Abstract
The unicellular green alga Acetabularia acetabulum has proven itself to be a superior model for studies of morphogenesis because of its large size and distinctive polar morphology. The giant cell forms an elongated tube (a stalk of up to 60 mm in length), which at its apical pole makes whorls of hairs, followed by one whorl of gametophores in the shape of a cap. At its basal pole, the cell extends into a rhizoid wherein the single nucleus is positioned. In this study, we have determined the level of specific messenger RNAs in the apical, middle, and basal regions using reverse transcriptase-PCR methodology. Four mRNA classes were distinguished: those that were uniformly distributed (small subunit of Rubisco, actin-1, ADP-glucose, centrin, and α- and β-tubulin), those that expressed apical/basal (calmodulin-4) or basal/apical gradients (calmodulin-2 and a Ran-G protein), and those with development-specific patterns of distribution (mitogen-activated protein kinase, actin-2, and UDP-glucose-epimerase). Restoration of the apical/basal calmodulin-4 mRNA gradient after amputation of the apical region of the cell requires the nucleus and was abolished by cytochalasin D. Accumulation of actin-1 mRNA in the vicinity of the wound set by the amputation needs, likewise, the presence of the nucleus and was also inhibited by cytochalasin. This suggests that actin microfilaments of the cytoskeleton are involved in directed transport and/or anchoring of these mRNAs.
In the early 1930s, Haemmerling (1931, 1932, 1934a) introduced the unicellular green alga Acetabularia acetabulum (formerly known as Acetabularia mediterranea) as a research object into cell biology primarily because of its large size and distinct polar morphology. The life cycle of A. acetabulum begins with the fusion of two isogametes to form a zygote, which, after attachment to a solid substratum, grows into a long, tube-shaped stalk. The stalk elongates at the tip, periodically forming lateral whorls of branched hairs. Morphogenesis at the apical pole is completed with formation of a whorl of gametophores known as the cap. In contrast, the basal pole ends in a short, convoluted rhizoid resembling the fingers of a gnarled hand wherein the nucleus is located during all vegetative stages of the cell cycle. Thus, both cellular poles of A. acetabulum execute very different morphogenetic programs (for details, see Berger and Kaever, 1992; Mandoli, 1998).
Grafting experiments led Haemmerling to the conclusion that “morphogenetic substances” are released from the nucleus into the cytoplasm where they elicit cellular morphogenesis. He considered them to be “gene products of the nucleus,” distinguishing between cap-, whorls of hair-, stalk-, and rhizoid-forming substances (Haemmerling, 1934b). A major feature of Haemmerling's morphogenetic substances was their extreme stability, because enucleated cells could perform stalk-, hair-, and cap-formation for several weeks after removal of the nucleus (Haemmerling, 1932). When these concepts were formulated, neither the chemical nature of these substances nor the molecular basis of the gene was known. Later, Haemmerling refined the definition of morphogenetic substances to those “carrying genetic information from the nucleus to the cytoplasm” (Haemmerling, 1963). Subsequent experiments using inhibitors of RNA and protein synthesis, UV irradiation, and isolation of long-lived (several weeks) poly(A) RNA from enucleated cells all favored the hypothesis that the morphogenetic substances of A. acetabulum are messenger RNAs (Haemmerling, 1963; Zetsche, 1964, 1966a; Kloppstech and Schweiger, 1975b, 1982). Haemmerling also proposed (based on the differential morphogenetic capacity of anucleate apical, middle, and basal cell fragments) the existence of apical/basal gradients of cap-, hair-, and stalk-forming substances and of basal/apical gradients of rhizoid-forming substances (Haemmerling, 1934a). Biochemical parameters, such as specific activities of enzymes being localized predominantly in the apical region of the cell, gave further credence to the existence of such gradients in A. acetabulum (Zetsche, 1969; Bachmann and Zetsche, 1979; Mandoli, 1998, and refs. therein).
Although several reports indicate that polar distribution of mRNAs plays an important role in animal cell development (for review, see Bashirullah et al., 1998), much less is known about mRNA gradients in plant cells. Quatrano's group, studying the brown alga Fucus sp., found accumulation of actin mRNA at the thallus pole of the zygote upon polar axis fixation (Bouget et al., 1996), and most recently, the subcellular location of a homeobox-like and a carbonic anhydrase mRNA in A. acetabulum was reported to be under developmental control (Serikawa and Mandoli, 1999; Serikawa et al., 2001).
It was against this backdrop that the current study with A. acetabulum was performed, examining subcellular distribution of a set of specific mRNAs during different developmental stages and in response to cellular injury. Here, we report evidence that differential mRNA gradients exist abundantly in this giant cell and that establishment of such polarity involves the microfilament system of the cytoskeleton.
RESULTS
Selection of Nuclear Genes
For this study, we generated cDNA probes for genes representing multiple levels of cellular functions: housekeeping genes; photosynthesis-related genes; genes of the cytoskeleton; genes involved in cellular signaling, transport, and differentiation; and genes related to cell wall synthesis (Table I). To ascertain that these molecular probes were specific for nuclear genes, we verified that all cDNAs conformed with the unusual codon usage of A. acetabulum. In the nuclear genes of A. acetabulum, TGA is the only stop codon, and TAG and TAA code for Gln (Schneider et al., 1989; Jukes, 1996, and refs. therein); whereas in all chloroplast genes analyzed so far, the universal code is used (H. Vogel and K. Zetsche, unpublished data). Upon phylogenetic analysis, the selected nuclear gene sequences clustered with the corresponding genes of green alga and/or terrestrial plants rather than with those of prokaryotes such as cyanobacteria and bacteria (Vogel, 1998).
Table I.
Survey of the distribution of analyzed mRNAs in the A. acetabulum cell and characterization of the PCR fragments used in the investigation
| Gene Name | mRNA Localization | PCR Fragment Length | No. of Unusual Codons | Function of Protein Product |
|---|---|---|---|---|
| bp | ||||
| UDP-Glc epimerase | Young cells: basalAdult cells: apical | 650 | 2 (TAA/TAG⩵Q) | Conversion of UDP-Glc to UDP-Gal |
| ADP-Glc-pyrophosphorylase | Evenly | 500 | 1 (TAA⩵Q) | Synthesis of activated sugar molecules (ADP-Glc) |
| Actin-1 | Evenly (but accumulates at wounded sites) | 700 | 4 (TAA/TAG⩵Q) | Cytoskeletal protein, involved in plasma streaming |
| Actin-2 | Young cells: evenlyAdult cells: apical | 750 | 1 (TAG⩵Q) | |
| Calmodulin-2 | Basal | 180 | 1 (TAA⩵Q) | Central mediator of Ca2+ signals |
| Calmodulin-4 | Apical | 180 | 2 (TAA/TAG⩵Q) | |
| Ran-G protein | Basal | 160 | 1 (TAG⩵Q) | Transport of RNA and proteins across the nuclear membrane |
| RbcS | Evenly | 250 | 2 (TAA/TAG⩵Q) | Small subunit of Rubisco enzyme |
| 18SrRNA | Evenly | 850 | None | 18S ribosomal rRNA |
| Centrin | Evenly | 300 | 1 (TAA⩵Q) | Ca2+-binding protein (e.g. basal body, centrosome) |
| MAP-kinase | Young cells: basalAdult cells: apical | 300 | 1 (TAA⩵Q) | Ser-Thr kinase, transduction of extracellular signals to intracellular targets |
| α- and β-Tubulin | Evenly | α: 650β: 750 | Not applicable | Cytoskeletal proteins |
Use of 18S Ribosomal RNA as Reference
Ribosomes are nearly equally distributed throughout Acetabularia cells (Kloppstech and Schweiger, 1975a), with their RNA comprising more than 80% of total cellular RNA. Reverse transcriptase-PCR (RT-PCR) using specific 18S rRNA primers with total RNA as a template showed comparable levels of this RNA species throughout the cell (Fig. 1). This pattern held whether the RNA was derived from cells of different developmental stages or whether alternate input RNA concentrations and cycle numbers per RT-PCR were used. Therefore, throughout this study, the distribution of specific mRNAs is compared with 18S rRNA levels as an internal reference, unless otherwise indicated.
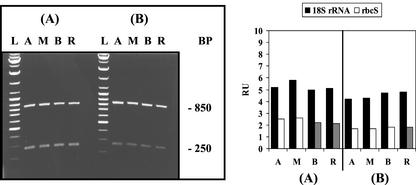
Figure 1.
Distribution of 18S rRNA and rbcS mRNA in different A. acetabulum cell fragments: evaluation by semiquantitative RT-PCR analysis. Young cells of 20 to 30 mm in length without caps (A) and adult cells of 40 to 50 mm in length with nearly full-sized caps (B) were cut into four fragments corresponding to apical stalk fragment (A), middle stalk fragment (M), basal stalk fragment (B), and rhizoidal fragment (R). Equal amounts of total RNA, isolated from these cell fragments, were used as templates in RT-PCR with appropriate specific primers (see “Materials and Methods”). Left panel, Separation of RT-PCR products by agarose gel electrophoresis. The 850-bp piece corresponds to 18S rRNA and the 250-bp piece to rbcS mRNA. (L) indicates a DNA size ladder. Right panel, Densitometric record of the individual signals after blotting and hybridization with fluorescein-labeled A. acetabulum cDNA probes, expressed as relative units (RU).
Other Uniformly Distributed Messenger RNAs
In green algae, the product of the nuclear small subunit of Rubisco (rbcS) gene functions exclusively in the chloroplasts. During the day, chloroplasts are evenly distributed in the A. acetabulum cytoplasm. Figure 1 shows the similarity of A. acetabulum rbcS mRNA signals, generated by semiquantitative RT-PCR analysis, whether the template RNA was derived from apical, middle, basal, or rhizoid-containing fragments, a pattern that was found in both young and adult cells. The uniform spatial distribution of the rbcS signals parallels that found for the RT-PCR products corresponding to 18S rRNA.
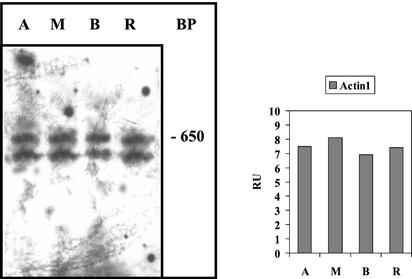
A host of other messenger RNAs also exhibited even cellular distribution, including actin-1 mRNA, α- and β-tubulin mRNA, ADP-Glc-P mRNA, and centrin mRNA (Table I). An RNA protection assay confirms the RT-PCR result for the actin isoform-1 mRNA (Fig. 2) and is, therefore, also a control for the reliability of our RT-PCR experiments.
Figure 2.
Cellular distribution of actin isoform-1 mRNA evaluated by RNase protection analysis. Cells of 40 to 50 mm in length with caps were used for cell fragmentation. Left panel, Electrophoretic separation of the products of an RNase protection assay of actin-1 mRNA (see “Materials and Methods”). The 650-bp band and the lower band are protected actin-1 mRNA fragments; they occur because the bacterial polymerases used often produce shorter by-products resulting in generation of two different antisense RNA probes. Right panel, Densitometric record of the combined bands after blotting. Abbreviations are the same as in Figure 1.
Messenger RNAs Forming Apical/Basal and Basal/Apical Gradients
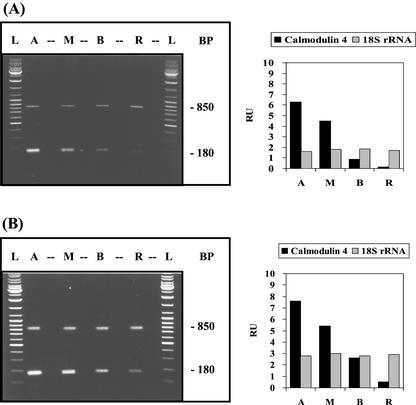
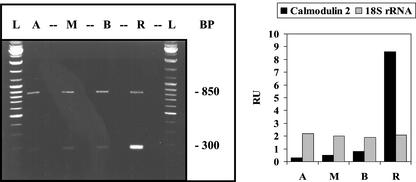
There are indications that, like other plant cells with tip growth, A. acetabulum exhibits an apical/basal calcium gradient (Reiss and Herth, 1979). We, therefore, examined the distribution of mRNA for calmodulin, a key protein in calcium metabolism. Four different A. acetabulum calmodulin genes were identified. The four calmodulin isoforms are 75% to 89% identical with one another, a range similar to that found for higher plants (Vogel, 1998). The distribution of mRNAs for two isoforms with the most divergent sequences were analyzed. As it turned out, the messenger RNAs both formed very steep gradients (50- to 100-fold enrichment) but in opposite directions: apical/basal isoform-4 and basal/apical isoform-2 (Figs. 3 and 4, respectively). These opposite distributions suggest very different functions.
Figure 3.
Apical/basal gradient of calmodulin-4 mRNA. Developmental stages, experimental conditions, and abbreviations used are similar to Figure 1. The 180-bp RT-PCR product corresponds to calmodulin-4 mRNA and the 850-bp RT-PCR product to 18S rRNA.
Figure 4.
Basal/apical distribution of calmodulin-2 mRNA. A. acetabulum cells with nearly full-size caps were used. Left, Electrophoretic separation of RT-PCR products. The 300-bp RT-PCR signal corresponds to calmodulin-2 mRNA and the 850-bp signal to 18S rRNA. Abbreviations and conditions are the same as in Figure 1.
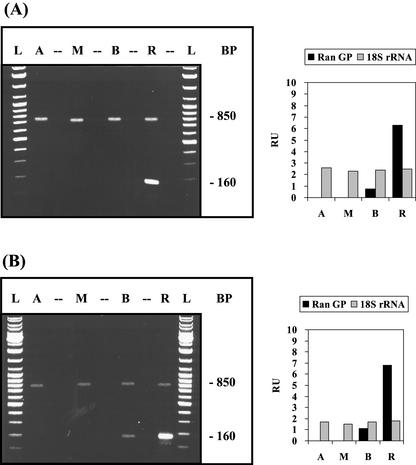
Because Ran-G protein, a highly conserved, small GTP-binding protein, is involved predominantly in protein and RNA transport across the nuclear envelope (for review, see Melchior and Gerace, 1998), we generated a specific cDNA probe for A. acetabulum to evaluate whether its mRNA has a close nuclear localization. RT-PCR analysis indicates that Ran-G mRNA is enriched in the basal region, particularly in the rhizoid, which contains the nucleus (Fig. 5), forming a basal/apical gradient similar to that of calmodulin-2 mRNA (Fig. 4).
Figure 5.
Basal/apical gradient of the mRNA for the small GTP-binding protein Ran. Cell stages, experimental conditions, and abbreviations are similar to Figure 1. The 160-bp RT-PCR product corresponds to the mRNA of the small GTP-binding protein Ran (Ran-GP) and the 850-bp RT-PCR product to 18S rRNA.
mRNAs That Change Distribution Pattern with Development
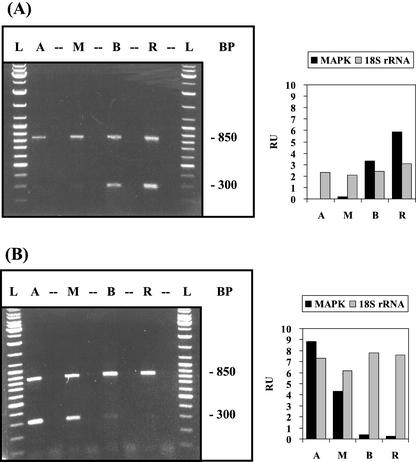
Mitogen-activated protein (MAP) kinases belong to a group of Ser/Thr protein kinases that, in eukaryotes, are part of the phosphorylation cascade in cellular signal transduction (for review, see Hirt, 1997). In immature A. acetabulum cells, the mRNA for MAP-kinase accumulated preferentially in the basal region. Upon cap formation, the gradient became reversed with a roughly 50-fold enrichment of MAP-kinase mRNA in the cap region (Fig. 6). A similar developmental reorganization was observed for the cellular distribution of UDP-Glc-epimerase mRNA and actin-2 mRNA (data not shown).
Figure 6.
Reversal of the gradient of MAP-kinase mRNA after cap formation. Distribution of MAP-kinase mRNA in cells without cap (A) and with nearly fully developed cap (B) was evaluated by RT-PCR analogous to the experiment of Figure 1. The 300-bp signal corresponds to MAP-kinase mRNA, the 850-bp signal to 18S rRNA.
Gradient Formation Is Dependent on the Presence of the Nucleus
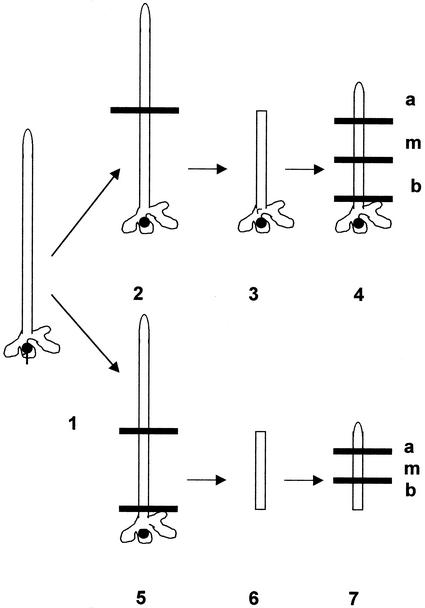
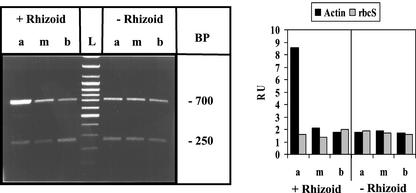
It is well known that the basal region of the A. acetabulum cell remaining after decapitation can regenerate a cap only if it includes a nucleus-containing rhizoid (Haemmerling, 1932). To investigate whether formation of mRNA gradients also relies on continued presence of a nucleus, the amputated basal fragments were cultured with and without rhizoid (see Fig. 7 for a schematic depiction of the amputation procedure, continued culture, and final fragmentation directly before analysis). As shown in Figures 8 and 9A, establishment of an actin-1 mRNA gradient in amputated cells and re-establishment of the calmodulin-4 mRNA gradient in the basal fragment of the A. acetabulum cell is completely dependent on the presence of a nucleus. It was only the fragments containing the nucleus that within 3 or 7 d of treatment, respectively, re-established strong apical/basal gradients, whereas in the fragments without rhizoids—and, therefore, without nucleus—actin-1 mRNA and calmodulin-4 mRNA, respectively, remained evenly dispersed.
Figure 7.
Schematic representation of A. acetabulum cell fragments: culture with and without rhizoid and in absence and presence of cytochalasin D. Cells are not drawn to scale; whorls of hairs are omitted. Amputation is indicated by a horizontal line. 1, Intact cell of 30 to 40 mm in length without cap; 2, amputation of apical cell-region only (5) generation of apical- and rhizoid-amputated fragment. The nucleated (3) and enucleated (6) cell fragments were cultured for 7 d in the absence and presence of cytochalasin D. The cultured cellular fragments were then redissected into apical (a), middle (m), and basal (b) fragments (4, 7) and immediately processed for analysis.
Figure 8.
Regeneration after injury induces accumulation of actin-1 mRNA at the apical wound site: the role of the nucleus. Basal cell fragments with or without rhizoid were generated and cultured (in the absence of inhibitor) for 3 d after amputation before processing occurred, as described schematically in Figure 7. Experimental conditions are similar to the one described in Figure 1; abbreviations as in Figure 7. The 700-bp RT-PCR product corresponds to actin-1 mRNA and the 250-bp product to rbcS mRNA.
Figure 9.
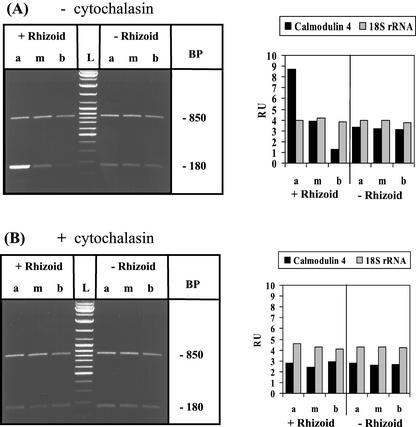
Restablishment of the apical/basal gradient of calmodulin-4 mRNA after decapitation: the role of actin microfilaments. Basal cell fragments (with or without rhizoid) were generated and cultured in the absence or presence of 10 μg mL−1 of cytochalasin D for 7 d after amputation before processing occurred, as described schematically in Figure 7. Experimental conditions are similar to those described in Figure 1; same abbreviations as in Figure 8. The 180-bp RT-PCR product corresponds to calmodulin 4 mRNA and the 850-bp RT-PCR product to 18S rRNA.
Induction of Gradient Formation after Cellular Injury
Microfilaments appear to play an important role in occlusion of wounds in A. acetabulum (Menzel, 1988). Actin-1 mRNA is uniformly distributed throughout the A. acetabulum cell (Fig. 2). However, removal of the apical region induced its accumulation at the tip of the regenerating amputated cell fragment (Fig. 8), a process that occurred within the 3-d culture period required for wound healing. Such a gradient re-formed only when the nucleus was present, i.e. in rhizoid-containing fragments. The original uniform distribution of actin-1 mRNA was re-established after several days of additional culture associated with continued cellular development (not shown).
The Cytoskeleton Is Required for mRNA Gradient Formation
It has been suggested that microfilaments may be generally involved in mRNA gradient formation (Bashirullah et al., 1998). Because cytochalasin D disrupts the distribution of actin microfilaments and works very efficiently with A. acetabulum cells in culture (Koop and Kiermayer, 1980), we tested this hypothesis by treating cell fragments with this potent inhibitor. Our experimental approach is schematically outlined in Figure 7, indicating which cell fragments were generated by amputation, when the inhibitor was added to the culture, and how analysis upon further fragmentation was performed. Cytochalasin D completely prevented the re-establishment of gradients upon amputation. This was true for all the mRNA species found in this study to exhibit apical/basal distribution (Fig. 9 and not shown). Figure 9 shows a representative example—the effect of the inhibitor on calmodulin-4 mRNA gradient formation. These findings strongly suggest that the microfilament system is involved in either the polar transport of these mRNAs and/or their local anchoring in the A. acetabulum cell.
DISCUSSION
Cell Polarity and the Distribution of mRNAs
This study demonstrates the existence of multiple mRNA gradients in the A. acetabulum cell (Table I). Cellular distribution was studied with cells at two developmental ages: young cells of 20 to 30 mm in length having not yet developed the cap and adult cells of 40 to 50 mm in length with nearly full-sized caps. Because at these cell stages, distribution is likely more a consequence of the existing cellular polarity than its cause, it is our suggestion that polarly distributed mRNAs not only stabilize cell polarity but facilitate the execution of specialized morphogenetic events at opposite ends. Our underlying assumption was that proteins would have to be synthesized locally close to where they were actually needed.
Because the cytoskeleton spans the entire cell, as does the distribution of the chloroplasts, it came as no surprise that mRNAs of their constituents were evenly dispersed also. RbcS mRNA is responsible for the synthesis of the small subunit of the plastidial enzyme Rubisco. ADP-Glc-P mRNA specifies ADP-Glc pyrophosphorylase, an enzyme involved in starch synthesis that is at least partly located in the plastids. A similar localization is expected for the actin isoform-1 mRNA, because actin microfilaments run through the entire cell (see Menzel, 1994). The distribution of tubulin mRNAs deserves some special consideration. Sawitzky (1997) established that tubulin mRNAs are present in the A. acetabulum cell at all developmental ages, and in this study, we found these mRNAs to be nearly ubiquitous in all parts of the cell. However, neither microtubules nor unassembled tubulin proteins were detected during vegetative development until after meiosis (Menzel, 1994; Sawitzky, 1997). These findings suggest that tubulin mRNAs are synthesized and stored in the A. acetabulum cytoplasm long before they are actually used for synthesis of the corresponding tubulin proteins. The mechanism for this exquisite case of translational control remains to be elucidated: e.g. What is it that activates these dormant messenger RNAs?
Like vascular plants (Yang et al., 1998; see also Zielinski, 1998), A. acetabulum contains several calmodulin isoforms. We studied the location of the two evolutionary most diverged calmodulins in further detail and found their messenger RNAs to behave antipodally: isoform-4 concentrated at the top and isoform-2 concentrated at the bottom of the cell. The calmodulin-4 gradient correlates well with the likely apical/basal calcium gradient (Reiss and Herth, 1979) and the gradient of calmodulin proteins that has been suggested based on measurement of fluophenacin fluorescence (Cotton and Vanden Driessche, 1987). Because amino acid replacements in the calcium binding site of the various isoforms affect not only calcium binding per se but also the affinity with which they bind to target proteins during signal transduction (Liu et al., 1998; Zielinski, 1998; Lee et al., 1999), it is reasonable to assume widely different functions for the various calmodulin isoforms (Yang et al., 1998). For quite some time, calmodulin was considered to be predominantly a cytosolic protein, but calmodulin has recently also been found in the nuclear compartment, associated with transcription factors, histone H1, and a specific nucleoside triphosphate phosphatase (Bachs et al., 1994; Zielinski, 1998). In yeast (Saccharomyces cerevisiae), a role for calmodulin in spindle organization during nuclear division has been suggested (Moser et al., 1997). Although calmodulin-2 mRNA formed a rather weak basal/apical gradient in young A. acetabulum cells (data not shown), older cells with nearly full-sized caps had massive accumulations in the rhizoid (Fig. 4). The high level of calmodulin-2 mRNA in the nuclear area of cells with mature caps at a time shortly before division of the primary nucleus may indicate a role for this isoform in the process.
In contrast, accumulation of Ran-G protein mRNA in the rhizoid was found to be similar in young and older cells with caps (Fig. 5). This suggests a constitutive requirement in the nuclear vicinity for Ran-G protein, which takes part in the general transport of proteins and RNAs across the nuclear membrane (for review, see Melchior and Gerace, 1998).
In addition we observed dramatic qualitative changes in mRNA distribution with development. A clear distributional shift was observed for MAP-kinase mRNA. In young cells, MAP-kinase mRNA was located preferentially in the basal region of the cell; whereas in cells with caps, the mRNA accumulates in the apical region (Fig. 6). Ser/Thr kinases in plant cells act as processing units, converting information from receptors via specific changes in gene expression into appropriate responses, which eventually result in cell growth or cell division (Hirt, 1997). The question of whether the observed developmental change to apical accumulation of MAP-kinase mRNA in cells with caps has to do with the transition from stalk growth to cap formation needs further investigations. The mRNA of UDP-Glc-epimerase, an enzyme that has been implicated in the transition of cell wall formation from stalk to cap (Zetsche, 1966b), followed a similar developmental pattern as MAP kinase mRNA (Fig. 6).
How Does mRNA Travel?
Because polar distribution of mRNAs turned out to be a common phenomenon in A. acetabulum, the question did arise of how such gradients are established in the giant cell. In animal cells, microtubules and microfilaments are involved (Bashirullah et al., 1998), whereas in Fucus sp. the spatial redistribution of poly(A) RNA appears to be dependent only on the presence of microfilaments (Bouget et al., 1995). In A. acetabulum, microtubules or tubulin proteins are undetectable during the vegetative growth, but microfilaments and intermediate filaments form a well-developed network (Menzel, 1994; Berger et al., 1998). We found that, without exception, cytochalasin D completely prevented gradient formation of all the mRNAs exhibiting apical/basal distribution (Fig. 9; data not shown).
Kloppstech and Schweiger (1975b) showed that poly(A) mRNA was transported to the apex of A. acetabulum at about 0.2 μm s−1. The time it would take to move a particular mRNA to the top of the cell (approximately 60 mm) is, therefore, much less than what could be achieved by simple diffusion. What distinguishes mRNAs that are retained around the nucleus, are transported to the tip, are evenly distributed, or are sequestered and temporarily silenced (such as tubulin mRNA) is completely unknown. It remains to be determined what specific RNA motifs and/or ribonucleo-proteins in concert with corresponding binding sites of the cytoskeleton are responsible for stop and go.
The direction of transport may be set by the polarity of the filament system. We know that the nucleus in some way is involved in this process, because in enucleated cell fragments, it is the position of a newly implanted nucleus that determines at which end (apical or basal) the morphogenetic events (stalk or rhizoid formation) will take place (Haemmerling, 1955; Zetsche, 1963). Similarly, either one or both of the filament systems could be involved in the anchoring and/or storing of mRNAs in the apical regions of the cell.
MATERIALS AND METHODS
Culture, Cell Fragmentation, and Harvesting
Unialgal cultures of Acetabularia acetabulum L. Silva (Haemmerling's isolate) cells were grown in Erdschreiber-medium (Schweiger et al., 1977) at 21°C under 2,500 Lux (36 W/19 Daylight 5000 De Luxe, Osram L., Munich) with a light-dark interval of 12 h. Culture medium was changed every 2 weeks. Cytochalasin D (Sigma, St. Louis) was diluted from a 1 mg mL−1 stock solution in dimethyl sulfoxide to a final concentration of 10 μg mL−1; control cells were exposed to same concentration of solvent only.
A. acetabulum cells were fragmented by a simple amputation technique using scissors and scalpel; little or no leakage of cytoplasm was observed. Cell fragments were either returned to culture or harvested directly into liquid nitrogen. Gametangia were isolated from matured caps (derived from 16-week-old cells) that had been kept without light for several weeks in sea water before dissection.
RNA Isolation and Generation of A. acetabulum cDNA Clones
Cells, cell fragments (10–50), germinating zygotes, and gametangia that had been harvested into liquid nitrogen were ground with a mortar and pestle into a fine powder, and RNA was extracted using RNA-Clean according to the manufacturer's instructions (AGS, Heidelberg, Germany).
Specific A. acetabulum cDNA probes were cloned with PCR methodology. Multiple orthologous sequences of selected proteins (EMBL, GenBank) were aligned with CLUSTAL W. Mixed primers, corresponding to well-conserved amino acid clusters, were used in PCR with oligo(dT)-primed cDNA that had been reverse transcribed from RNA of either gametangia or germinating zygotes of A. acetabulum. The resulting amplified fragments were isolated from agarose gels, purified, cloned into pBluescript II KS+ (Stratagene, La Jolla, CA) or pCR 2.1-TOPO (Invitrogen, Carlsbad, CA), and sequenced. Standard procedures were used (Sanger et al., 1977; Sambrook et al., 1989).
Gene-Specific A. acetabulum Primers Used in RT-PCR
Calmodulin-2: 180 bp [forward, ATGGCAAGGAGACATCGAACTTCGGC; reverse, CACCAAGGTACGTCATCACGTG]; calmodulin-4: 180 bp [forward, ATGGTAATG GTACTATAGACTTCCCT; reverse, CACCAAGGTACGTCATCACGTG]; Ran G-protein: 160 bp [forward, AAAGGCACTTGACGGGCGAG; reverse, TTG TAG TCT TCT TGG GTG CGG]; actin-1: 700 bp [forward, CCCAGCATCGTCGGCAAACCC; reverse, GTGTGTCCAAACAATCAGAAGC]; MAP-kinase: 300 bp [forward, TTACGTATTGTTGAGCGATAG; reverse, TGTTACCGTTGGGGATGGG]; rbcS: 250 bp [forward, TTTGCTGCTTCGGACTAGGC; reverse, TAGTCGGTAGCAGAGGGAGG]; 18S rRNA: 850 bp [forward, AAGTCTGGTGCCAGCAGCCGCG; reverse, AAGGCGCGCACCTCTACCGAGGC].
Semiquantitative Reverse Transcriptase PCR (RT-PCR)
cDNA was reverse transcribed from either 1 to 2 μg of total RNA or 100 ng of poly(A) RNA as template, with oligo(dT) or random primers, respectively, according to the manufacturer's instructions (Promega, Madison, WI), and after dilution, PCR with A. acetabulum gene-specific primers followed. For each individual primer pair, linear standard curves were established by varying the amount of input cDNA in conjunction with optimizing thermal cycling parameters. To examine two gene transcripts in the same assay, conditions were chosen to achieve optimal amplification of test and internal reference cDNAs, either 18S rRNA or rbcS.
Electrophoresis, Blotting, and Densitometry
RT-PCR products (20-μl aliquots per lane corresponding to 40 ng of total RNA in the RT-PCR approach) were separated on agarose gels and stained with ethidium bromide. Gels were then blotted onto nylon membranes (Hybond N+, Amersham, Buckinghamshire, UK) and hybridized with specifically cloned A. acetabulum cDNA probes. The plasmid cDNA, which had been labeled with fluorescein-11-dUTP, was detected by enhanced chemiluminescence (ECL-Kit RNP 3001, Amersham-Buchler, Braunschweig, Germany), and its binding pattern was captured on film (Amersham). The intensity of scanned signals was analyzed with the TINA software package (version 2.07 d, Raytest Isotopenmessgeraete, Straübenhardt, Germany) and expressed as relative units from the linear standard curves established for individual primer pairs.
In Vitro Transcription and RNase Protection Assay
Plasmid DNA was used for SP6 or T7 polymerase-directed RNA synthesis, with labeling of the RNA probes by incorporating fluorescein-conjugated UTP as recommended by the manufacturer (Roche Molecular Biochemicals, Mannheim, Germany). Total RNA (1–20 μg) was coprecipitated with fluorescein-labeled antisense RNA and digested with RNase T1. The protected RNAs were separated on 4% (w/v) SDS-PAGE, blotted on a Hybond N+ membrane, and detected by densitometry as described above.
All experiments were repeated at least three times. In each case, comparable results were observed. The data presented are from individual experiments representative for the respective results.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for non-commercial research.
Footnotes
This study was supported by the Deutsche Forschungsgemeinschaft (grant nos. Ze 71/27–1 and Ze 71/27–2).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010983.
LITERATURE CITED
- Bachmann P, Zetsche K. A close temporal and spatial correlation between cell growth, cell wall synthesis and the activity of enzymes of mannan synthesis in Acetabularia mediterranea. Planta. 1979;145:331–337. doi: 10.1007/BF00388357. [DOI] [PubMed] [Google Scholar]
- Bachs O, Agell N, Carafoli E. Calmodulin and calmodulin binding proteins in the nucleus. Cell Calcium. 1994;16:289–296. doi: 10.1016/0143-4160(94)90092-2. [DOI] [PubMed] [Google Scholar]
- Bashirullah A, Cooperstock RL, Lipshitz HD. RNA localization in development. Annu Rev Biochem. 1998;67:335–394. doi: 10.1146/annurev.biochem.67.1.335. [DOI] [PubMed] [Google Scholar]
- Berger S, Kaever MJ. Dasycladales. Stuttgart, Germany: Georg Thieme Verlag; 1992. [Google Scholar]
- Berger S, Wittke W, Traub P. Occurrence of proteinaceous 10 nm filaments throughout the cytoplasm of algae of the order Dasycladales. Exp Cell Res. 1998;240:176–186. doi: 10.1006/excr.1997.3924. [DOI] [PubMed] [Google Scholar]
- Bouget FY, Gerttula AS, Quatrano RS. Spatial redistribution of poly(A)+ RNA during polarization of the Fucus zygote is dependent on microfilaments. Dev Biol. 1995;171:258–261. doi: 10.1006/dbio.1995.1277. [DOI] [PubMed] [Google Scholar]
- Bouget FY, Gerttula S, Shaw SL, Quatrano RS. Localization of actin mRNA during the establishment of cell polarity and early cell division in Fucus embryos. Plant Cell. 1996;8:189–201. doi: 10.1105/tpc.8.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton G, Vanden Driessche T. Identification of calmodulin in Acetabularia: its distribution and physiological significance. J Cell Sci. 1987;87:337–347. [Google Scholar]
- Haemmerling J. Entwicklung und Formbildungsvermögen von Acetabularia mediterranea. Biol Zentbl. 1931;51:633–647. [Google Scholar]
- Haemmerling J. Entwicklung und Formbildungsvermögen von Acetabularia mediterranea. Biol Zentbl. 1932;52:42–61. [Google Scholar]
- Haemmerling J. Ueber formbildende Substanzen bei Acetabularia mediterranea, ihre raeumliche und zeitliche Verteilung und ihre Herkunft. Wilhelm Roux's Arch Entwicklungsmech Org. 1934a;131:1–81. doi: 10.1007/BF00649809. [DOI] [PubMed] [Google Scholar]
- Haemmerling J. Ueber Genomwirkungen und Formbildungsfähigkeit bei Acetabularia. Wilhelm Roux's Arch Entwicklungsmech Org. 1934b;132:424–462. doi: 10.1007/BF00577051. [DOI] [PubMed] [Google Scholar]
- Haemmerling J. Neuere Versuche ueber Polarität und Differenzierung bei Acetabularia. Biol Zentbl. 1955;74:545–554. [Google Scholar]
- Haemmerling J. Nucleo-cytoplasmic interactions in Acetabularia and other cells. Annu Rev Plant Physiol. 1963;14:65–92. [Google Scholar]
- Hirt H. Multiple roles of MAP kinases in plant signal transduction. Trends Plant Sci. 1997;1:11–15. [Google Scholar]
- Jukes TH. Neutral changes and modifications of the genetic code. Theor Popul Biol. 1996;49:143–145. doi: 10.1006/tpbi.1996.0008. [DOI] [PubMed] [Google Scholar]
- Kloppstech K, Schweiger HG. 80S ribosomes in Acetabularia major: distribution and transportation within the cell. Protoplasma. 1975a;83:27–40. doi: 10.1007/BF01289328. [DOI] [PubMed] [Google Scholar]
- Kloppstech K, Schweiger HG. Polyadenylated RNA from Acetabularia. Differentiation. 1975b;4:115–123. [Google Scholar]
- Kloppstech K, Schweiger HG. Stability of poly(A)+RNA in nucleate and anucleate cells of Acetabularia. Cell Rep. 1982;1:165–167. doi: 10.1007/BF00269189. [DOI] [PubMed] [Google Scholar]
- Koop HU, Kiermayer O. Protoplasmic streaming in the giant unicellular green alga Acetabularia mediterranea: II. Differential sensitivity of movement systems to substances acting on microfilaments and microtubules. Protoplasma. 1980;102:295–306. [Google Scholar]
- Lee SH, Kim MC, Heo WD, Kim JC, Chung WS, Park CY, Park HC, Cheong YH, Kim CY, Lee KJ et al. Competitive binding of calmodulin isoforms to calmodulin-binding proteins: implication for the function of calmodulin isoforms in plants. Biochim Biophys Acta. 1999;1433:56–67. doi: 10.1016/s0167-4838(99)00149-1. [DOI] [PubMed] [Google Scholar]
- Liu Z, Xia M, Poovaiah BW. Chimeric calcium/calmodulin-dependent protein kinase in tobacco differential regulation by calmodulin isoforms. Plant Mol Biol. 1998;38:889–897. doi: 10.1023/a:1006019001200. [DOI] [PubMed] [Google Scholar]
- Mandoli DF. Elaboration of body plan and phase change during development of Acetabularia: How is the complex architecture of a giant unicell built? Annu Rev Plant Physiol Plant Mol Biol. 1998;49:173–198. doi: 10.1146/annurev.arplant.49.1.173. [DOI] [PubMed] [Google Scholar]
- Melchior F, Gerace I. Two-way trafficking with Ran. Trends Cell Biol. 1998;8:157–179. doi: 10.1016/s0962-8924(98)01252-5. [DOI] [PubMed] [Google Scholar]
- Menzel D. How do giant plant cells cope with injury? The wound response in siphonous green alga. Protoplasma. 1988;144:73–91. [Google Scholar]
- Menzel D. Cell differentiation and cytoskeleton in Acetabularia: tansley review number 77. New Phytol. 1994;128:369–393. doi: 10.1111/j.1469-8137.1994.tb02984.x. [DOI] [PubMed] [Google Scholar]
- Moser MJ, Flory MR, Davis TM. Calmodulin localizes to the spindle pole body of Schizosaccharomyces pombe and performs an essential function chromosome segregation. J Cell Sci. 1997;110:1805–1812. doi: 10.1242/jcs.110.15.1805. [DOI] [PubMed] [Google Scholar]
- Reiss HD, Herth W. Calcium gradients in tip growing plant cells visualized by chlorotetracycline fluorescence. Planta. 1979;146:615–621. doi: 10.1007/BF00388841. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsche EF, Maniatis T. Molecular Cloning. A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain termination inhibitors. Proc Natl Acad Sci USA. 1977;74:5463. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawitzky H. Untersuchungen zur Rolle des Cytoskelettes bei der Hutmorphogenese der einzelligen Grünalge Acetabularia acetabulum. Inaugural Dissertation. Heidelberg: Ruprechts-Karls Universitaet; 1997. [Google Scholar]
- Schneider SU, Leible MB, Yang XP. Strong homology between the small subunit of ribulose-1, 5-bisphosphate carboxylase/oxygenase of two species of Acetabularia and the occurrence of unusual codon usage. Mol Gen Genet. 1989;218:445–452. doi: 10.1007/BF00332408. [DOI] [PubMed] [Google Scholar]
- Schweiger HG, Dehm P, Berger S. Culture conditions for Acetabularia. In: Woodcock CLF, editor. Progress in Acetabularia Research. New York: Academic Press; 1977. pp. 319–330. [Google Scholar]
- Serikawa KA, Mandoli DF. Aaknox1, a kn1-like homeobox gene in Acetabularia acetabulum, undergoes developmentally regulated subcellular localization. Plant Mol Biol. 1999;41:785–793. doi: 10.1023/a:1006387107071. [DOI] [PubMed] [Google Scholar]
- Serikawa KA, Porterfield DM, Mandoli DF. Asymmetric subcellular mRNA distribution correlates with carbonic anhydrase activity in Acetabularia acetabulum. Plant Physiol. 2001;125:900–911. doi: 10.1104/pp.125.2.900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel H. Untersuchungen zu spezifischen mRNA-Gradienten in der einzelligen, siphonalen Grünalge Acetabularia acetabulum (L) Silva. Inaugural Dissertation. Giessen: Justus Liebig Universität; 1998. [Google Scholar]
- Yang T, Lev-Yadun S, Feldman M, Fromm H. Developmentally regulated organ-, tissue-, and cell-specific expression of calmodulin genes in common wheat. Plant Mol Biol. 1998;37:109–120. doi: 10.1023/a:1005902905512. [DOI] [PubMed] [Google Scholar]
- Zetsche K. Das morphologische und physiologische Verhalten implantierter Zellkerne bei Acetabularia mediterranea. Planta. 1963;60:331–338. [Google Scholar]
- Zetsche K. Hemmung der Synthese morphogenetischer Substanzen im Zellkern von Acetabularia mediterranea durch Actinomycin. Z Naturforsch. 1964;19b:751–759. [PubMed] [Google Scholar]
- Zetsche K. Regulation der zeitlichen Aufeinanderfolge von Differenzierungsvorgängen bei Acetabularia. Z Naturforsch. 1966a;21b:375–379. [Google Scholar]
- Zetsche K. Regulation der UDPG-4-Epimerase Synthese in kernhaltigen und kernlosen Acetabularien. Biochim Biophys Acta. 1966b;124:332–338. doi: 10.1016/0304-4165(66)90196-6. [DOI] [PubMed] [Google Scholar]
- Zetsche K. Regulation der UDPG-Pyrophosphorylaseaktivität in Acetabularia: II. Unterschiedliche Synthese des Enzyms in verschiedenen Zellregionen. Planta. 1969;89:244–253. doi: 10.1007/BF00385029. [DOI] [PubMed] [Google Scholar]
- Zielinski RE. Calmodulin and calmodulin-binding proteins in plants. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:697–725. doi: 10.1146/annurev.arplant.49.1.697. [DOI] [PubMed] [Google Scholar]