Abstract
Phosphoinositides (PIs) constitute a minor fraction of total cellular lipids in all eukaryotic cells. They fulfill many important functions through interaction with a wide range of cellular proteins. Members of distinct inositol lipid kinase families catalyze the synthesis of these phospholipids from phosphatidylinositol. The hydrolysis of PIs involves phosphatases and isoforms of PI-specific phospholipase C. Although our knowledge of the roles played by plant PIs is clearly limited at present, there is no doubt that they are involved in many physiological processes during plant growth and development. In this review, we concentrate on inositol lipid-metabolizing enzymes from the model plant Arabidopsis for which biochemical characterization data are available, namely the inositol lipid kinases and PI-specific phospholipase Cs. The biochemical properties and structure of characterized and genome-predicted isoforms are presented and compared with those of the animal enzymes to show that the plant enzymes have some features clearly unique to this kingdom.
Phosphatidylinositol (PtdIns) is a major phospholipid in eukaryotic cells. Three of the five free hydroxyl groups of PtdIns can be phosphorylated in cells in different combinations. In total, seven phosphorylated derivatives of PtdIns have been detected, one of which, PtdIns 3,4,5-trisphosphate [PtdIns(3,4,5)P3], has not been found in plant cells (Fig. 1). These inositol phospholipids are collectively referred to as phosphoinositides (PIs). In animal cells, PIs and their derivatives operate in signal transduction pathways triggered by stimuli as diverse as growth factors, hormones, neurotransmitters, and light (Berridge, 1993).
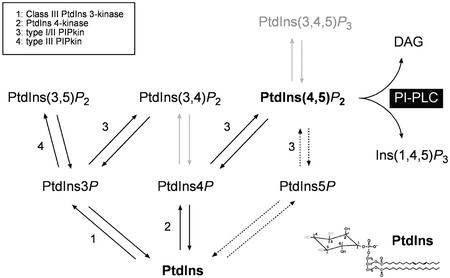
Figure 1.
PI metabolism. The different steps in the synthesis of PIs and the lipid kinases catalyzing the different reactions are indicated. PtdIns(3,4,5)P3 is present in animal cells but has not been detected in plant tissues, so far. In animal cells, PtdIns(3,4)P2 can be generated from PtdIns4P by a PtdIns 3-kinase or by an as-yet-unidentified PIPkin from PtdIns3P. Plant cells do not contain any homolog of the heterodimeric inositol lipid 3-kinases that are able to phosphorylate PtdIns4P to PtdIns(3,4)P2 and PtdIns(4,5)P2 to PtdIns(3,4,5)P3. PtdIns(4,5)P2 can be synthesized by type I and type II PIPkins from PtdIns4P and PtdIns5P, respectively. On the basis of sequence comparison, plants cells do not possess type II PIPkins. PtdIns5P is present in plants, but an enzyme capable of producing it has not been identified.
Historically, the first major insight into the importance of PIs was the discovery that the two PtdIns 4,5-bisphosphate [PtdIns(4,5)P2]-derived second messengers inositol 1,4,5-trisphosphate [Ins(1,4,5)P3] and diacylglycerol promote Ca2+ release from internal stores and activate protein kinase C, respectively (Berridge and Irvine, 1989; for review, see Berridge, 1993). The production of these two second messengers from PtdIns(4,5)P2 is catalyzed by PI-specific phospholipase C (PI-PLC) isoforms. During the last decade, it has become evident that in addition to serving as precursors to Ins(1,4,5)P3 and diacylglycerol, PIs actively participate in several other cellular processes. They have been shown to regulate the dynamics of the actin cytoskeleton through interaction with actin-binding proteins (Lassing and Lundberg, 1985; Janmey and Stossel, 1987; Brill et al., 2000), and to potentiate the activation of protein kinase C (Oh et al., 1998), PI-PLC (Bae et al., 1998; Falasca et al., 1998), and phospholipase D (Pappan et al., 1997). In addition, PIs phosphorylated at the D3-hydroxy group of the inositol head group are required for specific vesicle trafficking steps (De Camilli et al., 1996; Wurmser et al., 1999) and are able to activate the protein kinase Akt/PKB and PI-dependent kinases (Wymann and Pirola, 1998). A 3-phosphorylated PI, PtdIns 3,5-bisphosphate [PtdIns(3,5)P2], was recently identified and shown to accumulate in yeast cells subjected to hyperosmotic or NaCl stress (Dove et al., 1997). In addition, mutations in many of the proteins involved in the PI system cause various diseases or severe defects (Bloomquist et al., 1988; Yamamoto et al., 1995; Carvajal et al., 1996; Maehama et al., 2001).
The PIs clearly constitute a group of lipids with very important, diverse functions. It is therefore critical that the levels of these phospholipids are tightly regulated. The lipid kinases catalyzing the synthesis of the phosphorylated derivatives of PtdIns, and isoforms of PI-PLC have been extensively studied and characterized; some of them have also been crystallized and their structure determined.
In comparison with the wealth of information available for the components, regulation, and function of the animal PI system, and despite the identification of plant homologs of many of these animal components, our knowledge of the plant PI system is, at present, quite limited. However, it has been demonstrated that micro-injected “caged” Ins(1,4,5)P3 can release Ca2+ from internal stores (Alexandre et al., 1990) and is also able to trigger stomatal closure (Blatt et al., 1990; Gilroy et al., 1990). There is also evidence that PIs may participate in the regulation of cytoskeletal structures in plant cells (Yang et al., 1993; Drøbak et al., 1994) and in polar pollen tube growth (Kost et al., 1999). A number of reports also suggest that a wide range of signals, such as light, hormones, and stress, may mediate their effect through PI-dependent processes (Munnik et al., 1998a; Drøbak et al., 1999), although the effects reported were often limited and the identity of the lipid or inositol phosphate species affected not always clearly demonstrated. More recently, hyperosmotic stress was shown to induce an increase in two distinct PtdIns bisphosphate (PtdInsP2) isomers, PtdIns(4,5)P2 in Arabidopsis cells (Pical et al., 1999) and PtdIns(3,5)P2 in Chlamydomonas moewusii and in some higher plant cells (Dove et al., 1997; Meijer et al., 1999). It is also now clear that, as in other organisms, vesicle trafficking in plant cells is dependent on PIs (Matsuoka et al., 1995; Kim et al., 2001). A PtdIns 3-kinase was recently found associated with nuclear transcriptionally active sites, suggesting a potential role for this lipid kinase in the regulation of transcription in the nucleus (Bunney et al., 2000). It is also interesting that chloroplasts contain both PtdIns 3- and 4-kinase activities (Bovet et al., 2001).
Additional evidence that a functional PI system operates in plant cells has been provided by the identification of homologs of the components of the animal PI system, such as PI-metabolizing enzymes (Hirayama et al., 1995; Shi et al., 1995; Mikami et al., 1998; Kopka et al., 1998; Xue et al., 1999, 2000), PI-regulated enzymes (Deak et al., 1999), and PIs themselves (Parmar and Brearley, 1993; Munnik et al., 1994; Brearley and Hanke, 1995; Pical et al., 1999). Of the seven phosphorylated derivatives of PtdIns identified in animal systems, only PtdIns(3,4,5)P3 has never been observed in plant cells (or in yeast). The presence of PtdIns5P in higher plants has only recently been unambiguously demonstrated (Meijer et al., 2001).
Previous reviews on the metabolism and function of PIs in plant cells have provided overviews of some aspects of this field but were published before the availability of a complete plant genome sequence and lack a thorough description of some of the enzymes involved in the metabolism of these lipids (Drøbak, 1992; Munnik et al., 1998a; Drøbak et al., 1999; Stevenson et al., 2000). The aim of the present review is to provide a detailed summary of the information available on the enzymes involved in the synthesis and hydrolysis of PIs, i.e. the PI kinases and PI-PLCs. It includes a description of the structure, and of the molecular and biochemical properties of these enzymes, with new information gleaned from sequence comparison and analysis. When possible, in vivo functions are also discussed.
PtdIns KINASES
As shown in Figure 1, PtdIns can be phosphorylated on three of the five free hydroxyl groups of the inositol ring in reactions catalyzed by PtdIns kinases. The enzymes catalyzing the phosphorylation of the hydroxyl groups at positions 3 and 4 have been identified and characterized. There is an animal enzyme capable of generating PtdIns5P from PtdIns in vitro (Sbrissa et al., 2000); however, whether this enzyme is responsible for the synthesis of PtdIns5P in vivo is not known.
PtdIns 3-Kinase
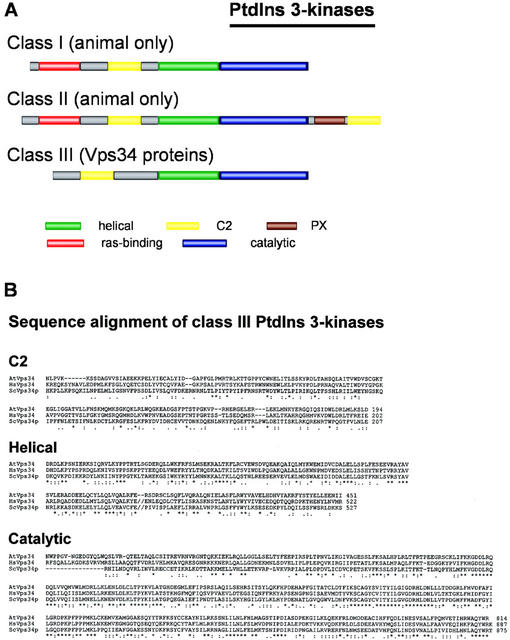
Multiple PtdIns 3-kinase isoforms have been identified. On the basis of their structure and substrate specificity, they can be divided into three classes (Vanhaesebroeck and Waterfield, 1999). All PtdIns 3-kinases share three conserved domains: a catalytic domain preceded by a helical domain and a C2 domain (a Ca2+-dependent or -independent lipid-binding domain originally identified in protein kinase C; Rizo and Südhof, 1998; Fig. 2). Class I PtdIns 3-kinases are heterodimeric enzymes that can phosphorylate PtdIns, PtdIns 4-phosphate (PtdIns4P), and PtdIns(4,5)P2 in vitro, but preferentially phosphorylate PtdIns(4,5)P2 in vivo. These PtdIns 3-kinases are subdivided into class IA and class IB. The catalytic subunit of class IA enzymes associates with one of three different adaptor proteins, whereas the class IB catalytic subunit (only one has been identified) associates with a p101 adaptor protein. Class II PtdIns 3-kinases preferentially use PtdIns and PtdIns4P as substrates. Class I and II PtdIns 3-kinases possess a ras-binding domain at their N terminus, and class II enzymes are characterized by the presence of a Phox homology (PX) domain (Ellson et al., 2002) and a second C2 domain at their carboxy terminus (see Fig. 2). Finally, class III PtdIns 3-kinases include the only PtdIns 3-kinase identified in Brewer's yeast (Saccharomyces cerevisiae), ScVps34p, and all of its orthologs from other organisms. Class III PtdIns 3-kinases phosphorylate only PtdIns and are believed to represent the ancestral form of PtdIns 3-kinase. The Vps34 protein exists as a complex with a Ser/Thr protein kinase both in Brewer's yeast (Stack and Emr, 1994) and animals (Volinia et al., 1995). This protein kinase, encoded by the VPS15 gene in Brewer's yeast, phosphorylates and activates Vps34. A putative ortholog of this protein kinase is also present in Arabidopsis (At4g29380), suggesting that some of the processes regulating protein trafficking may be conserved in animals, yeast, and plants.
Figure 2.
Domain structure representation of PtdIns 3-kinases. A, The domains conserved in the catalytic subunits of the three PtdIns 3-kinase classes are represented by colored boxes. B, Vps34 proteins from Brewer's yeast (Vps34p; accession no. X53531), Arabidopsis (AtVps34; accession no. U10669), and human (HsVps34; accession no. Z46973) were aligned using the ClustalW program from the MacVector package. Identical, conserved, and semiconserved residues are indicated below the alignment by *, :, and ., respectively. // indicates a large gap in the sequence.
The only PtdIns 3-kinase identified so far in Arabidopsis, AtVps34, is a homolog of ScVps34p. The VPS34 gene was identified in a screen for Brewer's yeast mutants altered in the sorting of vacuolar proteins (Herman and Emr, 1990), and was shown to encode a PtdIns kinase that phosphorylates PtdIns exclusively at the D-3 position of the inositol ring (Schu et al., 1993). Although the substrate specificity of AtVps34 has not been fully determined, it is reasonable to assume that like ScVps34p, it can only phosphorylate PtdIns, not PtdIns4P and PtdIns(4,5)P2. In both Brewer's yeast and fission yeast (Schizosaccharomyces pombe), the VPS34 gene is not essential but is necessary for normal growth (Herman and Emr, 1990; Takegawa et al., 1995). Point mutations in the catalytic domain of Vps34p or deletion of the VPS34 gene lead to dramatic decreases in cellular levels of PtdIns 3-phosphate (PtdIns3P) in Brewer's yeast and severe defects in vacuolar protein sorting (Herman and Emr, 1990; Schu et al., 1993; Stack et al., 1995). Down-regulation of the expression of the AtVPS34 gene in transgenic Arabidopsis plants resulted in severe inhibition of growth and development (Welters et al., 1994), indicating that AtVps34 is required for normal plant development. The Arabidopsis AtVPS34 cDNA was unable to complement a VPS34 yeast deletion mutant. However, a chimeric gene consisting of Brewer's yeast VPS34 in which the region coding for the catalytic domain had been replaced by the corresponding region of the Arabidopsis gene could complement the yeast mutant. This also suggests that regions other than the catalytic domain are required for the function of Vps34p in vivo, and that the regulation mechanisms of Vps34 enzymes differ between species. A distinctive feature of Vps34 proteins versus other PtdIns 3-kinases is the presence of a longer insert between the amino-terminal C2 domain and the helical domains, and of an extended insert (approximately 60 amino acid residues) in the helical domain. The latter insert is not present in AtVps34 (Fig. 2B).
The two most commonly used PtdIns 3-kinase inhibitors are wortmannin (a fungal metabolite) and LY294002 (a synthetic molecule; Walker et al., 2000). In vitro, their IC50 values are around 5 nm for wortmannin and 1 μm for LY294002 (Vanhaesebroeck et al., 2001). Surprisingly, ScVps34p is much less sensitive to these compounds (IC50 of 3 μm for wortmannin and 50 μm for LY294002; Stack and Emr, 1994), whereas the human Vps34 ortholog exhibits sensitivities similar to those of the most sensitive PtdIns 3-kinases. In addition, wortmannin inhibits other enzymes including PtdIns 4-kinases (e.g. Meyers and Cantley, 1997; Xue et al., 1999) and PtdIns phosphate (PtdInsP) kinases (PIPkins; Vanhaesebroeck et al., 2001), but with IC50 values 20- to 1,000-fold higher than for PtdIns 3-kinase inhibition. Fifty micromolar LY294002 does not inhibit a range of animal protein kinases (Vlahos et al., 1994). However other enzymes, such as the PtdIns 3-kinase-related protein kinases TOR and DNA-dependent protein kinase, and to a lesser extent casein kinase-2, are sensitive to LY294002 (Vanhaesebroeck et al., 2001). Casein kinase-2 is not inhibited by wortmannin (Davies et al., 2000). One major problem with using wortmannin is that it is rather unstable in solution, and incubation times longer than a couple of hours should be avoided (Vanhaesebroeck and Waterfield, 1999). In summary, in animal systems, wortmannin and LY294002 are, at low concentrations (up to 1 μm for wortmannin and up to 50 μm for LY294002), specific inhibitors of PtdIns 3-kinase; but at higher concentrations, they lose specificity.
Wortmannin has been shown to inhibit PtdIns 3-kinase activity in crude tobacco extracts, with calculated IC50 values of 98 nm in vitro and approximately 1 μm in vivo (Matsuoka et al., 1995). In this plant system, wortmannin also inhibited the sorting of one type of protein to the vacuole, with an IC50 of approximately 7 μm, a value close to the IC50 of 9 μm for the synthesis of all phospholipids examined, except PtdIns3P (Matsuoka et al., 1995). The effects of wortmannin on vacuolar sorting in these studies are difficult to interpret in terms of the involvement of PIs and/or other phospholipids because extremely high concentrations of wortmannin and long incubation times were used, and the specificity of wortmannin has not been tested with plant enzymes. Before using wortmannin and LY294002 to assess the possible involvement of PtdIns3P in physiological processes in plant systems, it must first be established that they specifically inhibit PtdIns 3-kinase and do not significantly affect other enzymes in the same concentration range.
Wortmannin was demonstrated to covalently bind to a Lys residue in the ATP-binding site of a class I PtdIns 3-kinase, inducing a conformational change in the catalytic domain of the enzyme (Walker et al., 2000). This Lys residue is conserved in all PtdIns 3-kinases. The difference in sensitivity to wortmannin of the different PtdIns 3-kinases is believed to be attributable to differences in amino acid residues surrounding the conserved Lys residue in the ATP-binding site of these isoforms. However, it remains unknown which residues determine wortmannin sensitivity.
As the sole PtdIns 3-kinase present in Arabidopsis, AtVps34 is probably responsible for the synthesis of the majority of the PtdIns3P found in plants. Class I and II PtdIns 3-kinases are activated by G-protein-coupled receptors and Tyr kinase receptors, which are represented by a large number of distinct members in animal cells. Plants, on the other hand, possess few putative G-protein-coupled receptors and no receptor Tyr kinases. It is therefore not surprising that class I and II PtdIns 3-kinases are absent from plants. Bunney et al. (2000) recently showed that the homolog of AtVps34 is associated with active transcription sites in soybean (Glycine max) cell nucleoli. As in yeast, the product of Vps34 activity, PtdIns3P, participates in the trafficking of proteins to the plant vacuole (Kim et al., 2001). In that study, Kim et al. also demonstrated that PtdIns3P is present in several compartments, including the Golgi apparatus and vacuole networks. PtdIns3P-dependent trafficking involves proteins with a FYVE domain (named after four proteins that it has been found in: Fab1, YOTB/ZK632.12, Vac1, and EEA1; Stenmark et al., 1996), a domain that confers highly specific binding to PtdIns3P (Stenmark and Aasland, 1999). In Arabidopsis, more than 10 proteins contain a FYVE domain (see the SMART database: http://smart.embl-heidelberg.de/browse.shtml), only one of which has been characterized (Jensen et al., 2001). The Brewer's yeast genome encodes fewer than 10 FYVE domain-containing proteins; the human genome encodes more than 50. Two of the plant FYVE proteins probably represent putative homologs of the PtdIns3P 5-kinase protein Fab1p from Brewer's yeast. Several PX domains, including all of the PX domains present in Brewer's yeast, have been shown to bind PtdIns3P (Ellson et al., 2002). Arabidopsis contains up to nine putative proteins with a PX domain. Fifteen have been identified in Brewer's yeast, whereas humans have more than 50 (SMART database). This indicates that the functions of PtdIns 3-kinases are much more diverse in animals than in plants and yeast.
PtdIns 4-Kinases
Biochemical Characterization
PtdIns 4-kinases catalyze the phosphorylation of PtdIns to PtdIns4P [a lipid believed to be the major precursor of PtdIns(4,5)P2], and therefore represents a potentially crucial point in the regulation of the PI-dependent pathways. Two major types of PtdIns 4-kinase, II and III, differing in size and sensitivity to detergents and adenosine, have been identified in a wide range of tissues and cellular compartments in animals (Endemann et al., 1987; Pike, 1992). Type II PtdIns 4-kinase is a membrane-bound, 55-kD enzyme that is readily renaturable after SDS-PAGE. It has comparatively low Km values for PtdIns and ATP, is strongly inhibited by adenosine and Ca2+ (Carpenter and Cantley, 1990), and is insensitive to wortmannin (Endemann et al., 1991). Two forms of type III PtdIns 4-kinase have been detected in membrane and soluble fractions of animal tissues. One is 110 kD in size, and the other is 200 to 230 kD. Type III enzymes have 3- to 7-fold higher Km values for PtdIns and ATP than the type II enzyme and are not inhibited by adenosine and Ca2+. In addition, type III enzymes are inhibited by wortmannin (at concentrations significantly higher than those inhibiting PtdIns 3-kinases), whereas type II enzymes are insensitive (Balla, 1998).
PtdIns 4-kinase activity is present in many different compartments in plant cells, and has been partially purified from the soluble fraction of the unicellular alga Dunaliella parva (Steinert et al., 1994) and from carrot (Daucus carota) cells (Okpodu et al., 1995). It has also been found in plasma membranes from Catharanthus roseus (Hanenberg et al., 1995) and spinach (Spinacia oleracea; Westergren et al., 1999). The size of the partially purified plant PtdIns 4-kinases was estimated to be 80 kD for the soluble activity from carrot cells, 500 kD for the activities purified from D. parva and C. roseus, and 65 and 125 kD for the two distinct activities purified from spinach plasma membranes. A soluble 49-kD protein, PIK-A49, was able to stimulate a PtdIns 4-kinase activity solubilized from carrot plasma membranes (Yang et al., 1993). PIK-A49 was shown to bind and bundle actin and was identified as an elongation factor 1 (Yang et al., 1993). Stimulation of PtdIns 4-kinase was dependent on the phosphorylation status of PIK-A49 (Yang and Boss, 1994). Using antibodies raised against a recombinant PtdIns 4-kinase, Stevenson et al. (1998) were able to immunoaffinity purify an active PtdIns 4-kinase from Arabidopsis microsomes. Wortmannin was recently shown to inhibit the two PtdIns 4-kinase activities partially purified from spinach plasma membranes (Westergren et al., 1999).
Genes, Structure, and Function
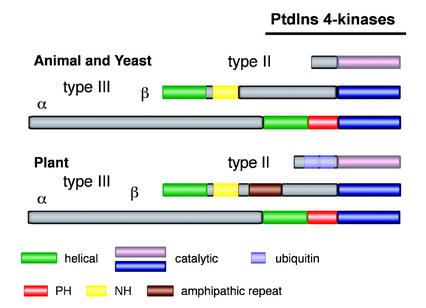
cDNAs encoding functional PtdIns 4-kinases have been isolated from animals, Brewer's yeast, and Arabidopsis, using either peptide sequences from purified proteins or homologous cloning using previously available sequences. Genes encoding type III enzymes have been grouped into two distinct subfamilies, α and β, based on sequence and structure similarities (Balla, 1998). Subfamily β is represented by proteins of 68 to 122 kD, and subfamily α by proteins of 200 to 230 kD (with the exception of human PI4Kα, which is a 97-kD protein [Wong and Cantley, 1994]). Several domains are conserved in these proteins, including a catalytic domain of about 230 amino acid residues within the C-terminal part of these proteins, and a helical domain (previously referred to as PIKa or LKU domain), the location of which varies among the different isoforms (Fig. 3). Members of the α subfamily all contain a pleckstrin homology (PH) domain that separates the helical and catalytic domains in these proteins. Another conserved domain, novel homology (NH) is conserved in all of the isoforms belonging to the β subfamily (Fig. 3; Xue et al., 1999). NH domains were first identified in yeast Pik1p and in a soluble PtdIns 4-kinase from rat (Nakagawa et al., 1996).
Figure 3.
Domain structure representation of animal, yeast, and plant PtdIns 4-kinases. The various conserved domains are represented by colored boxes. The catalytic domains of type II and III PtdIns 4-kinase share no homology and are therefore shown in different colors. Some plant type II PtdIns 4-kinases contain one or two ubiquitin-like domains.
Although PtdIns 4-kinase activity was first detected, characterized, and partially purified from higher plants many years ago, the isolation and functional expression of a full-length plant PtdIns 4-kinase cDNA was first reported in 1999 (Xue et al., 1999). The corresponding 126-kD protein from Arabidopsis, AtPI4Kβ1 (previously AtPI4Kβ), is similar in size and overall primary structure to the PtdIns 4-kinase β isoforms from yeast and animals. Another cDNA from Arabidopsis, AtPI4Kα1 (previously AtPI4Kα), encoding a PtdIns 4-kinase has been described (Stevenson et al., 1998). The deduced amino acid sequence corresponding to this partial cDNA comprised a helical, a PH, and a catalytic domain (Stevenson et al., 1998). The gene corresponding to this partial cDNA has now been identified and deposited in the GenBank/EMBL databases (Table I). The predicted protein is 227 kD in size and structurally similar to the α-isoforms from animal and yeast (Fig. 3). No peptide sequences for any of the partially purified plant PtdIns 4-kinases are available, making it impossible to verify whether any of them correspond to one of the Arabidopsis PtdIns 4-kinase genes. The subcellular localization of these two Arabidopsis type III PtdIns 4-kinases is at present not known, but AtPI4Kα1 was shown to be present in a microsomal fraction and an actin-enriched fraction (Stevenson et al., 1998).
Table I.
List of characterized and predicted PtdIns 4-kinases from Arabidopsis
| Type | Name | Molecular Mass | Amino Acid Residues | Accession No./Locus | mRNA/ESTa | E Valueb |
|---|---|---|---|---|---|---|
| kD | ||||||
| II | AtPI4Kγ1 | 61 | 561 | AC002409/At2g40850 | No | 5xe−8 (mouse) |
| AtPI4Kγ2 | 56 | 505 | AC009519/At1g64460 + At1g64470 | W43386 | 4xe−6 (mouse) | |
| AtPI4Kγ3 | 64 | 574 | AB006701/At5g24240 | AQ965429 | 3xe−11 (human) | |
| AtPI4Kγ4 | 63 | 566 | AC006418/At2g46500 | AF419566 | 1xe−9 (mouse) | |
| AtPI4Kγ5 | 70 | 630 | AC079829/At1g26270 | AY035014 | 6xe−16 (human) | |
| AtPI4Kγ6 | 69 | 620 | AC027656/At1g13640 | AY060574 | 3xe−15 (human) | |
| AtPI4Kγ7 | 71 | 638 | AC007196/At2g03890 | AY056101 | 6xe−11 (mouse) | |
| AtPI4Kγ8 | 58 | 533 | AL163972/At3g56600 | No | 3xe−7 (rat) | |
| III | AtPI4Kα1 | 227 | 2051 | AC007504/At1g49340 | AF035936 | |
| AtPI4Kα2 | 59 | 526 | AC079828/At1g51040 | No | ||
| AtPI4Kβ1 | 126 | 1121 | AB008266/At5g64070 | AJ002685 | ||
| AtPI4Kβ2 | 127 | 1120 | AL391712/At5g09350 | No |
mRNA or EST accession numbers are provided, when available.
For each putative type II PtdIns 4-kinase, the E value for the closest animal type II PtdIns 4-kinase determined from a BlastP search is given.
AtPI4Kβ1 possesses the two conserved domains present in all type III PtdIns 4-kinases, namely the helical and catalytic domains, but lacks a PH domain, features typical of PtdIns 4-kinase α isoforms. A conserved domain previously only identified in the yeast PIK1 protein and a rat soluble PtdIns 4-kinase (Nakagawa et al., 1996) is also conserved in AtPI4Kβ1 (domain NH in Fig. 3). Surprisingly, AtPI4Kβ1 is the only known PtdIns 4-kinase to possess a unique repetitive motif constituted of 11 repeats of a charged core unit (Xue et al., 1999). No function has yet been assigned to the conserved helical, NH, and repetitive motif domains. Because PtdIns 4-kinase activities have been detected in most cellular compartments, it is possible that the domains identified in PtdIns 4-kinases are involved in the targeting of these enzymes to various cellular compartments through interactions with other proteins or lipids.
The PH domain of the carrot homolog of AtPI4Kα1 was shown, using a lipid-protein overlay, to bind PtdIns4P but not PtdIns in vitro (Stevenson et al., 1998). It, thus, appears likely that the PH domain of type α PtdIns 4-kinases is not responsible for binding the lipid substrate. It is consequently possible that substrate binding in PtdIns 4-kinases is controlled by the helical domain, or a domain conserved structurally but not at the sequence level.
As recently observed with other PtdIns 4-kinases, AtPI4Kβ1 is inhibited by wortmannin at concentrations similar to those affecting the yeast and mammalian enzymes (Balla et al., 1997; Meyers and Cantley, 1997; Xue et al., 1999)—concentrations that are significantly higher than those required to inhibit PtdIns 3-kinases. The residue corresponding to the Lys residue of PI3Kα at which wortmannin binds covalently is conserved in AtPI4Kβ1 as it is in all other type III PtdIns 4-kinases, including AtPI4Kα1.
Two PtdIns 4-kinase genes, PIK1 and STT4, are present in the genome of Brewer's yeast, both of which are essential for yeast viability (Audhya et al., 2000). The PtdIns 4-kinases corresponding to these two genes are responsible for the synthesis of distinct pools of PtdIns4P (Audhya et al., 2000). Using temperature-conditional mutants, the authors demonstrated that Pik1p is essential for normal secretion, Golgi and vacuole membrane dynamics, and endocytosis, and that Stt4p is required for maintenance of vacuole morphology, cell wall integrity, and actin cytoskeleton organization.
Intriguingly, Arabidopsis is, so far, the only organism found to possess two genes encoding type IIIβ PtdIns 4-kinases (Table I). The two corresponding protein sequences share 80% identity, and AtPI4Kβ2 also possesses the NH domain and the repetitive motif. The genome of Arabidopsis contains large segments of duplicated DNA that can be grouped into 103 blocks (Vision et al., 2000). The two AtPI4Kβ genes do not belong to any of the duplicated blocks present in the genome, but they may have occurred from a duplication event not identified by Vision et al. (2000). Arabidopsis also contains a gene encoding a putative protein, AtPI4Kα2, with significant similarity to AtPI4Kα1 (Table I). However, this putative protein is much smaller than AtPI4Kα1 and consists of a catalytic domain and a region corresponding to the 114 first amino acid residues of AtPI4Kα1. AtPI4Kα2 does not contain any other domain, i.e. the PH and helical domains are lacking. No expressed sequence tag (EST) corresponding to this gene is present in the databases, suggesting that it may not be expressed.
The conservation of the structure of PtdIns 4-kinases in the different phyla contrasts markedly with the current data available for PI-PLC. On the basis of protein structure and biochemical properties, all plant PI-PLC isoforms appear to belong to a single family closely related to the δ-type of mammalian PI-PLCs, but lacking a PH domain.
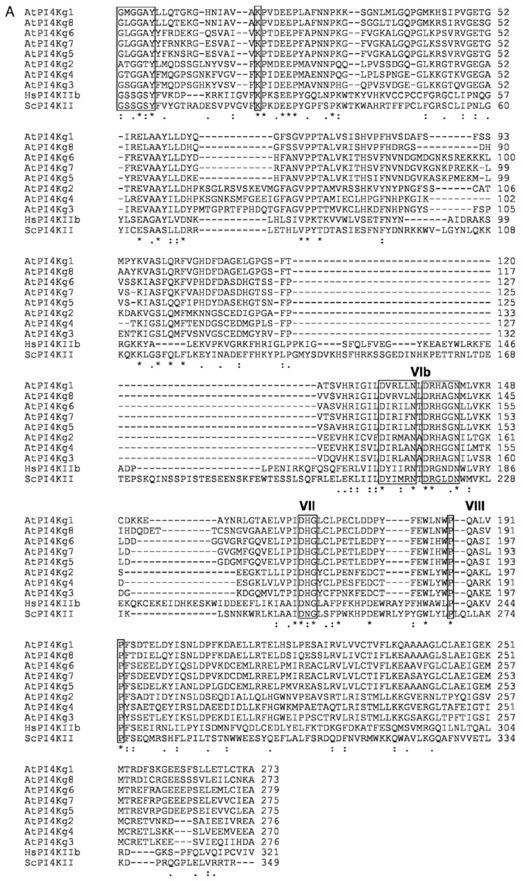
The identification of genes encoding type II PtdIns 4-kinases was achieved only recently (Barylko et al., 2001; Minogue et al., 2001). The corresponding proteins from rat and human form a novel class of PtdIns 4-kinases that is unrelated to previously identified inositol lipid kinases (Fig. 3). Potential homologs of these animal type II PtdIns 4-kinases are present in yeast, fruitfly (Drosophila melanogaster), and Arabidopsis. Surprisingly perhaps, there is only one gene in Brewer's yeast, but search of databases with the entire sequence of human PI4K IIα (Minogue et al., 2001) identified eight putative genes (AtPI4Kγ1–8) in Arabidopsis (Table I). The predicted Arabidopsis proteins show low overall identity with human PI4K IIα, but such is also the case for the putative type II PtdIns 4-kinase from Brewer's yeast (28% identical and 10% similar residues). However, Minogue et al. (2001) identified several candidate kinase motifs in type II PtdIns 4-kinases in more than 10 different putative type II PtdIns 4-kinases. Alignment of the Arabidopsis protein sequences with human PI4K IIα and the sequence from Brewer's yeast clearly shows that the kinase motifs identified in the animal proteins, which correspond to residues found in subdomains I, II, VIb, VII of protein kinases (Hanks and Quinn, 1991), are also present in the Arabidopsis sequences (Fig. 4A). Although, as noted by Minogue et al. (2001), there is no close similarity between type II PtdIns 4-kinases and other PtdIns kinases, a number of residues conserved in the candidate catalytic domain of type II PtdIns 4-kinases are also conserved in other PtdIns kinases and/or PIPkins. The GXXG motif of the candidate subdomain I is found in PIPkins (Rao et al., 1998), including all Arabidopsis PIPkins, except AtPIPK11 (see Fig. 7). The FK motif of the candidate subdomain II, the DRH motif of the candidate subdomain VIb of the Arabidopsis sequences, and the DXG motif of the candidate subdomain VII are all present in PtdIns 3-kinases and PtdIns 4-kinases (Rao et al., 1998). These observations strongly suggest that the Arabidopsis putative type II PtdIns 4-kinases may well be functional homologs of the animal enzymes. However, none of the Arabidopsis genes have yet been shown to encode functional PtdIns 4-kinases. Most of the Arabidopsis putative type II PtdIns 4-kinases are significantly larger than the animal enzymes: Six of them are 61 kD or greater in size (Table I), instead of 55 kD for the animal enzymes. This is because of the presence of an N-terminal extension in the plant enzymes. Sequence analysis reveals that six of the putative plant type II PtdIns 4-kinases contain one or two ubiquitin domains (Fig. 4B). No ubiquitin domain is present in non-plant type II PtdIns 4-kinases.
Figure 4.
Comparison of putative Arabidopsis type II PtdIns 4-kinases with the catalytic domain of type II PtdIns 4-kinases from human and Brewer's yeast (A) and with ubiquitin (B). Sequences were aligned with the ClustalW program and adjusted by hand. Identical, conserved, and semiconserved residues are indicated below the alignment by *, :, and ., respectively. A, Alignment of predicted catalytic domains. The location of candidate kinase subdomains I, II, VIb, and VII, identified by Minogue et al. (2001), are indicated in roman numerals above the alignment. A candidate subdomain VIII is also indicated. The kinase motifs in subdomains I, II, VIb, and VII identified by Minogue et al. (2001) are boxed. The two P residues in the PXXXXP motif in the candidate subdomain VIII are also boxed. B, Alignment of the N-terminal domains of six of the Arabidopsis putative type II PtdIns 4-kinases with ubiquitin 7 from Arabidopsis (AtUBQ7; accession no. NM_129118). AtUBQ7, AtPI4Kγ2, AtPI4Kγ3, and AtPI4Kγ4 contain two ubiquitin domains, whereas AtPI4Kγ5, AtPI4Kγ6, and AtPI4Kγ7 contain only one. Amino acid residues identical or conserved at identical positions in the two ubiquitin domains of AtUBQ7 and the ubiquitin-like domains of Arabidopsis type II PtdIns 4-kinases are boxed.
Figure 7.
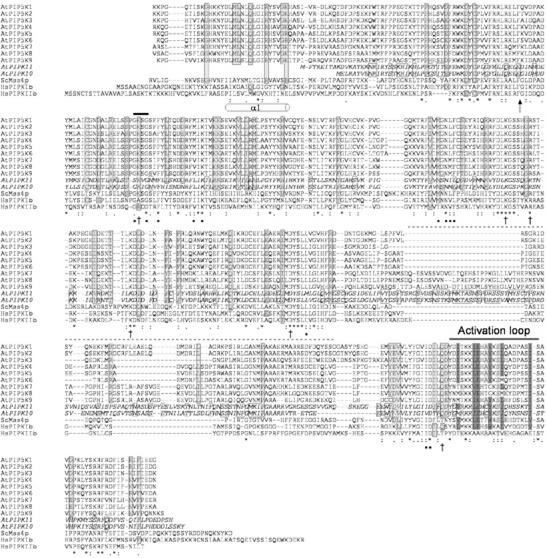
Comparison of the catalytic domain of type I and II PIPkins from animal, yeast, and Arabidopsis. The PIPkin catalytic domain of the nine Arabidopsis PIPkins, Brewer's yeast Mss4p and human PIPKIβ and PIPKIIβ were aligned using the ClustalW program. Arabidopsis subfamily A sequences are in italics. Residues identical in all sequences are indicated by *. The residues conserved or semiconserved are marked with : and ., respectively. Residues identical or conserved in Arabidopsis sequences only are boxed and have gray backgrounds. Residues identical in AtPIPK10 and AtPIPK11 only, and not conserved in other proteins, are boxed. The activation loop is indicated. Residues in HsPIPKIIb proposed to interact with ATP and PtdIns5P are indicated with ● and †, respectively (Rao et al., 1998). Residues in the activation loop that are conserved among type I or II PIPkins are shaded in dark and light gray, respectively, only when they are also conserved in Arabidopsis sequences. The position of the first α-helix of the catalytic domain of human PIPKIIβ (Rao et al., 1998) is shown under the sequence alignment, and the location of the end of the N-terminal dimerization domain is indicated by an arrow. The inserts in the catalytic domains are marked by a dotted line on top of the alignment. The GXXG motif conserved in the catalytic domain of protein kinases and type II PtdIns 4-kinases is indicated by a black bar over the alignment.
PtdInsP KINASES
The synthesis of the three PtdInsP2 isomers from PtdInsP isomers is catalyzed by PIPkins (see Fig. 1). These enzymes form a family of lipid kinases distinct from PtdIns 3-kinases and PtdIns 4-kinases, and are classified into three subfamilies (type I–III; Table II), depending on their substrate specificity (Hinchliffe et al., 1998). Type I enzymes are PtdIns4P 5-kinase, and type II enzymes are PtdIns5P 4-kinase (Hinchliffe et al., 1998). In vitro, type I PIPkins can make PtdIns5P from PtdIns, and PtdIns(3,5)P2 from PtdIns3P (Tolias et al., 1998), and type II PIPkins can synthesize PtdIns 3,4-bisphosphate [PtdIns(3,4)P2] from PtdIns3P (Tolias et al., 1998). In yeast, Mss4p is the only type I PIPkin identified and no type II members appear to be present. Last, type III enzymes are PtdIns3P 5-kinases and are represented by the Brewer's yeast FAB1 gene product and its homologs from other species (McEwen et al., 1999). In vitro, type III PIPkins can make PtdIns5P from PtdIns (Sbrissa et al., 2000).
Table II.
List of characterized and predicted PIPkins from Arabidopsis
| Type | Subfamily | Name | Molecular Mass | Amino Acid Residues | Accession No./Locus | mRNA/ESTa |
|---|---|---|---|---|---|---|
| kD | ||||||
| I/II | A | AtPIPK10 | 46 | 401 | AF007269/At1g01460 | No |
| AtPIPK11 | 48 | 421 | Y12776/At4g01190 | No | |
||
| I/II | B | AtPIP5K1 | 86 | 752 | AC013482/At1g21980 | AF19380 |
| AtPIP5K2 | 86 | 754 | AC012193/At1g77740 | Z34594 | ||
| AtPIP5K3 | 80 | 705 | AC002505/At2g26420 |
AV553828 | |
||
| AtPIP5K4 | 89 | 779 | AL138655/At3g56960 | No | ||
| AtPIP5K5 | 88 | 772 | AC005662/At2g41210 | AF260903 | ||
| AtPIP5K6 | 81 | 715 | AC013483/At3g07960 | No | |
||
| AtPIP5K7 | 98 | 859 | U95973/At1g10900 | AJ009782 | ||
| AtPIP5K8 | 88 | 769 | AC018908/At1g60890 | AY074507 | ||
| AtPIP5K9 | 92 | 815 | AC015985/At3g09920 | AI997830 | ||
| III | AtFab1a | 196 | 1757 | AL161583/At4g33240 | AV529335 | |
| AtFab1b | 201 | 1791 | AB022220/At3g14270 | AV554884 | ||
| AtFab1c | 181 | 1609 | NM_105770/At1g71010 | AV554569 | ||
| AtFab1d | 165 | 1456 | NM_103148/At1g34260 | AV548076 |
The vertical lines on the right-hand side indicate genes found in duplicated regions of the genome. Two mRNA sequences representing two alternative splice variants of the gene encoded by the At1g10900 locus (AtPIP5K7) have been reported. The accession number for the second mRNA, which encodes an 86-kD protein, is AY062718 (see also Figure 5).
mRNA, or EST, accession numbers are provided, when available.
All three PtdInsP2 isomers are present in plant cells, but their relative abundance varies depending on cell type and/or growth conditions (Parmar and Brearley, 1993; Meijer et al., 1999; Pical et al., 1999), suggesting that the lipid kinases involved in the synthesis of these lipids and/or the transcription of genes encoding these enzymes are tightly regulated. A plant enzyme capable of phosphorylating PtdIns4P to a PtdInsP2 isomer has long been known to be present in the plasma membrane (Sommarin and Sandelius, 1988), and both PtdIns(3,4)P2 and PtdIns(3,5)P2 can be generated from endogenous PIs when plant plasma membranes are incubated with ATP (S.K. Dove and B.K. Drøbak, personal communication). In animal cells, PtdIns(3,4)P2 synthesis can proceed via two distinct pathways, involving either 3-phosphorylation of PtdIns4P by heterodimeric inositol lipid kinases or a new PIPkin capable of phosphorylating PtdIns3P on the 4-OH group (Banfic et al., 1998). As mentioned previously, the only PtdIns 3-kinase present in plants is a homolog of the yeast Vps34p protein, which only phosphorylates PtdIns. This suggests that a distinct enzyme is responsible for the synthesis of PtdIns(3,4)P2 in plant cells. The first gene encoding a functional plant PIPkin was recently identified (Mikami et al., 1998) and characterized in more detail (Elge et al., 2001; Westergren et al., 2001). When expressed as a recombinant protein in Escherichia coli, AtPIP5K1 was able to phosphorylate both PtdIns3P and PtdIns4P to generate PtdIns(3,4)P2 and PtdIns(4,5)P2, respectively (Westergren et al., 2001). When expressed in insect cells, the enzyme was shown to preferentially stimulate the synthesis of PtdIns(4,5)P2 and, surprisingly, PtdIns(3,4,5)P3 but not PtdIns(3,4)P2 via 5-phosphorylation of endogenous precursors (Elge et al., 2001).
It is difficult to explain the discrepancy in the lipids generated by the same enzyme in the two heterologous systems. Regulation of AtPIP5K1 may include phosphorylation of one or several residues, because its activity was inhibited upon phosphorylation by cAMP-dependent protein kinase and because a soluble protein kinase activity from Arabidopsis plants was able to phosphorylate recombinant AtPIP5K1 (Westergren et al., 2001). Which PIs are generated by AtPIP5K1 in vivo is at present not known, but the enzyme must be responsible for the production of part of the PtdIns(4,5)P2 pool in cells where it is present. Furthermore, AtPIP5K1 is believed to play an important function in the response of plants to drought and salt stress because its expression is low in non-stressed cells and induced upon stress (Mikami et al., 1998). A confirmation of the probable implication of a PtdIns4P 5-kinase in the response of plant cells to osmotic and salt stress was provided by the clear demonstration that the level of PtdIns(4,5)P2 increases dramatically within a few minutes of subjecting Arabidopsis cells to such stress (Pical et al., 1999; DeWald et al., 2001).
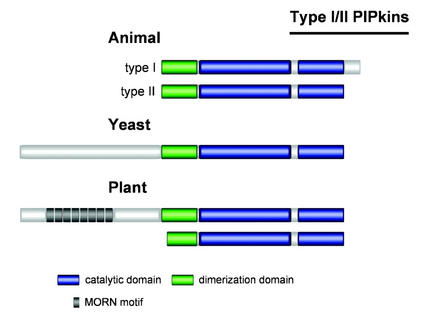
Type I/II AtPIPkins: Genes, Structure, and Function
The Arabidopsis genome contains a number of genes encoding proteins with significant similarity to AtPIP5K1, all of which have a PIPkin catalytic domain (Table II; Fig. 5). These additional putative AtPIPkins have not yet been characterized. Fifteen distinct Arabidopsis genes encode proteins with a PIPkin catalytic domain. Four of these genes encode putative homologs of the Fab1p protein from Brewer's yeast and have therefore been classified as type III PIPkins. The remaining 11 genes code for proteins that, based on sequence comparison alone, cannot be assigned to either type I or type II PIPkins (see below). They can, however, be classified into two distinct subfamilies, designated A and B in Table II based on differences in overall structure (see Fig. 5). Subfamily A is represented by two members, whereas subfamily B is represented by nine members including AtPIP5K1 (Table II).
Figure 5.
Domain structure representation of animal, yeast, and plant type I/II PIPkins. The conserved domains are indicated by boxes of different colors.
Two domains, the dimerization and catalytic domains, are conserved in all type I and II PIPkins (Figs. 5 and 6; Rao et al., 1998). The PIPkin catalytic domain is highly conserved in the 11 type I/II AtPIPkins, and like in all other type I and type II PIPkins, this domain contains a highly variable insert. This insert is significantly longer in AtPIPkins than in non-plant enzymes (Fig. 7). The function of this insert in PIPkins is not understood. In addition, a significant number of residues are specifically conserved in the catalytic domains of the Arabidopsis proteins (Fig. 7). Most of the residues in HsPIPKIIβ proposed to participate in binding ATP and the lipid substrate are conserved in AtPIPkins (Fig. 7).
Figure 6.
Alignment of the N-terminal MORN domain of type I/II PIPkins from Arabidopsis. The alignment was generated with the ClustalW program and adjusted by hand. Identical, conserved, and semiconserved residues are indicated below the alignment by *, :, and ., respectively. The eight MORN motifs (consensus sequence: Y-Q/E-G-E/Q-T-X-N-G-K-X-H-G-Y-G) are indicated by black lines on top of the alignment. Residues within the MORN consensus sequence that are conserved in only a few of the sequences are boxed.
Subfamily A AtPIPkins are composed of the PIPkin catalytic domain and of a shorter dimerization domain in which the first α-helix appears to be missing, as compared with other type I and II PIPkins (Figs. 5 and 7). In addition, a number of residues are identically conserved in the catalytic domain of the two class A AtPIPkins but not in any other type I or type II PIPkins (Fig. 7). Whether these two putative AtPIPkins are expressed is at present not known, because no mRNA or EST sequences have been reported for any of them.
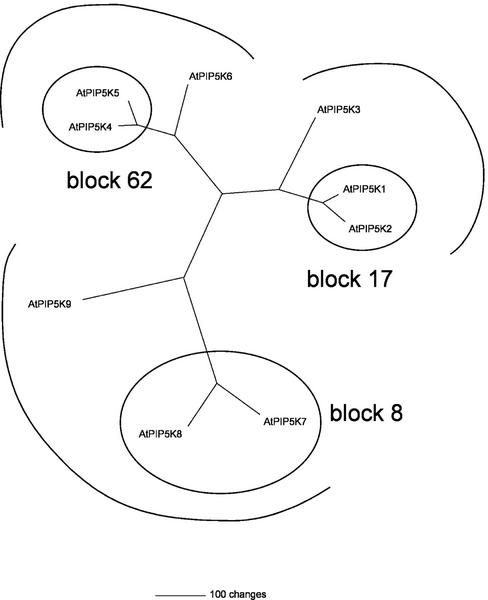
Subfamily B AtPIPkins are composed of the conserved PIPkin catalytic domain and a dimerization domain. In addition, they all have a third conserved domain at their N-terminal end (Figs. 5 and 6). A comparison of the nine subfamily B AtPIPkins shows that six of them form three pairs of two closely related proteins (Figs. 7 and 8). An identical tree is obtained if the catalytic domains only are compared. For each of the three AtPIPkin pairs, the two corresponding genes appear to have occurred from a duplication event because each of them is found in one of the two regions forming one duplicated block of the Arabidopsis genome, whereas the other gene belongs to the other region of the same duplicated block. Two of the PIPkin pairs belong to two duplicated blocks (8 and 62, respectively) that have been assigned to the same age class, i.e. the two blocks were duplicated at the same time, approximately 50 million years ago (Vision et al., 2000). The third pair of duplicated AtPIPkin genes belongs to an older age class, which occurred approximately 100 million years ago (block 17). Surprisingly, the two AtPIPkin genes belonging to block 8 show a significantly lower degree of identity (77%) than the two AtPIPkin genes belonging to block 62 (87%). This suggests that distinct physiological functions are fulfilled by the two pairs of AtPIPkins. It is also interesting to note that AtPIP5K6 and AtPIP5K3 are very closely related to the two pairs from block 62 and block 17, respectively (Fig. 8). Subfamily B AtPIPkins are characterized by the presence of a conserved domain at their N terminus, preceding the dimerization domain (Figs. 5 and 6). This domain is found in no other PIPkin from any organism and consists of eight repeats of a conserved 23-residue long motif. The presence of a repeated motif was noticed in the first PIPkin sequence (AtPIP5K1) obtained from Arabidopsis (Mikami et al., 1998), but at that time, no other protein with this domain had been identified. The repeated motif was subsequently identified as the membrane occupation and recognition nexus (MORN) motif by the SMART sequence analysis program (http://smart.embl-heidelberg.de/).
Figure 8.
A phylogenic tree of subfamily B AtPIPkins. Full-length protein sequences were aligned and the tree was constructed using the PAUP software (Phylogenetic Analysis Using Parsimony, version 4.0b4, Sinauer Associates, Sunderland, MA) from 12,000 replicates. An identical tree is obtained if the sequences of the catalytic domains alone are compared. The three pairs of duplicated genes are circled, and the duplicated blocks they belong to are indicated (Vision et al., 2000).
MORN repeats are present in a relatively small number of proteins from various organisms, fungi being one exception lacking proteins with such a motif. The best-characterized non-PIPkin proteins with such repeats are proteins called junctophilins, which have so far only been described in animal cells. These proteins are components of the junctional complexes present between the plasma membrane and the endoplasmic reticulum (Takeshima et al., 2000). In junctophilins, the eight MORN motifs are arranged in two groups of six and two motifs each (Nishi et al., 2000). Using green fluorescent protein fusion proteins, it was demonstrated that the MORN motifs are necessary for junctophilin-1 binding to the plasma membrane (Takeshima et al., 2000). The target of the MORN domain in the plasma membrane is thought to be phospholipids rather than proteins (Takeshima et al., 2000). Whether the MORN domain of AtPIPkins plays a role in the subcellular localization of these proteins is not known, but this is a plausible hypothesis. Membrane-binding in human PIPKIIβ is believed to be provided by a region of the catalytic domain (Rao et al., 1998). HsPIPKIIβ contains a protein kinase ATP-binding core that is believed to be present in all PI kinases (Rao et al., 1998). One main difference between the structures of the catalytic domain of HsPIPKIIβ and PKA is the absence in the former of structures that might hinder their association with the membrane (Rao et al., 1998). It is possible that, in AtPIPkins and PIPkins from plants in general, the MORN domain is involved in membrane binding, not the catalytic domain, and that animal type I and II PIPkins have evolved from an ancestral enzymes including a MORN domain.
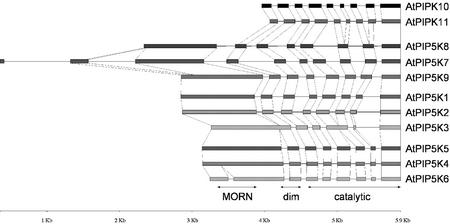
A comparison of the structure of the type I/II AtPIPkin genes is presented in Figure 9. The two class A AtPIPkin genes have identical structures, consisting of nine exons and eight introns. The structure of class B AtPIPkin genes is conserved within the three groups defined in Figure 8. In particular, the three genes encoding AtPIP5K4-6 are almost identical in their structure except for one additional intron present in AtPIP5K6. The group consisting of AtPIP5K1-3 also features two genes with identical structures. The third gene in this group lacks two of the introns, and one of its introns is shorter than those of the two other genes. The structure of the three genes composing the third group of AtPIPkins (AtPIP5K7-9) is identical except that the first exon of AtPIP5K8 and AtPIP5K9 is interrupted by an intron in AtPIP5K7, and that AtPIP5K7 contains an additional exon at its 5′ end. This exon is apparently absent from all the other type I/II AtPIPkin genes. AtPIP5K1 expression using promoter-GUS fusion and single-cell RT-PCR is strongest in the cells of the procambial tissues of leaves, flowers, and roots (Elge et al., 2001). This suggests that AtPIP5K1 may play a role in cell proliferation.
Figure 9.
Schematic representation of the genes encoding characterized and putative type I/II PIPkins from Arabidopsis. The introns and exons for AtPIP5K1, AtPIP5K5, AtPIP5K7, and AtPIP5K8 were determined by comparing the mRNA and gene sequences, whereas for the other genes, they were deduced by comparison with the four known gene structures and examination of the exon-intron splice junctions. The regions coding for the different conserved domains are indicated by double arrows. Dim, Dimerization domain.
The molecular basis for the difference in substrate specificity between type I and II PIPkins was recently shown to implicate the so-called activation loop of these enzymes. The activation loop is a short segment, 22 to 27 amino acid residues long, located close to the C-terminal end of the catalytic domain of all PIPkins, including type III (Fig. 7; Kunz et al., 2000). When the activation loop of a human type I PIPkin was exchanged with that of a type II PIPkin, the resulting chimera exhibited type II substrate specificity, whereas a chimera consisting of a type II backbone with only the activation loop from a type I enzyme exhibited type I specificity (Kunz et al., 2000). A number of residues of the activation loop are specifically conserved in all members of each PIPkin type, including type III, and are therefore thought to confer substrate specificity. However, these type-specific residues are not specifically conserved in any PIPkins from Arabidopsis, but rather AtPIPkins possess in their activation loops both type I- and type II-specific residues. The substrate specificity of animal and plant PIPkins has not been determined in vivo. So far, the yeast PIPkin Mss4p is the only PIPkin whose substrate specificity has been studied in vivo, and it was shown that it produces mainly PtdIns(4,5)P2, only minor amounts of PtdIns(3,4)P2, and no PtdIns(3,5)P2 (Desrivières et al., 1998).
Fab1 Homologs
Type III PIPkins were recently proposed as a name for the group of enzymes consisting of the Brewer's yeast Fab1p protein and its homologs from other species (McEwen et al., 1999). These enzymes are PIPkins that only use PtdIns3P as a substrate and produce PtdIns(3,5)P2 (Cooke et al., 1998; Gary et al., 1998; McEwen et al., 1999). Yeast cells lacking a functional FAB1 gene do not contain any PtdIns(3,5)P2, and a mammalian type I PIPkin gene cannot restore this phenotype (McEwen et al., 1999). This demonstrates that, in vivo, the enzyme responsible for the synthesis of PtdIns(3,5)P2 is Fab1p, and that the ability of type I PIPkins to produce PtdIns(3,5)P2 from PtdIns3P in vitro (Rameh et al., 1997; Tolias et al., 1998) does not represent their biologically relevant activity. PIKfyve, a mammalian Fab1, generates PtdIns5P from PtdIns in vitro (Sbrissa et al., 2000). However, when expressed in yeast, it did not induce the formation of PtdIns5P, nor did any other type III PIPkin (McEwen et al., 1999), confirming that in vivo type III PIPkins are so far the only PIPkin able to synthesize PtdIns(3,5)P2 and that they are most likely not responsible for the synthesis of PtdIns5P. In yeast, animal, and plant cells, salt stress stimulates the production of PtdIns(3,5)P2 (Dove et al., 1997). The mechanism(s) involved in the regulation of Fab1 enzymes are at present not known. Deletion or mutation of yeast FAB1 results in temperature-sensitive growth and massive enlargement of the vacuole caused by a defective membrane flux (Yamamoto et al., 1995).
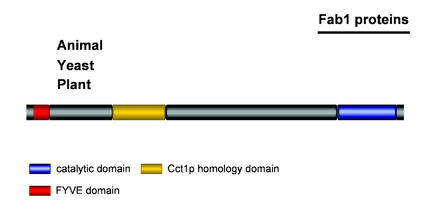
Four Arabidopsis genes encode putative Fab1p homologs, and all four genes are expressed in plant cells, because at least one EST for each has been reported (Table II). These four genes do not belong to any of the duplicated blocks defined by Vision et al. (2000), but they may well have arisen from an older duplication event. However, none of the four putative AtFab1 enzymes has yet been characterized in vitro or in vivo. The proteins encoded by AtFab1c and AtFab1d lack a putative FYVE domain. Surprisingly, only one FAB1 gene appears to be present in the human genome as well as in yeast. The structure of most Fab1 proteins is similar and is composed of three conserved domains: an N-terminal PtdIns3P-binding FYVE domain (this domain is apparently absent from AtFab1c and AtFab1d), a central domain with similarities to a conserved motif present in one of the subunits, Cct1p of the chaperonin complex and its homologs, and a C-terminal PIPkin catalytic domain (Fig. 10; McEwen et al., 1999). They are much larger than other PIPkins, most of them having a molecular mass of over 200 kD.
Figure 10.
Schematic representation of Fab1 proteins. The conserved domains are represented by blocks of different colors. The intervals between the conserved domains vary among species. Two of the four Arabidopsis genes encoding putative Fab1 proteins do no contain a FYVE domain.
PI-PLC
In vitro, PI-PLC isoforms catalyze the hydrolysis of PtdIns, PtdIns4P, and PtdIns(4,5)P2, and are Ca2+-dependent enzymes. Bacteria such as Bacillus cereus and Listeria monocytogenes contain a PtdIns-specific PLC that cannot use PtdIns4P or PtdIns(4,5)P2 as substrates.
Biochemical Characterization
PI-PLC activity has been detected in a number of different plant species and tissues. On the basis of in vitro assays, two types of activity were distinguished: One predominantly present in the soluble fraction of plant cells prefers PtdIns as a substrate and requires millimolar Ca2+ concentrations, and a second present in the plasma membrane that prefers PtdIns4P and PtdIns(4,5)P2 and requires much lower (0.1–10 μm) Ca2+ concentrations (Drøbak, 1992). It was also demonstrated early on that PI-PLC activity is present in both soluble (Melin et al., 1987; Yotsushima et al., 1992; Huang et al., 1995) and particulate (Melin et al., 1987; Yotsushima et al., 1993; Huang et al., 1995) fractions of plant cells. In most cases, the particulate fractions studied were purified plasma membranes. In the instances when substrate specificity was examined, it was found that the PI-PLC activity in plasma membranes showed a net preference for PtdIns4P and PtdIns(4,5)P2, the exception being a plasma membrane fraction from suspension-cultured rice (Oryza sativa) cells (Yotsushima et al., 1993), which showed an apparent preference for PtdIns.
Although PI-PLC has been purified from several plant tissues, no amino acid sequence has ever been obtained for any of these purified proteins. It is, therefore, still not known whether any of the PI-PLC genes identified so far correspond to any of the activities detected in or partially purified from plant extracts. However, Shi et al. (1995) isolated a cDNA encoding a biochemically active PI-PLC from soybean by screening an expression library with an antiserum raised against total proteins from purified plasma membranes and demonstrated that when expressed in transgenic tobacco plants, the fusion protein was present in the plasma membrane but also in the cytosol. Antibodies raised against a peptide specific for one PI-PLC from Arabidopsis recognize a single polypeptide of approximately 66 kD that is significantly enriched in plasma membranes purified from Arabidopsis plants and cannot be detected in the cytosol (Otterhag et al., 2001).
Although it is known that plant PI-PLC is activated by Ca2+, its regulation remains unresolved. When tested, guanine nucleotides have not stimulated PI-PLC activity at all in most cases. Even when activity has been stimulated, inositol phosphate production increased only 2- to 3-fold (not 20- to 30-fold as seen in animal systems) and these increases were accompanied by similar decreases in PtdInsP and PtdInsP2 (Harden et al., 1987; Einspahr et al., 1989). Clear, marked stimulatory effects of guanine nucleotides, or any other compounds, on plant PI-PLC activity have never been described. Several plant studies have reported that mastoparan (a potent G-protein activator) and/or its analogs induced Ins(1,4,5)P3 formation (Quarmby et al., 1992; Legendre et al., 1993; Yueh and Crain, 1993; Drøbak and Watkins, 1994; Cho et al., 1995; van Himbergen et al., 1999; Kuin et al., 2000) and suggested that plant PI-PLC may be regulated by a G-protein-dependent pathway. However, it was recently demonstrated that mastoparan and Mas-7 induce the formation of pores in the plasma membrane of human cells (Suh et al., 1998a). These pores allowed small molecules, such as ethidium bromide, lucifer yellow, but not Evan's blue, to penetrate the cells. Treatment with mastoparan and Mas-7, thus, resulted in a rise in the intracellular Ca2+ concentration. Chlamydomonas spp. was shown to respond to non-permeabilizing concentrations of mastoparan (i.e. concentrations that did not result in cells becoming permeable to Evan's blue) and its analogs by increasing Ins(1,4,5)P3 levels (Munnik et al., 1998b; Kuin et al., 2000). It was also demonstrated that these non-permeabilizing concentrations did not induce an influx of Evan's blue inside the cells, however, the influx of smaller molecules such as Ca2+ was not examined. Increases in [Ca2+]i upon mastoparan treatment have been observed in a higher plant system (Tucker and Boss, 1996). It is therefore possible, as also suggested by van Himbergen et al. (1999), that the Ins(1,4,5)P3 increase and simultaneous PtdIns(4,5)P2 decrease detected in Chlamydomonas spp. upon treatment with mastoparan and its analogs are attributable to a stimulation of PI-PLC activity via an increase in [Ca2+]i, rather than through activation of a G-protein. Further doubt to the possible regulation of plant PI-PLCs by G-proteins is raised by several facts. First, Arabidopsis contains only one gene coding for each of the three subunits forming heterotrimeric G-proteins, whereas in animal cells, several genes for each subunit exist. Second, and as mentioned earlier, only a few genes encoding putative G-protein-coupled receptors are present in Arabidopsis, whereas hundreds have been identified in animals. Third, as described below, the only type of PI-PLC identified in plants is not of the β- or ε-type, the only type of PI-PLC activated by heterotrimeric G-proteins (Rebecchi and Pentyala, 2000).
Genes, Structure, and Function
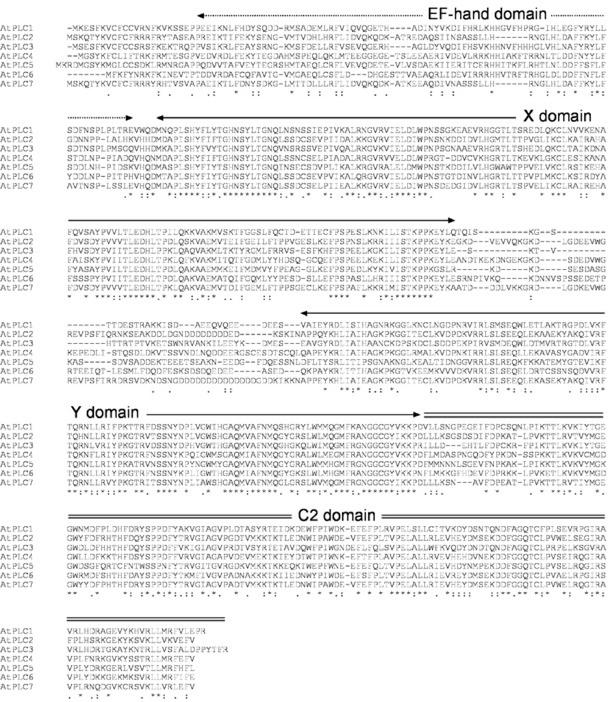
The first cDNA clones encoding functional plant PI-PLCs were reported in 1995 for Arabidopsis (Hirayama et al., 1995) and soybean (Shi et al., 1995), 7 years after the first animal PI-PLC clones were described (Bristol et al., 1988; Katan et al., 1988; Stahl et al., 1988; Suh et al., 1988b). Plant cDNA clones encoding active PI-PLC isozymes have also been obtained from potato (Solanum tuberosum; Kopka et al., 1998) and wild tobacco (Nicotiana rustica; Pical et al., 1997). Genes encoding additional putative PI-PLCs have been identified in Arabidopsis, garden pea (Pisum sativum), cowpea (Vigna unguiculata; accession no. U85250), rape seed (Brassica napus), soybean, and hairy finger-grass (accession no. AJ291467). In Arabidopsis, one cDNA clone, AtPLC1 (accession no. D38544), encoding a protein with demonstrated PI-PLC activity has been partially characterized (Hirayama et al., 1995). It showed a marked preference for PtdIns(4,5)P2 versus PtdIns, with a specific activity near the optimal free Ca2+ concentration of each substrate (approximately 100 times higher with PtdIns(4,5)P2 than with PtdIns). Maximal hydrolysis rates against PtdIns(4,5)P2 were reached at 1 to 50 μm free Ca2+, whereas Ca2+ concentrations above 1 mm were required to observe substantial PtdIns hydrolysis. Similar characteristics have been described for recombinant PI-PLCs from potato (Kopka et al., 1998) and soybean (Shi et al., 1995). Arabidopsis contains six additional genes with deduced protein sequences showing high similarity to AtPLC1 (Fig. 11; Table III).
Figure 11.
Alignment of the seven PI-PLCs from Arabidopsis. The accession numbers for the different proteins are given in Table II. The sequences were aligned using the ClustalW program. The four conserved domains are indicated. The X and Y domains together form the catalytic domain of the enzymes. Because no mRNA or EST sequences for AtPLC6 have been obtained, the sequence shown here is the predicted one that fits best with the other sequences.
Table III.
List of characterized and predicted PI-PLCs from Arabidopsis
| Name | Molecular Mass | Amino Acid Residues | Accession No./Locus | mRNA/ESTa |
|---|---|---|---|---|
| kD | ||||
| AtPLC1 | 64 | 561 | AB020755/At5g58670 | D38544 |
| AtPLC2 | 66 | 581 | AC074395/At3g08510 | D50804 |
| AtPLC3 | 65 | 564 | · · · · · · · · /At4g38530 | U13203 |
| AtPLC4 | 65 | 571 | · · · · · · · · /At5g58700 | AY053422 |
| AtPLC5 | 66 | 573 | · · · · · · · · /At5g58690 | AY062681 |
| AtPLC6 | 66 | 576 | AC018721/At2gxxxxx | No |
| AtPLC7 | 67 | 584 | AL163832/At3g55940 | No |
The AtPLC6 gene is located on chromosome 2 but is not annotated in the complete sequence of the chromosome, and therefore no locus has been assigned to it.
mRNA or EST accession numbers are provided, when available.
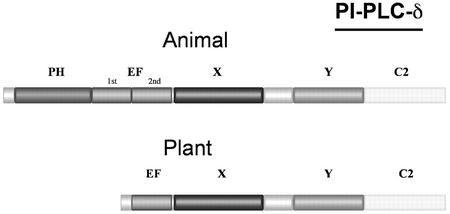
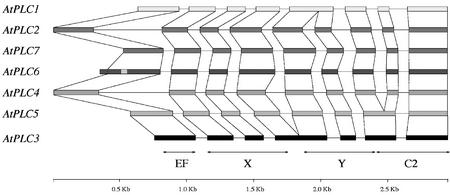
PI-PLCs are classified in four distinct subfamilies: β, γ, δ (Rebecchi and Pentyala, 2000), and ε (Lopez et al., 2001; Song et al., 2001). Five domains, a PH domain, an EF-hand domain, an X and a Y domain (together forming the catalytic domain), and a lipid-binding C2 domain, are conserved in PI-PLC enzymes and represent the core sequence of all known PI-PLCs from animals and yeast. This core sequence constitutes the structure of δ-isozymes (Fig. 12). The β-, γ-, and ε-isozymes contain specific insertions in the core sequence and are found only in animals. PI-PLC-βs are activated by G-protein-coupled receptors, PI-PLC-γs by receptor Tyr kinases (Rebecchi and Pentyala, 2000), and PI-PLC-εs by heterotrimeric G-proteins and Ras (Rhee, 2001). Sequence analysis indicates that all plant PI-PLC sequences are structurally identical and composed of four conserved domains, three of which correspond to the X, Y, and C2 domains conserved in non-plant PI-PLCs (Figs. 11 and 12). The structure of plant PI-PLCs resembles most closely that of PLC-δ fisoforms. The fourth conserved domain, preceding the X domain in plant PI-PLCs, is less conserved than the rest of the proteins, apart from the linker between the X and Y domains. Secondary structure predictions for this region show a clear resemblance to that of the secondary structure determined for the second loop of the EF-hand domain present in PLC-δ1, as determined by x-ray crystallography (Otterhag et al., 2001). Helices corresponding to the four helices forming the second loop of the EF-hand domain of PLC-δ1 are predicted in all plant PI-PLCs at sequence positions that are also conserved between plant isozymes and PLC-δ1. In PLC-δ1, the second loop of the EF-hand domain is believed to serve a critical structural role, through interaction with the C-terminal C2 domain. Deletion variants of animal PLC-δ1 and PLC-γ lacking this second loop are inactive (Emori et al., 1989; Nakashima et al., 1995). We have recently shown that in AtPLC2, a deletion variant lacking the 36 first amino acids (i.e. including a helix corresponding to helix E3α of PLC-δ1), is inactive, whereas a variant lacking the 22 first amino acids is fully active (Otterhag et al., 2001). This strongly suggests that plant PI-PLCs probably contain an EF-hand domain corresponding to the second loop of the EF-hand domain of animal PI-PLCs. Interestingly, all Arabidopsis PI-PLC genes contain an intron immediately after the region encoding the predicted EF-hand domain (Fig. 13). The seven Arabidopsis PI-PLC genes are composed of seven to nine exons: AtPLC3 contains only seven, AtPLC1 and AtPLC5 contain eight, and the remaining four genes have nine exons. Despite having the same number of exons and introns, AtPLC1 and AtPLC5 do not have the same structure: They differ by two introns. AtPLC2 and AtPLC4 are characterized by a much longer intron following the first exon. Because no cDNA sequences have been reported for AtPLC6 and AtPLC7, it is not possible to ascertain their structure.
Figure 12.
Representation of the modular domain arrangements of PI-PLC δ isozymes from animals and plants. The conserved domains are represented by blocks of different colors. The EF-hand domain of plant PI-PLCs corresponds to the second loop of the EF-hand domain of animal PI-PLCs. The X and Y domains constitute together the catalytic domain of PI-PLCs.
Figure 13.
Schematic representation of the seven PI-PLC genes from Arabidopsis. The introns and exons for AtPLC1, AtPLC2, AtPLC3, AtPLC4, and AtPLC5 were determined by comparing the mRNA and gene sequences, whereas for the other three genes, they were deduced by comparison with AtPLC1-5 and examination of the exon-intron splice junctions. In AtPLC6, two putative start codons can be identified. In addition, an insert in the first exon of this gene is present, and indicated by a gray box. The regions coding for the different conserved domains are indicated by double arrows.
Plant PI-PLCs do not contain a PH domain. In animal PI-PLC-δ, this domain is required for interaction with the plasma membrane (Paterson et al., 1995) and is involved in the binding of the lipid substrate and in processive catalysis (Essen et al., 1996; Katan and Williams, 1997). The C2 domain is also involved in membrane interaction but is not sufficient to position the enzyme in a catalytically active orientation (Katan and Williams, 1997). A thorough investigation of the lipid-binding and cellular localization of the C2 domains from the four mammalian PLC-δ isoforms recently showed that two of them bind specifically and in a Ca2+-dependent manner to phosphatidyl-Ser localized in the plasma membrane (Ananthanarayanan et al., 2001). Because plant PI-PLCs lack a PH domain, their interaction with the plasma membrane and PtdIns(4,5)P2 clearly differs from PI-PLC-δ. The C2 domain of plant PI-PLCs may be sufficient for membrane-binding. Apart from their C2 domain, other regions in plant PI-PLCs may be required for plasma membrane association, such as hydrophobic moieties, with or without posttranslational modification of the protein.
The function of plant PI-PLCs in vivo has been addressed, but is still obscure—in particular, the expression and subcellular distribution of the seven Arabidopsis isoforms has not been addressed. Using the aminosteroid compound U-73122, which inhibited the in vitro activity of a PI-PLC from Nicotiana rustica guard cells, it was demonstrated that stomatal guard cell responses to abscisic acid (ABA), aperture regulation, and cytosolic Ca2+ oscillations are inhibited by U-73122 at concentrations that also inhibited the recombinant PI-PLC. This suggests that ABA may activate a PI-PLC leading to cytosolic Ca2+ alterations and ultimately to stomatal closure (Staxén et al., 1999). Using sense and antisense transgenic Arabidopsis plants, Sanchez and Chua (2001) demonstrated that AtPLC1 is necessary for the inhibition of germination by ABA but that overexpression of AtPLC1 did not result in the induction of ABA-regulated genes, demonstrating that AtPLC1 may be involved in secondary ABA responses. It was also demonstrated that a second PI-PLC gene from Arabidopsis, AtPLC2 is not involved in these responses, in agreement with the fact that AtPLC1 is expressed only when plants are treated with ABA or subjected to drought or salt stress, whereas AtPLC2 expression is not affected by ABA (Hirayama et al., 1995, 1997).
CONCLUDING REMARKS AND PERSPECTIVES
Progress in the understanding of the function of PIs and their derivatives in plant cells has been slow, especially when compared with the animal field. A number of reasons can explain this lag. The identification of PIs from plant tissues has proved to be a challenge in plants. Attempts at purifying enzymes involved PI metabolism have failed to provide protein sequences. It has consequently been difficult to study the regulation of these enzymes, manipulate PI metabolism in vivo, and identify possible targets of these lipids. However, sequencing of the Arabidopsis genome has enabled the isolation and characterization of a number of cDNAs encoding PI-metabolizing enzymes and the identification of many genes coding for putative orthologs of other PI-metabolizing animal enzymes.
The enzymes catalyzing the removal of phosphate from PIs and/or inositol (poly) phosphate have not been discussed in the present review, mainly because only inositol (poly) phosphate phosphatases, but no PI phosphatase, from plants have been characterized. The phosphatases catalyzing these reactions form a large family, and can be categorized in four groups according to the position of the phosphoester bond on the inositol group they hydrolyze (i.e. 1-, 3-, 4-, and 5-phosphatases; for review, see Woscholski and Parker, 2000). Some of these phosphatases can hydrolyze inositol (poly) phosphates only, others PIs only, whereas others can use both as substrates. 1-Phosphatases cannot use PIs as a substrate. One Arabidopsis gene (At5g63980) encoding an inositol polyphosphate 1-phosphatase has been identified, characterized, and shown to be involved in ABA and stress responses (Quintero et al., 1996; Xiong et al., 2001), and three from tomato have also been characterized (Gillaspy et al., 1995). The Arabidopsis enzyme possesses catalytic bifunctional activity, namely a 3′(2′),5′-bisphosphate nucleotidase activity, and an inositol polyphosphate phosphatase activity. In Arabidopsis, five additional genes encode putative inositol polyphosphate phosphatase, two of which are located immediately after At5g63980 on chromosome 5, and another two genes are also found on chromosome 5.
3-Phosphatases can use either inositol polyphosphate or PIs as substrates, but not both. None have been biochemically characterized from plants. However, the Arabidopsis contains a number of putative orthologs of animal 3-phosphatases. Most interesting is the presence of genes encoding putative members of the two families of PI 3-phosphatases, phosphatase and tensin homolog (PTEN) and myotubularin (Maehama et al., 2001). Mutated forms of PTEN were identified in numerous human tumors. Originally thought to be a Tyr phosphatase, PTEN was demonstrated to have only weak phosphatase activity toward phospho-Tyr-containing peptides. Instead, its physiological substrate is believed to be PtdIns(3,4,5) P3, with other 3-phosphorylated PIs being poorer substrates in vitro. It is, therefore, intriguing that Arabidopsis contains two genes encoding putative orthologs of enzymes whose preferred substrate is a lipid that has never been detected in plant cells. Mutations in the MTM1 gene cause the neonatal disorder X-linked myotubular myopathy, which in severe cases is often fatal (Maehama et al., 2001). MTM1 and MTM-related genes encode myotubularin isoforms that are highly similar to each other. Similar to PTEN, myotubularin was first thought to be a protein phosphatase because it contains the active site present in members of the protein Tyr phosphatase family of proteins. However, it too has low activity in the presence of artificial protein substrates. It was then observed that the catalytic domain of myotubularin shows significant similarity with the active site of another PI phosphatase, the SacIp protein (see below) from Brewer's yeast. Myotubularin is indeed a PI phosphatase, showing a clear preference for PtdIns3P as substrate (Taylor et al., 2000). Two genes in Arabidopsis encode putative myotubularin-like proteins. Both genes appear to be expressed because one EST sequence for each gene has been deposited in the GenBank database. Finally, one enzyme, multiple inositol polyphosphate phosphatase, has been demonstrated to hydrolyze various inositol polyphosphates (Craxton et al., 1995). One Arabidopsis gene encodes a potential ortholog of the animal enzyme. Only two genes encoding 4-phosphatases have identified from animal tissues. They encode phosphatases that can hydrolyze PtdIns(3,4)P2, Ins 1,3,4-trisphosphate, and Ins 3,4-bisphosphate. No putative orthologs of these two proteins are present in Arabidopsis.
The largest group of inositol (poly) phosphates and/or PI phosphatase is constituted by enzymes hydrolyzing the 5-phosphate group of these inositol-containing molecules. They all contain a 5-phosphatase catalytic domain and can be divided into type I and type II enzymes. Type I enzymes do not hydrolyze PIs, whereas the larger type II can. Ins(1,4,5)P3 and Ins 1,3,4,5-tetrakisphosphate are the favored substrates of type I enzymes. One cDNA clone from Arabidopsis encoding type I 5-phosphatases with substrate specificities similar to the animal enzymes have been characterized (Berdy et al., 2001). Ten additional genes encoding putative type I 5-phosphatases are present in Arabidopsis (Berdy et al., 2001). This multiplicity is intriguing, especially in view of the fact that Brewer's yeast does not appear to possess a 5-phosphatase with type I substrate specificity (Wiradjaja et al., 2001), although the Inp54p protein is more similar in size to type I than type II 5-phosphatases. This suggests that the metabolism of inositol (poly) phosphates in plant cells is highly complex. Type II 5- phosphatases can hydrolyze PIs, and some enzymes are also able to hydrolyze Ins polyphosphates (Woscholski and Parker, 2000). They all possess a type II domain, except for Inp54p in which this domain is truncated (Hughes et al., 2000). A variety of protein modules, e.g. Sac, SH2, and GTPase activating protein domains, are found in individual members. In Brewer's yeast, the four different 5-phosphatases Inp51p, Inp52p, Inp53p, and Inp54p belong to the type II group (Hughes et al., 2000). No type II 5-phosphatase from plants has been characterized. However, the genome of Arabidopsis encodes four putative type II 5-phosphatases (Berdy et al., 2001). The predicted proteins do not contain the Sac domain present in some phosphatases, including three of the type II 5-phosphatases from Brewer's yeast. The Sac domain confers phosphatase activity toward PIs, but not Ins polyphosphates (Hughes et al., 2000). Of all the existing PIs, PtdIns(4,5)P2 and PtdIns(3,4)P2 are not hydrolyzed by Sac-containing enzymes, whereas PtdIns5P has not been tested as a substrate. As just mentioned, the Sac domain is present in some type 5-phosphatases. There are, in addition, proteins with a Sac domain but no 5-phosphatase domain (two such proteins exist in Brewer's yeast; Hughes et al., 2000). None have been characterized in plants, but Arabidopsis contains nine genes encoding putative proteins with a domain highly similar to the Sac domain.
From these studies and observations, it is obvious that plant PI-metabolizing enzymes are related to their animal counterparts both in their primary structure and substrate specificity. However, the pathways involved in PI metabolism in plant cells present some unique features, including a higher number of type I and type III PIPkin isoforms than in animal cells, an apparent lack of animal-like type II PIPkins, an absence of class I and II PtdIns 3-kinases and PtdIns(3,4,5)P3, and only one type of PI-PLC. The subcellular distribution of these different enzymes and the PIs has also been only superficially examined, because it is still not simple to obtain some of the subcellular compartments with adequate purity; in particular, there is at present no protocol allowing the isolation of highly purified Golgi and endoplasmic reticulum membranes from plant extracts.
A number of major questions regarding plant cell PI metabolism now clearly need to be addressed. Which of the various isoforms of the different inositol lipid kinases are responsible for the synthesis of the different inositol lipid isomers in vivo? How are these kinases and the PI-PLC isoforms regulated? What is the function of the different isoforms of PI-PLC, PIPkins? As pointed out above, Ins (poly) phosphates and/or PI phosphatases may form a family of more than 35 proteins in Arabidopsis. This virtually unexplored and exciting area of plant PI research undoubtedly deserves much attention; the properties and functions of these proteins await characterization.
An even more important challenge is to determine the physiological role of PIs in plant cells. One approach could involve the characterization of the numerous predicted proteins from Arabidopsis containing PI-binding domains, such as the PH, PX, FYVE, and epsin N-terminal homology domains (Cullen et al., 2001). Arabidopsis knock-out mutants should also provide useful information when they result in a clear phenotype.
Note Added in Proof
A fifth subfamily of PI-PLC, designated ζ, has been identified in animals. One member from this new subfamily of PI-PLC has been characterized. It is smaller in size, 74 kD, than any other PI-PLC from animals, and lacks an N-terminal PH domain (Saunders et al., 2002).
ACKNOWLEDGMENTS
We thank Marianne Sommarin (Lund University) for helpful comments on the manuscript, Megan McKenzie (MPI of Molecular Plant Physiology, Golm) for critical comments on the manuscript and language, and Harold Meijer and Teun Munnik (University of Amsterdam) for sharing results before publication.
Footnotes
This work was supported by the Swedish Council for Forestry and Agricultural Research (grant to C.P.), by the Lund Fysiografiska Sällskapet (grant to C.P.), by the Crafoord Foundation (grant to C.P.), and by the Max-Planck Society (to B.M.-R.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.004770.
LITERATURE CITED
- Alexandre J, Lassalles JF, Kado RD. Opening of Ca2+channels in isolated red beet vacuole membrane by inositol 1,4,5-trisphosphate. Nature. 1990;343:567–570. [Google Scholar]
- Ananthanarayanan B, Das S, Rhee SG, Murray D, Cho W. Membrane targeting of C2 domains of phospholipase C-δ isoforms. J Biol Chem. 2001;277:3568–3575. doi: 10.1074/jbc.M109705200. [DOI] [PubMed] [Google Scholar]
- Audhya A, Foti M, Emr SD. Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol Biol Cell. 2000;11:2673–2689. doi: 10.1091/mbc.11.8.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae YS, Cantley LG, Chen CS, Kim SR, Kwon KS, Rhee SG. Activation of phospholipase C-γ by phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:4465–4469. doi: 10.1074/jbc.273.8.4465. [DOI] [PubMed] [Google Scholar]
- Balla T. Phosphatidylinositol 4-kinases. Biochim Biophys Acta. 1998;1436:69–85. doi: 10.1016/s0005-2760(98)00134-9. [DOI] [PubMed] [Google Scholar]
- Balla T, Downing GJ, Jaffe H, Kim S, Zólyomi A, Catt KJ. Isolation and molecular cloning of wortmannin-sensitive bovine type III phosphatidylinositol 4-kinases. J Biol Chem. 1997;272:18358–18366. doi: 10.1074/jbc.272.29.18358. [DOI] [PubMed] [Google Scholar]
- Banfic H, Downes CP, Rittenhouse SE. Biphasic activation of PKBalpha/Akt in platelets: evidence for stimulation both by phosphatidylinositol 3,4-bisphosphate, produced via a novel pathway, and by phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:11630–11637. doi: 10.1074/jbc.273.19.11630. [DOI] [PubMed] [Google Scholar]
- Barylko B, Gerber SH, Binns DD, Grichine N, Khvotchev M, Südhof TC, Albanesi JP. A novel family of phosphatidylinositol 4-kinases conserved from yeast to humans. J Biol Chem. 2001;276:7705–7708. doi: 10.1074/jbc.C000861200. [DOI] [PubMed] [Google Scholar]
- Berdy SE, Kudla J, Gruissem W, Gillaspy GE. Molecular characterization of At5Ptase1, an inositol phosphatase capable of terminating inositol trisphosphate signaling. Plant Physiol. 2001;126:801–810. doi: 10.1104/pp.126.2.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge MJ. Inositol trisphosphate and calcium signalling. Nature. 1993;361:315–325. doi: 10.1038/361315a0. [DOI] [PubMed] [Google Scholar]
- Berridge MJ, Irvine RF. Inositol phosphates and cell signaling. Nature. 1989;341:197–205. doi: 10.1038/341197a0. [DOI] [PubMed] [Google Scholar]
- Blatt MF, Thiel G, Trentham DR. Reversible inactivation of K+ channels of Viciastomatal guard cells following the photolysis of caged inositol 1,4,5-trisphosphate. Nature. 1990;346:766–769. doi: 10.1038/346766a0. [DOI] [PubMed] [Google Scholar]
- Bloomquist BT, Shortridge RD, Schneuwly S, Perdew M, Montell C, Steller H, Rubin G, Pak WL. Isolation of a putative phospholipase C gene of Drosophila, norpA, and its role in phototransduction. Cell. 1988;54:723–733. doi: 10.1016/s0092-8674(88)80017-5. [DOI] [PubMed] [Google Scholar]
- Bovet L, Müller M-O, Siegenthaler P-A. Three distinct lipid kinase activities are present in spinach chloroplast envelope membranes: phosphatidylinositol phosphorylation is sensitive to wortmannin and not dependent on chloroplast ATP. Biochem Biophys Res Commun. 2001;289:269–275. doi: 10.1006/bbrc.2001.5969. [DOI] [PubMed] [Google Scholar]
- Brearley CA, Hanke DE. Evidence for substrate-cycling of 3-, 3,4-, 4-, and 4,5-phosphorylated phosphatidylinositols in plants. Biochem J. 1995;311:1001–1007. doi: 10.1042/bj3111001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill JA, Hime GR, Scharer-Schuksz M, Fuller MT. A phospholipid kinase regulates actin organization and intercellular bridge formation during germline cytokinesis. Development. 2000;127:3855–3864. doi: 10.1242/dev.127.17.3855. [DOI] [PubMed] [Google Scholar]
- Bristol A, Hall SM, Kriz RW, Stahl ML, Fan YS, Byers MG, Eddy RL, Shows TB, Knopf JL. Phospholipase C-148: chromosomal location and deletion mapping of functional domains. Cold Spring Harb Symp Quant Biol. 1988;53:915–920. doi: 10.1101/sqb.1988.053.01.105. [DOI] [PubMed] [Google Scholar]
- Bunney TD, Watkins PAC, Beven AF, Shaw PJ, Hernandez LE, Lomonossoff GP, Shanks M, Peart J, Drøbak BK. Association of phosphatidylinositol 3-kinase with nuclear transcription sites in higher plants. Plant Cell. 2000;12:1679–1687. doi: 10.1105/tpc.12.9.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter CL, Cantley LC. Phosphoinositide kinases. Biochemistry. 1990;29:11147–11156. doi: 10.1021/bi00503a001. [DOI] [PubMed] [Google Scholar]
- Carvajal JJ, Pook MA, dos Santos M, Doudney K, Hillermann R, Minogue S, Williamson R, Hsuan JJ, Chamberlain S. The Friedreich's ataxia gene encodes a novel phosphatidylinositol-4-phosphate 5-kinase. Nat Genet. 1996;14:157–162. doi: 10.1038/ng1096-157. [DOI] [PubMed] [Google Scholar]
- Cho MH, Tan Z, Erneux C, Shears SB, Boss WF. The effects of mastoparan on the carrot cell plasma membrane polyphosphoinositide phospholipase C. Plant Physiol. 1995;107:845–856. doi: 10.1104/pp.107.3.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke FT, Dove SK, McEwen RK, Painter G, Holmes AB, Hall MN, Michell RH, Parker PJ. The stress-activated phosphatidylinositol 3-phosphate 5-kinase Fab1p is essential for vacuole function in S. cerevisiae. Curr Biol. 1998;9:1219–1222. doi: 10.1016/s0960-9822(07)00513-1. [DOI] [PubMed] [Google Scholar]
- Craxton A, Ali N, Shears SB. Comparison of the activities of a multiple inositol polyphosphate phosphatase obtained from several sources: a search for heterogeneity in this enzyme. Biochem J. 1995;305:491–498. doi: 10.1042/bj3050491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ, Cozier GE, Banting G, Mellor H. Modular phosphoinositide-binding domains: their role in signalling and membrane trafficking. Curr Biol. 2001;11:882–893. doi: 10.1016/s0960-9822(01)00523-1. [DOI] [PubMed] [Google Scholar]
- Davies SP, Reddy H, Caivano M, Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deak M, Casamayor A, Currie RA, Downes CP, Alessi DR. Characterisation of a plant 3-phosphoinositide-dependent protein kinase-1 homologue which contains a pleckstrin homology domain. FEBS Lett. 1999;451:220–226. doi: 10.1016/s0014-5793(99)00556-6. [DOI] [PubMed] [Google Scholar]
- De Camilli P, Emr SD, McPherson PS, Novick P. Phosphoinositides as regulators in membrane traffic. Science. 1996;271:1533–1539. doi: 10.1126/science.271.5255.1533. [DOI] [PubMed] [Google Scholar]
- Desrivières S, Cooke FT, Parker PJ, Hall MN. MSS4, a phosphatidylinositol-4-phosphate 5-kinase required for organization of the actin cytoskeleton in Saccharomyces cerevisiae. J Biol Chem. 1998;273:15787–15793. doi: 10.1074/jbc.273.25.15787. [DOI] [PubMed] [Google Scholar]
- DeWald DB, Torabinejad J, Jones CA, Shope JC, Cangelosi AR, Thompson JE, Prestwich GD, Hama H. Rapid accumulation of phosphatidylinositol 4,5-bisphosphate and inositol 1,4,5-trisphosphate correlates with calcium mobilization in salt-stressed Arabidopsis. Plant Physiol. 2001;126:759–769. doi: 10.1104/pp.126.2.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove SK, Cooke FT, Douglas M, Sayers L, Parker PJ, Michell RH. Osmotic stress activates phosphatidylinositol-3,5-bisphosphate synthesis. Nature. 1997;390:187–192. doi: 10.1038/36613. [DOI] [PubMed] [Google Scholar]
- Drøbak BK. The plant phosphoinositide system. Biochem J. 1992;288:697–712. doi: 10.1042/bj2880697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drøbak BK, Dewey RE, Boss WF. Phosphoinositide kinase and the synthesis of polyphosphoinositides in higher plant cells. Int Rev Cytol. 1999;189:95–130. doi: 10.1016/s0074-7696(08)61386-8. [DOI] [PubMed] [Google Scholar]
- Drøbak BK, Watkins PAC. Inositol(1,4,5) trisphosphate production in plant cells: stimulation by the venom peptides, melittin and mastoparan. Biochem Biophys Res Commun. 1994;205:739–745. doi: 10.1006/bbrc.1994.2727. [DOI] [PubMed] [Google Scholar]
- Drøbak BK, Watkins PAC, Valenta R, Dove SK, Lloyd CW, Staiger CJ. Inhibition of plant plasma membrane phosphoinositide phospholipase C by the actin-binding protein, profiling. Plant J. 1994;6:389–400. [Google Scholar]
- Einspahr KJ, Peeler TC, Thompson GA., Jr Phosphatidylinositol 4,5-bisphosphate phospholipase C and phosphomonoesterase in Dunaliella salinamembranes. Plant Physiol. 1989;90:1115–1120. doi: 10.1104/pp.90.3.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elge S, Brearley C, Xia H-J, Kehr J, Xue H-W, Müller-Röber B. An Arabidopsis inositol phospholipid kinase strongly expressed in procambial cells: synthesis of PtdIns(4,5) P2 and PtdIns(3,4,5) P3in insect cells by 5-phosphorylation of precursors. Plant J. 2001;26:561–571. doi: 10.1046/j.1365-313x.2001.01051.x. [DOI] [PubMed] [Google Scholar]
- Ellson CD, Andrews S, Stephens LR, Hawkins PT. The PX domain: a new phosphoinositide-binding module. J Cell Sci. 2002;115:1099–1115. doi: 10.1242/jcs.115.6.1099. [DOI] [PubMed] [Google Scholar]
- Emori Y, Homma Y, Sorimachi H, Kawasaki H, Nakanishi O, Suzuki K, Takenawa T. A second type of rat phosphoinositide-specific phospholipase C containing a src-related sequence not essential for phosphoinositide-hydrolyzing activity. J Biol Chem. 1989;254:21885–21890. [PubMed] [Google Scholar]
- Endemann G, Dunn SN, Cantley LC. Bovine brain contains two types of phosphatidylinositol kinase. Biochemistry. 1987;26:6845–6852. doi: 10.1021/bi00395a039. [DOI] [PubMed] [Google Scholar]
- Endemann GC, Graziani A, Cantley LC. A monoclonal antibody distinguishes two types of phosphatidylinositol 4-kinase. Biochem J. 1991;273:63–66. doi: 10.1042/bj2730063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essen L-O, Perisic O, Cheung M, Katan M, Williams RL. Crystal structure of a mammalian phosphoinositide-specific phospholipase Cδ. Nature. 1996;380:595–602. doi: 10.1038/380595a0. [DOI] [PubMed] [Google Scholar]
- Falasca M, Logan SK, Lehto VP, Baccante G, Lemmon MA, Schlessinger J. Activation of phospholipase C gamma by PI 3-kinase-induced PH domain-mediated membrane targeting. EMBO J. 1998;17:414–422. doi: 10.1093/emboj/17.2.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary JD, Wurmser AE, Bonangelino CJ, Weisman LS, Emr SD. Fab1p is essential for PtdIns(3) P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J Cell Biol. 1998;143:65–79. doi: 10.1083/jcb.143.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillaspy GE, Keddie JS, Oda K, Gruissem W. Plant inositol monophosphatase is a lithium-sensitive enzyme encoded by a multigene family. Plant Cell. 1995;7:2175–2185. doi: 10.1105/tpc.7.12.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy S, Read ND, Trewavas AJ. Elevation of cytoplasmic calcium by caged calcium or caged inositol trisphosphate initiates stomatal closure. Nature. 1990;346:769–771. doi: 10.1038/346769a0. [DOI] [PubMed] [Google Scholar]
- Hanenberg A, Wissing JB, Wagner KG. Properties of phosphatidyl 4-kinase I from suspension cultured Catharanthus roseuscells. Plant Sci. 1995;112:53–63. [Google Scholar]
- Hanks SK, Quinn AM. Protein kinase catalytic domain sequence database: identification of conserved features of primary structure and classification of family members. Methods Enzymol. 1991;200:38–62. doi: 10.1016/0076-6879(91)00126-h. [DOI] [PubMed] [Google Scholar]
- Harden TK, Stephens L, Hawkins PT, Downes CP. Turkey erythrocyte membranes as a model for regulation of phospholipase C by guanine nucleotides. J Biol Chem. 1987;262:9057–9061. [PubMed] [Google Scholar]
- Herman PK, Emr SD. Characterization of VPS34, a gene required for vacuolar protein sorting and vacuole segregation in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:6742–6754. doi: 10.1128/mcb.10.12.6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchliffe KA, Ciruela A, Irvine RF. PIPkins, their substrates and their products: new functions for old enzymes. Biochim Biophys Acta. 1998;1436:87–104. doi: 10.1016/s0005-2760(98)00140-4. [DOI] [PubMed] [Google Scholar]
- Hirayama T, Mitsukawa N, Shibata D, Shinozaki K. AtPLC2, a gene encoding phosphoinositide-specific phospholipase C, is constitutively expressed in vegetative and floral tissues in Arabidopsis thaliana. Plant Mol Biol. 1997;34:175–180. doi: 10.1023/a:1005885230896. [DOI] [PubMed] [Google Scholar]
- Hirayama T, Ohto C, Mizoguchi T, Shinozaki K. A gene encoding a phosphatidylinositol-specific phospholipase C is induced by dehydration and salt stress in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1995;92:3903–3907. doi: 10.1073/pnas.92.9.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C-H, Tate BF, Crain RC, Coté GG. Multiple phosphoinositide-specific phospholipases C in oat roots: characterization and partial purification. Plant J. 1995;8:257–267. [Google Scholar]
- Hughes WE, Cooke FT, Parker PJ. Sac phosphatase domain proteins. Biochem J. 2000;350:337–352. [PMC free article] [PubMed] [Google Scholar]
- Janmey PA, Stossel TP. Modulation of gelsolin function by phosphatidylinositol 4,5-bisphosphate. Nature. 1987;325:362–364. doi: 10.1038/325362a0. [DOI] [PubMed] [Google Scholar]
- Jensen RB, La Cour T, Albrethsen J, Nielsen M, Skriver K. FYVE zinc-finger proteins in the plant model Arabidopsis thaliana: identification of PtdIns3P-binding residues by comparison of classic and variant FYVE domains. Biochem J. 2001;359:165–173. doi: 10.1042/0264-6021:3590165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katan M, Kriz RW, Totty N, Philp R, Meldrum E, Aldape RA, Knopf J, Parker PJ. Primary structure of PLC-154. Cold Spring Harbor Symp Quant Biol. 1988;53:921–926. doi: 10.1101/sqb.1988.053.01.106. [DOI] [PubMed] [Google Scholar]
- Katan M, Williams RL. Phosphoinositide-specific phospholipase C: structural basis for catalysis and regulatory interactions. Semin Cell Dev Biol. 1997;8:287–296. doi: 10.1006/scdb.1997.0150. [DOI] [PubMed] [Google Scholar]
- Kim DH, Eu Y-J, Yoo CM, Kim Y-W, Pih KT, Jin JB, Kim SJ, Stenmark H, Hwang I. Trafficking of phosphatidylinositol 3-phosphate from the trans-Golgi network to the lumen of the central vacuole in plant cells. Plant Cell. 2001;13:287–301. doi: 10.1105/tpc.13.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopka J, Pical C, Gray JE, Müller-Röber B. Molecular and enzymatic characterization of three phosphoinositide-specific phospholipase C isoforms from potato. Plant Physiol. 1998;116:239–250. doi: 10.1104/pp.116.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost B, Lemichez E, Spielhofer P, Hong Y, Tolias K, Carpenter C, Chua NH. Rac homologues and compartmentalized phosphatidylinositol 4,5-bisphosphate act in a common pathway to regulate polar pollen tube growth. J Cell Biol. 1999;145:317–330. doi: 10.1083/jcb.145.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuin H, Koerten H, Ghijsen WE, Munnik T, van den Ende H, Musgrave A. Chlamydomonascontains calcium stores that are mobilized when phospholipase C is activated. Planta. 2000;210:286–294. doi: 10.1007/PL00008136. [DOI] [PubMed] [Google Scholar]
- Kunz J, Wilson MP, Kisseleva M, Hurley JH, Majerus PW, Anderson RA. The activation loop of phosphatidylinositol phosphate kinases determines signaling specificity. Mol Cell. 2000;5:1–11. doi: 10.1016/s1097-2765(00)80398-6. [DOI] [PubMed] [Google Scholar]
- Lassing I, Lundberg U. Specific interaction between phosphatidylinositol 4,5-bisphosphate and profilactin. Nature. 1985;314:472–474. doi: 10.1038/314472a0. [DOI] [PubMed] [Google Scholar]
- Legendre L, Yueh YG, Crain R, Haddock N, Heinstein PF, Low PS. Phospholipase C activation during elicitation of the oxidative burst in cultured plant cells. J Biol Chem. 1993;268:24559–24563. [PubMed] [Google Scholar]
- Lopez I, Mak EC, Ding S, Hamm HE, Lomasney JW. A novel bifunctional phospholipase c that is regulated by Galpha 12 and stimulates the Ras/mitogen-activated protein kinase pathway. J Biol Chem. 2001;276:2758–2765. doi: 10.1074/jbc.M008119200. [DOI] [PubMed] [Google Scholar]
- Maehama T, Taylor GS, Dixon JE. PTEN and myotubularin: novel phosphoinositide phosphatases. Annu Rev Biochem. 2001;70:247–279. doi: 10.1146/annurev.biochem.70.1.247. [DOI] [PubMed] [Google Scholar]
- Matsuoka K, Bassham DC, Raikhel NV, Nakamura K. Different sensitivity to wortmannin of two vacuolar sorting signals indicates the presence of distinct sorting machineries in tobacco cells. J Cell Biol. 1995;130:1307–1318. doi: 10.1083/jcb.130.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen RK, Dove SK, Cooke FT, Painter GF, Holmes AB, Shisheva A, Ohya Y, Parker PJ, Michell RH. Complementation analysis in PtdInsP kinase-deficient yeast mutants demonstrates that Schizosaccharomyces pombeand murine Fab1p homologues are phosphatidylinositol 3-phosphate 5-kinases. J Biol Chem. 1999;274:33905–33912. doi: 10.1074/jbc.274.48.33905. [DOI] [PubMed] [Google Scholar]
- Meijer HJ, Berrie CP, Iurisci C, Divecha N, Musgrave A, Munnik T. Identification of a new polyphosphoinositide in plants, phosphatidylinositol 5-monophosphate (PtdIns5P), and its accumulation upon osmotic stress. Biochem J. 2001;360:491–498. doi: 10.1042/0264-6021:3600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer HJG, Divecha N, van den Ende H, Musgrave A, Munnik T. Hyperosmotic stress induces rapid synthesis of phosphatidyl-d-inositol 3,5-bisphosphate in plant cells. Planta. 1999;208:294–298. [Google Scholar]
- Melin P-M, Sommarin M, Sandelius AS, Jergil B. Identification of Ca2+-stimulated polyphosphoinositide phospholipase C in isolated plant plasma membranes. FEBS Lett. 1987;223:87–91. doi: 10.1016/0014-5793(87)80515-x. [DOI] [PubMed] [Google Scholar]
- Meyers R, Cantley LC. Cloning and characterization of a wortmannin-sensitive human phosphatidylinositol 4-kinase. J Biol Chem. 1997;272:4384–4390. doi: 10.1074/jbc.272.7.4384. [DOI] [PubMed] [Google Scholar]
- Mikami K, Katagiri T, Luchi S, Yamaguchi-Shinozaki K, Shinozaki K. A gene encoding phosphatidylinositol-4-phosphate 5-kinase is induced by water stress and abscisic acid in Arabidopsis thaliana. Plant J. 1998;15:563–568. doi: 10.1046/j.1365-313x.1998.00227.x. [DOI] [PubMed] [Google Scholar]
- Minogue S, Anderson JS, Waugh MG, dos Santos M, Corless S, Cramer R, Hsuan JJ. Cloning of a human type II phosphatidylinositol 4-kinase reveals a novel lipid kinase family. J Biol Chem. 2001;276:16635–16640. doi: 10.1074/jbc.M100982200. [DOI] [PubMed] [Google Scholar]
- Munnik T, Irvine RF, Musgrave A. Phospholipid signalling in plants. Biochim Biophys Acta. 1998a;1389:222–272. doi: 10.1016/s0005-2760(97)00158-6. [DOI] [PubMed] [Google Scholar]
- Munnik T, Musgrave A, de Vrije T. Rapid turnover of polyphosphoinositides in carnation flower petals. Planta. 1994;193:89–98. [Google Scholar]
- Munnik T, van Himbergen JAJ, ter Riet B, Braun F-J, Irvine RF, van den Ende H, Musgrave A. Detailed analysis of the turnover of polyphosphoinositides and phosphatidic acid upon activation of phospholipases C and D in Chlamydomonascells treated with non-permeabilizing concentrations of mastoparan. Planta. 1998b;207:133–145. [Google Scholar]
- Nakagawa T, Goto K, Kondo H. Cloning and characterization of a 92 kDa soluble phosphatidylinositol 4-kinase. Biochem J. 1996;320:643–649. doi: 10.1042/bj3200643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima S, Banno Y, Watanabe T, Nakamura Y, Mizutani T, Sakai H, Zhao Y, Sugimoto Y, Nozawa Y. Deletion and site-directed mutagenesis of EF-hand domain of phospholipase C-delta 1: effects on its activity. Biochem Biophys Res Commun. 1995;211:364–369. doi: 10.1006/bbrc.1995.1822. [DOI] [PubMed] [Google Scholar]
- Nishi M, Mizushima A, Nakagawara K-i, Takeshima H. Characterization of human junctophilin subtype genes. Biochem Biophys Res Commun. 2000;273:920–927. doi: 10.1006/bbrc.2000.3011. [DOI] [PubMed] [Google Scholar]
- Oh ES, Woods A, Lim St, Theibert AW, Couchman JR. Syndecan-4 proteoglycan cytoplasmic domain and phosphatidylinositol 4,5-bisphosphate coordinately regulate protein kinase C activity. J Biol Chem. 1998;273:10624–10629. doi: 10.1074/jbc.273.17.10624. [DOI] [PubMed] [Google Scholar]
- Okpodu CM, Gross W, Burkhart W, Boss WF. Purification and characterization of a soluble phosphatidylinositol 4-kinase from carrot suspension culture cells. Plant Physiol. 1995;107:491–500. doi: 10.1104/pp.107.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otterhag L, Sommarin M, Pical C. N-Terminal EF-hand-like domain is required for phosphoinositide-specific phospholipase C activity in Arabidopsis thaliana. FEBS Lett. 2001;497:165–170. doi: 10.1016/s0014-5793(01)02453-x. [DOI] [PubMed] [Google Scholar]
- Pappan K, Zheng S, Wang X. Identification and characterization of a novel phospholipase D that requires polyphosphoinositide and submicromolar calcium for activity in Arabidopsis. J Biol Chem. 1997;272:7048–7054. doi: 10.1074/jbc.272.11.7048. [DOI] [PubMed] [Google Scholar]
- Parmar PN, Brearley CA. Identification of 3- and 4-phosphorylated phosphoinositides and inositol phosphates in stomatal guard cells. Plant J. 1993;4:255–263. [Google Scholar]
- Paterson HF, Savopoulos JW, Perisic O, Cheung R, Ellis MV, Williams RL, Katan M. Phospholipase C delta 1 requires a pleckstrin homology domain for interaction with the plasma membrane. Biochem J. 1995;312:661–666. doi: 10.1042/bj3120661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pical C, Kopka J, Müller-Röber J, Hetherington AM, Gray JE. Isolation of 2 cDNA clones for phosphoinositide-specific phospholipase C from epidermal peels (accession no. X95877) and guard cells (accession no. Y11931) of Nicotiana rustica (PGR 97-086) Plant Physiol. 1997;114:747. [Google Scholar]
- Pical C, Westergren T, Dove SK, Larsson C, Sommarin M. Salinity and hyperosmotic stress induce rapid increases in phosphatidylinositol 4,5-bisphosphate, diacylglycerol pyrophosphate, and phosphatidylcholine in Arabidopsis thalianacells. J Biol Chem. 1999;274:38232–38240. doi: 10.1074/jbc.274.53.38232. [DOI] [PubMed] [Google Scholar]
- Pike L. Phosphatidylinositol 4-kinases and the role of polyphosphoinositides in cellular regulation. Endocr Rev. 1992;13:692–706. doi: 10.1210/edrv-13-4-692. [DOI] [PubMed] [Google Scholar]
- Quarmby LM, Yueh YG, Cheshire JL, Keller LR, Snell WJ, Crain RC. Inositol phospholipid metabolism may trigger flagellar excision in Chlamydomonas reinhardtii. J Cell Biol. 1992;116:737–744. doi: 10.1083/jcb.116.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero FJ, Garciadeblas B, Rodriguez-Navarro A. The SAL1 gene of Arabidopsis, encoding an enzyme with 3′(2′),5′-bisphosphate nucleotide and inositol polyphosphate 1-phosphatase activities, increases salt tolerance in yeast. Plant Cell. 1996;8:529–537. doi: 10.1105/tpc.8.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rameh LE, Tolias KF, Duckworth BC, Cantley LC. A new pathway for synthesis of phosphatidylinositol-4,5-bisphosphate. Nature. 1997;390:192–196. doi: 10.1038/36621. [DOI] [PubMed] [Google Scholar]
- Rao VD, Misra S, Boronenkov IV, Anderson RA, Hurley JH. Structure of type II beta phosphatidylinositol phosphate kinase: a protein kinase fold flattened for interfacial phosphorylation. Cell. 1998;94:829–839. doi: 10.1016/s0092-8674(00)81741-9. [DOI] [PubMed] [Google Scholar]
- Rebecchi MJ, Pentyala SN. Structure, function, and control of phosphoinositide-specific phospholipase C. Physiol Rev. 2000;80:1291–1335. doi: 10.1152/physrev.2000.80.4.1291. [DOI] [PubMed] [Google Scholar]
- Rhee SG. Regulation of phosphoinositide-specific phospholipase C. Annu Rev Biochem. 2001;70:281–312. doi: 10.1146/annurev.biochem.70.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizo J, Südhof TC. C2-domains, structure and function of a universal Ca2+-binding domain. J Biol Chem. 1998;273:15879–15882. doi: 10.1074/jbc.273.26.15879. [DOI] [PubMed] [Google Scholar]
- Sanchez JP, Chua NH. Arabidopsis plc1 is required for secondary responses to abscisic acid signals. Plant Cell. 2001;13:1143–1154. doi: 10.1105/tpc.13.5.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders CM, Larman MG, Parrington J, Cox LJ, Royse J, Blayney LM, Swann K, La FA. PLCζ: a sperm-specific trigger of Ca2+oscillations in eggs and embryo development. Development. 2002;129:3533–3544. doi: 10.1242/dev.129.15.3533. [DOI] [PubMed] [Google Scholar]
- Sbrissa D, Ikonomov OC, Shisheva A. PIKfyve lipid kinase is a protein kinase: downregulation of 5′-phosphoinositide product formation by autophosphorylation. Biochemistry. 2000;39:15980–15989. doi: 10.1021/bi001897f. [DOI] [PubMed] [Google Scholar]
- Schu PV, Takegawa K, Fry MJ, Stack JH, Waterfield MD, Emr SD. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260:88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- Shi J, Gonzales RA, Bhattacharyya MK. Characterization of a plasma membrane-associated phosphoinositide-specific phospholipase C from soybean. Plant J. 1995;8:381–390. doi: 10.1046/j.1365-313x.1995.08030381.x. [DOI] [PubMed] [Google Scholar]
- Sommarin M, Sandelius AS. Phosphatidylinositol and phosphatidylinositol phosphate kinases in plant plasma membranes. Biochim Biophys Acta. 1988;958:268–278. [Google Scholar]
- Song C, Hu C-D, Masago M, Kariya K-i, Yamawaki-Kataoka Y, Shibatohge M, Wu D, Satoh T, Kataoka T. Regulation of a novel human phospholipase C, PLCε, through membrane targeting by Ras. J Biol Chem. 2001;276:2752–2757. doi: 10.1074/jbc.M008324200. [DOI] [PubMed] [Google Scholar]
- Stack JH, DeWald DB, Takegawa K, Emr SD. Vesicle-mediated protein transport: regulatory interactions between the Vps15 protein kinase and the Vps34 PtdIns 3-kinase essential for protein sorting to the vacuole in yeast. J Cell Biol. 1995;129:321–334. doi: 10.1083/jcb.129.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack JH, Emr SD. Vps34p required for yeast vacuolar protein sorting is a multiple specificity kinase that exhibits both protein kinase and phosphatidylinositol-specific PI 3-kinase activities. J Biol Chem. 1994;269:31552–31562. [PubMed] [Google Scholar]
- Stahl ML, Ferenz CR, Kelleher KL, Kriz RW, Knopf JL. Sequence similarity of phospholipase C with the non-catalytic region of src. Nature. 1988;332:269–272. doi: 10.1038/332269a0. [DOI] [PubMed] [Google Scholar]
- Staxén I, Pical C, Montgomery LT, Gray JE, Hetherington AM, McAinsh MR. Abscisic acid induces phosphoinositide-specific phospholipase C-dependent oscillations in guard cell cytosolic free calcium. Proc Natl Acad Sci USA. 1999;96:1779–1784. doi: 10.1073/pnas.96.4.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert P, Wissing JB, Wagner KG. Manganese-stimulated phosphatidylinositol 4-kinase from Dunaliella parva: purification and characterization. Plant Sci. 1994;101:105–114. [Google Scholar]
- Stenmark H, Aasland R. FYVE-finger proteins: effectors of an inositol lipid. J Cell Sci. 1999;112:4175–4183. doi: 10.1242/jcs.112.23.4175. [DOI] [PubMed] [Google Scholar]
- Stenmark H, Aasland R, Toh BH, D'Arrigo A. Endosomal localization of the autoantigen EEA1 is mediated by a zinc-binding FYVE finger. J Biol Chem. 1996;271:24048–24054. doi: 10.1074/jbc.271.39.24048. [DOI] [PubMed] [Google Scholar]
- Stevenson JM, Perera IY, Boss WF. A phosphatidylinositol 4-kinase pleckstrin homology domain that binds phosphatidylinositol 4-monophosphate. J Biol Chem. 1998;273:22761–22767. doi: 10.1074/jbc.273.35.22761. [DOI] [PubMed] [Google Scholar]
- Stevenson JM, Perera IY, Heilmann I, Persson S, Boss WF. Inositol signaling and plant growth. Trends Plant Sci. 2000;5:252–258. doi: 10.1016/s1360-1385(00)01652-6. [DOI] [PubMed] [Google Scholar]
- Suh BC, Lee IS, Chae HD, Han S, Kim KT. Characterization of Mas-7-induced pore formation in SK-N-BE(2) C human neuroblastoma cells. Mol Cells. 1998a;8:162–168. [PubMed] [Google Scholar]
- Suh PG, Ryu SH, Moon KH, Suh HW, Rhee SG. Cloning and sequence of multiple forms of phospholipase C. Cell. 1988b;54:161–169. doi: 10.1016/0092-8674(88)90548-x. [DOI] [PubMed] [Google Scholar]
- Takegawa K, DeWald DB, Emr SD. Schizosaccharomyces pombeVps34p, a phosphatidylinositol-specific PI 3-kinase essential for normal cell growth and vacuole morphology. J Cell Sci. 1995;108:3745–3756. doi: 10.1242/jcs.108.12.3745. [DOI] [PubMed] [Google Scholar]
- Takeshima H, Komazaki S, Nishi M, Iino M, Kangawa K. Junctophilins: a novel family of junctional membrane complex proteins. Mol Cell. 2000;6:11–22. doi: 10.1016/s1097-2765(00)00003-4. [DOI] [PubMed] [Google Scholar]
- Taylor GS, Maehama T, Dixon JE. Myotubularin, a protein tyrosine phosphatase mutated in myotubular myopathy, dephosphorylates the lipid second messenger, phosphatidylinositol 3-phosphate. Proc Natl Acad Sci USA. 2000;97:8910–8915. doi: 10.1073/pnas.160255697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolias KF, Rameh LE, Ishihara H, Shibasaki Y, Chen J, Prestwich GD, Cantley LC, Carpenter CL. Type I phosphatidylinositol-4-phosphate 5-kinases synthesize the novel lipids phosphatidylinositol 3,5-bisphosphate and phosphatidylinositol 5-phosphate. J Biol Chem. 1998;273:18040–18046. doi: 10.1074/jbc.273.29.18040. [DOI] [PubMed] [Google Scholar]
- Tucker EB, Boss WF. Mastoparan-induced intracellular Ca2+fluxes may regulate cell-to-cell communication in plants. Plant Physiol. 1996;111:459–467. doi: 10.1104/pp.111.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Leevers SJ, Ahmadi K, Timms J, Katso R, Driscoll PC, Woscholski R, Parker PJ, Waterfield MD. Synthesis and function of 3-phosphorylated inositol lipids. Annu Rev Biochem. 2001;70:535–602. doi: 10.1146/annurev.biochem.70.1.535. [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Waterfield MD. Signaling by distinct classes of phosphoinositide 3-kinases. Exp Cell Res. 1999;253:239–254. doi: 10.1006/excr.1999.4701. [DOI] [PubMed] [Google Scholar]
- van Himbergen JAJ, ter Riet B, Meijer HJG, van den Ende H, Musgrave A, Munnik T. Mastoparan analogues stimulate phospholipase C- and phospholipased-activity in Chlamydomonas: a comparative study. J Exp Bot. 1999;50:1735–1742. [Google Scholar]
- Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis. Science. 2000;290:2114–2117. doi: 10.1126/science.290.5499.2114. [DOI] [PubMed] [Google Scholar]
- Vlahos CJ, Matter WF, Hui KY, Brown RF. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002) J Biol Chem. 1994;269:5241–5248. [PubMed] [Google Scholar]
- Volinia S, Dhand R, Vanhaesebroeck B, MacDougall LK, Stein R, Zvelebil MJ, Domin J, Panaretou C, Waterfield MD. A human phosphatidylinositol 3-kinase complex related to the yeast Vps34p-Vps15p protein sorting system. EMBO J. 1995;14:3339–3348. doi: 10.1002/j.1460-2075.1995.tb07340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker EH, Pacold ME, Perisic O, Stephens L, Hawkins PT, Wymann MP, Williams RL. Structural determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercitin, myricetin, and staurosporine. Mol Cell. 2000;6:909–919. doi: 10.1016/s1097-2765(05)00089-4. [DOI] [PubMed] [Google Scholar]
- Welters P, Takegawa K, Emr SD, Chrispeels MJ. AtVPS34, a phosphatidylinositol 3-kinase of Arabidopsis thaliana, is an essential protein with homology to a calcium-dependent lipid binding domain. Proc Natl Acad Sci USA. 1994;91:11398–11402. doi: 10.1073/pnas.91.24.11398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergren T, Dove SK, Sommarin M, Pical C. AtPIP5K1, an Arabidopsis thaliana phosphatidylinositol phosphate kinase, synthesizes PtdIns(3,4)P2 and PtdIns(4,5)P2 in vitroand is inhibited by phosphorylation. Biochem J. 2001;359:583–589. doi: 10.1042/0264-6021:3590583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergren T, Ekblad L, Jergil B, Sommarin M. Phosphatidylinositol 4-kinase associated with spinach plasma membranes: isolation and characterization of two distinct forms. Plant Physiol. 1999;121:507–516. doi: 10.1104/pp.121.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiradjaja F, Ooms LM, Whisstock JC, McColl B, Helfenbaum L, Sambrook JF, Gething M-J, Mitchell CA. The yeast inositol polyphosphate 5-phosphatase Inp54p localizes to the endoplasmic reticulum via a C-terminal hydrophobic anchoring tail. J Biol Chem. 2001;276:7643–7653. doi: 10.1074/jbc.M010471200. [DOI] [PubMed] [Google Scholar]
- Wong K, Cantley LC. Cloning and characterization of a human phosphatidylinositol 4-kinase. J Biol Chem. 1994;269:28878–28884. [PubMed] [Google Scholar]
- Woscholski R, Parker PJ. Inositol phosphatases: constructive destruction of phosphoinositides and inositol phosphates. In: Cockcroft S, editor. Biology of Phosphoinositides. Oxford: Oxford University Press; 2000. pp. 320–338. [Google Scholar]
- Wurmser AE, Gary JD, Emr SD. Phosphoinositide 3-kinases and their FYVE domain-containing effectors as regulators of vacuolar/lysosomal membrane trafficking pathways. J Biol Chem. 1999;274:9129–9132. doi: 10.1074/jbc.274.14.9129. [DOI] [PubMed] [Google Scholar]
- Wymann MP, Pirola L. Structure and function of phosphoinositide 3-kinases. Biochim Biophys Acta. 1998;1436:127–150. doi: 10.1016/s0005-2760(98)00139-8. [DOI] [PubMed] [Google Scholar]
- Xiong L, Lee B-H, Ishitani M, Lee H, Zhang C, Zhu J-K. FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev. 2001;15:1971–1984. doi: 10.1101/gad.891901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue H-W, Hosaka K, Plesch G, Müller-Röber B. Cloning of Arabidopsis thalianaphosphatidylinositol synthase and functional expression in the yeast pis mutant. Plant Mol Biol. 2000;42:757–764. doi: 10.1023/a:1006308909105. [DOI] [PubMed] [Google Scholar]
- Xue H-W, Pical C, Brearley C, Elge S, Müller-Röber B. A plant 126-kDa phosphatidylinositol 4-kinase with a novel repeat structure: cloning and functional expression in baculovirus-infected insect cells. J Biol Chem. 1999;274:5738–5745. doi: 10.1074/jbc.274.9.5738. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, DeWald DB, Boronenkov IV, Anderson RA, Emr SD, Koshland D. Novel PI(4) P 5-kinase homologue, Fab1p, essential for normal vacuole function and morphology in yeast. Mol Biol Cell. 1995;6:525–539. doi: 10.1091/mbc.6.5.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Boss WF. Regulation of phosphatidylinositol 4-kinase by the protein activator PIK-A49: activation requires phosphorylation of PIK-A49. J Biol Chem. 1994;269:3852–3857. [PubMed] [Google Scholar]
- Yang W, Burkhart W, Cavallius J, Merrick WC, Boss WF. Purification and characterization of a phosphatidylinositol 4-kinase activator in carrot cells. J Biol Chem. 1993;268:392–398. [PubMed] [Google Scholar]
- Yotsushima K, Mitsui T, Takaoka T, Hayakawa T, Igaue I. Purification and characterisation of membrane-bound inositol phospholipid-specific phospholipase C from suspension-cultured rice (Oryza sativaL.) cells: identification of a regulatory factor. Plant Physiol. 1993;102:165–172. doi: 10.1104/pp.102.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yotsushima K, Nakamura K, Mitsui T, Igaue I. Purification and characterization of phosphatidylinositol-specific phospholipase C in suspension-cultured cells of rice (Oryza sativaL.) Biosci Biotech Biochem. 1992;56:1247–1251. [Google Scholar]
- Yueh YG, Crain RC. Deflagellation of Chlamydomonas reinhardtii follows a rapid transitory accumulation of inositol 1,4,5-trisphosphate and requires Ca2+entry. J Cell Biol. 1993;123:869–875. doi: 10.1083/jcb.123.4.869. [DOI] [PMC free article] [PubMed] [Google Scholar]