Abstract
Background
Defining measures of linkage disequilibrium (LD) that have good small sample properties and are applicable to multiallelic markers poses some challenges. The potential of volume measures in this context has been noted before, but their use has been hampered by computational challenges.
Results
We design a sequential importance sampling algorithm to evaluate volume measures on I × J tables. The algorithm is implemented in a C routine as a complement to exhaustive enumeration. We make the C code available as open source. We achieve fast and accurate evaluation of volume measures in two dimensional tables.
Conclusion
Applying our code to simulated and real datasets reinforces the belief that volume measures are a very useful tool for LD evaluation: they are not inflated in small samples, their definition encompasses multiallelic markers, and they can be computed with appreciable speed.
Background
Linkage disequilibrium (LD) is the term used in genetics to indicate association between the qualitative random variables corresponding to alleles at different polymorphic sites. Measuring the levels of linkage disequilibrium is important for gene mapping and increasing our understanding of genome architecture. The current literature documents agreement only on the definition of measures of LD for biallelic markers. Consider two markers, with alleles A, a and B, b. Their haplotype distribution can be synthetically described as:
Fixing the marginals p and q, the distribution π is completely identified by the probability x of the haplotype (A, B). The discrepancy of a generic π from the distribution under linkage equilibrium, can be quantified simply by D = (x - pq). Measures of LD are defined as the standardized values of D. Two common such measures are and , where Dmax is min(p(1 - q), q(1 - p)) when the numerator is positive, and min(pq, (1 - p) (1 - q)) otherwise. The definition of R2 can be understood by considering the alleles as realizations of quantitative random variables (with values 0 and 1), among which we calculate a correlation coefficient. The measure R2 ranges between 0 and 1, and it is equal to 1 only when two entries of the table in (1) are equal to 0. The measure D' ranges, by definition, between -1 and 1, and its absolute value is equal to 1 whenever one entry of the table in (1) is equal to 0. There is a large literature discussing the choice of these measures (see, for example, [1]). Typically, R2 is preferred when the focus is on the predictability of one polymorphism given the other (and hence it is often used in power studies for association designs). D', instead, is the measure of choice to assess recombination patterns (haplotypes blocks have often been defined on the basis of D'). Despite their effectiveness, these measures suffer from two limitations: (a) they are not easily generalizable to multiallelic markers; (b) they are defined on the population haplotype distribution, and their performance can be rather unsatisfactory when applied to the empirical distribution derived from a finite sample.
With regard to point (a), it is clear that the definitions of R2 and D' are based on properties of the joint distribution of two biallelic markers and their generalization is not immediate. A partial solution is to evaluate R2 or D' on all 2 × 2 sub-tables obtainable from the joint distribution of two multiallelic markers and then summarize these results in one value. However, this measure is not easily interpretable and does not have good small sample properties (see the discussion in the following paragraphs).
Finally, let us remark how the problem of defining measures of disequilibrium generalizable to I × J tables remains actual, even if the current high density genotyping efforts are focused on biallelic markers as SNPs. Often we are interested in studying the relation between SNPs haplotypes at different loci: these can be considered as qualitative variables with multiple levels, just as multiallelic markers.
With regard to point (b), R2 and D' are defined and studied assuming that the population haplotype distribution is known [2]. In practice, this is rarely (if ever) the case: the sample haplotype frequencies provide an estimate of the population frequencies, and these estimates are used, following the plug-in principle, to obtain estimates of R2 and D'. This approach encounters some difficulties in the case of D'. If a SNP has a low minor allele frequency, it is quite possible that the rare haplotype that carries it, is not observed in a small sample. This leads to a D' being equal to 1, irrespective of the level of linkage disequilibrium. Detailed analysis of this phenomena is available in [3] and [4]. To avoid spurious results, researchers often calculate empirical confidence intervals for D' using resampling schemes (see, for example, [5]). While this certainly takes care of the variability of D', it does not result in a "correct" measure of linkage disequilibrium and it clearly comes with a substantial computational cost. Moreover, the fact that the values of D' are inflated in the presence of rare alleles makes it difficult to obtain a multiallelic version of D' based on pooling statistics: as mentioned above, it is quite likely for a particular haplotype of multiallelic markers to have very low population frequency, resulting in zero observed counts. Volume measures [6-8] both take effectively into account the variability due to sample size and are immediately applicable to multiallelic markers. Let us first recall the main idea of volume measures. For distributions like (1), volume measures can be described as a different strategy for normalizing D with reference to the class of distributions with the same marginals p and q. Rather than dividing the observed D by its maximum value over distributions in , one evaluates the proportion of distributions in that have a smaller difference from the distribution under independence than D. In general, we can consider any quantification of the discrepancy between a generic distribution π and the distribution under independence. When π is known, a volume measure is defined as the probability that a distribution selected uniformly among all possible ones with the same margins as π results in a lower discrepancy from equilibrium. If, however, the population haplotype distribution π is unknown, and a sample of size n is available, volume measures are defined directly on the contingency table summarizing the data, avoiding spurious effects due to the sample size. To clarify this point, let us consider again the case of two biallelic markers. If the population distribution (1) is known, we define Dvol, the volume measure equivalent to D', as the ratio of two volumes V1/V2. V1 is the volume of the space of all distributions with marginals equal to p and q, and Pr(0, 0) = z such that (x - pq) (z - pq) ≥ 0 and |x - pq| > |z - pq|. V2 is the volume of the space of distributions that satisfy all but the last one of the constraints for V1. A simple geometric argument shows that Dvol = D'. (See Additional file 1 for a graphical illustration). Suppose now, instead, that we do not know the population distribution (1), but we obtain a sample of size n from it, leading to the contingency table F
with row sums equal to r1, r2 and column sums c1, c2. Consider now the set Ω of tables T, with row and column sums r = (r1, r2) and c = (c1, c2). We define Dvol(F) as the fraction of contingency tables T in Ω that lead to a value |t11 - c1r1/n| smaller than |f11 - c1r1/n| among those for which (t11 - r1c1/n) (f11 - r1c1/n) > 0.
This Dvol value will be different from D', calculated by treating fij/n as population frequencies. For example, the fact that one table entry is equal to zero will not be sufficient to guarantee Dvol = 1. To make this point clearer, however, it is appropriate to consider a more general and precise definition. In the remainder of the paper, we will only concern ourselves with volume measures defined on haplotype sample frequencies.
Implementation
Consider the table of observed haplotypes counts F:
where Bi represent alleles at marker B and Ai alleles at marker A. Let Ω be the set of all tables T with row and column sums equal to r1, ..., rI and c1, ..., cJ, respectively. Given a criterion to quantify the discrepancy between F and the table expected under independence (linkage equilibrium), a volume measure is defined as the proportion of tables T ∈ Ω that lead to a smaller discrepancy value. If the recorded discrepancy is the biggest possible, then the volume measure will have value close to 1 (the exact value 1 will be attained as the sample size increases to ∞). Conversely, if all other tables lead to larger discrepancies, the volume measure will be zero.
One may notice that this definition of volume measure is similar to one minus the p-value for a test of independence. Indeed, volume measures are related to the "volume test," an original notion introduced by Hotelling [8], and the effect of sample size on the measures is very much the same as its effect on a p-value. The key difference between volume measures and variants of the commonly used Fisher's exact test for independence is that in the case of volume measures, the relevant proportion of tables is evaluated assuming that all tables with the same margins are equally probable, while in the case of Fisher's exact test tables are generated under the hypothesis of independence. Because of this, volume measures and Fisher's exact tests answer two very different questions: the first compares the observed table to all tables with the same margins, while the second one assesses the likelihood of the observed table under independence. A thorough discussion of the different interpretations and uses of these two approaches can be found in [7]. In order to concretely evaluate volume measures, one has to choose a criterion for discrepancy and be able to explore the space of tables with fixed margins to evaluate the required proportions. We start illustrating the first point by focusing on three specific measures: a) Dvol, which is defined only on 2 × 2 tables and coincides with D' when the population haplotype distribution is known; b) Mvol, which is a generalization of Dvol to multiallelic markers; c) Hvol, which is based on expected homozygosity and captures information that is close to the one described by R2, although it can be defined on tables with any number of entries.
When I = J = 2, let Ω1 = {T : ti+ = ri, t+j = cj, (t11 - r1c1/n)(f11 - r1c1/n) > 0}. We then define Dvol as
where .
For general I × J tables, recall that Ω denotes the set of all contingency tables with the same row and column sums as F: Ω = {T : ti+ = ri, t+j = cj}. Then, we define
The definition above should clarify how Mvol is closely related to Dvol, and the difference between the two is that Mvol does not consider the "sign" of the association, a notion that is undefined in generic I × J tables.
Letting , we can define the measure Hvol:
We have mentioned how Hvol captures information closely related to that of R2. A careful discussion of the interpretation of LD measures based on homozygosity can be found in [9]. Here it suffices to recall that joint homozygosity relates to a measure of agreement between the two markers and excess in homozygosity indicates that knowledge of the allele value at one marker increases predictive accuracy of the allele values at the other marker. The results of a recent empirical study conducted using homozygosity-based measures are documented in [10].
Note that all the above definitions use the strict inequality sign. The choice of this over ≤ is irrelevant for large n, but it makes a difference in the case of small n, where strict inequality allows us to better discriminate against apparent association due to small samples.
To evaluate these measures, we need to explore the space of all tables with the same margins. In the case of I = J = 2, this can be done by simple enumeration. For multiallelic tables enumeration is impractical. An obvious alternative is to restrict one's attention to a sample of possible tables. However, obtaining a sample of tables according to the uniform distribution among all tables with fixed margin (as opposed to according to the Fisher-Yates distribution) is not easy. It is indeed the computational difficulty associated with volume tests [7] and measures [6] that has substantially hindered their wide-spread application. Previous solutions have been proposed with Markov chain Monte Carlo algorithms in [11], as well as rejection sampling (see [12] for a review). The main contribution of this paper is that we have successfully implemented a sequential importance sampling (SIS) algorithm, originally introduced in [12], to evaluate volume measures accurately and in a timely manner. This implementation makes volume measures applicable to high throughput analysis.
To enumerate all tables in Ω1 (I = J = 2), it is useful to notice that t11 must satisfy
max(0, r1 + c1 - n) ≤ t11 ≤ min(r1, c1), (2)
and after t11 is chosen, we can fill in other entries of the 2 × 2 table by the marginal sum constraints. Therefore we can enumerate tables in Ω1 by assigning all possible integers satisfying (2) to t11, and keeping those tables such that (t11 - r1c1/n) has the same sign as in F.
We now consider the SIS procedure for I × J tables. Let u(T) be the uniform distribution over all tables in Ω. Then Mvol(F) can be treated as the expectation of the indicator function 1{m(T)<m(F)} with respect to u(T). It is hard to sample directly from u(T). The idea of importance sampling is to sample tables from another proposal distribution g(T), and then estimate Mvol(F) by
where T1, ..., TL are L independent and identically distributed (i.i.d.) samples from g(T). SIS generates a table cell by cell by decomposing the proposal distribution g(T) as
g(T)= g(t11)g(t21|t11) ... g(tIJ|tI-1, J, ..., t11).
Notice that the support for the first entry t11 is max(0, r1 + c1 - n) ≤ t11 ≤ min(r1, c1). We sample an integer uniformly from the above range for in, i.e., g(t11) is the uniform distribution on the support of t11.
Recursively, suppose we have chosen ti1 = for i = 1, ..., k - 1. Then the support for tk1 is . We sample an integer uniformly from the above range for tk1. The procedure is continued until all the entries in the first column have been considered. Then we update the row sums by subtracting the realization of the first column from the original row sum, and sample the second column of the table in the same way.
The computing time and precision of the algorithm are different for 2 × 2 or larger size tables. For 2 × 2 tables, our algorithm simply lists all possible tables with fixed margins. CPU time is then proportional to the total number of tables: usually their enumeration takes a fraction of a second. The algorithm is exact and we do not have approximation errors in the output. For the general case of I × J tables, the CPU time depends on the number of generated uniform random variables: I × J × L for L Monte Carlo samples. It is important to keep in mind that the output of the algorithm is not exact, but an estimate of the true volumes ratio (so different runs will give slightly different results). The precision of the final estimate depends on the number L of Monte Carlo samples and how well the proposal distribution in the SIS algorithm approximates the target distribution for a given table. Indeed, the value of the parameter L has to be specified by the user. It is advisable to conduct multiple trial runs to estimate the precision of the estimate and select a value of L that assures an acceptable precision.
Results
We now illustrate the performance of our algorithm and the relevance of volume measures with three examples.
The effect of small sample size on D' and Dvol
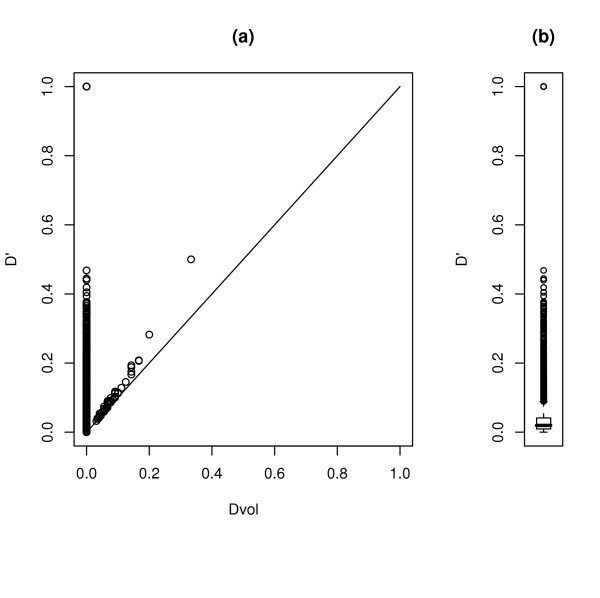
It has been noted that D' tends to be biased upwards in small samples [3,4]. We conducted a simulation study to illustrate how this problem is less severe when using Dvol. We generated 100 two-markers haplotype tables with 200 observations, each under the hypothesis of linkage equilibrium between the markers. The distribution of the frequency of the minor alleles of the simulated SNPs matched a random sample of markers on chromosome 22 that were used in [13]. In a situation where the true population value of D' is equal to zero, any sample based estimator is going to be upward biased, since 0 is the minimum value that D' can achieve. The point of our investigation was to compare the severity of this bias. Figure 1 illustrates the results: D' is always larger than Dvol, and it is occasionally equal to 1; Dvol is actually equal to zero in the majority of cases.
Figure 1.
Comparison of D' and Dvol. Comparison of D' and Dvol on tables generated under linkage equilibrium, (a) Scatterplot of the values of D' and Dvol. (b) Boxplot of the values of D'.
Patterns of LD between multiallelic markers
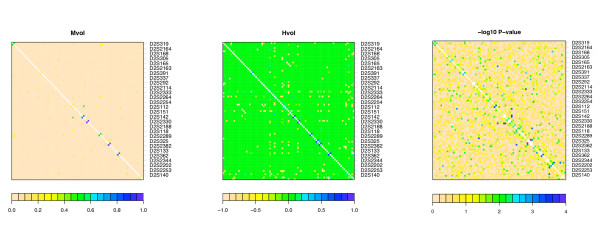
Our next example focuses on the application of volume measures to multiallelic markers. The data consists of 157 phase-known non-transmitted chromosomes 2 of parents of BP-I persons from the Central Valley of Costa Rica. The chromosomes were typed with 85 markers in the course of the study by [14]. Using volume measures Mvol and Hvol we were able to evaluate the level of disequilibrium between all the possible marker pairs in this sample. Figure 2 gives a graphical representation of the values of Mvol and Hvol in this data set as well as the negative of log10 p-value for a Fisher exact test of independence. This last one is reported for comparison purposes, as it is often used as a measure of dependence, despite the fact that it is rather inappropriate with this goal [7]. Volume measures make it unnecessary to resort to this unsatisfactory surrogate when comparing multiallelic markers. To analyze these tables, we used L = 1, 000, 000 Monte Carlo sample. The average time to evaluate the measures on one table was 48 seconds on a Dell desktop with 2.19 GHz CPU and 384 MB ram.
Figure 2.
Measuring LD between multiallelic markers. Measure of disequilibrium between microsatellites. Each square in this symmetric picture corresponds to a marker pair (the same markers are reported on both rows and columns). The three panels report, from left to right, Mvol, Hvol, and the negative of the log10 of the p-value for a Fisher's exact test of independence.
Consistency of LD patterns on chr 22 in 12 populations
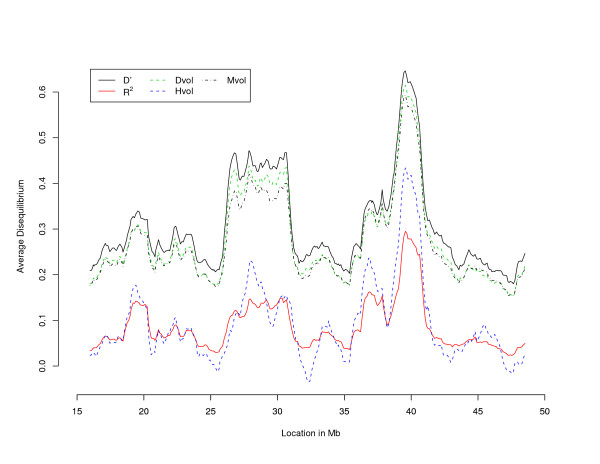
We have used the measures D', |R|, Dvol, Mvol and Hvol to assess the distribution and extent of linkage disequilibrium on chromosome 22 in samples of 200 persons from each of eleven population isolates and in an out-bred Caucasian sample, using 2486 SNP markers spaced at a density of approximately one marker every 13.8 kb. [10,13]. To conduct a complete analysis of the linkage disequilibrium patterns in the 12 population samples, we restricted our attention to the SNPs with sample minor allele frequencies larger than 0.1. We did so for uniformity with previous studies (for example, [15]) and to make sure that our results were not strongly influenced by the rare markers with exceptionally high homozygosity. This leads us to work with 1920 SNPs. Phase was unknown and the two markers haplotypes counts necessary to evaluate pair-wise disequilibrium measures were reconstructed using EM [16]. Five measures, D', Dvol, Mvol, R2 and Hvol were calculated for each of the 1,842,240 pairs of SNPs. The results were summarized by averaging the measured disequilibrium within windows of 1.7 Mb sliding along chromosome 22. Figure 3 reports the values of the five measures in the Costa Rica population. The observed relation between the measures is consistent across populations. In particular, it can be noted that the average values of Dvol are lower than the ones of D', while clearly exhibiting very similar patterns. This testifies that even if the sample size is moderately large (200 individuals) and only markers with minor allele frequency >0.1 are considered, D' is inflated, making a strong case for the use of Dvol over D'. The values of Mvol are very close to the one of Dvol, even if Mvol are often smaller as expected given the differences in definitions. As far as Hvol, one can notice that its values are closer to those of R2 than to those of any other measure. Finally, let us observe that the computational time required for evaluating all the volume measures above amounts to an average of 5 minutes for each population on a Dell desktop with 2.19 GHz CPU and 384 MB ram (this is after pairwise haplotypes counts were reconstructed, which required approximately the same amount of time). The substantial difference in computational time with the results reported in the previous subsection is due to the fact that here we are dealing with 2 × 2 tables.
Figure 3.
LD pattern on Chr 22 in a Costa Rican population. Linkage disequilibrium of chromosome 22 in Costa Rica according to five different measures. D', R2, Dvol, Mvol and Hvol are represented, respectively, with a solid yellow, a broken green, a solid blue, a broken magenta, and a solid red line. The average value of the measures, between markers that are within a 1.7 Mb window, is plotted against the middle point of the window, with the x axis representing the length of chromosome 22.
Conclusion
We describe a novel implementation of a sequential importance sampling algorithm to evaluate volume measures of linkage disequilibrium. We focus on three measures. Dvol corresponds conceptually to D', but we show that Dvol is not inflated for small sample size. Mvol represents a generalization of Dvol that can be evaluated on generic I × J tables. Hvol is based on expected homozygosity and measures agreement between markers, so that it captures information similar to that of R2. However, unlike R2, Hvol can be evaluated on generic I × J tables.
Availability and requirements
The source code for evaluating the volume measures described in this paper is available at the following url: http://www.stat.uiuc.edu/~yuguo/software/volume/.
It is a C program that can be compiled with gcc and requires the libraries math.h, stdlib.h, malloc.h, time.h, and stdio.h.
Authors' contributions
YC has been responsible mainly for the algorithm and code development. CL has contributed mainly to data analysis. CS has supervised the project.
Supplementary Material
Measures of disequilibrium between biallelic markers. This .pdf file contains a detailed description of measures of disequilibrium for population haplotype distributions for biallelic markers. We recall the definition of D' and R2 as well of Dvol and Mvol. We graphically illustrate the difference in normalization procedure between all of these measures. This makes it easy to see the identity of D' and Dvol when the population haplotype frequency is known.
Acknowledgments
Acknowledgements
We thank Nelson Freimer and his lab for making their data available to us. We also thank Hui Wang for help in drawing figures. Y. Chen is partially supported by NSF grant DMS-0503981. C. Sabatti acknowledges support from NSF grant DMS0239427, NIH grants R01NS037484 and R01MH049499, and USPHS grant GM53275.
Contributor Information
Yuguo Chen, Email: yuguo@uiuc.edu.
Chia-Ho Lin, Email: chiaholi@ucla.edu.
Chiara Sabatti, Email: csabatti@mednet.ucla.edu.
References
- Pritchard JK, Przeworski M. Linkage disequilibrium in humans: models and data. American Journal of Human Genetics. 2001;69:1–14. doi: 10.1086/321275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B, Risch N. A Comparison of Linkage Disequilibrium Measures for Fine-Scale Mapping. Genomics. 1995;29:311–322. doi: 10.1006/geno.1995.9003. [DOI] [PubMed] [Google Scholar]
- Teare M, Dunning A, Durocher F, Rennart G, Easton DF. Sampling distribution of summary linkage disequilibrium measures. Annals of Human Genetics. 2002;66:223–233. doi: 10.1046/j.1469-1809.2002.00108.x. [DOI] [PubMed] [Google Scholar]
- Tenesa A, et al. Extent of linkage disequilbirum in a Sardinian sub-isolate: sampling and methodological considerations. Human Molecular Genetics. 2004;13:25–33. doi: 10.1093/hmg/ddh001. [DOI] [PubMed] [Google Scholar]
- Gabriel S, Schaffner S, Nguyen H, Moore J, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero S, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander E, Daly M, Altshuler D. The Structure of Haplotype Blocks in the Human Genome. Science. 2002;21:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Sabatti C. Measuring dependence with volume tests. The American Statistician. 2002;50:191–195. doi: 10.1198/000313002128. [DOI] [Google Scholar]
- Diaconis P, Efron B. Testing for Independence in a Two-Way Table: New Interpretations of the Chi-Square Statistics. The Annals of Statistics. 1985;13:845–874. [Google Scholar]
- Hotelling H. Tubes and Spheres in n-spaces, and a Class of Statistical Problems. American Journal of Mathematics. 1939;61:440–460. doi: 10.2307/2371512. [DOI] [Google Scholar]
- Sabatti C, Risch N. Homozygosity and linkage disequilibrium. Genetics. 2002;160:1707–1719. doi: 10.1093/genetics/160.4.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Lin C, Service S, The international collaborative group on isolated populations . Chen Y, Freimer N, Sabatti C. Linkage disequilibrium and haplotype homozygosity in population samples genotyped at a high marker density. Hum Hered. 2006;62:175–89. doi: 10.1159/000096599. [DOI] [PubMed] [Google Scholar]
- Diaconis P, Sturmfels B. Algebraic Algorithms for Sampling from Conditional Distributions. The Annals of Statistics. 1997;26:363–397. [Google Scholar]
- Chen Y, Diaconis P, Holmes S, Liu J. Sequential Monte Carlo Methods for Statistical Analysis of Table. Journal of the American Statistical Association. 2005;100:109–120. doi: 10.1198/016214504000001303. [DOI] [Google Scholar]
- Service S, DeYoung J, Karayiorgou M, Louw Roos J, Pretorious H, Bedoya G, Ospina J, Ruiz-Linares A, Macedo A, Palha J, Heutink P, Aulchenko Y, Oostra B, van Duijn C, Jarvelin M, Varilo T, Peddle L, Rahman P, Piras G, Monne M, Murray S, Galver L, Peltonen L, Sabatti C, Collins A, Freimer N. Magnitude and distribution of linkage disequilibrium in population isolates and implications for genome-wide association studies. Nature Genetics. 2006;38:556–560. doi: 10.1038/ng1770. [DOI] [PubMed] [Google Scholar]
- Ophoff R, Escamilla M, Service S, Spesny M, Meshi D, Poon W, Molina J, Fournier E, Gallegos A, Mathews C, Neylan T, Batki S, Roche E, Ramirez M, Silva S, De Mille M, Dong P, Leon P, Reus V, Sandkuijl L, Freimer N. Genomewide linkage disequilibrium mapping of severe bipolar disorder in a population isolate. American Journal of Human Genetics. 2002;71:565–74. doi: 10.1086/342291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds D, Stuve L, Nilsen G, Halperin E, Eskin E, Ballinger D, Frazer K, Cox D. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Slatkin Maximum likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol. 1995;12:921–7. doi: 10.1093/oxfordjournals.molbev.a040269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Measures of disequilibrium between biallelic markers. This .pdf file contains a detailed description of measures of disequilibrium for population haplotype distributions for biallelic markers. We recall the definition of D' and R2 as well of Dvol and Mvol. We graphically illustrate the difference in normalization procedure between all of these measures. This makes it easy to see the identity of D' and Dvol when the population haplotype frequency is known.