Abstract
The spontaneous occurrence of resistance to the herbicide glyphosate in weed species has been an extremely infrequent event, despite over 20 years of extensive use. Recently, a glyphosate-resistant biotype of goosegrass (Eleusine indica) was identified in Malaysia exhibiting an LD50 value approximately 2- to 4-fold greater than the sensitive biotype collected from the same region. A comparison of the inhibition of 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) activity by glyphosate in extracts prepared from the resistant (R) and sensitive (S) biotypes revealed an approximately 5-fold higher IC50(glyphosate) for the (R) biotype. Sequence comparisons of the predicted EPSPS mature protein coding regions from both biotypes revealed four single-nucleotide differences, two of which result in amino acid changes. One of these changes, a proline to serine substitution at position 106 in the (R) biotype, corresponds to a substitution previously identified in a glyphosate-insensitive EPSPS enzyme from Salmonella typhimurium. Kinetic data generated for the recombinant enzymes suggests that the second substitution identified in the (R) EPSPS does not contribute significantly to its reduced glyphosate sensitivity. Escherichia coli aroA− (EPSPS deficient) strains expressing the mature EPSPS enzyme from the (R) biotype exhibited an approximately 3-fold increase in glyphosate tolerance relative to strains expressing the mature EPSPS from the (S) biotype. These results provide the first evidence for an altered EPSPS enzyme as an underlying component of evolved glyphosate resistance in any plant species.
Spontaneous resistance is generally thought to occur within weed populations as a consequence of the intense selective pressure exerted by a lack of diversity in weed management practices (Gressel and Segel, 1978). Factors such as extended residual soil activity, lack of rotation to other herbicidal modes of action, and specific managerial practices further discriminate between resistant and susceptible individuals within a population (Powles and Holtum, 1994). In addition, the rate and severity at which resistant weed infestations occur is influenced by genetic and ecophysiological determinants such as the mode of inheritance of a given resistance mechanism, the absence or presence of fitness penalties associated with resistance, as well as the reproductive habit of a given weed species (Gressel and Segel, 1978; Jasieniuk et al., 1996; Gardner et al., 1998). To date, there exist more than 249 herbicide-resistant weed biotypes distributed among 52 different countries, involving at least 17 different herbicide modes of action (Heap, 2001).
Goosegrass (Eleusine indica) is an annual, self-pollinating, diploid grass species of undetermined origin possessing a relatively small genome size of approximately 8.03 × 108 bp (Ganeshaiah and Umashaanker, 1982; Mysore and Baird, 1997). The species' habitat is wide-ranging and includes south Asia, eastern and southern Africa, and North America. Although goosegrass is used as animal feed and a source of grain in some regions, it is considered one of the five “world's worst weeds” and has been reported to be a problem weed in 46 different crop species in more than 60 countries (Holm et al., 1977). Compounding this problem is the fact that the species has demonstrated the capacity to evolve resistance against dinitroanilines, acetohydroxyacid synthase inhibitors such as imazapyr, and acetyl-CoA carboxylase inhibitors such as fluazifop, all important herbicides for maintaining its control within crops (Mudge et al., 1984; Marshall et al., 1994; Heap, 1997).
Glyphosate [N-(phosphonomethyl) Gly] is the active ingredient of the most extensively used foliar-applied, broad-spectrum herbicide, Roundup (Malik et al., 1989). This herbicide has demonstrated efficacy against the majority of annual and perennial grasses and broad-leaved weeds (Bradshaw et al., 1997). The primary mode of action in planta for glyphosate is competitive inhibition of the enzyme 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS; E.C. 2.5.1.19), which catalyzes the penultimate step of the prechorismate part of the plastid-localized shikimate pathway (Steinrücken and Amrhein, 1980; Franz et al., 1997; Gruys and Sikorski, 1999). In plants, as much as 20% of all fixed carbon flows through the shikimate pathway leading to the formation of the aromatic amino acids Tyr, Phe, and Trp, as well as tetrahydrofolate, ubiquinone, and vitamins K and E (Haslam, 1993; Franz et al., 1997). The aromatic amino acids, in turn, serve as precursors for an array of secondary metabolites including lignin, flavonoids, and alkaloids (Herrmann, 1995). The shikimate pathway occurs exclusively in plants and microorganisms, and this, coupled with the specificity of glyphosate as an inhibitor of EPSPS, contributes in large part to glyphosate's lack of toxicity to animals.
Evaluation of resistance risk criteria along with current and historical use data have led to the suggestion that glyphosate is a herbicide at low risk for the evolution of weed resistance (e.g. Benbrook, 1991; Bradshaw et al., 1997). This notion has been supported in theory by saturation mutagenesis experiments conducted with Arabidopsis, as well as in vitro mutagenesis experiments performed using a petunia (Petunia hybrida) EPSPS cDNA (Haughn and Somerville, 1987; Padgette et al., 1996; Bradshaw et al., 1997; Baerson et al., 1999). Nevertheless, recent reports have documented the appearance of glyphosate-resistant Lolium rigidum in Australia (Pratley et al., 1996; Powles et al., 1998), and glyphosate-resistant biotypes of goosegrass in Malaysia (Tran et al., 1999; Lee and Ngim, 2000). In all but one instance (Lee and Ngim, 2000), resistant weed biotypes occurred in areas subjected to repeated applications of glyphosate for at least 10 years before their emergence.
In this paper, we examine and compare expression levels, sensitivity to glyphosate, as well as describe cloning experiments and kinetic analysis for the herbicide target site, EPSPS, in glyphosate-resistant and -sensitive goosegrass biotypes collected in Malaysia. We have obtained evidence that a simple amino acid substitution in the EPSPS enzyme expressed in the resistant biotype contributes to the underlying basis for resistance. This is the first report to our knowledge for a glyphosate resistance mechanism involving an altered EPSPS enzyme to have evolved in any plant species.
RESULTS AND DISCUSSION
EPSPS Expression Levels and Genomic Characterization of the Sensitive (S) and Resistant (R) Goosegrass Biotypes
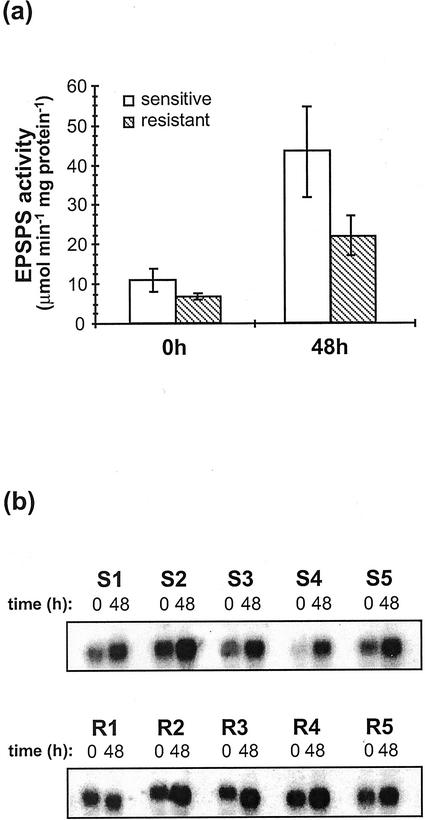
To examine the possibility that the glyphosate resistance mechanism of goosegrass involves overexpression of EPSPS, both enzyme-specific activity and steady-state mRNA accumulation levels were determined for (R) and (S) biotype individuals (Fig. 1, a and b). Because target site overexpression could potentially involve differences in basal expression levels as well as differences in the capacity to respond to herbicide challenge via feedback regulation, both possibilities were examined. Two clones each were first generated from representative (R) and (S) individuals by removing approximately 10 to 20 tillers and subtending root mass from mature plants, which were then repotted. All clones were then subjected to sublethal glyphosate spray applications equivalent to 0.5 kg a.e. ha−1 for (S) clones, and 2.0 kg a.e. ha−1 for (R) clones. Crown tissues, which contained the highest levels of extractable EPSPS activity among tissues examined (data not shown), were then harvested at t = 0 and t = 48 h postapplication for analysis. In addition, control experiments involving clones generated from both biotypes were performed using surfactant-only treatments to ensure that any differences observed were not due to abiotic stress imposed by the surfactant or other variables not specific to glyphosate-treated plants (not shown).
Figure 1.
Basal and induced EPSPS expression levels in glyphosate-sensitive and -resistant goosegrass individuals. Two clones were generated from each representative plant; (S) clones were spray-treated with 0.5 kg acid equivalent (a.e.) ha−1 glyphosate and (R) clones were treated with 2 kg a.e. ha−1 glyphosate. Clones were harvested immediately (time [t] = 0 h) or 48 h post-treatment (t = 48 h), then independently analyzed for EPSPS activity and mRNA levels. a, EPSPS activity levels. Extracts prepared from crown tissues were radiometrically assayed for EPSPS activity (see “Materials and Methods”). Specific activities were calculated based on extract protein concentration and all assays were performed in triplicate. Each bar represents the mean activity observed within five individuals from each biotype. Error bars indicate sds. b, RNA-blot analysis. Ten micrograms of total RNA, isolated from crown tissues, was loaded per lane, then size fractionated on 1.0% (w/v) agarose gels containing 0.66 m formaldehyde and transferred to nylon membranes. Blots were hybridized with an L. rigidum 32P-labeled EPSPS cDNA. S1 through S5 samples were isolated from duplicate clones derived from five different (S) individuals; R1 through R5 samples were isolated from duplicate clones derived from five different (R) individuals.
The comparison of EPSPS activity levels between the two biotypes indicated that resistance was not associated with target enzyme overexpression (Fig. 1a). In fact, basal activity levels (t = 0 h) appeared slightly higher (approximately 35%) in the (S) biotype as compared with the (R) biotype. In response to sublethal glyphosate applications, significant increases in EPSPS activity levels were observed in both biotypes at t = 48 h. The fold induction observed was similar for (S) and (R) individuals, increasing approximately 4-fold in (S) plants and slightly less than 3-fold in the (R) biotype. Collectively, these results indicate that the resistance phenotype is not associated with increased levels of either basal or glyphosate-induced EPSPS activity.
Northern analyses were also performed using the same crown tissue samples analyzed for determining EPSPS enzyme activity levels (Fig. 1b). Total cellular RNAs were prepared from five (S) and five (R) individuals (S1–S5 and R1–R5, Fig. 1b), then probed using a radiolabeled L. rigidum EPSPS cDNA (see “Materials and Methods”). As was the case for enzyme activity levels, these data did not indicate elevated basal or glyphosate-induced EPSPS steady-state mRNA levels in the (R) as compared with the (S) biotype. Although basal (t = 0 h) mRNA levels were slightly higher in the (R) plants analyzed for this study, this was not observed in all cases. Increases in EPSPS mRNA levels were seen for both biotypes 48 h after exposure to glyphosate; however, overall levels were similar in (R) and (S) individuals.
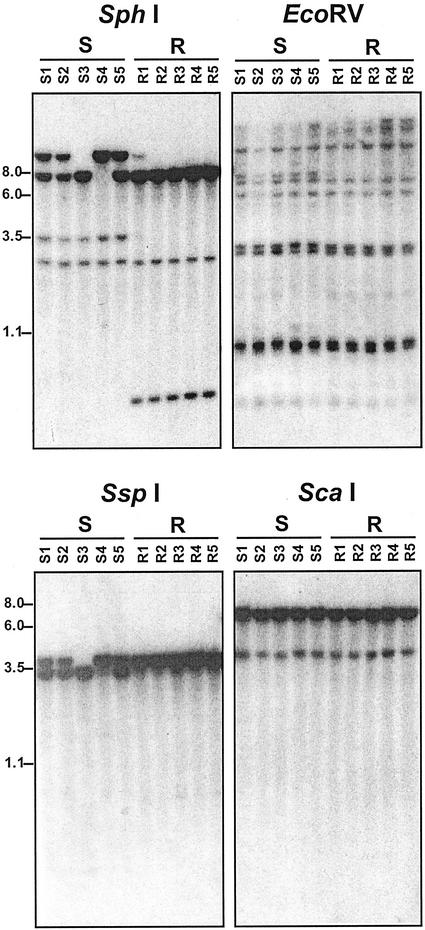
Because specific amplification of EPSPS genes has frequently been associated with resistance to glyphosate in selected cell lines (e.g. Shah et al., 1986; Suh et al., 1993; Jones et al., 1996), Southern analyses were also performed using genomic DNAs isolated from five (S) and five (R) individuals (Fig. 2). In all individuals analyzed, the data indicate that the EPSPS gene families from the two biotypes are of similar complexity, comprising either small gene families or possibly a single gene (Fig. 2). In particular, the data generated from SspI and ScaI digests are consistent with EPSPS being represented by a single locus in goosegrass, with two alleles segregating within the (S) population. This is also consistent with the data generated from SphI digests, which reveal EPSPS-related polymorphisms among different (S) individuals, but not (R) individuals. Furthermore, the lack of detectable polymorphisms from ScaI digests allows for unambiguous comparison of (R) and (S) plants, revealing that the two biotypes contain identical EPSPS gene copy numbers.
Figure 2.
DNA-blot analysis of glyphosate-sensitive and -resistant goosegrass individuals. Ten micrograms of Genomic DNA from five sensitive individuals (samples S1–S5) and five resistant individuals (samples R1–R5) were digested with either EcoRV, SphI, SspI, or ScaI as indicated above, then size fractionated on 0.8% (w/v) agarose gels and transferred to nylon membranes. Blots were then hybridized with an L. rigidum 32P-labeled EPSPS cDNA, washed at high stringency (see “Materials and Methods”), then subjected to autoradiography.
Comparison of EPSPS Sensitivity to Glyphosate for the (S) and (R) Biotypes
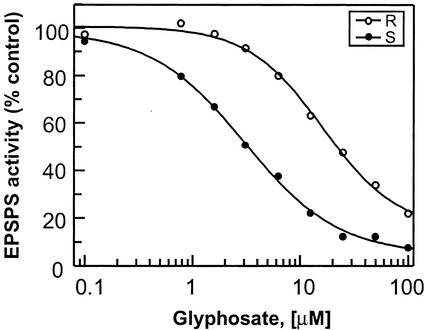
Previous mutagenesis studies have demonstrated the ability to generate glyphosate-insensitive EPSPS variants that differ from their wild-type counterparts by only a single amino acid (for review, see Padgette et al., 1996). Therefore, the possibility could not be excluded that a similar mutation occurred spontaneously in goosegrass, leading to the emergence of a new glyphosate-resistant biotype. To address this question, crown tissue extracts prepared from (R) and (S) individuals were compared for inhibition of [14C]phosphoenolpyruvate (PEP) conversion to [14C]5-enolpyruvylshikimate-3-phosphate (EPSP) in the presence of varying amounts of glyphosate (Fig. 3). At 0.1 mm glyphosate concentrations, [14C]PEP turnover in extracts prepared from (S) biotype plants was inhibited approximately 92%, whereas the (R) biotype extracts retained approximately 22% activity at the same concentrations. The glyphosate concentrations required to inhibit [14C]PEP turnover by 50% (IC50 values) were also calculated based on nonlinear regression analysis of the inhibition curves (see “Materials and Methods”). By this analysis, the IC50 values for EPSPS activities from the (S) and (R) biotypes were determined to be approximately 3.0 and 16.0 μm, respectively. These data indicate the presence of an EPSPS enzyme expressed in the (R) biotype exhibiting reduced sensitivity to glyphosate.
Figure 3.
IC50 (glyphosate) determinations for the (R) and (S) biotype EPSPS activities. Radiometric EPSPS assays were performed on (R) and (S) biotype crown tissue extracts in the presence of glyphosate at concentrations ranging from 0.1 to 100 μm. Inhibition curves and IC50 (inflection point) values were generated by nonlinear regression analysis using the GraFit software package (Leatherbarrow, 1998).
Comparison of EPSPS cDNAs for the (S) and (R) Goosegrass Biotypes
To examine the molecular basis for the differential glyphosate sensitivity of the activities detected in crude extracts (Fig. 3), EPSPS cDNAs were isolated from both biotypes by reverse transcriptase (RT)-PCR, then compared by sequence analysis (Fig. 4). Degenerate oligonucleotides were designed for amplification of EPSPS coding regions based on a comparison of known amino acid sequences for plant EPSPS enzymes (see “Materials and Methods”). In plants, EPSPS is synthesized as a cytosolic precursor possessing an N-terminal chloroplast transit peptide that is removed by proteolysis during translocation, resulting in the mature form (Della-Cioppa et al., 1986). The oligonucleotides Ei2a and Ei9a (Fig. 4) were initially chosen for these experiments because the approximately 1.0-kb RT-PCR products generated would represent the majority of the mature protein coding sequence (minus 63 N-terminal and 36 C-terminal amino acids; based on the published full-length corn EPSPS sequence), and encompass critical residues identified in all known glyphosate-resistant EPSPS variants (Padgette et al., 1996; Franz et al., 1997; Lebrun et al., 1997).
Figure 4.
Sequence comparison of EPSPS cDNAs isolated from the (S) and (R) goosegrass biotypes. The nucleotide sequence of the (S) EPSPS is indicated in the line below only where nucleotide differences occur between the two biotypes. The translation termination codon (TAA) is marked with an asterisk. Single nucleotide differences and corresponding amino acid substitutions are indicated in bold face. PCR primer sites are indicated by arrows. The deduced amino acid sequence of the (R) EPSPS mature protein coding region is shown above the nucleotide sequence, which has approximately 98% identity with the EPSPS enzyme from corn (Zea mays; GenBank accession no. X63374) and a predicted Mr of 47,402. A motif conserved in all plant and most bacterial EPSPS enzymes is boxed in black. Amino acid assignments referred to in the text are based on the plant EPSPS numbering system used by Padgette et al. (1996). EMBL accession numbers for the (R) and (S) EPSPS sequences are AJ417033 and AJ417034, respectively.
To minimize ambiguity due to potential thermostable polymerase-induced errors, RT-PCR products were cloned from multiple, pooled reactions, then clones were randomly selected for sequence analysis. In addition, a proofreading polymerase mixture was used for all amplifications. Single nucleotide polymorphisms within a biotype that occurred in only one of the 20 clones analyzed were not included because these could potentially have arisen during amplification. Twenty clones were sequenced for each biotype and the comparison is summarized in Figure 4.
Four single-nucleotide differences were identified between the (R) and (S) biotype, which were consistent among all the cDNAs analyzed. No other polymorphisms were detected that occurred in more than one clone, indicating that a single mRNA species predominates in goosegrass crown tissues. Two of the nucleotide differences detected, A225 (S biotype) → G (R biotype) and A651 (S biotype) → G (R biotype), correspond to silent changes in codon wobble positions for Ala-74 and Leu-216, respectively. The remaining two nucleotide changes result in amino acid substitutions within the predicted mature EPSPS protein from the (R) biotype. C319 (S biotype) → T (R biotype) replaces Pro-106 with Ser-106 in the (R) biotype, substituting a polar residue (Ser) for a helix-destabilizing, nonpolar residue (Pro). C1145 (S biotype) → T (R biotype) replaces Pro-381 with Leu-381 in the (R) biotype, causing a nonconservative change of Pro for the hydrophobic Leu residue. The identification of a Pro-106 → Ser substitution in the (R) biotype EPSPS is of particular significance because the same amino acid substitution at the corresponding residue in the Salmonella typhimurium aroA gene (EPSPS) was previously determined to be the genetic basis of glyphosate resistance in an ethyl methanesulfonate (EMS)-mutagenized strain (Comai et al., 1983; Stalker et al., 1985). More recent studies have shown that the analogous change made to the petunia EPSPS enzyme via site-directed mutagenesis results in an approximately 7.5-fold increase in Ki(app)(glyphosate), reflecting a significant reduction in affinity for glyphosate (Padgette et al., 1991).
Differential Growth Response to Glyphosate of Escherichia coli aroA− Cells Expressing the (R) or (S) Biotype EPSPS
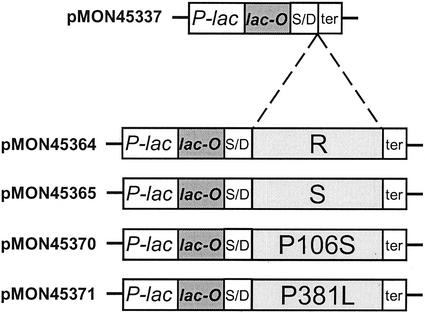
To determine if the EPSPS enzyme from the (R) biotype is able to confer glyphosate resistance in intact cells, liquid phase growth assays were utilized to directly compare the effects of glyphosate on E. coli transformants expressing the (R) or (S) EPSPS. To generate expression cassettes, the remainder of the mature EPSPS coding sequence from goosegrass was first determined using 5′- and 3′-RACE, and RT-PCR (Fig. 4; see also “Materials and Methods”). Gene-specific primers were designed based on sequence data obtained from the initial RT-PCR products (described above), then the remainder of the mature protein sequence was deduced from the resulting 5′- and 3′-RACE products. The high degree of sequence identity observed between goosegrass and corn EPSPS (Lebrun et al., 1997) allowed for the identification of a homologous Ala residue as the putative N terminus for the mature goosegrass enzyme. Two oligonucleotides were then designed for additional RT-PCR reactions (EiLFT1 and EiRT1; Fig. 4), which generated products from both the (R) and (S) biotype cDNA preparations suitable for direct cloning into the Plac-based E. coli expression vector pMON45337 (Fig. 5). To ensure accuracy of the cloned sequences, RT-PCR products were generated using a proofreading thermostable polymerase mixture, then cloned from multiple, pooled reactions. The resulting E. coli expression constructs, pMON45364 and pMON45365 (Fig. 5), contain the predicted mature EPSPS open reading frames from the (R) and (S) biotypes, respectively (preceded by an ATG start codon), controlled by the native promoter and operator sequences of the E. coli lac operon. Both plasmids were subsequently transferred into an E. coli aroA− host deficient in EPSPS activity (SR481; Padgette et al., 1987).
Figure 5.
DNA cassettes containing different EPSPS mature protein coding regions for E. coli expression studies. P-lac and lac-O represent the native promoter and operator sequence from the E. coli lac operon. The Shine/Dalgarno sequence is indicated by the letters S/D. The predicted EPSPS mature coding region from the resistant biotype is indicated by (R) and the sensitive biotype EPSPS by (S). P106S is an engineered variant of (R) containing a Pro at position 381; P381L is a an engineered variant of (R) containing a Pro at position 106; “ter” represents the rho-independent transcriptional terminator element of the E. coli trpA gene (Sato et al., 1987).
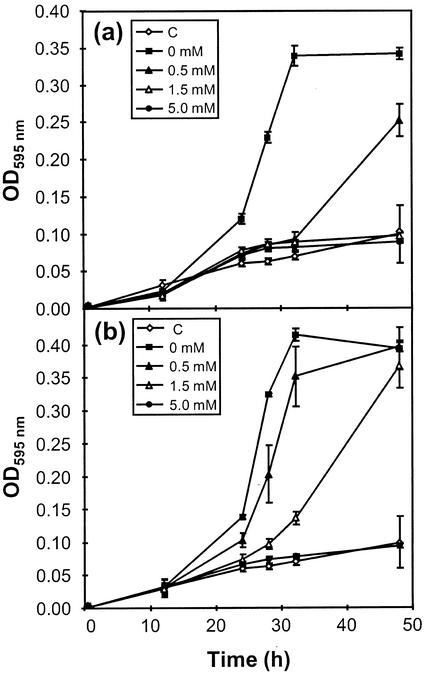
Fresh overnight cultures of SR481 cells harboring pMON45364 (R-EPSPS) and pMON45365 (S-EPSPS) were used to inoculate culture tubes containing minimal media supplemented with ampicillin, isopropylthio-β-galactoside (IPTG), and either 0, 0.5, 1.5, or 5.0 mm glyphosate. At various time points, aliquots were removed and cell densities were monitored spectrophotometrically. The results of these experiments are shown in Figure 6. Both the (S) and (R) biotype EPSPS complemented the deficiency in SR481, which normally requires exogenously supplied aromatic amino acids for growth (Fig. 6, a and b). In the absence of glyphosate, pMON45364 and pMON45365 transformants both grew to saturation within 32 h. In contrast, cells transformed with an “empty” control plasmid (pMON45337) showed little growth over the entire 48-h time course, and the small increase in cell density that was observed is most likely attributable to aromatic amino acid carryover from the inoculum (see “Materials and Methods”). Cell lines expressing the (S) EPSPS from pMON45365 were extremely sensitive to added glyphosate, and failed to reach saturation in the presence of 0.5 mm glyphosate even after 48 h of incubation at 37°C (Fig. 6a). The same glyphosate concentrations showed little or no inhibitory effect on pMON45364 transformants expressing the (R) EPSPS enzyme (Fig. 6b). At 1.5 mm glyphosate concentrations, pMON45365 transformants showed no detectable growth above control levels by 48 h (Fig. 6a), whereas for pMON45364 transformants, growth above control levels was observable within 28 h, and cultures had reached saturation by the time the 48-h experimental period was completed (Fig. 6b). Growth was completely inhibited for both cell lines at 5.0 mm glyphosate concentrations. These data indicate that the SR481 cells expressing the EPSPS cDNA isolated from the (R) biotype exhibit greater than 3-fold higher levels of glyphosate resistance than cells expressing the EPSPS cDNA from the (S) biotype. It is interesting to note that the level of resistance conferred by the (R) EPSPS in the present study is on par with greenhouse experiments where approximately 2- to 4-fold higher glyphosate levels were necessary to achieve equivalent weed control for the goosegrass (R) biotype as were necessary for the (S) biotype (Tran et al., 1999). Taken together, these data provide compelling evidence that the genetic basis for glyphosate resistance in goosegrass involves the resistant EPSPS variant described in this report.
Figure 6.
Glyphosate sensitivity of pMON45364- and pMON45365-transformed cell lines. Overnight cultures of E. coli SR481 (aroA−) cell lines harboring pMON45364 [containing the EPSPS cDNA from the (R) biotype], and pMON45365 [containing the EPSPS cDNA from the (S) biotype], were used to inoculate 3.0-mL cultures containing minimal media supplemented with either 0.0, 0.5, 1.5, or 5.0 mm glyphosate. Aliquots were removed at t = 0, 24, 28, 32, and 48 h and optical densities were monitored at 595 nm. Before inoculation, optical density measurements were taken from the original overnight cultures to confirm similar cell densities. Each data point represents the mean of three replicates; error bars indicate sd. Where not visible, the error is enclosed within the data symbol. C, Growth rates of cells harboring pMON45337 (“empty” vector control). a, Growth rates of cells expressing the mature (S) EPSPS from pMON45365. b, Growth rates of cells expressing the mature (R) EPSPS from pMON45364.
Kinetic Characterization of E. coli-Expressed EPSPS Variants
Glyphosate inhibition of EPSPS is competitive with respect to its substrate PEP (Boocock and Coggins, 1983); thus, it is not surprising that most efforts to engineer glyphosate resistance in plant enzymes by one or two amino acid substitutions result in perturbations in PEP binding (Padgette et al., 1996). This has been proposed as the mechanistic basis for the lack of evolved target-based resistance to glyphosate in weed species because resistant enzymes may not provide sufficient EPSP to support aromatic amino biosynthesis under conditions of limited PEP availability (Bradshaw et al., 1997). Previous work performed by Padgette et al. (1991) using the cloned petunia enzyme has shown that substitution of Gly-101 with Ala, as well as Pro-106 with Ser, results in an apparent reduced affinity for PEP as well as glyphosate. Given that, at least in crown tissues, a resistant EPSPS, which includes a Pro-106 → Ser substitution, appears to be the predominant enzyme species expressed in the goosegrass (R) biotype, it is of considerable interest to determine if perturbations in substrate binding also occur in this enzyme. It is possible that the second substitution identified (Pro-381 → Leu; Fig. 4) could modulate the effect of the Pro-106 → Ser substitution, allowing the enzyme to bind more tightly to PEP, thus lowering or eliminating fitness penalties associated with the expression of a glyphosate-resistant EPSPS enzyme. Although not specifically implicated in catalysis, Pro-381 lies in proximity to other putative active site residues (Shuttleworth et al., 1999). To address these questions, two additional constructs were prepared from pMON45364 (R-EPSPS expression vector; Fig. 5) via site-directed mutagenesis to determine the effect each substitution has on PEP and glyphosate interactions with the (R) EPSPS enzyme. In pMON45370, Leu-381 is converted to a Pro, and in pMON45371, Ser-106 is converted to a Pro, resulting in single-substitution (R) EPSPS variants, containing only the Pro-106 substitution or Leu-381 substitution, respectively (Fig. 5). The apparent affinity for PEP [Km(app)] and glyphosate [Ki(app)] were then determined for these variants as well as the native (R) and (S) biotype EPSPS enzymes expressed in E. coli. The results of these experiments are shown in Table I.
Table I.
Kinetic parameters for E. coli-expressed enzymes
| Construct | EPSPS | IC50 | Km(app)(PEP) | Ki(app)(Glyphosate) |
|---|---|---|---|---|
| μm | nm | |||
| pMON45364 | Resistant | 30.4 ± 2.7 | 7.0 ± 1.0 | 759 ± 140 |
| pMON45370 | P106S | 38.2 ± 7.6 | 8.9 ± 1.0 | 1,040 ± 138 |
| pMON45365 | Sensitive | 6.3 ± 0.9 | 3.8 ± 1.1 | 47.8 ± 16 |
| pMON45371 | P381L | 8.6 ± 1.3 | 3.2 ± 0.5 | 29.2 ± 5.1 |
Kinetic constants were determined as described previously (Padgette et al., 1991) using triplicate assays for all experiments. Calculations were performed using the GraFit software package (Leatherbarrow, 1998). IC50, Km(app)(PEP), and Ki(app)(glyphosate) values are expressed as ± se derived from the best fit.
The Km(app)(PEP) for the (S) EPSPS obtained was 3.8 μm (Table I), similar to results obtained for other plant enzymes, which generally fall within the 5 to 14 μm range (Franz et al., 1997). The Ki(app)(glyphosate) versus PEP was found to be 47.8 nm, which is lower than values reported for other plant enzymes, which generally fall between 80 and 400 μm. However, the majority of these studies involved dicot sources that tend to be slightly less sensitive to glyphosate than monocot enzymes (Franz et al., 1997). In comparison, the Ki(app)(glyphosate) versus PEP for the (R) biotype EPSPS was determined to be 759 nm, indicating an approximately 16-fold decrease in glyphosate sensitivity as compared with the (S) biotype EPSPS. The difference in sensitivity at saturating substrate levels as indicated by IC50 values (Table I) was approximately 5-fold, similar to the differences observed in crude extracts prepared from (R) and (S) biotype plants (Fig. 3).
The Km(app)(PEP) for the (R) EPSPS was determined to be 7.0 μm, which is surprisingly similar (less than 2-fold higher) to the Km(app)(PEP) for the sensitive enzyme. When the corresponding substitution (Pro-106 → Ser) was made in the wild-type petunia enzyme by Padgette and coworkers (1991), Km(app)(PEP) increased from 5.0 to 44 μm, and Ki(app)(glyphosate) increased approximately 8-fold. These data further support the notion that the (R) EPSPS could represent a component or all of the underlying genetic basis for the observed resistance to glyphosate in goosegrass because the expected loss of enzyme substrate affinity associated with reduced glyphosate inhibition appears to be minimized in this case.
The kinetic data for the (R) EPSPS single mutation derivatives strongly suggest that the observed differences in substrate and inhibitor affinity between the (R) and (S) EPSPS enzymes can be primarily attributed to the Pro to Ser substitution at position 106 (Table I). The P106S variant expressed from pMON45370 shows similar Km(app)(PEP) and Ki(app)(glyphosate) values to the (R) EPSPS expressed from pMON45365, and furthermore, the kinetic parameters for the P381L variant from pMON45371 are close to those for the (S) EPSPS enzyme. Using the three-dimensional structure of the E. coli EPSPS/shikimate-3-phosphate/glyphosate ternary complex as a guide (Schönbrunn et al., 2001), the residue at position 381 would likely reside on the enzyme's outer surface within a turn between two beta-sheets, away from the active site. Thus, it is unlikely to be directly involved in either catalysis, substrate binding, or domain closure (J. Evans, personal communication). Based on these data, the Pro-381 → Leu substitution may represent a neutral change in the primary sequence of the (R) enzyme, occurring during an interval in which the two biotypes had evolved in isolation. Determination of the relative catalytic efficiencies using purified enzyme preparations will be required to definitively resolve this question.
Taken together, these studies suggest that an altered EPSPS provides a significant component of the glyphosate resistance mechanism in goosegrass, and represents the first example for target-based resistance to glyphosate occurring in any plant species. The potential for the existence of other factors that contribute to resistance cannot be eliminated at the present time; however, recent studies indicate that metabolic inactivation of glyphosate and differential herbicide uptake or translocation are not associated with this biotype (T.L. Reynolds, unpublished data). Cosegregation of the (R) EPSPS allele with the resistance phenotype as well as the demonstration of monogenic inheritance for this trait will be necessary to demonstrate that the resistant EPSPS is the sole determinant involved. Given that a simple C → T nucleotide transition in the (R) EPSPS allele could have created a resistant weed, it is surprising that no other biotypes have evolved with an altered EPSPS enzyme, and that Arabidopsis screens employing the mutagen EMS have been unsuccessful in recovering glyphosate-resistant mutants (Haughn and Somerville, 1987; Baerson et al., 1999). GC → AT transitions are the predominant substitutions caused by EMS (Kreig, 1963); thus, the corresponding Pro-106 → Ser EPSPS substitution would have been an expected outcome from such mutagenesis screens. It is possible that goosegrass may be predisposed to this type of mechanism due to species-specific genetic or physiological characteristics that remain obscure at present.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seed samples of glyphosate-resistant and susceptible goosegrass (Eleusine indica) biotypes were collected from orchards located in the Jahor region of Malaysia. The orchards from which these biotypes were collected have been treated an average of eight times per year with 0.9 to 1.8 kg a.e. ha−1 glyphosate for the past 10 years (Tran et al., 1999). Plants of both biotypes were greenhouse grown in 10-cm square pots filled with prefertilized potting soil (1:1 [w/w] US10:Metro Mix 200), with an approximately 1-cm-thick overlay of Metro Mix 350. Greenhouse conditions were maintained at approximate day/night temperatures of 28°C and 20°C, respectively, relative humidity from 30% to 70%, and supplemental lighting added as needed to maintain a 14-h photoperiod and a minimum of 500 μmol m−2 s−1. For analysis of EPSPS enzyme activity and steady-state mRNA levels, duplicate clones were generated from representative (R) and (S) biotype individuals by removing approximately 10 to 20 tillers and subtending root mass per plant, which were then repotted in 10-cm2 pots filled with Metro Mix 510 potting media. Before harvest, (S) and (R) clones were challenged with sublethal spray applications of the isopropylamine salt of glyphosate, as described by Pratley et al. (1999).
EPSPS Protein Extraction and Enzymatic Assays
Crown regions (compressed stem tissue near soil level where new tillers arise) were dissected from whole plants, pulverized under liquid nitrogen with a mortar and pestle, then stored at −80°C before extraction. Homogenates were prepared from 0.5 g of tissue per sample in 25 mL of extraction buffer [100 mm Tris-HCl, 10% (v/v) glycerol, 1 mm EDTA, 1 mm benzamidine, 1 mm dithiothreitol, 1 mm 4-(2-aminoethyl)-benzenesulfonyl fluoride HCl, and 0.1 mm leupeptin, pH 7.4] at 4°C using a model PT3000 Polytron homogenizer (Brinkman Instruments Inc., Westbury, NY). Debris was then removed by 0.2-μm filtration, then the resulting supernatants were concentrated and desalted using an Ultrafree-15 centrifugal filtration unit (catalog no. UFV2-BGC-10, Millipore Corp., Bedford, MA). Final sample volumes were approximately 0.5 mL. Protein concentrations of the extracts were determined spectrophotometrically using the method of Bradford (1976) with bovine serum albumin as the standard.
EPSPS activity levels of the extracts were determined using a radiometric HPLC-based assay essentially as described by Padgette et al. (1987, 1988). Assays were performed using 10 μL of extract incubated at 25°C for 5 to 15 min (50-μL reactions included 50 mm HEPES, 5 mm potassium fluoride, 1 mm shikimate-3-phosphate, 0.5 mm [1-14C]PEP, and 1.073 GBq mmol−1 cyclohexylammonium salt; catalog no. CFQ10004, Amersham Life Science, Inc., Arlington Heights, IL, and 0.1 mm ammonium molybdate, pH 7.0). Reactions were then quenched by the addition of 50 μL of an ethanol:0.1 n acetic acid, pH 4.5 (9:1, v/v) mixture before loading. Thirty microliters of the quenched reactions was then injected onto a Synchropak AX100 anion-exchange column (catalog no. 942804, P.J. Cobert Associates, Inc., St. Louis) equilibrated with 0.235 m potassium phosphate buffer, pH 6.5, and eluted isocratically with the same buffer. A model D525 radioactive flow detector (Packard Instrument Co., Downer's Grove, IL) was used to monitor the production of [14C]EPSP in the reactions.
For IC50 determinations, assays were performed in the presence of glyphosate at concentrations ranging from 0.1 to 100 μm, then inhibition curves and IC50 (inflection point) values were generated from the data by nonlinear regression analysis using the GraFit software package (Leatherbarrow, 1998).
RT-PCR, 5′-RACE, and 3′-RACE for Isolation of EPSPS Mature Protein Coding Sequences
For the isolation of EPSPS coding sequences by RT-PCR, total RNAs were first extracted from flash-frozen, pulverized crown tissues using the RNeasy Plant Mini Kit (catalog no. 74904, Qiagen Inc., Valencia, CA) per manufacturer's instructions. Oligo(dT)-primed first-strand cDNAs were prepared from 5-μg samples of total RNA using the Superscript Pre-Amplification System (catalog no. 18089-011, Life Technologies, Rockville, MD) per manufacturer's instructions. Two microliters of first-strand cDNA was used to generate partial goosegrass EPSPS cDNAs via PCR using a modification of the “touchdown PCR” technique (Don et al., 1991). Degenerate oligonucleotides were designed based on published plant EPSPS sequences: Ei2a (22 mer) 5′-T(ACGT)(AT) (CG)(ACGT) G T(ACGT) G A(AG) G C(ACGT) G A(CT) A A(AG) G T-3′, and Ei9a (23 mer) 5′-GCC AT(ACGT) GCC AT(ACGT) C(TG)(AG) TG(AG) TC(AG) TC-3′, and added to 50 μL of RT-PCR reactions at a final concentration of 25 μm each. PCR amplifications were performed using the Expand High Fidelity PCR System (catalog no. 1732 641, Roche Molecular Biochemicals, Indianapolis) per manufacturer's instructions. A thermal profile of 94°C for 20 s, followed by 60°C for 1 min, then 72°C for 1 min 30 s was used for the initial 30 cycles with a 0.5°C decrease in annealing temperature per cycle, followed by 10 additional cycles of 94°C for 20 s, 45°C for 1 min, then 72°C for 1 min 30 s. RT-PCR products were then gel purified, and directly cloned into the pCR2.1-TOPO vector (catalog no. K4500-40, Invitrogen, Carlsbad, CA). The identity of cloned RT-PCR products was confirmed by automated DNA sequence analysis using the Prism reagent system (Applied Biosystems, Foster City, CA).
The remainder of the 3′ end of EPSPS coding regions was generated using the 3′-RACE System for Rapid Amplification of cDNA Ends (catalog no. 18373-027, Life Technologies), with the gene-specific oligonucleotide: Ei3′gsp1 (24 mer) 5′-GTGAAAGCAGAGCATTCTGATAGC-3′, per manufacturer's instructions. The resulting 3′-RACE products were then gel purified and directly sequenced using internal, gene-specific oligonucleotides.
The remainder of the 5′ end of the predicted mature protein coding regions were generated using the SMART RACE cDNA Amplification Kit (catalog no. K1811-1, CLONTECH Laboratories Inc., Palo Alto, CA), using the gene-specific oligonucleotides Ei5′gsp1 (29 mer) 5′-GGCTGCTGTCAATGTCGCATTGCAGTTCC-3′ and Ei5′gsp2 (32 mer) 5′-CTCTTTCGCATCCTTCTCAACTGGGAACTTGC-3′, per manufacturer's instructions. First strand cDNAs for 5′-RACE reactions were prepared from 150 ng of poly(A+) mRNA isolated from crown tissues using an Oligotex mRNA Midi Kit (catalog no. 28704, Qiagen Inc.). PCR reactions were conducted as recommended by the manufacturer, except that the Expand High Fidelity PCR System (catalog no. 1732641, Roche Molecular Biochemicals) was used, and dimethyl sulfoxide was included in all reactions at a final concentration of 5% (v/v) to facilitate the amplification of GC-rich sequences. The oligonucleotide Ei5′gsp1 (described above) was used in primary PCR amplifications, then second round (“nested”) amplifications were performed using the oligonucleotide Ei5′gsp2 (described above), with a 1-μL aliquot of a 1:100 dilution of primary PCR reactions. The resulting 5′-RACE products were then gel purified and directly sequenced using internal, gene-specific oligonucleotides. Sequences generated by RT-PCR, 3′-RACE, and 5′-RACE were assembled into continuous DNA sequences containing the entire open reading frame for the mature goosegrass EPSPS protein using the SEQ-Man II software package (DNASTAR Inc., Madison, WI).
Vector Construction
DNA manipulations and transformations of Escherichia coli were performed according to standard procedures (Sambrook et al., 1989). An E. coli, lac promoter-based heterologous expression vector, pMON45337, was employed for the inducible expression of different EPSPS cDNAs. pMON45337 was derived from pMON34610 (Valentin et al., 2000) to facilitate cloning using NdeI. Two complementary oligonucleotides, 34610Nde1a (79 mer) 5′-GATCTCCTAGGGCTTAATTAATTAAGCACTAGTCACACAGGAGGTAATTCATATGGGCATGCAGTACTGGTACCGAGCT-3′ and 34610Nde1b (71 mer) 5′-CGGTACCAGTACTGCATGCCCATATGAATTACCTCCTGTGTGACTAGTGCTTAATTAATTAAGCCCTAGGA-3′, were annealed together, then the resulting double-stranded oligonucleotide was ligated with SacI- and BglII-digested pMON34610 DNA, resulting in the plasmid pMON45337.
To generate cDNAs containing the predicted mature EPSPS coding sequences from the (S) and (R) goosegrass biotypes, the oligonucleotides EiRT1 (36 mer) 5′-GCAATTCCATATGGCGGGCGCGGAGGAGGTGGTGCT-3′ and EiLFT1 (48 mer) 5′-GACTAGGAATTCTTAGTTCTTTTGACGAAAGTGCTCAGCACGTCGAAG-3′ were employed in RT-PCR reactions using 2 μL of the first strand cDNAs described above for the initial isolation of (S) and (R) biotype-specific EPSPS coding sequences. Oligonucleotides EiRT1 and EiLFT1 were added in 50-μL RT-PCR reactions at a final concentration of 0.4 μm each. PCR amplifications were performed using the Expand High Fidelity PCR System (catalog no. 1 732 641, Roche Molecular Biochemicals) per manufacturer's instructions, using a thermal profile of 94°C for 30 s, then 57°C for 2 min, followed by 75°C for 3 min, for a total of 35 cycles. The resulting PCR products were then gel purified, digested with NdeI and EcoRI, then ligated with NdeI- and EcoRI-digested pMON45337, resulting in the plasmids pMON45364 and pMON45365, which contain the predicted mature protein coding region for goosegrass EPSPS isolated from the (R) and (S) biotype, respectively. To ensure accuracy of the cloned EPSPS sequences, PCR products were pooled from a minimum of three independent RT-PCR reactions before ligation into pMON45337, then five randomly selected isolates per construct were confirmed by DNA sequence analysis.
Two additional constructs, pMON45370 and pMON45371, were derived from pMON45364 using the QuikChange Site-Directed Mutagenesis Kit (catalog no.200518, Stratagene, La Jolla, CA) per manufacturer's instructions. pMON45370 was created from pMON45364 using the complementary mutagenic oligonucleotides EiP106Sa (37 mer) 5′-CGTCGGTCGAGGAAGGACCGGACTACTGCATTATCAC-3′ and EiP106Sb (37 mer) 5′-GTGATAATGCAGTAGTCCGGTCCTTCCTCGACCGACG-3′. pMON45371 was created by mutagenesis of pMON45364 using the complementary mutagenic oligonucleotides EiP328La (31 mer) 5′-GGAACTGCAATGCGACCATTGACAGCAGCCG-3′ and EiP328Lb (31 mer) 5′-CGGCTGCTGTCAATGGTCGCATTGCAGTTCC-3′. After mutagenesis, the resulting plasmids were confirmed by DNA sequence analysis.
Heterologous Expression in E. coli
For enzyme kinetic studies, pMON45337, pMON45364, pMON45365, pMON45370, and pMON45371 were all transformed into the E. coli strain SR481, an aroA minus strain which lacks endogenous EPSPS activity (Padgette et al., 1987). For preparation of bacterial extracts, fresh overnight cultures were grown at 37°C in Terrific Broth (Sambrook et al., 1989), supplemented with 50 μg mL−1 ampicillin and 100 μg mL−1 each of l-Phe, l-Tyr, and l-Trp. Overnight cultures were used to inoculate large-scale cultures containing the same media at a 1:100 dilution, then grown at 37°C to an optical density at 595 nm of 0.6. IPTG was then added to a final concentration of 1.0 mm, and incubations were continued for an additional 4 h at 37°C. Cells were pelleted by centrifugation at 10,000g for 5 min, then washed twice in ice-cold 0.9% (w/v) NaCl. Cell pellets were flash frozen in liquid nitrogen and stored at −80°C before use. Homogenates were prepared by resuspending 1 g of pelleted cells in 3 mL of extraction buffer, followed by lysis using a French press (model no. J5-598A, American Instrument Co., Silver Springs, MD). Cell debris was subsequently removed by centrifugation at 14,000g for 10 min at 4°C. Supernatants were then desalted using a PD-10 column (catalog no. 17-0851-01, Amersham Pharmacia Biotech Inc., Piscataway, NJ). EPSPS radiometric assays were then performed as described above for plant-derived extracts.
Northern and Southern Analyses
Total RNA was isolated from flash-frozen, pulverized crown tissues as described above. Genomic DNA was extracted from the same material as for RNA isolations, using the Plant DNAzol Reagent (Life Technologies). Approximately 1 g of powdered tissue was mixed with 3.0 mL of Plant DNAzol reagent supplemented with RNase A at a final concentration of 1.0 mg mL−1, then incubated at room temperature for 10 min with gentle shaking. The remainder of the extraction procedure was carried out per manufacturer's instructions, with an additional chloroform:isoamyl alcohol (24:1, v/v) extraction step performed before ethanol precipitation. Restriction endonuclease digestions and northern and Southern blotting procedures were performed according to standard protocols (Sambrook et al., 1989) using a partial Lolium rigidum EPSPS cDNA as the probe (GenBank accession no. AF349754). The integrity of all purified RNA and DNA samples was confirmed by agarose gel electrophoresis, and the concentration and purity of each preparation was determined spectrophotometrically.
E. coli Growth Comparisons
For comparative growth studies, fresh overnight cultures of E. coli SR481 cell lines harboring pMON45337, pMON45364, and pMON45365 were grown in Terrific Broth (Sambrook et al., 1989), supplemented with 1.0 mm IPTG, 50 μg mL−1 ampicillin, and 100 μg mL−1 each of l-Phe, l-Tyr, and l-Trp, at 37°C. One hundred-microliter aliquots of each overnight culture were then used to inoculate 3.0 mL of M9 minimal media (Sambrook et al., 1989), supplemented with 50 μg mL−1 ampicillin, 1.0 mm IPTG, and either 0.0, 0.5, 1.5, or 5.0 mm glyphosate. Before inoculation, optical density measurements at 595 nm were taken on all of the original overnight cultures to confirm similar cell densities. Growth rates were monitored by removing 100-μL aliquots from each tube at indicated timepoints, then monitoring optical densities at 595 nm.
ACKNOWLEDGMENTS
We thank Ron Brinker, Laura Casagrande, Matthew Faletti, Margie Nemeth, David Schafer, Doug Sammons, Yeew-Thai Teng, and Jinsong You for providing materials and excellent technical assistance. We are also grateful to Drs. Jeremy Evans, Kenneth Gruys, Stephen Padgette, David Stalker, and Jonathan Gressel for critical reviews, numerous helpful discussions, and support of this work.
Footnotes
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.001560.
LITERATURE CITED
- Baerson SR, Tran M, Rodriguez DJ, Gonzalez KA, Schafer DE, Krupa DM, Gruys KJ, Stalker DM, Taylor NA, Dill GM. Proceedings, 10th International Conference on Arabidopsis Research. Melrose Park, Australia: Manning Printers PTY LTD; 1999. Comparison of frequencies of individuals resistant to imazethapyr, chlorsulfuron, and glyphosate in EMS-mutagenized populations of Arabidopsis thaliana (cv. Col-0) pp. 5–7. [Google Scholar]
- Benbrook C. Racing against the clock. Agrichem Age. 1991;35:30–33. [Google Scholar]
- Boocock MR, Coggins JR. Kinetics of 5-enolpyruvylshikimate-3-phosphate synthase inhibition by glyphosate. FEBS Lett. 1983;154:127–133. doi: 10.1016/0014-5793(83)80888-6. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bradshaw LD, Padgette SR, Kimball SL, Wells BH. Perspectives on glyphosate resistance. Weed Technol. 1997;11:189–198. [Google Scholar]
- Comai L, Sen LC, Stalker DM. An altered aroA gene product confers resistance to the herbicide glyphosate. Science. 1983;221:370–371. doi: 10.1126/science.221.4608.370. [DOI] [PubMed] [Google Scholar]
- Della-Cioppa G, Bauer SC, Klein BK, Shah DM, Fraley RT, Kishore GM. Translocation of the precursor of 5-enolpyruvylshikimate-3-phosphate synthase into chloroplasts of higher plants in vitro. Proc Natl Acad Sci USA. 1986;83:6873–6877. doi: 10.1073/pnas.83.18.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS. “Touchdown” PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz JE, Mao MK, Sikorski JA. Glyphosate: A Unique Global Herbicide. Washington, DC: American Chemical Society; 1997. Glyphosphate's molecular mode of action; pp. 521–642. [Google Scholar]
- Ganeshaiah KN, Umashaanker R. Evolution of reproductive behavior in the genus Eleusine. Euphytica. 1982;31:397–404. [Google Scholar]
- Gardner SN, Gressel J, Mangel M. A revolving dose strategy to delay the evolution of both quantitative vs. major monogene resistances to pesticides and drugs. Int J Pest Manag. 1998;44:161–180. [Google Scholar]
- Gressel J, Segel LA. The paucity of plants evolving genetic resistance to herbicides: possible reasons and implications. J Theor Biol. 1978;75:349–371. doi: 10.1016/0022-5193(78)90340-5. [DOI] [PubMed] [Google Scholar]
- Gruys KJ, Sikorski JA. Inhibitors of tryptophan, phenylalanine, and tyrosine biosynthesis as herbicides. In: Singh B, editor. Plant Amino Acids. New York: Marcel Dekker, Inc.; 1999. pp. 357–384. [Google Scholar]
- Haslam E. Shimikic Acid: Metabolism and Metabolites. Chichester, UK: John Wiley and Sons Inc; 1993. [Google Scholar]
- Haughn GW, Somerville CR. In Selection for herbicide resistance at the whole plant level. In: Lebaron H, Mumma RO, Honeycutt RC, Duesing JH, editors. Applications of Biotechnology to Agricultural Chemistry. Washington, DC: American Chemical Society; 1987. pp. 98–108. [Google Scholar]
- Heap IM. The occurrence of herbicide-resistant weeds worldwide. Pest Sci. 1997;51:235–243. [Google Scholar]
- Heap IM (2001) International survey of herbicide resistant weeds. WeedScience.com. http://www.weedscience.com (December 1, 2001)
- Herrmann KM. The shikimate pathway: early steps in the biosynthesis of aromatic compounds. Plant Cell. 1995;7:907–919. doi: 10.1105/tpc.7.7.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm LG, Plucknett DL, Pancho JV, Herberger JP. The World's Worst Weeds. Distribution and Biology. University Press of Hawaii, Honolulu. 1977. Eleusine indica (L.) Gaertn; pp. 47–53. [Google Scholar]
- Jasieniuk M, Brule-Babel AL, Morrison IN. The evolution and genetics of herbicide resistance in weeds. Weed Sci. 1996;44:176–193. [Google Scholar]
- Jones JD, Goldsbrough PB, Weller SC. Stability and expression of amplified EPSPS genes in glyphosate resistant tobacco cells and plantlets. Plant Cell Rep. 1996;15:431–436. doi: 10.1007/BF00232070. [DOI] [PubMed] [Google Scholar]
- Kreig D. Ethyl methanesulfonate-induced reversion of bacteriophage T4r II mutants. Genetics. 1963;48:561–580. doi: 10.1093/genetics/48.4.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatherbarrow RJ. GraFit Version 4.0. Staines, UK: Erithacus Software Ltd; 1998. [Google Scholar]
- Lebrun M, Sailland A, Freyssinet G, inventors. February 6, 1997. Mutated 5-enol pyruvylshikimate-3-phosphate synthase, gene coding for said protein and transformed plants containing said gene. World Patent Application No. WO9704103.
- Lee LJ, Ngim J. A first report of glyphosate-resistant goosegrass (Eleusine indica (L) Gaertn) in Malaysia. Pest Manag Sci. 2000;56:336–339. [Google Scholar]
- Malik J, Barry G, Kishore G. The herbicide glyphosate. Biofactors. 1989;2:17–25. [PubMed] [Google Scholar]
- Marshall G, Kirkwood RC, Leach GE. Comparative studies on graminicide resistant and susceptible biotypes of Eleusine indica. Weed Res. 1994;34:177. [Google Scholar]
- Mudge LC, Gossett BJ, Murphy TR. Resistance of goosegrass (Eleusine indica) to dinitroaniline herbicides. Weed Sci. 1984;32:591–594. [Google Scholar]
- Mysore KS, Baird V. Nuclear DNA content in species of Eleusine (Graminae): a critical re-evaluation using laser flow cytometry. Plant Syst Evol. 1997;207:1–11. [Google Scholar]
- Padgette SR, Huynh QK, Aykent S, Sammons RD, Sikorski JA, Kishore GM. Identification of the reactive cysteines of Escherichia coli 5-enolpyruvylshikimate-3-phosphate synthase and their nonessentiality for enzymatic catalysis. J Biol Chem. 1988;263:1798–1802. [PubMed] [Google Scholar]
- Padgette SR, Huynh QK, Borgmeyer J, Shah DM, Brand LA, Re DB, Bishop BF, Rogers SG, Fraley RT, Kishore GM. Bacterial expression and isolation of Petunia hybrida 5-enol-pyruvylshikimate-3-phosphate synthase. Arch Biochem Biophys. 1987;258:564–573. doi: 10.1016/0003-9861(87)90378-x. [DOI] [PubMed] [Google Scholar]
- Padgette SR, Re DB, Barry GF, Eichholtz DE, Delannay X, Fuchs RL, Kishore GM, Fraley RT. New weed control opportunities: development of soybeans with a Roundup Ready gene. In: Duke S, editor. Herbicide Resistant Crops: Agricultural, Economic, Environmental, Regulatory, and Technological Aspects. Boca Raton, FL: CRC Press; 1996. pp. 53–84. [Google Scholar]
- Padgette SR, Re DB, Gasser CS, Eichholtz DA, Frazier RB, Hironaka CM, Levine EB, Shah DM, Fraley RT, Kishore GM. Site-directed mutagenesis of a conserved region of the 5-enolpyruvylshikimate-3-phosphate synthase active site. J Biol Chem. 1991;266:22364–22369. [PubMed] [Google Scholar]
- Powles SB, Holtum JAM. Herbicide Resistance in Plants: Biology and Biochemistry. Boca Raton, FL: Lewis Publishers; 1994. [Google Scholar]
- Powles SB, Lorraine-Colwill DF, Dellow JJ, Preston C. Evolved resistance to glyphosate in rigid ryegrass (Lolium rigidum) in Australia. Weed Sci. 1998;46:604–607. [Google Scholar]
- Pratley J, Baines P, Eberbach P, Incerti M, Broster J. Proceedings of the 11th Annual Conference of the Grassland Society of NSW. Orange, NSW, Australia: The Grassland Society of NSW Inc.; 1996. Glyphosate resistance in annual ryegrass; p. 122. [Google Scholar]
- Pratley JE, Urwin NAR, Stanton RA, Baines PR, Broster JC, Cullis K, Schafer DE, Bohn JA, Krueger RW. Resistance to glyphosate in Lolium rigidum: I. Bioevaluation Weed Sci. 1999;47:405–411. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sato T, Matsui H, Shibahara S, Kobayashi T, Morinaga Y, Kashima N, Yamasaki S, Hamuro J, Taniguchi T. New approaches for the high-level expression of human interleukin-2 cDNA in Escherichia coli. J Biochem. 1987;101:525–534. doi: 10.1093/oxfordjournals.jbchem.a121940. [DOI] [PubMed] [Google Scholar]
- Schönbrunn E, Eschenburg S, Shuttleworth WA, Schloss JV, Amrhein N, Evans JNS, Kabsch W. Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail. Proc Natl Acad Sci USA. 2001;98:1376–1380. doi: 10.1073/pnas.98.4.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah DM, Horsch RB, Klee HJ, Kishore GM, Winter JA, Tumer NE, Hironaka CM, Sanders PR, Gasser CS, Aykent S et al. Engineering herbicide tolerance in transgenic plants. Science. 1986;233:478–481. doi: 10.1126/science.233.4762.478. [DOI] [PubMed] [Google Scholar]
- Shuttleworth WA, Pohl ME, Helms GL, Jakeman DL, Evans JNS. Site-directed mutagenesis of putative active site residues of 5-enoylpyruvylshikimate-3-phosphate synthase. Biochemistry. 1999;38:296–302. doi: 10.1021/bi9815142. [DOI] [PubMed] [Google Scholar]
- Stalker DM, Hiatt WR, Comai L. A single amino acid substitution in the enzyme 5-enolpyruvylshikimate-3-phosphate synthase confers resistance to the herbicide glyphosate. J Biol Chem. 1985;260:4724–4728. [PubMed] [Google Scholar]
- Steinrücken H, Amrhein N. The herbicide glyphosate is a potent inhibitor of 5-enolpyruvylshikimic acid-3-phosphate synthase. Biochem Biophys Res Commun. 1980;94:1207–1212. doi: 10.1016/0006-291x(80)90547-1. [DOI] [PubMed] [Google Scholar]
- Suh H, Hepburn AG, Kriz AL, Widholm JM. Structure of the amplified 5-enolpyruvylshikimate-3-phosphate synthase gene in glyphosate-resistant carrot cells. Plant Mol Biol. 1993;22:195–205. doi: 10.1007/BF00014928. [DOI] [PubMed] [Google Scholar]
- Tran M, Baerson S, Brinker R, Casagrande L, Faletti M, Feng Y, Nemeth M, Reynolds T, Rodriguez D, Schafer D et al. Proceedings 1(B) 17th Asian-Pacific Weed Science Society Conference, The Asian-Pacific Weed Science Society, Los Banos, Philippines. 1999. Characterization of glyphosate resistant Eleusine indica biotypes from Malaysia; pp. 527–536. [Google Scholar]
- Valentin HE, Reiser S, Gruys KJ. Poly(3-hydroxybutyrate-co-4-hydroxybutyrate) formation from γ-aminobutyrate and glutamate. Biotechnol Bioeng. 2000;67:291–299. [PubMed] [Google Scholar]