Abstract
A full-length cDNA encoding a metallothionein (MT)-like polypeptide, designated GmarMT1, was identified in an expressed sequence tag collection from germinated spores of the arbuscular mycorrhizal fungus Gigaspora margarita (BEG34). The GmarMT1 gene is composed of two exons separated by an 81-bp intron. It codes for a 65-amino acid polypeptide comprising a plant type 1 MT-like N-terminal domain and a C-terminal domain that is most closely related to an as-yet-uncharacterized fungal MT. As revealed by heterologous complementation assays in yeast, GmarMT1 encodes a functional polypeptide capable of conferring increased tolerance against Cd and Cu. The GmarMT1 RNA is expressed in both presymbiotic spores and symbiotic mycelia, even in the absence of metal exposure, but is significantly less abundant in the latter stage. An opposite pattern was observed upon Cu exposure, which up-regulated GmarMT1 expression in symbiotic mycelia but not in germinated spores. Together, these data provide the first evidence, to our knowledge, for the occurrence in an arbuscular mycorrhizal fungus of a structurally novel MT that is modulated in a metal and life cycle stage-dependent manner and may afford protection against heavy metals (and other types of stress) to both partners of the endomycorrhizal symbiosis.
Contaminated soils and waters are a major threat for the environment and for human health. Within the framework of emerging bioremediation technologies, much attention has been given lately to natural biotools capable of removing, containing, or detoxifying various environmental pollutants (Gadd, 2000; Barkay and Schaefer, 2001). Basic and applied research has been mainly directed toward plants (Baker et al., 1999; Meagher, 2000; Reeves and Baker, 2000) and bacteria (Pieper and Reineke, 2000; Lloyd and Lovley, 2001; Watanabe, 2001), whereas the kingdom of fungi, which represents a very versatile microbial entity, has remained largely unexplored. The extraordinary genetic and physiological diversity of fungi—particularly the richness of species found in soil—makes them key components of almost all ecosystems, with a great potential for bioremediation purposes. Especially interesting within this kingdom are mycorrhizal fungi, which represent direct links between plants and soil and which are often needed to ensure plant survival in heavily polluted areas. Arbuscular mycorrhizal (AM) fungi (Glomeromycota, Schüßler et al., 2001), which are ubiquitous root symbionts, are particularly attractive in this regard. In natural and agricultural environments, they significantly contribute to plant growth not only by improving mineral nutrition (Harrison, 1999; Gadkar et al., 2001), but also by protecting plants against a variety of biotic and abiotic stresses, including heavy metal (HM) stress (Leyval et al., 1997). Several studies indicate that an increased HM tolerance and other beneficial effects are conferred to the host plant by the AM symbiosis (Shetty et al., 1994; Hildebrandt et al., 1999; Tonin et al., 2001). High levels of mycorrhizal colonization have accordingly been documented in agricultural soils with different kinds of HM contamination (Weissenhorn et al., 1995a, 1995b). Although AM fungi are presently the object of an increasing attention and many international projects are aimed at exploiting their ecological potentialities for bioremediation purposes, no information about the molecular HM tolerance mechanisms operating in these organisms is currently available.
A complex network of transport, chelation, and extracellular and intracellular sequestration processes operates to maintain essential metal (e.g. Cu) homeostasis and to minimize the damage caused by nonessential metals (e.g. Cd; Sanità di Toppi et al., 2002). A variety of membrane transporters controlling the trafficking of both types of metal ions (e.g. ATP-binding cassette-type transporters and the so-called cation diffusion facilitators) have been identified recently in plants and microorganisms (Clemens, 2001). By comparison, only a fairly limited number of intracellular metal chelators have been identified so far. Citrate and His have been shown to act as major Ni chelators, whereas phytochelatins (PCs) and metallothioneins (MTs) are of primary importance in buffering the intracellular concentration of free thiophilic metal ions, such as Cu, Zn, and Cd (Clemens, 2001; Sanità di Toppi et al., 2002, and refs. therein). The latter two are Cys-rich polypeptides that chelate metal ions through the formation of tetrahedrally coordinated metal-thiolate clusters. However, at variance with PCs, which are synthesized through the ribosome-independent polymerization of reduced glutathione-derived γ-glutamyl-Cys units, MTs are gene-encoded (Robinson et al., 1993; Rauser, 1999; Cobbett, 2000).
Fungal MTs and PCs have been characterized almost exclusively in yeasts. Brewer's yeast (Saccharomyces cerevisiae) contains a multigene MT (CUP1) family, which is mainly involved in Cu detoxification (Hamer et al., 1985; Ecker et al., 1986), and the single-copy MT gene CRS5 (Jensen et al., 1996), but no canonical PC synthase gene (Mewes et al., 1997). On the other hand, only one sequence annotated as a putative MT (accession no. CAB57404) has been reported so far in fission yeast (Schizosaccharomyces pombe), which, however, produces HM-chelating PC peptides through a plant-like PC-synthase enzyme (SpPCS; Clemens et al., 1999; Ha et al., 1999). Budding yeast cup1 (Hamer et al., 1985; Ecker et al., 1986) and fission yeast SpPCS (Clemens et al., 1999) disruptants are both metal hypersensitive, as if distinct HM detoxification strategies were predominantly used by these two organisms. Some fungi, however, are able to synthesize both MTs and PCs. This is the case of Candida glabrata, for example, which produces MTs when exposed to toxic concentrations of Cu but produces mainly PCs in response to a Cd stress (Mehra et al., 1988, 1989). Only a few data are available for filamentous fungi (Munger et al., 1987; Singh and Ashby, 1998; Averbeck et al., 2001).
The occurrence of MTs, PCs, or both, in mycorrhizal fungi is still a matter of debate (Leyval et al., 1997). There are a few reports on MT-like sequences, obtained in the frame of expressed sequence tag projects. cDNAs coding for putative MT-like polypeptides have been identified in the ectomycorrhizal basidiomycete Pisolithus tinctorius (Voiblet et al., 2001) and in the AM fungi Gigaspora rosea (Stommel et al., 2001) and Glomus intraradices (accession no. BI451899; M.J. Harrison, personal communication). However, the metal sequestration capacity of these three predicted polypeptides and, thus, their actual MT-like nature have not yet been determined. Here, we describe the identification and functional characterization of GmarMT1, a MT-encoding gene from the AM fungus Gigaspora margarita. Besides documenting for the first time, to our knowledge, the existence of an MT-based HM detoxification machinery in AM fungi, we also show that the GmarMT1 mRNA is differentially expressed in symbiotic versus presymbiotic life cycle stages of G. margarita. These data raise interesting questions as to the possible role of MTs in metal detoxification and/or more general stress responses in AM fungi.
RESULTS
Identification and Sequence Analysis of the GmarMT1 cDNA
A full-length cDNA similar to previously described MTs was identified in an expressed sequence tag collection from G. margarita germinated spores. This cDNA, named GmarMT1, codes for a 6.9-kD polypeptide with a predicted pI of 8.6. As shown in Figure 1, other diagnostic features of GmarMT1 besides its small size are the presence of 14 cysteines, with only one aromatic residue (Tyr-23), on a total of 65 amino acids. Furthermore, all but two of such Cys residues are part of the characteristic MT motif C-X-C (underlined in Fig. 1). Also shown in Figure 1, is the genomic sequence of GmarMT1, obtained from PCR experiments carried out with oligonucleotide primers designed on the 5′- and 3′-untranslated regions of the GmarMT1 cDNA. As revealed by comparison between genomic and cDNA sequences, a small (81-bp) intron, delimited by consensus splice junctions, is present in the GmarMT1 gene at position 200.
Figure 1.
Nucleotide and deduced amino acid sequence of GmarMT1. The intron sequence is shown in lowercase letters; Cys residues are in bold; C-X-C motifs and GmarMT1-specific primers (MT1/MT2) are underlined.
Interestingly, the GmarMT1 polypeptide could not be assigned to any of the six, presently classified subfamilies of fungal MTs (Binz and Kägi, 1999). Additional insight into the putative G. margarita MT was, thus, obtained from an extended similarity search using the sequence of the predicted GmarMT1 polypeptide as a query. Quite surprisingly, all but one of the 20 most similar sequences identified by this analysis were type 1 MTs from plants. Besides a partial polypeptide sequence from the related fungus G. rosea (Stommel et al., 2001), the only other non-plant GmarMT1 homolog within this set of sequences is a predicted (but as-yet-unclassified) MT-like polypeptide from the basidiomycete Agaricus bisporus (Eastwood et al., 2001). As revealed by the multiple sequence alignment reported in Figure 2, the GmarMT1 polypeptide can be subdivided more specifically into two distinct N- and C-terminal domains, each bearing three C-X-C motifs and separated by an approximately 28-amino acid spacer. The first of such domains best fits the sequence pattern of the corresponding domain of plant type 1 MTs even at amino acid residues other than the cysteines. The C-terminal domain and part of the predicted spacer, instead, most closely resemble the corresponding regions of the Agaricus MT-like polypeptide (Fig. 2).
Figure 2.
Alignment of GmarMT1 with MT-like polypeptides from other organisms. The polypeptide sequence of GmarMT1 (boxed) was aligned with the partial sequence of a predicted polypeptide from G. rosea (Stoffel et al., 2001) and with seven of the best scoring sequences identified by BLAST analysis: Arabidopsis MT1A (National Center for Biotechnology Information [NCBI] accession no. P43392; Yeh et al., 1995; Zhou and Goldsbrough, 1995); Arabidopsis MT1C (NCBI accession no. Q38804; Zhou and Goldsbrough, 1995) canola (Brassica napus) MT-like (NCBI accession no. P43402; Buchanan-Wollaston, 1994); rice (Oryza sativa) MT1 (NCBI accession no. Q40633; Hsieh et al., 1995); barley (Hordeum vulgare) MT1 (NCBI accession no. P26571; Okumura et al., 1991); Mimulus guttatus MT1 (NCBI accession no. P20238; de Miranda et al., 1990); and A. bisporus MT like (NCBI accession no. CAB85689; Eastwood et al., 2001). Amino acid residues that are identical in at least seven of the nine sequences are shown on a black background, and residues shared by all three fungal sequences are shaded gray; these two types of conserved residues are indicated with uppercase and lowercase letters, respectively, in the consensus pattern shown below the alignment. Gaps introduced to optimize the alignment are indicated by dots.
Functional Complementation Assays
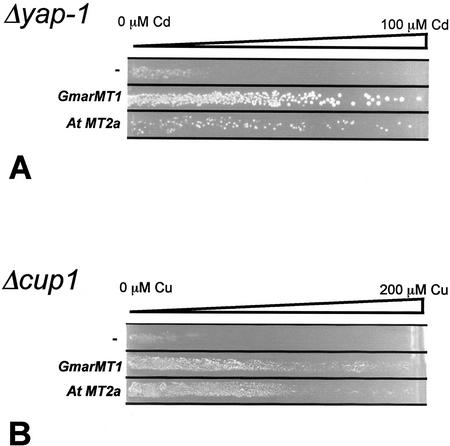
yAP-1 is a transcription factor related to the mammalian AP-1 complex that positively controls various genes involved in HM and, more generally, oxidative stress tolerance in yeast (Kuge and Jones, 1994; Toone et al., 2001). Although yeast MT genes are not direct targets of such an activator, Δyap-1 mutants are particularly sensitive to Cd and are, thus, suitable to highlight tolerant phenotypes induced by exogenous cDNAs (Wu et al., 1993). To test whether GmarMT1 expression confers an increased tolerance to HMs, we transformed a Δyap-1 yeast mutant strain with the complete open reading frame of the GmarMT1 cDNA placed under the control of a constitutive yeast promoter provided by the expression vector pFL61 (Minet et al., 1992). The empty pFL61 vector and the same vector carrying the MT2a cDNA from Arabidopsis (Zhou and Goldsbrough, 1994, 1995; A. Bolchi and S. Ottonello, unpublished data) were used as negative and positive controls, respectively. The various yeast transformants were streaked onto synthetic dextrose (SD)-agar plates containing a linear 0 to 100 μm gradient of CdSO4. As shown in Figure 3A, cells carrying the pFL61 vector alone grew only a very short distance into the Cd concentration gradient. In contrast, both positive control (MT2a) and GmarMT1 transformed yeast cells grew up to the highest Cd concentration. No difference between the three yeast transformants (pFL61-GmarMT1, pFL61-MT2a, and pFL61) was observed when similar assays were conducted on CuSO4 or NiSO4 gradients (both ranging from 0 to 3 mm, data not shown). The HM protection capacity of GmarMT1, evidenced by the above experiments, was further verified by similar but functionally more direct complementation assays carried out in a yeast mutant (Δcup1) that is highly sensitive to HMs (especially Cu) because of the complete disruption of the MT-encoding locus CUP1 (Hamer et al., 1985; Ecker et al., 1986). Confirming and extending the results obtained with the Δyap-1 mutant, an increased tolerance to CuSO4 (0–200 μm gradient; Fig. 3B) and to CdSO4 (0–10 μm gradient; data not shown) was conferred to Δcup1 cells by the GmarMT1 cDNA. Once again, no differential sensitivity was observed when the three yeast transformants were plated onto 0 to 3 mm NiSO4 gradients (data not shown).
Figure 3.
Increased HM tolerance conferred by GmarMT1 in metal-hypersensitive yeast mutants. Δyap-1 (A) or Δcup1 (B) yeast mutants harboring the pFL61-GmarMT1 plasmid (GmarMT1), the positive control plasmid pFL61-Mt2a (AtMt2a), or the empty pFL61 vector (−) were grown on SD-agar (−uracil) plates with the indicated linear gradients of CdSO4 (A) or CuSO4 (B).
GmarMT1 mRNA Expression Analysis
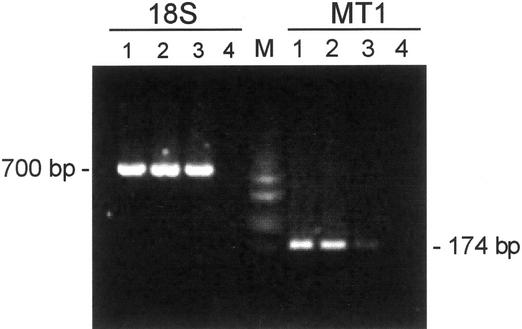
GmarMT1 mRNA expression levels were next analyzed by reverse transcriptase (RT)-PCR. Sequence-specific amplification primers designed on the GmarMT1 sequence (MT1/MT2; underlined in Fig. 1) were initially tested on total DNA extracted from the host plant white clover (Trifolium repens). The negative results obtained from such control amplifications (data not shown) allowed us to exclude any cross-hybridization with the plant genome. Total RNA extracted from quiescent spores, germinated spores, and mycorrhizal roots was then reverse-transcribed and used for RT-PCR analysis. The amount of cDNA obtained from different fungal samples was first quantified by amplifying a small aliquot with fungus-specific 18S rRNA primers (R1/R2) that did not recognize any sequence in the plant genome (data not shown). Balanced amounts of each cDNA sample were finally amplified with GmarMT1-specific oligonucleotide primers. As shown in Figure 4, an amplified fragment of the expected size (174 bp) was obtained from all cDNA samples, thus, indicating that the GmarMT1 gene is expressed in all three stages of the G. margarita life cycle. The amount of amplified product, however, was considerably higher (7- to 8-fold) in quiescent and germinated spores than in symbiotic mycelia (Fig. 4). Such a difference in GmarMT1 mRNA abundance was reproducibly observed regardless of the extent of AM colonization, which in different experiments varied from 30% to 60% of colonized segments.
Figure 4.
RT-PCR analysis of GmarMT1 mRNA levels in presymbiotic and symbiotic life cycle stages of G. margarita. Balanced amounts of cDNA from quiescent spores (lane 1), germinated spores (lane 2), or mycorrhizal roots (lane 3) were amplified with GmarMT1-specific oligonucleotide primers (MT1). 18S rDNA amplicons, obtained from parallel control reactions and loaded in the same order as above, were used as internal standards (18S). The sizes of GmarMT1 and 18S rDNA amplicons are indicated. No cDNA template was added to reaction mixtures run in lane 4; DNA size markers (HaeIII-digested pUC18) were run in lane M.
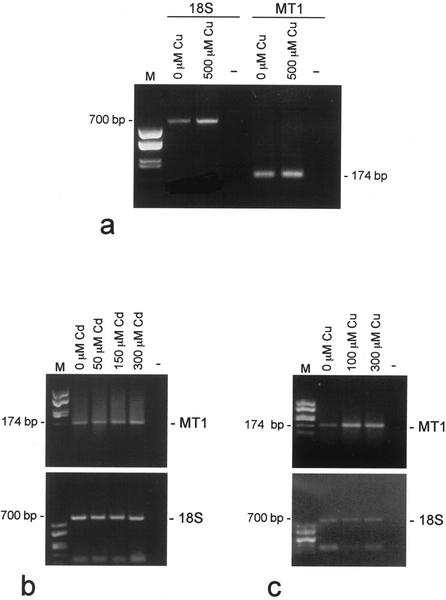
The metal responsiveness of GmarMT1 was then investigated by measuring mRNA levels in germinated spores and symbiotic mycelia exposed for 24 h to increasing concentrations of either CuSO4 or CdSO4. Data reported in Figure 5a show that Cu did not exert any appreciable effect on GmarMT1 expression in germinated spores, and the same result was obtained after Cd exposure (data not shown). GmarMT1 mRNA levels in symbiotic mycelia were similarly not affected by a Cd treatment (Fig. 5b). When exposed to Cu, instead, the same symbiotic mycelia exhibited a significant increase in GmarMT1 mRNA abundance (approximately 4-fold) even at the lowest Cu concentration (100 μm; Fig. 5c).
Figure 5.
GmarMT1 mRNA expression levels after metal exposure. Balanced amounts of total RNA extracted from germinated spores (a) and mycorrhizal roots (b and c), either untreated or exposed to the indicated concentrations of CuSO4 or CdSO4, were reverse-transcribed and amplified with GmarMT1-specific primers (MT1). The 18S rRNA was used as an internal calibration standard for all reactions (18S). The sizes and migration positions of GmarMT1 (MT1) and 18S rDNA (18S) amplicons are indicated. Template RNA was omitted from reaction mixtures shown in lane −; DNA size markers (HaeIII-digested pUC18) were run in lane M.
DISCUSSION
MTs are ubiquitous Cys-rich metalloproteins with a characteristically low (or null) aromatic amino acid content (Robinson et al., 1993). The possible occurrence of MT-like polypeptides in AM fungi was first suggested by electron energy loss spectroscopic analyses, which revealed the presence of polyphosphate granules containing S and N together with Al, Fe, Ti, and B in the fungal cytoplasm (Turnau et al., 1993). However, neither the potential metal-binding polypeptides contained in such granules nor the genes coding for them were identified thereafter. The results of this work thus provide the first direct evidence for the occurrence of a functional MT gene in the genome of an AM fungus.
GmarMT1 Codes for a Functional MT
Standardized protocols for the genetic transformation of Glomalean fungi are not yet available (Harrier and Millam, 2001). We, thus, resorted to functional complementation assays in yeast (Thiele et al., 1986; Minet et al., 1992; Zhou and Goldsbrough, 1994) to unambiguously demonstrate that the GmarMT1 gene product can sequester metal ions, thereby conferring in vivo protection against HMs. Two distinct metal hypersensitive mutants (Δyap-1 and Δcup1) and three different HMs (Cd, Cu, and Ni) were used for these assays. Only a Cd tolerance phenotype was observed in GmarMT1-transformed, Δyap-1 mutants. The Δyap-1 mutant is impaired in reduced glutathione/glutaredoxin and thioredoxin but not in MT synthesis (Kuge and Jones, 1994; Grant, 2001; Toone et al., 2001). Therefore, it is likely endowed with a residual tolerance against Cu (the preferred ligand of yeast MTs) that is much too high to allow the detection of an increased Cu tolerance phenotype conferred by a transgene. Such a phenotype, in fact, was observed in an MT-deficient, Δcup1 mutant (Hamer et al., 1985; Ecker et al., 1986), which upon GmarMT1 transformation became more resistant to both Cu and Cd. As furthermore expected for a thionein metal-binding protein, GmarMT1 failed to confer to either mutant an increased resistance against Ni, a metal with an exceedingly low affinity for thiolate groups.
GmarMT1 Identifies a Novel Class of Fungal MT-Like Polypeptides
The G. margarita MT displays fairly unique sequence features that distinguish it from known homologs of either fungal or plant origin (Binz and Kägi, 1999). Its N-terminal domain is most similar to type 1 MTs from plants (Rauser, 1999), whereas a much closer resemblance with a predicted fungal MT from A. bisporus (Eastwood et al., 2001) is observed at the level of its C-terminal domain. The GmarMT1 polypeptide, thus, appears to comprise two evolutionarily distinct domains, one of which is more closely related to plant MTs than to any known fungal MT. Because of this peculiar structural organization and region-specific similarity with an as-yet-unclassified fungal MT, GmarMT1 could not be assigned to any of the presently categorized fungal MT subfamilies (Binz and Kägi, 1999). Additional proof of the authentic AM fungal origin of GmarMT1 was provided by the isolation of its corresponding genomic sequence. This also revealed the presence of a short intron separating the region coding for the predicted C-terminal domain (exon 2) from the rest of the coding sequence (exon 1). Besides the N-terminal domain, the first exon of GmarMT1 encodes a putative intervening spacer that comprises the only aromatic residue found in the entire protein. Interestingly, a similar two-exon organization with a centrally located intron and a conserved Tyr residue within the spacer is also found in the genes for Arabidopsis (and other plants) MTs. At variance with GmarMT1, however, Arabidopsis exon 1 only encodes for the N-terminal domain, whereas the spacer and the C-terminal domain are both encoded by exon 2.
In many eukaryotes, including Brewer's yeast, MT genes are organized into tandemly repeated multigene clusters (Hamer et al., 1985; Mehra et al., 1990). So far, the extremely limited amount of genomic DNA that can be extracted from the spores of G. margarita (an obligate biotroph) has unfortunately precluded a direct examination of MT gene multiplicity in this organism.
Differential Expression of the GmarMT1 mRNA
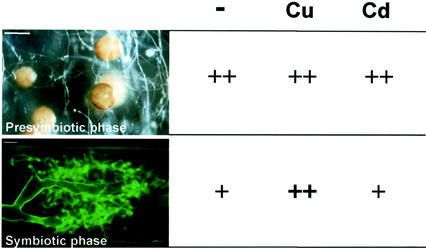
GmarMT1 is expressed in presymbiotic spores and in symbiotic mycelia. Lower expression levels, however, were consistently measured in mycorrhizal roots (sampled at different colonization densities) compared with spores (Fig. 6). Considering that MTs are known to respond to a variety of stresses besides HM exposure (Liu and Thiele, 1997; Kondoh et al., 2001), one may imagine that such an expression pattern somehow reflects a generalized stress situation occurring within the “free-living” spores of an obligate biotrophic organism. This situation, in which the fungus suffers from both C and N starvation (Bago et al., 1999), contrasts with the more favorable conditions experienced inside the roots by the symbiotic mycelium (Smith, 1979). Interestingly, Glc starvation is known to activate MT (CUP1) genes in Brewer's yeast (Tamai et al., 1994), and various other metal-unrelated stresses (e.g. wounding, pathogen infection, and senescence) similarly up-regulate MT expression in both plants and fungi (Buchanan-Wollaston, 1994; Coupe et al., 1995; Choi et al., 1996; Butt et al., 1998; Averbeck et al., 2001). Two features shared by these seemingly diverse stress conditions are an increased production of activated oxygen species and a frequent occurrence of cell death events. Because of their limited life span (and compromised metabolic situation), it is likely that similar events also occur in germinating AM spores. After germination, their hyphae elongate for about 15 to 20 d, after which many morphological modifications occur, including cytoplasmic retraction, production of septa, development of lateral branches, and swollen apices. All of these events correlate with a compromised metabolic situation and the arrest in hyphal growth (Bianciotto and Bonfante, 1999).
Figure 6.
Scheme of the differential expression and HM-induction of GmarMT1 in two phases of the G. margarita life cycle. The steps illustrated are germinating spores of G. margarita as seen under the stereomicroscope (presymbiotic phase; bar = 300 μm) and an arbuscule visualized by fluorescence microscopy (symbiotic phase; bar = 5 μm). The GmarMT1 mRNA is selectively up-regulated by Cu ions in the symbiotic mycelium.
In keeping with this view, oxidative stress response genes, such as those coding for superoxide dismutase and glutathione S-transferase, were found to be expressed at high levels in germinated spores of G. margarita (Lanfranco et al., 2000; L. Lanfranco, unpublished data). A generalized, and preexisting activation of stress response genes in spores might also explain the lack of metal-induced GmarMT1 mRNA up-regulation in this particular stage of the fungus life cycle. Metal-dependent up-regulation, in fact, was only observed in symbiotic mycelia (Fig. 6), as if a basal GmarMT1 expression state, such as the one associated to this more favorable growth condition, were a necessary prerequisite for responding to the stress ensuing from metal exposure.
An alternative explanation is that GmarMT1 up-regulation in symbiotic mycelia might be caused indirectly by a secondary stress resulting from metal-induced damage to the host root.
Similarly to what has been reported for other unicellular and multicellular fungi relying on MTs to control Cu homeostasis (Mehra et al., 1988; Thorvaldsen et al., 1995; Liu and Thiele, 1997), GmarMT1 responded to Cu, but not to Cd exposure (Fig. 6). This selectivity of response, which contrasts with the joint protective effect observed upon constitutive overexpression of GmarMT1 in metal-hypersensitive yeast mutants, likely reflects the Cu specificity of the trans-acting factors regulating GmarMT1 expression in its natural host. It will, thus, be interesting to identify putative metal (and other stress) response elements in the GmarMT1 promoter and to test their role in controlling MT expression by either homologous or heterologous reporter gene experiments.
MTs and HM Tolerance in AM Fungi
In both Brewer's yeast (Hamer et al., 1985; Ecker et al., 1986; Jensen et al., 1996) and C. glabrata (Mehra et al., 1988; Thorvaldsen et al., 1995), MTs have been shown to be the chief effectors of Cu tolerance. By contrast, a plant-like PC synthase (Clemens et al., 1999; Ha et al., 1999) and a P1-type ATPase (Riggle and Kumamoto, 2000) seem to play major roles in the detoxification of Cu in fission yeast and C. albicans, respectively. This variety of metal protection strategies, which sometimes coexist in a single organism, raises a question as to the actual role of GmarMT1 in Cu detoxification and to the possible existence in AM fungi of additional HM protection mechanisms, besides MTs. A second, more general question, prompted by the obligate symbiotic and endomycorrhizal nature of AM fungi and by the metal protective effects that are thought to be exerted by plant MTs (Murphy and Taiz, 1995; van Hoof et al., 2001), is whether mycobiont-derived MTs, such as GmarMT1, can confer an increased HM tolerance to host plants. What is becoming increasingly clear, however, is that mycorrhizal fungi contribute not only to plant nutrition, but also to the uptake and detoxification of various environmental pollutants, including HMs (Perotto and Martino, 2001). Understanding the molecular bases of HM tolerance in this symbiotic system will, thus, also aid the selection of the most effective AM isolates (Tonin et al., 2001; Turnau et al., 2001) and plant-fungus combinations for bioremediation and soil protection purposes.
MATERIALS AND METHODS
Biological Materials
Spores of Gigaspora margarita (BEG 34) were collected from pot cultures of mycorrhizal white clover (Trifolium repens) and sterilized with 4% (w/v) chloramine T/0.004% (w/v) streptomycin, plus four rounds of sonication. To induce germination, spores were incubated in water at 24°C for 2 weeks. Mycorrhizal roots were obtained from pot cultures of white clover. The percentage of total infected root lengths was evaluated with the grid-line intersect method of Giovannetti and Mosse (1980). Mycorrhizal plants were collected from pot cultures, and cleaned roots were submerged for 24 h in water solutions containing increasing amounts of CuSO4 (100 and 300 μm) or CdSO4 (50, 150, and 300 μm). Germinated spores were exposed for the same length of time to 500 μm CuSO4 or 300 μm CdSO4 dissolved in water. The G. margarita (germinated spores) cDNA library, with an estimated complexity of 50,000 recombinant clones, was constructed into the λTriplEx II vector using the SMART cDNA library construction kit (CLONTECH Laboratories, Palo Alto, CA). Individual clones were randomly selected and sequenced (Lanfranco et al., 2000).
PCR Amplifications
Genomic DNA was extracted from spores, mycorrhizal roots, or leaves as described by Lanfranco et al. (1999). PCR reactions were carried out in a final volume of 50 μL containing 10 mm Tris-HCl, pH 8.3, 50 mm KCl, 1.1 mm MgCl2, 0.01% (w/v) gelatin, 200 μm of each dNTP, 1 μm of each primer, 50 to 100 ng of genomic DNA, and 2 units of REDTaq DNA polymerase (Sigma, St. Louis). The PCR program was as follows: 95°C for 3 min (1 cycle), 92°C for 45 s, 45 s annealing at temperatures indicated below, 72°C for 45 s (30 cycles), and 72°C for 5 min (1 cycle).
The oligonucleotide primers MTG1 (5′-ATCAAATAAGTATATCTCTC) and MTG2 (5′-TTTGAACCCAATATACAACG) were employed for genomic GmarMT1 amplifications at an annealing temperature of 48°C. The primers MT1 (5′-TGTGGTTCCGCTTGTCAATG) and MT2 (5′-TTTACAGTTGCCTTTGGTGC) were used to amplify a specific subregion of the GmarMT1 coding sequence at an annealing temperature of 60°C. R1 (5′-GAATTTCTACCTTCTGGGGAAC) and R2 (5′-TCGACCATTCAAAAGAATAGCCTG), two oligonucleotide primers specifically recognizing the G. margarita 18S ribosomal gene were used at an annealing temperature of 60°C. PCR products were separated on 1.2% (w/v) agarose gels and visualized by ethidium bromide staining. Negative controls for all PCR experiments consisted of reaction mixtures from which template DNA was omitted.
Cloning and Sequencing
The PCR product amplified from genomic DNA was extracted and purified from agarose gels using the QIAEX II Gel Extraction Kit (Qiagen USA, Valencia, CA) and directly cloned into the pGEM-T vector (Promega, Madison, WI). XL-2 Blue ultracompetent cells (Stratagene, La Jolla, CA) were transformed and plated onto selective medium following the manufacturer's instructions. Plasmid DNAs were prepared with the Qiagen Plasmid Mini Kit. DNA sequences were determined by MWG Biotech (Ebersberg, Germany) using T7 and Sp6 primers. The genomic sequence of GmarMT1 has been submitted to the DNA data bank of Japan/EMBL/GenBank databases under accession number AJ421527. DNA sequence analyses were performed with the PC/gene software (IntelliGenetics, Mountain View, CA) and the BLASTX software available through the NCBI.
RT-PCR Analyses
RNA was extracted from about 100 spores or 100 mg (fresh weight) of mycorrhizal roots using the SV Total RNA Isolation System kit (Promega). Before RT-PCR experiments, RNA was treated (30 min, 37°C) with RNase-free DNase (Amersham, Buckinghamshire, UK), extracted once with phenol:chloroform:isoamylalcohol (25:24:1, v/v), precipitated with 2 m LiCl, and resuspended in 20 μL of sterile diethyl pyrocarbonate-treated, double-distilled water. All of the above RNA samples were routinely checked for DNA contamination by RT-PCR analyses conducted with the 18S rRNA universal primers NS1/NS2 (White et al., 1990), in the presence or absence of RT. When appropriate, reverse transcription reactions were performed in a final volume of 20 μL containing 50 mm Tris-HCl, pH 8.3, 75 mm KCl, 3 mm MgCl2, 10 mm dithiothreitol, 1 μm dNTPS, 1 unit of RNaseOUT (Invitrogen, Carlsbad, CA), 500 ng of the ribosomal primer NS2 (White et al., 1990), 1 μL of total RNA, and 200 units of SuperScript II RT (Invitrogen). After 50 min at 42°C, a PCR mix was added consisting of 10 mm Tris-HCl, pH 8.3, 50 mm KCl, 1.1 mm MgCl2, 0.01% (w/v) gelatin, 200 μm of each dNTP, 500 ng of the NS1 primer, and 2 units of REDTaq DNA polymerase (Sigma). The same PCR mix, but with both NS primers, was added to parallel samples not subjected to reverse transcription. The PCR program was as follows: 95°C for 3 min (1 cycle), 92°C for 45 s, 50°C for 45 s, 72°C for 45 s (30 cycles), and 72°C for 5 min (1 cycle).
An essentially identical RT-PCR protocol, except for the use of random primers (Invitrogen) and a higher amount (8 μL) of total RNA in the initial reverse transcription step, was used for GmarMT1 mRNA determinations. The amount of reverse-transcribable RNA contained in the different samples was first determined by RT-PCR using oligonucleotide primers (R1/R2; see above) specific for the G. margarita 18S rRNA. Internally balanced amounts of RNA and the sequence-specific primers MT1/MT2 (see above) were then used to amplify the GmarMT1 cDNA. PCR reactions (annealing temperature of 60°C) were allowed to proceed for different number of cycles to determine the exponential phase of amplification.
The One-Step RT-PCR kit (Qiagen USA) was used for RT-PCR experiments conducted on RNA extracted from HM-treated samples. Reactions were carried out in a final volume of 25 μL containing 5 μL of 5× buffer, 5 μL of Q-solution, 400 μm dNTPs, 0.6 μm of each primer, 1 μL of One-Step RT-PCR enzyme mix, and 0.1 to 2 μL of total RNA. Samples were incubated for 30 min at 50°C, followed by a 15 min incubation at 95°C. Amplification reactions (92°C for 45 s, 60°C for 45 s, and 72°C for 45 s) were run for a maximum of 26 cycles.
RT-PCR products were quantified by densitometric analysis of ethidium bromide-stained bands (Multi-Analyst/PC software, Bio-Rad, Hercules, CA); RT-PCR experiments were conducted in triplicate on three independent samples.
Yeast Complementation Assays
The full-length GmarMT1 sequence was first amplified under standard PCR conditions using the NotI site-containing primers MTA (5′-TGACATTGCGGCCGCAAAATGTGCCAAAATTGTA) and MTB (5′-ACTTCGAGCGGCCGCAGAAATTAGCATTTACAGT) at an annealing temperature of 50°C. The resulting product was digested with NotI and cloned into the dephosphorylated NotI site of the yeast expression vector pFL61 (Minet et al., 1992). The pFL61-GmarMT1 construct, the empty pFL61 vector, and a positive control pFL61 derivative carrying the cDNA for the Arabidopsis MT2a MT (A. Bolchi and S. Ottonello, unpublished data) were then transformed (Rose et al., 1990) into chemically competent Δyap-1 (strain WYT; Kuge and Jones, 1994) or Δcup1 (strain DTY 113; Tamai et al., 1994) yeast mutants. The various transformants were grown at 30°C for 3 d on selective (−uracil) SD-agar medium, before being transferred to SD-agar plates containing linear concentration gradients of Cd, Cu, or Ni (Cunningham et al., 1986). Gradient plate assays, performed at the HM concentrations specified in “Results,” were conducted in triplicate.
ACKNOWLEDGMENTS
We thank Shusuke Kuge (Department of Microbiology, University of Tokyo) and Dennis Thiele (Department of Biological Chemistry, University of Michigan) for Δyap-1 and Δcup1 yeast mutant strains and Michéle Minet (Centre National de la Recherche Scientifique, Gif-sur-Yvette) for the gift of the pFL61 plasmid and the yeast expressible Arabidopsis cDNA library. We thank Riccardo Percudani for assistance with sequence analysis and Chiara Chiapponi and Roberta Ruotolo (Dipartimento di Biochimica e Biologia Molecolare, Universita di Parma) for help with the setting up of HM gradient plate assays.
Footnotes
This work was supported by the European Union GENOMYCA project (grant no. QLK5–CT–2000–01319) and by the Italian Project Biotechnology (subproject 2: Environmental Biotechnology), National Research Council of Italy.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.003525.
LITERATURE CITED
- Averbeck NB, Borghouts C, Hamann A, Specke V, Osiewacz HD. Molecular control of copper homeostasis in filamentous fungi: increased expression of a metallothionein gene during aging of Podospora anserina. Mol Gen Genet. 2001;264:604–612. doi: 10.1007/s004380000346. [DOI] [PubMed] [Google Scholar]
- Bago B, Pfeffer PE, Douds DD, Brouillette J, Bécard G, Shachar-Hill Y. Carbon metabolism in spores of the arbuscular mycorrhizal fungus Glomus intraradicesas revealed by nuclear magnetic resonance spectroscopy. Plant Physiol. 1999;121:263–271. doi: 10.1104/pp.121.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker AJM, McGrath SP, Reeves RD, Smith JAC. Metal hyperaccumulator plants: a review of the ecology and physiology of a biological resource for phytoremediation of metal-polluted soils. In: Terry N, Banuelos GS, editors. Phytoremediation of Contaminated Soil and Water. Boca Raton, FL: CRC Press; 1999. pp. 85–107. [Google Scholar]
- Barkay T, Schaefer J. Metal and radionuclide bioremediation: issues, considerations and potentials. Curr Opin Microbiol. 2001;4:318–323. doi: 10.1016/s1369-5274(00)00210-1. [DOI] [PubMed] [Google Scholar]
- Bianciotto V, Bonfante P. Presymbiotic versus symbiotic phase in arbuscular endomycorrhizal fungi. In: Varma A, Hock B, editors. Mycorrhiza. Ed 2. Berlin: Springer-Verlag; 1999. pp. 229–251. [Google Scholar]
- Binz PA, Kägi JHR. Metallothionein: molecular evolution and classification. In: Klaassen C, editor. Metallothionein IV. Basel: Birkhäuser Verlag; 1999. pp. 7–13. [Google Scholar]
- Buchanan-Wollaston V. Isolation of cDNA clones for genes that are expressed during leaf senescence in Brassica napus. Plant Physiol. 1994;105:839–846. doi: 10.1104/pp.105.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt A, Mousley C, Morris K, Beynon J, Can C, Holub E, Greenberg JT, Buchanan-Wollaston V. Differential expression of a senescence-enhanced metallothionein gene in Arabidopsis in response to isolates of Peronospora parasitica and Pseudomonas syringae. Plant J. 1998;16:209–221. doi: 10.1046/j.1365-313x.1998.00286.x. [DOI] [PubMed] [Google Scholar]
- Choi D, Kim HM, Yun HK, Park JA, Kim WT, Bok SH. Molecular cloning of a metallothionein-like gene from Nicotiana glutinosaL. and its induction by wounding and tobacco mosaic virus infection. Plant Physiol. 1996;112:353–359. doi: 10.1104/pp.112.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens S. Molecular mechanisms of plant metal tolerance and homeostasis. Planta. 2001;212:475–486. doi: 10.1007/s004250000458. [DOI] [PubMed] [Google Scholar]
- Clemens S, Kim EJ, Neumann D, Schroeder JI. Tolerance to toxic metals by a gene family of phytochelatin synthases from plants and yeast. EMBO J. 1999;18:3325–3333. doi: 10.1093/emboj/18.12.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbett CS. Phytochelatins and their roles in heavy metal detoxification. Plant Physiol. 2000;123:825–832. doi: 10.1104/pp.123.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coupe SA, Taylor JE, Roberts JA. Characterization of an mRNA encoding a metallothionein-like protein that accumulates during ethylene-promoted abscission of Sambucus nigraL. leaflets. Planta. 1995;197:442–447. doi: 10.1007/BF00196665. [DOI] [PubMed] [Google Scholar]
- Cunningham RP, Saporito SM, Spitzer SG, Weiss B. Endonuclease IV (nfo) mutant of Escherichia coli. J Bacteriol. 1986;168:1120–1127. doi: 10.1128/jb.168.3.1120-1127.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Miranda JR, Thomas MA, Thurman DA, Tomsett AB. Metallothionein genes from the flowering plant Mimulus guttatus. FEBS Lett. 1990;260:277–280. doi: 10.1016/0014-5793(90)80122-y. [DOI] [PubMed] [Google Scholar]
- Eastwood DC, Kingsnorth CS, Jones HE, Burton KS. Genes with increased transcript levels following harvest of the sporophore of Agaricus bisporushave multiple physiological roles. Mycol Res. 2001;105:1223–1230. [Google Scholar]
- Ecker DJ, Butt TR, Sternberg EJ, Neeper MP, Debouck C, Gorman JA, Crooke ST. Yeast metallothionein function in metal ion detoxification. J Biol Chem. 1986;261:16895–16900. [PubMed] [Google Scholar]
- Gadd GM. Bioremedial potential of microbial mechanisms of metal mobilization and immobilization. Curr Opin Biotechnol. 2000;11:271–279. doi: 10.1016/s0958-1669(00)00095-1. [DOI] [PubMed] [Google Scholar]
- Gadkar V, David-Schwartz, Kunik T, Kapulnik Y. Arbuscular mycorrhizal fungal colonization: factors involved in host recognition. Plant Physiol. 2001;127:1493–1499. [PMC free article] [PubMed] [Google Scholar]
- Giovannetti M, Mosse B. An evaluation of techniques for measuring vesicular arbuscular mycorrhizal infection in roots. New Phytol. 1980;84:489–500. [Google Scholar]
- Grant CM. Role of the glutathione/glutaredoxin and thioredoxin systems in yeast growth and response to stress conditions. Mol Microbiol. 2001;39:533–541. doi: 10.1046/j.1365-2958.2001.02283.x. [DOI] [PubMed] [Google Scholar]
- Ha SB, Smith AP, Howden R, Dietrich WM, Bugg S, O'Connell MJ, Goldsbrough PB, Cobbett CS. Phytochelatin synthase genes from Arabidopsis and the yeast Schizosaccharomyces pombe. Plant Cell. 1999;11:1153–1164. doi: 10.1105/tpc.11.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer DH, Thiele DJ, Lemontt JE. Function and autoregulation of yeast copperthionein. Science. 1985;228:685–690. doi: 10.1126/science.3887570. [DOI] [PubMed] [Google Scholar]
- Harrier LA, Millam S. Biolistic transformation of arbuscular mycorrhizal fungi: progress and perspectives. Mol Biotechnol. 2001;18:25–33. doi: 10.1385/MB:18:1:25. [DOI] [PubMed] [Google Scholar]
- Harrison MJ. Molecular and cellular aspects of the arbuscular mycorrhizal symbiosis. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:361–389. doi: 10.1146/annurev.arplant.50.1.361. [DOI] [PubMed] [Google Scholar]
- Hildebrandt U, Kaldorf M, Bothe H. The Zn violet and its colonization by arbuscular mycorrhizal fungi. J Plant Physiol. 1999;154:709–717. [Google Scholar]
- Hsieh HM, Liu WK, Huang PC. A novel stress-inducible metallothionein-like gene from rice. Plant Mol Biol. 1995;28:381–389. doi: 10.1007/BF00020388. [DOI] [PubMed] [Google Scholar]
- Jensen LT, Howard WR, Strain JJ, Winge DR, Culotta VC. Enhanced effectiveness of copper ion buffering by CUP1 metallothionein compared with CRS5 metallothionein in Saccharomyces cerevisiae. J Biol Chem. 1996;271:18514–18519. doi: 10.1074/jbc.271.31.18514. [DOI] [PubMed] [Google Scholar]
- Kondoh M, Inoue Y, Atagi S, Futakawa N, Higashimoto M, Sato M. Specific induction of metallothionein synthesis by mitochondrial oxidative stress. Life Sci. 2001;69:2137–2146. doi: 10.1016/s0024-3205(01)01294-2. [DOI] [PubMed] [Google Scholar]
- Kuge S, Jones N. YAP1 dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiaeto oxidative stress by hydroperoxides. EMBO J. 1994;13:655–664. doi: 10.1002/j.1460-2075.1994.tb06304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfranco L, Gabella S, Bonfante P. EST as a useful tool for studying gene expression in arbuscular mycorrhizal fungi. In: Weber H, Imhof S, Zeuske D, editors. Abstract and Papers of the Third International Congress on Symbiosis. Germany: Philipps University of Marburg; 2000. pp. 108–114. [Google Scholar]
- Lanfranco L, Vallino M, Bonfante P. Differential expression of chitin synthase genes in the arbuscular mycorrhizal fungus Gigaspora margarita. New Phytol. 1999;142:347–354. [Google Scholar]
- Leyval C, Turnau K, Haselwandter K. Effects of heavy metal pollution on mycorrhizal colonization and function: physiological, ecological and applied aspects. Mycorrhiza. 1997;7:139–153. [Google Scholar]
- Liu XD, Thiele DJ. Yeast metallothionein gene expression in response to metals and oxidative stress. Methods. 1997;11:289–299. doi: 10.1006/meth.1996.0423. [DOI] [PubMed] [Google Scholar]
- Lloyd JR, Lovley DR. Microbial detoxification of metals and radionuclides. Curr Opin Biotechnol. 2001;12:248–253. doi: 10.1016/s0958-1669(00)00207-x. [DOI] [PubMed] [Google Scholar]
- Meagher RB. Phytoremediation of toxic elemental and organic pollutants. Curr Opin Plant Biol. 2000;3:153–162. doi: 10.1016/s1369-5266(99)00054-0. [DOI] [PubMed] [Google Scholar]
- Mehra RK, Garey JR, Butt TR, Gray WR, Winge DR. Candida glabratametallothioneins: cloning and sequence of the genes and characterization of proteins. J Biol Chem. 1989;264:19747–19753. [PubMed] [Google Scholar]
- Mehra RK, Garey JR, Winge DR. Selective and tandem amplification of a member of the metallothionein gene family in Candida glabrata. J Biol Chem. 1990;265:6369–6375. [PubMed] [Google Scholar]
- Mehra RK, Tarbet EB, Gray WR, Winge DR. Metal-specific synthesis of two metallothioneins and γ-glutamil peptides in Candida glabrata. Proc Natl Acad Sci USA. 1988;85:8815–8819. doi: 10.1073/pnas.85.23.8815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mewes HW, Albermann K, Rahr M, Frishman D, Gleissner A, Hani J, Heumann K, Kleine K, Maierl A, Oliver SG et al. Overview of the yeast genome. Nature Suppl. 1997;387:7–65. doi: 10.1038/42755. [DOI] [PubMed] [Google Scholar]
- Minet M, Dufour ME, Lacroute F. Complementation of mutants by Arabidopsis thalianacDNA. Plant J. 1992;32:417–422. doi: 10.1111/j.1365-313x.1992.00417.x. [DOI] [PubMed] [Google Scholar]
- Munger K, Germann UA, Lerch K. The Neurospora crassametallothionein gene: regulation of expression and chromosomal location. J Biol Chem. 1987;262:7363–7367. [PubMed] [Google Scholar]
- Murphy A, Taiz L. Comparison of metallothionein gene expression and non-protein thiols in ten Arabidopsisecotypes. Plant Physiol. 1995;109:945–954. doi: 10.1104/pp.109.3.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura N, Nishizawa NK, Umehara Y, Mori S. An iron deficiency-specific cDNA from barley roots having two homologous cysteine-rich MT domains. Plant Mol Biol. 1991;17:531–533. doi: 10.1007/BF00040651. [DOI] [PubMed] [Google Scholar]
- Perotto S, Martino E. Molecular and cellular mechanisms of heavy metal tolerance in mycorrhizal fungi: what perspectives for bioremediation? Minerva Biotecnol. 2001;13:55–63. [Google Scholar]
- Pieper DH, Reineke W. Engineering bacteria for bioremediation. Curr Opin Biotechnol. 2000;11:262–270. doi: 10.1016/s0958-1669(00)00094-x. [DOI] [PubMed] [Google Scholar]
- Rauser WE. Structure and function of metal chelators produced by plants: the case of organic acids, amino acids, phytin and metallothioneins. Cell Biochem Biophys. 1999;31:18–48. doi: 10.1007/BF02738153. [DOI] [PubMed] [Google Scholar]
- Reeves RD, Baker AJM. Metal-accumulating plants. In: Raskin I, Ensley BD, editors. Phytoremediation of Toxic Metals: Using Plants to Clean Up the Environment. New York: John Wiley & Sons; 2000. pp. 193–229. [Google Scholar]
- Riggle PJ, Kumamoto CA. Role of a Candida albicansP1-type ATPase in resistance to copper and silver ion toxicity. J Bacteriol. 2000;182:4899–4905. doi: 10.1128/jb.182.17.4899-4905.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NJ, Tommey AM, Kuske C, Jackson PJ. Plant metallothioneins. Biochem J. 1993;295:1–10. doi: 10.1042/bj2950001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. pp. 122–123. [Google Scholar]
- Sanità di Toppi L, Prasad MNV, Ottonello S. Metal chelating peptides and proteins in plants. In: Prasad MNV, Strzalka K, editors. Physiology and Biochemistry of Metal Toxicity and Tolerance in Plants. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2002. pp. 59–94. [Google Scholar]
- Schüßler A, Schwarzott D, Walker C. A new phylum, the Glomeromycota: phylopenyanol evolution. Mycol Res. 2001;105:1413–1421. [Google Scholar]
- Shetty KG, Banks MK, Hetrick BA, Schwab AP. Biological characterization of a southeast Kansas mining site. Water Air Soil Pollut. 1994;78:169–177. [Google Scholar]
- Singh G, Ashby AM. Cloning of the mating type loci from Pyrenopeziza brassicaereveals the presence of a novel mating type gene within a discomycete MAT 1–2 locus encoding a putative metallothionein-like protein. Mol Microbiol. 1998;30:799–806. doi: 10.1046/j.1365-2958.1998.01112.x. [DOI] [PubMed] [Google Scholar]
- Smith DC. From extracellular to intracellular: the establishment of a symbiosis. In: Richmond MH, Smith DC, editors. The Cell as a Habitat. London: Royal Society; 1979. pp. 115–130. [DOI] [PubMed] [Google Scholar]
- Stommel M, Mann P, Franken P. EST-library construction using spore RNA of the arbuscular mycorrhizal fungus Gigaspora rosea. Mycorrhiza. 2001;10:281–285. [Google Scholar]
- Tamai KT, Liu X, Silar P, Sosinowski T, Thiele DJ. Heat shock transcription factor activates yeast metallothionein gene expression in response to heat and glucose starvation via distinct signalling pathways. Mol Cell Biol. 1994;14:8155–8165. doi: 10.1128/mcb.14.12.8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele DJ, Walling MJ, Hamer DH. Mammalian metallothionein is functional in yeast. Science. 1986;231:854–856. doi: 10.1126/science.3080806. [DOI] [PubMed] [Google Scholar]
- Thorvaldsen JL, Mehra RK, Yu W, Sewell AK, Winge DR. Analysis of copper-induced metallothionein expression using autonomously replicating plasmids in Candida glabrata. Yeast. 1995;11:1501–1511. doi: 10.1002/yea.320111505. [DOI] [PubMed] [Google Scholar]
- Tonin C, Vandenkoornhuyse P, Joner EJ, Straczek J, Leyval C. Assessment of arbuscular mycorrhizal fungi diversity in the rizosphere of Viola calaminariaand effect of these fungi on heavy metal uptake by clover. Mycorrhiza. 2001;10:161–168. [Google Scholar]
- Toone WM, Morgan BA, Jones N. Redox control of AP-1-like factors in yeast and beyond. Oncogene. 2001;20:2336–2346. doi: 10.1038/sj.onc.1204384. [DOI] [PubMed] [Google Scholar]
- Turnau K, Kottke I, Oberwinkler F. Element localization in mycorrhizal roots of Pteridium aquilinum(L.) Kuhn collected from experimental plots with cadmium dust. New Phytol. 1993;123:313–324. [Google Scholar]
- Turnau K, Ryszka P, Gianinazzi-Pearson V, van Tuinen D. Identification of arbuscular mycorrhizal fungi in soils and roots of plants colonizing zinc wastes in southern Poland. Mycorrhiza. 2001;10:169–174. [Google Scholar]
- van Hoof NALM, Hassinen VH, Hakvoort HWJ, Ballintijn KF, Shact H, Verkleij JAC, Ernst WHO, Karenlampi SO, Tervahauta AI. Enhanced copper tolerance in Silene vulgaris(Moench) Garcke populations from copper mines is associated with increased transcript levels of a 2b-type metallothionein gene. Plant Physiol. 2001;126:1519–1526. doi: 10.1104/pp.126.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voiblet C, Duplessis S, Encelot N, Martin F. Identification of symbiosis-regulated genes in Eucalyptus globulus-Pisolithus tinctoriusectomycorrhiza by differential hybridization of arrayed cDNA. Plant J. 2001;25:181–191. doi: 10.1046/j.1365-313x.2001.00953.x. [DOI] [PubMed] [Google Scholar]
- Watanabe K. Microorganisms relevant to bioremediation. Curr Opin Biotechnol. 2001;12:237–241. doi: 10.1016/s0958-1669(00)00205-6. [DOI] [PubMed] [Google Scholar]
- Weissenhorn I, Leyval C, Berthelin J. Bioavailability of heavy metals and arbuscular mycorrhiza in a soil polluted by atmospheric deposition from a smelter. Biol Fertil Soils. 1995a;19:22–28. [Google Scholar]
- Weissenhorn I, Mench M, Leyval C. Bioavailability of heavy metals and arbuscular mycorrhiza in a sewage sludge amended sandy soil. Soil Biol Biochem. 1995b;27:287–296. [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols. A Guide to Methods and Applications. San Diego: Academic Press; 1990. pp. 315–322. [Google Scholar]
- Wu A, Wemmie JA, Edgington NP, Goebl M, Guevara JL, Moye-Rowley WS. Yeast bZip proteins mediate pleiotropic drug and metal resistance. J Biol Chem. 1993;268:18850–18858. [PubMed] [Google Scholar]
- Yeh SC, Hsieh HM, Huang PC. Transcripts of metallothionein genes in Arabidopsis thaliana. DNA Seq. 1995;5:141–144. doi: 10.3109/10425179509029353. [DOI] [PubMed] [Google Scholar]
- Zhou J, Goldsbrough PB. Functional homologs of fungal metallothionein genes from Arabidopsis. Plant Cell. 1994;6:875–884. doi: 10.1105/tpc.6.6.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Goldsbrough PB. Structure, organization and expression of the metallothionein gene family in Arabidopsis. Mol Gen Genet. 1995;248:318–328. doi: 10.1007/BF02191599. [DOI] [PubMed] [Google Scholar]