Abstract
The flowering process in grapevine (Vitis vinifera) takes place in buds and extends for two consecutive growing seasons. To understand the genetic and molecular mechanisms underlying this process, we have characterized grapevine bud development, cloned the grapevine FLORICAULA/LEAFY (FLO/LFY) ortholog, VFL, and analyzed its expression patterns during vegetative and reproductive development. Flowering induction takes place during the first season. Upon induction, the shoot apical meristem begins to produce lateral meristems that will give rise to either inflorescences or tendrils. During the second season, after a winter dormancy period, buds reactivate and inflorescence meristems give rise to flower meristems. VFL is expressed in lateral meristems that give rise to inflorescence and flower meristems, consistent with a role in reproductive development. Furthermore, VFL is also detected in other meristematic regions such as the vegetative shoot apical meristem and the lateral meristems that will give rise to tendrils. VFL is also expressed in leaf primordia and in growing leaf margins until later stages of development. Accumulation of VFL transcripts in cell-proliferating regions suggests a role for VFL not only in flower meristem specification, but also in the maintenance of indeterminacy before the differentiation of derivatives of the apical meristem: flowers, leaves, or tendrils.
Genetic and molecular approaches in snapdragon (Antirrhinum majus) and Arabidopsis have allowed the identification of some of the key genes regulating flowering induction and reproductive development (Pidkowich et al., 1999; Simpson et al., 1999; Araki, 2001). Among them, FLORICAULA (FLO) from snapdragon (Carpenter and Coen, 1990; Coen et al., 1990) and its Arabidopsis ortholog, LEAFY (LFY; Schultz and Haughn, 1991; Weigel et al., 1992), seem to have a central role in the specification of flower meristem identity. Inactivation of FLO causes the development of indeterminate shoots in place of flowers (Coen et al., 1990), whereas lfy mutants show partial flower-to-shoot conversions (Weigel et al., 1992). Constitutive expression of LFY is sufficient to promote flower initiation and development from shoot apical and axillary meristems in Arabidopsis and has similar effects in other dicot and monocot species (Weigel and Nilsson, 1995; He et al., 2001; Peña et al., 2001), suggesting a conservation of LFY function across long phylogenetic distances within angiosperms.
Despite FLO/LFY sequence conservation among distantly related species (Frohlich and Parker, 2000), significant differences are emerging in relation to their expression patterns that could indicate the existence of a functional divergence. For instance, although in snapdragon expression of FLO is specific of the reproductive phase (bracts and young floral meristems; Coen et al., 1990), low levels of expression of FLO/LFY orthologs have been detected in leaf primordia during vegetative growth in Arabidopsis, tobacco (Nicotiana tabacum), Impatiens sp., pea (Pisum sativum), petunia (Petunia hybrida), and tomato (Lycopersicon esculentum; Kelly et al., 1995; Blázquez et al., 1997; Bradley et al., 1997; Hofer et al., 1997; Pouteau et al., 1997; Souer et al., 1998; Molinero-Rosales et al., 1999). Consistent with this evidence, a role in leaf development has been proposed for pea and tomato FLO/LFY orthologs based on the morphological alterations shown in the leaves of loss-of-function mutants. In monocots, expression of FLO/LFY orthologs further deviates from that of their dicots counterparts. In rice (Oryza sativa), the RLF gene is expressed in young panicles but not in mature florets or leaves, thus, making it unlikely its involvement in flower meristem initiation (Kyozuka et al., 1998). In Lolium temulentum, LtLFY is expressed later than the SQUA/AP1 ortholog, LtMADS2 (other genes involved in the initiation of floral development; Gocal et al., 2001). In contrast, FLO/LFY precedes SQUA/AP1 expression and LFY is required for AP1 up-regulation (Liljegren et al., 1999; Wagner et al., 1999).
Regulation of flowering in woody perennials shows remarkable differences with respect to herbaceous species, i.e. long juvenile phases, winter bud dormancy and the need of two consecutive growing seasons for flowering. Despite the interest of these processes for the management and improvement of woody species, very little is known about their underlying molecular mechanisms. FLO/LFY orthologs have been cloned and characterized in several woody species such as eucalyptus (Eucalyptus globulus; Southerton et al., 1998), Monterey pine (Pinus radiata; Mellerowicz et al., 1998; Mouradov et al., 1998), Populus trichocarpa (Rottmann et al., 2000), and kiwifruit (Actinidia deliciosa; Walton et al., 2001). However, its specific role in the characteristic features of tree reproductive development is still being elucidated. Furthermore, partial or total FLO/LFY-like sequences have been reported from other basal angiosperms and gymnosperms (Frohlich and Meyerowitz, 1997; Frohlich and Parker, 2000), although in these cases functional information is not available.
We are interested in the reproductive development of grapevine (Vitis vinifera), an important fruit crop in temperate regions that belongs to the family of Vitaceae, a basal family within the eudicots (Judd et al., 1999). Grapevine plants grown from seeds go through 2 to 5 years of juvenile phase before they start to flower. In grapevine, flowering requires two growing seasons: flowering is induced in latent buds during the summer but flower initiation and development takes place the following spring (Mullins et al., 1992). Grapevine, similar to other Vitaceae species, produces either inflorescences or tendrils opposite to leaves. Interestingly, inflorescences and tendrils can substitute for each other depending on the environmental conditions or hormonal treatments (Srinivasan and Mullins, 1976, 1981), with gibberellins inhibiting inflorescence and promoting tendril development (Boss and Thomas, 2002). To understand the genetic and molecular mechanisms underlying the flowering process in grapevine, we have analyzed by scanning electron microscopy (SEM) the development of buds during two growing seasons and have related their development with the temporal and spatial expression patterns of VFL, the grapevine FLO/LFY ortholog gene. Cloning and characterization of VFL indicate that it is a single-copy gene, as in other angiosperm species. In situ hybridization experiments shows that VFL is expressed in lateral meristems irrespective of meristematic fate. These results could suggest that VFL is involved not only in flower initiation and development but also in inflorescence, leaf, and tendril development.
RESULTS
Flower Initiation in Grapevine
In grapevine, bud development, from the time of bud initiation until the development of flowers, extends during two growing seasons (Mullins et al., 1992). This process has been described using SEM in the grapevine var Shiraz (Srinivasan and Mullins, 1976, 1981). We have used a similar strategy to monitor the development of grapevine var Tempranillo buds during two consecutive seasons. Buds collected at each stage were both analyzed by SEM and used to study VFL expression by in situ hybridization.
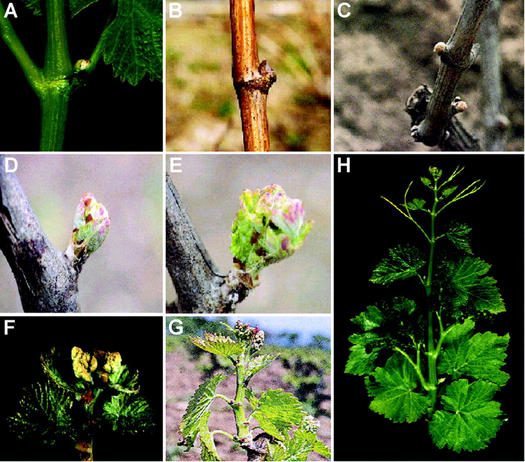
During the first season, buds are first detectable around March in the axils of current year's young leaves (Fig. 1A). They are formed by several shoot apical meristems (SAM) protected by bracts. The earliest-formed meristem usually develops as a lateral shoot during this first season. The others remain latent and one of them will give rise to next season's crop of grapes. The whole process described below takes place within this bud (Fig. 1, A–E).
Figure 1.
Stages of development in grapevine. A, Newly formed latent bud in the axil of a young leaf. B, Winter bud; phenological stage A according to Baggiolini (1952). C, Swelling bud; phenological stage B. D, Sprouting bud; phenological stage C. E, Phenological stage D. F, Outgrowing shoot; phenological stage E. G, The inflorescences are clearly visible and separated; phenological stage G. H, General view of a growing cane bearing inflorescences with developing flowers at phenological stage H.
During the first months of development (March–May; Fig. 1A), the SAM goes through a vegetative phase producing three to four leaf primordia with distichous phyllotaxis (Fig. 2A). A pair of scales flanks each leaf primordium. Around May to June, the SAM begins to produce lateral meristems opposite to leaf primordia (Fig. 2B). Under normal conditions, the first two to three lateral meristems will give rise to inflorescences and the rest to tendrils (Fig. 2, C and D; Srinivasan and Mullins, 1976, 1981). The SAM continues to produce two consecutive nodes containing opposed leaf primordia and lateral meristems, which alternate with one node bearing a solitary leaf primordium (Fig. 2D; see also Figs. 1H and 6). Inflorescence meristems grow rapidly during the summer (June–August). First, a bract is formed at the region farthest from the apex (Fig. 2D). Then, the inflorescence meristem splits into two meristems (not shown) and each new inflorescence branch meristem begins to produce additional branch meristems subtended by bracts in a spiral phyllotaxy (Fig. 2C). During this first season, there is no internode elongation, so at the end of summer, the bud encloses a compressed shoot with inflorescence meristems and developing tendrils and leaves (Fig. 2D). These buds are protected from desiccation and freezing by bracts, scales, and epidermal hairs and enter dormancy around September (Fig. 1B).
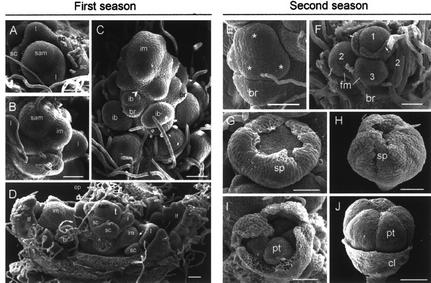
Figure 2.
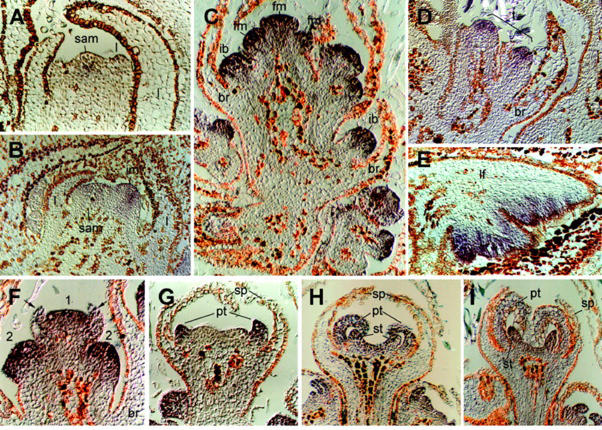
Development of a grapevine var Tempranillo bud and its derivatives as revealed by SEM. A through D, First season; E through J, second season. A, Detail of an April latent bud. The vegetative shoot apical meristem is forming leaf primordia flanked by scales (sc) in spiral phyllotaxis. B, Detail of a June bud. The shoot apical meristem has undergone flowering transition and begins to form lateral inflorescence meristem (im) opposite to leaf primordia. C, Inflorescence meristem in August, around the end of the first season. Notice the spiral phyllotaxis of inflorescence branch meristem (ib). One bract (br) subtends each branch. Only some of the ib and br are indicated. D, General view of a July bud showing the derivatives formed by the SAM (sam) during the 1st year. At this stage, the bud encloses developing leaves (lf), inflorescence meristems (im), newly formed leaf primordia (l), and tendril primordia (t). E, Detail of an inflorescence branch in a bud of phenological stage B–C (second season). The inflorescence branch meristem has divided into three to four flower meristems (asterisks). F, Flower meristems derived from an inflorescence branch in a bud of phenological stage C–D. The terminal flower meristem, labeled as 1, is more advanced in development than flanking 2 and 3. G, Developing flower in a bud of phenological stage E. The sepals (sp) grow to enclose the inner part of the flower. H and I, Developing flower at the end of stage E. In I, sepal primordia have been partially removed to show the petal primordia (pt). J, Flower from phenological stage G shoots. The petals have overgrown the calyx (cl). ep, Epidermal hair. All bars represent 50 μm.
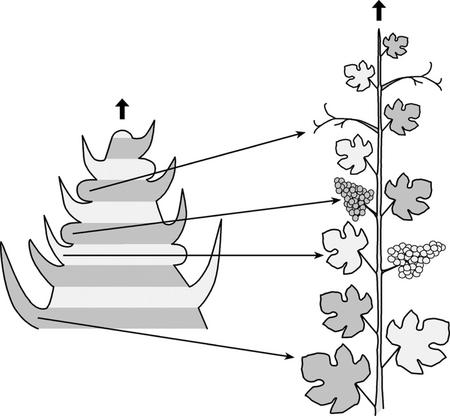
Figure 6.
Bud derivatives in grapevine. Left, Schematic representation of a latent bud during the first season showing the phyllotaxis of meristems and primordia at this stage. Right, derivatives formed from those meristems and primordia during the second season. Lateral meristems giving rise to inflorescences or tendrils are indistinguishable in morphology and position at the time they are formed.
During the second season, bud growth resumes when the environmental conditions become permissive, around February or March (Fig. 1C). Upon reactivation, the SAM produces further leaf and tendril primordia. The inflorescence branch meristems form additional inflorescence meristems in a spiral phyllotaxis until mid-March. At the end of March, each racemose inflorescence is formed by many branches that prefigure the conical bunch of grapes. At this stage, each branch meristem divides into a cluster of three to four flower meristems arranged as a dicasium. The terminal flower develops first, then the lateral ones develop, and the basal-most develops the latest (Fig. 2, E and F). Flower development takes place during April to May (Fig. 1, D and E), when the bud swells and shoot internodes begin to elongate. Flower meristems sequentially form sepals, petals and stamens, and carpels. Sepal primordia form as a ring of tissue (Fig. 2G) that grows to enclose the flower bud (Fig. 2H). Afterward, petal and stamen primordia develop (Fig. 2I). Petals become fused together by the interlocking of epidermal cells at the petal margins and grow out of the sepal cover (Fig. 2J). At anthesis, petals cannot separate and they get detached at the base so that the corolla is lifted up and thrown off by the expansion of the filaments of the stamens (not shown, Srinivasan and Mullins, 1981; Weberling, 1989).
Cloning of the Grapevine FLO/LFY Ortholog Gene
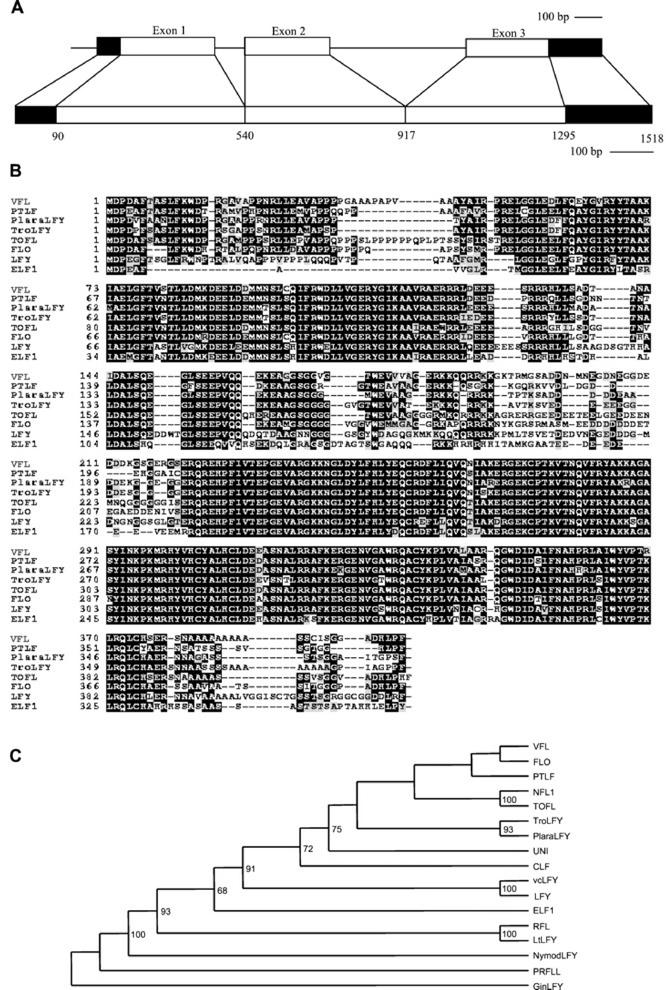
To analyze how bud development relates to the function of the FLO/LFY grapevine ortholog, we have cloned VFL. The VFL cDNA was isolated by a 3′/5′-RACE strategy. The longest (1,518 bp) cDNA clone (GenBank accession no. AF450278) has a 1,206-bp open reading frame preceded by a 5′-untranslated region of 89 bp. Considerable heterogeneity in length was found in the 3′-untranslated region revealing the use of four different polyadenylation sites at positions 1,393, 1,453, 1,484, and 1,518, respectively. Amplification of the corresponding genomic sequences by PCR showed that the VFL gene contains two introns of 153 and 662 bp (Fig. 3A) at positions conserved with those described in others species (Frohlich and Meyerowitz, 1997). Genomic DNA-blot hybridization analysis using either a 3′ or a 5′ probe under high stringency conditions detected a single hybridizing restriction fragment (data not shown). These results suggest the existence of a single VFL gene in the grapevine genome.
Figure 3.
The VFL gene and sequence comparison to FLO/LFY-like proteins. A, Genomic organization of VFL (top) and VFL cDNA (bottom). B, The deduced amino acid sequence of VFL was compared with (accession nos. in parentheses): PTLF from Populus balsamifera (U93196); PlaraLFY from Platanus racemosa (AF106842); TroLFY from Trochodendron aralioides (AF230078); TOFL from tomato (AF197934); FLO from snapdragon (M55525); LFY from Arabidopsis (M91208); and ELF1 from eucalyptus (AF34806). Black boxes indicate identical amino acids, shaded boxes similar residues, and dashed lines gaps introduced to optimize the alignment. Sequences were aligned using the ClustalW program. C, Phylogenetic relationship among FLO/LFY-like proteins. The protein sequences shown in B are included, together with: GinLFY from ginkgo (Ginkgo biloba; AF108228); PRFLL from Monterey pine (U92008); NymodLFY from Nymphaea odorata (AF105110); LtLFY from L. temulentum (AF321273); RFL from rice (AB005620); vcLFY from violet cress (Jonopsidium acaule; AF184589); CFL from cucumber (Cucumis sativus; AF059320); UNI from pea (AF010190); and NFL1 from tobacco (U16172). Bootstrap support values are indicated when over 50.
The 1,206-bp open reading frame is predicted to encode a 402-amino acid protein. This predicted protein aligns well with the sequences of other FLO/LFY-like proteins (Fig. 3B). The VFL protein shares two highly conserved regions with all the FLO/LFY proteins. Furthermore, it shares with other angiosperm FLO/LFY proteins specific regions such as an amino-terminal Pro-rich region (10 Pro residues between positions 14 and 50), a short putative Leu zipper (residues 84, 91, and 98), a basic region formed by a core of Arg and Lys residues from position 180 to 192, and an acidic region (nine aspartic/glutamic residues between positions 197 and 213). The Pro-rich, the acidic, and the basic regions are not so well conserved in gymnosperm FLO/LFY-like proteins.
To determine the evolutionary relationships between VFL and other FLO/LFY-like proteins, a phylogenetic tree was constructed using the predicted protein sequences (Fig. 3C). The general phylogenetic relationships found in this tree are consistent with the currently accepted species phylogenies (Judd et al., 1999): The VFL protein is more closely related to other FLO/LFY dicot proteins than to their monocot counterparts or to gymnosperm FLO/LFY-like proteins. Within the dicots, the highest level of identity was found between VFL and FLO/LFY ortholog proteins from P. racemosa (66% identity), T. aralioides (66% identity), P. balsamifera (65%), snapdragon (63%), and several Solanaceae spp. However, the high number of amino acid substitutions precluded a more accurate positioning of grapevine within this group.
Expression of VFL during Grapevine Development
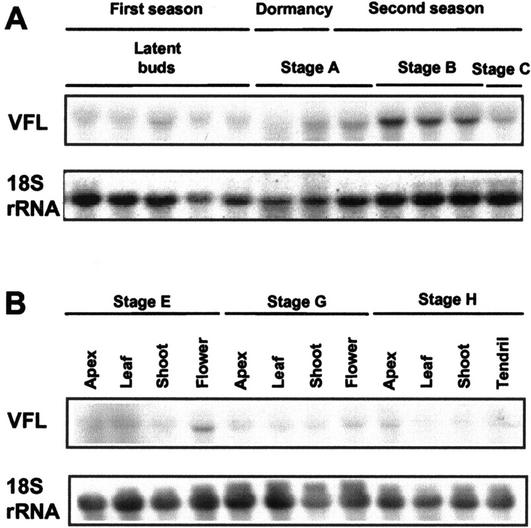
To determine the involvement of VFL in grapevine development, we have studied its temporal and spatial patterns of expression in buds and vegetative and reproductive structures. The expression patterns of VFL were first studied by RNA-blot hybridization. RNA levels of VFL were analyzed in axillary buds collected at equivalent branch positions during two consecutive growing seasons (Fig. 4A). This study revealed that VFL mRNA was already detected in latent buds during the first season (Fig. 4A, latent buds, lane 3), when inflorescence meristems are being initiated. Expression was barely detectable in dormant buds (Fig. 4A, stage A, lane 1). During the second season, expression was already detected in advanced stage A (Fig. 4A, stage A, lanes 2–3) and increased significantly in stages B and C, when the buds start swelling (Fig. 4A, stages B and C). At these stages, the inflorescence meristems are dividing to generate new branch meristems and flower meristems. From stage E onward, when the shoot is growing out, VFL expression was analyzed in separate organs (Fig. 4B). Expression of VFL was mainly detected in flowers of stages E and G (Fig. 4B). When the branches elongate, during stage H, VFL expression could also be detected in the shoot apex (Fig. 4B).
Figure 4.
Expression of VFL during grapevine development. A, VFL expression in latent buds (from June to August) in the first growing season, in winter buds (phenological stage A) and in buds from phenological stages B and C during the spring of the second growing season (see “Materials and Methods” for further details). B, VFL expression in different tissues during cane development (phenological stages E, G, and H). For each lane, 25 μg of total RNA was loaded, blotted, and hybridized with a VFL probe. Filters were also hybridized with 18S rRNA as a quantitative control of loading and blotting.
In situ hybridizations were carried out to localize the VFL transcripts during bud development. Expression of VFL was detected in meristematic tissues throughout development. In agreement with the RNA-blot hybridization results, the mRNA levels detected varied during the year and were maximum at the time of flowering induction during the first growing season and at the time of bud reactivation and flower initiation during the second growing season. During the first season, VFL mRNA was detected as early as April at very low levels in the vegetative SAM and at higher levels in leaf primordia (Fig. 5A). Expression was maintained in the leaf margins until later stages of leaf development (Fig. 5E). Around May to June, when the SAM begins to produce lateral meristems (Fig. 2B), VFL levels rose and a strong signal was detected in the dome of those meristems that would become either inflorescences or tendrils (Fig. 5B). From July to August, VFL was expressed in the inflorescence branch meristems but not in the subtending bract primordia as illustrated in Figure 5C for second season inflorescences. VFL also accumulated in the apex of developing tendrils (Fig. 5D).
Figure 5.
VFL expression patterns during bud development. A, Vegetative bud comparable with that in Figure 2A. VFL accumulates in the inner layers of the SAM and in leaf primordia. B, June bud during floral transition similar to that in Figure 2B. VFL is expressed in the SAM and in the lateral meristem that will develop an inflorescence. C, Inflorescence of a bud of phenological stage B-C, second season, showing accumulation of VFL in inflorescence branches and in newly formed flower meristems. D, Tendril primordia during the second season. VFL accumulates in the apical region of the tendril that is likely to have meristematic activity. E, Developing leaf. VFL accumulates at the growing tips of the leaf. F, Close up of an inflorescence branch comparable with that in Figure 2F where flower meristems have just formed. VFL is strongly expressed in newly formed flower meristems and begins to disappear at the regions where sepal primordia form (arrows). G, Flower comparable with that in Figure 2G. VFL is not detected in sepal primordia but accumulates in the inner part of the flower preferentially in petal primordia. H, Flower comparable with those in Figure 2, H and I. I, Flower corresponding to a stage slightly earlier than the one shown in Figure 2J. VFL is detected at very low levels in petals and at higher levels in stamens. VFL mRNA is absent from the bracts throughout development. Nomenclature is the same as in Figure 2.
During the second season, VFL transcripts were already detectable in stage A buds (Fig. 1B) in the inflorescence branch meristems (Fig. 5C). However, the expression dramatically increased in the newly formed flower meristems (stages B and C buds; Fig. 5, C and F). VFL was also expressed in developing floral organs but at lower levels than in inflorescence or flower meristems. When sepal primordia began to grow (Fig. 2G), VFL expression became restricted to the inner part of the flower meristem that will form petals, stamens, and carpel (Fig. 5G). Later, VFL was expressed in petal primordia until the petal fuse and, at lower levels, in stamen primordia (Fig. 5, H and I).
DISCUSSION
The basic function of FLO/LFY floral meristem identity genes seems to be conserved in several annual dicot species based on the common aspects of the corresponding mutant phenotypes (Hofer et al., 1997; Souer et al., 1998; Molinero-Rosales et al., 1999). However, their role in monocots and basal dicots is not so well established. Moreover, the role of FLO/LFY-like genes in woody perennial species has only started to be established due to the difficulties of the genetic analysis and transgenic approaches in those species. Although the available information suggests that overexpression of LFY is sufficient to promote the conversion of shoots into flowers in species like Populus spp. (Weigel and Nilsson, 1995) and Citrus spp. (Peña et al., 2001), the role of the endogenous FLO/LFY homologs and their function during meristem development are poorly understood.
As a first step to analyze flowering induction and flower initiation in grapevine, we have characterized bud development during this process, cloned the grapevine FLO/LFY ortholog gene and analyzed its expression during vegetative and reproductive development. cDNA cloning and genomic DNA-blot hybridization studies indicate that VFL is a single-copy gene in grapevine as it has been reported for other diploid angiosperms (Frohlich and Parker, 2000). The number and position of introns found in VFL are identical to those reported for other FLO/LFY-like genes (Frohlich and Meyerowitz, 1997). No alternative splicing sites for the first intron were detected in VFL, in contrast to what has been described for FLO and LFY (Coen et al., 1990; Weigel et al., 1992). Furthermore, analysis of the deduced VFL protein shows the presence of the typical motives found in most FLO/LFY-like proteins (Frohlich and Meyerowitz, 1997).
VFL expression is associated to reproductive meristems. During the first season, VFL is expressed in the lateral meristems giving rise to inflorescences and in the developing inflorescence meristems. Winter dormant buds show reduced levels of VFL transcripts, which could be due to either a reduced expression or to a low detectability of the transcripts in these hardy buds. During the second season, the expression levels increase in the proliferating inflorescence meristems generating inflorescence branches, with the highest levels detected in young floral meristems. VFL is also expressed in petal and stamen primordia, and its expression declines as organs expand, as described for other species (Weigel et al., 1992; Hofer et al., 1997; Southerton et al., 1998). In summary, expression of VFL in reproductive meristems and developing floral organs suggests that VFL plays an important role during reproductive development as it has been suggested for most FLO/LFY-like genes studied. Similar temporal patterns of expression spanning two seasons have also been described for the FLO/LFY ortholog in kiwifruit, another woody perennial with winter bud dormancy (Walton et al., 2001). However, in the case of kiwifruit, LFY expression levels are higher during the first growing season corresponding to the time of initiation of flower meristems. The expression levels during the second season are lower and coincide with the initiation and development of flower organs. Therefore, in grapevine and kiwifruit, the highest levels of FLO/LFY expression correspond to the time of flower meristem formation (first season in the case of kiwifruit and second season in the case of grapevine), supporting a role for these genes in this process.
Expression of VFL is not restricted to reproductive tissues. Low transcription levels are also detectable in the inner cell layers of the vegetative shoot apical meristem and in leaf and tendril primordia. Expression in the vegetative SAM has been reported in violet cress (Shue et al., 2000) and several Solanaceae species such as tobacco (Kelly et al., 1995), Impatiens sp. (Pouteau et al., 1997), and tomato (Molinero-Rosales et al., 1999), but its role in this meristematic region is still unknown. Leaf primordia and developing leaves also accumulate VFL. Most analyzed FLO/LFY orthologs have been reported to be expressed in developing leaves (Kelly et al., 1995; Blázquez et al., 1997; Bradley et al., 1997; Hofer et al., 1997; Pouteau et al., 1997; Souer et al., 1998; Southerton et al., 1998; Molinero-Rosales et al., 1999; Rottmann et al., 2000; Walton et al., 2001). In some cases, FLO/LFY expression is restricted to the adaxial side of the developing leaf as in tobacco, eucalyptus, and kiwifruit (Kelly et al., 1995; Southerton et al., 1998; Walton et al., 2001), but this is not the case for VFL mRNA, which is expressed dorsally and ventrally in leaf primordia. In developing leaves, VFL accumulates at the growing margins. At this region, VFL could be involved in maintaining proliferation and, thus, helping generate the palmate shape of the grapevine leaves. A similar role for FLO/LFY-like genes has been shown in pea where UNIFOLIATA is required to generate the wild-type compound leaves (Hofer et al., 1997) and in tomato where falsiflora mutants have leaves with fewer leaflets than wild-type plants (Molinero-Rosales et al., 1999).
A unique feature of grapevine is the occurrence of lateral meristems, indistinguishable in morphology and position, that will give rise to inflorescences, tendrils, or even lateral shoots (Srinivasan and Mullins, 1981; this work). Although inflorescence meristems are only seasonally formed as a result of flower induction, tendril meristems are continuously formed together with leaves following the pattern shown in Figure 6. Poor environmental conditions (e.g. low growing temperatures) can cause lateral meristems to develop as tendrils or even as lateral shoots (Srinivasan and Mullins, 1976). In addition, gibberellin and cytokinins have antagonistic effects on the development of these meristems: Gibberellins cause inflorescence meristems to develop as tendrils (Boss and Thomas, 2002), and cytokinins can result in the production of inflorescences from tendril meristems. VFL is expressed in all lateral meristems independently of the structure they will form, suggesting that VFL is not sufficient to confer reproductive fate. Thus, the specification of reproductive characteristics may require the observed increase in VFL expression, unknown posttranscriptional regulatory mechanisms or the activity of additional factors.
In summary, VFL transcripts accumulate in regions such as meristems and growing primordia that maintain part of its undifferentiated state. Thus, VFL could be involved in maintaining a transient phase of indeterminacy that precedes differentiation of lateral derivatives of the apical meristem: flowers, leaves, or tendrils. A similar role has been previously proposed (Hofer et al., 1997; Mouradov et al., 1998) based on results from other species (Kelly et al., 1995; Hofer et al., 1997; Pouteau et al., 1997; Mouradov et al., 1998). This extended expression of FLO/LFY orthologs can be more easily followed in woody perennials such as grapevine than in annual herbaceous plants. Functional studies of the role of VFL in grapevine will be required to demonstrate its function in the development of vegetative and reproductive primordia.
MATERIALS AND METHODS
Plant Material
Grapevine (Vitis vinifera L. var Tempranillo) plants were collected in the fields of Instituto Madrileño de Investigaciones Agrarias (Alcalá de Henares, Madrid). Young expanding leaves were used for isolation of genomic DNA. RNA-blot and in situ hybridization analyses were performed on plant tissues collected and fixed in different developmental stages during two growing seasons. Parallel samples were also analyzed by SEM.
Cloning of VFL
Cloning of VFL was performed using a 3′/5′-RACE strategy (Frohman et al., 1988), following the instructions of a commercial RACE kit (Roche Diagnostics GmbH, Mannheim, Germany). 3′-RACE was performed with oligo(dT)-primed single-stranded cDNA synthesized from total RNA of developing buds (stage C). Further PCR amplification was performed with anchored primers to the 3′-end and degenerated primers specific to highly conserved regions of FLO/LFY-like genes. These primers were 5′-CAA/GAGGGAGCAT/CCCCTTCATAGTA/GAC-3′ for the first amplification and 5′-TACATA/CAACAAGCCGAAA/GATG-3′ for the re-amplification. Amplified fragments from two independent experiments were cloned in pGEM-T easy vector (Promega, Madison, WI). Thirty clones corresponding to the 3′ region of the gene were sequenced to analyze the sequence diversity at the 3′ end. For 5′-RACE, specific single-stranded cDNA was synthesized using a gene-specific primer (5′-AGCAGCAAGGGCGACAAGAGGCTTGTAACA-3′). Then, terminal transferase was used to add a homopolymeric A-tail to the 3′ end of the cDNA. Tailed cDNA was amplified with a gene-specific primer (5′-CCAACGTTCTCTCCTCTCTCCTT-3′) and an oligo(dT)-anchor primer. The resulting product was re-amplified in a second PCR using a nested, specific primer (5′-GCTCTCCTTAAGGCATTGGAAGCCTCCTCA-3′) and an anchor primer. The gene sequence was completed after two successive 5′-RACE experiments, which allowed the isolation of the middle part and the 5′ end of the gene, respectively. The complete coding region of the cDNA was obtained by reverse transcriptase-PCR with primers flanking this zone (5′-AGAGATAGAGAGGCAATCAGCAGGATGGAT-3′ and 5′-GGGGAATACGTTAAGTTCAGAATGGCAAGT-3′). This cDNA was cloned in pGEM-T easy. To avoid PCR-based errors, six clones, obtained from three independent PCR reactions, were completely sequenced and compared.
To analyze the genomic structure of the gene, two overlapping genomic fragments were amplified by PCR on genomic DNA. Amplification products were cloned, and six clones from each fragment were completely sequenced and compared. For sequencing, the Big Dye Terminator Cycle Sequencing kit and a sequencer (Prism 377, ABI, Sunnyvale, CA) were used. Comparisons of sequences of six cDNA clones and six genomic clones revealed a complete identity between them. Only a single-base change that resulted in a synonymous codon (base 221 is G or T) was detected in both the cDNA and the genomic clones.
RNA-Blot Hybridization Analyses
For RNA-blot hybridization, plant material was collected at different developmental stages. In the first growing season, young buds in the axils of the leaves (latent buds) were collected every 15 d from June to August (Fig. 4A, latent buds, lanes 1–5). For phenological stage A, bud samples were taken in November (dormancy period), February, and March (Fig. 4A, stage A, lanes 1–3, respectively). In the second growing season, swelling buds were collected. These samples correspond to buds of early (lanes 1) or advanced (lanes 2) phenological stages B and C (Fig. 4A). Further in development, when the new cane begins to grow out (phenological stages E, F, G, and H), the different organs of the plant were independently analyzed: shoot apex, young leaves, shoots, tendrils, and flowers. Total RNA extraction was performed following the protocol of Chang et al. (1993). For RNA-blot hybridization analyses, 25 μg of total RNA was loaded per lane on agarose/formaldehyde gels, electrophoretically separated, and transferred to Hybond-N+ membranes. Filters were hybridized with a 32P-radiolabeled VFL 5′-probe, comprising 290 amino acids and the untranslated 5′ region. Hybridization was performed overnight at 65°C in Church's buffer (Church and Gilbert, 1984). Filters were washed several times in 2× SSC and 0.1% (w/v) SDS and once in 0.1× SSC and 0.1% (w/v) SDS for 20 min at 65°C.
In Situ Hybridization
Digoxigenin labeling of RNA probes, tissue preparation, and hybridization were performed as described by Coen et al. (1990). The template for the VFL digoxigenin-labeled riboprobes was the 1,249-bp fragment, containing the complete coding region, obtained by reverse transcriptase-PCR with the primers indicated above and cloned in pGEM-T easy vector. The hybridized sections were visualized with Nomarski optics in a microscope (DMR, Leica, Wetzlar, Germany).
SEM
Samples were collected once every month for 1 year. Samples were dissected in the field and collected in ice-cold fixation buffer (phosphate-buffered saline, 4% [w/v] paraformaldehyde, 0.1% [v/v] Triton, and 0.1% [v/v] Tween 20), vacuum-infiltrated in fresh fixation buffer, and left overnight in the cold room. They were then washed in saline solution and incubated for 24 h in a phosphate-buffered saline solution with 2% (w/v) osmium tetroxide. After washing them once with saline solution, samples were dehydrated through an ethanol series (50%, 70%, 85%, 95%, and 100% [v/v]) and critical-point dried. They were then covered with gold particles and observed under the SEM.
Phylogenetic Analyses
To construct the phylogenetic tree, predicted proteins were aligned with ClustalW. Using this original data set, 100 data sets were generated by bootstrap resampling using the SEQBOOT program. Distance matrices were made for each bootstrap data set using the PROTDIST program-Dayhoff PAM matrix algorithm. The distance matrices obtained were used to construct 100 un-rooted trees by the neighbor-joining method using the NEIGHBOR program. A consensus tree was obtained using CONSENSE. SEQBOOT, PROTDIST, NEIGHBOR, and CONSENSE programs belong to the PHYLIP program (Phylogeny Inference Package, version 3.57c, Department of Genetics, University of Washington, Seattle).
ACKNOWLEDGMENTS
We thank Félix Cabello and the Instituto Madrileño de Investigaciones Agrarias (Alcalá de Henares) for providing plant material for this research and some of the photographs of Figure 1, Esperanza Salvador and Laura Tormo from the SEM service of the Universidad Autónoma de Madrid for assistance with the microphotography work, and Joaquín García and Christiane Germonprez for help with the artwork.
Footnotes
This work was supported by the Comunidad Autónoma de Madrid (grant no. 07B/0035/1999) and the Ministerio de Educación Ciencia, Spain (postdoctoral fellowship to P.C.). Support to research activity at Centro Nacional de Biotecnología is provided through specific agreement of Consejo Superior de Investigaciones Científicas-Instituto Nacional de Investigacion y Tecnologia Agraria y Alimentaria.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.002428.
LITERATURE CITED
- Araki T. Transition from vegetative to reproductive phase. Curr Opin Plant Biol. 2001;4:63–68. doi: 10.1016/s1369-5266(00)00137-0. [DOI] [PubMed] [Google Scholar]
- Baggiolini M. Les stades repères dans le developpement annuel de la vigne et leur utilisation pratique. Rev Romande Agric Vitic Arbor. 1952;8:4–6. [Google Scholar]
- Blázquez MA, Soowal LN, Lee I, Weigel D. LEAFY expression and flower initiation in Arabidopsis. Development. 1997;124:3835–3844. doi: 10.1242/dev.124.19.3835. [DOI] [PubMed] [Google Scholar]
- Boss PK, Thomas MR. Association of dwarfism and floral induction with a grape “green revolution” mutation. Nature. 2002;416:847–850. doi: 10.1038/416847a. [DOI] [PubMed] [Google Scholar]
- Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E. Inflorescence commitment and architecture in Arabidopsis. Science. 1997;275:80–83. doi: 10.1126/science.275.5296.80. [DOI] [PubMed] [Google Scholar]
- Carpenter R, Coen ES. Floral homeotic mutations produced by transposon mutagenesis in Antirrhinum majus. Genes Dev. 1990;4:1483–1493. doi: 10.1101/gad.4.9.1483. [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep. 1993;11:113–116. [Google Scholar]
- Church JM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen ES, Romero JM, Doyle S, Elliot R, Murphy G, Carpenter R. Floricaula: a homeotic gene required for flower development in Antirrhinum majus. Cell. 1990;63:1311–1322. doi: 10.1016/0092-8674(90)90426-f. [DOI] [PubMed] [Google Scholar]
- Frohlich MW, Meyerowitz EM. The search for flower homeotic gene homologous in basal angiosperms and gnetales: a potential new source of data on the evolutionary origin of flowers. Int J Plant Sci. 1997;158:S131–S142. [Google Scholar]
- Frohlich MW, Parker DS. The mostly male theory of flower evolutionary origins: from genes to fossils. Syst Bot. 2000;25:155–170. [Google Scholar]
- Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNA from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocal FW, King RW, Blundell CA, Schwartz OM, Andersen CH, Weigel D. Evolution of floral meristem identity genes: analysis of Lolium temulentum genes related to APETALA1 and LEAFY of Arabidopsis. Plant Physiol. 2001;125:1788–1801. doi: 10.1104/pp.125.4.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Zhu Q, Dabi T, Li D, Weigel D, Lamb C. Transformation of rice with the Arabidopsis floral regulator LEAFY causes early heading. Transgenic Res. 2001;9:223–227. doi: 10.1023/a:1008992719010. [DOI] [PubMed] [Google Scholar]
- Hofer J, Turner L, Hellens R, Ambrose M, Matthews P, Michaels A, Ellis N. UNIFOLIATA regulates leaf and flower morphogenesis in pea. Curr Biol. 1997;7:581–587. doi: 10.1016/s0960-9822(06)00257-0. [DOI] [PubMed] [Google Scholar]
- Judd WS, Campbell CS, Kellogg E, Stevens PF. Plant Systematics. A Phylogenetic Approach. Sunderland, MA: Sinauer Associates; 1999. [Google Scholar]
- Kelly AJ, Bonnlander MB, Meeks-Wagner RD. NFL, the tobacco homolog of FLORICAULA and LEAFY, is transcriptionally expressed in both vegetative and floral meristems. Plant Cell. 1995;7:225–234. doi: 10.1105/tpc.7.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyozuka J, Konishi S, Nemoto K, Izawa T, Shimamoto K. Down-regulation of RFL, the FLO/LFY homolog of rice, accompanied with panicle branch initiation. Proc Natl Acad Sci USA. 1998;95:1979–1982. doi: 10.1073/pnas.95.5.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren SJ, Gustafson-Brown C, Pinyopich A, Ditta GS, Yanofsky MF. Interactions among APETALA1, LEAFY, and TERMINAL FLOWER1 specify meristem fate. Plant Cell. 1999;11:1007–1018. doi: 10.1105/tpc.11.6.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellerowicz EJ, Horgan K, Walden A, Coker A, Walter C. PRFLL: a Pinus radiata homologue of FLORICAULA and LEAFY is expressed in buds containing vegetative shoot and undifferentiated male cone primordial. Planta. 1998;206:619–629. doi: 10.1007/s004250050440. [DOI] [PubMed] [Google Scholar]
- Molinero-Rosales N, Jamilena M, Zurita S, Gomez P, Capel J, Lozano R. FALSIFLORA, the tomato orthologue of FLORICAULA and LEAFY, controls flowering time and floral meristem identity. Plant J. 1999;20:685–693. doi: 10.1046/j.1365-313x.1999.00641.x. [DOI] [PubMed] [Google Scholar]
- Mouradov A, Glassick T, Hamdorf B, Murphy L, Fowler B, Marla S, Teasdale RD. NEEDLY, a Pinus radiata ortholog of FLORICAULA/LEAFY genes, expressed in both reproductive and vegetative meristems. Proc Natl Acad Sci USA. 1998;95:6537–6542. doi: 10.1073/pnas.95.11.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins MG, Bouquet A, Williams LE. Biology of the Grapevine. Cambridge, UK: Cambridge University Press; 1992. [Google Scholar]
- Peña L, Martin-Trillo M, Juárez J, Pina JA, Navarro L, Martínez-Zapater JM. Constitutive expression of Arabidopsis LEAFY or APETALA1 genes in citrus reduces their generation time. Nat Biotechnol. 2001;19:263–267. doi: 10.1038/85719. [DOI] [PubMed] [Google Scholar]
- Pidkowich MS, Klenz JE, Haughn GW. The making of a flower: control of floral meristem identity in Arabidopsis. Trends Plant Sci. 1999;4:64–70. doi: 10.1016/s1360-1385(98)01369-7. [DOI] [PubMed] [Google Scholar]
- Pouteau S, Nicholls D, Tooke F, Coen E, Battey N. The induction and maintenance of flowering in Impatiens. Development. 1997;124:3343–3351. doi: 10.1242/dev.124.17.3343. [DOI] [PubMed] [Google Scholar]
- Rottmann WH, Meilan R, Sheppard LA, Brunner AM, Skinner JS, Ma C, Cheng S, Jouanin L, Pilate G, Strauss SH. Diverse effects of overexpression of LEAFY and PTLF, a poplar (Populus) homolog of LEAFY/FLORICAULA, in transgenic poplar and Arabidopsis. Plant J. 2000;22:235–245. doi: 10.1046/j.1365-313x.2000.00734.x. [DOI] [PubMed] [Google Scholar]
- Schultz EA, Haughn GW. LEAFY, a homeotic gene that regulates inflorescence development in Arabidopsis. Plant Cell. 1991;3:771–781. doi: 10.1105/tpc.3.8.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shue G, Amaral W, Hileman LC, Baum DA. LEAFY and the evolution of rosette flowering in violet cress (Jonopsidium acaule, Brassicaceae) Am J Bot. 2000;87:634–641. [PubMed] [Google Scholar]
- Simpson GG, Gendall AR, Dean C. When to switch to flowering. Annu Rev Cell Dev Biol. 1999;99:519–550. doi: 10.1146/annurev.cellbio.15.1.519. [DOI] [PubMed] [Google Scholar]
- Souer E, van der Krol A, Kloos D, Spelt C, Bliek M, Mol J, Koes R. Genetic control of branching pattern and floral identity during Petunia inflorescence development. Development. 1998;125:733–742. doi: 10.1242/dev.125.4.733. [DOI] [PubMed] [Google Scholar]
- Southerton SG, Strauss SH, Olive MR, Harcourt RL, Decroocq V, Zhu X, Llewellyn DJ, Peacock WJ, Dennis E. Eucalyptus has a functional equivalent of the Arabidopsis floral meristem identity gene LEAFY. Plant Mol Biol. 1998;37:897–910. doi: 10.1023/a:1006056014079. [DOI] [PubMed] [Google Scholar]
- Srinivasan C, Mullins MG. Reproductive anatomy of the grapevine (Vitis vinifera L.): origin and development of the anlage and its derivatives. Ann Bot. 1976;38:1079–1084. [Google Scholar]
- Srinivasan C, Mullins MG. Physiology of flowering in the grapevine: a review. Am J Enol Vitic. 1981;32:47–63. [Google Scholar]
- Wagner D, Sablowski RWM, Meyerowitz EM. Transcriptional activation of APETALA1 by LEAFY. Science. 1999;285:582–584. doi: 10.1126/science.285.5427.582. [DOI] [PubMed] [Google Scholar]
- Walton EF, Podivinsky E, Wu RM. Bimodal pattern of floral gene expression over the two seasons that kiwifruit flowers develop. Physiol Plant. 2001;111:396–404. doi: 10.1034/j.1399-3054.2001.1110318.x. [DOI] [PubMed] [Google Scholar]
- Weberling F. Morphology of flowers and inflorescences. Cambridge, UK: Cambridge University Press; 1989. [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. LEAFY controls floral meristem identity in Arabidopsis. Cell. 1992;69:843–859. doi: 10.1016/0092-8674(92)90295-n. [DOI] [PubMed] [Google Scholar]
- Weigel D, Nilsson O. A developmental switch sufficient for flower initiation in diverse plants. Nature. 1995;377:495–500. doi: 10.1038/377495a0. [DOI] [PubMed] [Google Scholar]