Abstract
The glycosylated forms of abscisic acid (ABA) have been identified from many plant species and are known to be the forms of ABA-catabolism, although their (physiological) roles have not yet been elucidated. ABA-glucosyltransferase (-GTase) is thought to play a key role in the glycosylation of ABA. We isolated an ABA-inducible GTase gene from UDP-GTase homologs obtained from adzuki bean (Vigna angularis) seedlings. The deduced amino acid sequence (accession no. AB065190) showed 30% to 44% identity with the known UDP-GTase homologs. The recombinant protein with a glutathione S-transferase-tag was expressed in Escherichia coli and showed enzymatic activity in an ABA-specific manner. The enzymatic activity was detected over a wide pH range from 5.0 to 9.0, the optimum range being between pH 6.0 and 7.3, in a citrate and Tris-HCl buffer. The product from racemic ABA and UDP-d-glucose was identified to be ABA-GE by gas chromatography/mass spectrometry. The recombinant GTase (rAOG) converted 2-trans-(+)-ABA better than (+)-S-ABA and (−)-R-ABA. Although trans-cinnamic acid was slightly converted to its conjugate by the GTase, (−)-PA was not at all. The mRNA level was increased by ABA application or by water stress and wounding. We suggest that the gene encodes an ABA-specific GTase and that its expression is regulated by environmental stress.
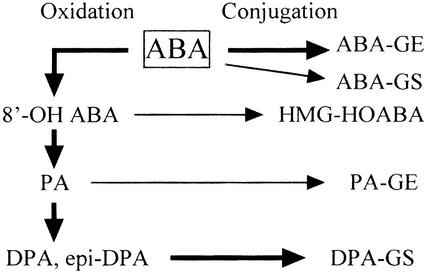
Abscisic acid (ABA) is a plant hormone that regulates plant growth, development, seed maturation or dormancy, and germination (Fincher, 1989; McCarty, 1995; Leung and Giraudat, 1998) and that mediates such stress responses as environmental stress adaptation to salinity, low temperature, UV irradiation, and water deficiency, including the procedure for rapid stomatal closure by ion efflux from guard cells (Chandler and Robertson, 1994; Ingram and Bartels, 1996; Albinsky et al., 1999). In these phenomena, ABA induces or regulates corresponding gene expression in the biochemical and physiological processes (Leung and Giraudat, 1998; Rock, 2000; Söderman et al., 2000). The ABA level is simultaneously regulated by catabolism and/or biosynthesis in these processes (Bray, 1997; Zeevaart, 1999). Stress not only stimulates ABA biosynthesis to increase its level, but also promotes ABA catabolism to increase phaseic acid (PA), dihydrophaseic acid (DPA), etc. (Zeevaart, 1980, 1983; Creelman et al., 1987). Various findings from structure-activity relationships have demonstrated that the activities of ABA were markedly decreased or even lost when its side chain was modified (Walton, 1983). In general, active ABA can be rapidly metabolized to some inactive structures in higher plants through two main routes (Zeevaart and Creelman, 1988; Zeevaart, 1999; Barthe et al., 2000): One is the hydroxylation pathway, and the other is conjugation (Fig. 1). The former route involves active ABA first being converted to 8′-hydroxy ABA (HOABA) and then further metabolized to other inactive structures such as hydroxymethyl glutaryl (HMG)-HOABA, methyl ester (Me)HMG-HOABA, PA, DPA, etc. The latter is the simple conjugation process of ABA to ABA-glucosyl ester (-GE) or ABA-glucosyl ether (-GS). This route also converts ABA catabolites on the hydroxylation pathway to corresponding conjugate forms such as HMG-HOABA, PA-GE, and DPA-GS. It is, therefore, important to obtain information about the enzymes involved in these reactions, including the regulation of enzyme activities, during the ABA-mediating process.
Figure 1.
Two major pathways for ABA metabolism. The bold arrows indicate the major pathways that have been reported to function in plants. The normal arrows indicate the minor pathways (Zeevaart and Creelman, 1988; Zeevaart, 1999; Barthe et al., 2000).
ABA-GE and ABA-GS have been isolated from many plant species (Zeevaart, 1999), although the physiological significance of ABA conjugations and especially of ABA glycosylation in plants remains unclear. The glycosylation of plant hormones such as zeatin and indole-3-acetic acid (IAA) has been confirmed to be catalyzed by glucosyltransferase (GTase) and is believed to play an important role in hormonal transport, protecting hormones against peroxidation, their storage in seed, and hormonal homeostasis (Leznicki and Bandurski, 1988a, 1988b; Dixon et al., 1989; Szerszen et al., 1994; Martin et al., 1999). It is believed that ABA-GE is not likely to be directly transported in phloem. When exogenous ABA-GE was applied to a mature castor bean (Ricinus communis) leaf, it was hydrolyzed to free ABA before it entered the phloem and then translocated out of the leaf (Zeevaart and Boyer, 1984). At the cellular level, ABA-GE mainly exists in vacuoles (Bray and Zeevaart, 1985; Lehmann and Glund, 1986). There are reports contradicting the idea that ABA-GE takes part in hormonal homeostasis by its hydrolysis. The level of ABA-GE increased when a Xanthium strumarium leaf was repeatedly subjected to water stress (Zeevaart, 1983). The increase in free ABA due to a water stress treatment was greater than that of endogenous ABA-GE under normal conditions (Neill et al., 1983), ABA-GE was not hydrolyzed under water stress conditions (Milborrow, 1978), and the activity of the ABA-Glc-splitting enzymes was not increased (Lehmann and Vlasov, 1988). Based on these results, Zeevaart (1999) has suggested that the endogenous conjugation of ABA to ABA-GE is irreversible. In contrast to these results, some reports suggest that the hydrolysis of ABA conjugates forms free ABA. The level of conjugated ABA decreased and the level of free ABA increased in needles of Pseudotsuga menziesii during water stress (Johnson and Ferrell, 1982). Exogenous ABA-GE showed about one-half of the inhibitory activity toward the growth of rice seedlings that (+)-ABA showed (Koshimizu et al., 1966), although it did not show activity toward stomatal closure in broad bean (Vicia faba; Hornberg and Weiler, 1984). This inhibitory activity may have originated from free ABA produced by the hydrolysis of exogenous ABA-GE. These reports may not be contradictory, because the results were obtained from different species of plants, and from some specific organs, tissues or cells. To address the physiological meaning of ABA conjugation, it is important to know how the formation and hydrolysis of ABA conjugates are regulated at the molecular level.
The present study is focused on GTase involved in the formation of glycosylated ABA. GTases are thought to play an important role in the biosynthesis of many plant secondary metabolites. They transfer nucleotide diphosphate-activated sugars to low-molecular-weight substrates. They belong to a superfamily of over 100 members in plants, and native GTases in plants generally have a molecular mass of between 45 and 60 kD (Vogt and Jones, 2000). Several results have indicated that ABA-GE could be synthesized from ABA and UDP-d-Glc (UDPG) by a GTase (Lehmann and Schütte, 1980; Schwarzkopf and Miersch, 1992). In these reports, ABA-GTase was only partially purified by gel-affinity or gel-chromatography on Sephadex. The genes of the zeatin- and IAA-GTase have been cloned and characterized after purifying their protein (Szerszen et al., 1994; Martin et al., 1999). However, the isolation of pure ABA-GTase has not been reported until recently. As described by Vogt and Jones (2000), GTases are generally labile, which renders their purification as difficult and cumbersome as the purification of other superfamilies of modifying enzymes. In this article, we report the isolation of an ABA-GTase gene (accession no. AB065190) by molecular biological methods. By using the recombinant protein bacterially expressed, we characterize the substrate specificity for the enzyme. We also examine the effects of exogenous ABA and such stress treatment as wounding and water deficiency on gene expressions, based on which the role of ABA-GE in plants is discussed in this report.
RESULTS
ABA Metabolites in Adzuki Bean (Vigna angularis) Seedlings
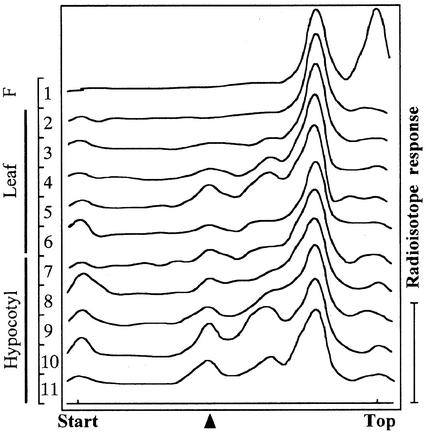
ABA-GE showed an RF of 0.45 by thin-layer chromatography (TLC) in the solvent employed. The [3H]ABA metabolites extracted from adzuki bean plants that had been treated with [3H]ABA were separated by TLC. Radioactivity was detected by an imaging analyzer. When [3H]ABA was taken up into the plants without roots by leaf transpiration, it was metabolized in the plants to compounds with higher polarity than ABA. As shown in Figure 2, ABA metabolites more polar than ABA appeared in a range of RF 0.38 to 0.66. Among them, peaks at RF 0.45 were also detected (Fig. 2, lanes 4–6 and lanes 7–11), which suggested the existence of ABA-GE as one of the ABA metabolites. In the hypocotyls, the products were detectable within 2 h (Fig. 2, lane 7) and accumulated with increasing time (Fig. 2, lanes 7–10). When the plants that had been previously treated with non-labeled ABA for 6 h and then with the [3H]ABA solution for 2 h, the signals of products were stronger (about 2-fold) than those from the plants only treated in the [3H]ABA solution for 2 h (Fig. 2, lanes 7 and 11). On the other hand, although [3H]ABA was translocated into the leaves within 2 h (Fig. 2, lane 2), only faint signals from the products were detectable until at least 4 to 6 h had elapsed (Fig. 2, lanes 3 and 4). In the leaves that had been pretreated with non-labeled ABA for 6 h and then with [3H]ABA for 2 h (Fig. 2, lane 6), a product-peak with a weak signal at RF = 0.45 could be detected, whereas no corresponding signal was detectable from the leaf treated only with [3H]ABA for 2 h (Fig. 2, lane 2).
Figure 2.
Autoradiographic profile of the [3H]ABA metabolites produced in adzuki. Lane 1 is free [3H]ABA. Lanes 2 to 6 are the extracts from leaves, and lanes 7 to 11, from hypocotyls. Plants were treated with [3H]ABA for 2 h (lanes 2 and 7), 4 h (lanes 3 and 8), 6 h (lanes 4 and 9), and 8 h (lanes 5 and 10). Lanes 6 and 11 are for the extracts from plants pretreated with 5 × 10−5 m non-labeled ABA for 6 h and then incubated in a [3H]ABA solution for 2 h.
Isolation and Sequence of GTase
We presumed from the foregoing results that GTases catalyzing ABA conjugation may be present in adzuki seedlings. We, therefore, tried to isolate the gene responsible for this ABA conjugation. Using the designed degenerate primers, 16 different PCR fragments of GTase homologs with the enzymatic signature were cloned from the cDNA library. Based on the assumption that ABA-GTase would be inducible by ABA treatment, northern-blot hybridization experiments were performed to select the ABA-inducible GTase genes. Among the 16 PCR fragments, two showed ABA inducibility (clones 105G and 109G). Their full-length cDNAs were cloned by the PCR procedure described in “Materials and Methods.” Only clone 105G showed clear ABA-GTase activity in successive continuous analyses using recombinant protein, so the description of clone 109G is omitted.
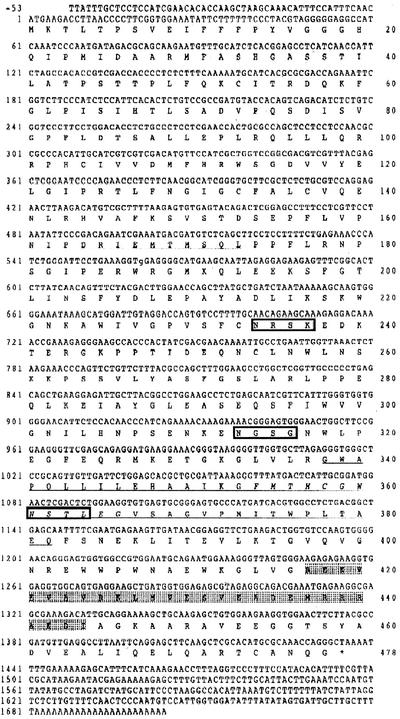
The cDNA of 105G was 1.75 kb long. Its open reading frame configuration encoded a polypeptide of 478 amino acid residues (Fig. 3) with a calculated molecular mass of 53.35 kD. A search of the databases located several important sequence motifs in the deduced amino acid sequence (Fig. 3). The UDP-GTase signature was at 338–382 (underlined); the second peroxisomal targeting signal was located at 164–173 (broken line), the presence of this signal showing the possibility that the GTase can enter peroxisomes; three N-glycosylation motifs, each with the pattern N[∧P][ST][∧P] ([∧P] means any amino acid residue except for Pro), were found at 233–237, 313–316, and 361–364 (box), this motif indicating that the GTase was possibly a glycoprotein; a coiled-coil region was at 417–444 by Lupas's algorithm for detecting coiled-coil regions (shaded); although these features can be found in such structural proteins as myosins and DNA-binding proteins, we do not know how to interpret the structure of the GTase. According to BLAST and FASTA searches of the international databases, the deduced amino acid sequence shows only 30% to 44% identity with known UDP-GTase homologs from plants.
Figure 3.
cDNA and deduced amino acid sequences of ABA-GTase from adzuki bean seedlings. The broken line indicates the second peroxisomal targeting signal. The boxes indicate the N-glycosylation motif with a pattern N[∧P][ST][∧P]. The underlining shows the UDP-glycosyltransferase signature. The shaded area is the coiled-coil region detected by Lupas's algorithm.
Authentication of the GTase Activity
A GTase is generally categorized in a transferase which catalyzes the transfer of a glucosyl group from α-d-Glc-1-phosphate (G-1-P), NDP-Glc, or other glucosyl donors to an acceptor. An ABA-GTase (designated as AOG) assay was first performed at pH 8.0 according to the methods used for the IAA-GTase, zeatin-GTase, or other recombinant GTase assays. The recombinant protein (Fig. 4A, lane 1) was used in the assay to catalyze the glycosylation of plant hormones.
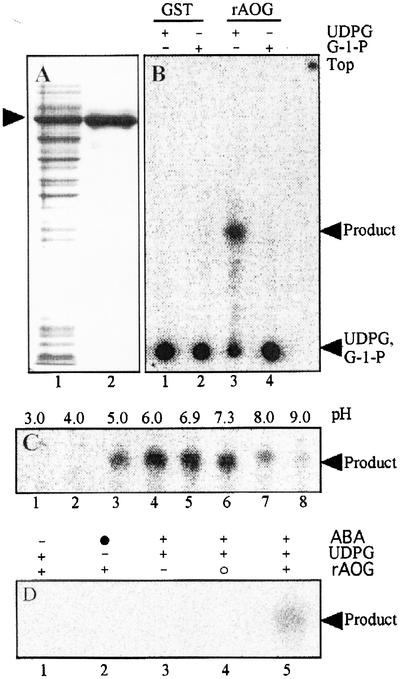
Figure 4.
Results of enzyme assays using recombinant protein. A, SDS-PAGE pattern of the recombinant protein. Lane 1 is crude protein prepared from the cell lysate after inducing rAOG by 0.2 mm IPTG for 4 h at 22°C. Lane 2 shows the fusion protein purified by affinity chromatography. B, Reaction products from ABA and UDPG or G-1-P by rAOG. Lanes 1 and 2 and lanes 3 and 4, respectively, indicate the patterns from GST and rAOG. + and −, The presence and absence, respectively, of UDPG or G-1-P in a reaction mixture. C, The pH dependence of ABA-GTase activity expressed in E. coli. The arrowhead indicates the product. D, The signal intensities of the products under different reaction conditions. Reactions were performed without ABA (lane 1), UDPG (lane 2), or rAOG (lane 3), respectively, and with rAOG denatured by boiling for 10 min (lane 4). Lane 5 shows the product using the native rAOG as a positive control. + and −, The presence and absence, respectively, of ABA, UDPG, or rAOG in a reaction mixture. ●, The [3H]ABA. ○, The boiled rAOG.
cDNA containing the open reading frame configuration was cloned in the expression vector, pGEX-4T-2, and a glutathione S-transferase (GST)-GTase fusion protein of 80 kD was expressed in Escherichia coli (Fig. 4A). When the control plasmid pGEX-4T-2 was used for preparing the protein expected to express GST, no product signal could be detected (Fig. 4B, lanes 1 and 2). To the contrary, in the reactions using the protein extract containing AOG fused in-frame to the GST-tag (namely recombinant AOG, designated as rAOG), a product signal could be detected in the presence of UDPG (Fig. 4B, lane 3), but not in the presence of G-1-P (Fig. 4B, lane 4). Assuming that the product was ABA-GE, the pH dependence of the rAOG activity was determined with a crude protein solution (Fig. 4C). In these experiments, reaction mixtures were prepared with pH 3.0 to 6.9 sodium citrate and pH 7.3 to 9.0 Tris-HCl buffers. Although the AOG activity was detectable in a wide pH range from 5.0 to 9.0 (Fig. 4C, lanes 3–8), high enzymatic activity was observed in the pH range from 6.0 to 7.3 (Fig. 4C, lanes 4–6). Because Tris-HCl (pH 7.3) was used as an extraction buffer and clear AOG activity was observed with this buffer, the same buffer was adopted in the subsequent experiments.
The product was not detected in the absence of ABA, UDPG, or rAOG (Fig. 4D, lanes 1–3). The reaction did not occur when the boiled rAOG was used (Fig. 4D, lanes 4), whereas a clear signal of the product was obtained with native rAOG (Fig. 4D, lanes 5). The results indicated that the product at RF 0.45 was synthesized from ABA and UDPG, being catalyzed by rAOG. To decrease the glucosidase activities or non-specific reactions by proteins of E. coli cells, a fusion protein with the GST-tag was purified with an affinity column (Fig. 4A, lane 2). When either purified GST or the purified recombinant protein (designated as prAOG) was used, the products show similar patterns by TLC to those observed for each of their crude proteins in Figure 4B, except that the enzymatic activity of prAOG was much higher (about 100- to 300-fold per milligram of protein) than that of the crude protein (rAOG).
Identification of the Product
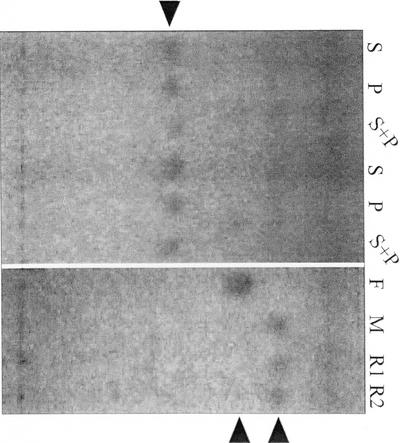
To prove that the product was from ABA and UDPG, [3H]ABA was also used as the substrate together with non-labeled UDPG, in addition to the combination of non-labeled ABA and [14C]UDPG. rAOG was used in this experiment. The products of both combinations showed the same RF value (Fig. 5A, lanes 2 and 3), suggesting that both products were the glycosylated ABA. In the reaction with prAOG, enzymatic activity was only found in the presence of free ABA (Fig. 5B, lane 1) and not in the presence of ABA-Me (Fig. 5B, lane 2), which shows that glycosylation did not occur at the 1′-hydroxyl group. To characterize the ABA-GTase, the non-isotope-labeled product was used for identification experiments. The conjugated ABA was prepared from free ABA and UDPG by using prAOG and then purified by TLC. When the purified product and authentic ABA-GE were developed by TLC, both showed an identical RF value of 0.45 (Fig. 6A, lanes S and P). Furthermore, when a mixture of the product and authentic ABA-GE were cochromatographed by TLC, only a single spot was detected (Fig. 6A, lane S+P). When treated with 0.2 m NaOMe, the enzymatically formed product and authentic ABA-GE afforded compounds with an RF value identical with that of ABA-Me by TLC (Fig. 6B, lanes R1, R2, and M). These results demonstrate that the compound enzymatically formed from ABA and UDPG was the GE of ABA (ABA-GE).
Figure 5.
Identification of the substrate for recombinant GTase. A, Confirmation of the substrate. Lane 1 is free [3H]ABA. Lane 2 is the product from UDPG and [3H]ABA. Lane 3 shows the reaction product from ABA and [14C]UDPG. The reaction mixture was incubated for 2 h at 30°C. B, Reactivity of ABA and the ABA-Me derivative. The mixture was incubated for 4 h at 30°C. F, ABA; M, ABA-Me. The arrowhead indicates the products.
Figure 6.
Identification of the product. A, Comparison of RF values between the ABA-GE standard and the product. S, P, and S+P indicate the ABA-GE standard, the product, and the equimolar mixture of the ABA-GE standard and the product, respectively. An arrowhead indicates the position of ABA-GE separated in a solvent of CHCl3:MeOH:AcOH:H2O (40:15:3:2, v/v). B, Pattern for standard ABA-GE and the product treated with 0.2 m NaOMe, having been separated in a solvent of EtOAc:CHCl3:AcOH (25:15:1, v/v). F and M are free ABA and ABA-Me, respectively. R1 and R2, respectively, show the solvolysis products from the enzyme and standard ABA-GE. The upper arrowhead indicates the position of ABA-Me. The lower arrowhead shows the position of free ABA.
Identity of the GTase product was further confirmed by a gas chromatography/mass spectrometry analysis. Trimethylsilyl derivatives of the product and authentic ABA-GE showed the identical retention time (15.2 min) on gas chromatography, and the same fragmentation: m/z 786 [M+] (7.4%), 768 (5.1%), 450 (40%), 331 (36%), 318 (26%), and 217 (100%). These data clearly indicated that the product of the enzyme reaction was ABA-GE.
Substrate Specificity of ABA-GTase
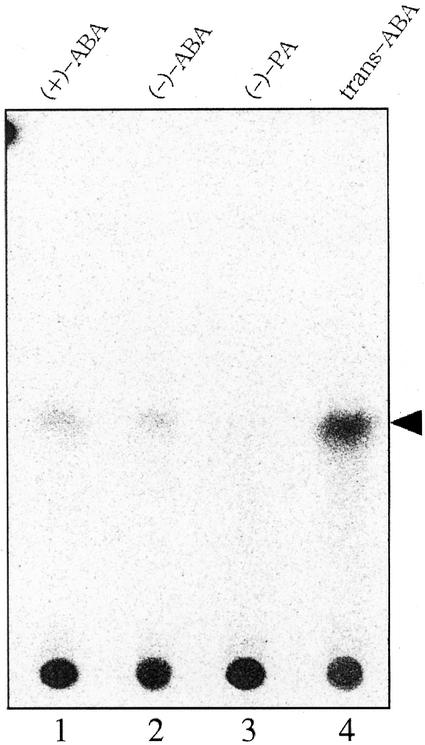
The specificity of prAOG toward the substrates was examined by observing the glycosylation of compounds such as plant hormone or potential organic acids. Although salicylic acid did not induce any signal from the glycosylated products after incubation with [14C]UDPG and prAOG (Fig. 7, lane 1), trans-cinnamic acid gave a weak signal at RF 0.5 (Fig. 7, lane 2). 2-trans-ABA gave the strongest signal of a product at RF 0.45 (Fig. 7, lane 3), overwhelming the clear signal of the product from RS-ABA (Fig. 7, lane 4). The signal intensity of the former product was about three to four times higher than that of the latter. Other hormones such as GA3 (Fig. 7, lane 5), IAA (Fig. 7, lane 6), JA (Fig. 7, lane 7), and zeatin (Fig. 7, lane 8) did not show any glycosylated products. These results indicate that the GTase isolated from adzuki bean seedlings was highly specific for ABA. On the other hand, the reactivity of prAOG toward PA, an immediate metabolite of ABA, and the enantiomer of ABA was likewise examined. Although both (+)-S-ABA and (−)-R-ABA gave clear signals of the glucosylated products after incubation with [14C]UDPG and prAOG (Fig. 8, lane 1 and 2), (−)-PA did not give a detectable signal (Fig. 8, lane 3). The higher reactivity of the ABA-GTase to 2-trans-ABA than to cis-ABA was confirmed in this experiment (Fig. 8, lane 4). The signal intensity of the product from 2-trans-ABA was about 12 times higher than that of the (−)-R-ABA product. The signal intensity of the (+)-S-ABA product (Fig. 8, lane 1) was about two times higher than that of (−)-R-ABA product (Fig. 8, lane 2).
Figure 7.
Substrate specificity of ABA-GTase. Lanes 1 to 8 show the products of prAOG incubated with [14C]UDPG and salicylic acid, trans-cinnamic acid, trans-ABA, cis-ABA, GA3, IAA, JA, and zeatin, respectively. The upper and lower arrowheads indicate the putative trans-cinnamic acid-glucosyl ester and ABA-GE, respectively.
Figure 8.
Characterization of ABA-GTase specificity. Lanes 1 to 4 show the products of prAOG incubated with (+)-S-ABA, (−)-R-ABA, (−)-PA, and 2-trans-ABA, respectively, in the presence of [14C]UDPG. The arrowhead indicates the products discussed in the text.
Northern-Blot Analysis
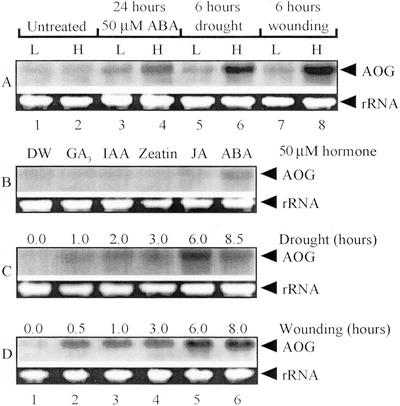
To determine whether the level of AOG gene expression was influenced by exogenous ABA and other stimuli such as drought stress and wounding, total RNA was isolated from leaves or hypocotyls of the intact treated plants. Exogenous ABA promoted the expression of the AOG gene in both the leaves (Fig. 9A, lane 3) and hypocotyls (Fig. 9A, lane 4). Drought and wounding also increased its expression level in leaves (Fig. 9A, lanes 5 and 7) and to a greater extent in hypocotyls (Fig. 9A, lanes 6 and 8). Apart from ABA as the positive control, none of the plant hormones, GA3, IAA, zeatin, or JA, significantly promoted the gene expression (Fig. 9B, lanes 2–5). On the other hand, the drought and wounding treatments rapidly increased the level of the AOG transcript (Fig. 9, C and D). The drought treatment slightly increased the mRNA level within 1 h (Fig. 9C, lane 2), reaching the maximum within 6 h (Fig. 9C, lanes 3–5), and then decreasing (Fig. 9C, lane 6). The wounding treatment increased the expression of the AOG gene, even more rapidly than the drought treatment (Fig. 9D, lanes 1–6). A marked increase in the mRNA level of the AOG gene was detected within 30 min (Fig. 9D, lane 2). After 6 h, the mRNA level reached its maximum and remained at the same level at least until 8 h had elapsed (Fig. 9D, lanes 5 and 6).
Figure 9.
Northern-blot analysis of AOG mRNA. A, mRNA level expressed in the leaves (L) and hypocotyls (H) with no treatment (lanes 1 and 2), and after being treated with 50 μm ABA for 24 h (lanes 3 and 4), drought for 6 h (lanes 5 and 6), and wounding for 6 h (lanes 7 and 8). B, mRNA levels expressed in the hypocotyls of plants treated with DW (lane 1), GA3 (lane 2), IAA (lane 3), zeatin (lane 4), JA (lane 5), and ABA (lane 6). C, mRNA levels expressed in the hypocotyls of plants subjected to drought for up to 8.5 h. D, mRNA levels expressed in the hypocotyls of plants subjected to wounding for 0.0 to 8.0 h. The upper or lower arrowheads in A through D indicate the AOG transcript and rRNA as an indicator of the total RNA quantity (7 μg/lane), respectively.
Southern-Blot Analysis
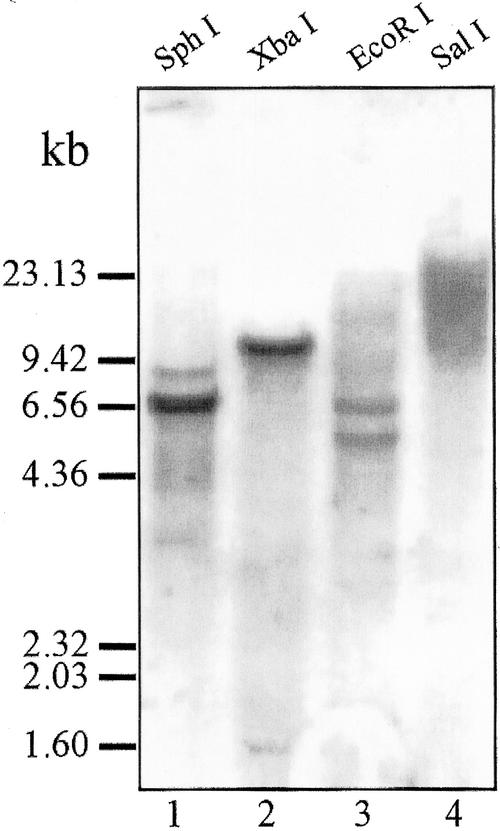
Restriction enzymes, whose recognition site was not found in the cDNA sequence, were used for genomic DNA digestion. In the hybridization process, two copies of the gene were detected under highly stringent conditions (Fig. 10, lanes 1–4).
Figure 10.
Southern-blot analysis of genomic DNA digested with SphI (lane 1), XbaI (lane 2), EcoRI (lane 3), and SalI (lane 4). The amount of DNA loaded was 20 μg/lane. Hybridization and washing were carried out at 65°C and 68°C, respectively.
DISCUSSION
It is widely accepted that ABA is transported in plants through the xylem and phloem (Zeevaart and Boyer, 1984; Wolf et al., 1990). Exogenous ABA could be slightly metabolized to DPA or PA during such transport (Everat-Bourbouloux, 1982). We have shown in the present work that exogenous ABA was partly metabolized in the plants to other forms with higher polarity (Fig. 2), when ABA was taken up by plants without roots through the xylem by leaf-transpiration. A comparison of the RF values for the products and authentic ABA-GE suggested that one of the ABA metabolites was ABA-GE. This result provided us with the basis to isolate an ABA-glucosyl transferase gene from adzuki been seedlings. The ABA metabolites were detectable in the hypocotyls within 2 h of the ABA treatment and in the leaves after 4 h at the earliest (Fig. 2). This difference in the metabolism time course between the hypocotyls and leaves cannot be explained solely by a greater delay in the translocation of absorbed ABA to the leaves than to the hypocotyls, because a substantial level of free ABA was already present in the leaves within 2 h of the ABA treatment. There could be other reasons: e.g. the basal and/or induced enzymatic activity was lower in the leaf cells than in the hypocotyl cells. We will discuss this point later in conjunction with some experimental results on ABA-GTase expression.
Expecting the presence of ABA-GTase in adzuki, and assuming that a high concentration of ABA would increase the expression level of this gene, we screened ABA-inducible GTase genes and isolated one encoding ABA-GTase (Fig. 3). This approach for selecting GTases from many homologous genes is reasonable, because glycosylation is usually involved in the rapid inactivation of bioactive compounds, which are toxic at a high concentration. It has been shown, for instance, that a GTase for salicylic acid from tobacco was substrate inducible (Lee and Raskin, 1999). The ABA-inducible GTase could use UDPG as a glucosyl-source, but not G-1-P (Fig. 4). This enzyme also showed specificity to free ABA as a glucosyl acceptor (Figs. 5 and 7). These findings demonstrate that the enzyme was an ABA-specific UDP-Glc-transferase. The substrate specificity is also supported by the results that the AOG gene expression was increased by the application of ABA but not by other plant hormones (Fig. 9B).
The optimum pH value for the enzyme activity of recombinant protein ranges from 6.0 to 7.4, whereas Lehmann and Schütte (1980) have reported that it was 5.0, and Schwarzkopf and Miersch (1992), 5.2. However, they did not necessarily determine the optimum pH of native ABA-GTase itself, because the β-glucosidase activities were not completely separated from the partially purified GTases, which may have affected the pH dependency of the native enzyme. We determined the pH dependency with a recombinant protein and avoided the affects of other contaminants. It should be noted, however, that the recombinant enzyme fused to GST may have had different pH dependency from that of the native enzyme due to a conformational difference.
The GTase did not catalyze the glucosylation of ABA-Me (Fig. 5B) that had been methylated at the carboxyl of ABA. This result suggests that the GTase could catalyze only the esterification of ABA to ABA-GE. To elucidate whether the GS formation at C-1′ of ABA was involved in ABA conjugation by ABA-GTase, we examined the enzymatic products to determine their structure. The behavior of the product on TLC was compared with that of the authentic ABA-GE, and both showed identical RF values (Fig. 6A). Solvolysis with NaOMe of both the product and authentic material afforded solvolysates showing RF values identical with that of ABA-Me by TLC (Fig. 6B). These results demonstrate that the glucosyl group on the GTase product was located at the C-1 position of ABA. The product was definitively identified by gas chromatography/mass spectrometry as ABA-GE.
Because the enzyme was active for conjugating ABA, but not other plant hormones, the substrate specificity was examined in more detail. 2-trans-ABA was found to be a better substrate than RS-ABA (Fig. 7). In young tomato (Lycopersicon esculentum) shoots, 2-trans-ABA was reportedly converted to 2-trans-ABA-GE 10-fold faster than RS-ABA to RS-ABA-GE, when an equimolar mixture of 2-trans- and RS-ABA was fed to the shoots (Milborrow, 1970). The higher 2-trans-ABA GTase activity may reflect the more effective conversion of 2-trans-ABA to 2-trans-ABA-GE in plants. The GTase also slightly catalyzed the glycosylation of trans-cinnamic acid (Fig. 7). This low reactivity may come from the structural similarity between the side chains of ABA and trans-cinnamic acid; i.e. both have an α,β-unsaturated carboxyl group. Although the reactivity of the GTase with trans-cinnamic acid was much lower than that with ABA, it has to be concluded that the recombinant GTase did not strictly recognize the ABA structure. Because trans-cinnamic acid and its derivatives are abundant in plants as precursors of lignin and other phenylpropanoids, it is important to consider how the glycosylation of ABA and of phenolic compounds is differentially regulated. One possible explanation is that the GTase expression is specifically tissue- and cell-type regulated so that the appropriate substrates are glycosylated. It is also possible that, because the GTase used in the experiment was a recombinant enzyme fused to GST and the reaction conditions were not necessarily identical to those in plants, the substrate specificity in vitro may not reflect that in vivo.
The recombinant GTase did not catalyze the glycosylation of PA (Fig. 8, lane 3). This finding indicates that the ABA-GTase can distinguish the ABA structure from that of PA. The ABA GTase not only catalyzed the glucosylation of (+)-S-ABA but also catalyzed that of (−)-R-ABA (Fig. 8, lane 1 and 2). Comparing the signal intensity between the products of (+)-S-ABA and (−)-R-ABA, the results indicate that the ABA-GTase has higher reactivity to (+)-S-ABA than to (−)-R-ABA. When (+)-S-ABA and/or (−)-R-ABA were fed to X. strumarium, tomato plants, or suspension-cultured maize (Zea mays) cells, the glucosylation rate of (+)-S-ABA was different from that of (−)-R-ABA (Vaughan and Milborrow, 1984; Boyer and Zeevaart, 1986; Balsevich et al., 1994). In these plants or cells, natural (+)-S-ABA was mainly converted to PA, whereas (−)-R-ABA was easily conjugated to form the GE. Similar results were obtained with bean (Phaseolus vulgaris; Zeevaart and Milborrow, 1976). Considering that the experiments were performed in an in vivo system, the different metabolism between (+)-S-ABA and (−)-R-ABA in plants or cells may be due to the higher affinity or the advantageous access of the enzyme catabolizing ABA to PA to (+)-S-ABA rather than to (−)-R-ABA.
The expression level of the AOG gene was very low under normal conditions (Fig. 9, lane 1). A high concentration of ABA (50 μm) and drought and wounding treatments increased its expression level in the plants (Fig. 9A, lanes 3–8) more effectively in the hypocotyls (Fig. 9A, lane H) than in the leaves (Fig. 9A, lane L). The differences among the mRNA levels in those tissues were especially conspicuous with the drought and wounding treatments (Fig. 9A). As already described, ABA taken up from the hypocotyl was rapidly translocated into the leaves, and ABA in the hypocotyls and leaves reached almost the same levels in 2 h (Fig. 2). Therefore, the difference in the GTase inducibility by exogenous ABA in those tissues could be attributable to the difference in the responsiveness to ABA and not in the arrival time and concentration of ABA. The result from drought stress further supports the supposition that the different patterns of ABA metabolism between the two tissues were due to different regulation of the metabolic enzymes, because the drought stress would have been equally applied to these tissues. According to the results from the Southern-blot analysis (Fig. 10), the gene had two copies. It is, therefore, probable that one of the gene promoters was hypocotyl specific and highly responsive to the stress treatment, whereas the other was leaf specific and less stress inducible.
Several studies have demonstrated that the endogenous ABA level is rapidly increased by severalfold or more when plants are under conditions that cause dehydration (Ilahi and Dörffling, 1982; Herde et al., 1996; Ingram and Bartels, 1996; Bray, 1997; Moons et al., 1997; Birkenmeier and Ryan, 1998; Qin and Zeevaart, 1999). The increased concentration of endogenous ABA produced during the response to stress may also promote the metabolism of ABA to ABA conjugates by increasing the corresponding enzymes. Thus, the increased AOG expression under stress may be considered to come from the induction of a high concentration of endogenous ABA. In fact, several reports have demonstrated that endogenous ABA-GE increased during stress (Zeevaart, 1980, 1983; Creelman et al., 1987), although there was also the contrary result that conjugated ABA was decreased and free ABA was increased in needles of P. menziesii during water stress (Johnson and Ferrell, 1982). A stress treatment was more effective for inducing AOG than application of ABA. One explanation for this is that the exogenous ABA is less effective than endogenous ABA for inducing AOG (Fig. 9A), because exogenous ABA could be partially catabolized before entering the cells (Everat-Bourbouloux, 1982; Zeevaart and Boyer, 1984). The increased level of endogenous ABA may more efficiently and directly induce the GTase expression during stress, even though it is local and small. An alternative pathway to enhance the AOG mRNA level not through ABA may exist, because a clear and rapid increase of the mRNA was observed in the plants whose roots were carefully cut off without apparent wilting (Fig. 9D).
Assuming that the effect of a stress treatment is manifested by the ABA produced, these reports and our findings on the AOG expression under stress conditions suggest that ABA-GTase takes part in hormonal homeostasis to reduce the excess amount of free ABA. According to other reports that free ABA was not released from ABA-GE during stress response in young tomato, silverbeet shoots, and X. strumarium leaves (Milborrow, 1978; Zeevaart, 1983), we may conclude that ABA-GE is not involved in supplying ABA. To confirm this, a detailed study will be needed on the fluctuation of ABA and ABA-GE, and on the expression of ABA biosynthetic enzymes and the glucosidase responsible for the hydrolysis of ABA-conjugates and GTase during response to stress.
MATERIALS AND METHODS
Plant Materials
Adzuki bean (Vigna angularis) seeds were washed and soaked in distilled water (DW) at 25°C ± 2°C in the dark overnight. The swollen seeds were planted and grown in a greenhouse at 25°C ± 2°C, with 7- to 8-d-old seedlings being used for the subsequent experiments.
Chemical Compounds
UDPG [d-Glc-14C (U), 254 μCi/mmol; Moravek], G-1-P [14C(U), 254 μCi/mmol; Moravek], RS-[G-3H]ABA (30 Ci/mmol; Amersham Pharmacia Biotech AB, Uppsala), (+)-S-ABA (Sigma, St. Louis), (−)-R-ABA (Sigma), and ABA-GE (Lancaster, Newgate, UK) were obtained from commercial sources. ABA-Me was prepared by treating ABA with ethereal diazomethane. Natural (−)-PA was a gift from Dr. Nobuhiro Hirai (Kyoto University).
Plant Treatments
Eight-day-old seedlings were used in the feeding experiments. The plants, whose roots had been removed, were placed in a 5 × 10−5 m [3H]ABA solution for 0 to 8 h or in a 5 × 10−5 m non-labeled ABA solution for 6 h and then in the [3H]ABA solution for 2 h. Total RNA was isolated after the roots of 7-d-old intact seedlings had been dipped into a hormone solutions containing 5 × 10−5 m RS-ABA, jasmonic acid (JA), IAA, gibberellic acid (GA3), or zeatin and incubated for 24 h. In the other experiments, the leaves of 8-d-old intact plantlets were sprayed with each of the hormone solutions and incubated for 8 h. Eight-day-old seedlings were also used for the wounding treatment. The hypogeal parts including their roots were removed in DW, and the hypocotyls were immediately transferred and placed in fresh DW for 0 to 8 h. The intact seedlings were transplanted into a dry molding for the drought treatment for 0 to 8 h.
Extraction and Detection of ABA-GE from the Plantlets
One gram fresh weight of the hypocotyl segments or leaves was homogenized in 5 mL of 85% (v/v) methanol. After filtration, the extract solution was concentrated under reduced pressure to remove the methanol, and the remaining aqueous solution was acidified to pH 4 to 5 with 1 m acetic acid, before being extracted with n-butanol. The butanol phase was concentrated under reduced pressure at 50°C and then dissolved in 10 μL of 60% (v/v) methanol. A 2-μL aliquot of the solution was applied to a silica gel plate, developed with a solvent of chloroform:methanol:acetic acid:water (40:15:3:2, v/v). The radioactivity was visualized on a Bas-Tr2040 imaging-plate, (Fuji Photo Film, Tokyo) and analyzed by a BAS-2000 imaging analyzer (Fuji Photo Film).
Preparation of Adzuki Hypocotyl cDNA
The hypocotyl segments (3 cm from an apical bud) of the seedlings that had been treated with 5 × 10−5 m RS-ABA for 6 h were collected and frozen at −80°C until needed. Total RNA was isolated according to the guanidine-hydrochloride method combined with phenol/chloroform extraction as described by Logemann et al. (1982) and then purified by the CsCl cushion centrifugation method. cDNA was prepared by a Marathon cDNA amplification kit (CLONTECH Laboratories, Palo Alto, CA) according to the recommended protocol for the kit, using the poly (A+) RNA purified by chromatography on oligo(dT)-cellulose type 7 (Amersham Pharmacia Biotech).
Cloning and Sequencing
To obtain fragments of the GTase genes, a nested-PCR was used comprising two-step PCR with two degenerate antisense primers. The degenerate primers were designed according to the consensus sequence (Fig. 3, italic-underlined) of UDP-GTases: DP-1 designed for 5′-CYYTCYARBRTHGARTTCCAHCCRC-3′, and DP-2 designed for 5′-GARTTCCAHCCRCWRTGBGTBAMRA-3′. The adaptor primer (AP-1) in the Marathon cDNA amplification kit (CLONTECH Laboratories) was used as the sense-primer. The PCR reaction was optimized for a 50 μL of reaction mixture per tube containing 220 μm dNTP mix, 0.2 μm AP-1, 4.0 μm DP-1 or DP-2, 5.0 of μL cDNA (5.0 ng) or 1.0 μL of the DNA solution from PCR, 0.5 μL of Advantage cDNA polymerase (CLONTECH Laboratories), and 5.0 μL of the PCR buffer in the polymerase kit by using a PCR thermal cycler (Takara, Kyoto). The DP-1 and AP-1 pair of primers was used for first-step PCR, the reaction temperature being set to 95°C 30 s, 55°C for 60 s, and 72°C for 2 min with 25 cycles. A 1-μL amount of the product mixture from the first step of the PCR reaction was added into the second-step PCR mix containing the primer pair of DP-2 and AP-1. The reaction conditions were the same as those of the first PCR, except that the annealing temperature was increased to 58.5°C. PCR fragments of 1.0 to 1.2 kb were collected after their separation on 1.0% (w/v) agarose, purified by Geneclean (Bio 101, Vista, CA) according to the methods described in the protocol, and then ligated into the pGEM T-easy plasmid vector (Promega, Madison, WI), which was transformed into JM109 Escherichia coli. The plasmids were purified with a Qiaprep Spin Miniprep kit (Qiagen USA, Valencia, CA). To obtain sequence information on full-length cDNAs, 5′- or 3′-RACE PCR reactions were performed by using primers based on the cDNA-fragment sequences. These were designed as 5′-GATGGAACATGTCGACGACGATGCAAT-3′ (for 5′-RACE PCR) and 5′-GAAGGGTTCGAGCAGAGGATGAAGGA-3′ (for 3′-RACE PCR).
The coding-region for the GTase gene was obtained by a PCR reaction with primers, which had been designed according to the full-length cDNA sequence. The sense-primer was 5′-GGAATTCACATGAAGACCTTAACCCCTTCGGTGG-3′, and the antisense-primer was 5′-GGAATTCTTAGCCCTGGTTTGCGCATGTGCGAG-3′. The EcoRI site was added at the 5′ end of each primer. The cDNA fragment, which contained the complete coding region encoding a GTase, was ligated in the EcoRI site of the pGEX-4T-2 plasmid vector of the GST fusion system (Amersham Pharmacia Biotech) and cloned in JM 109 E. coli. The in-frame connection of the GST-GTase fusion gene (designated as pGEX-GTase) was confirmed by sequencing according to the recommended sequencing primers in the GST fusion system. All the sequences were analyzed by a DNA sequence analyzer (model 373A, Applied Biosystems, Foster City, CA) with dye primers (-21 M13 and M13), or by a DSQ-2000L DNA sequencer (Shimadzu, Kyoto) with fluorescein isothiocyanate-labeling primers (M4 and RV-M).
Expression and Isolation of the Recombinant Proteins
The pGEX-GTase plasmid was transformed into E. coli BL21 (DE3) pLysS-competent cells (Stratagene, La Jolla, CA) according to the manufacturer's protocol, before selecting the colonies on an Luria Bertani (LB) plate containing 1.5% (w/v) agar and 100 μg mL−1 ampicillin. A single colony was grown overnight in 5.0 mL of LB broth containing 100 μg mL−1 ampicillin at 26°C. A 2.0-mL aliquot of the culture was inoculated into 100 mL of fresh LB broth containing 100 μg mL−1 ampicillin medium containing 2% (w/v) Glc and incubated while shaking at 22°C. After the culture had grown to a cell density of OD600 = 1.0–1.2, the fusion protein was induced by adding isopropyl-β-d-thiogalactoside at 0.2 mm. After incubating for 2 to 5 h, the cells were collected by centrifugation at 10,000g for 10 min, washed twice with a 10 mm Tris-HCl buffer (pH 7.3), and resuspended in 10 mL of the 10 mm Tris-HCl buffer (pH 7.3) containing 150 mm NaCl and 1 mg mL−1 lysozyme. After incubation for 10 min at room temperature, the suspension was frozen and left overnight at −80°C. The cell suspension was thawed in a 30°C water bath, chilled on ice, and then sonicated. Soluble proteins were collected by centrifugation. The recombinant protein was purified in a glutathione 4B affinity column according to the recommended method for the GST fusion system. The excess glutathione in the eluate was removed by ultrafiltration at 4°C. After being washed three times with a buffer containing 10 mm Tris-HCl (pH 7.3) and 100 mm NaCl, the purified protein was collected and stored at −80°C.
Enzyme Assays
Enzyme activity was determined by using either the affinity-purified or crude recombinant proteins. The reaction mixture was made up in 100 μL of 100 mm Tris-HCl at pH 7.3 containing 5 μg of purified protein or 40 μg of crude protein and either (a) 5.0 mm ABA, 3.0 mm UDPG, and 3.7 kBq of [14C]UDPG, or (b) 3.0 mm ABA, 5.0 mm UDPG, and 3.7 kBq of [3H]ABA. After incubation for 2 to 4 h at 30°C, the reaction was stopped by adding 20 μL of 1 m acetic acid. Instead of free ABA in reaction mixture a, 5.0 mm ABA-Me was also used to examine the substrate specificity of the enzyme. The product was extracted twice with 120 μL of n-butanol and then concentrated to 20 μL. A 2-μL aliquot of the concentrate was applied to a thin-layer plate and developed with the solvent system already described. Likewise, G-1-P and [14C]G-1-P were used, respectively, instead of UDPG and [14C]UDPG for examining the glucosyl source. The same method was used to determine the optimum pH value of the enzyme by using sodium citrate in the pH range from 4.0 to 6.9 and Tris-HCl from pH 7.3 to 9.0. The substrate specificity of the enzyme was also examined by using such possible glucosyl acceptors as salicylic acid, JA, IAA, GA3, and zeatin, each at a 5.0 mm concentration.
Chemical Analysis of the Reaction Product
The spots of the product showing UV absorption at 254 nm were recovered from the chromatogram, extracted with 60% (v/v) methanol, and evaporated to dryness. The purified product and authentic ABA-GE were treated with 0.2 m sodium methoxide in methanol at −10°C for 1 h. The reaction mixtures were then applied to a thin-layer plate and developed with ethyl acetate:chloroform:acetic acid (25:15:1, v/v) to compare their RF values.
The glucosylated ABA from the enzymatic reaction was purified by reversed phase partition chromatography (1 g, octadecylsilanized silica gel, Fuji-Davison, Tokyo) eluted step wise with 20%, 25%, and 28% (v/v) acetonitrile 1 mL each. The 25% and 28% fractions containing the ABA-conjugates were combined and concentrated to dryness and then trimethylsilylated in N-methyl-N-(trimethylsilyl)trifluoroacetamide by heating for 1 min. Authentic ABA-GE was likewise trimethylsilylated. The derivatives were analyzed with an HP-9800II gas chromatograph connected with a mass spectrometer (M-4100, Hitachi, Tokyo). A capillary column DB-1 (15-m × 0.3-mm i.d., 0.2 mm thick; J&W Scientific, Folsom, CA) was used under the following condition: initial oven temperature, 100°C (0–2 min) and then linear gradient to 300°C at 15°C min−1; He flow, 1 mL min−1; ionization, EI (70eV).
Northern-Blot Analysis
Total RNA was isolated from seedlings by several treatments according to the guanidine-hydrochloride system combined with phenol/chloroform extraction as already described. Total RNA (7 μg lane−1) was separated on 1% (w/v) formaldehyde agarose gel and then transferred to a Hybond N+ nylon membrane (Amersham Pharmacia Biotech) according to the manufacturer's instructions. To efficiently synthesize a digoxigenin-UTP-labeled RNA probe, pGEM T-easy plasmid DNA containing the PCR fragment (from 63 to 1,079 in Fig. 3) was used for preparing the DNA template by a PCR reaction with primers T7F (5′-CAGGGTTTTCCCAGTCACGACGTTG-3′) and SP6R (5′-CACACAGGAAACAGCTATGACCATG-3′) according to the DNA sequence of the pGEM T-easy plasmid. The T7 or SP6 promoter sequence was contained in the PCR product. The undesired sequence downstream of the T7 or SP6 promoter was cut out with the NdeI or NcoI restriction enzyme to leave 5′ overhanging ends, and purified by agarose gel. The digoxigenin-UTP-labeled RNA probe was prepared with the Dig RNA labeling kit (Roche Molecular Biochemicals, Summerville, NJ) according to the manufacturer's protocol. The membranes were prehybridized, hybridized, and washed at 68°C before being stained according to the manufacturer's instructions.
Southern-Blot Analysis
Genomic DNA was extracted from the budding adzuki beans without cotyledons by using the cetyl-trimethyl-ammonium bromide method (Rogers and Bendich, 1985) with some modifications. Genomic DNA (20 μg) was digested with restriction enzymes, separated on 1% (w/v) agarose gel, and transferred to a Hybond-N+ nylon membrane (Amersham Pharmacia Biotech). The membrane was prehybridized, hybridized at 65°C, and washed at 68°C. The other conditions were the same as those used for the northern-blot analysis.
ACKNOWLEDGMENT
We thank Dr. Nobuhiro Hirai (Kyoto University, Japan) for his generous gift of (−)-PA.
Footnotes
This work was supported in part by a grant from Bio-oriented Technology Research Advancement Institution.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.001784.
LITERATURE CITED
- Albinsky D, Masson JE, Bogucki A, Afsar K, Vaass I, Nagy F, Paszkowski J. Plant responses to genotoxic stress are linked to an ABA/salinity signaling pathway. Plant J. 1999;17:73–82. [Google Scholar]
- Balsevich JJ, Cutler AJ, Lamb N, Friesen LJ, Kurz EU, Perras MR, Abrams SR. Response of cultured maize cells to (+)-abscisic acid, (−)-abscisic acid, and their metabolite. Plant Physiol. 1994;106:135–142. doi: 10.1104/pp.106.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthe P, Garello G, Bianco-Trinchant J, le Page-Degivry MT. Oxygen availability and ABA metabolism in Fagus sylvatica seeds. Plant Growth Regul. 2000;30:185–191. [Google Scholar]
- Birkenmeier GF, Ryan CA. Wounding signaling in tomato plants: evidence that ABA is not a primary signal for defense gene activation. Plant Physiol. 1998;117:687–693. doi: 10.1104/pp.117.2.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer GL, Zeevaart JAD. 7′-Hydroxy (−)-R-abscisic acid: a metabolite of feeding (−)-R-abscisic acid to Xanthium strumarium. Phytochemistry. 1986;25:1103–1105. [Google Scholar]
- Bray EA. Plant responses to water deficit. Trends Plant Sci. 1997;2:48–54. [Google Scholar]
- Bray EA, Zeevaart JAD. The compartmentation of abscisic acid and β-d-glucopyranosyl abscisate in mesophyll cells. Plant Physiol. 1985;79:719–722. doi: 10.1104/pp.79.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler PM, Robertson M. Gene expression regulated by abscisic acid and its relation to stress tolerance. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:113–141. [Google Scholar]
- Creelman RA, Gage DA, Stults JT, Zeevaart JAD. Abscisic acid biosynthesis in leaves and roots of Xanthium strumarium. Plant Physiol. 1987;85:726–732. doi: 10.1104/pp.85.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon SC, Martin RC, Mok MC, Shaw G, Mok DWS. Zeatin glycosylation enzymes in Phaseolus: isolation of O-glucosyltransferase from P. lunatus and comparison to O-xylosyltransferase from P. vulgaris. Plant Physiol. 1989;90:1316–1321. doi: 10.1104/pp.90.4.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everat-Bourbouloux A. Transport and metabolism of labelled abscisic acid in broad-bean plants (Vicia faba L.) Physiol Plant. 1982;54:431–439. [Google Scholar]
- Fincher GB. Molecular and cellular biology associated with endosperm mobilization in germinating cereal grains. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:305–346. [Google Scholar]
- Herde O, Atzorn R, Fisahn J, Wasternack C, Willmitzer L, Peña-Cortés H. Localized wounding by heat initiates the accumulation of proteinase inhibitor II in abscisic acid-deficient plants by triggering jasmonic acid biosynthesis. Plant Physiol. 1996;112:853–860. doi: 10.1104/pp.112.2.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornberg C, Weiler EW. High-affinity binding sites for abscisic acid on the plasmalemma of Vicia faba guard cells. Nature. 1984;310:321–324. [Google Scholar]
- Ilahi I, Dörffling K. Changes in abscisic acid and proline levels in maize varieties of different drought resistance. Physiol Plant. 1982;55:129–135. [Google Scholar]
- Ingram J, Bartels D. The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:377–403. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- Johnson JD, Ferrell WK. The relationship of abscisic acid metabolism to stomatal conductance in Douglas fir during water stress. Physiol Plant. 1982;55:431–437. [Google Scholar]
- Koshimizu K, Fukui H, Mitsui T, Ogawa Y. Identity of lupin inhibitor with abscisin II and its biological activity on growth of rice seedling. Agric Biol Chem. 1966;30:941–943. [Google Scholar]
- Lee H, Raskin I. Purification, cloning, and expression of a pathogen inducible UDP-glucose: salicylic acid glucosyltransferase from tobacco. J Biol Chem. 1999;274:36637–36642. doi: 10.1074/jbc.274.51.36637. [DOI] [PubMed] [Google Scholar]
- Lehmann H, Glund K. Abscisic acid metabolism: vacuolar/extravacuolar distribution of metabolites. Planta. 1986;168:559–562. doi: 10.1007/BF00392276. [DOI] [PubMed] [Google Scholar]
- Lehmann H, Schütte HR. Purification and characterization of an abscisic acid glucosylating enzyme from cell suspension cultures of Macleaya microcarpa. Z Pflanzenphysiol. 1980;96:277–280. [Google Scholar]
- Lehmann H, Vlasov P. Plant growth and stress: the enzymic hydrolysis of an abscisic acid conjugate. J Plant Physiol. 1988;132:98–101. [Google Scholar]
- Leung J, Giraudat J. Abscisic acid signal transduction. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:199–222. doi: 10.1146/annurev.arplant.49.1.199. [DOI] [PubMed] [Google Scholar]
- Leznicki AJ, Bandurski RS. Enzymic synthesis of indole-3-acetyl-1-O-β-d-glucose: I. Partial purification and characterization of the enzyme from Zea mays. Plant Physiol. 1988a;88:1474–1480. doi: 10.1104/pp.88.4.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leznicki AJ, Bandurski RS. Enzymic synthesis of indole-3-acetyl-1-O-β-d-glucose: II. Metabolic characteristics of the enzyme. Plant Physiol. 1988b;88:1481–1485. doi: 10.1104/pp.88.4.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann J, Schell J, Willmitzer L. Improved method for the isolation of RNA from plant tissues. Anal Biochem. 1982;163:16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- Martin RC, Mok MC, Moc DWS. Isolation of a cytokinin gene, ZOG1, encoding zeatin O-glucosyltransferase from Phaseolus lunatus. Proc Natl Acad Sci USA. 1999;96:284–289. doi: 10.1073/pnas.96.1.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DR. Genetic control and integration of maturation and germination pathways in seed development. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:71–93. [Google Scholar]
- Milborrow BV. The metabolism of abscisic acid. J Exp Bot. 1970;21:17–29. [Google Scholar]
- Milborrow BV. The stability of conjugated abscisic acid during wilting. J Exp Bot. 1978;29:1059–1066. [Google Scholar]
- Moons A, Prinsen E, Bauw G, Montagu MV. Antagonistic effects of abscisic acid and jasmonates on salt stress-inducible transcripts in rice roots. Plant Cell. 1997;9:2243–2259. doi: 10.1105/tpc.9.12.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill SJ, Horgan R, Heald JK. Determination of the levels of abscisic acid-glucose ester in plants. Planta. 1983;157:371–375. doi: 10.1007/BF00397410. [DOI] [PubMed] [Google Scholar]
- Qin XQ, Zeevaart JAD. The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proc Natl Acad Sci USA. 1999;96:15354–15361. doi: 10.1073/pnas.96.26.15354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock CD. Pathways to abscisic acid-regulated gene expression. New Phytol. 2000;148:357–396. doi: 10.1046/j.1469-8137.2000.00769.x. [DOI] [PubMed] [Google Scholar]
- Rogers SO, Bendich AJ. Extraction of DNA from milligram amounts of fresh herbarium and mummified plant tissues. Plant Mol Biol. 1985;5:69–76. doi: 10.1007/BF00020088. [DOI] [PubMed] [Google Scholar]
- Schwarzkopf E, Miersch O. In vitro glucosylation of dihydrojasmonic acid and abscisic acid. Biochem Physiol Pflanz. 1992;188:57–65. [Google Scholar]
- Söderman EM, Brocard IM, Lynch TJ, Finkelstein RR. Regulation and function of the Arabidopsis ABA-insensitive 4 gene in seed and the abscisic acid response signaling network. Plant Physiol. 2000;124:1752–1765. doi: 10.1104/pp.124.4.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szerszen JB, Szczyglowski K, Bandurski RS. iaglu, a gene from Zea mays involved in the conjugation of growth hormone indole-3-acetic acid. Science. 1994;265:1699–1701. doi: 10.1126/science.8085154. [DOI] [PubMed] [Google Scholar]
- Vaughan GT, Milborrow BV. The resolution by HPLC of RS-[2-14C]Me 1′,4′-cis-diol of abscisic acid and metabolism of (−)-R- and (+)-S-abscisic acid. J Exp Bot. 1984;35:110–120. [Google Scholar]
- Vogt T, Jones P. Glycosyltransferases in plant natural product synthesis: characterization of a supergene family. Trends Plant Sci. 2000;5:380–386. doi: 10.1016/s1360-1385(00)01720-9. [DOI] [PubMed] [Google Scholar]
- Walton DC. Structure-activity relationships of abscisic acid analogs and metabolites. In: Addicott FT, editor. Abscisic Acid. New York: Praeger Publishers; 1983. , pp, 113–146. [Google Scholar]
- Wolf O, Jeschke WD, Hartung W. Long distance transport of abscisic acid in NaCl-treated intact plants of Lupinus albus. J Exp Bot. 1990;41:593–600. [Google Scholar]
- Zeevaart JAD. Changes in the levels of abscisic acid and its metabolites in excised leaf blades of Xanthium strumarium during and after water stress. Plant Physiol. 1980;66:672–678. doi: 10.1104/pp.66.4.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart JAD. Metabolism of abscisic acid and its regulation in Xanthium leaves during and after water stress. Plant Physiol. 1983;71:477–481. doi: 10.1104/pp.71.3.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart JAD. Abscisic acid metabolism and its regulation. In: Hooykaas PJJ, Haall MA, Libbenga KR, editors. Biochemistry and Molecular Biology of Plant Hormones. Amsterdam: Elsevier Science; 1999. pp. 189–207. [Google Scholar]
- Zeevaart JAD, Boyer GL. Accumulation and transport of abscisic acid and its metabolites in Ricinus and Xanthium. Plant Physiol. 1984;74:934–939. doi: 10.1104/pp.74.4.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart JAD, Creelman RA. Metabolism and physiology of abscisic acid. Annu Rev Plant Physiol Plant Mol Biol. 1988;39:439–473. [Google Scholar]
- Zeevaart JAD, Milborrow BV. Metabolism of abscisic acid and the occurrence of epi-dihydrophaseic acid in Phaseolus vulgaris. Phytochemistry. 1976;15:493–500. [Google Scholar]