Abstract
Functional analyses of a number of hydrolase gene promoters, induced by gibberellin (GA) in aleurone cells following germination, have identified a GA-responsive complex as a tripartite element containing a pyrimidine box motif 5′-CCTTTT-3′. We describe here that BPBF, a barley (Hordeum vulgare) transcription factor of the DOF (DNA-Binding with One Finger) class, previously shown to be an activator of reserve protein encoding genes during development, also has a role in the control of hydrolase genes following seed germination. Northern-blot, reverse transcriptase-polymerase chain reaction, and in situ hybridization analyses evidenced that the transcripts of the BPBF-encoding gene (Pbf), besides being present during endosperm development, are also expressed in aleurone cells of germinated seeds where they are induced by GA, an effect counteracted by abscisic acid. Electrophoretic mobility shift assays have shown that the BPBF protein binds specifically to the pyrimidine box motif in vitro within the different sequence contexts that naturally occur in the promoters of genes encoding a cathepsin B-like protease (Al21) and a low-isoelectric point α-amylase (Amy2/32b), both induced in the aleurone layers in response to GA. In transient expression experiments, BPBF repressed transcription of the Al21 promoter in GA-treated barley aleurone layers and reverted the GAMYB-mediated activation of this protease promoter.
In cereal seeds, the endosperm is the major organ for storage of the nutritional reserves that will support seedling growth before the onset of photosynthesis. The endosperm is differentiated into two predominant tissue types, the central starchy endosperm and the peripheral aleurone layer (Bosnes et al., 1992). During seed maturation, the starchy endosperm is committed to the synthesis and deposition of carbohydrates and storage proteins. When the seed desiccation phase initiates, it undergoes a degenerative process related to programmed cell death (Young and Gallie, 1999). By contrast, the aleurone cells remain viable in the mature seed, and, following germination, secrete a number of hydrolytic enzymes that mobilize the stored nutrients, mainly starch and proteins (Fincher, 1989; Skadsen, 1998). Gene regulation in these aleurone cells is under the control of phytohormones, mainly of the ratio of gibberellins (GA) to abscisic acid (ABA). GAs are synthesized by the embryo and diffuse into the aleurone (Appleford and Lenton, 1997), where they trigger the transcriptional activation of a number of hydrolase genes such as those encoding several classes of α-amylases, proteases, and β-glucanases (Fincher, 1989). The activating effect of GA is counteracted by ABA (Lovegrove and Hooley, 2000).
Although a GA receptor has not yet been isolated, many interesting findings have been documented related to the site of perception of GA, elegantly localized to the external face of the plasma membrane (Gilroy and Jones, 1994), and related to the transduction events that follow after the signal is perceived (for review, see Bethke and Jones, 1998; Lovegrove and Hooley, 2000).
However, little is known about the actual transcription factors involved in the regulation of the hydrolase encoding genes. Functional analysis of the α-amylase promoters from barley (Hordeum vulgare), wheat (Triticum aestivum), and rice (Oryza sativa) have identified a conserved cis-element required for GA induction, termed the GA responsive complex (GARC). Although this GARC may not always be tripartite, most often it includes three sequence motifs, the TAACAAA box or GA responsive element (GARE), the pyrimidine box CCTTTT, and the TATCCAC box (Skriver et al., 1991; Gubler and Jacobsen, 1992; Rogers et al., 1994). Mutation analysis of the GARC from low and high pI α-amylase genes (Amy2/32b and Amy1/pHV19, respectively) have shown that these three boxes are necessary for a full GA response in transiently transformed barley (Hordeum vulgare) aleurone layers (Lanahan et al., 1992; Jacobsen et al., 1995). Disruption of either of them resulted in a decreased level of promoter expression in GA-treated aleurone layers, but did not completely abolish the GA induction. Promoter regions closely resembling the GARC have also been described in other genes expressed in the aleurone such as those encoding the (1-3, 1-4)-β-glucanase isozyme II (Slakeski and Fincher, 1992; Wolf, 1992), and the thiol-proteases cathepsin B-like (Cejudo et al., 1992a, 1992b) and EPB1 (Cercós et al., 1999). In spite of the similarity of most GARC in different hydrolase promoters, significant differences are also noted that may explain the timing and level of expression controlled by them. In this regard, the EPB1 promoter contains a functional GARE, a pyrimidine box, and an upstream element in different orientations than those in α-amylase gene promoters. This has been proposed to be the reason why the EPB1 gene shows a lower level of expression than α-amylase genes in response to GA (Cercós et al., 1999).
Although the cis-motifs constitutive of the tripartite GARC element have been well defined, the isolation of the corresponding trans-acting factors have lagged far behind. The first such factor to be characterized was the GAMYB protein from barley (Gubler et al., 1995). GAMYB was induced by GA in aleurone cells and, through binding to the TAACAAA-like sequences, was able to trans-activate a number of hydrolase gene promoters induced in those cells upon germination (Gubler et al., 1995, 1999). A transcription repressor (HRT) has also been described associated with the GARE (Raventós et al., 1998). However, the trans-acting factors recognizing the pyrimidine and the TATCCAC boxes have not yet been identified.
In this context, we found relevant that the pyrimidine box, 5′-CTTTT-3′, contains in the anti-sense strand the 5′-AAAAG-3′ sequence defined as the core motif required for the binding of transcription factors of the DNA-binding with one finger (DOF) class (Yanagisawa, 1996). Protein BPBF (PBF; gene Pbf) is a DOF transcription factor expressed in the developing endosperm of maize (Zea mays), barley, and wheat where it is involved in the activation of seed storage protein genes (Vicente-Carbajosa et al., 1997; Mena et al., 1998; Yanagisawa and Schmidt, 1999). Moreover, Washio (2001) reported the isolation of five cDNAs encoding DOF proteins (OsDof1-5) from a rice aleurone library by southwestern screening with a pyrimidine box probe. In transient assays, OsDof3 was a repressor of the GA-induced activity of a type III carboxypeptidase promoter. These data and the fact that the transcription factor Viviparous-1 (VP1) in maize seems to be involved in the control of seed maturation and germination programs (Hoecker et al., 1995) led us to explore whether the barley gene Pbf might also have a role in seed germination.
In the present study, we have investigated the expression profile of the barley Pbf gene in seeds following germination and we tested the binding ability of BPBF toward the pyrimidine box sequence in promoters of GA-controlled hydrolase genes. More specifically, we show here that the recombinant PBF was able to bind in vitro the pyrimidine box sequence motif in the cathepsin B-type protease Al21 and in the α-amylase Amy2/32b promoter contexts. In transient expression experiments, BPBF repressed the GA-induced activity of the Al21 gene promoter and could largely revert its GAMYB-mediated trans-activation. This indicates that Pbf has a functional role as a transcriptional repressor in aleurone cells, beside being a transcription activator of prolamin genes during endosperm development, as previously described by us (Mena et al., 1998).
RESULTS
The Barley Pbf Gene Is Expressed in the Aleurone Cells following Germination
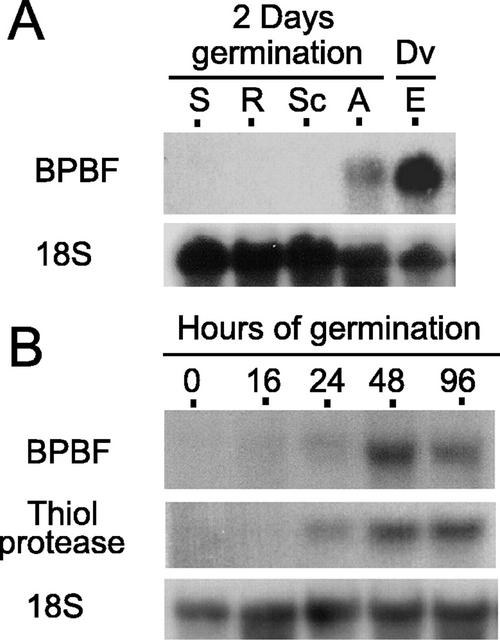
To explore the possibility of Pbf being expressed in germinating seeds, total RNA was prepared from dissected tissues (aleurone, scutellum, shoots, and roots) of 2-d-imbibed kernels of the barley cv Bomi. The Pbf transcript was detected in the aleurone sample upon northern-blot analysis (Fig. 1A). It should be noted that the hybridization signal for Pbf was approximately 50 times lower in the aleurone than in the developing endosperm sample run as a control. Our previous results had shown that Pbf was expressed in developing barley endosperms, but its mRNA was not detected in immature embryos or vegetative tissues (Mena et al., 1998). To discard the possibility that the Pbf mRNA was present following germination as a result of storage in the dormant aleurone of mRNAs previously synthesized during development, the kinetics of the mRNA accumulation was examined at different times after seed rehydration (Fig. 1B). The barley Pbf message was not detected in the mature dry seeds, and, upon imbibition, it was first detected in northern blots at 16 h, gradually increasing until 48 h, and decreasing thereafter (96 h). All blots were subsequently hybridized with a probe for a wheat cDNA encoding a cathepsin B-like thiol protease, which is highly expressed in the aleurone layer during germination (Cejudo et al., 1992b). The expression pattern of the thiol protease gene closely followed that found for Pbf, which was compatible with its being regulated by Pbf. The blots were finally hybridized with an 18S ribosomal probe as a loading control (Fig. 1).
Figure 1.
Northern-blot analysis of barley Pbf expression during germination. A, Ten micrograms of total RNA from different tissues of 2-d germinated kernels of the barley cv Bomi: shoots (S), roots (R), scutellum (Sc), and aleurone (A). For comparison, a sample of 4 μg of total RNA from developing endosperms (Dv/E) at 15 d after pollination (DAP) was included. B, Comparative accumulation of Pbf transcripts in germinating barley aleurones. Total RNA (10 μg) was extracted from aleurones rehydrated during 0, 16, 24, 48, and 96 h. The blot was first hybridized with the barley Pbf-specific probe (BPBF) and subsequently with a probe for the gene Al21 of the cathepsin-B type from wheat (Thiol protease) and/or a 18S rRNA probe as a loading control.
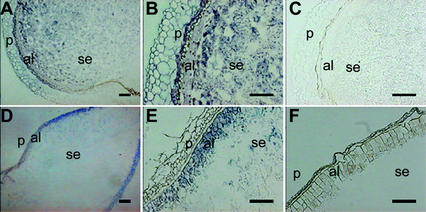
To determine the spatial expression of the Pbf mRNA, we did in situ hybridization experiments. This analysis was performed in barley seeds after 2 d of imbibition and in developing kernels at 15 DAP (Fig. 2). During development, a clear signal was detected not only in the starchy endosperm, but also in the aleurone layer (Fig. 2, A and B), which is clearly differentiated at this stage of development. No expression was observed in the pericarp or in the nucellar tissue. When longitudinal sections of 2-d rehydrated grains were hybridized with the barley Pbf antisense probe, a clear signal was observed that was exclusively localized to the aleurone layer (Fig. 2, D and E). No signal above background was detected when sections of developing or germinating seeds were hybridized to the sense probe (Fig. 2, C and F).
Figure 2.
Spatial expression of the Pbf mRNA in developing and germinating seeds of barley determined by in situ hybridization. A, B, and E are transversal sections of 15 DAP developing seeds. D through F are longitudinal sections of 2-d germinating seeds. Sections A, B, D, and E were hybridized with a Pbf antisense RNA probe. Hybridization with a Pbf sense RNA probe as negative controls C and F. al, Aleurone; se, starchy endosperm; p, pericarp. Bars = 100 μm in A, C, E, and F; 250 μm in A; and 500 μm in D.
The Expression of Barley Pbf in the Aleurone Is Hormonally Regulated
The observed accumulation pattern of the barley Pbf transcript upon seed imbibition suggested that it may be up-regulated by GA. We have used isolated aleurone layers as a system to study the hormonal control of the Pbf gene because they do not synthesize GA, but are able to respond to it. Aleurone layers were isolated from deembryonated grains of Himalaya barley after 4 d of imbibition and were incubated in the presence of 1 μm GA3 over different periods of time up to 24 h.
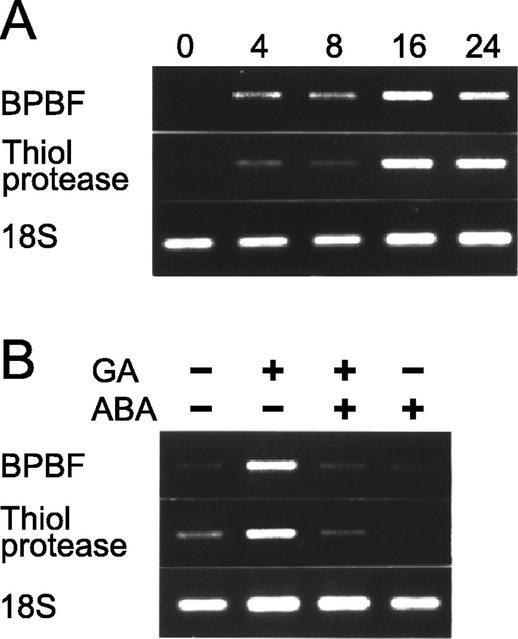
Total RNA was prepared from these samples and the time course of Pbf mRNA accumulation was analyzed using relative reverse transcriptase (RT)-PCR techniques. For comparative purposes, the induction profile of the barley homolog of the wheat gene encoding the cathepsin B-like thiol protease Al21 was also studied. As shown in Figure 3A, aleurone layers at the beginning of the incubation period (0 time) contained barely detectable levels of the Pbf mRNA. After 4 h of GA treatment, these levels sharply increased, reaching their maximum by 16 h and then declining slightly. As expected, incubation with GA3 induced the thiol protease transcript levels (Cejudo et al., 1992b). It is well documented that the GA effect on hydrolase gene transcription in aleurone layers can be reverted by an external concentration of 10 μm ABA. We analyzed the possible interaction between these two hormones in Pbf expression. Figure 3B shows the levels of the Pbf mRNA, as well as those of the thiol protease run as a control, in aleurone layers incubated for 16 h in the presence of GA3, ABA, and GA3 plus ABA. The treatment with ABA did not activate the expression of either of the genes tested, and was effective in antagonizing the induction of Pbf transcription promoted by GA3.
Figure 3.
RT-PCR analysis of hormone response of Pbf expression in rehydrated aleurone layers. Total RNA was isolated from aleurone layers after different hormone treatments and was reverse transcribed in the presence of random hexamers. The first strand cDNA was then amplified by PCR using gene-specific primers for the Pbf (BPBF) or the cathepsin B-like thiol protease (thiol protease) transcripts. Amplification of a region of the 18S RNA was used as the internal control (18S). The PCR products were fractionated in agarose gels and were visualized by ethidium bromide staining. A, Comparative Pbf expression pattern in aleurone layers incubated in the presence of 1 μm GA3 for 0, 4, 8, 16, or 24 h. B, Expression analysis of Pbf and of the cathepsin B-like thiol protease transcripts in aleurone layers incubated for 4 h with (+) or without (−) 1 μm GA3, 10 μm ABA, or both, as indicated above.
BPBF Binds to the Pyrimidine Box Element from Hydrolase Gene Promoters
Because the complementary strand of the pyrimidine box element (5′-CTTTT-3′) in GA-induced hydrolase gene promoters was identical to the core sequence (5′-AAAAG-3′) recognized by PBF in prolamin gene promoters (P-box: 5′-T/AAAAG-3′; Vicente-Carbajosa et al., 1997; Mena et al., 1998; Yanagisawa and Schmidt, 1999), it seemed plausible that the PBF might bind to the pyrimidine box motif.
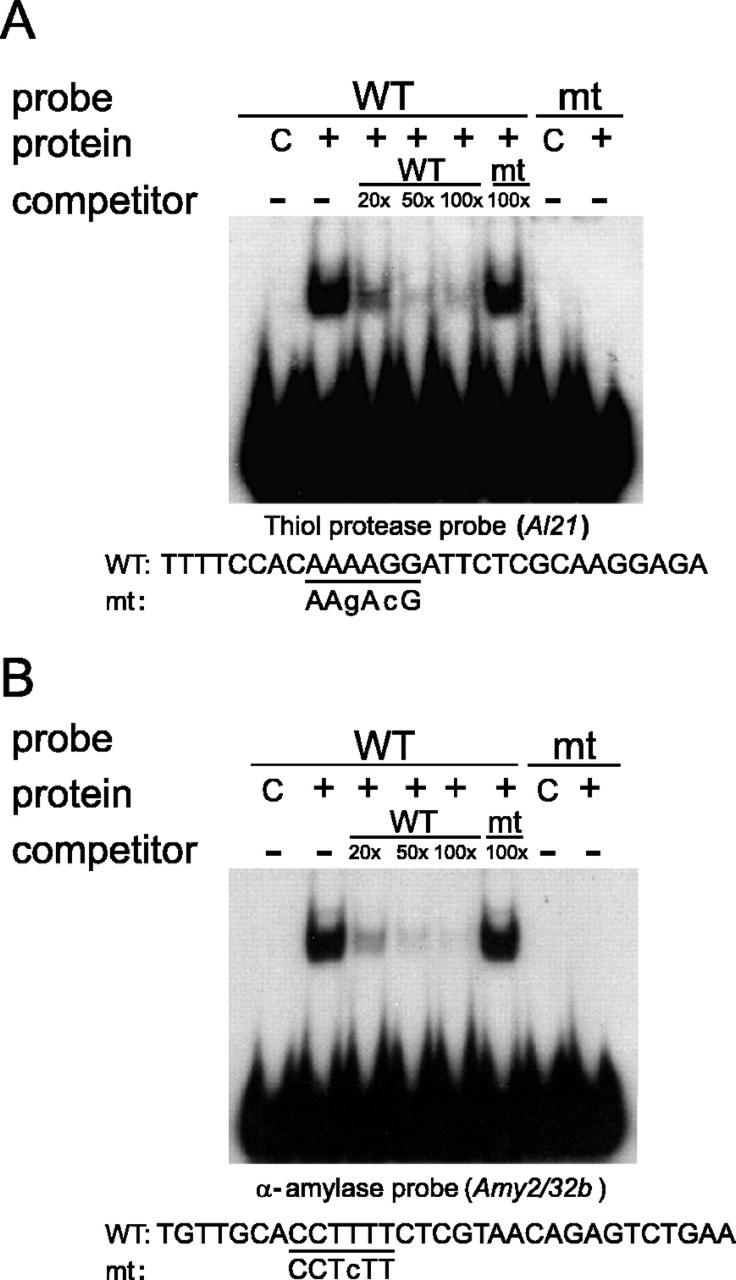
To test this possibility, two pyrimidine box elements placed in different contexts were assayed in vitro in electrophoretic mobility shift assays (EMSA; Fig. 4). A glutathione S-transferase (GST)-BPBF fusion protein was expressed and purified from Escherichia coli extracts and was used in the EMSA with appropriate radiolabeled DNA probes. The thiol protease probe was a 29-mer oligonucleotide containing the pyrimidine box motif 5′-AAAAGG-3′ (Fig. 5, D1) from the promoter of the gene Al21 (Cejudo et al., 1992b), considered a high-affinity binding site for PBF, according to Yanagisawa and Schmidt (1999). Two other pyrimidine boxes of lower affinity (our data not shown and Yanagisawa and Schmidt, 1999) were also present in the Al21 promoter (Fig. 5, D2 and D3).
Figure 4.
In vitro binding of the recombinant BPBF protein to the pyrimidine box sequence motif. EMSA performed with affinity purified GST-BPBF protein and oligonucleotides deduced from the thiol protease AL21 (A; D1 motif in Fig. 5) and the α-amylase Amy2/32b gene promoters (B). The sequences of the corresponding wild-type (WT) and mutant derivative (mt) oligonucleotides, used as probes, are shown at the bottom of each panel. The pyrimidine box motif is underlined and the point mutations are indicated by lowercase letters. Binding reactions were performed with the GST protein as a negative control (C) or with the GST-BPBF protein (+), in the absence (−) or in the presence of probe competitors at the indicated molar excesses (20×, 50×, and 100×).
Figure 5.
Schematic structure of effector and reporter constructs. A, Schematic representation of the effector constructs. The barley Pbf (BPBF protein) and GAMyb genes were under the control of the cauliflower mosaic virus (CaMV) 35S promoter (p35S) followed by the first intron of the maize AdhI gene (I-ADHI) and, downstream, flanked by the 3′-nopaline synthase (nos) sequences (nos). B, Diagrammatic structures of the reporter constructs used in this study: pCBG13, pCBG13.1, and pCBG13.5 as described in Cejudo et al. (1992a). The white and black boxes indicate, respectively, the location of putative binding sites for BPBF (D1, D2, and D3) and GAMYB (M) transcription factors, along with the cathepsin B-like thiol protease AL21 promoter sequences. C, Sequence of cis-motifs conforming the putative DOF- and MYB-binding sites in the AL21 gene promoter. Numbers at the left indicate the position from the translation start codon. Motifs containing the BPBF-binding sequence core (indicated by uppercase letters) are shown aligned with the consensus sequence for pyrimidine box elements.
The α-amylase probe was a 31-mer oligonucleotide derived from the promoter sequence of the low-pI α-amylase gene Amy2/32b (Whittier et al., 1987), which contains a canonical pyrimidine box element (5′-CCTTTT-3′). When these probes were incubated independently with the GST-BPBF protein, a shifted complex was observed that was competed out when the probes were incubated with a molar excess of the corresponding unlabeled oligonucleotides. As expected, this binding was not produced when the control GST protein was used in the assay (Fig. 4, A and B).
The specificity of the interaction was also confirmed using variants of these probes. In a previous study, we had shown that the binding of the BPBF protein to the prolamin-box motif in storage protein gene promoters was abolished when the core AAAG sequence was changed to AgAc (Mena et al., 1998). The same nucleotide substitutions were introduced at the pyrimidine box sequence in the thiol protease oligonucleotide (AAAAGG was changed to AAgAcG) to generate a mutant thiol protease probe. Even when a single base substitution was introduced, as in the mutated α-amylase probe (changing CCTTTT to CCTcTT), the binding of BPBF was abolished. As shown in Figure 4, none of these mutant versions of the probes were bound by the GST-BPBF protein or able to compete the binding of the corresponding wild-type probes, even at 100 molar excess.
BPBF Negatively Regulates the GA-Responsive Expression of a Thiol Protease Promoter in Cobombarded Aleurones
Because promoters containing a pyrimidine box motif are likely to be the target for BPBF in germinating aleurone nuclei, we addressed the question of whether BPBF modulates their transcriptional activity. Transient expression experiments were done by using particle bombardment into aleurone layers of cv Himalaya barley embryoless half-seeds.
The pBPBF effector construct expressed the whole barley Pbf cDNA under the control of the CaMV35S promoter followed by the first intron of the maize AdhI gene (Mena et al., 1998), and an equivalent construct with the GAMYB cDNA was used as the GAMYB effector; both constructs had the 3′-noncoding region of the nos gene (Fig. 5A).
As reporters, several fragments of the Al21 promoter fused to the β-glucuronidase (GUS) gene were chosen. The Al21 promoter constructs assayed had been described in an earlier study (Cejudo et al., 1992a) and consisted of three fragments spanning to positions −480, −381, and −355, respectively, from the translation initiation codon. Sequence analysis of the −480-bp promoter region in the pCBG13 construct showed (Fig. 5, B and C) that it contained a canonical pyrimidine box motif at position −401 (Fig. 5, B and C, “D1”), and two additional imperfect boxes at −434 and −235 (Fig. 5, B and C, “D2 and D3,” respectively). One putative GAMYB-binding site (Gubler et al., 1995) was found at position −367 (Fig. 5, B and C, “M”).
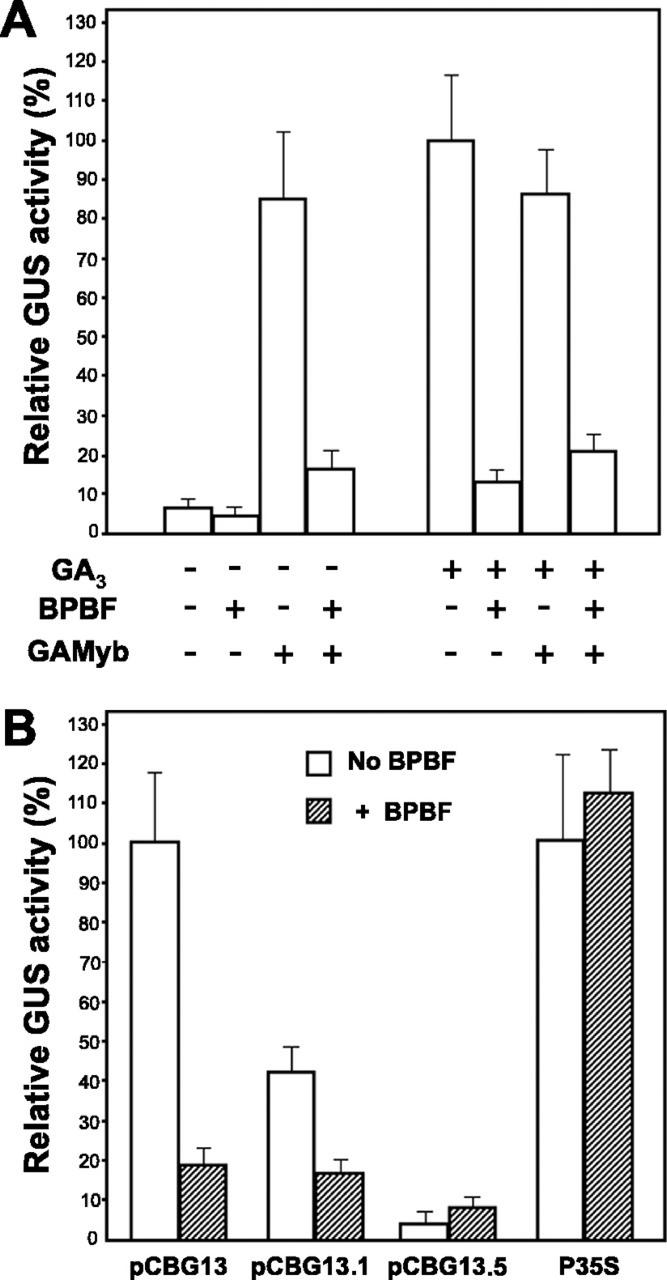
In a first set of assays, the pCBG13 reporter construct was bombarded into rehydrated aleurone cells, alone or with the pBPBF effector, at a 1:1 molar ratio. The GAMYB as effector (1:1) was used as a positive control (Gubler et al., 1999). The bombarded aleurone layers were then incubated in a solution with 1 μm GA3 or without this hormone. As represented in Figure 6A, the exogenous GA treatment resulted in ∼10-fold enhancement of the GUS activity over that found with the GA-untreated controls. Coexpression of BPBF largely reverted this GA-induced expression, reducing levels of GUS activity to ∼15% of that found in GA-treated aleurone layers. As expected, GUS activity from pCBG13 was highly induced by coexpression with GAMYB (Gubler et al., 1999), reaching comparable levels with those found with the GA-treatment alone. Cobombardment with BPBF greatly reduced the GAMYB trans-activation of pCBG13 in the presence and in the absence of exogenous GA3. These data indicate that BPBF functions in rehydrated aleurone cells as a negative regulator of the GA-dependent or GAMYB-inducible GUS activity driven by the Al21 thiol protease promoter. In the absence of GAMYB or when GA incubation was suppressed, GUS activity was very low, and the repression mediated by BPBF, if any, could not be evaluated (Fig. 6A).
Figure 6.
BPBF negative regulation of the transcription of AL21 thiol protease promoter. Intact aleurone layers were prepared from half-grains and were transfected by particle bombardment with the indicated combinations of reporter and effector plasmids at a 1:1 molar ratio. A, Effect of BPBF on the AL21 promoter activity of the reporter construct pCBG13. + indicates the presence and − indicates the absence of the effector constructs shown at the left (BPBF and/or GAMYB). When indicated, half-grains were incubated in the presence of 1 μm GA3. B, Effect of BPBF on the GA-induced transcription of three different AL21 promoter constructs. Aleurone cells were bombarded with the reporter construct indicated, alone (white bars) or in combination (1:1 molar ratio) with the BPBF effector (dashed bars), and were then incubated with 1 μm GA3. The p35S construct, where the GUS reporter gene was under the control of the CaMV35S promoter, was used as a control. Relative GUS activity is shown, considering expression levels of the pCBG13 construct in the presence of GA3 as 100%. Column height represents the mean value of 12 replicates from three independent experiments. Bars indicate ses.
To address the contribution of the various DOF motifs in the AL21 promoter to the BPBF transcriptional repression, the reporter constructs pCBG13.1 and pCBG13.5 (Fig. 5B), spanning, respectively, to positions −381 and −355 were assayed (Fig. 6B). The GA-induced GUS expression from pCBG13.1 was only ∼50% of that supported by the pCBG13 construct, but still retained its capacity to be repressed in trans by the BPBF transcription factor. Although the percentage of repression obtained relative to the respective background levels was smaller for pCBG13.1 (40% GUS) than for pCBG13 (18% GUS), the actual remaining activity after BPBF cotransformation was identical in both cases, probably indicating that interaction with D3 plays a major role in the repression mediated by BPBF, whereas promoter sequences between −480 and −381, namely the pyrimidine boxes D1 and D2 (Fig. 5, B and C), are positive elements. The GUS activity driven by the pCBG13.5 construct appears not to be GA inducible or repressible by BPBF. Considering that pCBG13.1 and pCBG13.5 differ only by the deletion of 26 bp of the promoter sequences, which removes the putative GAMYB-binding site (M), but not the D3 sequence likely to be recognized by BPBF, these results suggest that the repressing activity of BPBF in the aleurone cells may work through interaction with GAMYB. In this context, it is pertinent to mention that recent experiments from our laboratory have detected interaction between GAMYB and BPBF in the yeast two-hybrid system (Diaz et al., 2002). Overexpression of BPBF had no effect over the transcriptional activity of the constitutive promoter CaMV35S (Fig. 6B), indicating that BPBF does not act as a general negative regulator of transcription in the aleurone layers.
DISCUSSION
The two phases of the life cycle of cereal seeds, development and germination, are associated with the expression of different sets of genes, and the ratio of GA to ABA plays an essential role regulating gene expression in both programs.
The success of germination depends on the responses of aleurone cells to GA, which activates the expression of genes encoding hydrolytic enzymes, an effect that is counteracted by ABA (Lovegrove and Hooley, 2000). Initial analysis of GA-regulated gene promoters identified a cis-element (referred to as GARE) in cereal α-amylase promoters that was able to confer GA-regulated expression to truncated 35S promoters in transient assays (Skriver et al., 1991). However, it became clear that correct GA regulation of gene expression required additional elements that conformed the tripartite GARC (Lanahan et al., 1992; Jacobsen et al., 1995). So far, only transcription factors interacting with the GARE have been described are GAMYB, acting as an activator, and HRT, which is a repressor of transcription (Gubler et al., 1995, 1999; Raventós et al., 1998). However, to our knowledge, no transcription factors interacting with the other two cis-elements of the GARC have been characterized thus far.
Here, we describe that barley BPBF (gene Pbf), a transcription factor of the DOF class previously shown to be an activator of reserve protein genes in the developing barley endosperm (Mena et al., 1998), functions as a transcriptional repressor upon germination through interaction with the pyrimidine box of the GARC.
As shown by northern-blot analysis, RT-PCR, and in situ hybridization analysis, Pbf is expressed in the aleurone cells following germination in such a way that its mRNA is up-regulated by GA. This pattern of expression, together with in vitro binding assays of BPBF to the pyrimidine box in the context of promoters of α-amylase and protease-encoding genes is compatible with BPBF being a transcriptional regulator of hydrolase genes induced by GA in the aleurone cells upon germination. In transient expression assays, BPBF behaves as a transcription repressor of the GAMYB-transactivated or GA-inducible GUS reporter activity controlled by the thiol-protease Al21 gene promoter.
According to our results, the regulation of the Al21 gene by BPBF may be modeled under the possible scenario that BPBF can compete with another transcriptional activator(s) of the DOF class for binding to the pyrimidine box motifs. This view of BPBF as a competitor of another putative DOF transcription activator(s) is consistent with the observation that pyrimidine box motifs D1 and D2 are positive promoter elements for the response of the Al21 gene to GA (Fig. 6B; Cejudo et al., 1992a). It is also supported by similar results reported in the analysis of the Amy 2/32b (Lanahan et al., 1992), Amy 1/6-4 (Jacobsen et al., 1995), and EPB-1 (Cercós et al., 1999) genes. From this evidence, the pyrimidine boxes D1 and D2 are expected to be the site for binding of transcriptional activator(s). According to this model, the relative concentrations of BPBF and of such an activator(s), and their relative binding affinities, would direct the overall effect of the pyrimidine box elements upon GA induction. Although the nature of such an activator remains to be determined, it seems probable that it could also belong to the DOF class of transcription factors. In maize leaves, two DOF factors have been described as being involved in the light regulation of certain genes (Yanagisawa, 2000); modulation being achieved by competition for a common binding site between Dof1 acting as an activator, and Dof2 acting as a repressor (Yanagisawa and Sheen, 1998). Genetic evidence for two Arabidopsis DOF genes, DAG1 and DAG2, with opposite effects on germination has been recently reported (Papi et al., 2000; Gualberti et al., 2002), although their mechanism of action is not completely elucidated (Papi et al., 2002).
The repressor activity of BPBF might also operate indirectly through the interaction with other transcription factors associated with the AL21 promoter because BPBF contains no obvious repression domain and it is a good transcriptional activator itself in the yeast one-hybrid system and in the developing endosperm cells (Mena et al., 1998; Diaz et al., 2002). Transient expression in planta (this paper) and yeast two-hybrid assays (Diaz et al., 2002) indicate that BPBF interacts with GAMYB. This result opens the possibility that GAMYB function might be regulated by BPBF and, because the Pbf transcripts appear later than those of GAMYB and are simultaneous to downstream GAMYB-regulated genes such as those encoding hydrolytic enzymes, a possible role for BPBF might be to terminate Al21 GA-induced transcription as part of the start of programmed cell death in aleurone cells.
Coupled activator and repressor functions have been well documented for VP1 in the developing maize seeds, where it acts as a transcriptional activator of maturation-specific genes (McCarty et al., 1991) and as a repressor of α-amylase-encoding genes normally expressed after germination (Hoecker et al., 1995). Similar to VP1 in maize, during barley endosperm development, the PBF would act as an activator of hordein genes and as a transcriptional repressor of the protease gene Al21. Another transcription factor, expressed during seed development and in aleurone cells following germination, is GAMYB. Recent data from our laboratory indicate that GAMYB is a transcription factor involved in seed-storage protein gene regulation (Diaz et al., 2002), in addition to being a transcriptional activator of hydrolase-encoding genes in aleurone cells following germination (Gubler et al., 1995, 1999).
MATERIALS AND METHODS
RNA Isolation and Northern-Blot Analysis
Seeds of barley (Hordeum vulgare cv Bomi) were germinated at 22°C in the dark, and were used to collect samples of aleurone, scutellum, and root and shoot tissues. Developing endosperms were prepared from 15 DAP grains. Total RNA was extracted using the Chang et al. (1993) method for aleurone samples, or the Lagrimini et al. (1987) procedure for the other tissues analyzed.
RNA fractionation in formaldehyde-agarose gels, blotting, and hybridization with [32P]-labeled random-primed probes were performed as previously described (Mena et al., 1998). The barley Pbf-specific probe was obtained from an 896-bp SmaI-XhoI fragment of the barley Pbf cDNA clone, which spans from position 302 to 1,198 (Mena et al., 1998). Expression of a thiol protease gene of the cathepsin B-like class was analyzed using as a probe the 1,065-bp insert in the 2529 wheat (Triticum aestivum) cDNA clone, reported by Cejudo et al. (1992b). As control for loading, the blots were also hybridized with a 18S rDNA probe from wheat.
In Situ Hybridization
Barley seeds of the cv Bomi were collected at 15 DAP or after 2 d of germination and were fixed in 50% (v/v) ethanol, 5% (v/v) acetic acid, and 3.7% (w/v) formaldehyde with an occasional vacuum. After dehydration with ethanol and exchange with xylene, tissues were embedded in Paraplast Plus (Sigma, St. Louis). Sections (8 μm thick) were mounted in Poly-l-Lys and were hybridized overnight at 52°C with appropriate Pbf Biotin-labeled probes at a final concentration of 100 ng μL−1, according to the mRNA locator-Hyb and nonisotopic labeling kits (Ambion, Austin, TX). For probe preparation, the barley Pbf-specific fragment of 896 bp, used for northern-blot analysis, was subcloned in pBluescript SK+ (Stratagene, La Jolla, CA) and was amplified by PCR with standard M13 reverse and forward primers. About 1 μg of the PCR product was used as template for the synthesis of the Biotin-labeled, in vitro-transcribed RNA from the T7 (antisense probe) or the T3 (sense probe) promoter sequences. Hybridization signals were detected by color development with a Streptavidin alkaline phosphatase conjugate (Ambion), following the manufacturer's instructions. Sections were stained with 0.1% (w/v) calcofluor and were then photographed under UV light on an microscope (BX60; Olympus, Tokyo).
Hormone Treatments and RT-PCR Analysis
Cultivar Himalaya barley seeds (1992 harvest, Washington State University, Pullman) were deembryonated and sterilized in 1.7% (w/v) NaOCl for 10 min, treated with 0.01 m HCl for 5 min, and thoroughly washed with distilled water. Half seeds were placed for 48 h at 22°C in the dark on filter paper soaked with a buffer containing 20 mm Na succinate, pH 5.0, and 20 mm CaCl2 for 3 d. Aleurone layers were isolated under a dissecting microscope and were incubated in petri plates at 22°C in the dark, for various times, with the buffer described above, including no hormone, 1 μm GA3, 10 μm ABA, or a mixture of both hormones.
Total RNA for RT-PCR analysis was isolated from aleurone layers by the RNeasy Plant protocol (Qiagen, Valencia, CA). Contaminating genomic DNA in the RNA preparations was then digested by DNase treatment using the DNA-free system (Ambion). First-strand cDNA synthesis was primed with random hexamers and was catalyzed by M-MuLV Reverse Transcriptase according to the manufacturer's recommendations (Amersham Pharmacia Biotech, Piscataway, NJ). PCR amplification of a 460-bp portion of the Pbf cDNA was performed using the following oligonucleotides as primers: The forward BPBF14 was 5′-ACCCTTCGTTCACCTGATGG-3′, which spans an intron-exon boundary in the Pbf gene sequence (I. Isabel-Lamoneda, M. Mena, and P. Carbonero, unpublished data); the reverse BPBF15 was 5′-GACCCAAAAGTTCTCAGGGA-3′. A 650-bp portion of the cDNA encoding a cathepsin B-like thiol protease was amplified using the primers CB1, 5′-TCGCGAATTACACTATTGAGC-3′, and CB2, 5′-CACCGGTGATGTG-CTTGTA-3′. These primers were designed based on nucleotide sequences highly conserved between the different wheat cathepsin B-like cDNAs (Cejudo et al., 1992b). The resulting PCR products were cloned and sequenced, and this showed that we had amplified the desired target messages without detectable heterogeneity. As an internal control, the 18S amplicon was used, performing the PCR reaction with a mixture of 18S primers/competimers (Ambion) at a 2:8 and 3:7 molar ratio for Pbf and the thiol protease analysis, respectively. All amplifications were carried out for 35 cycles with AmpliTaq Gold DNA polymerase (Applied Biosystems, Norwalk, CT). The PCR products were analyzed by agarose gel (2%, w/v) electrophoresis and were visualized by ethidium bromide staining.
EMSA
Plasmid expression constructs and procedures for purification of the fusion proteins GST-BPBF and its mutant derivative GST-mtBPBF have been described previously (Mena et al., 1998). The oligonucleotide probes, described in Figure 4, were generated by annealing complementary oligonucleotides designed to create 5′ overhangs that were end-filled by treatment with Klenow DNA polymerase in the presence of [α-32P]dATP. Each binding reaction contained approximately 5 ng of GST-purified proteins, 1 ng of gel-purified radiolabeled probe (25,000 cpm), and the indicated excess of unlabeled double-stranded oligonucleotide as a competitor in 15 μL of binding buffer [10 mm HEPES, pH 7.9, 50 mm KCl, 10 mm dithiothreitol, 10% (w/v) glycerol, 1 μg d(I)-d(C), and 2 mg mL−1 bovine serum albumin]. After incubation for 30 min at room temperature, the DNA-protein complexes were analyzed by electrophoresis on 7% (w/v) acrylamide gels (29:1), prepared, and run in 0.5× Tris borate-EDTA, pH 8 (10 mm Tris-ClH, 90 mm boric acid, and 2.5 mm EDTA), at 150 V for 3.5 h at 4°C. Gels were dried under vacuum and were autoradiographed using film (X-OMAT S; Kodak, Rochester, NY).
Transient Expression Assays in Barley Aleurone Cells
Reporter constructs of the wheat cathepsin B-like protease promoter were obtained by linking different promoter fragments of the Al21 gene to the coding sequence for the GUS reporter gene, followed by the 3′-nos terminator. The pCBG13 reporter plasmid contained a −480-bp fragment upstream of the putative translational start site, and the pCBG13.1 and pCBG13.5 plasmids were deletions spanning up to −381 and −355 of the ATG, respectively (Cejudo et al., 1992a).
Effector constructs were derived from the plasmid p35IN harboring the CaMV35S promoter followed by the first intron of the maize (Zea mays) AdhI gene and the 3′-nos terminator in a pBluescript vector. Plasmids pBPBF and pGAMYB contained the full-length coding regions of the barley Pbf (Mena et al., 1998) and GAMyb cDNAs (Gubler et al., 1995), respectively.
Particle bombardment was performed with a biolistic Helium gun device (PSD-1000; DuPont, Wilmington, DE), basically as described in a previous study (Mena et al., 1998), but with the following modifications: Each shot delivered equal molar amounts (to a maximum of 175 ng) of the tested reporter plasmid on 0.15 mg of gold particles of 1 μm in size; for cotransfection, the reporter was combined with the effector plasmid(s) at 1:1 molar ratio after preliminary studies indicated that the same inhibition rates were obtained at three effector (BPBF) concentrations (0.5:1, 1:1, and 2:1); and rupture discs of 1,500 psi were used and the distance between macrocarrier and sample was set to 6 cm.
Cultivar Himalaya barley seeds (1992 harvest) were deembryonated and sterilized as described above. Half seeds were placed for 48 h at 22°C in the dark on filter paper soaked with a buffer (pH 5.0) containing 20 mm Na succinate and 20 mm CaCl2 before removing their pericarp and testa layers under a dissecting microscope. Exposed aleurone layers were shot in sets of eight. Four of them were then incubated separately in the buffer described above with no hormone or with 1 μm GA3 for 24 h at 22°C in the dark in a petri plate with gentle shaking. GUS expression was determined by histochemical staining for 24 h according to Jefferson (1987). Blue spots were counted under a dissecting microscope, and the GUS activity was expressed as the mean value of blue spots per half-grain of aleurone. This measure of GUS activity directly correlates with fluorimetrically quantitated GUS activity per milligram of protein with a correlation coefficient of 0.95 (data not shown).
Footnotes
This work was financed by Ministerio de Educación y Cultura (Spain; grant nos. PB97–0561 and PB97–0745), by Ministerio de Ciencia y Tecnología (Spain; grant no. BMC2000–1483), and by Junta de Andalucia (Spain; grant no. CVI–0182). M.M. and I.I.-L. were the recipients of a postdoctoral contract and a PhD scholarship, respectively, from Ministerio de Educación y Cultura.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.005561.
LITERATURE CITED
- Appleford NEJ, Lenton JR. Hormonal regulation of α-amylase gene expression in germinating wheat (Triticum aestivum) grains. Physiol Plant. 1997;100:534–542. [Google Scholar]
- Bethke PC, Jones RL. Gibberellin signaling. Curr Opin Plant Biol. 1998;1:440–446. doi: 10.1016/s1369-5266(98)80270-7. [DOI] [PubMed] [Google Scholar]
- Bosnes M, Weideman F, Olsen OA. Endosperm differentiation in barley wild-type and sex mutants. Plant J. 1992;2:661–674. [Google Scholar]
- Cejudo FJ, Ghose TK, Stabel P, Baulcombe DC. Analysis of the gibberellin-responsive promoter of a cathepsin B-like gene from wheat. Plant Mol Biol. 1992a;20:849–856. doi: 10.1007/BF00027156. [DOI] [PubMed] [Google Scholar]
- Cejudo FJ, Murphy G, Chinoy C, Baulcombe DC. A gibberellin-regulated gene from wheat with sequence homology to cathepsin B of mammalian cells. Plant J. 1992b;2:937–948. [PubMed] [Google Scholar]
- Cercós M, Gómez-Cadenas A, Ho T-HD. Hormonal regulation of a cystein proteinase gene, EPB-1, in barley aleurone layers: cis- and trans-acting elements involved in the coordinated gene expression regulated by gibberellins and abscisic acid. Plant J. 1999;19:107–118. doi: 10.1046/j.1365-313x.1999.00499.x. [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep. 1993;11:113–116. [Google Scholar]
- Diaz I, Vicente-Carbajosa J, Abraham Z, Martinez M, Isabel-LaMoneda I, Carbonero P. The GAMYB protein from barley interacts with the DOF transcription factor BPBF and activates endosperm-specific genes during seed development. Plant J. 2002;29:401–414. doi: 10.1046/j.0960-7412.2001.01230.x. [DOI] [PubMed] [Google Scholar]
- Fincher GB. Molecular and cellular biology associated with endosperm mobilisation in germinating cereal grains. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:305–346. [Google Scholar]
- Gilroy S, Jones RL. Perception of gibberellin and abscisic acid at the external face of the plasma membrane of barley (Hordeum vulgare L.) aleurone protoplasts. Plant Physiol. 1994;14:1185–1192. doi: 10.1104/pp.104.4.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualberti G, Papi M, Belluci L, Ricci I, Bouchez D, Camilleri C, Costantino P, Vittorioso P. Mutations in the DOF zinc finger genes DAG2 and DAG1 influence with opposite effects germination of Arabidopsis seeds. Plant Cell. 2002;14:1253–1263. doi: 10.1105/tpc.010491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Jacobsen JV. Gibberellin-responsive elements in the promoter of a barley high-pI α-amylase gene. Plant Cell. 1992;4:1435–1441. doi: 10.1105/tpc.4.11.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Kalla R, Roberts JK, Jacobsen JV. Gibberellin-regulated expression of a myb gene in barley aleurone cells: evidence for myb transactivation of a high-pI α-amylase gene promoter. Plant Cell. 1995;7:1879–1891. doi: 10.1105/tpc.7.11.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Raventós D, Keys M, Watts R, Mundy J, Jacobsen JV. Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. Plant J. 1999;17:1–9. doi: 10.1046/j.1365-313x.1999.00346.x. [DOI] [PubMed] [Google Scholar]
- Hoecker U, Vasil IK, McCarty DM. Integrated control of seed maturation and germination programs by activator and repressor functions of Viviparous-1 of maize. Genes Dev. 1995;9:2459–2469. doi: 10.1101/gad.9.20.2459. [DOI] [PubMed] [Google Scholar]
- Jacobsen JV, Gubler F, Chandler PM. Gibberellins action in germinating cereal grains. In: Davies PJ, editor. Plant Hormones: Physiology, Biochemistry, and Molecular Biology. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 246–271. [Google Scholar]
- Jefferson RA. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- Lagrimini LM, Burkhart W, Moyer M, Rothstein S. Molecular cloning of complementary DNA encoding the lignin forming peroxidases from tobacco: molecular analysis and tissue-specific expression. Proc Natl Acad Sci USA. 1987;84:7542–7546. doi: 10.1073/pnas.84.21.7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanahan MB, Rogers SW, Rogers JC. A gibberellin response complex in cereal α-amylase gene promoters. Plant Cell. 1992;4:203–211. doi: 10.1105/tpc.4.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovegrove A, Hooley R. Gibberellin and abscisic acid signalling in aleurone. Trends Plant Sci. 2000;5:102–110. doi: 10.1016/s1360-1385(00)01571-5. [DOI] [PubMed] [Google Scholar]
- McCarty DR, Hattori T, Carson CB, Vasil V, Lazar M, Vasil IK. The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell. 1991;66:895–905. doi: 10.1016/0092-8674(91)90436-3. [DOI] [PubMed] [Google Scholar]
- Mena M, Vicente-Carbajosa J, Schmidt RJ, Carbonero P. An endosperm-specific DOF protein from barley, highly conserved in wheat, binds to and activates transcription from the prolamin-box of a native B-hordein promoter in barley endosperm. Plant J. 1998;16:53–62. doi: 10.1046/j.1365-313x.1998.00275.x. [DOI] [PubMed] [Google Scholar]
- Papi M, Sabatini S, Altamura MM, Hennig L, Schäfer E, Costantino P, Vittorioso P. Inactivation of the phloem-specific DOF zinc finger gene DAG1 affects response to light and integrity of the testa of Arabidopsis seeds. Plant Physiol. 2002;128:411–417. doi: 10.1104/pp.010488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papi M, Sabatini S, Bouchez D, Camilleri C, Costantino P, Vittorioso P. Identification and disruption of an Arabidopsis zinc finger gene controlling seed germination. Genes Dev. 2000;14:28–33. [PMC free article] [PubMed] [Google Scholar]
- Raventós D, Skriver K, Schlein M, Karnahl K, Rogers SW, Rogers JC, Mundy J. HRT, a novel zinc finger, transcriptional repressor from barley. J Biol Chem. 1998;273:23313–23320. doi: 10.1074/jbc.273.36.23313. [DOI] [PubMed] [Google Scholar]
- Rogers JC, Lanahan MB, Rogers SW. The cis-acting gibberellin response complex in high-pI α-amylase gene promoters. Plant Physiol. 1994;105:151–158. doi: 10.1104/pp.105.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skadsen RW. Physiological and molecular genetic mechanisms regulating hydrolytic enzymes gene expression in cereal grains. Physiol Plant. 1998;104:486–502. [Google Scholar]
- Skriver K, Olsen FL, Rogers JC, Mundy J. Cis-acting DNA elements responsive to gibberellin and its antagonist abscisic acid. Proc Natl Acad Sci USA. 1991;88:7266–7270. doi: 10.1073/pnas.88.16.7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slakeski N, Fincher GB. Developmental regulation of (1–3, 1–4)-α-glucanase gene expression in barley. Plant Physiol. 1992;99:1226–1231. doi: 10.1104/pp.99.3.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Carbajosa J, Moose SP, Parsons RL, Schmidt RJ. A maize zinc-finger binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque-2. Proc Natl Acad Sci USA. 1997;94:7685–7690. doi: 10.1073/pnas.94.14.7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washio K. Identification of DOF proteins with implication in the gibberellin-regulated expression of a peptidase gene following the germination of rice grains. Biochim Biophys Acta. 2001;1520:54–62. doi: 10.1016/s0167-4781(01)00251-2. [DOI] [PubMed] [Google Scholar]
- Whittier RF, Dean DA, Rogers JC. Nucleotide sequence analysis of α-amylase and thiol protease genes that are hormonally regulated in barley aleurone cells. Nucleic Acids Res. 1987;15:2515–2535. doi: 10.1093/nar/15.6.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf N. Structure of the genes encoding Hordeum vulgare (1–3, 1–4)-α-glucanase isoenzymes I and II and functional analysis of their promoters in barley aleurone protoplasts. Mol Gen Genet. 1992;234:33–42. doi: 10.1007/BF00272342. [DOI] [PubMed] [Google Scholar]
- Yanagisawa S. DOF DNA binding proteins contain a novel zinc finger motif. Trends Plant Sci. 1996;1:213–214. [Google Scholar]
- Yanagisawa S. DOF1 and DOF2 transcription factors are associated with expression of multiple genes involved in carbon metabolism in maize. Plant J. 2000;21:281–288. doi: 10.1046/j.1365-313x.2000.00685.x. [DOI] [PubMed] [Google Scholar]
- Yanagisawa S, Schmidt RJ. Diversity and similarity among recognition sequences of DOF transcription factors. Plant J. 1999;17:209–214. doi: 10.1046/j.1365-313x.1999.00363.x. [DOI] [PubMed] [Google Scholar]
- Yanagisawa S, Sheen J. Involvement of maize DOF zinc finger proteins in tissue-specific and light-regulated gene expression. Plant Cell. 1998;10:75–89. doi: 10.1105/tpc.10.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young TE, Gallie DR. Analysis of programmed cell death in wheat endosperm reveals differences in endosperm development between cereals. Plant Mol Biol. 1999;39:915–916. doi: 10.1023/a:1006134027834. [DOI] [PubMed] [Google Scholar]