Abstract
We examined the effect of a single exercise session on the protein and mRNA expression of the monocarboxylate transporters MCT1 and MCT4 in rat soleus (SOL), and red (RG) and white gastrocnemius (WG) muscles. Muscle samples were obtained at rest before 2 h of treadmill exercise (21 m min−1, 15% grade) and immediately after exercise, as well as 5, 10 and 24 h after exercise. During the 2 h exercise bout, MCT1 proteins in RG (+60%) and WG (+56%) were increased (P < 0.05). MCT1 protein was further increased thereafter, with peak increments occurring 10 h after exercise in RG (+157%), WG (+193%) and SOL (+179%) (P < 0.05). Twenty-four hours after exercise, MCT1 protein was still up-regulated in WG (+100%) and SOL (+55%) (P < 0.05), but not in RG. MCT1 mRNA was up-regulated during exercise in RG (+53%) and WG (+98%) and remained elevated until 24 h post-exercise in RG (P < 0.05), but in WG, MCT1 mRNA decreased transiently to pre-exercise levels at 5 and 10 h after exercise, before increasing again at 24 h (+150%) (P < 0.05). MCT4 protein and mRNA were not increased in WG muscle during and after exercise (P > 0.05). In contrast, during exercise, in RG (+41%) and SOL (+98%) MCT4 protein was increased (P < 0.05). Peak increases in MCT4 protein were observed 10 h after exercise in RG (+131%) and SOL (+323%) (P < 0.05). MCT4 protein was still up-regulated 24 h after exercise (RG: +106%; SOL +225%) (P < 0.05). MCT4 mRNA in RG was not increased until 10 (+132%) and 24 h after exercise (+55%) (P < 0.05). These studies have shown that MCT1 and 4 proteins are transiently up-regulated by a single bout of exercise, involving post-transcriptional and transcriptional mechanisms. Thus, MCT1 and MCT4 belong to a class of selected metabolic genes that are very rapidly up-regulated with an exercise stimulus.
During exercise, lactate is transported out of the working muscle cell. This substrate can be taken up by neighbouring muscle cells or enter the circulation to be metabolized in other muscles or tissues, including the heart, kidney and liver. The transmembrane movement of lactate into and out of muscle involves a lactate–proton cotransport system that is pH dependent and stereo-specific for l-lactate (Roth & Brooks, 1990; Juel, 1997; Juel & Halestrap, 1999). Studies investigating the kinetics of lactate transport with small or giant vesicles have shown that the Km values for lactate transport rates are within the physiological ranges of circulating and intramuscular lactate concentration (5–40 mm) (Juel, 1997).
In the past decade a family of monocarboxylate transporters (MCTs) has been identified. Although 14 MCTs are known to exist (see Halestrap & Meredith, 2004), the kinetics and tissue distribution of only a few are known, while many remain orphan transporters. Among the MCTs that have been characterized, MCT1 is ubiquitously expressed in many tissues (Garcia et al. 1995; Jackson et al. 1997; Gerhart et al. 1998; Pellerin et al. 1998a,b; Bonen, 2001; Boussouar et al. 2003; Sepponen et al. 2003). Some other MCTs have a more restricted tissue distribution. MCT3 is expressed in the basal membrane of retinal pigment epithelium (Philp et al. 1998), while MCT4 is primarily expressed in skeletal muscle, along with MCT1 (Wilson et al. 1998; Pilegaard et al. 1999b; Bonen et al. 2000b; Dubouchaud et al. 2000; Bonen, 2001; Sepponen et al. 2003). In rodents, muscles that are composed of predominantly oxidative fibres (e.g. soleus) express considerable quantities of MCT1 and very little MCT4 (Bonen et al. 2000b), whereas muscles with a large proportion of fast-twitch, glycolytic fibres (e.g. white gastrocnemius) express primarily MCT4 and very little MCT1 (Bonen et al. 2000b). Muscles composed of fast-twitch oxidative glycolytic fibres (e.g. red gastrocnemius) express both MCT1 and MCT4 in appreciable quantities (Bonen et al. 2000b). MCT1 and 4 are also coexpressed in human muscle, with MCT1 being more prevalent in type I fibres and MCT4 in type II fibres (Pilegaard et al. 1999b). Kinetics data suggest that MCT1 (Km ∼5 mm) and MCT4 (Km ∼20 mm) are key transporters for lactate, the most prominent of the monocarboxylates, since their Km values are consistent with lactate concentrations that occur physiologically (Broer et al. 1998; Dimmer et al. 2000; Manning Fox et al. 2000).
The expression of MCT1 and 4 in muscle is regulated by a number of factors. Endocrine regulation of MCT1 and/or MCT4 expression has recently been reported (Enoki et al. 2003; Wang et al. 2003). Muscle activity also exerts a strong influence on the expression of MCTs. Muscle inactivity (denervation) reduces MCT1 and 4 expression (Wilson et al. 1998), while conversely, increased muscle activity induces the expression of MCT1 and MCT4 (McCullagh et al. 1996a; Baker et al. 1998; Bonen et al. 1998, 2000c; Pilegaard et al. 1999a; Dubouchaud et al. 2000). Reduction in post-exercise lactate accumulation is one of the very early adaptive responses, and appears to be associated with an enhanced rate of lactate clearance (Donovan & Brooks, 1983). Thus, it is possible that an exercise-induced, up-regulation of MCTs is an early response to exercise training.
It had long been thought that induction of protein expression was a relatively slow process. However, a number of studies have recently shown that a single exercise session transiently up-regulated a number of genes involved with substrate metabolism. For example, mRNAs of peroxisome proliferator activated receptor coactivator 1α (PGC-1α), uncoupling protein 3 (UCP3), lipoprotein lipase (LPL) and carnitine palmitoyl transferase 1 (CPT1) were rapidly up-regulated during exercise (Pilegaard et al. 2000, 2003; Zhou et al. 2000). In other studies, the mRNAs of the glucose transporter 4 (GLUT4), UCP3, LPL, PGC-1α, hexokinase II, peroxisome proliferator activated receptor α (PPARα), calcineurin Aα and Aβ, pyruvate dehydrogenase kinase 4 (PDK4), and haem-oxygenase 1 (HO-1) were increased within a few hours after a single bout of exercise in humans (Pilegaard et al. 2003) and rats (Ren et al. 1994; Hildebrandt et al. 2003). These increments were associated with increased rates of transcription of these genes (Hildebrandt et al. 2003; Pilegaard et al. 2003). Concomitant changes in protein expression have typically not been examined in these studies, but there are exceptions. Ren et al. (1994) found that along with a 200% increase in GLUT-4 mRNA, GLUT-4 protein was also markedly increased (+50%) in epitrochlearis muscle, 16 h after one prolonged exercise session. Similarly, Zhou et al. (2000) observed that both PGC-1α protein (+465%) and mRNA (+444%) were increased during exercise. Thus, it appears that for selected genes, exercise-induced up-regulation of their mRNA and protein expression can occur much more rapidly than had generally been recognized.
MCTs may also be rapidly up-regulated by exercise. Baker et al. (1998) observed that the training-induced increase in MCT1 occurred quite rapidly, as MCT1 was increased +30% after only a few exercise bouts. Others have shown qualitatively that MCT4 mRNA in rat muscle was transiently increased during exercise (Zhou et al. 2000). In human muscle, MCT1 and 4 proteins were increased by 20–40%, 2 and 4 days after a prolonged exercise bout (Green et al. 2002), although, presumably, earlier and larger changes in these MCTs had occurred shortly after exercise. Indeed, in pilot studies in our laboratory we have observed that MCT1 and 4 proteins were already considerably up-regulated 24 h after an exercise bout. Therefore, to determine whether MCTs belong to a unique class of metabolic genes that are very rapidly up-regulated by exercise (Pilegaard et al. 2000, 2002, 2003; Hildebrandt et al. 2003), we examined the exercise-induced changes in MCT1 and MCT4 at both the mRNA and protein levels within the first 24 h after a 2-h exercise bout. These studies were performed in three types of rat skeletal muscle (red (RG) and white gastrocnemius (WG), and soleus (SOL)), since (a) the rapid exercise-induced increase in gene expression can occur in a muscle fibre type-specific manner (Hildebrandt et al. 2003) and (b) metabolically heterogeneous muscles express different amounts of MCT1 and MCT4 (Bonen et al. 2000b).
Methods
Ethical approval for these studies was obtained from the Animal Care Committee at the University of Guelph. Female Sprague-Dawley rats were housed in an air-conditioned room on a 12: 12-h light–dark cycle. They were fed a diet of Purina rat chow and water ad libitum. Between 8 and 10 weeks of age, animals (204 ± 11 g) were familiarized with treadmill running at low speeds (15–20 m min−1, 8% grade) for approximately 5–10 min day−1 for 5 days.
For the purpose of the present study, exercise consisted of running on a treadmill at a moderate-intensity (21 m min−1, 15% grade). The rats completed 30 min of exercise followed by 30 min rest until 2-h of exercise had been completed. In preliminary experiments 2 h of exercise was shown to reduce glycogen concentrations by 18–40% (P < 0.05) in SOL, RG and WG muscle. In the experimental studies, animals were aneasthetized with an intraperitoneal injection of Somnotol (60 mg (100 g body wt)−1) before and immediately after 2 h of exercise, as well as at 5, 10 and 24 h after exercise. Anaesthesia was confirmed according to the Canadian Council on Animal Care standards, namely by the absence of foot and toe reflexes when pinched and the absence of eyelid reflexes when touched. While animals were under anaesthesia, SOL, RG and WG muscles were rapidly removed, frozen in liquid nitrogen, and stored at −80°C until analysed for MCT1 and MCT4 protein and mRNA. After obtaining the required tissues, animals were killed with an overdose of Somnotol (150 mg (100 g body wt)−1).
MCT1 and MCT4 proteins
Proteins were isolated from the muscles and were separated using SDS-PAGE, followed by Western blotting to detect the presence of MCT1 and 4 proteins. Equal quantities of protein were loaded into each lane for each muscle, and a common standard was included in all blots. These routine procedures have been previously described by us (Bonen et al. 2000b,c; Tonouchi et al. 2002; Wang et al. 2003). Antibodies for MCT1 and 4 were raised in rabbits against the oligopeptide corresponding to the C-terminus regions of each MCT (Qiagen, Tokyo, Japan). MCT1 and 4 blots were quantified using an enhanced chemiluminescence (ECL, Amersham, Oakville, Canada) detection method by exposing the membranes to film (Hyperfilm-ECL, Amersham) at room temperature. The film was developed in GBX developer and fixed in GBX fixer (Kodak). Films were scanned onto a computer and densitometric analysis was performed using Scion Image software (Scion Corporation, Frederick, MD, USA).
MCT1 and MCT4 mRNAs
MCT1 mRNA and MCT4 mRNA were determined in RG and WG muscles only, using Northern blotting procedures, as we have previously described (Bonen et al. 2000b,c; Wang et al. 2003). Briefly, total RNA was isolated from muscle tissues using the GIT/CsCl centrifugation method (Chirgwin et al. 1979), with some modifications. The tissues were homogenized in 10 ml of 4 m guanidine isothiocyanate, and layered on top of 3.3 ml of 5.7 m caesium chloride solution. The samples were centrifuged in an SW-41 Ti rotor (Beckman), at 27 500 g for 23 h. The RNA pellets were recovered and purified by two precipitations in ethanol. Three micrograms of total RNA was used for electrophoresis on 1.2% formaldehyde–agarose gels, and then transferred to positively charged nylon membrane (Boehringer Mannheim, Laval, Quebec, Canada). The Northern blots were UV cross-linked with a GS-Gene linker (Bio-Rad). Data were expressed relative to the 18S rRNA signal.
A 1.9 kb fragment containing the coding sequence of MCT1 cDNA was isolated from the full length (3.3 kb) MCT1 by digestion with EcoRI restriction enzyme from the full length (3.3 kb) MCT1 cDNA (Jackson et al. 1997) and subcloned into the EcoRI restriction enzyme site of pBluescript (KS). The orientation was checked by digestion with HindIII restriction endonuclease. Template DNA was linearized with XbaI restriction enzyme, and DIG-labelled MCT1 antisense riboprobe was generated by in vitro transcription with T3 RNA polymerase. MCT4 cDNA was originally subcloned into BamHI/ApaI restriction enzyme sites of pBluescript (Wilson et al. 1998). DIG-labelled MCT4 antisense riboprobe was generated by digestion of the template DNA with XbaI restriction enzyme, and in vitro transcription with T7 RNA polymerase.
The ingredients for RNA transcription included 1–2 μg of DNA template plus the NTP mix (2.5 mm CTP, 2.5 mm rGTP, 2.5 mm ATP, 1.625 mm UTP (Promega) and 0.875 mm Dig-11 UTP (Boehringer Mannheim)), 20 mm DTT (Promega), 1 U/1 μg template DNA of RNase inhibitor (Promega) and 1× RNA polymerase buffer (5× buffer: 400 mm Tris-HCl pH 7.5; 60 mm MgCl2 and 20 mm spermidine-HCl (Promega)) maintained at room temperature. The appropriate RNA polymerase (T3 or T7 RNA polymerases; Boehringer Mannheim) was added (at least 20 IU/1 μg of DNA template) and incubated for 2 h at 37°C. The DNA template was then digested for 10 min at 37°C with RNase-free DNase (1 IU/1 μg of DNA template; Promega). After precipitation in ethanol and centrifugation at 13 500 g for 15 min, the probe was resuspended in 10–20 ml DIG easy-hyb hybridization buffer (Boehringer Mannheim), or standard hybridization buffer with 50% formamide (5× SSC, 50% formamide, 0.1% sodium-lauroylsarcosine, 0.02% SDS and 2% Blocking reagent; Boehringer Mannheim).
After pre-hybridization of the membrane for at least 4 h at 68°C, the pre-hybridization buffer was replaced with the same buffer containing DIG-labelled antisense RNA probe and the membrane was incubated with the probe overnight at 68°C. High stringency washes and chemiluminescence detection were performed in accordance with the protocol supplied by the manufacturer (Boehringer Mannheim), and the membrane was exposed to Kodak BioMax film. After a 7–10 min exposure the film was developed in Kodak developer and fixed in Kodak fixer.
Statistics
The data were analysed using analysis of variance. Fisher's least squares difference (LSD) comparison was used for post hoc analysis. All data are reported as means ± s.e.m.
Results
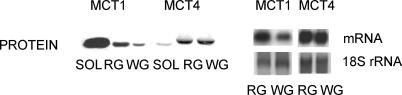
Concentrations of MCT1 and MCT4 protein and mRNA differ widely among the muscles examined (Fig. 1 and Table 1). MCT1 protein was most abundantly expressed in SOL with lower levels of expression in RG and WG (Table 1). In contrast, MCT4 protein expression is most abundant in WG and RG muscles, while SOL muscle expressed considerably less MCT4 protein (Table 1). Differences in MCT1 and 4 mRNA abundances in RG and WG are less than for their respective proteins (Fig. 1 and Table 1). In the present study, MCT protein and mRNA prior to exercise were set to 100 in each muscle, and the remaining data were expressed relative to this pre-exercise control.
Figure 1. Representative Western and Northern blots of MCT1 and 4 in skeletal muscle.
Equal quantities of protein (20 μg) and RNA (3 μg) were loaded for each muscle in order to make direct comparisons among muscles.
Table 1.
MCT1 and 4 protein and mRNA in rat hindlimb muscles (mean ±s.e.m.)
| Muscle | MCT1 protein (a.u./20 μg potein) | MCT4 protein (a.u./20 μg potein) | MCT1 mRNA (a.u./18S rRNA) | MCT4 mRNA (a.u./18S rRNA) |
|---|---|---|---|---|
| Soleus | 311 ± 36** | 100 ± 8.5 | ND | ND |
| Red gastrocnemius | 238 ± 27* | 298 ± 9.1*** | 100 ± 2.3* | 100 ± 5.0 |
| White gastrocnemius | 100 ± 3 | 315 ± 7.8*** | 85 ± 2.3 | 97.8 ± 6.1 |
Equal quantities of protein (20 μg) and RNA (3 μg) were loaded for each muscle so as to be able to make direct comparisons among muscles. Protein data are arbitrary units (a.u) per 20 μg protein, with the mean for muscles with the lowest content of MCT1 (white gastrocnemius) and MCT4 proteins (soleus) being set to 100. mRNA data are normalized to 18S rRNA, followed by a further transformation of the data so that the red gastrocnemius muscle ratio ± s.e.m. was set to 100 ± s.e.m., with the white gastrocnemius being set relative to this. ND, not determined. MCT1, n = 13; MCT4, n = 7; MCT1 mRNA, n = 3; MCT4 mRNA n = 3.
MCT1: RG versus WG, P < 0.05
MCT1: SOL versus RG, and SOL versus WG P < 0.05
MCT4: RG versus SOL and WG versus SOL, P < 0.05.
Effects of one exercise session on MCT1 protein
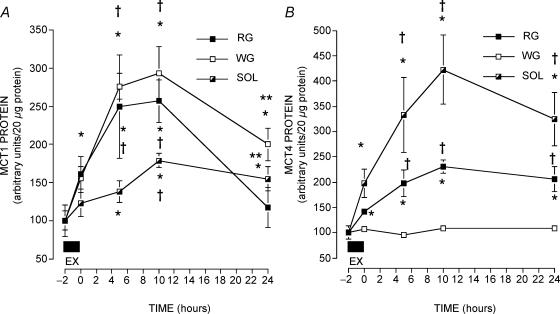
After a single bout of treadmill exercise, MCT1 protein levels increased in all muscles. In general, there was a significant change in RG MCT1 protein over the 24-h period (Fig. 2A). Already after 2 h of exercise there was a significant increase in MCT1 (+61%) (P < 0.05). Relative to the pre-exercise RG MCT1, there was a further increase at 5 h (+150%) and 10 h (+157%) after exercise (P < 0.05). Thereafter, RG MCT1 decreased sharply between 10 and 24 h (P < 0.05) (Fig. 2A). The pre-exercise and 24-h post exercise RG MCT1 levels did not differ (P > 0.05) (Fig. 2A).
Figure 2. MCT1 (A) and MCT4 (B) protein in Soleus (SOL) and in red (RG) and white gastrocnemius (WG) muscles before and after a 2 h bout of exercise.
Data are means ± s.e.m.; n = 4–9 muscles at each point. Equal quantities of protein (20 μg) were loaded in each well. Protein data are arbitrary units (a.u) per 20 μg protein. In each muscle the pre-exercise ratio ±s.e.m. was set to 100 ±s.e.m., with the remaining data in each muscle set relative to the pre-exercise data. *P < 0.05 compared to pre-exercise; †P < 0.05 compared to immediate post-exercise (t = 0); **P < 0.05 24 h post-exercise versus 10 h post-exercise.
In WG, MCT1 protein was significantly increased over the 24-h period following exercise (Fig. 2A). An increase in MCT1 (+56%) was already present at the end of 2 h of exercise. WG MCT1 was further increased at 5 h (+176%) and 10 h (+193%) after exercise (P < 0.05). MCT1 protein levels in WG were still elevated (+100%) 24 h after exercise (P < 0.05) (Fig. 2A).
In SOL muscle, MCT1 protein levels were not increased during exercise. However, SOL MCT1 protein was increased significantly at 10 h (+79%) and 24 h (+55%) after exercise (Fig. 2A) (P < 0.05).
Effects of one exercise session on MCT4 protein expression
During the 24 h observation period, RG MCT4 was significantly increased (Fig. 2B). At the end of exercise there was already a significant increase in RG MCT4 (+41%) (P < 0.05), and there was a further increase 5 h after exercise (+98%) (P < 0.05). This increase remained stable at 10 h (+131%) and 24 h (+106%) after exercise (P < 0.05, Fig. 2B). In contrast to RG, WG MCT4 protein expression was not changed during the 24-h period after exercise (P > 0.05) (Fig. 2B).
SOL MCT4 was significantly up-regulated by the 2 h exercise bout (Fig. 2B). During exercise MCT4 was already increased (+98%) (P < 0.05), and there was an additional increase 5 h after exercise (+233%) (P < 0.05). This MCT4 up-regulation remained stable at 10 h (+323%) and 24 h (+225%) after exercise (P < 0.05).
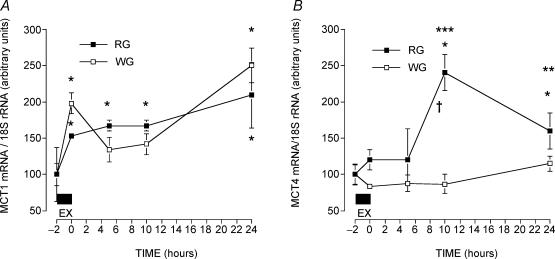
Effects of one exercise session on MCT1 mRNA abundance
After 2 h of exercise, RG MCT1 mRNA was significantly increased (+53%) (P < 0.05) and remained increased at 5 h (+67%), 10 h (+68%) and 24 h (+108%) after exercise (P < 0.05) (Fig. 3A). There was also a significant increase in WG MCT1 mRNA (+98%) during exercise (P < 0.05) (Fig. 3A), and at 5 h (+34%), 10 h (+42%) and 24 h (+150%) after exercise (P < 0.05, Fig. 3A).
Figure 3. MCT1 mRNA (A) and MCT4 mRNA (B) in red (RG) and white gastrocnemius (WG) muscles before and after a 2 h bout of exercise.
Data are means ± s.e.m. n = 3–4 muscles at each point. Equal quantities of RNA (3 μg) were loaded in each well and data were normalized to 18S rRNA. This was followed by a further transformation of the data, so that in each muscle the pre-exercise ratio ± s.e.m. was set to 100 ± s.e.m., with the remaining data in each muscle set relative to the pre-exercise data. *P < 0.05 compared to pre-exercise; †P < 0.05 compared to immediate postexercise (t = 0); **P < 0.05 24 h post-exercise versus 10 h post-exercise; ***P < 0.05 10 h post-exercise versus 5 h post-exercise.
Effects of one exercise session on MCT4 mRNA abundance
MCT4 mRNA accumulation in RG did not differ from control muscles either immediately after exercise or 5 h after exercise (P > 0.05). There was, however, a significant increase in MCT4 mRNA at 10 h (+132%) and 24 h (+55%) after exercise (P < 0.05) (Fig. 3B).
Although WG MCT4 mRNA abundance was slightly reduced during the first 10 h after exercise (−16 to −12%), this decrease was not significant (P > 0.05) (Fig. 3B). Similarly, the slight increase observed 24 h after exercise in WG MCT4 mRNA (+15%) was not significant (P > 0.05) (Fig. 3B).
Discussion
The present study has shown that MCT1 and 4 belong to a class of metabolic genes that can be very rapidly up-regulated by a bout of exercise. The novel observations in this study are as follows: (a) there were large changes in both MCT1 and MCT4 proteins during the first 24 h after a single exercise bout in most muscles examined, (b) the changes in MCT1 and 4 had in most instances already been initiated during the 2 h exercise period, (c) the increases in MCT1 were maximal 5–10 h after exercise, after which MCT1 began to decrease to pre-exercise levels in RG and WG, but not SOL, (d) maximal, exercise-induced MCT4 increments also occurred 5–10 h after exercise, but these MCT4 protein levels remained elevated up to 24 h after exercise, and (e) based on the post-exercise changes in MCT1 and 4 mRNA, the changes in MCT1 and 4 protein expression appeared to be mediated by transcriptional and post-transcriptional mechanisms.
Previous studies have shown that MCT1 and 4 proteins are increased by training (Baker et al. 1998; Bonen et al. 1998; Pilegaard et al. 1999a; Dubouchaud et al. 2000). Already at the end of five exercise sessions there was a marked increase in MCT1 protein (Baker et al. 1998). An increase in MCT4 mRNA has recently been observed within the first 100 min of exercise, with a decrease in this transcript occurring during the next 100 min of exercise (Zhou et al. 2000). Others have reported modest, exercise-induced increments in MCT1 and 4 protein (Green et al. 2002), although it was not clear why these exercise-induced increments were examined 2–6 days after completing exercise, rather than in the immediate hours or first day after exercise. From the present studies it is clear that the exercise-induced increments in MCT1 and 4 protein had at times already occurred during exercise, and attained a maximum 5–10 h after exercise. The increases in the MCT proteins were quite large. With the exception of WG MCT4, which was not up-regulated, increases in MCT1 and 4 in the SOL and RG ranged from +23% to +98% at the end of exercise, and from +79% to +323% at their peak, 10 h after exercise, depending on the muscle examined. Changes in UCP3 protein up-regulation during exercise (i.e. at min 30 (+∼100%), at min 100 (+∼400%) and at min 200 (+∼550%)) appear to be of a similar magnitude (Zhou et al. 2000), although post-exercise changes in UCP3 protein expression were not examined. We are not aware of other studies that have examined changes in protein expression during or immediately after exercise.
Several studies have shown that mRNA abundance of some, but not all, metabolic genes is increased during exercise and/or in the first 4–6 h after exercise. This has been attributed to increased rates of transcription of selected genes (Pilegaard et al. 2000; Hildebrandt et al. 2003; Pilegaard et al. 2003). The intensity and duration of exercise, as well as the muscle fibre composition, can influence the increased rates of transcription in the post-exercise period (Hildebrandt et al. 2003). In the present study, the exercise-induced increases in MCT1 and 4 protein expression appeared to be mediated by both transcriptional and post-transcriptional mechanisms, as the increase in MCT proteins was not always accompanied by concurrent changes in their transcripts. We (Bonen et al. 2000c) have previously shown that increments in MCT1 protein during 7 days of chronic muscle contraction are mediated by post-transcriptional mechanisms. In the present study, the patterns of change in the mRNAs after exercise differed somewhat within and between muscles (see Results). Others have shown that MCT4 mRNA can increase and decrease during the same prolonged exercise bout (Zhou et al. 2000). Thus, the variable MCT mRNA patterns in muscle, induced by exercise, may not be unusual, particularly as studies by Hildebrandt, Pilegaard and Neufer (Hildebrandt et al. 2003) have recently shown that gene-specific and muscle fibre type-specific responses to exercise can occur. For example, the rate of transcription for a given gene can be greater in red than in white muscle (e.g. HO-1), or alternatively, the rate of transcription can be greater in white than in red muscle (e.g. uncoupling protein 3 (UCP3)) (Hildebrandt et al. 2003). In addition, light prolonged exercise (180 min) provides the greatest stimulus for up-regulating HO-1 in red muscle while comparable changes in this gene are observed in white muscle with heavy short duration exercise (45 min) (Hildebrandt et al. 2003). Although low muscle glycogen content enhances the transcriptional activation of some metabolic genes in response to exercise (Pilegaard et al. 2002), this was probably not a determining factor for the differing MCT1 and 4 mRNA responses in RG and WG muscles, as in preliminary experiments the exercise-induced glycogen loss in RG and WG was very similar. Collectively, the present study and others (Pilegaard et al. 2000, 2002, 2003; Zhou et al. 2000; Hildebrandt et al. 2003) illustrate that the exercise-induced regulation of protein expression is complex. Explanatory models describing the mechanisms that regulate gene expression have undergone a number of revisions in the past 50 years. A recent series of reviews have addressed the current understandings of the complexities involved in regulating gene expression (Baker & Parker, 2004; Chambeyron & Bickmore, 2004; Dahlberg & Lund, 2004; Grewal & Rice, 2004; Huang & Richter, 2004; Murchison & Hannon, 2004; Proudfoot, 2004; Van De Bor & Davis, 2004). These reviews clearly show that there is not a simple concordance between the rate of gene transcription, mRNA abundance and the changes in protein levels.
In studies by Pilegaard and Neufer (Pilegaard et al. 2000, 2002, 2003), it was clear that not all genes are up-regulated by exercise. Recently, they (Hildebrandt et al. 2003) have postulated that genes, which are rapidly up-regulated by exercise, may not be expressed in sufficient quantities to meet the needs of the contracting muscle. Hence, these types of genes are thought to be the most susceptible to exercise-activated signalling/regulatory mechanisms that acutely activate transcription. This principle at first glance appears to apply to the MCTs in the present study. The muscles with the lowest quantity of MCT1 (WG) and MCT4 (SOL) showed the greatest relative increases in MCT1 (WG: +193%) and MCT4 (SOL: +323%). However, the ‘low expression–rapid induction’ principle (Hildebrandt et al. 2003) does not appear to apply to RG muscle. This muscle expresses MCT1 protein in greater amounts than WG, yet the peak increase in MCT1 (+157%) was quite comparable to that of WG (+193%). Moreover, the quantity of MCT4 in RG is quite comparable to that of WG (Table 1 and Bonen et al. 2000b), yet there was a large increase in RG MCT4 protein (+131% at 10 h post-exercise) while concomitantly there was no change in WG MCT4 protein. Thus, it will require further study to ascertain whether the attractive ‘low expression–rapid induction’ hypothesis (Hildebrandt et al. 2003) is robust. Our present results do not fully support this scheme.
We have previously shown that the rate of lactate flux into and out of muscle is correlated with the content of MCT1 and MCT4 in muscle (McCullagh et al. 1996a,b, 1997; Bonen et al. 2000b,c). Thus, the increase in MCT1 and 4 in the present study may well be expected to affect the rates of lactate flux. However, in one study (Green et al. 2002) the small increments in MCT1 and MCT4, measured 2–6 days after exercise, were only crudely associated with muscle lactate concentrations, as the reductions in muscle lactate did not fully mirror the small increases in muscle MCT1 and 4 (Green et al. 2002). It may well be important to examine changes in lactate concentrations or flux in relation to the plasmalemmal content of MCTs, as MCT1 and 4 are also located within intracellular sites such as mitochondria (Brooks et al. 1999; Benton et al. 2004), and MCT4 is also present in low density microsomes and t-tubules (Bonen et al. 2000b). Thus, just as for other substrate transporters, such as GLUT4 (Goodyear & Kahn, 1998; Lemieux et al. 2000) and fatty acid translocase (FAT/CD36) (Bonen et al. 2000a; Steinberg et al. 2002), it is the quantity of MCT 1 and 4 at the plasma membrane, not necessarily their expression, that regulates the rate of lactate flux. We do believe that the observed increases in MCT1 and 4 expression in the present study will result in increased rates of lactate flux, as this has been observed in previous studies when MCT1 proteins have been experimentally up-regulated (McCullagh et al. 1997; Baker et al. 1998; Bonen et al. 2000c).
The present study indicates that care may be required when examining the training-induced alterations in lactate uptake and MCTs in skeletal muscle. Specifically, experiments will now need to be designed to rule out the possibility that the observed effects in these parameters after training do not simply reflect the effects of the last exercise bout. Such care has for many years been required in studies examining glucose metabolism, as the effects of an exercise bout on up-regulating glucose uptake is well known to last for many hours. Whether such care is always necessary in studies examining exercise- or training-induced alterations in lactate uptake and metabolism is not known. The results observed in the present study may well have occurred because the exercise was prolonged. Less prolonged and/or less intense exercise may not provoke changes in MCTs and lactate metabolism after a single exercise bout, whereas repetition of such bouts (i.e. training) may over time increase MCT protein expression and alter lactate metabolism. Clearly, until we have a better understanding as to what type of exercise up-regulates MCT protein expression rapidly, future studies may have to distinguish between acute, exercise-induced effects and training-induced effects on MCTs and lactate metabolism.
Summary
We have shown that a single bout of treadmill exercise increased the expression of both MCT1 and MCT4 in SOL and RG, and MCT1 in WG. Notably, the protein levels had already increased during exercise in RG and WG. Peak MCT1 and MCT4 protein expression occurred in all muscles 10 h after exercise, except in WG MCT4. MCT1 and MCT4 protein expression appeared to be regulated by both transcriptional and post-transcriptional mechanisms. We speculate that these rapid exercise-induced increments in MCTs could contribute to the well known reductions in the post-exercise muscle lactate and circulating lactate concentrations that are one of the earliest adaptive responses observed with the onset of an exercise training programme. However, it must be acknowledged that changes in lactate transport cannot always be accounted for by concomitant changes in MCT1 or 4 (e.g. Bonen et al. 2000b,c; Tonouchi et al. 2002; Wang et al. 2003). Finally, it appears that MCT1 and MCT4 belong to a class of metabolic genes that are very rapidly up-regulated by a single exercise session.
Acknowledgments
These studies were supported by a grant from the Natural Sciences and Engineering Research Council of Canada. A. Bonen is Canada Research Chair in Metabolism and Health.
References
- Baker SK, McCullagh KJA, Bonen A. Training intensity dependent and tissue specific increases in lactate uptake and MCT1 in heart and muscle. J Appl Physiol. 1998;84:987–994. doi: 10.1152/jappl.1998.84.3.987. 10.1063/1.368165. [DOI] [PubMed] [Google Scholar]
- Baker KE, Parker R. Nonsense-mediated mRNA decay: terminating erroneous gene expression. Curr Opin Cell Biol. 2004;16:293–299. doi: 10.1016/j.ceb.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Benton CR, Campbell SE, Tonouchi M, Hatta H, Bonen A. Monocarboxylate transporters in subsarcolemmal and intermyofibrillar mitochondria. Biochem Biophys Res Comm. 2004;323:249–253. doi: 10.1016/j.bbrc.2004.08.084. [DOI] [PubMed] [Google Scholar]
- Bonen A. Expression of lactate transporters (MCT1, MCT4) in heart and muscle. Europ J Appl Physiol. 2001;86:6–11. doi: 10.1007/s004210100516. [DOI] [PubMed] [Google Scholar]
- Bonen A, Luiken JJFP, Arumugam Y, Glatz JFC, Tandon NN. Acute regulation of fatty acid uptake involves the cellular redistribution of fatty acid translocase. J Biol Chem. 2000a;275:14501–14508. doi: 10.1074/jbc.275.19.14501. [DOI] [PubMed] [Google Scholar]
- Bonen A, McCullagh KJA, Putman CT, Hultman E, Jones NL, Heigenhauser GJF. Short-term training increases human muscle MCT1 and femoral venous lactate in relation to muscle lactate. Am J Physiol Endocrinol Metab. 1998;274:E102–E107. doi: 10.1152/ajpendo.1998.274.1.E102. [DOI] [PubMed] [Google Scholar]
- Bonen A, Miskovic D, Tonouchi M, Lemieux K, Wilson MC, Marette A, et al. Abundance and subcellular distribution of MCT1 and MCT4 in heart and fast-twitch skeletal muscles. Am J Physiol Endocrinol Metab. 2000b;278:E1067–E1077. doi: 10.1152/ajpendo.2000.278.6.E1067. [DOI] [PubMed] [Google Scholar]
- Bonen A, Tonuchi M, Miskovic D, Heddle C, Heikkila JJ, Halestrap AP. Isoform-specific regulation of the lactate transporters MCT1 and MCT4 by contractile activity. Am J Physiol Endocrinol Metab. 2000c;279:E1131–E1138. doi: 10.1152/ajpendo.2000.279.5.E1131. [DOI] [PubMed] [Google Scholar]
- Boussouar F, Mauduit C, Tabone E, Pellerin L, Magistretti PJ, Benahmed M. Developmental#and hormonal regulation of the monocarboxylate transporter 2 (MCT2) expression in the mouse germ cells. Biol Reprod. 2003;69:1069–1078. doi: 10.1095/biolreprod.102.010074. [DOI] [PubMed] [Google Scholar]
- Broer S, Schneider HP, Broer A, Rahman B, Hamprecht B, Deitmer JW. Characterization of the monocarboxylate transporter 1 expressed in Xenopus laevis oocytes by changes in cytosolic pH. Biochem J. 1998;333:167–174. doi: 10.1042/bj3330167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks GA, Brown MA, Butz CE, Sicurello JP, Dubouchaud H. Cardiac and skeletal muscle mitochondria have a monocarboxylate transporter MCT1. J Appl Physiol. 1999;87:1713–1718. doi: 10.1152/jappl.1999.87.5.1713. [DOI] [PubMed] [Google Scholar]
- Chambeyron S, Bickmore WA. Does looping and clustering in the nucleus regulate gene expression? Curr Opin Cell Biol. 2004;16:256–262. doi: 10.1016/j.ceb.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Chirgwin J, Przbyla A, MacDonald R, Rutter W. Isolation of biologically active ribonucleic acid from source enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Dahlberg JE, Lund E. Does protein synthesis occur in the nucleus? Curr Opin Cell Biol. 2004;16:308–313. doi: 10.1016/j.ceb.2004.03.006. 10.1016/j.ceb.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Dimmer K-S, Friedrich B, Lang F, Deitmer JW, Broer S. The low affinity monocarboxylate transporter MCT4 is adapted to the export of lactate in highly glycolytic cells. Biochem J. 2000;350:219–227. 10.1042/0264-6021:3500219. [PMC free article] [PubMed] [Google Scholar]
- Donovan C, Brooks GA. Endurance training affects lactate clearance, not lactate production. Am J Physiol. 1983;244:83–92. doi: 10.1152/ajpendo.1983.244.1.E83. [DOI] [PubMed] [Google Scholar]
- Dubouchaud H, Butterfield GE, Wolfel EE, Bergman BC, Brooks GA. Endurance training, expression, and physiology of LDH, MCT1, and MCT4 in human skeletal muscle. Am J Physiol Endocrinol Metab. 2000;278:E571–E579. doi: 10.1152/ajpendo.2000.278.4.E571. [DOI] [PubMed] [Google Scholar]
- Enoki T, Yoshida Y, Hatta H, Bonen A. Exercise training alleviates MCT1 and 4 reductions in heart and skeletal muscles of STZ-induced diabetic rats. J Appl Physiol. 2003;94:2433–2438. doi: 10.1152/japplphysiol.01155.2002. [DOI] [PubMed] [Google Scholar]
- Garcia CK, Brown MS, Pathak RK, Goldstein JL. cDNA cloning of MCT2, a second monocarboxylate transporter expressed in different cells than MCT1. J Biol Chem. 1995;270:1843–1849. doi: 10.1074/jbc.270.4.1843. [DOI] [PubMed] [Google Scholar]
- Gerhart DZ, Enerson BE, Zhdankina OY, Leino RL, Drewes LR. Expression of the monocarboxylate transporter MCT2 by rat brain glia. Glia. 1998;22:272–281. 10.1002/(SICI)1098-1136(199803)22:3<272::AID-GLIA6>3.3.CO;2-T. [PubMed] [Google Scholar]
- Goodyear LJ, Kahn BB. Exercise, glucose transport, and insulin sensitivity. Ann Rev Med. 1998;49:235–261. doi: 10.1146/annurev.med.49.1.235. 10.1146/annurev.med.49.1.235. [DOI] [PubMed] [Google Scholar]
- Green H, Halestrap A, Mockett C, O'Toole D, Grant S, Ouyang J. Increases in muscle MCT are associated with reductions in muscle lactate after a single exercise session in humans. Am J Physiol Endocrinol Metab. 2002;282:E154–E160. doi: 10.1152/ajpendo.2002.282.1.E154. [DOI] [PubMed] [Google Scholar]
- Grewal SIS, Rice JC. Regulation of heterochromatin by histone methylation and small RNAs. Curr Opin Cell Biol. 2004;16:230–238. doi: 10.1016/j.ceb.2004.04.002. 10.1016/j.ceb.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Halestrap AP, Meredith D. The SLC16 gene family – from monocarboxylate transporters (MCTs) to aromatic amino acid transporters and beyond. Pflugers Arch. 2004;447:619–628. doi: 10.1007/s00424-003-1067-2. 10.1007/s00424-003-1067-2. [DOI] [PubMed] [Google Scholar]
- Hildebrandt AL, Pilegaard H, Neufer PD. Differential transcriptional activation of select metabolic genes in response to variations in exercise intensity and duration. Am J Physiol Endocrinol Metab. 2003;285:E1021–E1027. doi: 10.1152/ajpendo.00234.2003. [DOI] [PubMed] [Google Scholar]
- Huang Y-S, Richter JD. Regulation of local mRNA translation. Curr Opin Cell Biol. 2004;16:308–313. doi: 10.1016/j.ceb.2004.03.002. 10.1016/j.ceb.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Jackson VN, Price NT, Halestrap AP. Cloning of the monocarboxylate transporter isoform MCT2 from rat testis provides evidence that the expression is species specific and may involve post-transcriptional regulation. Biochem J. 1997;324:447–453. doi: 10.1042/bj3240447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juel C. Lactate – proton cotransport in skeletal muscle. Physiol Rev. 1997;77:1–37. doi: 10.1152/physrev.1997.77.2.321. [DOI] [PubMed] [Google Scholar]
- Juel C, Halestrap AP. Lactate transport in skeletal muscle – role and regulation of the monocarboxylate transporter. J Physiol. 1999;517:633–642. doi: 10.1111/j.1469-7793.1999.0633s.x. 10.1111/j.1469-7793.1999.0633s.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux K, Han X-X, Dombrowski L, Bonen A, Marette A. The transferrin receptor defines two distinct contraction-responsive GLUT4 vesicle populations. Diabetes. 2000;49:183–189. doi: 10.2337/diabetes.49.2.183. [DOI] [PubMed] [Google Scholar]
- McCullagh KJA, Juel C, O'Brien M, Bonen A. Chronic muscle stimulation increases lactate transport in rat skeletal muscle. Mol Cell Biochem. 1996a;156:51–57. doi: 10.1007/BF00239319. [DOI] [PubMed] [Google Scholar]
- McCullagh KJA, Poole RC, Halestrap AP, O'Brien M, Bonen A. Role of the lactate transporter (MCT1) in skeletal muscles. Am J Physiol Endocrinol Metab. 1996b;271:E143–E150. doi: 10.1152/ajpendo.1996.271.1.E143. [DOI] [PubMed] [Google Scholar]
- McCullagh KJA, Poole RC, Halestrap AP, O'Brien M, Bonen A. Chronic electrical stimulation increases MCT1 and lactate uptake in red and white skeletal muscle. Am J Physiol Endocrinol Metab. 1997;273:E239–E246. doi: 10.1152/ajpendo.1997.273.2.E239. [DOI] [PubMed] [Google Scholar]
- Manning Fox JE, Meredith D, Halestrap AP. Characterisation of human monocarboxylate transporter 4 substantiates its role in lactic acid efflux from skeletal muscle. J Physiol. 2000;529:285–293. doi: 10.1111/j.1469-7793.2000.00285.x. 10.1111/j.1469-7793.2000.00285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Hannon GJ. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr Opin Cell Biol. 2004;16:223–229. doi: 10.1016/j.ceb.2004.04.003. 10.1016/j.ceb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Pellerin L, Pellegri G, Bittar PG, Charnay Y, Bouras C, Martin JL. Evidence supporting the existence of an activity-dependent astrocyte-neuron lactate shuttle. et al. 1998a;20:291–299. doi: 10.1159/000017324. Dev Neurosci. [DOI] [PubMed] [Google Scholar]
- Pellerin L, Pellegri G, Martin JL, Magistretti PJ. Expression of monocarboxylate transporter mRNAs in mouse brain: support for a distinct role of lactate as an energy substrate for the neonatal vs. adult brain. Proc Natl Acad Sci U S A. 1998b;95:3990–3995. doi: 10.1073/pnas.95.7.3990. 10.1073/pnas.95.7.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philp NJ, Yoon H, Grollman EF. Monocarboxylate transporter MCT1 is located in the apical membrane and MCT3 in the basal membrane of rat RPE. Am J Physiol Regulatory, Integrative Comp Physiol. 1998;274:R1824–R1828. doi: 10.1152/ajpregu.1998.274.6.R1824. [DOI] [PubMed] [Google Scholar]
- Pilegaard H, Domino K, Noland T, Juel C, Hellsten Y, Halestrap AP, et al. Effect of high-intensity exercise training on lactate/H+ transport capacity in human skeletal muscle. Am J Physiol Endocrinol Metab. 1999a;276:E255–E261. doi: 10.1152/ajpendo.1999.276.2.E255. [DOI] [PubMed] [Google Scholar]
- Pilegaard H, Keller C, Steensberg A, Helge JW, Pedersen BK, Saltin B, et al. Influence of pre-exercise muscle glycogen content on exercise-induced transcriptional regulation of metabolic genes. J Physiol. 2002;541:261–271. doi: 10.1113/jphysiol.2002.016832. 10.1113/jphysiol.2002.016832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilegaard H, Ordway G, Saltin B, Neufer PD. Transcripional regulation of gene expression in human skeletal muscle during recoverey from exercise. Am J Physiol Endocrinol Metab. 2000;279:E806–E814. doi: 10.1152/ajpendo.2000.279.4.E806. [DOI] [PubMed] [Google Scholar]
- Pilegaard H, Saltin B, Neufer PD. Exercise induces transient transcriptional activation of the PGC-1a gene in human skeletal muscle. J Physiol. 2003;546:851–858. doi: 10.1113/jphysiol.2002.034850. 10.1113/jphysiol.2002.034850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilegaard H, Terzis G, Halestrap A, Juel C. Distribution of the lactate/H+ transporter isoforms MCT1 and MCT4 in human skeletal muscle. Am J Physiol Endocrinol Metab. 1999b;276:E843–E848. doi: 10.1152/ajpendo.1999.276.5.E843. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. New perspectives on connecting messenger RNA-3′ end formation to transcription. Curr Opin Cell Biol. 2004;16:272–278. doi: 10.1016/j.ceb.2004.03.007. 10.1016/j.ceb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Ren JM, Semenkovich CF, Gulve EA, Gao J, Holloszy JO. Exercise induces rapid increases in Glut4 expression, glucose transport capacity, and insulin-stimulated glycogen storage in muscle. J Biol Chem. 1994;269:14396–14401. [PubMed] [Google Scholar]
- Roth DA, Brooks GA. Lactate and pyruvate transport is dominated by a pH gradient-sensitive carrier in rat skeletal muscle sarcolemmal vesicles. Arch Biochem Biophys. 1990;279:386–394. doi: 10.1016/0003-9861(90)90506-t. 10.1016/0003-9861(90)90506-T. [DOI] [PubMed] [Google Scholar]
- Sepponen K, Koho N, Puolanne E, Ruusunen M, Poso AR. Distribution of monocarboxylate transporter isoforms MCT1, MCT2 and MCT4 in porcine muscles. Acta Physiol Scand. 2003;177:79–86. doi: 10.1046/j.1365-201X.2003.01051.x. 10.1046/j.1365-201X.2003.01051.x. [DOI] [PubMed] [Google Scholar]
- Steinberg GR, Dyck DJ, Calles-Escandon J, Tandon NN, Luiken JJFP, Glatz JF, et al. Chronic leptin administration decreases fatty acid uptake and fatty acid transporters in rat skeletal muscle. J Biol Chem. 2002;277:8854–8860. doi: 10.1074/jbc.M107683200. 10.1074/jbc.M107683200. [DOI] [PubMed] [Google Scholar]
- Tonouchi M, Hatta H, Bonen A. Muscle contraction increases lactate transport while reducing sarcolemmal MCT4, but not MCT1. Am J Physiol Endocrinol Metab. 2002;282:E1062–E1069. doi: 10.1152/ajpendo.00358.2001. [DOI] [PubMed] [Google Scholar]
- Van De Bor M, Davis I. mRNA localisation gets more complex. Curr Opin Cell Biol. 2004;16:300–307. doi: 10.1016/j.ceb.2004.03.008. 10.1016/j.ceb.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Wang Y, Tonouchi M, Miskovic D, Hatta H, Bonen A. T3 increases lactate transport and the expression of MCT4, but not MCT1, in rat skeletal muscle. Am J Physiol Endocrinol Metab. 2003;285:E622–E628. doi: 10.1152/ajpendo.00069.2003. [DOI] [PubMed] [Google Scholar]
- Wilson MC, Jackson VN, Hedle C, Price NT, Pilegaard H, Juel C, et al. Lactic acid efflux from white skeletal muscle is catalyzed by the monocarboxylate transporter MCT3. J Biol Chem. 1998;273:15920–15926. doi: 10.1074/jbc.273.26.15920. 10.1074/jbc.273.26.15920. [DOI] [PubMed] [Google Scholar]
- Zhou M, Lin B-Z, Coughlin S, Vallega G, Pilch PF. UCP-3 expression in skeletal muscle: effects of exercise, hypoxia and AMP-activated protein kinase. Am J Physiol Endocrinol Metab. 2000;279:E622–E629. doi: 10.1152/ajpendo.2000.279.3.E622. [DOI] [PubMed] [Google Scholar]