Abstract
In black cherry (Prunus serotina Ehrh.) seed homogenates, (R)-amygdalin is degraded to HCN, benzaldehyde, and glucose by the sequential action of amygdalin hydrolase (AH), prunasin hydrolase (PH), and mandelonitrile lyase. Leaves are also highly cyanogenic because they possess (R)-prunasin, PH, and mandelonitrile lyase. Taking both enzymological and molecular approaches, we demonstrate here that black cherry PH is encoded by a putative multigene family of at least five members. Their respective cDNAs (designated Ph1, Ph2, Ph3, Ph4, and Ph5) predict isoforms that share 49% to 92% amino acid identity with members of glycoside hydrolase family 1, including their catalytic asparagine-glutamate-proline and isoleucine-threonine-glutamate-asparagine-glycine motifs. Furthermore, consistent with the vacuolar/protein body location and glycoprotein character of these hydrolases, their open reading frames predict N-terminal signal sequences and multiple potential N-glycosylation sites. Genomic sequences corresponding to the open reading frames of these PHs and of the previously isolated AH1 isoform are interrupted at identical positions by 12 introns. Earlier studies established that native AH and PH display strict specificities toward their respective glucosidic substrates. Such behavior was also shown by recombinant AH1, PH2, and PH4 proteins after expression in Pichia pastoris. Three amino acid moieties that may play a role in conferring such aglycone specificities were predicted by structural modeling and comparative sequence analysis and tested by introducing single and multiple mutations into isoform AH1 by site-directed mutagenesis. The double mutant AH ID (Y200I and G394D) hydrolyzed prunasin at approximately 150% of the rate of amygdalin hydrolysis, whereas the other mutations failed to engender PH activity.
O-Glycoside hydrolases (EC 3.2.1.x) constitute a widespread group of enzymes that hydrolyze the glycosidic bond between two or more carbohydrates or between a carbohydrate and a non-carbohydrate moiety. These enzymes have been classified into distinct families based on amino acid sequence similarities (Henrissat, 1991; Henrissat and Bairoch, 1996). One of these, family 1, includes many β-glucosidases (EC 3.2.1.21) that play diverse and important roles in prokaryotes and eukaryotes. In bacteria and fungi, the cellulolytic β-glucosidases are key enzymes in biomass conversion (Béguin, 1990; Fowler, 1993), whereas in animals, lysosomal glucocerebrosidase, for example, is critical to glycosphingolipid metabolism (Grabowski et al., 1993). In higher plants, β-glucosidases have been implicated in such fundamental processes as chemical defense against herbivores and pathogens (Conn, 1979), lignification (Dharmawardhana et al., 1995), and regulation of the biological activity of phytohormones by hydrolysis of their inactive hormone-glucoside conjugates (Falk and Rask, 1995).
In recent years, our laboratory has been investigating the nature and regulation of β-glucosidases involved in large-scale cyanogenesis (HCN production) in plants. As an experimental system, we have used black cherry (Prunus serotina Ehrh.), a species grown for its fruit and high-value hardwood. Like other rosaceous stone fruits, black cherry accumulates the cyanogenic monoglucoside (R)-prunasin [the O-β-d-glucoside of (R)-mandelonitrile] in its leaves and immature fruits (Kingsbury, 1964). The related diglucoside (R)-amygdalin [the β-gentiobioside of (R)-mandelonitrile] is found in high concentrations in mature seeds of these crops. Upon seed disruption, amygdalin is hydrolyzed to mandelonitrile by a two-step process. First, amygdalin hydrolase (AH; EC 3.2.1.117) hydrolyzes the β-1,6-glycosidic bond of amygdalin, yielding prunasin and d-Glc. Prunasin is then hydrolyzed to mandelonitrile and d-Glc by prunasin hydrolase (PH; EC 3.2.1.118). Because AH and PH show very pronounced specificities toward their respective glucosidic substrates (Kuroki and Poulton, 1986, 1987), both enzymes are required in cherry macerates for complete hydrolysis of amygdalin. Finally, mandelonitrile dissociates either enzymatically in the presence of the α-hydroxynitrile lyase mandelonitrile lyase (MDL; EC 4.1.2.10) or spontaneously, generating HCN and benzaldehyde (Hu and Poulton, 1999). In disrupted black cherry leaves, prunasin is degraded to HCN in similar fashion by PH and MDL.
In previous studies, AH and PH were purified to homogeneity from mature seeds, and their major physicochemical properties were characterized (Poulton, 1993). Subsequently, we demonstrated by immunocytochemistry and tissue printing that tissue level compartmentation prevents premature cyanogenesis in undamaged seeds. Whereas amygdalin accumulates in cotyledonary parenchyma cells, AH and PH are located in the procambium, where each β-glucosidase occurs as multiple forms (Li et al., 1992; Swain et al., 1992; Poulton and Li, 1994). Four isoforms of AH, designated AH I, AH I′, AH II, and AH II′, were recognized. All are monomeric glycoproteins with similar kinetic properties but differing in their pI and N-terminal sequences. The sequencing of a near full-length AH cDNA that encodes AH I showed that this β-glucosidase belongs to glycoside hydrolase family 1 (Zheng and Poulton, 1995). Far less is known about PH, because no authentic cDNA clone has as yet been isolated from any species. However, in black cherry seeds, PH is a glycoprotein that exists as three isoforms, designated PH I, IIa, and IIb, that were separable by hydroxyapatite and Sephacryl S-200 chromatography (Kuroki and Poulton, 1987). Whether PH displays microheterogeneity in vegetative tissues was unknown until now, because its purification from such sources has not been reported.
To gain a better understanding of cyanogenesis in rosaceous stone fruits, we have now cloned and characterized a full-length cDNA (designated Ph1) that encodes seed isoform PH I. Using both biochemical and molecular approaches, we also investigated PH microheterogeneity in black cherry seedling shoots. Four novel PH cDNAs (named sequentially Ph2 through Ph5) were isolated and characterized, of which three were shown to encode specific PH isoforms purified here from the same source. Sequence analysis has indicated that the five PHs, like AH, belong to glycoside hydrolase family 1 and are probably encoded by a multigene family. Genomic sequences corresponding to the open reading frames (ORFs) of Ah1 and Ph1 through Ph5 were amplified from black cherry genomic DNA by long distance (LD)-PCR, allowing analysis of their exon-intron organization. Finally, contributing to efforts to engineer the substrate specificities of β-glucosidases for particular biotechnological purposes, we have used computer modeling, sequence comparison, and site-directed mutagenesis to predict and alter amino acid residues that may confer upon AH and PH their characteristic aglycone specificities.
RESULTS AND DISCUSSION
In recent years, we have been investigating the biochemistry and molecular biology of cyanogenesis in rosaceous stone fruits using black cherry as an experimental system. One intriguing aspect of this species, which is shared by several other stone fruits (Haisman et al., 1967; Gerstner et al., 1971), is the microheterogeneity displayed by AH, PH, and MDL (Poulton, 1993). Possible sources of enzyme multiplicity include allelic differences at the structural loci for the polypeptides (i.e. allozymes), posttranslational modifications (e.g. N- and C-terminal processing, glycosylation, and phosphorylation), and gene duplications leading to multigene families (Weeden, 1983). In higher plants, polyploidy is perhaps the most conspicuous mechanism for wholesale gene duplication. Several rosaceous stone fruits are polyploid (Darlington, 1928), including black cherry, which is a tetraploid species (2n = 4x = 32) of ancient origin whose chromosomes pair as bivalents during meiosis (Maynard et al., 1991). Thus, in some Prunus spp., it is possible that allozymes could contribute significantly to the observed multiplicity of the cyanogenic enzymes.
In previous work, we demonstrated that MDL, which occurs in black cherry seed and shoot homogenates as several closely related isoforms, is encoded by a gene family of approximately eight members that exhibit differential expression (Hu and Poulton, 1999). The sources of AH and PH microheterogeneity remain less clear (Poulton, 1993). The four AH isoforms purified from homogenates of mature black cherry seeds exhibited similar kinetic properties but differed in their N-terminal sequences and pI, suggesting the existence of an AH multigene family or perhaps multiple alleles (Li et al., 1992). Three PH isoforms, designated PH I, PH IIa, and PH IIb, were purified from the same source and shown to be highly active toward prunasin but inactive toward amygdalin (Kuroki and Poulton, 1987). Whereas PH I and PH IIb were monomeric (68 kD), PH IIa was dimeric (140 kD). However, sequencing their N termini revealed no unequivocal differences among them, nor did it shed light upon the physical relationship between PH IIa and the monomeric isoforms (Li et al., 1992). In the present study, we have utilized both enzymological and molecular approaches to reexamine PH microheterogeneity in black cherry, focusing principally, for the first time (to our knowledge) in rosaceous stone fruits, on vegetative tissues.
Identification of Three Novel PH Isoforms in Black Cherry Seedling Shoots
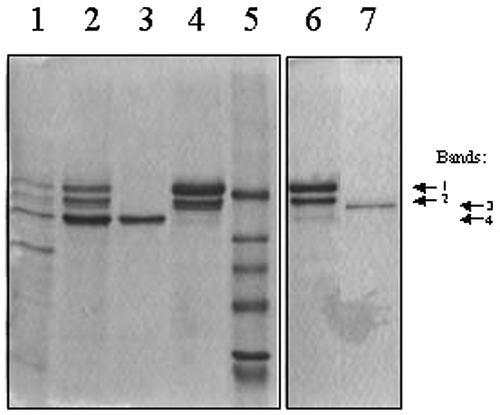
To purify PH from black cherry seedling shoots, we developed a protocol that began with Concanavalin A (Con A)-Sepharose 4B chromatography, because this approach had proven extremely successful in earlier purifications of AH, PH, and MDL from black cherry seeds (Poulton, 1993). SDS-PAGE analysis showed that most proteins failed to bind to this affinity matrix, when the crude preparation was applied in 10 mm His-HCl buffer, pH 6.0 (Fig. 1). However, the matrix became yellow due to the binding of the flavoprotein MDL (Hu and Poulton, 1999). After elution by α-methyl-d-glucoside, the bound proteins exhibited four polypeptide bands on SDS-PAGE (Fig. 1). Designated bands 1 through 4, all lay within the range of 52 to 67 kD. Previous N-terminal sequencing had established that band 1 was MDL (Hu and Poulton, 1999). To resolve these polypeptides, the Con A-Sepharose eluate was subjected to DEAE-cellulose chromatography at pH 5.0 in 20 mm sodium acetate-HCl buffer. Unbound proteins were removed by extensive washing with this buffer, before eluting bound proteins with a linear NaCl gradient. Substantial β-glucosidase activity was found in both unbound and bound fractions, suggesting that ion-exchange chromatography had resolved two distinct PH isoforms. This outcome was confirmed by SDS-PAGE, which showed that the unbound PH appeared as a single polypeptide of approximately 55 kD (Fig. 1, band 4). This band was submitted for N-terminal sequencing without further purification. Because the polypeptides represented by bands 1 through 3 were not fully resolved by DEAE-cellulose chromatography, they were applied to a Reactive Red 120-agarose column. Band 2 failed to bind to this column and co-eluted with MDL, whereas band 3 bound and could be eluted in homogeneous form by a salt gradient. Bands 2 and 3 were subsequently submitted for N-terminal sequencing.
Figure 1.
SDS-PAGE analysis of purification of PH isoforms from black cherry seedling shoots. PH preparations at various stages of purification were subjected to SDS-PAGE with Coomassie Brilliant Blue staining: crude homogenate (lane 1), Con A-Sepharose eluate (lane 2), unbound (lane 3) and bound (lane 4) fractions from DEAE-cellulose chromatography, and unbound (lane 6) and bound (lane 7) fractions from Reactive Red-agarose chromatography. Lane 5 indicates the position (from top to bottom) of the molecular mass markers bovine serum albumin (66 kD), egg albumin (45 kD), glyceraldehyde- 3-phosphate dehydrogenase (36 kD), carbonic anhydrase (29 kD), trypsinogen (24 kD), and soybean trypsin inhibitor (20.1 kD). Protein bands: 1, MDL4 isoform (Hu and Poulton, 1999); 2, PH4; 3, PH5; 4, PH3.
N-Terminal sequencing of bands 2 through 4 provided an initial estimate of the minimum number of PH genes expressed in black cherry shoots. As Table I shows, their N termini are similar but not identical, thus ruling out the possibility that these isoforms represent different posttranslational modifications of a single gene product. Furthermore, their N-terminal sequences also differed from those reported previously for the seed isoforms PH I, IIa, and IIb (Li et al., 1992).
Table I.
Comparison of the experimentally determined N-terminal amino acid sequences of black cherry AH I and PHs
| Band 4 (PH3) | D P P S H X P V L L R R S F . . . . . . . . |
| Band 2 (PH4) | T D P P G V V T T L X R T H F D T X F P G F . |
| Band 3 (PH5) | T D P X I V X A T L X R T H F D X L F P G . . |
| PH Iab | X X T Y P P V V X A T L X R T H . . . . . . . . . |
| PH IIaa | A G T Y P P V V X A T L X R T H . . . . . . . . . |
| PH IIba | X G T Y P P V V L A T L X R T H . . . . . . . . . |
| AH Iab | A K T D P P I H X A S L X R S S . . . . . . . . . |
Amino acids are given in the one-letter code, with unassigned residues indicated by X. Sequences have been aligned to maximize similarity.
In the present study, PHI and AHI are renamed PH1 and AH1, respectively.
During the purification of PH isoforms from black cherry shoots, β-glucosidase activity was routinely monitored using the facile p-nitrophenyl-β-d-glucosidase assay (Kuroki and Poulton, 1986). To confirm that these enzymes are PHs and not, for example, AHs, it was important that specific assays be used to ascertain their activities toward physiological substrates. This goal was achieved here using specific AH and PH assays that quantitate the d-Glc released upon hydrolysis of these cyanogenic glucosides (Kuroki and Poulton, 1986). These assays unequivocally showed that each of the three highly purified PH isoforms exhibits high activity toward (R)-prunasin but is completely inactive toward (R)-amygdalin (data not shown).
Isolation and Characterization of Black Cherry PH cDNAs
To gain a better understanding of the biochemical nature and possible physiological significance of PH multiplicity in black cherry, we isolated cDNAs encoding PHs expressed in leaves and/or seeds. To obtain a PH probe for screening leaf and seed cDNA libraries, we first undertook reverse transcriptase (RT)-PCR. Given the high amino acid identity (approximately 50%) shared by the N termini of AH and PH isoforms isolated from black cherry seeds (Li et al., 1992), we decided to maximize our probability of isolating PH clones by selecting leaves, which lack AH activity, as RNA source. Poly(A+) RNA from leaves was reverse transcribed using a degenerate primer based on the active site Ile-Thr-Glu-Asn-Gly (ITENG) motif characteristic of glycoside hydrolase family 1 members (Esen, 1993). Subsequent amplification using nested primers based on known peptide sequences of seed isoform PH I yielded an 834-bp fragment that was cloned and double-strand sequenced. Designated “partial-length Ph2,” this cDNA represents a single ORF that shares 92% amino acid identity (95% similarity) with the Prunus avium mesocarp/exocarp β-glucosidase, including its catalytic Asn-Glu-Pro (NEP) motif (Wiersma and Fils-Lycaon, 1995). The N terminus of the deduced PH2 protein was extremely similar to, but did not exactly match, those of the PH isoforms already purified from black cherry seeds and shoots.
Isolation and Characterization of Ph1 cDNA
To obtain cDNAs corresponding to PH genes expressed in black cherry seeds, we used the partial-length Ph2 cDNA as probe to screen a λgt11 cDNA library previously constructed from mRNA from immature seeds (Zheng and Poulton, 1995). A cDNA insert exhibiting high homology to known PH sequences was double-strand sequenced and assigned accession number U50201 (Table II). Designated Ph1, this cDNA is 2,056 nucleotides in length and contains two potential translational start points that lie within a tandem repeat of CAGTTATGGCAT. Assuming that the first ATG codon is the most likely translation start point, the Ph1 cDNA has 34 nucleotides of 5′-UTR, a 1,650-nucleotide ORF (beginning at position 35 and ending at position 1,681 before a TGA stop codon), and a 3′-UTR of 372 nucleotides that includes a presumptive polyadenylation (AATAAA) signal and a short poly(A+) tail. The ORF encodes a polypeptide of 549 amino acids with a predicted molecular mass of 62,080 D and a pI of 5.75.
Table II.
Comparison of major features of black cherry PH and AH cDNAs and their encoded polypeptides
| cDNA | Accession No. | Encoded Protein | Sequence Length | 5′-Untranslated Region (UTR) | 3′-UTR | Polypeptide Length | Features |
|---|---|---|---|---|---|---|---|
| bp | amino acids | ||||||
| Ph1 | U50201 | PH1a | 2,056 | 34 | 372 | 549 | NEP and ITENG active site motifsN-terminal signal sequenceEight N-glycosylation sitesPoly(A+) tail |
| Ph2 | AF221527 | PH2 | 2,059 | 37 | 387 | 544 | NEP and ITENG active site motifsLikely N-terminal signal sequenceTwo N-glycosylation sitesPoly(A+) tail |
| Ph3 | AF221526 | PH3 | 1,960 | 31 | 315 | 537 | NEP and ITENG active site motifsN-terminal signal sequenceSix N-glycosylation sitesPoly(A+) tail |
| Ph4 | AF411009 | PH4 | 1,911 | 38 | 235 | 545 | NEP and ITENG active site motifsN-terminal signal sequenceSeven N-glycosylation sitesPoly(A+) tail |
| Ph5 | AF411131 | PH5 | 1,819 | 29 | 161 | 542 | NEP and ITENG active site motifsN-terminal signal sequenceSeven N-glycosylation sitesPoly(A+) tail |
| Ah1 | AF411130 | AH1b | 1,915 | 29 | 224 | 553 | NEP and ITENG active site motifsN-terminal signal sequenceFive N-glycosylation sitesPoly(A+) tail |
Formerly designated PH I (Li et al., 1992).
Formerly designated AH I (Li et al., 1992).
Considerable evidence supports our contention that the Ph1 cDNA codes for seed isoform PH I (henceforth designated PH1). First, the amino acid sequence deduced from this cDNA (AGTYPPVVCATLNRTH) matches almost perfectly the reported N-terminal sequence of PH1 (XXTYPPVVXATLXRTH; Li et al., 1992). We believe that our earlier failure to assign residues 9 and 13 was due to the inability of the Edman degradation to detect Cys and glycosylated Asn, respectively. Second, the sequence (GLDAYRFSISXSRLLPXGTLSGGIN) of an internal peptide generated by CNBr cleavage of isoform PH1 is essentially identical to that deduced from the Ph1 cDNA sequence (GLDAYRFSISWSRLLPNGTLSGGIN). Here again, we assume that the observed discrepancies reflect the inability of the sequenator to identify Trp and glycosylated Asn residues. Third, because the N terminus of the mature PH1 protein corresponds to residue 31 of the polypeptide deduced from the Ph1 cDNA, it seems likely that the preceding 30 residues serve as a signal peptide; this correlates well with the known protein body location of PH in seeds (Swain et al., 1992). Finally, the presence of eight potential N-glycosylation sites (Asn-X-Thr/Ser) in the Ph1 cDNA sequence is consistent with the glycoprotein character of PH1 (Kuroki and Poulton, 1987).
Isolation and Characterization of PH cDNAs from Vegetative Tissues
Several cDNAs corresponding to PH transcripts expressed in seedling leaves were obtained by RT-PCR and library screening. In addition to Ph1, four novel PH cDNAs (Ph2 through Ph5) were isolated, whose major features, together with those of the deduced proteins that they encode, are summarized in Table II. As detailed below, three of the cDNAs (Ph3, Ph4, and Ph5) were verified as authentic PH clones by matching their deduced polypeptide sequences with the N-terminal sequences of the purified seedling shoot isoforms (bands 4, 2, and 3, respectively). The Ph2-encoded isoform was not detected during our purification studies here and therefore is assumed to be a minor component. However, to establish its identity as a PH, this protein was heterologously expressed in Pichia pastoris and shown to exhibit PH, but not AH, activity (see below). Tables II and III illustrate that the five deduced PH proteins are of similar length (537–549 amino acids) and share 71% to 93.4% amino acid identity among themselves. Furthermore, consistent with their vacuolar localization (Swain and Poulton, 1994) and their recognition by Con A-Sepharose 4B (this work), the shoot PH isoforms exhibit N-terminal signal sequences and multiple (two–seven) potential N-glycosylation sites.
Table III.
Percentage identity of the deduced amino acid sequences of black cherry AH and PHs and several β-glucosidases belonging to glycoside hydrolase family 1
| Enzyme | PH2 | PH3 | PH4 | PH5 | AH1 | U39228 | X56733 | S35175 |
|---|---|---|---|---|---|---|---|---|
| PH1 | 85.9 | 73.2 | 93.4 | 91.5 | 69.7 | 89.6 | 59.7 | 50.3 |
| PH2 | – | 71.0 | 83.3 | 86.1 | 66.9 | 92.3 | 57.9 | 50.0 |
| PH3 | – | – | 71.9 | 74.1 | 71.0 | 74.0 | 63.2 | 49.2 |
| PH4 | – | – | – | 87.4 | 68.8 | 86.3 | 57.5 | 49.7 |
| PH5 | – | – | – | – | 69.2 | 88.5 | 59.3 | 50.8 |
Percentage amino acid identity was determined by the GAP program. GenBank accession nos.: U39228, P. avium β-glucosidase (Wiersma and Fils-Lycaon, 1995); X56733, Trifolium repens linamarase (Barrett et al., 1995); and S35175, cassava (Manihot esculenta Crantz) linamarase (Hughes et al., 1992).
Authenticity of the Ph3 cDNA was established by recognition that nucleotides 110 through 151 encode the known N terminus (DPPSHXPVLLRRSF) of band 4 (Table I, PH3). Since the Ph3 cDNA encodes Cys as the sixth residue, this readily explains the failure of the Edman degradation to assign this moiety. Because the N-terminal Asp residue of the PH3 isoform corresponds to residue 27, as predicted by the cDNA, it strongly suggests that the initial 26 amino acids act as a signal peptide to facilitate intracellular movement of this polypeptide to the vacuole (Swain and Poulton, 1994) via the endoplasmic reticulum. Since the N-terminal sequence of band 2 (TDPPGVVTTLXRTHFDTXFPGF) almost perfectly matches the amino acid sequence predicted by the Ph4 cDNA (TDPPGVCTTLNRTNFDTLFPGF) beginning at residue 29, we suggest that this polypeptide is encoded by the Ph4 cDNA. Finally, the amino acid sequence (TDPPIVCATLNRTHFDTLFPG) deduced from the Ph5 cDNA perfectly matches the N-terminal sequence of band 3 (TDPXIVXATLXRTHFDXLFPG).
Phylogenetic Analysis
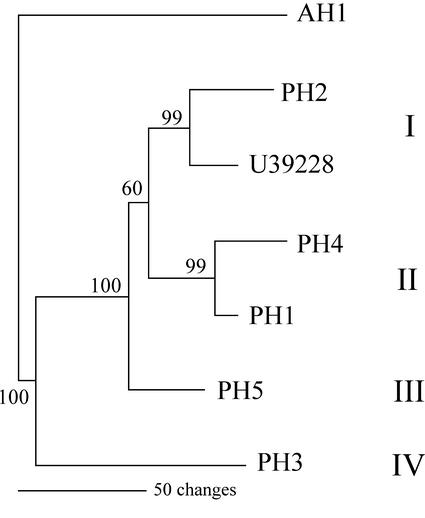
The amino acid sequences of the black cherry PHs and AH I (henceforth designated AH1), together with that of the P. avium β-glucosidase, were aligned using the Clustal V program. Phylogenetic analysis of the aligned sequences was then performed using the heuristic option of PAUP* (Phylogenetic Analysis Using Parsimony, version 4.0b2, Sinauer Associates, Sunderland, MA), with support for the branches being gathered by bootstrap analysis. As Figure 2 illustrates, the known PHs fell into four apparent clades. One clade contains PH2 and U39228, a likely ortholog from P. avium; both proteins are expressed in vegetative tissues. The second clade contains PH1 and PH4. The remaining clades are represented by PH3 and PH5, respectively. In this analysis, AH1 appears as a distinct branch. In future work, we intend to isolate cDNAs corresponding to other members of the putative AH multigene family predicted by our earlier enzymological studies (Li et al., 1992).
Figure 2.
Phylogenetic tree of the alignment of the deduced amino acid sequences of black cherry PHs and AH1. P. avium β-glucosidase (U39228), which shows 74% to 92% amino acid identity to the black cherry PHs, is also included. Amino acid sequences were aligned using the Clustal V program. Phylogenetic analysis was performed using the heuristic option of PAUP* (version 4.0 b2). Support for the branches was determined by bootstrap analysis (1,000 replicates). This tree is arbitrarily rooted with the midpoint method and does not necessarily reflect the order of divergences.
Because of the tetraploid nature of black cherry, and because our experimental material is obtained from natural populations and thus not genetically defined, at this time, we cannot unequivocally distinguish whether the PH gene sequences described here represent multiple alleles or members of a gene family. This would require extensive crosses between lines differing with respect to their zymogram profiles, followed by backcrosses to parental lines to determine whether there is linkage or independent assortment between any two isoforms. Considering that black cherry has a generation time of 10 to 15 years, this would clearly be a long-term project. In the absence of such information, the observed high level of sequence divergence (71%–93.4% amino acid identity) of the black cherry PH genes is viewed as evidence of a multigene family, although we are aware that this criterion may not always be valid (e.g. the Brassica campestris S locus [Watanabe et al., 2000]). The contribution of polyploidy to the putative PH gene family remains unknown.
Black Cherry AH and PH Belong to Glycoside Hydrolase Family 1
In 1991, Henrissat introduced a novel classification system for glycoside hydrolases to complement the International Union of Biochemistry and Molecular Biology (1984) enzyme nomenclature for this class of enzymes. Based on amino acid sequence similarities, this system better reflects the structural features of these enzymes than do systems based solely on their substrate specificities. Over 950 sequences of glycoside hydrolases and related enzymes were subsequently grouped into 82 families (Henrissat and Davies, 2000); this compilation is being continuously updated at http://afmb.cnrs-mrs.fr/~cazy/CAZY/index.html. According to sequence information, most known β-glucosidases belong to glycoside hydrolase family 1 (also referred to as the BGA enzyme family by Béguin, 1990). This large family includes β-glucosidases, 6-phospho-β-galactosidases, 6-phospho-β-glucosidases, and myrosinases from organisms as diverse as archae, bacteria, and eukaryotes. Typified by the Agrobacterium faecalis and T. repens β-glucosidases (Wakarchuk et al., 1988; Hughes, 1993), family 1 members utilize two Glu residues, embedded in highly conserved ITENG (nucleophile) and NEP (acid/base catalyst) motifs, to catalyze glycosidic bond cleavage with net retention of anomeric configuration (McCarter and Withers, 1994). In earlier work (Zheng and Poulton, 1995), we concluded that black cherry AH1 belonged to glycoside hydrolase family 1, as evidenced by its high homology (37%–65% identity) to other family members as well as its possession of the active site NEP and ITENG motifs. The present study reveals that the five deduced PHs also share high sequence identity (49%–92%) with members of glycoside hydrolase family 1, including the NEP, ITENG, and many other motifs characteristic of this family (Tables II and III). Therefore, we conclude that the black cherry PH isoforms, like AH, belong to this family. This conclusion concurs with the assignment to this family of sweet almond (Prunus dulcis var. sativa) β-glucosidase (presumably a mixture of AH and PH; He and Withers, 1997) and bitter almond (Prunus dulcis) β-glucosidase (Legler and Harder, 1978), based on partial sequences gained by irreversible inactivator studies.
Exon-Intron Organization of PH and AH Genes
Nothing is known about the evolution of PH and AH genes in Prunus spp. A plausible hypothesis is that the extant PH and AH multigene families are derived from two ancestral PH and AH genes, which in turn arose by gene duplication and divergence of a single ancestral β-glucosidase gene. Supporting this hypothesis are the high sequence identities between individual members of the PH multigene family (71%–93%), and between these same members and AH1 (67%–71%). Further support was sought here by investigating the exon-intron organization of the PH and AH genes.
Although relatively little information is available about the exon-intron organization of plant β-glucosidase genes, certain patterns are being delineated as more sequences become available. Whereas the barley (Hordeum vulgare) bgp60 and maize (Zea mays) Glu1 genes exhibit nine and 11 introns, respectively (Leah et al., 1995; Esen and Bandaranayake, 1998), the O-β-glucosidase genes from the dicots cassava (accession no. X94986; Liddle et al., 1998) and Arabidopsis (Malboobi and Lefebvre, 1997) possess 12 introns. In addition, six myrosinase genes from three crucifers consist of 12 exons separated by 11 introns (Rask et al., 2000). Closer analysis has shown that the O- and S-β-glucosidases share a similar exon-intron organization, although certain exons are fused or split (Rask et al., 2000). All the aforementioned introns lie within the ORFs of their respective genes.
In our work, we obtained genomic sequences corresponding to the ORFs of the black cherry cDNAs Ah1 and Ph1 through Ph5 by LD-PCR amplification of genomic DNA using gene-specific primers (Table IV). Comparison of cDNA and genomic sequences revealed that the ORFs of these six genes are interrupted at identical positions by 12 AT-rich (63%–84%) introns, each of which shows significant sequence similarity between genes. These findings are consistent with our hypothesis that AH and PH are derived from a common ancestral gene. The intron lengths, which average 165 bp, lie within the range of 83 to 666 bp. All exon-intron junction sequences of the black cherry β-glucosidase genes conform to the GT-AG rule for RNA splicing with the exceptions of intron 1 of Ah1 and Ph3 and intron 8 of Ah1, which have GC-AG junction sequences (Brown, 1986). Interestingly, a GC donor splice site was noted for intron 1 of the Arabidopsis tgg1 and tgg2, and probably also tgg3, myrosinase genes (Xue and Rask, 1995).
Table IV.
Compilation of primers used to generate PH and AH genomic sequences by PCR amplification
| Gene | Accession No. | Primers |
|---|---|---|
| Ph1 | AF414608 | CTGTTGTTTGTGCAACTCTCAACAGGACCC (sense) |
| CACGGTACGTTATCCATCATTGGGATTCAC (antisense) | ||
| Ph2 | AF414607 | GATTACAGGACAGATCCACCCGTTGTTTGC (sense) |
| GACGGGAACCATCCTTTGGGATTCAAATTC (antisense) | ||
| Ph3 | AF413214 | CAATGCAGTTAGGACAGATCCACCCTCTCA (sense) |
| GGCGTATGGGATTCAAATTTCATACCCAGC (antisense) | ||
| Ph4 | AF411928 | AATTCGGGACAGATCCACCCGGTGTTTGTA (sense) |
| CGCATGTGGTACACAAATTTGGTAGCCCTG (antisense) | ||
| Ph5 | AF413213 | TTGTCAATGCTGCCAGAACAGATCCACCCA (sense) |
| AGCCCGATACACAAATTTGGTAGCCCTAC (antisense) | ||
| Ah1 | AF414606 | GCGAAAACAGATCCACCCATTCACT (sense) |
| TCACACGACGACTGCTAGAGAGCTT (antisense) |
Identification of Amino Acid Residues That May Confer Aglycone Specificity on AH and PH
Although exceptions exist (e.g. Selmar et al., 1987), most plant β-glucosidases display high aglycone specificities (Hösel and Conn, 1982; Conn, 1993). Prime examples include β-glucosidases exhibiting pronounced specificities toward such endogenous substrates as cyanoglucosides (Hösel and Nahrstedt, 1975; Hösel et al., 1987; Poulton, 1993), coumarinyl acid glucoside (Kosuge and Conn, 1961), cinnamyl alcohol glucosides (Hösel and Todenhagen, 1980; Dharmawardhana et al., 1995), isoflavone glycosides (Hösel and Barz, 1975), saponin glycosides (Inoue and Ebizuka, 1996), hydroxamic acid glucosides (Babcock and Esen, 1994), and cytokinin glucosides (Falk and Rask, 1995). This high degree of aglycone specificity raises the question as to which amino acid residues confer such selectivity. Understanding this aspect of enzyme behavior might ultimately allow us to efficiently engineer the substrate specificities of β-glucosidases for particular purposes such as biomass conversion.
Black cherry AH and PH are ideal candidates for addressing the above question because: (a) They show strict specificities toward their respective substrates (Poulton, 1993), and (b) their cDNAs predict 60% to 70% amino acid identity not only to each other but also to T. repens linamarase, a cyanogenic β-glucosidase of known crystal structure belonging to glycoside hydrolase family 1 (Barrett et al., 1995). The linamarase backbone folds into a single, large (β/α)8 barrel consisting of a core of eight twisted parallel β-strands with the connecting α-helices lying on the outside of the barrel. At its C terminus is a solvated pocket that holds the catalytic NEP (acid/base catalyst) and ITENG (nucleophile) motifs characteristic of family 1 members. This pocket is created by eight loops that connect the carboxy end of the β-strands with the amino ends of the adjacent α-helices (Branden and Tooze, 1991). These loops, which do not contribute to the structural stability of the barrel “scaffold” but instead participate in substrate binding, were the principal focus of our efforts to recognize aglycone specificity-conferring residues in PH and AH. Starting from the primary structures of black cherry AH1 and the five PHs encoded by the cDNAs Ph1 through Ph5, candidate residues were selected based on the following criteria: (a) They should occur in the aforementioned “loop regions” and lie within 20Å of the active site Glu residues, and (b) these residues should be identical (or at least a conservative replacement) in the five PH proteins but differ significantly in nature in AH1. Using structural modeling (data not shown) and sequence analysis (Fig. 3), three candidate residues were thereby identified, each of which lies near the entrance to or within the side wall of a channel leading down to the active site pocket. For AH1, these residues are Ser-216, Tyr-220, and Gly-394.
Figure 3.
Alignment of the deduced amino acid sequences of black cherry PHs and AH1. Alignment was performed using the PILEUP program. Where necessary, periods are inserted to obtain maximal homology. Three residues, whose predicted role in conferring aglycone specificity upon AH and PH was assessed here by site-directed mutagenesis, are highlighted by black boxes. The active site NEP and ITENG motifs are indicated by horizontal bars.
To assess whether the three candidate residues confer substrate specificity upon AH and PH requires expression of active recombinant AH and PH in a suitable system. Reasoning that, because certain monocotyledonous β-glucosidases are unglycosylated (Esen, 1993), the carbohydrate side chains of AH1 and PH1 may be inessential for catalytic activity, we first attempted to express these black cherry hydrolases in Escherichia coli. Unfortunately, the recombinant proteins were essentially inactive and were sequestered largely in insoluble form in bacterial inclusion bodies (data not shown). Because glycosylation of AH and PH may be required for hydrolase activity, as has been reported for other β-glucosidases (Grace and Grabowski, 1990; McMahon et al., 1995), we next attempted to express AH1, PH2, and PH4 in the eukaryotic expression system P. pastoris, which displays most of the posttranslational modification pathways typically associated with higher eukaryotes (Cregg et al., 1993). As Table V illustrates, all three recombinant proteins were secreted into the culture supernatants, were catalytically active, and maintained the strict aglycone specificity exhibited by native AH and PH.
Table V.
Retention by recombinant AH1, PH2, and PH4 of the characteristic aglycone specificities shown by the native AH and PH
| Recombinant Enzyme | AH Activity | PH Activity |
|---|---|---|
| rAH1 | 6.8 | 0 |
| rPH2 | 0 | 24.2 |
| rPH4 | 0 | 18.5 |
Activities are expressed in micromoles per milliliter enzyme per hour and are the average of three separate measures.
After successful expression of wild-type AH1 in P. pastoris, we investigated whether the three candidate residues identified above are important in determining the aglycone specificity of black cherry AH and PH. Site-directed mutagenesis was employed to introduce several single, double, and triple mutations into the AH1 cDNA, thereby replacing the candidate residues of recombinant AH1 with those found in PH1. The desired mutations were verified by sequencing. The β-glucosidase activities of the mutated recombinant enzymes, which were also secreted into the culture medium, were tested against amygdalin and prunasin (Table VI). In summary, all mutant forms showed AH activities ranging from 2.1 to 14.1 μmol h−1 mL−1 enzyme (compare with WT-AH1, 6.0 μmol h−1 mL−1 enzyme) but, with one exception, they lacked PH activity. The obvious exception was the double mutant AH ID (Y220I, G394D) that hydrolyzed prunasin at approximately 150% of the rate of amygdalin hydrolysis (i.e. AH and PH activities of 3.9 and 6.1 μmol h−1 mL−1 enzyme, respectively). To our knowledge, this is the first report of an AH exhibiting PH activity. Future research will include the purification of the AH ID enzyme before thorough kinetic analysis. In addition, we are also introducing specific mutations into the Ph4 cDNA that we predict may elicit AH activity.
Table VI.
AH, PH, and p-nitrophenyl-β-d-glucosidase (PNPGase) activities of recombinant AH1 (rAH1) and its mutant forms in P. pastoris culture supernatants
| Recombinant Enzyme | Mutation(s) | AH Activity | PH Activity | PNPGase Activity |
|---|---|---|---|---|
| rAH1 | – | 6.0 | 0 | 0.7 |
| AH SH | S 216 H | 9.4 | 0 | 1.3 |
| AH YI | Y 220 I | 10.9 | 0 | 1.7 |
| AH GD | G 394 D | 13.8 | 0 | 2.4 |
| AH HI | S 216 H | 14.1 | 0 | 1.8 |
| Y 220 I | ||||
| AH HD | S 216 H | 3.5 | 0 | 1.2 |
| G 394 D | ||||
| AH ID | Y 220 I | 3.9 | 6.1 | 14.2 |
| G 394 D | ||||
| AH HID | S 216 H | 2.1 | 0 | 0.2 |
| Y 220 I | ||||
| G 394 D |
The following single, double, and triple mutations were introduced into the Ah1 cDNA by site-directed mutagenesis and verified by sequencing. The substrate specificities of the mutated recombinant AH1 toward amygdalin, prunasin, and PNPG were compared with that of unmutated, recombinant AH1. Activities are expressed in micromoles substrate hydrolysed per milliliter enzyme per hour and are the average of three separate measures.
Our findings correlate well with data obtained by Czjzek et al. (2000), who recently undertook x-ray crystallography of maize β-glucosidase isozyme 1 using the catalytically inactive mutant Glu1E191D in complexes with its natural substrate (2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one [DIMBOA]-glucoside),the corresponding aglycone (DIMBOA), and a competitive inhibitor dhurrin. Found in all family 1 β-glucosidases studied to date, the highly conserved residues Q38, H142, E191(D), E406, E464, and W465 were shown to contribute to this isozyme's glycone-binding pocket, forming hydrogen bonds to the hydroxyl groups of the glucopyranoside ring. These residues are also present in the black cherry PHs and AH1. The bulky aryl group of DIMBOA was sandwiched within a hydrophobic aglycone-binding pocket that consisted of W378 on one side and F198, F205, and F466 on the other side. Whereas W378 is highly conserved among plant family 1 β-glucosidases (including the black cherry hydrolases described here), sites homologous to the three Phe residues are not conserved among β-glucosidases that differ in substrate specificities, leading to the conclusion that these residues might play an important role in aglycone recognition and binding. In our work, we demonstrated that the substrate specificity of black cherry AH1 might be broadened to include prunasin by simultaneous mutation of residues Y220 and G394, which were selected on the basis of comparative sequence analysis and computer modeling. It should be noted that Y220 lies within a narrow 19-amino acid region between the highly conserved NEP and Ala-Pro-Gly motifs that includes sites homologous to the maize F198 and F205 residues. Furthermore, G394 lies adjacent to W395, which is the homologous site in AH1 of the highly conserved W378 that forms one side of the maize Glu1 aglycone-binding pocket. In contrast, our site-directed mutagenesis studies suggest that S216, which is homologous in AH1 to maize Glu1 F198, does not play a significant role in conferring aglycone specificity upon this black cherry hydrolase.
MATERIALS AND METHODS
Plant Materials
Unexpanded leaves, young leaves, and immature seeds of black cherry (Prunus serotina Ehrh.) were harvested from a single tree on the University of Iowa campus. Samples were collected into liquid N2 and stored at −70°C until used. Black cherry seeds were germinated by soaking them overnight in aerated distilled water before planting them in Jiffy-mix Plus (Jiffy Products of America, Batavia, IL). Unexpanded leaves (for mRNA isolation) or seedling shoots (for PH protein isolation) were harvested after 6 weeks in the greenhouse.
Purification of Black Cherry Shoot PH Isoforms
All steps were undertaken at 4°C. Seedling shoots (125 g) were harvested approximately 1 cm above soil level using a razor blade and homogenized in a blender with 440 mL of buffer A (100 mm His-HCl, pH 6.0) and 9 g of polyvinylpolypyrrolidone. The homogenate was filtered through four layers of cheesecloth, and the resulting filtrate was centrifuged at 10,000g for 30 min. The supernatant was dialyzed overnight against 4 L of buffer B (10 mm His-HCl, pH 6.0, containing 0.17 m NaCl). After clarification by centrifugation, the dialysate was applied to a Con A-Sepharose 4B column (1.6 × 10 cm) pre-equilibrated with buffer B. Unbound proteins were removed by extensive washing with buffer B, before eluting bound proteins with 125 mL of the same buffer containing 0.5 m α-methyl-d-glucoside. Fractions showing highest β-glucosidase activity were combined, dialyzed overnight against 4 L of buffer C (20 mm sodium acetate-HCl, pH 5.0), and applied to a DEAE-cellulose column (1.6 × 12 cm) pre-equilibrated with buffer C. After extensive washing with buffer C, bound proteins were eluted with a linear 0 to 350 mm NaCl gradient (200-mL total volume) in the same buffer. The active fractions were pooled, dialyzed overnight against buffer C, and applied to a Reactive Red 120-agarose column (1.0 × 7.5 cm), pre-equilibrated with buffer C. The column was washed with 100 mL of buffer C, before eluting bound proteins with a linear 0 to 0.5 m NaCl gradient (200-mL total volume) in the same buffer.
SDS-PAGE and Protein Microsequencing
Protein samples were fractionated on 12.5% (w/v) SDS-polyacrylamide gels according to the method of Laemmli (1970) using a minigel system (Bio-Rad Laboratories, Richmond, CA). Gels were stained with Coomassie Brilliant Blue G. For microsequencing, resolved polypeptides were electroblotted onto Trans-Blot polyvinylidene difluoride membranes (Bio-Rad Laboratories) and visualized by Coomassie Brilliant Blue R-250 staining (Matsudaira, 1993). N-Terminal sequences were obtained by automated Edman degradation on an Applied Biosystems 475A protein sequencer at the University of Iowa Protein Structure Facility.
Enzyme Assays
AH, PH, and PNPGase activities were assayed as described previously (Kuroki and Poulton, 1986).
Isolation and Sequencing of PH and AH cDNAs
Partial-Length Ph2 cDNA
After preparing total RNA from arborescent leaves by previously described methods (Zheng and Poulton, 1995), poly(A+) RNA was isolated utilizing magnetic beads (PolyATract mRNA isolation kit, Promega, Madison, WI). The poly(A+) RNA was reverse-transcribed using Stratascript RNase H− RT (Stratagene, La Jolla, CA) and the primer 5′-CCITTITCIGTIAT-3′ (I = inosine), whose sequence was deduced from the highly conserved ITENG region of glycoside hydrolase family 1 members (Esen, 1993). Subsequent PCR amplification involved two primers based on N-terminal and internal peptide sequences of isoform PH1 (E. Swain, unpublished data); these were 5′-GGNACNTAYCCNCCNGTGGT-3′ (sense) and 5′-CATRAACCANCCRTACAT-3′ (antisense), respectively. The resulting 834-bp PCR product (designated “partial-length Ph2”) was purified and cloned into pGEM-T (Promega) for transformation into Escherichia coli XL1-Blue competent cells (Stratagene). Plasmids were isolated by standard protocols (Sambrook et al., 1989), and their inserts were sequenced manually in both directions by the dideoxy chain termination method (Sanger et al., 1977) using Sequenase version 2.0 (United States Biochemical, Cleveland).
Full-Length Ph2 and Ph4 cDNAs
An unamplified λTriplEx2 cDNA library (105 plaque-forming units [pfu]) was constructed from seedling leaf total RNA using a SMART cDNA Library Construction Kit (BD Biosciences Clontech, Palo Alto, CA). This library was screened with a 237-bp BamHI/HindIII restriction fragment of the Ph5 cDNA labeled with digoxigenin by random priming (Roche Applied Science, Indianapolis). Upon restriction analysis, 100 putative clones fell into three distinct groups. For each group, five clones were selected at random for plasmid rescue and automated double-strand sequencing at the University of Iowa DNA Facility. Sequencing was undertaken at the University of Iowa DNA Facility with a 373S Fluorescent Automated Sequencer (Perkin-Elmer Applied Biosystems, Foster City, CA). This approach yielded a novel cDNA, designated Ph4, and a full-length Ph2 cDNA.
Ph3 cDNA
An unamplified λgt11 library (5 × 104 pfu), constructed from seedling shoots total RNA as described previously (Hu and Poulton, 1999), was screened with radiolabeled partial-length Ph2 cDNA. After two rounds of plaque hybridization, phage DNA was isolated from positive clones by a plate-lysate method. Inserts were excised with SalI, gel purified, and ligated into SalI-digested pBluescript SK(−) for transformation into E. coli DH5α competent cells (Gibco-BRL, Rockville, MD) and automated double-strand sequencing.
Ph5 cDNA
Starting with leaf total RNA, a SMART cDNA Library Construction Kit (BD Biosciences Clontech) was used to obtain double-stranded cDNA bearing adaptors. With this cDNA serving as template, a 1,715-bp product was obtained by PCR amplification using the sense primer 5′-ACAGATCCACCCATTGTTTGTGCA-3′ (based principally on the N-terminal sequence of band 3 [PH5]) and the CDS III/3′ PCR primer (BD Biosciences Clontech) as antisense primer. An overlapping fragment of 990 bp in length was subsequently generated by PCR amplification utilizing the following primers: 5′-PCR primer (BD Biosciences Clontech) as sense primer and 5′-ACAATAGATCGCATGGTCTGCGGG-3′ as antisense primer. Finding that the two cDNAs matched perfectly in the 900-bp overlap region, a contig was constructed and designated Ph5.
Ph1 cDNA
The partial-length Ph2 cDNA was radiolabeled with [α-32P]dCTP by random priming (Roche Applied Science) and used to screen a λgt11 cDNA library constructed from mRNA isolated from mid-maturation black cherry seeds (Zheng and Poulton, 1995). Prehybridization and hybridization were carried out at 65°C in 6× SSC, 1× Denhardt's solution (Sambrook et al., 1989), 0.5% (w/v) SDS, 100 mg mL−1 denatured salmon sperm DNA, and 0.05% (w/v) sodium pyrophosphate. Filters were washed with 6× SSC and 0.05% (w/v) sodium pyrophosphate for 5 min at 40°C and then for 10 min each at 45°C, 50°C, 55°C, and 60°C. Three successive rounds of screening yielded four positive clones, whose inserts were recovered by in vivo excision into pBluescript SK(−) (Stratagene). When partial sequencing revealed that one clone exhibited high sequence identity with known PH internal peptide sequences, its insert (designated Ph1) was subjected to automated double-strand sequencing.
Isolation of Full-Length Ah1 cDNA
An unamplified λTriplEx2 cDNA library (105 pfu), constructed from mRNA isolated from immature black cherry seeds, was screened with a 488-bp AH cDNA fragment (Zheng and Poulton, 1995) previously labeled with digoxigenin by random priming (Roche Applied Science). Two rounds of screening yielded 32 positive clones with inserts larger than 1.8 kb. These clones were selected, converted into pTriplEx2 plasmids, and sequenced in both directions. This approach yielded a full-length Ah1 cDNA clone, whose insert was 1,915 bp in length.
Sequence Analyses and Molecular Modeling
Homology searches were undertaken using the BLAST network server of the National Center for Biotechnology Information (Altschul et al., 1990). Nucleotide and deduced amino acid sequences were analyzed with the University of Wisconsin Genetics Computer Group software package (Devereux et al., 1984). Amino acid sequences were aligned by the Clustal V program. Phylogenetic analysis of the aligned sequences was performed using the heuristic option of PAUP* (Saitou and Nei, 1987). Support for the branches was determined by bootstrap analysis (1,000 replicates). Computer modeling of PH1 and AH1 was undertaken with the RasMol V2.6 program (Sayle, 1996) using data generated by the Swiss Model server (http://www.expasy.ch/swissMod/SWISS-MODEL.html).
Isolation and Characterization of Genomic Sequences Corresponding to cDNA Clones Ah1 and Ph1 through Ph5
Genomic DNA was isolated from young cherry leaves using the Floraclean Kit following manufacturer's instructions (Qbiogene, Carlsbad, CA). Genomic sequences corresponding to the cDNAs Ah1 and Ph1 through Ph5 were obtained by LD-PCR amplification in 50-μL reactions containing 1 μg of genomic DNA template, 0.2 mm dNTP Mix, 50× Advantage cDNA Polymerase Mix (BD Biosciences Clontech), 0.1 μm each of the respective gene-specific primers (Table IV), and 1× PCR buffer. Reactions were cycled 18 times for 94°C at 30 s and 5 min at 68°C and, in each case, yielded a single product of approximately 3.5 kb. After GENECLEAN II purification (Qbiogene), the PCR products were ligated into the pCR-4 TOPO vector before transformation into E. coli Top10 competent cells (Invitrogen, Carlsbad, CA) and sequencing. The intron-exon organization of these PH and AH genes was determined by comparison of their respective genomic and cDNA sequences.
Functional Expression in Pichia pastoris of β-Glucosidases Encoded by Ah1, Ph2, and Ph4 cDNAs
Construction of Expression Vectors and P. pastoris Transformation
The full-length Ah1 cDNA clone (in pTriplEx2) was used as template in PCR amplification using the SMART 3 Oligonucleotide and CDS III/3′ PCR primers (BD Biosciences Clontech) as forward and reverse primers under conditions previously described for LD-PCR from genomic DNA. The amplified product was purified using the GENECLEAN II kit and cloned into the pCR4-TOPO plasmid (Invitrogen) following the supplier's instructions. After partial sequencing using T7 and T3 primers, the full-length Ah1 cDNA was excised by EcoRI digestion (1U EcoRI, 37°C, 10 min), purified, and cloned into the E. coli/P. pastoris shuttle vector pPICZA (Invitrogen; previously digested with EcoRI and dephosphorylated). The ligation product was transformed into competent E. coli Top10 cells that were cultured on Luria-Bertani broth plates containing zeocin. Plasmids isolated from 10 zeocin-resistant transformants by standard methods were analyzed by restriction enzyme digestion and partial sequencing to identify clones having the desired insert direction. Subsequently, the Ah1-pPICZA plasmid DNA was isolated, purified, and linearized with the restriction enzyme Pme1 to allow integration of the vector DNA into the P. pastoris chromosome by homologous recombination. After transforming P. pastoris host cells X-33 and KM71H using the Easycomp kit (Invitrogen), transformants were cultured in the dark on yeast extract peptone dextrose plates containing 100 μg mL−1 Zeocin for 2 to 4 d at 30°C. Construction of pPICZA vectors containing full-length Ph2 or Ph4 cDNAs and their transformation into competent P. pastoris KM71H cells were accomplished by identical means.
Expression of Recombinant AH1, PH2, and PH4 Proteins
Single zeocin-resistant colonies were selected to inoculate 10 mL of buffered glycerol-complex medium in 50-mL conical tubes. Cultures were grown in a shaking incubator (300 rpm) at 30°C until the OD600 reached 2 to 6 (approximately 18 h). Cells were harvested by centrifugation at 3,000 rpm for 5 min and resuspended in buffered methanol-complex medium at an OD600 of 1.0. Cultures were maintained under the same conditions except for addition of methanol every 24 h to induce expression. At 0, 24, 48, 72, 96, and 120 h after induction, aliquots (1 mL) were removed for analysis. Supernatants and cell pellets obtained by centrifugation for 5 min at 14,000 rpm were stored at −80°C for subsequent enzyme assays. Control cultures (KM71H cells transformed with empty pPICZA vector) were inoculated and induced by identical methods. Culture supernatants were used without further purification in assays of PNPGase, AH, and PH activities as previously described (Kuroki and Poulton, 1986). Cell lysates were obtained by extracting cells by shearing with 0.5-mm glass beads followed by centrifugation.
Site-Directed Mutagenesis of AH1
Mutagenesis was performed by the QuikChange Site-Directed Mutagenesis kit (Stratagene) using the synthetic mutagenic oligonucleotides 5′-GCCATATACCTTTAGTAGCCATGGTTATGCATACGGGGTCC-3′, 5′-GTAGCAGTGGTTATGCAATCGGGGTCCATGCACCAGGACG-3′, and 5′-GGTCCAATGGCTGCTTCAGACTGGTTATATGTTTATCCC-3′ for construction of AH1 mutants SH, YI, and GD, respectively. The double mutants HI, HD, and ID, as well as the triple mutant HID, were obtained using the same primers by mutagenesis of cDNAs from appropriate single mutants. Desired mutations were verified by partial sequencing. The culture, induction, and enzymatic assays of the mutant lines were undertaken as described for wild-type recombinant AH1 expression.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from third party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
ACKNOWLEDGMENTS
We thank Drs. Asim Esen, Chi-Lien Cheng, Erin Irish, and Ming-Che Shih for their helpful suggestions and careful reviews of the manuscript. We are grateful to Dr. Debashish Bhattacharya for assistance with the phylogenetic analysis. We acknowledge the expert technical assistance of the University of Iowa Protein Structure and DNA Sequencing Facilities.
Footnotes
This work was supported by the National Science Foundation (grant nos. IBN 9630935 and MCB 9723302).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010863.
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Babcock GD, Esen A. Substrate specificity of maize β-glucosidase. Plant Sci. 1994;101:31–39. [Google Scholar]
- Barrett T, Suresh CG, Tolley SP, Dodson EJ, Hughes MA. The crystal structure of a cyanogenic β-glucosidase from white clover, a family 1 glycosyl hydrolase. Structure. 1995;3:951–960. doi: 10.1016/s0969-2126(01)00229-5. [DOI] [PubMed] [Google Scholar]
- Béguin P. Molecular biology of cellulose degradation. Annu Rev Microbiol. 1990;44:219–248. doi: 10.1146/annurev.mi.44.100190.001251. [DOI] [PubMed] [Google Scholar]
- Branden C, Tooze J. Introduction to Protein Structure. New York: Garland Publishing, Inc.; 1991. [Google Scholar]
- Brown JW. A catalogue of splice junction and putative branch point sequences from plant introns. Nucleic Acids Res. 1986;14:9549–9559. doi: 10.1093/nar/14.24.9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn EE. Cyanide and cyanogenic glycosides. In: Rosenthal GA, Janzen DH, editors. Herbivores, Their Interactions with Secondary Plant Metabolites. New York: Academic Press; 1979. pp. 387–412. [Google Scholar]
- Conn EE. β-Glycosidases in plants: substrate specificity. In: Esen A, editor. β-Glucosidases: Biochemistry and Molecular Biology, ACS Symposium Series 533. Washington, DC: American Chemical Society; 1993. pp. 15–26. [Google Scholar]
- Cregg JM, Vedvick TS, Raschke WC. Recent advances in the expression of foreign genes in Pichia pastoris. Bio/Technology. 1993;11:905–910. doi: 10.1038/nbt0893-905. [DOI] [PubMed] [Google Scholar]
- Czjzek M, Cicek M, Zamboni V, Bevan DR, Henrissat B, Esen A. The mechanism of substrate (aglycone) specificity in β-glucosidases is revealed by crystal structures of mutant maize β-glucosidase-DIMBOA, -DIMBOAGlc, and -dhurrin complexes. Proc Natl Acad Sci USA. 2000;97:13555–13560. doi: 10.1073/pnas.97.25.13555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlington CD. Studies in Prunus I, II. J Genet. 1928;19:213–256. [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmawardhana DP, Ellis BE, Carlson JE. A β-glucosidase from lodgepole pine specific for the lignin precursor coniferin. Plant Physiol. 1995;107:331–339. doi: 10.1104/pp.107.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esen A. β-Glucosidases: Biochemistry and Molecular Biology, ACS Symposium Series 533. Washington, DC: American Chemical Society; 1993. [Google Scholar]
- Esen A, Bandaranayake H. Insertional polymorphism in introns 4 and 10 of the maize beta-glucosidase gene glu1. Genome. 1998;41:597–604. doi: 10.1139/g98-061. [DOI] [PubMed] [Google Scholar]
- Falk A, Rask L. Expression of a zeatin-O-glucoside-degrading β-glucosidase in Brassica napus. Plant Physiol. 1995;108:1369–1377. doi: 10.1104/pp.108.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler T. Deletion of the Trichoderma reesei β-glucosidase gene, bgl1. In: Esen A, editor. β-Glucosidases: Biochemistry and Molecular Biology, ACS Symposium Series 533. Washington, DC: American Chemical Society; 1993. pp. 56–65. [Google Scholar]
- Gerstner E, Steffen A, Pfeil E. Zur Entwicklung der multiplen Formen des Flavin-Enzyms d-Oxynitrilase bei Prunus spinosa. Naturwissenschaften. 1971;58:269–270. [Google Scholar]
- Grabowski GA, Berg-Fussman A, Grace M. Molecular biology and enzymology of human acid β-glucosidase. In: Esen A, editor. β-Glucosidases: Biochemistry and Molecular Biology, ACS Symposium Series 533. Washington, DC: American Chemical Society; 1993. pp. 66–82. [Google Scholar]
- Grace ME, Grabowski GA. Human β-glucosidase: glucosylation is required for catalytic activity. Biochem Biophys Res Commun. 1990;168:771–777. doi: 10.1016/0006-291x(90)92388-g. [DOI] [PubMed] [Google Scholar]
- Haisman DR, Knight DJ, Ellis MJ. The electrophoretic separation of the β-glucosidases of almond “emulsin.”. Phytochem. 1967;6:1501–1505. [Google Scholar]
- He S, Withers SG. Assignment of sweet almond β-glucosidase as a family 1 glycosidase and identification of its active site nucleophile. J Biol Chem. 1997;272:24864–24867. doi: 10.1074/jbc.272.40.24864. [DOI] [PubMed] [Google Scholar]
- Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B, Bairoch A. Updating the sequence-based classification of glycosyl hydrolases. Biochem J. 1996;316:695–696. doi: 10.1042/bj3160695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B, Davies GJ. Glycoside hydrolases and glycosyltransferases. Families, modules, and implications for genomics. Plant Physiol. 2000;124:1515–1519. doi: 10.1104/pp.124.4.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hösel W, Barz W. β-Glucosidases from Cicer arietinum L. Purification and properties of isoflavone-7-O-glucoside-specific β-glucosidases. Eur J Biochem. 1975;57:607–616. doi: 10.1111/j.1432-1033.1975.tb02336.x. [DOI] [PubMed] [Google Scholar]
- Hösel W, Conn EE. The aglycone specificity of plant β-glycosidases. Trends Biochem Sci. 1982;7:219–221. [Google Scholar]
- Hösel W, Nahrstedt A. Spezifische Glucosidasen fuer das Cyanoglucosid Triglochinin: Reinigung und Charakterisierung von β-Glucosidasen aus Alocasia macrorrhiza Schott. Hoppe-Seyler's Z Physiol Chem. 1975;356:1265–1275. [PubMed] [Google Scholar]
- Hösel W, Tober I, Eklund SH, Conn EE. Characterization of β-glucosidases with high specificity for the cyanogenic glucoside dhurrin in Sorghum bicolor (L.) Moench seedlings. Arch Biochem Biophys. 1987;252:152–162. doi: 10.1016/0003-9861(87)90019-1. [DOI] [PubMed] [Google Scholar]
- Hösel W, Todenhagen R. Characterization of a β-glucosidase from Glycine max which hydrolyses coniferin and syringin. Phytochemistry. 1980;19:331–339. [Google Scholar]
- Hu Z, Poulton JE. Molecular analysis of (R)-(+)-mandelonitrile lyase microheterogeneity in black cherry. Plant Physiol. 1999;119:1535–1546. doi: 10.1104/pp.119.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes MA. Molecular genetics of plant cyanogenic β-glucosidases. In: Esen A, editor. β-Glucosidases: Biochemistry and Molecular Biology, ACS Symposium Series 533. Washington, DC: American Chemical Society; 1993. pp. 153–169. [Google Scholar]
- Hughes MA, Brown K, Pancoro A, Murray BS, Oxtoby E, Hughes J. A molecular and biochemical analysis of the structure of the cyanogenic beta-glucosidase (linamarase) from cassava (Manihot esculenta Crantz) Arch Biochem Biophys. 1992;295:273–279. doi: 10.1016/0003-9861(92)90518-2. [DOI] [PubMed] [Google Scholar]
- Inoue K, Ebizuka Y. Purification and characterization of a beta-glucosidase which converts furostanol glycosides to spirostanol glycosides from Costus speciosus. Adv Exp Med Biol. 1996;404:57–69. doi: 10.1007/978-1-4899-1367-8_6. [DOI] [PubMed] [Google Scholar]
- Kingsbury JM. Poisonous Plants of the United States and Canada. Englewood Cliffs, NJ: Prentice-Hall; 1964. pp. 365–366. [Google Scholar]
- Kosuge T, Conn EE. The metabolism of aromatic compounds in higher plants: III. The β-glucosides of o-coumaric, coumarinic, and melilotic acids. J Biol Chem. 1961;236:1617–1621. [PubMed] [Google Scholar]
- Kuroki GW, Poulton JE. Comparison of kinetic and molecular properties of two forms of amygdalin hydrolase from black cherry (Prunus serotina Ehrh.) seeds. Arch Biochem Biophys. 1986;247:433–439. doi: 10.1016/0003-9861(86)90603-x. [DOI] [PubMed] [Google Scholar]
- Kuroki GW, Poulton JE. Isolation and characterization of multiple forms of prunasin hydrolase from black cherry (Prunus serotina Ehrh.) seeds. Arch Biochem Biophys. 1987;255:19–26. doi: 10.1016/0003-9861(87)90290-6. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leah R, Kigel J, Svendsen I, Mundy J. Biochemical and molecular characterization of a barley seed β-glucosidase. J Biol Chem. 1995;270:15789–15797. doi: 10.1074/jbc.270.26.15789. [DOI] [PubMed] [Google Scholar]
- Legler G, Harder A. Amino acid sequence at the active site of β-glucosidase A from bitter almonds. Biochim Biophys Acta. 1978;524:102–108. doi: 10.1016/0005-2744(78)90108-0. [DOI] [PubMed] [Google Scholar]
- Li CP, Swain E, Poulton JE. Prunus serotina amygdalin hydrolase and prunasin hydrolase: purification, N-terminal sequencing and antibody production. Plant Physiol. 1992;100:282–290. doi: 10.1104/pp.100.1.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddle S, Keresztessy Z, Hughes J, Hughes MA. A genomic cyanogenic beta-glucosidase gene from cassava (accession no. X94986) Plant Physiol. 1998;117:1526. [Google Scholar]
- Malboobi MA, Lefebvre DD. A phosphate-starvation inducible beta-glucosidase gene (psr3.2) isolated from Arabidopsis thaliana is a member of a distinct subfamily of the BGA family. Plant Mol Biol. 1997;34:57–68. doi: 10.1023/a:1005865406382. [DOI] [PubMed] [Google Scholar]
- Matsudaira P. A Practical Guide to Protein and Peptide Purification for Microsequencing. Ed 2. San Diego: Academic Press; 1993. [Google Scholar]
- Maynard CA, Kavanagh K, Fuerukranz H, Drew AP. Black cherry (Prunus serotina Ehrh.) In: Bajaj YPS, editor. Biotechnology in Agricultural Forestry. Vol. 16. Berlin: Springer-Verlag; 1991. pp. 3–22. [Google Scholar]
- McCarter JD, Withers SG. Mechanisms of enzymatic glycoside hydrolysis. Curr Opin Struct Biol. 1994;4:885–892. doi: 10.1016/0959-440x(94)90271-2. [DOI] [PubMed] [Google Scholar]
- McMahon JM, White WLB, Sayre RT. Cyanogenesis in cassava (Manihot esculenta Crantz) J Exp Bot. 1995;46:731–741. [Google Scholar]
- Poulton JE. Enzymology of cyanogenesis in rosaceous stone fruits. In: Esen A, editor. β-Glucosidases: Biochemistry and Molecular Biology, ACS Symposium Series No. 533. Washington, DC: American Chemical Society; 1993. pp. 170–190. [Google Scholar]
- Poulton JE, Li CP. Tissue level compartmentation of (R)-amygdalin and amygdalin hydrolase prevents large-scale cyanogenesis in undamaged Prunus seeds. Plant Physiol. 1994;104:29–35. doi: 10.1104/pp.104.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rask L, Andréasson E, Ekbom B, Eriksson S, Pontoppidan B, Meijer J. Myrosinase: gene family evolution and herbivore defense in Brassicaceae. Plant Mol Biol. 2000;42:93–113. [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbour-joining method: a new method for constructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayle R. RasMol V2.6. Uxbridge, UK: GlaxoSmithKline; 1996. [Google Scholar]
- Selmar D, Lieberei R, Biehl B, Vogt J. Hevea linamarase: a nonspecific glucosidase. Plant Physiol. 1987;83:557–563. doi: 10.1104/pp.83.3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain E, Li CP, Poulton JE. Tissue and subcellular localization of enzymes catabolizing (R)-amygdalin in mature Prunus serotina seeds. Plant Physiol. 1992;100:291–300. doi: 10.1104/pp.100.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain E, Poulton JE. Immunocytochemical localization of prunasin hydrolase and mandelonitrile lyase in stems and leaves of Prunus serotina. Plant Physiol. 1994;106:1285–1291. doi: 10.1104/pp.106.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakarchuk WW, Greenberg NM, Kilburn DG, Miller RC, Jr, Warren RAJ. Structure and transcription analysis of the gene encoding a cellobiase from Agrobacterium sp. strain ATCC 21400. J Bacteriol. 1988;170:301–307. doi: 10.1128/jb.170.1.301-307.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Ito A, Takada Y, Ninomiya C, Kakizaki T, Takahata Y, Hatakeyama K, Hinata K, Suzuki G, Takasaki T et al. Highly divergent sequences of the pollen self-incompatibility (S) gene in class-I S haplotypes of Brassica campestris (syn. rapa) L. FEBS Lett. 2000;473:139–144. doi: 10.1016/s0014-5793(00)01514-3. [DOI] [PubMed] [Google Scholar]
- Weeden NF. Evolution of plant isozymes. In: Tanksley SD, Orton TJ, editors. Developments in Plant Genetics and Breeding: Isozymes in Plant Genetics and Breeding, Part A. New York: Elsevier; 1983. pp. 175–205. [Google Scholar]
- Wiersma PA, Fils-Lycaon BR. Molecular cloning and nucleotide sequence (accession no. U39228) of a beta-glucosidase cDNA from ripening sweet cherry fruit. Plant Gene Register PGR95–127. Plant Physiol. 1996;110:337. [Google Scholar]
- Xue J, Rask L. The unusual 5′ splicing border GC is used in myrosinase genes of the Brassicaceae. Plant Mol Biol. 1995;29:167–171. doi: 10.1007/BF00019128. [DOI] [PubMed] [Google Scholar]
- Zheng L, Poulton JE. Temporal and spatial expression of amygdalin hydrolase and (R)-(+)-mandelonitrile lyase in black cherry (Prunus serotina Ehrh.) seeds. Plant Physiol. 1995;109:31–39. doi: 10.1104/pp.109.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]