Abstract
Regulatory elements that control tetracycline resistance in Escherichia coli were previously converted into highly specific transcription regulation systems that function in a wide variety of eukaryotic cells. One tetracycline repressor (TetR) mutant gave rise to rtTA, a tetracycline-controlled transactivator that requires doxycycline (Dox) for binding to tet operators and thus for the activation of Ptet promoters. Despite the intriguing properties of rtTA, its use was limited, particularly in transgenic animals, because of its relatively inefficient inducibility by doxycycline in some organs, its instability, and its residual affinity to tetO in absence of Dox, leading to elevated background activities of the target promoter. To remove these limitations, we have mutagenized tTA DNA and selected in Saccharomyces cerevisiae for rtTA mutants with reduced basal activity and increased Dox sensitivity. Five new rtTAs were identified, of which two have greatly improved properties. The most promising new transactivator, rtTA2S-M2, functions at a 10-fold lower Dox concentration than rtTA, is more stable in eukaryotic cells, and causes no background expression in the absence of Dox. The coding sequences of the new reverse TetR mutants fused to minimal activation domains were optimized for expression in human cells and synthesized. The resulting transactivators allow stringent regulation of target genes over a range of 4 to 5 orders of magnitude in stably transfected HeLa cells. These rtTA versions combine tightness of expression control with a broad regulatory range, as previously shown for the widely applied tTA.
The repressor of the Tn10 tetracycline (Tc) resistance operon of Escherichia coli (TetR) recognizes its genuine operator (tetO) with unusual specificity (1). The interaction between repressor and operator is efficiently prevented by Tc, particularly by doxycycline (Dox) that binds to TetR with high affinity (2). These parameters, as well as the fact that Dox, a nontoxic compound widely used in medicine, readily traverses cell membranes, have made the elements of the tet resistance operon attractive for the development of a transcription control system that would function in higher eukaryotic cells. It was expected that, because of their evolutionary distance, the prokaryotic regulatory elements would not interfere with the metabolism of, e.g., a mammalian cell. Accordingly, it appeared feasible to superimpose onto the complex regulatory network of a cell an independent control circuit that could be governed from outside at will. Indeed, by fusing TetR with transcription activation domains, Tc controlled transactivators (tTAs) were obtained that efficiently activate Ptet, minimal promoters fused downstream of an array of tetO sequences (3, 4). The presence of Dox would prevent this activation.
A TetR mutant containing four amino acid exchanges of which three are located in the protein core, where inducer is bound and triggers the conformational change necessary for induction and where dimerization takes place (5, 6), exhibits a reverse phenotype when fused to a transcription activator (C-terminal portion of VP16 of herpes simplex virus) and examined in mammalian cells (7). This mutant, called rtTA, requires Dox or anhydrotetracycline for activation of Ptet. Both Dox-controlled transcription activation systems, which operate in a complementary way, have been widely used in studies of gene function in various cellular systems, as well as in whole organisms including yeast, plants, Drosophila, mice, and rats (for review see ref. 8).
Despite numerous successful applications, the currently available Tet regulatory systems show some limitations. Here, we focus on the previously described rtTA, which requires Dox or anhydrotetracycline for binding to and activating Ptet. Controlling genes via rtTA is of particular advantage in transgenic animals whenever a gene has to be kept silent, e.g., during development, before it is activated at a defined point in time. Obviously, an animal is more rapidly “saturated” with an inducer than it is depleted of it. For kinetic reasons, rtTA is, therefore, often preferable over the tTA system (9). Detailed analysis of our previously described rtTA in cell culture as well as in transgenic mice has, however, revealed some properties that limit its application. For example, full activation of Ptet-1 (PhCMV*-1 in ref. 1) via rtTA is achieved only at 1–2 μg/ml of Dox, a concentration that cannot be readily obtained, e.g., in the brain of mice. Moreover, rtTA exhibits some residual affinity to tetO in absence of Dox, which is sometimes recognized as low intrinsic background activity. Finally, despite repeated attempts, it was not possible to generate mouse lines that would produce sufficient amounts of rtTA in certain cell types, although analogous tTA lines could be established (unpublished results). The latter results may be due to the reduced stability of rtTA, as observed in, e.g., HeLa cells (unpublished results). In addition, instability, particularly of the prokaryotic portion of the mRNA of rtTA, may also contribute to the failure of establishing this system in certain cell types.
Here, we describe several novel rtTAs that were discovered after random and directed mutagenesis followed by screening in a properly adapted yeast system. Surprisingly, some new rtTAs contain only mutations in the DNA binding domain and the α-helix that connects the DNA reading head with the core of the protein (5, 6). These mutations were embedded in a synthetic sequence background designed to optimize expression in mammals. When compared with the originally described rtTA, these transactivators show greatly improved properties with respect to all of the parameters discussed above. Together with a tTA version that is also encoded in a synthetic gene, they open up possibilities for the study of gene function in vivo.
Materials and Methods
Plasmid Constructions.
DNA encoding the enhanced green fluorescent protein (GFP) derivative GFP+ was obtained from Michael Niederweis (10). pCM190-GFP+ was derived by inserting the blunt ended GFP+ cDNA into the unique and blunt ended BamHI site of the yeast vector pCM190 (11).
To obtain pUHrT10-1 encoding rtTA-S2 or pUHrT12-1 encoding rtTA-S3, the mutagenized rtetR fragments were excised from pCM190-GFP+ with XbaI and BsiWI and ligated to equally restricted pUHD15-1 (3) to substitute wild-type tetR. pUHrT13-1 encoding rtTA-19/56R was obtained by excising an XhoI/EcoNI fragment (harboring the E19 → G19, A56 → P56 mutations) from the rtTA-S2 gene and ligating it to the equally restricted vector pUHD15-1.
pUHrT16-1 encoding rtTA-M2 was constructed by excising an XbaI/NsiI fragment (harboring the S12 → G12, E19 → G19, A56 → P56 exchanges) from rtTA-M1 (encoded by plasmid pUHrT15-1) and ligating it to the equally restricted vector pUHrT10-1.
Generation of Synthetic tTA2, rtTA2-S2, and rtTA2-M2 Genes.
The amino acid sequences of wild-type TetR, fused to three minimal activation domains of the “F”-type (4) was translated into a DNA sequence with an average codon composition similar to that found in human coding sequences and devoid of putative splice donor/acceptor sites, according to methods described previously (12). We used a set of 10 overlapping oligonucleotides anywhere from 90–109 nucleotides in length for synthesis. In addition, oligonucleotides were synthesized carrying specific mutations corresponding to amino acid positions 12, 19, 56, 148, and 179. The final PCR cocktail was analyzed on a 1% agarose gel, and a DNA fragment of an expected length was identified. To produce larger amounts of the full-length synthetic gene product, the PCR cocktail was used as a template for further amplification using outside oligonucleotide primers 1 and 10. The amplified material of the correct length was subcloned as an EcoRI/XmaCI fragment into mammalian expression vector pUHD141-1/X (4) cleaved with the same restriction enzymes. Positive recombinants for synthetic transactivators were identified by transfecting plasmid DNA into HeLa X1/6 cells, which contain a stably integrated tet-inducible luciferase reporter gene construct. Those recombinant plasmid clones that showed robust transactivation potential in the luciferase assay were verified by sequence analysis. The resulting plasmids encoding synthetic tTA2, synthetic rtTA2-S2, and rtTA2-M2 were designated pUHT61-1, pUHrT61-1, and pUHrT62-1, respectively.
PCR Mutagenesis of TetR.
The yeast expression vector pCM190 (11) was used as a template for PCR under conditions designed to induce frequent misincorporation of nucleotides (13). The N terminus (5′-gaccgatccagcctccgcgg) and the C terminus primers (5′-cgtgtgtcccgcggggagaa) overlap the SacII restriction sites, respectively, and allow the amplification of the tetR-encoding DNA region of tTA. The resulting mutagenized PCR products were digested with XbaI and BsiWI and cloned into pCM190-GFP+ to substitute the wild-type sequence for the mutagenized one.
Yeast Screening Assay.
The mutant tTA library was transformed into Saccharomyces cerevisiae strain RS453 (MATa ade2-1 trp1-1 can1-100 leu2-3,112 his3-1 ura3-52) by the lithium acetate method (14). Yeast cells were routinely grown in rich medium (15) at 28°C. When transformed with pCM190-derived plasmids, cells were grown on synthetic medium (SM) agar plates or in liquid SM (15) lacking uracil to maintain plasmids. After 4 days of incubation at 28°C, resulting colonies were replica-plated to SM agar plates supplemented with 10 μg per ml Dox or Tc. After 2 additional days, the colonies were screened under long range UV light (365 nm) for reverse phenotypes.
Fluorometric Determination of GFP+ Expression in Yeast.
Reverse mutants and control strains were grown in 3 ml of SM (containing no inducer, or 10 μg/ml Dox or Tc) to an OD600 of about 1, harvested, and washed once with 1 ml of PBS. Yeast cells were resuspended in 2 ml of PBS at an OD600 of 2. Fluorescence intensity of GFP+ in intact yeast cells was determined at 512 nm with an excitation wavelength of 490 nm at 22°C.
Transient Transfections.
HeLa X1/6 cells (4) harbor chromosomally integrated copies of the luciferase reporter construct pUHC13-3 (3). Transfections of HeLa cells were performed at 50%–60% confluency with 2 μg of DNA and 7 μl of Lipofectamine (Gibco Life Technologies, Basel) in 35-mm dishes, according to the instructions of the producer. The respective DNA mixtures consisted of 0.5 μg of transactivator plasmid [with the exception of pUHD17-1neo (7), which was added at 0.8 μg because of its higher molecular weight], 0.6 μg lacZ expression vector pUHD16-1 (for normalization of transfection efficiency) (4), and pWH802 as unspecific DNA. For induction, Dox (0 to 5 μg/ml; Sigma) was added. Cells were harvested 24 h after induction, and cell lysates were prepared.
Luciferase Assay.
The transfected cells were harvested from the 35-mm dishes and washed once with 1 ml of PBS. Cell lysis was performed by resuspension in 100 mM lysis buffer [100 mM potassium phosphate (pH 7.8)/2 mM DTT] and three subsequent freeze and thaw cycles. Luciferase activity of 50 μl of HeLa cell lysates was determined with 100 mM potassium phosphate (pH 7.8), 15 mM MgSO4, 5 mM ATP, and 0.2 mM d-luciferin (Boehringer Mannheim). Protein concentrations were measured with the Bio-Rad protein assay kit. Relative light units were controlled for β-galactosidase activity (16) as a measure of transfection efficiency.
Characterization of Transactivators Transiently Produced in HeLa Cells by Western Blot Analysis.
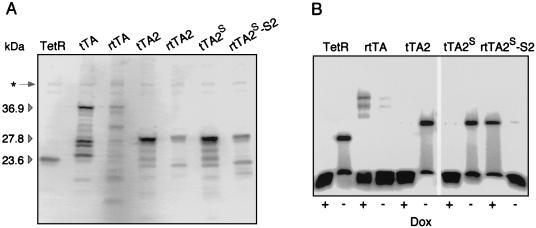
HeLa cells were grown in 10 cm dishes to 50% confluency and transiently transfected with 20 μg of plasmid DNA encoding TetR or the transactivators indicated. The transfection mixture contained also plasmid DNA conferring constitutive expression of the luciferase gene for normalization of transfection efficiency. Cell extracts were prepared after 30 h as described previously (4) and aliquots of the proteins (normalized to luciferase activity) were subjected to Western blot analysis. For immunochemical detection of TetR and fusion proteins thereof, a polyclonal TetR serum from rabbit was used (17). Microdensitometric measurements were normalized using the ubiquitous protein species indicated by an asterisk in Fig. 2A.
Figure 2.
In vitro characterization of transactivators. (A) Western blot analysis of tTA/rtTAs produced in HeLa cells. Cells that had been grown in 10-cm dishes to 40% confluency were transiently cotransfected with a plasmid mixture containing pUHD131-1, constitutively producing luciferase as well as DNA encoding TetR, tTA, rtTA, tTA2, rtTA2, tTA2S, or rtTA2S-S2, respectively. After 36 h, total cell extracts were prepared and separated by SDS/PAGE. Gel loading was normalized to the luciferase activity of the extracts. Western blot analysis with a polyclonal rabbit serum reactive against TetR revealed proteins of the expected molecular weight as well as numerous degradation products. The molecular weights of TetR, tTA, and rtTA, as well as of tTA2 and rtTA2, are indicated. The protein marked by an asterisk served as internal standard for estimating the abundancies of the various tTA and rtTA species. (B) Analysis of the tetO-binding properties of various Tc-controlled transactivators in DNA retardation assays. HeLa cells were grown and transfected as in A. Each transfection mixture contained plasmid DNA encoding TetR, rtTA, tTA2, tTA2S, or rtTA2S-S2, respectively. After 36 h, cell extracts were prepared and incubated with radiolabeled tetO DNA in the absence or presence of 5 μg/ml Dox. Protein-DNA complexes were electrophoretically separated and detected by using a PhosphorImager (Molecular Dynamics).
DNA Retardation Experiment.
Total extracts from HeLa cells transiently producing the various transactivators were prepared, and DNA retardation experiments were performed as described previously (4).
Generation of Stable HeLa Cell Lines.
Linearized plasmid DNA pUHrT61-1 and pUHrT62-1 were chromosomally integrated into the HeLa cell line X1/6. Positive cell clones stably producing rtTA2-S2 and rtTA2-M2 were identified as described previously (4).
Results
Screening for Novel tTA and rtTA Alleles in S. cerevisiae.
We have developed an efficient screening procedure for the identification of tTA/rtTA variants with novel properties in eukaryotic cells. This screen is based on the tTA/rtTA-dependent expression of the GFP from Aequorea victoria in S. cerevisiae. The fluorescence of GFP-producing yeast colonies is conveniently detected on appropriate agar plates under UV light and can be quantified by fluorescence spectroscopy or in 96-well plate readers, and individual cells can be isolated by FACS (Becton Dickinson) (18).
We made use of the S. cerevisiae expression plasmid pCM190 (11) harboring Ptet adapted to the yeast system upstream of a multiple cloning site, as well as the tTA gene, which is constitutively transcribed under the control of the cytomegalovirus IE promoter (PCMV). In addition, pCM190 carries the 2 μ replicon and the URA3 marker for selection in appropriate yeast strains. In a first step, the cDNA encoding an optimized enhanced GFP (GFP+; refs. 10 and 18) was inserted into the multiple cloning site of pCM190, resulting in pCM190GFP+. Because of the high activity levels of GFP+, yeast cells transformed with the latter plasmid and grown on appropriate plates were readily identified when illuminated with long-range UV light (365 nm). Positive colonies were grown in liquid culture and examined in a fluorometric assay. Excitation at 490 nm generated an emission (maximum at 512 nm) that was reduced to background levels by Tc or Dox (data not shown).
In a second step, the tetR gene was amplified by PCR under mutagenic conditions (13). The resulting PCR fragments were then used to substitute the wild-type tetR allele within the tTA gene of pCM190GFP+. Approximately 20,000 recombinant E. coli clones were cocultured, and their plasmid pools were isolated. Transformed into S. cerevisiae strain RS453 via lithium acetate (14), this plasmid preparation generated uracil-prototroph yeast clones. Around 1,000 clones were replicated on SM agar plates containing either no effector substance or 10 μg/ml Dox. After growth for two days at 30°C, GFP+ fluorescence was scored on agar plates under long wave length UV light.
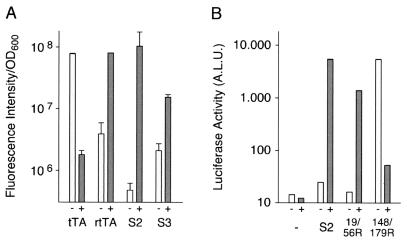
Our first screen led to two yeast clones designated S2 and S3, each harboring a new tTA allele characterized by a reverse phenotype because the clones showed strong fluorescence only in the presence of Dox. The light emission of both clones in the absence and presence of Dox was quantified by fluorometry (Fig. 1A). Comparison with the fluorescence of yeast clones carrying the unmodified tTA (or the original rtTA) revealed that rtTA-S2 has nearly the same maximal activation potential as the previously described rtTA; however, its background activity was considerably lower. Accordingly, the regulatory range of rtTA-S2 appeared about 10-fold enhanced. The sequence of the rtTA-S2 gene revealed four altered codons that result in amino acid exchanges E19 → G19, A56 → P56, D148 → E148, and H179 → R179. Interestingly, the amino acids at positions 71, 95, 101, and 102, whose alteration gave rise to the previously described rtTA (7), remain unaltered. Two of the mutations of rtTA-S2 are located in regions that are intimately involved in the interaction with DNA: E19 in the DNA binding domain itself and A56 in α-helix 4, which connects the DNA binding head with the core of TetR (5). By contrast, D148 and H179 are located in the protein core close to positions involved in Tc binding and in dimerization of the two TetR monomers. The rtTA-S3 allele (Table 1) contains only two mutations, of which one, A26, is again located within the DNA binding head, whereas the other, G95, affects α-helix 6 at a position that is also mutated in the original rtTA allele. Although the reverse phenotype of the S3 allele is not as pronounced as those of rtTA or rtTA-S2 (Fig. 1A), these findings demonstrate that just two mutations can be sufficient for the conversion of the phenotype from tTA to rtTA. Interestingly, such mutations can be located in different functional domains of TetR.
Figure 1.
Dox-dependent gene activation by novel rtTA alleles. (A) rtTA-mediated expression of GFP+ in yeast cells transformed with plasmid pCM190-GFP+ carrying the coding sequence of tTA, rtTA, or rtTA-S2 and -S3, the newly identified alleles, respectively, were grown overnight in the absence or the presence of 10 μg/ml Dox. Suspensions of yeast cells at an OD600 of 2 were exited at 490 nm, and fluorescence emission was measured at 512 nm. (B) Characterization of rtTA-S2 and its derivatives. HeLa cells were cotransfected with plasmid pUHC13-3 carrying the luciferase gene under Ptet-1 control and plasmids containing the coding sequence of transactivators rtTA-S2, -19/56R, or -148/179R, respectively. Cells were grown in the absence or in the presence of 5 μg/ml Dox. After 24 h, luciferase activity from the cell extracts was determined. Values shown are arbitrary light units.
Table 1.
Mutations contributing to the reverse phenotype of tTA
| Transactivator | Amino acid exchanges
|
||||
|---|---|---|---|---|---|
| rtTA | EK71 | DN95 | LS101 | GD102 | |
| rtTA-S2 | EG19 | AP56 | DE148 | HR179 | |
| rtTA-S3 | TA26 | DG95 | |||
| rtTA-19/56R | EG19 | AP56 | |||
| tTA-148/179R | DE148 | HR179 | |||
| rtTA-M1 | SG12 | EG19 | AP56 | ||
| rtTA-M2 | SG12 | EG19 | AP56 | DE148 | HR179 |
The table summarizes the amino acid exchanges within the TetR moiety of tTA discovered in the various tTA/rtTA alleles. The mutations were detected by DNA sequence analysis of alleles functionally identified in the yeast screening system.
Characterization of the rtTA Variants in Mammalian Cells.
To examine the contributions of the different mutated amino acids to the phenotype of rtTA-S2, we separated the mutations at positions 19 and 56 from those at 148 and 179. The resulting alleles designated 19/56R and 148/179 were generated by substitution of the corresponding region of the tTA gene in plasmid pUHD15-1 (3). The activation potential of the different transactivators was examined in X1/6 HeLa cells that contain the stably integrated luciferase gene controlled by Ptet-1. The results of transient expression experiments show (Fig. 1B) that the two mutations at positions 19 and 56 are sufficient for a stringent reverse phenotype, whereas the mutations at position 148 and 179 appear to primarily enhance the activation potential, but are not involved in the conversion of the phenotype. Thus, the dependence of rtTA on Dox for DNA binding can be achieved with mutations not directly affecting the inducer binding site. These experiments confirm furthermore that rtTA-S2 and also rtTA-19/56R cause only minute background activities when compared with mock transfected cells. The low elevation of background over the machine blank seen under these conditions is likely due to the high expression levels of the transactivators in transiently transfected cells.
rtTA-M2, a Transactivator with Increased Sensitivity Toward Dox.
As shown in Fig. 1B, the two mutations G19 and P56 are sufficient to convey a reverse phenotype to TetR because the removal of the mutations E148 and R179 merely lowers the level of induction. rtTA-19/56 was, therefore, subjected to a second round of mutagenesis and screening. A plasmid pool prepared from 6500 E. coli clones was transfected into S. cerevisiae strain RS453, and 2000 yeast transformants were screened for GFP+ fluorescence. The SM agar plates contained either no inducer or 10 ng/ml Dox, a concentration at which rtTA-19/56-containing cells ceased to show a GFP+ signal. Three clones showed a bright fluorescence under these conditions. Sequence analysis of the respective rtTA genes revealed that all three clones had acquired the same additional mutation: a serine to glycine exchange at position 12 in the DNA reading head of the transactivator. The three point mutations in “rtTA-M1” lead to a transactivator with an increased sensitivity toward Dox and a stringent reverse phenotype. To examine whether the two mutations at positions 148 and 179, which had been eliminated from rtTA-S2 to obtain rtTA-19/56, would also elevate the level of induction that can be achieved by rtTA-M1, the respective amino acid exchanges were introduced into the rtTA-M1 gene. Remarkably, the resulting rtTA-M2 combines indeed the advantageous contributions of all five mutations. Examination in HeLa cells (Table 2) reveals that this transactivator has maintained the stringent reverse phenotype, the low background expression properties, and higher sensitivity toward Dox of rtTA-M1. In addition, it has acquired a higher activation potential for Ptet-1.
Table 2.
Response of rtTA and of the new rtTA variants toward Dox
| Dox, ng/ml | Response in arbitrary light units of rtTA alleles
|
|||||
|---|---|---|---|---|---|---|
| — | rtTA | S2 | 19/56R | M1 | M2 | |
| 0 | 0.67 ± 0.28 | 1.24 ± 0.41 | 0.62 ± 0.48 | 2.54 ± 2.05 | 1.02 ± 1.16 | 3.02 ± 1.26 |
| 5 | 0.52 ± 0.06 | 57.27 ± 6.66 | 1.26 ± 0.17 | 3.24 ± 2.72 | 1.24 ± 1.54 | 17.07 ± 5.58 |
| 50 | 0.64 ± 0.55 | 229.72 ± 35.34 | 26.52 ± 12.4 | 0.91 ± 0.32 | 30.94 ± 25.75 | 1,517.37 ± 169.94 |
| 500 | 0.94 ± 0.63 | 696.27 ± 70.3 | 1,726.75 ± 110.64 | 112.68 ± 6.98 | 2,428.19 ± 254.42 | 10,746.33 ± 1,272.97 |
Dox-dependent gene activation of various rtTAs was measured in HeLa X1/6 cells that were transiently transfected with plasmids containing the genes for the transactivators rtTA and rtTA-S2, -19/56R, -M1, and -M2, all of which were controlled by PCMV. For standardization of transfection efficiencies, each transfection mixture contained also a vector that constitutively produces β-galactosidase. Cells were grown in absence of or in presence of 5, 50, and 500 ng/ml Dox. Twenty-four hours post transfection, luciferase activity was measured from cell extracts and normalized to β-galactosidase activity. Values given are arbitrary light units.
Optimization of the Coding Sequences of Transactivators.
For a number of reasons, it appeared advantageous to design a synthetic DNA sequence encoding the most promising transactivators. For example, previous experiments have shown that, depending on the sequence context, mRNA encoding TetR sequences fused to VP16 could be spliced via a number of potential splice-donor and acceptor sites located within the transactivator gene. Reduction or even elimination of functional mRNA was the consequence (unpublished data). Moreover, prokaryotic DNA sequences can be preferred targets of methylation, with concomitant negative effects on transcription. Finally, the VP16 activation domain is for a number of reasons not the optimal choice, as has been discussed previously (4). Therefore, the amino acid sequences of TetR, rTetR-S2, and rTetR-M2 each fused to 3 minimal activation domains were translated into DNA sequences that were based on codon frequencies as found in human genes. Potential splice-donor and acceptor sites, several motifs for endonuclease cleavage sites, and sequences inducing hairpin structures with -ΔG > 9 kcal at the level of mRNA were removed by additional bp changes. The genes were synthesized via an asymmetric PCR method described previously (12) and inserted into plasmid pUHD141-1/X (4). The synthetic transactivators are designated tTA2S, rtTA2S-S2, and rtTA2S-M2, respectively.
The intracellular stability and the binding to tetO sequences of some of these new transactivators were examined. For that purpose, HeLa cell extracts were subjected to SDS/PAGE followed by Western blot analysis with anti-TetR Abs and to DNA retardation experiments. As shown in Fig. 2A, the original VP16 fusions, tTA and rtTA, undergo extensive degradation in HeLa cells. Degradation is most pronounced in the case of rtTA. Fusion of the TetR moieties to minimal activation domains stabilizes both transactivators, which now represent the most abundant species detected by anti-TetR Abs. Estimating the abundancy of the various molecular species suggests furthermore that the synthetic genes encoding tTA2s and rtTA2s-S2 give rise to an approximately 2-fold increased intracellular concentration as compared with the nonsynthetic genes.
When the various transactivators were exposed to a high excess of radioactively labeled tetO DNA in presence and absence of Dox and analyzed by gel electrophoresis, the residual affinity of the original rtTA in the absence of Dox becomes obvious by the mobility change of tetO DNA (Fig. 2B). By contrast, only minute amounts of bound tetO DNA are found with TetR and tTA. As already suggested by the transcription activation experiments in yeast and HeLa cells, rtTA2s-S2 has a highly reduced affinity to tetO DNA in absence of Dox. Indeed, its affinity to tetO is now comparable to that of TetR and tTA in presence of Dox; i.e., it is negligible. Thus, the fusion of the new alleles to minimal activation domains and the generation of optimized coding sequences have further improved the properties of these transactivators.
Characterization of rtTA2s-S2 and rtTA2S-M2 in HeLa Cell Lines.
The synthetic coding sequences of the new transactivators were introduced into HeLa X1/6 cells (4). This cell line contains the luciferase gene under Ptet-1 control stably integrated into a chromosomal site where Ptet-1 is silent unless it is activated by tTA or rtTA. Cell lines that stably synthesize rtTA2S-S2 or rtTA2S-M2, respectively, were derived by subcloning as described previously (4). Two of the resulting cell lines, HrTAS2-1 and HrTAM2-1, were used to characterize these transactivators, particularly in comparison to the original rtTA that is stably produced in the analogous cell line h5-CL11 (7).
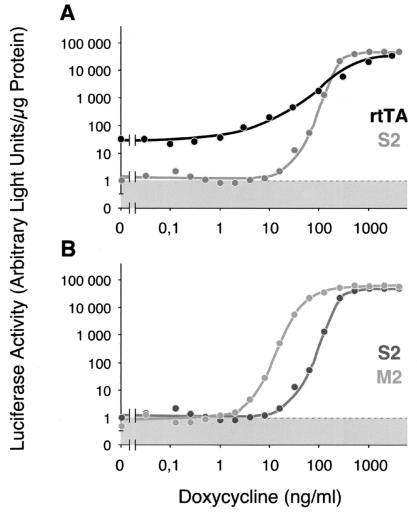
When the activations of Ptet-1 by rtTA and rtTA2S-S2 are compared (Fig. 3A), two remarkable differences between the two transactivators are revealed: (i) Whereas rtTA causes a low but defined background of luciferase activity in the absence of Dox, no such activity is found in cells harboring rtTA2S-S2, because the HrTAS2-1 cell line shows only instrumental background. (ii) When Ptet-1 activation is monitored at different Dox concentrations, full induction of luciferase by rtTA2S-S2 is achieved in a relatively narrow and well defined concentration window when compared with rtTA. However, both transactivators induce Ptet-1 to about the same level at Dox concentrations between 1 and 2 μg/ml. On the other hand, when rtTA2S-S2 and rtTA2S-M2 are compared (Fig. 3B), both resemble each other closely with respect to the lack of background activity in absence of Dox, to the maximal level of induction and to the dose-response profile. However, rtTA2S-M2 shows a highly increased sensitivity toward Dox. Full activation is achieved at a 10-fold lower Dox concentration, namely around 0.1 μg/ml, when compared with the other reverse transactivators. Thus, the combination of the five mutations identified in the various alleles of the Tet repressor has resulted in a reverse phenotype with highly improved properties.
Figure 3.
Induction characteristics of various rtTAs in stably transfected HeLa cell lines. X1/6 HeLa cells were transfected with linearized DNA encoding rtTA2S-S2 or rtTA2S-M2, respectively, under PCMV control, and cell lines constitutively expressing the respective transactivator gene were derived (HrTAS2-1, HrTAM2-1). The respective cell lines, as well as cell line h5CL11 harboring the original rtTA gene also under PCMV control, were grown in medium containing Dox at the concentrations indicated. After 3 days, luciferase activity was measured from the cell extracts and normalized to the protein content. The figure shows comparison (A) of cell lines h5CL11 and HrTAS2-1 and (B) of HrTAS2-1 and HrTAM2-1.
Discussion
The success of the Tet regulatory systems in studies of gene function is based on the unusual specificity of the TetR/tetO interaction and the availability of well characterized nontoxic and highly effective inducers, Tc and many of its derivatives, particularly Dox. Moreover, genetic and more recently detailed structural information on TetR (5, 6) and its interaction with DNA and effector molecules has allowed the modification of various TetR fusion proteins, which led not only to a transcriptional activator with reverse phenotype (7) but also to fusion proteins with novel DNA binding and dimerization specificities (19). These findings have suggested a large sequence space for TetR that could possibly be further exploited. It therefore appeared promising to develop a eukaryotic system for the genetic dissection of fusions between TetR and various functional domains. We have adapted S. cerevisiae for a screening procedure that would permit the direct assessment of the transcription activation potential of a fusion protein. This eukaryotic screening system circumvents inconsistencies observed previously when TetR alleles identified in E. coli would not necessarily maintain their phenotype when fused to the various eukaryotic domains and transferred into the eukaryotic environment. Moreover, in contrast to the approach in E. coli, the screen would allow use of antibiotically active Tcs such as Dox for the identification of TetR alleles. Screening directly for the function of TetR fusions allows also extension of the genetic dissection to the eukaryotic domains as well. Because the screening procedure described herein can be adapted to high throughput protocols, it is now feasible to explore in great detail the sequence space leading to reverse versions of TetR and its fusions with various eukaryotic domains.
Our primary goal was to search for novel alleles with a reverse phenotype because, on closer examination, the originally described rtTA revealed several limitations. They included (i) a low but distinct residual affinity toward tetO and concomitantly an intrinsic background activity of Ptet-1 in the noninduced state whenever rtTA was present in relatively high intracellular concentrations; (ii) a rather high instability in vivo, which may have prevented the establishment of the rtTA system in some cell lines and tissues; and (iii) a relatively low sensitivity toward Dox, which is of no relevance in studies carried out in vitro but which has hampered regulation in organs with limited accessibility for inducer, e.g., in the brain of transgenic animals.
Random mutagenesis of the tetR gene and a first direct screening for alleles with reverse transactivator function yielded two surprising results. First, it was unexpected that, within a rather small population of mutants, two alleles with reverse phenotypes could be identified, and second, none of the new rtTA alleles resembled in its mutation pattern the originally described quadruple mutant rtTA. In fact, it turned out that a stringent reverse phenotype can be obtained with only two mutations. Interestingly, in the 19/56R allele, one of the mutations is found in the DNA reading head and the other within α-helix 4, which connects the DNA binding domain with the core of TetR (5) and plays a crucial role in the conformational changes associated with induction (6). Furthermore, considering rtTA-M1, it is neither obvious nor can it presently be explained why an additional mutation in the DNA reading head (G12) increases the sensitivity for the inducer. In principle, because inducer binding allosterically affects DNA binding, there must, in turn, also be an influence of DNA binding on inducer recognition. We know of no other rigorous observation of such a functional connection, the mechanics of which deserve attention in the future. Surprising as well was the finding that the combination of five mutations, of which only two are essential for the reverse phenotype, led to rtTA-M2 that exhibits superior properties with respect to inducibility and specificity. Interestingly, the newly identified rtTA alleles allowed replacement of the VP16 activation domain by three minimal activation domains without affecting specificity and inducibility, in contrast to our experience with the original rtTA, where the corresponding changes resulted in drastically increased affinity of the transactivators to tetO in absence of Dox (20). Moreover, the combination of the new TetR alleles with three minimal activation domains has yielded transactivators with greatly enhanced stability in HeLa cells, when compared with the original rtTA. These favorable properties of the transactivators themselves are augmented by the features conveyed through the synthetic coding sequences. All potential splice donor and acceptor sites that could be identified by sequence analysis, as well as potentially stable hairpin structures of the mRNA, were eliminated, and human codon frequencies were introduced. An increased expression in HeLa cells is the consequence of this gene optimization, which may become even more obvious in other cell types. Together, the various modifications have abrogated obstacles that have limited the application of the rtTA system in the past. The wider applicability is supported by a well functioning hepatocyte-specific rtTA2-S2 mouse line that we have recently generated after several attempts with the original rtTA gene had failed (K. Schönig, unpublished data). The two novel rtTAs we have described herein, rtTA2S-S2 and rtTA2S-M2, differ in one important feature, their susceptibility toward Dox. rtTA2S-M2 is fully induced at an about 10-fold lower concentration of Dox than rtTA2S-S2. In principle, this feature can be exploited to turn on two genes sequentially by varying the Dox concentration. The modifications required for such a double switch system were recently shown to be feasible (19).
The findings described here open up interesting perspectives: (i) The S. cerevisiae-based screen can be used to obtain a saturating set of mutations leading to rtTAs, thus defining all residues involved in the switch of inducer action; this will be of value for our understanding of the mechanism of action of TetR as a paradigm of an allosteric protein that interacts with a defined DNA sequence. (ii) A large set of sensitivity mutations like rtTA-M2 may shed light on the reversibility of functional interplay between DNA and inducer binding. (iii) The exploration of the TetR sequence space responsible for the interaction with inducer molecules may lead to novel pairs of inducers and TetR fusion proteins that can be used independently from each other. (iv) The potential to efficiently screen or select for modified eukaryotic activator or repressor domains may lead to TetR-based regulatory proteins, adapted to fulfill specific functions. Such developments will not only further insights into repressor structure function relationship, but also enhance our capabilities in the study of gene function in higher organisms by providing new tools like the ones described here.
Acknowledgments
We thank Christiane Schirra-Müller for excellent technical assistance and Sibylle Reinig for help in preparing the manuscript. This work was supported by the Bayrische Forschungsstiftung through Forschungsverbund Grundlagen Gentechnischer Verfahren (FORGEN), by the Deutsche Forschungsgemeinschaft, by the Volkswagen-Stiftung, and by the Fonds der Chemischen Industrie Deutschlands.
Abbreviations
- Tc
tetracycline
- TetR
the repressor of the Tc resistance operon
- tTa
Tc-controlled transactivator
- rtTA
reverse tTa
- Dox
doxycycline
- SM
synthetic medium
- GFP
green fluorescent protein
- CMV
cytomegalovirus
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.130192197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.130192197
References
- 1.Hillen W, Berens C. Annu Rev Microbiol. 1994;48:345–369. doi: 10.1146/annurev.mi.48.100194.002021. [DOI] [PubMed] [Google Scholar]
- 2.Degenkolb J, Takahashi M, Ellestad G A, Hillen W. Antimicrob Agents Chemother. 1991;35:1591–1595. doi: 10.1128/aac.35.8.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gossen M, Bujard H. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baron U, Gossen M, Bujard H. Nucleic Acids Res. 1997;25:2723–2729. doi: 10.1093/nar/25.14.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hinrichs W, Kisker C, Duvel M, Müller A, Tovar K, Hillen W, Saenger W. Science. 1994;264:418–420. doi: 10.1126/science.8153629. [DOI] [PubMed] [Google Scholar]
- 6.Orth P, Cordes F, Schnappinger D, Hillen W, Saenger W, Hinrichs W. J Mol Biol. 1998;279:439–447. doi: 10.1006/jmbi.1998.1775. [DOI] [PubMed] [Google Scholar]
- 7.Gossen M, Freundlieb S, Bender G, Müller G, Hillen W, Bujard H. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 8.Baron, U. & Bujard, H. (2000) Methods Enzymol., in press. [DOI] [PubMed]
- 9.Kistner A, Gossen M, Zimmermann F, Jerecic J, Ullmer C, Lübbert H, Bujard H. Proc Natl Acad Sci USA. 1996;93:10933–10938. doi: 10.1073/pnas.93.20.10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scholz O, Thiel A, Hillen W, Niederweis M. Eur J Biochem. 2000;267:1565–1570. doi: 10.1046/j.1432-1327.2000.01170.x. [DOI] [PubMed] [Google Scholar]
- 11.Garí E, Piedrafita L, Aldea M, Herrero E. Yeast. 1997;13:837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 12.Pan W, Ravot E, Tolle R, Frank R, Mosbach R, Türbachova I, Bujard H. Nucleic Acids Res. 1999;27:1094–1103. doi: 10.1093/nar/27.4.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leung D W, Chen C, Goeddel D V. Technique. 1989;1:11–15. [Google Scholar]
- 14.Gietz R D, Schiestl R H, Willems A R, Woods R A. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 15.Rose A B, Broach J R. Methods Enzymol. 1990;185:234–279. doi: 10.1016/0076-6879(90)85024-i. [DOI] [PubMed] [Google Scholar]
- 16.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 17.Freundlieb S, Schirra-Müller C, Bujard H. J Gene Med. 1999;1:4–12. doi: 10.1002/(SICI)1521-2254(199901/02)1:1<4::AID-JGM4>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 18.Niedenthal R K, Riles L, Johnston M, Hegemann J H. Yeast. 1996;12:773–786. doi: 10.1002/(SICI)1097-0061(19960630)12:8%3C773::AID-YEA972%3E3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 19.Baron U, Schnappinger D, Helbl V, Gossen M, Hillen W, Bujard H. Proc Natl Acad Sci USA. 1999;96:1013–1018. doi: 10.1073/pnas.96.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baron U. Ph.D. thesis. Heidelberg, Germany: Universität Heidelberg; 1998. [Google Scholar]