Abstract
The expression of CHALCONE SYNTHASE (CHS) expression is an important control step in the biosynthesis of flavonoids, which are major photoprotectants in plants. CHS transcription is regulated by endogenous programs and in response to environmental signals. Luciferase reporter gene fusions showed that the CHS promoter is controlled by the circadian clock both in roots and in aerial organs of transgenic Arabidopsis plants. The period of rhythmic CHS expression differs from the previously described rhythm of chlorophyll a/b-binding protein (CAB) gene expression, indicating that CHS is controlled by a distinct circadian clock. The difference in period is maintained in the wild-type Arabidopsis accessions tested and in the de-etiolated 1 and timing of CAB expression 1 mutants. These clock-affecting mutations alter the rhythms of both CAB and CHS markers, indicating that a similar (if not identical) circadian clock mechanism controls these rhythms. The distinct tissue distribution of CAB and CHS expression suggests that the properties of the circadian clock differ among plant tissues. Several animal organs also exhibit heterogeneous circadian properties in culture but are believed to be synchronized in vivo. The fact that differing periods are manifest in intact plants supports our proposal that spatially separated copies of the plant circadian clock are at most weakly coupled, if not functionally independent. This autonomy has apparently permitted tissue-specific specialization of circadian timing.
Light is a key environmental signal for plants, regulating gene expression and development (Neff et al., 2000). Changes in fluence rate and light quality can occur unpredictably and rapidly during the day but have an underlying day-night cycle. Plants have evolved a circadian timing system that allows the anticipation of this predictable rhythm. When plants are deprived of environmental time cues and placed in constant (“free running”) environmental conditions, circadian rhythms persist with a period of around 24 h, often for many days (Millar, 1999; McClung, 2000; Murtas and Millar, 2000; Johnson, 2001). Within the circadian system of the whole organism, the term “circadian oscillator” has been used to denote the parts of the system responsible for rhythm generation. Light-dark signals entrain the oscillator via input phototransduction pathways, synchronizing its phase with the environmental light-dark cycle and also affecting its period. Rhythmic output from the oscillator controls a large number of physiological processes in plants (Lumsden and Millar, 1998). The abundance of 2% to 6% of RNA transcripts in Arabidopsis plants was scored as circadian-regulated in two recent microarray analyses, for example (Harmer et al., 2000; Schaffer et al., 2001).
The rhythmic expression of chlorophyll a/b-binding protein (CAB or Light-Harvesting Complex [LHCB]) genes has often been used as a marker for circadian regulation in plants (for review, see Fejes and Nagy, 1998), especially using firefly (Photinus pyralis) luciferase (LUC) reporter fusions (Millar et al., 1995a). CAB genes are strongly expressed in the mesophyll cells of photosynthetically active organs and in epidermal guard cells. However, physiological analysis shows that the plant circadian system comprises many copies of the circadian clock, with at least one clock in all the major plant organs and possibly one in most cells (Gorton et al., 1989; Kim et al., 1993; Mayer and Fischer, 1994). We use the term “circadian clock” to denote the smallest complete timing unit (comprising an oscillator with light input and output to overt rhythms). CAB rhythms, thus, reveal only a subset of clocks in the plant circadian system. The distributed circadian clocks are thought to function autonomously, because many circadian rhythms persist in isolated tissue explants (for example, Vaadia, 1960; Simon et al., 1976; Engelmann and Johnsson, 1978; Thain et al., 2000). Within a single plant, we have shown that various organs can maintain rhythmic expression of the same gene set to different phases (Thain et al., 2000). Central circadian pacemakers and communicated rhythmic signals, therefore, have little influence on plant rhythms, at least for the gene expression rhythms tested (Thain et al., 2000). This is in contrast to their well-documented involvement in mammalian rhythms (van Esseveldt et al., 2000; Yamazaki et al., 2000).
The autonomy of plant clocks implies that, if the timing properties of the circadian clocks varied among tissues, those differences would be reflected in each tissue's circadian rhythms. Such specialization in timing might be advantageous (Roenneberg and Mittag, 1996). It is unclear how much circadian timing actually varies among plant tissues and what are the molecular causes of such variation. Differences in circadian period between plant rhythms have been reported (Hennessey and Field, 1992; Millar et al., 1995a; Fowler et al., 1999; Sai and Johnson, 1999). The rhythms in question were not only expressed in different cells but also had overtly unrelated mechanisms (leaf movement and CAB gene expression, for example).
To investigate the heterogeneity of plant circadian clocks, we required rhythmic markers that can be tested for period under constant conditions, in broader spatial domains than the CAB genes. Here, we characterize the rhythmic expression of a CHS promoter:reporter gene fusion (CHS:LUC). CHS is expressed predominantly in epidermal cells of aerial organs and in roots, in Arabidopsis (Chory and Peto, 1990; Kaiser and Batschauer, 1995) as in other species (Schmelzer et al., 1988; Ehmann et al., 1991; Haussuhl et al., 1996). We show that its circadian rhythm has a significantly different period than rhythmic CAB expression, implying that CHS is controlled by a different circadian clock. To test whether the same molecular components are required for circadian control of CHS and CAB, we assayed the two markers in mutant backgrounds that are known to alter CAB rhythms by different mechanisms. The mutations had very similar effects upon both CHS and CAB rhythms, indicating that the circadian clocks share similar molecular components. The heterogeneity among circadian rhythms of plant gene expression is likely attributable to tissue-specific modifiers of a common biochemical oscillator, which is present in many if not all Arabidopsis cells.
RESULTS
Expression of CHS:LUC in Roots and Leaves
Fusions of firefly LUC to the promoters of the white mustard (Sinapis alba) chalcone synthase (mCHS) and Arabidopsis chalcone synthase (CHS) genes were transformed into Arabidopsis plants of the Landsberg erecta (Ler) and C24 strains, respectively. Figure 1B shows that luminescence driven by the CHS promoter was evident in the leaves, hypocotyl, and roots of 12-d-old seedlings. Particularly high expression occurred in the shoot apical region and in young lateral roots. Younger, 5-d-old seedlings showed no CHS expression in the hypocotyl (data not shown), consistent with previous reports (Kaiser and Batschauer, 1995; Kaiser et al., 1995). The mCHS promoter had an identical pattern of expression (data not shown). In contrast, CAB:LUC activity was largely confined to green tissues, with a gradient down the hypocotyl and no detectable activity in the roots (Fig. 1D).
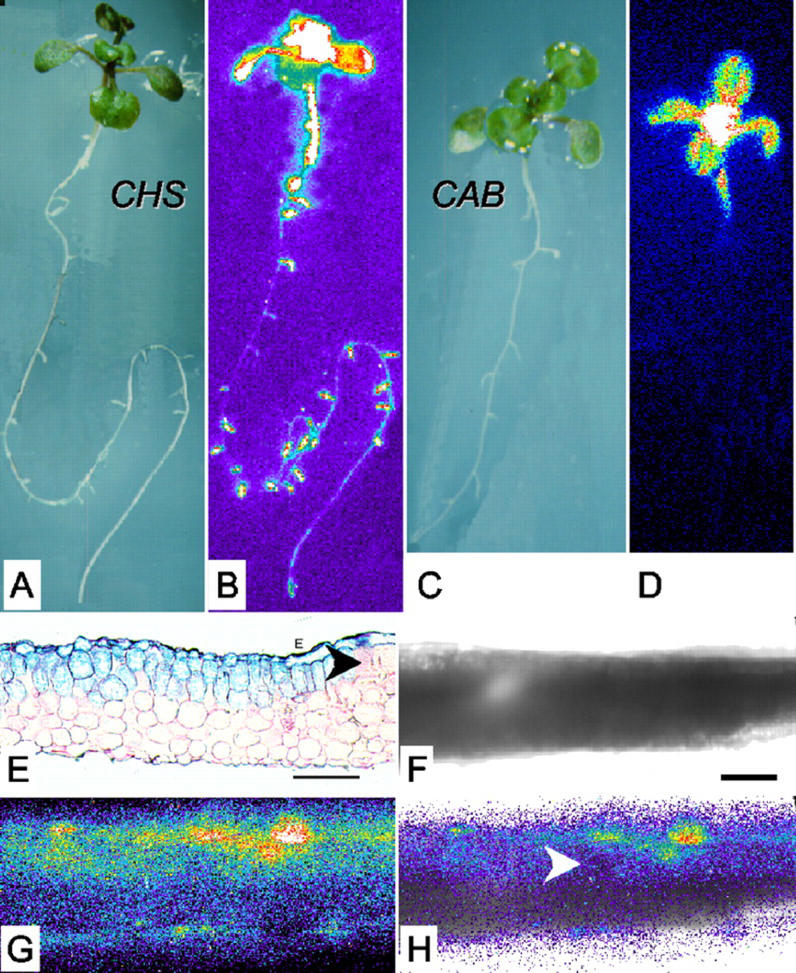
Figure 1.
Tissue-specific expression of the CHS promoter. Reflected-light (A and C) and luminescence (B and D) images of CHS:LUC (A and B) and CAB:LUC (C and D) seedlings after 12 d of growth in LD (12, 12) of 150 μmol m−2 s−1. Luminescence video images were processed in false color (see “Materials and Methods”): White and red shades represent the highest photon counts, and darker blues, the lowest. A reflected-light image of the plant is shown to the left of each false color image. E through H, Leaf sections were prepared from plants grown in LD (12, 12) of 150 μmol m−2 s−1 for 7 d and then transferred to 250 μmol m−2 s−1 for 5 d. E, Microtome section of a CHS:β-glucuronidase (GUS) leaf, stained for GUS activity and counter-stained with ruthenium red. F through H, Thick hand section of a CHS:LUC leaf: bright field (F), luminescence (G), and overlaid (H) images. E and F, Scale bars represent 60 μm. E and H, Arrowheads indicate areas where CHS expression is very weak or absent from the mesophyll.
The spatial pattern of CHS promoter activity was examined in greater detail in leaf sections from CHS:LUC plants and plants carrying a CHS-GUS fusion transgene. The CHS:LUC signal was highest in the upper epidermis, but both reporters showed patchy expression also in the palisade mesophyll layer (Fig. 1, E and G). The epidermal expression of CHS:LUC was about 2-fold higher than the mesophyll expression (in Fig. 1G, for example, average counts per pixel per minute were 0.36 for epidermis and 0.19 for upper mesophyll). A minor contribution from rhythmic CHS expression in the mesophyll would not be distinguished in our assays. Luminescence from the mesophyll is appreciably reduced by passage through the epidermis (Wood et al., 2001), indicating that the CHS:LUC rhythm preferentially reflects epidermal luminescence. CHS-GUS expression was also evident in all cell layers of root cross sections (data not shown).
The CHS Expression Rhythm Is Distinct from the CAB Expression Rhythm
The strong CHS:LUC activity in the root might make this a useful marker for circadian rhythms specifically in this organ, where root pressure and ion fluxes are the only previously reported, rhythmic markers (Vaadia, 1960; Parsons and Kramer, 1974; Gorr et al., 1995; Henzler et al., 1999). The circadian rhythm of CHS:LUC activity was tested under several fluence rates of constant light, because CHS mRNA levels increase in response to higher fluence rates (Feinbaum and Ausubel, 1988; Peter et al., 1991; Fuglevand et al., 1996). Transgenic seedlings were grown for 12 d on vertical agar plates under white light of 60, 150, or 250 μmol m−2 s−1 fluence rate. Seedlings grown and assayed in white light of 60 μmol m−2 s−1 showed low expression of CHS:LUC and mCHS:LUC (data not shown). Luminescence rhythms were measured after the plants were transferred to constant light of either the same fluence rates (Fig. 2, A and C), or of an increased fluence rate (from 150 to 250 μmol m−2 s−1; Fig. 2B). CHS:LUC activity showed circadian rhythmicity in aerial organs and roots under all conditions. The peak of activity on the 1st d in constant light occurred before predicted dawn under all conditions (at zeitgeber time [ZT] 20–22; ZT is defined as the number of hours since lights-on), 6 or 7 h before the peak of CAB expression. Transgenic plants carrying the mCHS:LUC fusion in the Ler background showed very similar luminescence rhythms (Fig. 2D).
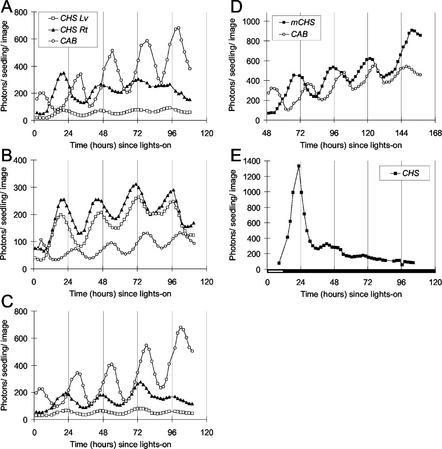
Figure 2.
Rhythmic luminescence in roots and aerial organs of Arabidopsis seedlings expressing CHS:LUC. CHS:Luc (A, B, C, and E) or mCHS:LUC (D) expression was assayed by video imaging (see “Materials and Methods”) at the times indicated. A, B, D, and E, Seedlings were grown on vertical agar plates, in 7 d of LD (12, 12) at 150 μmol m−2 s−1. Seedlings in A and D were transferred (at 0 h) to continuous light of 150 μmol m−2 s−1; seedlings in B were transferred to continuous light of 250 μmol m−2 s−1; and seedlings in E were transferred first to 250 μmol m−2 s−1 for 12 h and then to constant darkness. Seedlings in C were both grown and tested at 250 μmol m−2 s−1. The data are representative of at least five independent experiments. White box on time axis, Light interval; black box, dark interval. Lv, Luminescence from aerial organs; Rt, luminescence from roots.
The lighting conditions affected the amplitude of rhythmic CHS expression. The high-amplitude rhythm in the 1st d of constant light was followed by a reduction (damping) in the amplitude of the CHS:LUC circadian rhythm within two to three cycles. mCHS:LUC was less affected (Fig. 2D), possibly because of a difference between the C24 and Ler genetic backgrounds. CAB:LUC activity continued to cycle with high amplitude under constant light of all fluence rates. In plants transferred from lower to higher fluence rates of light (Fig. 2B), CAB:LUC luminescence levels were reduced compared with CHS:LUC. When 12-d-old plants expressing CHS:LUC were transferred to continuous darkness, transcription from the CHS promoter was greatly attenuated (Fig. 2E). A single, high-amplitude peak at the expected phase was followed by rapid damping of CHS:LUC activity, with a complete loss of rhythmic amplitude by 72h.
The CAB and CHS rhythms had different free-running periods under constant light. The lag (phase angle) between the rhythms, therefore, changed progressively during our experiments, most obviously when Ler plants were imaged on the 3rd to 7th d under constant light (Fig. 2D). The altered period was clear: The first peak of mCHS expression shown (at 68 h) occurred 10 h before the CAB peak (at 78 h), but by 147 h, the mCHS peak occurred with or slightly after that for CAB. Period estimates were derived from luminescence rhythms measured in the aerial tissues, where both CHS:LUC and CAB:LUC are expressed (Table I). The period of the CHS rhythm (25.4 h) was significantly longer than the period of the CAB rhythm (23.7 h). A similar period difference was observed between CHS and CAB expression rhythms in the Ler background (as in Fig. 2D). In contrast, the period of rhythmic CHS expression in the root was not significantly different from that in the leaf, in either genetic background (Fig. 2; Table II; data not shown). These results suggested that the circadian system that controlled CHS in aerial organs was distinct from that controlling CAB.
Table I.
Circadian period of gene expression in the aerial organs of det1
| LUC Marker | Genotype | Background | Period | se | n |
|---|---|---|---|---|---|
| h | |||||
| CAB | + | C24 | 23.7a | 0.36 | 40 |
| CHS | + | C24 | 25.4b | 0.38 | 40 |
| CHS | + | C24/Col | 25.0c | 0.41 | 20 |
| CHS | detI-1 | C24/Col | 21.8d | 0.48 | 15 |
| CAB | detI-1 | C24/Col | 20.7 | 0.39 | 14 |
Plants were maintained as described in the legend to Figure 2B: grown for 7 d under LD (12,12) of 150 μmol m−2 s−1 and transferred to constant light of 250 μmol m−2 s−1 for rhythm assays. Period estimation and statistical comparisons were performed as described in “Materials and Methods.” se are based on the analysis of seven experiments with 129 period estimates (119 degrees of freedom). Genetic backgrounds: C24, Columbia (Col); C24 × Columbia F2 (C24/Col). The significance levels of t tests comparing the mean periods were as follows: aCHS (+) versus CAB (+), P < 0.0001. bCHS (C24) versus CHS (C24/Col), P > 0.4. cCHS (+)versus CHS (det1), P < 0.0001. dCHS (det1) versus CAB (det1), P < 0.001.
Table II.
Circadian period of LUC activity rhythms in toc1
| LUC Marker | Genotype | Organs | Period | se | n |
|---|---|---|---|---|---|
| h | |||||
| CAB | + | Aerial | 24.3a | 0.08 | 4 |
| CHS | + | Aerial | 25.1 | 0.37 | 5 |
| CAB | toc1-1 | Aerial | 19.2b | 0.25 | 6 |
| CHS | toc1-1 | Aerial | 20.6 | 0.23 | 5 |
| CHS | + | Root | 24.8 | 0.21 | 5 |
| CHS | toc1-1 | Root | 21.2 | 0.26 | 4 |
Plants were maintained as described for Figure 2B and Table I. Period estimation and statistical comparisons were performed as described in “Materials and Methods.” All comparisons of toc1 versus wild type for a given marker and tissue, P < 0.0001; CHS in aerial tissues versus roots, P > 0.15 in both wild type and toc1. All lines are in the C24 background. The significance levels of t tests comparing the mean periods were as follows: aCHS (+) versus CAB (+), P = 0.09. bCAB (toc1) versus CHS (toc1) in aerial organs, P < 0.005.
de-etiolated 1 (det1) and timing of CAB expression 1 (toc1) Mutations Shorten the Period of Both CHS and CAB Rhythms
The det1 mutant has a severe short-period phenotype for CAB expression (Millar et al., 1995b), along with other phenotypes principally related to light signaling (Chory and Peto, 1990; Pepper et al., 1994). toc1 is a short-period mutant that affects only clock-regulated processes (Millar et al., 1995a; Kreps and Simon, 1997; Somers et al., 1998b; Dowson-Day and Millar, 1999); TOC1 encodes one of the best candidates for a plant circadian oscillator component (Strayer et al., 2000; Alabadi et al., 2001). The CHS:LUC construct was crossed into these mutant backgrounds to test whether these genes are involved in the circadian systems that control both the CAB and CHS markers. det1 and toc1 mutant seedlings expressed CHS:LUC rhythmically in both leaves and roots under constant light (Fig. 3A; data not shown). Light-grown det1 seedlings express the CHS promoter in all leaf cell layers, and express CAB genes inappropriately in roots and in aerial tissue (Chory and Peto, 1990). CAB:LUC activity in det1 roots was barely detectable and was too low for us to measure circadian rhythms (data not shown). The det1 mutation prevents the damping of rhythmic CAB expression in darkness (Fig. 3B; Millar et al., 1995b). It had little effect on CHS expression, which damped out in the dark similarly in the mutant (Fig. 3B) and wild type (Fig. 2E). Consistent with previous data, the toc1 mutation did not affect the mean expression level or damping of CAB and CHS expression rhythms (data not shown; Millar et al., 1995a).
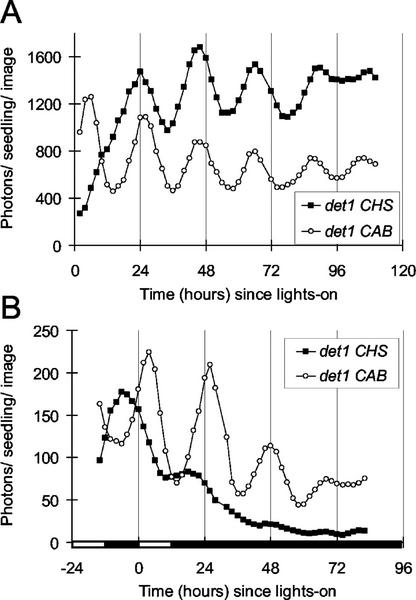
Figure 3.
Circadian rhythms of CHS:LUC activity in det1. Expression was assayed as in Figure 2. Seedlings were grown in LD (8, 16) at 20 μmol m−2 s−1 to confirm the det1 phenotype. Seedlings were transferred to LD (12, 12) at 150 μmol m−2 s−1, 2 d before the start of the experiment and to constant light of 250 μmol m−2 s−1 at 0 h (A) or to 250 μmol m−2 s−1 at 0 h and constant darkness at 12 h (B). Luminescence data were analyzed from whole seedlings, without separate analysis of roots and aerial tissues. The data are representative of at least four independent experiments. White box on time axis, Light interval; black box, dark interval.
Quantitative comparisons showed that the period of CHS expression was longer than the period of CAB expression in both mutant backgrounds (Tables I and II), reinforcing the conclusion that separate clocks control these two promoters. However, both rhythmic markers were very similarly affected by the mutations (Tables I and II), with much shorter periods in mutant progeny than in wild-type controls. This result indicates that TOC1 and DET1 function similarly in the circadian clocks that regulate CAB and CHS.
DISCUSSION
Chalcone synthase is one of the key biosynthetic enzymes controlling anthocyanin formation. These flavonoid pigments function to protect plant cells from UV radiation and from pathogen attack, act as insect repellents, and are involved in plant-microbe and pollen-pistil signaling (Hahlbrock and Scheel, 1989). We used the noninvasive LUC reporter gene in fusions with the CHS promoter (CHS:LUC), to monitor the dynamic pattern of CHS expression (Fig. 1). The rhythmic luminescence of CHS:LUC seedlings showed that the circadian clock regulates CHS expression at the level of transcription (Fig. 2). CHS:LUC represents the first noninvasive molecular marker for circadian rhythms in root tissue.
Regulation of CHS
The expression of CHS exhibited a very similar circadian rhythm in all tissues (Fig. 2). The phase of the circadian rhythm of CHS was earlier than CAB, with a peak occurring in the late subjective night (ZT 20–22). Chalcone synthase enzyme activity and mRNA abundance have previously been reported to exhibit diurnal and circadian regulation (Peter et al., 1991; Deikman and Hammer, 1995; Harmer et al., 2000; Schaffer et al., 2001). The reported phase of peak enzyme activity lags slightly behind the peaks of mRNA abundance and of transcriptional activity, suggesting that the circadian system principally regulates CHS expression at the transcriptional level. It may be advantageous for the plant to accumulate photoprotective pigments in advance of the daily photoperiod (Harmer et al., 2000); the timing of CHS transcription before dawn is consistent with this notion.
Increasing the fluence rate of white light from 150 to 250 μmol m−2 s−1 had little effect on the phase or period of CHS or CAB expression (Fig. 2; data not shown). However, mean expression levels were differentially affected by the lighting conditions. Plants entrained at 150 μmol m−2 s−1 but assayed at 250 μmol m−2 s−1 showed reduced levels of CAB:LUC activity compared with CHS:LUC activity (Fig. 2B), perhaps reflecting a requirement for increased photoprotection and reduced light-harvesting capacity. High-fluence rate light was required to maintain high expression levels of CHS transcription in wild-type plants: CHS:LUC activity was very low in plants assayed at 60 μmol m−2 s−1 (data not shown) or in darkness (Fig. 2E). The det1 mutation increases CAB expression levels in dark-adapted plants but had little effect on the level of CHS expression (Fig. 3B; Chory and Peto, 1990; Millar et al., 1995b). This presumably reflects a differential involvement of DET1 in the phototransduction pathways that regulate these promoters (Mustilli et al., 1999; Jenkins et al., 2001).
Differences in the Circadian Regulation of CHS and CAB
The rhythm of CHS expression was very similar in roots and in aerial organs under constant light, indicating that the clocks controlling CHS in these organs do not differ significantly. The period of CHS expression in aerial tissues was approximately 1.5 h longer than the period of CAB expression. This difference was maintained in two wild-type accessions and in the det1 and toc1 mutants (Tables I and II). Distinct circadian clocks, therefore, control the rhythms of CAB and CHS expression, although both are nuclear genes that could in principle respond to the same regulator. The phase of a single rhythm (free calcium concentration) has recently been shown to vary among tissues of transgenic tobacco (Nicotiana tabacum; Wood et al., 2001). The authors point out that a phase difference could result from tissue-specific clocks or from tissue-specific responses to a single, common clock. Where period differences are observed, the latter interpretation can be ruled out (Sai and Johnson, 1999).
It is not surprising that the period difference is small. Studies on cyanobacteria indicate that a clock with a period that matches the environmental light-dark cycle provides a competitive advantage (Ouyang et al., 1998). Balancing selection is, therefore, likely to maintain periods in a narrow range around 24 h. Consistent with this notion, we have previously shown that the similar periods of several Arabidopsis accessions are the result of balancing long- and short-period alleles at multiple loci (Swarup et al., 1999).
Period differences under constant conditions are relatively easy to measure. Under light-dark cycles, however, clocks with different periods will entrain to different phases (for example, Ouyang et al., 1998; Somers et al., 1998b). A clock with a longer period (such as that controlling CHS) will be set to a later phase, all else being equal. The longer period moves the peak of CHS expression away from midnight and toward dawn. If CAB was controlled by the same, longer-period clock, then the peak of CAB expression would also be later in the day, all else being equal. The delay between the peak of CHS expression and the peak of CAB expression would thus be greater than we observe. Independent clocks allow the temporal sequence of metabolic processes to be fine-tuned: A smaller delay between the peaks of CHS and CAB expression might be one example of this. Such a flexible timing system has potential selective advantages (Roenneberg and Mittag, 1996), although these remain to be demonstrated experimentally.
Differential Regulation of Circadian Period
The circadian clocks that control CAB and CHS might differ fundamentally in their oscillator mechanism, or they might alternatively be separate copies of a common biochemical mechanism with only minor modifications leading to the period difference (Millar, 1998). det1 and toc1 mutations are thought to affect the circadian rhythm of CAB expression via the input pathway and the oscillator, respectively (Millar et al., 1995a, 1995b; Strayer et al., 2000; Alabadi et al., 2001). Each mutation shortens the circadian period of CHS in aerial organs in parallel with CAB (Tables I and II). This result shows that both circadian clocks share at least the TOC1 and DET1 functions, so the clocks are not radically different. A parsimonious explanation is that the CHS and CAB promoters are controlled by separate copies of the same clock mechanism (Fig. 4). The characteristics of circadian timing also vary among rodent organs, outside the brain (Yamazaki et al., 2000). Differences in input pathways might be particularly important in that case (Damiola et al., 2000; Stokkan et al., 2001). The same canonical clock genes seem to be involved in diverse anatomical locations (Ripperger et al., 2000).
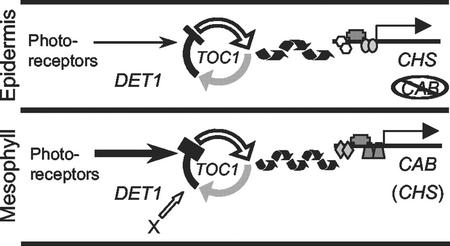
Figure 4.
Autonomy and specialization of the circadian clock. A simplified model of the circadian clock is shown in the mesophyll and in the epidermis, with the output markers and clock-related genes tested in this work. The clock is depicted as containing the same components in each case; TOC1 is thought to function in the oscillator, and DET1 is thought to affect the light input pathway from the photoreceptors. Rhythmic output from the oscillator is shown regulating transcription factors at the CAB or CHS promoter. Two hypothetical tissue-specific factors are shown: modification of the light input pathway and an unknown tissue-specific factor (X). One or both modify the function of an oscillator component, resulting in a longer period in the epidermis.
CHS is expressed principally in the epidermal cell layer of wild-type aerial organs, whereas CAB is expressed in the mesophyll layers. It is most likely that their different circadian periods reflect tissue-specific modifications of the clock in epidermal and mesophyll cells, respectively. In support of this conclusion, we have recently shown that the PHYTOCHROME B gene, which is expressed in the epidermis, also has a longer period than CAB (A. Hall, L. Kozma-Bognar, F. Nagy, and A.J. Millar, unpublished data). These data imply that epidermal circadian clocks are not tightly coupled to clocks in the mesophyll of the same leaf. We have previously demonstrated that several autonomous clocks exist within the mesophyll of a single leaf (Thain et al., 2000). Taken together, our results suggest that circadian control is local to one or a few cells, not widely coupled within or between tissues, at least for gene expression rhythms in the leaf.
The molecular cause of the observed period difference is unclear. Any circadian input pathway might contribute (Fig. 4). Photoreceptor genes are differentially expressed in the epidermal and mesophyll cell layers (Somers and Quail, 1995), and photoreceptors are known to alter circadian period in a dose-dependent manner (Somers et al., 1998b; Devlin and Kay, 2000). The light input pathways might thus control circadian period in a tissue-specific fashion. Observed distinctions between circadian rhythms suggest that further specialization of the circadian clock might be present in the cells that sense or respond to photoperiodic signals (for example, Salisbury and Denney, 1971; Fowler et al., 1999) or control rhythmic leaf movement (Hennessey and Field, 1992; Fowler et al., 1999; Park et al., 1999). Rhythmic reporters that peak at various phases in restricted spatial patterns are required to determine how much the circadian clock is modulated for specialized timing functions. Recent microarray experiments suggest many candidate promoters (Harmer et al., 2000; Schaffer et al., 2001), which can now be tested in detail using LUC fusions.
MATERIALS AND METHODS
Plant Material, Media, and Growth Conditions
Experiments were performed with transgenic Arabidopsis seedlings carrying the native firefly (Photinus pyralis) LUC gene under the control of one of two chalcone synthase promoters. Arabidopsis plants of the C24 accession carrying the native Arabidopsis CHS promoter fused to the LUC gene have been described (Michelet and Chua, 1996). The Columbia transgenic line carrying a fusion of the same Arabidopsis CHS promoter fragment to GUS was also described previously (Hartmann et al., 1998). Transgenic plants of the Ler accession carried the promoter of white mustard (Sinapis alba) CHS1 from −907 to +26 bp (Batschauer et al., 1991), fused to the LUC gene in the pMON 721 binary vector backbone: Four independently transformed lines were tested with very similar results (data not shown). The CAB:LUC transgene introgressed from C24 into the Ler background has been described (Somers et al., 1998b). CHS:LUC from the C24 accession was crossed with the det1-1 mutant in the Columbia background (from Dr. Joanne Chory, Salk Institute). det1-1 mutant and wild-type plants were selected from the F2 population on the basis of their morphology; mutant scoring was confirmed in the F3 generation. The line carrying CAB:LUC in the det1-1 background has been described (Millar et al., 1995b). CHS:LUC in C24 was crossed to the toc1-1 mutant that carries CAB:LUC in the C24 background (Millar et al., 1995a). Homozygous mutant F2 progeny were selected by scoring short-period CAB:LUC luminescence in leaves; among these, seedlings carrying CHS:LUC were selected by scoring luminescence in roots. The CAB:LUC reporter was removed by segregation in the F3, resulting in greatly reduced luminescence in leaves. The presence of CHS:LUC and absence of CAB:LUC was confirmed by PCR using transgene-specific primers designed from published sequences (data not shown).
Seedlings were grown at 22°C in 12:12-h light-dark cycles (unless otherwise indicated) on agar medium containing 3% (w/v) Suc under cool-white fluorescent lights of the fluence rates indicated for each figure. Our standard conditions (approximately 60 μmol m−2 s−1) gave low CHS expression (data not shown); higher fluence rates of 150 to 250 μmol m−2 s−1 increased expression from these constructs, so all subsequent experiments were conducted within this range.
Localization of CHS Expression
Histochemical localization of GUS (Fig. 1E) was performed as described (Jefferson et al., 1987). Ruthenium red counterstaining was carried out by standard techniques. Tissue sections were prepared for light microscopy using a Technovit 7100 embedding kit (Kulzer, Wehrheim, Germany). High-resolution imaging of LUC activity was performed essentially as described (Hall et al., 2001). The image in Figure 1, G and H, was captured through a Fluar 20x objective (Zeiss, Jena, Germany) by a 30-min exposure on a back-thinned CCD in a liquid-nitrogen cooled camera (LN/CCD-512-TKB with ST133 controller, Roper Scientific Ltd., Marlow, UK). Camera background has been removed from Figure 1G. The specific signal in this image ranges from 2 (dark blue) to 24 (white) counts per pixel. Dark current was undetectable and readout noise was 1.8 counts (sd). The lowest luminescence levels have been clipped in Figure 1H to reveal the tissue outline.
Imaging of LUC Bioluminescence Rhythms and Statistical Analysis
After luciferin pretreatment (Millar et al., 1992), seedlings were sprayed before each image with 1 mm d-luciferin (Promega, Madison, WI) in 0.01% (w/v) Triton X-100 and left in darkness for 5 min to allow chlorophyll chemiluminescence to decay. Bioluminescence was detected by ultra-low-light cameras (VIM C2400–47, Hamamatsu, Bridgewater, NJ; and Roper Scientific LN/CCD-512-TKB with ST138 controller), as described (Millar et al., 1992; Michelet and Chua, 1996; Hall et al., 2001). The data from both cameras were similar, although absolute luminescence counts are not directly comparable because of the different image acquisition methods (for example, in Figs. 2 and 3). Data are expressed as detected photons per plant per 25-min image, derived from data from groups of 10 to 20 plants. Data in Figures 2 and 3 have been processed with a three-point, boxcar filter. Period estimates from the raw data were produced by the fast Fourier transform-nonlinear least squares method (FFT-NLLS, Plautz et al., 1997). The first 24 h of each time course was excluded from the period estimation to exclude any transient effects of the transfer to continuous light.
To derive mean period estimates from the results of seven experiments, the data of Table I were analyzed using residual maximum likelihood (REML) (Patterson and Thompson, 1971) in the statistical package Genstat 5 (Payne et al., 1993). REML can be thought of as a generalization of analysis of variance to unbalanced designs. Data were weighted for analysis by the reciprocal of the estimated variance of the circadian period for the trace, which was derived from FFT-NLLS (Millar et al., 1995a). The data were analyzed with each line taken as a fixed effect and experiment and trace within experiment as random effects. The significance of differences between pairs of treatments were assessed using t tests, based on se of the differences derived from REML, rather than the se of individual means presented in Table I (Patterson and Thompson, 1971). The mean periods and se of Table II are derived from the same variance-weighting procedure on period estimates from FFT-NLLS for replicated samples in a separate experiment.
Waveforms in the data that are unusually close to a cosine wave can give very low estimated ses from FFT-NLLS and, thus, gain disproportionate weight in the variance-weighted means. The analysis was repeated with revised weights that were derived by adding 0.1 to the original, estimated ses to reduce the effects of such rare estimates. The conclusions of the analysis were not altered by this procedure, and the data presented are weighted using the original estimates.
ACKNOWLEDGMENTS
We thank Profs. Gareth Jenkins and Nam-Hai Chua for helpful discussions and materials and members of the chronobiology group at Warwick for discussions and assistance with imaging experiments.
Footnotes
This work was supported by the Biotechnology and Biological Science Research Council (graduate studentship to S.C.T. and grant no. BI11209 to A.J.M.). The Warwick imaging facility is supported by the Gatsby Charitable Foundation, the Royal Society, and the Biotechnology and Biological Science Research Council (grants to A.J.M.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.005405.
LITERATURE CITED
- Alabadi D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA. Reciprocal regulation between TOC1 and LHY/CCA1. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- Batschauer A, Ehmann B, Schafer E. Cloning and characterization of a chalcone synthase gene from mustard and its light-dependent expression. Plant Mol Biol. 1991;16:175–185. doi: 10.1007/BF00020550. [DOI] [PubMed] [Google Scholar]
- Chory J, Peto CA. Mutations in the DET1 gene affect cell-type-specific expression of light-regulated genes and chloroplast development in Arabidopsis. Proc Natl Acad Sci USA. 1990;87:8776–8780. doi: 10.1073/pnas.87.22.8776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deikman J, Hammer PE. Induction of anthocyanin accumulation by cytokinins in Arabidopsis thaliana. Plant Physiol. 1995;108:47–57. doi: 10.1104/pp.108.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Kay SA. Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell. 2000;12:2499–2509. doi: 10.1105/tpc.12.12.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowson-Day MJ, Millar AJ. Circadian dysfunction causes aberrant hypocotyl elongation patterns in Arabidopsis. Plant J. 1999;17:63–71. doi: 10.1046/j.1365-313x.1999.00353.x. [DOI] [PubMed] [Google Scholar]
- Ehmann B, Ocker B, Schafer E. Development-dependent and light-dependent regulation of the expression of two different chalcone synthase transcripts in mustard cotyledons. Planta. 1991;183:416–422. doi: 10.1007/BF00197741. [DOI] [PubMed] [Google Scholar]
- Engelmann W, Johnsson A. Attenuation of the petal movement rhythm in Kalanchoe with light pulses. Physiol Plant. 1978;43:68–76. [Google Scholar]
- Feinbaum RL, Ausubel F. Transcriptional regulation of the Arabidopsis thaliana chalcone synthase gene. Mol Cell Biol. 1988;8:1985–1992. doi: 10.1128/mcb.8.5.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fejes E, Nagy F. Molecular analysis of circadian clock-regulated gene expression in plants: features of the “output” pathways. In: Lumsden PJ, Millar AJ, editors. Biological Rhythms and Photoperiodism in Plants. Oxford: BIOS Scientific; 1998. pp. 99–118. [Google Scholar]
- Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Coupland G, Putterill J. GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 1999;18:4679–4688. doi: 10.1093/emboj/18.17.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglevand G, Jackson JA, Jenkins GI. UV-B, UV-A and blue light signal transduction pathways interact synergistically to regulate chalcone synthase expression in Arabidopsis. Plant Cell. 1996;8:2347–2357. doi: 10.1105/tpc.8.12.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorr G, Obst G, Doring O, Bottger M. Light-dependent proton excretion of wheat (Triticum aestivum L) and maize (Zea mays L) roots. Bot Acta. 1995;108:351–357. [Google Scholar]
- Gorton HL, Williams WE, Binns ME, Gemmell CN, Leheny EA, Shepherd AC. Circadian stomatal rhythms in epidermal peels from Vicia faba. Plant Physiol. 1989;90:1329–1334. doi: 10.1104/pp.90.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahlbrock K, Scheel D. Physiology and molecular biology of phenylpropanoid metabolism. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:347–369. [Google Scholar]
- Hall A, Kozma-Bognar L, Toth R, Nagy F, Millar AJ. Conditional circadian regulation of PHYTOCHROME A gene expression. Plant Physiol. 2001;127:1808–1818. [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- Hartmann U, Valentine WJ, Christie JM, Hays J, Jenkins GI, Weisshaar B. Identification of UV/blue light-response elements in the Arabidopsis thaliana chalcone synthase promoter using a homologous protoplast transient expression system. Plant Mol Biol. 1998;36:741–754. doi: 10.1023/a:1005921914384. [DOI] [PubMed] [Google Scholar]
- Haussuhl K, Rohde W, Weissenbock G. Expression of chalcone synthase genes in coleoptiles and primary leaves of Secale cereale L after induction by UV radiation: evidence for a UV-protective role of the coleoptile. Bot Acta. 1996;109:229–238. [Google Scholar]
- Hennessey TL, Field CB. Evidence of multiple circadian oscillators in bean plants. J Biol Rhythms. 1992;7:105–113. doi: 10.1177/074873049200700202. [DOI] [PubMed] [Google Scholar]
- Henzler T, Waterhouse RN, Smyth AJ, Carvajal M, Cooke DT, Schaffner AR, Steudle E, Clarkson DT. Diurnal variations in hydraulic conductivity and root pressure can be correlated with the expression of putative aquaporins in the roots of Lotus japonicus. Planta. 1999;210:50–60. doi: 10.1007/s004250050653. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GI, Long JC, Wade HK, Shenton MR, Bibikova TN. UV and blue light signalling: pathways regulating chalcone synthase gene expression in Arabidopsis. New Phytol. 2001;151:121–131. doi: 10.1046/j.1469-8137.2001.00151.x. [DOI] [PubMed] [Google Scholar]
- Johnson CH. Endogenous timekeepers in photosynthetic organisms. Annu Rev Physiol. 2001;63:695–728. doi: 10.1146/annurev.physiol.63.1.695. [DOI] [PubMed] [Google Scholar]
- Kaiser T, Batschauer A. cis-Acting elements of the CHS1 gene from white mustard controlling promoter activity and spatial patterns of expression. Plant Mol Biol. 1995;28:231–243. doi: 10.1007/BF00020243. [DOI] [PubMed] [Google Scholar]
- Kaiser T, Emmler K, Kretsch T, Weisshaar B, Schafer E, Batschauer A. Promoter elements of the mustard CHS1 gene are sufficient for light regulation in transgenic plants. Plant Mol Biol. 1995;28:219–229. doi: 10.1007/BF00020242. [DOI] [PubMed] [Google Scholar]
- Kim HY, Coté GG, Crain RC. Potassium channels in Samanea saman protoplasts controlled by phytochrome and the biological clock. Science. 1993;260:960–962. doi: 10.1126/science.260.5110.960. [DOI] [PubMed] [Google Scholar]
- Kreps JA, Simon AE. Environmental and genetic effects on circadian clock-regulated gene expression in Arabidopsis. Plant Cell. 1997;9:297–304. doi: 10.1105/tpc.9.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumsden PJ, Millar AJ, editors. Biological rhythms and photoperiodism in plants. Oxford: BIOS Scientific; 1998. [Google Scholar]
- Mayer WE, Fischer C. Protoplasts from Phaseolus coccineus L. pulvinar motor cells show circadian volume oscillations. Chronobiol Int. 1994;11:156–164. doi: 10.3109/07420529409057235. [DOI] [PubMed] [Google Scholar]
- McClung CR. Circadian rhythms in plants: a millennial view. Physiol Plant. 2000;109:359–371. [Google Scholar]
- Michelet B, Chua NH. Improvement of Arabidopsis mutant screens based on luciferase imaging in planta. Plant Mol Biol Rep. 1996;14:320–329. [Google Scholar]
- Millar AJ. The cellular organization of circadian rhythms in plants: not one but many clocks. In: Lumsden PJ, Millar AJ, editors. Biological Rhythms and Photoperiodism in Plants. Oxford: BIOS Scientific; 1998. pp. 51–68. [Google Scholar]
- Millar AJ. Tansley review no. 103: biological clocks in Arabidopsis thaliana. New Phytol. 1999;141:175–197. doi: 10.1046/j.1469-8137.1999.00349.x. [DOI] [PubMed] [Google Scholar]
- Millar AJ, Carré IA, Strayer CA, Chua NH, Kay SA. Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science. 1995a;267:1161–1163. doi: 10.1126/science.7855595. [DOI] [PubMed] [Google Scholar]
- Millar AJ, Short SR, Hiratsuka K, Chua N-H, Kay SA. Firefly luciferase as a reporter of regulated gene expression in higher plants. Plant Mol Biol Rep. 1992;10:324–337. [Google Scholar]
- Millar AJ, Straume M, Chory J, Chua N-H, Kay SA. The regulation of circadian period by phototransduction pathways in Arabidopsis. Science. 1995b;267:1163–1166. doi: 10.1126/science.7855596. [DOI] [PubMed] [Google Scholar]
- Murtas G, Millar AJ. How plants tell the time. Curr Opin Plant Biol. 2000;3:43–46. doi: 10.1016/s1369-5266(99)00034-5. [DOI] [PubMed] [Google Scholar]
- Mustilli AC, Fenzi F, Ciliento R, Alfano F, Bowler C. Phenotype of the tomato high pigment-2 mutant is caused by a mutation in the tomato homolog of DEETIOLATED1. Plant Cell. 1999;11:145–157. doi: 10.1105/tpc.11.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM, Fankhauser C, Chory J. Light: an indicator of time and place. Genes Dev. 2000;14:257–271. [PubMed] [Google Scholar]
- Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci USA. 1998;95:8660–8664. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh MS, Kim HJ, Kay SA, Nam HG. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science. 1999;285:1579–1582. doi: 10.1126/science.285.5433.1579. [DOI] [PubMed] [Google Scholar]
- Parsons L, Kramer PJ. Diurnal cycling in root resistance to water movement. Physiol Plant. 1974;30:19–23. [Google Scholar]
- Patterson HD, Thompson R. Recovery of inter-block information when block sizes are unequal. Biometrika. 1971;58:545–554. [Google Scholar]
- Payne RW, Lane P, Digby P, Harding S, Leech P, Morgan G, Todd A, Thompson R, Tunnicliffe W, Welham S et al. Genstat 5 Release 3 Reference Manual. Oxford: Oxford University Press; 1993. [Google Scholar]
- Pepper A, Delaney T, Washburn T, Poole D, Chory J. DET1, a negative regulator of light-mediated development and gene expression in Arabidopsis, encodes a novel nuclear-localized protein. Cell. 1994;78:109–116. doi: 10.1016/0092-8674(94)90577-0. [DOI] [PubMed] [Google Scholar]
- Peter H-J, Kroger-Alef C, Knogge W, Brinkmann K, Weissenbock G. Diurnal periodicity of chalcone-synthase activity during the development of oat primary leaves. Planta. 1991;183:409–415. doi: 10.1007/BF00197740. [DOI] [PubMed] [Google Scholar]
- Plautz JD, Straume M, Stanewsky R, Jamison CF, Brandes C, Dowse HB, Hall JC, Kay SA. Quantitative analysis of Drosophila period gene transcription in living animals. J Biol Rhythms. 1997;12:204–217. doi: 10.1177/074873049701200302. [DOI] [PubMed] [Google Scholar]
- Ripperger JA, Shearman LP, Reppert SM, Schibler U. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev. 2000;14:679–689. [PMC free article] [PubMed] [Google Scholar]
- Roenneberg T, Mittag M. The circadian program of algae. Semin Cell Dev Biol. 1996;7:753–763. [Google Scholar]
- Sai J, Johnson CH. Different circadian oscillators control Ca2+ fluxes and Lhcb gene expression. Proc Natl Acad Sci USA. 1999;96:11659–11663. doi: 10.1073/pnas.96.20.11659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury FB, Denney A. Separate clocks for leaf movements and photoperiodic flowering in Xanthium strumarium L. In: Menaker M, editor. Biochronometry. Washington, DC: National Academy of Sciences; 1971. pp. 292–311. [Google Scholar]
- Schaffer R, Landgraf J, Monica A, Simon B, Larson M, Wisman E. Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell. 2001;13:113–123. doi: 10.1105/tpc.13.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzer E, Jahnen W, Hahlbrock K. In situ localization of light-induced chalcone synthase messenger-RNA, chalcone synthase, and flavonoid end products in epidermal cells of parsley leaves. Proc Natl Acad Sci USA. 1988;85:2989–2993. doi: 10.1073/pnas.85.9.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon E, Satter RL, Galston AW. Circadian rhythmicity in excised Samanea pulvini: II. Resetting the clock by phytochrome conversion. Plant Physiol. 1976;58:421–425. doi: 10.1104/pp.58.3.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Devlin PF, Kay SA. Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science. 1998a;282:1488–1490. doi: 10.1126/science.282.5393.1488. [DOI] [PubMed] [Google Scholar]
- Somers DE, Quail PH. Temporal and spatial expression patterns of PHYA and PHYB genes in Arabidopsis. Plant J. 1995;7:413–427. doi: 10.1046/j.1365-313x.1995.7030413.x. [DOI] [PubMed] [Google Scholar]
- Somers DE, Webb AAR, Pearson M, Kay SA. The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development. 1998b;125:485–494. doi: 10.1242/dev.125.3.485. [DOI] [PubMed] [Google Scholar]
- Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Mas P, Panda S, Kreps JA, Kay SA. Cloning of the Arabidopsis clock cone TOC1, an autoregulatory response regulator homolog. Science. 2000;289:768–771. doi: 10.1126/science.289.5480.768. [DOI] [PubMed] [Google Scholar]
- Swarup K, Alonso-Blanco C, Lynn JR, Michaels SD, Amasino RM, Koornneef M, Millar AJ. Natural allelic variation identifies new genes in the Arabidopsis circadian system. Plant J. 1999;20:67–77. doi: 10.1046/j.1365-313x.1999.00577.x. [DOI] [PubMed] [Google Scholar]
- Thain SC, Hall A, Millar AJ. Functional independence of circadian clocks that regulate plant gene expression. Curr Biol. 2000;10:951–956. doi: 10.1016/s0960-9822(00)00630-8. [DOI] [PubMed] [Google Scholar]
- Vaadia Y. Autonomic diurnal fluctuations in rate of exudation and root pressure of decapitated sunflower plants. Physiol Plant. 1960;13:701–717. [Google Scholar]
- van Esseveldt LKE, Lehman MN, Boer GJ. The suprachiasmatic nucleus and the circadian time-keeping system revisited. Brain Res Rev. 2000;33:34–77. doi: 10.1016/s0165-0173(00)00025-4. [DOI] [PubMed] [Google Scholar]
- Wood NT, Haley A, Viry-Moussaid M, Johnson CH, van der Luit AH, Trewavas AJ. The calcium rhythms of different cell types oscillate with different circadian phases. Plant Physiol. 2001;125:787–796. doi: 10.1104/pp.125.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]