Abstract
To begin the functional dissection of light signal transduction pathways of maize (Zea mays), we have identified and characterized the light-sensing mutant elm1 (elongated mesocotyl1). Seedlings homozygous for elm1 are pale green, show pronounced elongation of the mesocotyl, and fail to de-etiolate under red or far-red light. Etiolated elm1 mutants contain no spectrally active phytochrome and do not deplete levels of phytochrome A after red-light treatment. High-performance liquid chromatography analyses show that elm1 mutants are unable to convert biliverdin IXα to 3Z-phytochromobilin, preventing synthesis of the phytochrome chromophore. Despite the impairment of the phytochrome photoreceptors, elm1 mutants can be grown to maturity in the field. Mature plants retain aspects of the seedling phenotype and flower earlier than wild-type plants under long days. Thus, the elm1 mutant of maize provides the first direct evidence for phytochrome-mediated modulation of flowering time in this agronomically important species.
The phytochrome family of photoreceptors mediates many of the responses that plants display to changes in their light environment (Smith, 2000). The basis of phytochrome action is a reversible photoconversion between a red light (R)-absorbing form (Pr) and a far-red light (FR)-absorbing form (Pfr; Quail, 2002). In lower plants, the family is represented by a small number of nuclear genes (Schneider-Poetsch et al., 1998). However, gene duplication and evolutionary divergence have resulted in the formation of functionally diverse multigene families in flowering plants. In Arabidopsis, the phytochrome family consists of five genes: PHYA, PHYB, PHYC, PHYD, and PHYE (Clack et al., 1994), whereas the grasses have three phytochromes: PhyA, PhyB, and PhyC (Mathews and Sharrock, 1996). In maize (Zea mays), an ancestral genomic duplication has increased the total family size to at least six: PhyA1, PhyA2, PhyB1, PhyB2, PhyC1, PhyC2, and possibly PhyC3 (Christensen and Quail, 1989; Childs et al., 1997; Basu et al., 2000). Although loss-of-function phy mutants have been characterized in a broad range of plants, including Arabidopsis (for review, see Whitelam et al., 1998), sorghum (Sorghum bicolor; Childs et al., 1997), barley (Hordeum vulgare; Hanumappa et al., 1999), and rice (Oryza sativa; Takano et al., 2001), no phytochrome gene mutants have been characterized in maize. Gene duplication within the maize phytochrome family and the accompanying potential for functional redundancy may have obscured genetic screens for phy mutants.
The photoactive holoprotein (phy) consists of a PHY apoprotein (PHY) covalently attached to a linear tetrapyrrole (bilin) chromophore, 3E-phytochromobilin (PΦB; Terry, 1997). The first committed step in the synthesis of PΦB is the conversion of heme to biliverdin (BV) IXα by the enzyme heme oxygenase (Weller et al., 1996). BV IXα is then reduced to 3Z-PΦB by PΦB synthase and subsequently isomerized to 3E-PΦB (Terry et al., 1995). Of these three activities, genes encoding the first two have now been cloned (Davis et al., 1999; Muramoto et al., 1999; Kohchi et al., 2001). The HO1 (HY1) gene encodes heme oxygenase, which is targeted to the plastid (Muramoto et al., 1999). The HY2 gene encodes PΦB synthase, a ferredoxin-dependent BV reductase, which is also plastid localized (Kohchi et al., 2001). It is not yet known whether the isomerization of 3Z-PΦB to 3E-PΦB is enzyme mediated or whether it occurs spontaneously.
Although phytochrome apoproteins are encoded by a multigene family, it is likely that all plant apophytochromes bind the same chromophore. Therefore, genetic disruption of linear tetrapyrrole synthesis offers a way to specifically inactivate the entire phytochrome system. There are a number of known mutants in which linear tetrapyrrole synthesis is disrupted. These include the hy1 and hy2 mutants of Arabidopsis (Koornneef et al., 1980; Muramoto et al., 1999; Davis et al., 1999; Kohchi et al., 2001), the pew1 (partially etiolated-in-white-light1) and pew2 mutants of Nicotiana plumbaginifolia (Kraepiel et al., 1994), the pcd1 (phytochrome chromophore-deficient1) and pcd2 mutants of pea (Pisum sativum; Weller et al., 1996, 1997), the au (aurea) and yg-2 (yellow-green2) mutants of tomato (Lycopersicon esculentum; Koornneef et al., 1985; Terry and Kendrick, 1996), and the se5 (photoperiodic sensitive5) mutant of rice (Yokoo and Okuno, 1993; Izawa et al., 2000). All these mutants have lesions in either heme oxygenase or PΦB synthase and show a reduction in light responsiveness. However, a common characteristic of these mutants is that they show a partial recovery of light sensitivity during development (López-Juez et al., 1990; Weller et al., 1996), suggesting that other enzymes are present that can partially complement these mutations. Support for such an idea has recently come from Davis et al. (2001), who have shown that heme oxygenase is encoded by a small gene family that may be functionally redundant.
Although many molecular characterizations of phytochrome signaling have focused on seedling responses, a number of studies have demonstrated the importance of phytochrome in mature, field-grown plants (Robson et al., 1996; Schmitt, 1997; Shlumukov et al., 2001). In several cases, the mutation of a single phy gene has dramatically changed the mature plant phenotype. Two such examples are the early flowering ma3R line of sorghum (Childs et al., 1997) and the photoperiod-insensitive BMDR1 line of barley (Hanumappa et al., 1999). Overexpression of an oat (Avena sativa) PHYA in tobacco (Nicotiana tabacum) resulted in dramatic morphological changes that increased harvest index (Robson et al., 1996). Characterization of the se5 heme-oxygenase mutant of rice has also demonstrated a significant contribution of the phytochrome system to the regulation of flowering time in this crop species (Izawa et al., 2000).
In this study, the isolation and initial characterization of the elongated mesocotyl1 (elm1) mutant of maize is presented. We show that the elm1 mutant has a reduced accumulation of active phytochrome. Under long-day (LD) growth conditions, elm1 mutants flower earlier than near-isogenic wild-type plants, indicating that phytochrome signaling can modulate flowering time in maize.
RESULTS
Isolation of the elm1 Mutant
The elm1 mutation was identified in seedling screens of an Ac-mutagenized population. All lines in this population were maintained in a standard W22 inbred, enabling near-isogenic comparisons between any alleles recovered. To identify light-signaling mutants, Ac transpositions were selected from several donor elements located throughout the maize genome. F1 plants were grown and self-pollinated to generate approximately 100 F2 families. Approximately 20 kernels from each ear were then screened in greenhouse sandbenches to identify elongated pale-green seedlings. A similar screen was previously used to identify long-hypocotyl mutants of Arabidopsis (Koornneef et al., 1980) and tomato (Koornneef et al., 1985). A single line was identified that segregated pale-green seedlings with elongated mesocotyls as a simple recessive trait. The mutation was designated elm1. Southern-blot analysis has failed to detect linkage of the transposable element Ac to the elm1 locus (data not shown); thus, it is unlikely that the elm1 allele contains an Ac insertion.
Mature, field-grown elm1 plants have elongated internodes (Fig. 1), pale-green leaves, and display a tendency to lodge (fall down). Under LD growth conditions in the field (14–16 h of light, Ithaca, NY), elm1 plants flowered approximately 5 d earlier than wild type (Elm1, n = 60, mean = 79.1 d after planting [dap], se = 0.22; elm1, n = 63, mean = 74.4 dap, se = 0.38; Wilcoxon two-sample, non-paired, rank test, U = 3,639.5, P < 0.01). The similarity of elm1 seedling and mature plant phenotypes to previously characterized mutants of Arabidopsis and tomato suggested that the elm1 mutant is impaired in light perception or signal transduction.
Figure 1.
Mature plant phenotypes of wild-type (Elm1) and mutant (elm1) plants. Wild-type (left) and elm1 mutant (right) plants grown at summer field site (LD conditions). elm1 mutants are taller, have pale internodes, and flower earlier than near-isogenic siblings.
elm1 Mutant Seedlings Show a Disruption of the De-Etiolation Response
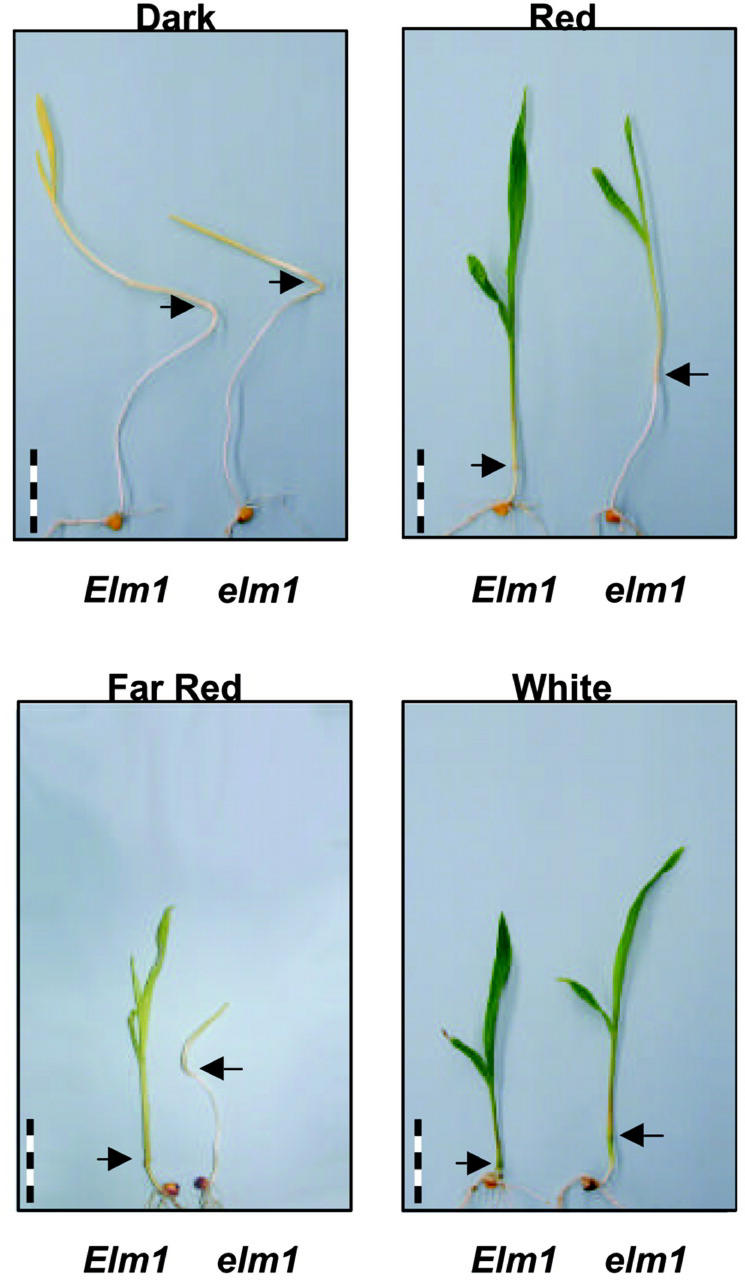
The phenotype of elm1 seedlings grown under white light (W) is shown in Figure 2. Under these conditions, elm1 showed a moderately elongated phenotype (Fig. 2), lower levels of chlorophyll (Chl), and an increased Chl a:b ratio (Table I). Carotenoid levels were also reduced in elm1 compared with wild-type seedlings (Table I). Detailed genetic analyses in Arabidopsis have indicated that phyB is the primary photoreceptor mediating de-etiolation in response to red (R), whereas phyA is the primary photoreceptor mediating responses to far-red (FR) (Quail, 2002). To further examine the light signal transduction pathway in elm1 mutants, we examined the inhibitory effect of R and FR on mesocotyl elongation. The mesocotyl can be considered functionally analogous to the dicot hypocotyl and, in wild-type seedlings, is greatly elongated in the absence of light stimuli (see Fig. 2). Wild-type and elm1 plants were grown in dark (D), R (3 μmol m−2 s−1), or FR (3 μmol m−2 s−1) and mesocotyl lengths measured 10 dap. As shown in Figure 2, etiolated (D) wild-type and elm1 seedlings showed similar elongation of the mesocotyl. In wild-type plants, elongation was strongly inhibited by both R and FR. In contrast, mesocotyl length was similar in elm1 seedlings under all growth conditions tested (Fig. 3). This morphology is indicative of a lack of responsiveness to either R or FR.
Figure 2.
Seedling phenotypes of wild-type (Elm1) and mutant (elm1) plants. Representative seedlings were photographed after 10 d of growth in D, R (3.0 μmol m−2 s−1), FR (1.2 μmol m−2 s−1), or W (100 μmol m−2 s−1). Arrows indicate boundary between mesocotyl and first internode. Scale bar divisions are in centimeters.
Table I.
Pigment measurements
| Pigment | Elm1 | elm1 |
|---|---|---|
| Total Chl (mg g−1 fresh wt)a | 1.16 ± 0.16 | 0.48 ± 0.07 |
| Chl a:b ratio | 3.73 ± 0.17 | 5.55 ± 1.03 |
| Carotenoids (mg g−1 fresh wt) | 0.27 ± 0.04 | 0.15 ± 0.01 |
Chl and carotenoids were measured in acetone extracts from 2-week-old W-grown seedlings (values shown are the mean ± se).
Figure 3.
De-etiolation responses of wild-type (Elm1) and mutant (elm1) plants. Mean (±se) mesocotyl length measurements in wild-type (Elm1) and mutant (elm1) seedlings grown for 10 d in D, R (3.0 μmol m−2 s−1), or FR (2.0 μmol m−2 s−1) growth conditions. Sample size is 15 to 20 seedlings per treatment/genotype.
elm1 Seedlings Do Not Contain Spectrally Active Phytochrome Pools
The nonresponsiveness of elm1 seedlings to both R and FR suggests disruption of both phyA- and phyB-mediated responses. To further investigate the activity of phytochrome in elm1 mutants, spectrophotometrically active pools of phytochrome were directly measured in etiolated elm1 and wild-type seedlings. Using in vivo spectrophotometry, the signal from elm1 seedlings was below the level of detection, whereas the signal from etiolated wild-type seedlings was 4.9 ± 0.1 (n = 2) units (1 unit is 1 × 10−3 ΔΔA730–800 nm). The level of detection is <0.3 units; therefore, elm1 seedlings contain <6% of the spectrally active phytochrome present in wild-type plants.
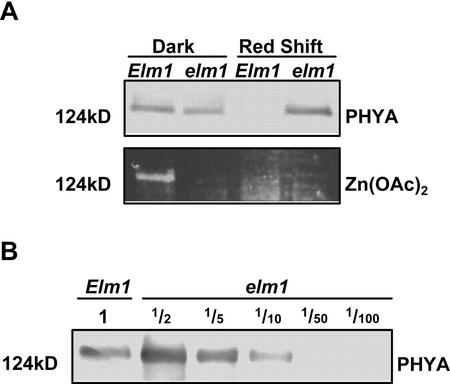
As an additional assay of phytochrome activity, PHYA accumulation was examined in elm1 seedlings. phyA is the most abundant phytochrome in etiolated tissue, but is rapidly degraded upon illumination. Because this degradation requires Pfr formation, the change in phyA stability after a light treatment can be used to assay the degree of photoconversion (Parks et al., 1989; Weller et al., 1996, 1997). As shown in Figure 4A, wild-type and elm1 seedlings accumulate PHYA in D. After 4 h of R (10 μmol m−2 s−1), phyA pools are rapidly depleted in wild-type but not in elm1 seedlings. Figure 4B shows that levels of PHYA were approximately 5- to 10-fold higher in elm1 seedlings relative to wild-type plants after a 4-h R treatment. This suggests that although PHYA accumulates in elm1 seedlings, it is not bound to PΦB.
Figure 4.
Immunoblot analysis of PHYA stability in wild-type (Elm1) and mutant (elm1) plants. A, Top, Immunodetection of PHYA apoprotein after SDS-PAGE and western blotting of crude protein extracts from wild-type (Elm1) and mutant (elm1) seedlings grown in dark (D) or given a 4-h R treatment (Red Shift; 10.0 μmol m−2 s−1). Lanes were loaded on an equivalent fresh weight basis and loading confirmed by Coomassie Blue stain (data not shown). Molecular masses (kD) were determined using prestained markers (see “Materials and Methods”). Bottom, Detection of bound PΦB by zinc-induced fluorescence after gel staining with Zn(OAc)2. Lanes as above. B, Immunodetection of PHYA apoprotein in crude protein extracts of R shift (4 h, 10.0 μmol m−2 s−1) wild-type (Elm1) and mutant elm1 seedlings. Extracts of elm1 seedlings were diluted from 2-fold (½) to 100-fold (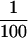 ) to allow semiquantitative determination of relative PHYA levels.
) to allow semiquantitative determination of relative PHYA levels.
To confirm that elm1 is deficient in holophytochrome, zinc-induced fluorescence was used to visualize the covalently bound chromophore (Berkelman and Lagarias, 1986). In wild-type plants, the presence of bound chromophore was detected as a fluorescent band in zinc-stained SDS-PAGE gels that comigrated with PHYA (Fig. 4A). However, although elm1 plants accumulate high levels of PHYA, no bound chromophore could be detected in extracts of either etiolated or red-shifted seedlings. The absence of chromophore associated with PHYA in etiolated elm1 seedlings suggests that there is either a disruption in holoenzyme assembly or PΦB synthesis in elm1 mutants.
elm1 Mutants Are Unable to Convert BV IXα to 3Z-PΦB
All phytochrome chromophore-deficient mutants characterized to date are blocked in one of two steps: heme to BV IXα or BV IXα to 3Z-PΦB (Terry, 1997). To examine the accumulation of heme oxygenase in elm1, we used an antibody raised to the HO1 (HY1) protein of Arabidopsis (Muramoto et al., 1999). Etiolated elm1 seedlings had an identical level of immunodetectable HO1 protein as wild-type seedlings (data not shown). This result suggests that heme oxygenase is unaffected in elm1 seedlings and that elm1 may be deficient in PΦB synthase. To directly assay PΦB synthase activity in elm1, we isolated plastids from dark-grown wild-type and elm1 seedlings, incubated these with BV IXα and heme, and analyzed the products by HPLC. Figure 5 shows that incubation of wild-type plastids with BV IXα results in the synthesis of two products, identified by their absorbance maxima and co-injection of authentic standards isolated from pea (data not shown), as 3Z-PΦB (peak 2, trace B) and 3E-PΦB (peak 3, trace B), respectively. In contrast, incubation of elm1 plastids with BV IXα produced no PΦB peaks (trace C). The major peak under these conditions was confirmed as the substrate BV IXα by its absorbance maximum and co-injection of authentic BV IXα (data not shown). To confirm that the elm1 mutation specifically affects PΦB synthase, we also assayed the same etioplast samples for heme oxygenase activity. Wild-type etioplasts were capable of converting heme to both BV IXα and 3Z-PΦB. In contrast, incubation of heme with elm1 etioplasts resulted in a small and reproducible increase in BV IXα synthesis, but no synthesis of PΦB (data not shown). These data strongly suggest that elm1 is specifically deficient in the enzyme PΦB synthase.
Figure 5.
HPLC analysis of BV metabolism by isolated plastids from wild-type (WT) and mutant (elm) seedlings. HPLC analysis of BV metabolism by isolated plastids from wild-type (WT) and mutant (elm) seedlings. A, Control incubation with BV IXα and all reaction components except plastids. B and C, Bilin products obtained after incubation of WT or elm1 mutant plastids with BV IXα. Peaks identified as 1 BV IXα, 2 3Z-PΦB, and 3 3E-PΦB are indicated.
Photosynthetic Transcripts Accumulate to Reduced Levels in elm1 Seedlings
To investigate the requirement of phytochrome for photosynthetic development, the levels of transcripts encoding several plastidic proteins were examined in elm1 seedlings. Wild-type and elm1 seedlings were grown in D, under two fluences of R and in W. RNA gel-blot analysis was used to assay the accumulation of transcripts encoded by Cab, RbcS, rbcL, and psbA genes (Fig. 6). Cab and RbcS are nuclear transcripts encoding the light-harvesting Chl a/b protein (LHCPII) and the small subunit of Rubisco, respectively. The transcription of Cab and RbcS genes is regulated by both phytochrome and blue light-mediated signaling (Fluhr et al., 1986; Tobin and Silverthorne, 1986). rbcL and psbA are chloroplast genes encoding the large subunit of Rubisco and the D1 peptide of photosystem II, respectively. As observed with Cab and RbcS, transcripts encoded by rbcL and psbA accumulate to higher levels after illumination (Bedbrook et al., 1978; Crossland et al., 1984). Under D growth conditions, wild-type and elm1 mutants showed similar low-level accumulation of photosynthetic transcripts. As expected, all photosynthetic transcripts examined in wild-type seedlings accumulated to much higher levels under R relative to D growth conditions. In contrast, there was a relatively modest accumulation of all photosynthetic transcripts examined in elm1 seedlings grown under low- or high-R conditions. Under W, rbcL and psbA transcripts accumulated to similar levels in wild-type and elm1 mutant seedlings. RbcS transcripts accumulated to slightly lower levels in W-grown elm1 relative to wild type, but Cab transcripts were significantly lower in elm1 plants relative to W-grown wild-type seedlings. These data show that the R-mediated nuclear (Cab and RbcS) and plastid (rbcL and psbA) transcript accumulation is impaired in elm1 seedlings.
Figure 6.
Northern-blot analysis of photosynthetic transcript accumulation in wild-type and elm1 seedlings. Total RNA was extracted from wild-type and elm1 mutant seedlings grown under D, low R (3 μmol m−2 s−1), high R (30 μmol m−2 s−1), or W (100 μmol m−2 s−1) light conditions. Filters were hybridized to gene-specific fragments for the Cab, RbcS, psbA, and rbcL genes as described in “Materials and Methods.” Approximately 5 μg of total RNA was loaded per lane. The 26S ribosomal band was visualized by ethidium bromide staining and was used as a loading control. Approximate transcript sizes (kb) are shown on the left.
DISCUSSION
The data presented here show that the elm1 mutant of maize is severely deficient in photoreversible phytochrome and responds only weakly to both R and FR irradiation. These results indicate that elm1 lacks multiple phytochromes and is consistent with a deficiency in phytochrome chromophore synthesis or assembly. Mutants disrupted in the synthesis of the phytochrome chromophore have been characterized in a number of species (Terry, 1997) and are disrupted at one of two loci encoding heme oxygenase or PΦB synthase. To date, no locus has been implicated in the regulation of chromophore synthetic enzymes, post-synthetic chromophore processing or holophytochrome assembly. Measurement of PΦB synthesis in isolated etioplasts demonstrated that elm1 was unable to synthesize 3Z-PΦB from BV IXα. This result, together with the retention of heme oxygenase protein and activity, suggest that elm1 is specifically deficient in PΦB synthase. Therefore, the elm1 mutant is similar to the hy2 mutant of Arabidopsis (Koornneef et al., 1980; Kohchi et al., 2001), pcd2 of pea (Weller et al., 1997), and the au mutant of tomato (Koornneef et al., 1985; Terry and Kendrick, 1996), but is not equivalent to any known mutants from monocot species. In Arabidopsis, the HY2 gene has recently been cloned and shown to encode PΦB synthase (Kohchi et al., 2001). Unfortunately, searches of public maize expressed sequence tag collections have failed to identify putative maize orthologs of HY2. This may be due to the relatively low abundance of HY2-like transcripts in maize or may reflect a high degree of sequence divergence between the maize and Arabidopsis gene sequences. The rapid progress being made in the sequencing of the rice genome could soon help to provide a bridge to the isolation of a maize HY2 ortholog and possibly to the cloning of Elm1 in maize.
elm1 mutants are pale green both as seedlings and as mature field-grown plants, a phenotypic trait observed in most chromophore-deficient mutants (Terry, 1997). Investigation of greening in elm1 seedlings has demonstrated a reduction in the accumulation of transcripts encoding a number of chloroplast components. Under R, Cab, RbcS, rbcL, and psbA transcripts accumulated to reduced levels in elm1 seedlings. Under W, rbcL and psbA transcripts accumulated to similar levels in elm1 and wild-type seedlings, suggesting that R and possibly blue light-signaling systems function redundantly to promote the accumulation of chloroplast-encoded transcripts. In contrast to rbcL and psbA, Cab and RbcS transcripts did not accumulate to similar levels in wild-type and elm1 seedlings grown in W. This result suggests that the blue-light signal transduction pathway is unable to compensate for impaired phytochrome signaling in elm1 mutants. However, it does not exclude the possibility that the reduced levels of Cab and RbcS transcripts reflect an altered physiology directly or indirectly responsible for lower Chl and carotenoid levels in elm1 plants.
Although non-plastidic photoreceptor systems may signal directly to chloroplasts, the probable targets of light regulation are nuclear factors required for the accumulation of plastid-encoded transcripts. The identification of nuclear-encoded plastid-localized RNA polymerase (Greenberg et al., 1984; Lerbs-Mache, 1993; Young et al., 1998) and plastid-targeted sigma factors (Tiller et al., 1991; Lahiri et al., 1999) has revealed the importance of transcriptional control in the regulation in plastid gene expression. The accumulation of both nuclear-encoded plastid-localized RNA polymerase and plastid-localized sigma factors is normally light regulated (Chang et al., 1999; Lahiri et al., 1999) and, therefore, may be disrupted in elm1 plants. Furthermore, genetic analyses have revealed a number of nuclear loci required for the processing of chloroplast mRNAs and highlight the importance of posttranscriptional regulation of transcript abundance (Stern et al., 1997). Thus, although we observed dramatic decreases in levels of psbA and rbcL transcripts in elm1 mutants, it is unclear if this reflects a decreased rate of transcription, an increased rate of transcript degradation, or a combination of both.
Although it is clear from the above discussion that deficiencies in light signaling are likely to be important in determining the pale-green phenotype of elm1, the analysis of chromophore-deficient mutants from other species has indicated that other factors may play a role (Terry, 1997). Inconsistencies between the degree of Chl deficiency and the level of functional phytochrome have led to the proposal that feedback inhibition of Chl synthesis results from the block in plastidic heme degradation (Terry and Kendrick, 1999). Consistent with this hypothesis, dark-grown au and yg-2 mutants have a reduced level of the Chl precursor protochlorophyllide (Terry and Kendrick, 1999) and this phenomenon has also been observed in chromophore-deficient mutants of pea and Arabidopsis (Terry et al., 2001). The pale-green phenotype and reduced Chl accumulation in elm1 plants, therefore, may result in part from a similar negative feedback of Chl biosynthesis. Examination of protochlorophyllide levels in D-grown elm1 seedlings should resolve this issue.
The elm1 mutant represents the first light-signaling lesion to be characterized in maize and presents evidence that phytochrome influences flowering time in this species. Flowering time in many species is modulated by the relative duration of light and darkness during a daily cycle (the photoperiod; Thomas and Vince-Prue, 1997). In some plants, flowering is promoted by short days (SD), whereas in others, flowering is promoted by LD. Genetic analyses, notably of Arabidopsis (an LD plant), have indicated that both phytochrome and blue light-signaling pathways interact in the perception of photoperiod and in the regulation of flowering. As a generalization, phyA acts to promote flowering (Johnson et al., 1994), whereas phyB is required to inhibit flowering (Goto et al., 1991; Reed et al., 1993). The blue light-sensing cryptochromes act to promote flowering, both in a phytochrome-independent manner and by antagonism of phyB signaling (Koornneef et al., 1991; Bagnall et al., 1996; Guo et al., 1998; Mockler et al., 1999).
Although accessions of maize grown in the United States are generally considered day neutral, the early flowering phenotype of elm1 shows that flowering is repressed under LD in the W22 inbred. The response is similar to that seen in the se5 mutant of rice (an SD plant), although the magnitude of the effect in rice is greater (under LD wild-type rice flowered after 100.8 ± 0.8 d, se5 flowered after 46.6 ± 0.6 d; Izawa et al., 2000). The expansion of cultivated maize beyond semitropical regions of early domestication has required the selection of day-neutral lines from ancestrally SD stocks. Nevertheless, the elm1 mutant suggests that standard U.S. inbreds retain a weak response to photoperiod. The elm1 mutant, the se5 mutant of rice, the BMDR-1 mutant of barley, and the ma3R line of sorghum collectively show that the establishment of early flowering under nonpermissive photoperiods can be achieved by selection for reduced phytochrome signaling. The utility of loss-of-function alleles in components of the phytochrome pathway has made this an efficient target for past selection and an attractive candidate for future genetic modification.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Homozygous elm1 mutants and near-isogenic wild-type seed stocks were maintained in a standard maize (Zea mays) W22 inbred line. Seeds were surface sterilized for 15 min in a 10% (v/v) commercial bleach solution containing 0.1% (v/v) Tween 20, rinsed five times in deionized water, and imbibed overnight with shaking at room temperature. For mesocotyl measurements, seeds were grown in Rootrainers (http://www.hummert.com) containing vermiculite. On d 10, mesocotyl length was measured to the nearest millimeter using digital calipers. All seedlings were grown under continuous light or in constant darkness at 28°C in Percival Scientific (Boone, IA) model E-30LED light chambers with the exception of white-light treatments. LED light modules provided red and far-red light sources with narrow wavebands and peak emissions at 664 and 736 nm, respectively. White-light growth used a combination of incandescent and cool-white fluorescent lighting. Plants were grown for 10 d under light treatments before photography, RNA isolation, and mesocotyl measurements.
Measurement of Photosynthetic Pigments
Wild-type and elm1 plants were grown for 2 weeks for 16 h in 180 μmol m−2 s−1 W and 8 h in D at 23°C. Three leaf discs were taken from the third leaf of four different plants and Chl and carotenoids were extracted into 80% (v/v) acetone and quantified according to Lichtenthaler (1987).
Protein Gel-Blot Analysis
Seedlings were frozen in liquid nitrogen, ground to a fine powder, and suspended at 2 mL g−1 in extraction buffer (37.5% [v/v] ethylene glycol, 75 mm Tris-HCl [pH 8.3], 7.5 mm Na4EDTA, 15 mm NaS2O5, 0.11% [v/v] polyethylenimine, and 1.5 mm phenylmethylsulfonyl fluoride; Davis et al., 2001). Extracts were clarified by centrifugation at 3,000g for 30 min at 4°C, fractionated by SDS-PAGE (7.5% [w/v] acrylamide gel) and transferred to nitrocellulose membrane (Schleicher & Schull, Keene, NH). Gels were loaded by mass of starting tissue and equal loading confirmed by Coomassie Blue staining. Samples were equivalent to approximately 50 μg of total protein as determined using the Bio-Rad DC protein assay antibody (Bio-Rad, Hercules, CA). PHYA protein was detected using the monoclonal antibody O73D (Boylan and Quail, 1991), horseradish peroxidase-conjugated goat-anti-mouse secondary antibody and the Bio-Rad Opti-4CN substrate kit. To detect PΦB, acrylamide gels were incubated for 2 h in 1 m zinc acetate and visualized under UV light (Berkelman and Lagarias, 1986). Molecular masses (kD) were determined using prestained markers (SeeBlue Plus 2, Invitrogen, Carlsbad, CA).
Spectrophotometric Assay for Phytochrome
Phytochrome levels in wild-type and elm1 seedlings were assayed by in vivo spectroscopy as described previously (Weller et al., 1996). Seedlings were grown for 7 d in D at 25°C and the top 1.5 cm of eight seedlings were used for each sample.
Assays for PΦB Synthesis
Maize seeds were sown in wet vermiculite (washed to remove fine particles before use), cold treated for 24h at 4°C, and then grown in the dark at 23°C for a further 8 to 10 d before etioplast isolation. Maize etioplasts were isolated as described previously (Weller et al., 1996) and PΦB synthesis from BV IXα was assayed essentially as described before for pea (Pisum sativum; Weller et al., 1996; Terry, 2001), but with the following modifications; PΦB assays were performed in 1 mL of reaction buffer, 20 mm TES, 10 mm HEPES-NaOH (pH 7.7) containing 500 mm sorbitol, 1 mm phenylmethylsulfonyl fluoride, 2 μm leupeptin, and 0.5 mm dithiothreitol, and an NADPH regenerating system (1.2 mm NADP+, 10 mm Glc-6-phosphate, and 2.5 units mL−1 Glc-6-phosphate dehydrogenase), 3,000 U mL−1 catalase, 1 mm desferrioxamine, and 5 mm ascorbate. The reaction was initiated by the addition of 10 μL of BV IXα (Porphyrin Products Inc., Logan, UT) to give a final substrate concentration of 10 μm. Bilins were recovered and concentrated using a C18 cartridge (SepPak Plus, Waters Corporation, Milford, MA) as described previously (Terry et al., 1995; Terry, 2001). HPLC analysis was performed using an LC-10 system (Shimadzu Corp., Kyoto) running VP-5 software and using an SPD-M10A photodiode array detector. An LC-18 column (5 μm; 250 × 4.6 mm; Supelco UK, Poole, UK) was used with a mobile phase of acetone:ethanol:100 mm formic acid (25:65:10 [v/v]) at an isocratic flow rate of 1 mL min−1 (Weller et al., 1996; Terry, 2001). The photodiode array detector was used to monitor spectra between 300 and 800 nm over 60 min.
RNA Gel-Blot Analysis
Seedling tissue was harvested at the base of the coleoptile and flash frozen in liquid nitrogen. Total RNA was extracted from approximately 1 g of tissue as previously described (Van Tunen et al., 1988). Approximately 5 μg of total RNA was fractionated on 1.5% (w/v) agarose gels containing 6.8% (v/v) formaldehyde and photographs taken of ethidium bromide-stained gels to visualize ribosomal bands. RNA was transferred to GeneScreen Plus nylon membrane (NEN, Boston, MA) through capillary transfer in 20× SSC. Digoxygenin (DIG)-labeled DNA probes were synthesized using the PCR DIG Probe Synthesis kit (Roche, Indianapolis, IN) according to the manufacturer's recommendations, using T7 and T3 primers. Gene-specific fragments for RbcS (pJL12), rbcL (pJL12), psbA (pSD7), and Cab (LHCP1020) were described previously (Roth et al., 1996; Hall et al., 1998). Hybridizations were performed using the Roche DIG Easy Hyb solution. Hybridization buffers and conditions were according to the manufacturer's recommendation (DIG Easy Hyb, Roche). In brief, membranes were prehybridized in 15 to 25 mL of DIG Easy Hyb buffer (Roche) for 30 min to 1 h. The prehybridization buffer was removed and 5 mL of hybridization buffer was added to 3 μL of labeled probe. Hybridization was performed overnight at 43°C. Membranes were washed twice in 2× SSC and 0.1% (w/v) SDS at room temperature for 5 min and twice in 0.1× SSC and 0.1% (w/v) SDS at 68°C for 15 min each. Membranes were then washed in 1× maleic acid buffer (0.1 m maleic acid and 0.15 m NaCl, pH 7.5) for 3 min followed by 1 to 2 h of shaking in blocking solution (10% [w/v] casein, 0.1 m maleic, and 0.15 NaCl) acid before addition of 5 μL of Anti-DIG-AP Fab fragments (Roche). Blots were incubated for 30 min at room temperature with gentle shaking, washed for 15 min twice in 1× Washing buffer (3% [v/v] Tween 20, 0.1 m maleic acid, and 0.15 m NaCl). Blots were incubated with detection buffer (0.1 m Tris and 0.1 m NaCl, pH 9.5) for 3 min and placed in plastic sheet protectors. Excess liquid was removed and 2 mL of CDP-Star solution (20 μL of CDP-Star reagent + 2 mL of detection buffer) were applied directly to membranes. Blots were incubated for 5 min and exposed on an image station 440 CF (Eastman-Kodak, Rochester, NY). Eastman-Kodak 1D 3.5.4 Image Analysis software was used to determine relative transcript abundance.
ACKNOWLEDGMENTS
We would like to thank Dr. James L. Weller (University of Tasmania, Australia) for help with the in vivo spectrophotometric experiments (performed in the Laboratory for Photoperception and Signal Transduction, Frontier Research Program, RIKEN, Wako, Japan), Dr. Takayuki Kohchi (Nara Institute of Science and Technology, Nara, Japan) for providing us with the HO1 antibody, and Dr. Peter Quail (Plant Gene Expression Center, Albany, CA) for providing us with the PHYA antibody. We would also like to thank Julie Batley (University of Cambridge, UK) for help with protein extractions.
Footnotes
This work was supported by the National Science Foundation (grant no. IBN–0110297 to T.P.B.), by the UK Biotechnology and Biological Sciences Research Council (grant no. 51/P10948 to M.J.T.), and by the Human Frontier Science Short-Term Fellowship Program (grant no. SF0085/1999–M to T.P.B. and M.J.T.). M.J.T. is a Royal Society University Research Fellow.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.006411.
LITERATURE CITED
- Bagnall DJ, King RW, Hangarter RP. Blue-light promotion of flowering is absent in hy4 mutants of Arabidopsis. Planta. 1996;200:278–280. doi: 10.1007/BF00208319. [DOI] [PubMed] [Google Scholar]
- Basu D, Dehesh K, Schneider-Poetsch HJ, Harrington SE, McCouch SR, Quail PH. Rice PhyC gene: structure, expression, map-position and evolution. Plant Mol Biol. 2000;44:27–42. doi: 10.1023/a:1006488119301. [DOI] [PubMed] [Google Scholar]
- Bedbrook JR, Link G, Coen DM, Bogorad L, Rich A. Maize plastid gene expressed during photoregulated development. Proc Natl Acad Sci USA. 1978;75:3060–3064. doi: 10.1073/pnas.75.7.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkelman TR, Lagarias JC. Visualization of bilin-linked peptides and proteins in polyacrylamide gels. Anal Biochem. 1986;156:194–201. doi: 10.1016/0003-2697(86)90173-9. [DOI] [PubMed] [Google Scholar]
- Boylan MT, Quail PH. Phytochrome A overexpression inhibits hypocotyl elongation in transgenic Arabidopsis. Proc Natl Acad Sci USA. 1991;88:10806–10810. doi: 10.1073/pnas.88.23.10806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Sheen J, Bligny M, Niwa Y, Lerbs-Mache S, Stern DB. Functional analysis of two maize cDNAs encoding T7-like RNA polymerases. Plant Cell. 1999;11:911–926. doi: 10.1105/tpc.11.5.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs KL, Miller FR, Cordonnier-Pratt MM, Pratt LH, Morgan PW, Mullet JE. The sorghum photoperiod sensitivity gene, Ma3, encodes a phytochrome B. Plant Physiol. 1997;113:611–619. doi: 10.1104/pp.113.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen AH, Quail PH. Structure and expression of a maize phytochrome-encoding gene. Gene. 1989;85:381–390. doi: 10.1016/0378-1119(89)90431-9. [DOI] [PubMed] [Google Scholar]
- Clack T, Mathews S, Sharrock RA. The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Mol Biol. 1994;25:413–427. doi: 10.1007/BF00043870. [DOI] [PubMed] [Google Scholar]
- Crossland LD, Rodermel SR, Bogorad L. Single gene for the large subunit of ribulosebisphosphate carboxylase in maize yields two differentially regulated mRNAs. Proc Natl Acad Sci USA. 1984;81:4060–4064. doi: 10.1073/pnas.81.13.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis SJ, Bhoo SH, Durski AM, Walker JM, Vierstra RD. The heme-oxygenase family required for phytochrome chromophore biosynthesis is necessary for proper photomorphogenesis in higher plants. Plant Physiol. 2001;126:656–669. doi: 10.1104/pp.126.2.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis SJ, Kurepa J, Vierstra RD. The Arabidopsis thaliana HY1 locus, required for phytochrome-chromophore biosynthesis, encodes a protein related to heme oxygenases. Proc Natl Acad Sci USA. 1999;96:6541–6546. doi: 10.1073/pnas.96.11.6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluhr S, Kuhlemeier C, Nagy F, Chua N-H. Organ-specific and light-induced expression of plant genes. Science. 1986;232:1106–1112. doi: 10.1126/science.232.4754.1106. [DOI] [PubMed] [Google Scholar]
- Goto N, Kumagai T, Koornneef M. Flowering responses to light-breaks in photomorphogenic mutants of Arabidopsis thaliana, a long-day plant. Physiol Plant. 1991;83:209–215. [Google Scholar]
- Greenberg BM, Narita JO, DeLuca-Flaherty C, Gruissem W, Rushlow KA, Hallick RB. Evidence for two RNA polymerase activities in Euglena gracilis chloroplasts. J Biol Chem. 1984;259:14880–14887. [PubMed] [Google Scholar]
- Guo H, Yang H, Mockler TC, Lin C. Regulation of flowering time by Arabidopsis photoreceptors. Science. 1998;279:1360–1363. doi: 10.1126/science.279.5355.1360. [DOI] [PubMed] [Google Scholar]
- Hall LN, Rossini L, Cribb L, Langdale JA. GOLDEN 2: a novel transcriptional regulator of cellular differentiation in the maize leaf. Plant Cell. 1998;10:925–936. doi: 10.1105/tpc.10.6.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanumappa M, Pratt LH, Cordonnier-Pratt MM, Deitzer GF. A photoperiod-insensitive barley line contains a light-labile phytochrome B. Plant Physiol. 1999;119:1033–1040. doi: 10.1104/pp.119.3.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T, Oikawa T, Tokutomi S, Okuno K, Shimamoto K. Phytochromes confer the photoperiodic control of flowering in rice (a short-day plant) Plant J. 2000;22:391–399. doi: 10.1046/j.1365-313x.2000.00753.x. [DOI] [PubMed] [Google Scholar]
- Johnson E, Bradley M, Harberd NP, Whitelam GC. Photoresponses of light-grown phyA mutants of Arabidopsis. Plant Physiol. 1994;105:141–149. doi: 10.1104/pp.105.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohchi T, Mukougawa K, Frankenberg N, Masuda M, Yokota A, Lagarias JC. The Arabidopsis Hy2 gene encodes phytochromobilin synthase, a ferredoxin-dependent biliverdin reductase. Plant Cell. 2001;13:425–436. doi: 10.1105/tpc.13.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Cone JW, Dekens RG, O'Herne-Robers EG, Spruitt CJP, Kendrick RE. Photomorphogenic responses of long-hypocotyl mutants of tomato. J Plant Physiol. 1985;120:153–165. [Google Scholar]
- Koornneef M, Hanhart CJ, van der Veen JH. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet. 1991;229:57–66. doi: 10.1007/BF00264213. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Rolff E, Spruitt CJP. Genetic control of light-induced hypocotyl elongation in Arabidopsis thaliana L. Z Pflanzenphysiol. 1980;100:147–160. [Google Scholar]
- Kraepiel Y, Jullien M, Cordonnier-Pratt MM, Pratt L. Identification of two loci involved in phytochrome expression in Nicotiana plumbaginifolia and lethality of the corresponding double mutant. Mol Gen Genet. 1994;242:559–565. doi: 10.1007/BF00285279. [DOI] [PubMed] [Google Scholar]
- Lahiri SD, Yao J, McCumbers C, Allison LA. Tissue-specific and light-dependent expression within a family of nuclear-encoded sigma-like factors from Zea mays. Mol Cell Biol Res Commun. 1999;1:14–20. doi: 10.1006/mcbr.1999.0102. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol. 1987;148:350–382. [Google Scholar]
- Lerbs-Mache S. The 110-kDa polypeptide of spinach plastid DNA-dependent RNA polymerase: single-subunit enzyme or catalytic core of multimeric enzyme complexes? Proc Natl Acad Sci USA. 1993;90:5509–5513. doi: 10.1073/pnas.90.12.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Juez E, Nagatani A, Buurmeijer WF, Peters JL, Furuya M, Kendrick RE, Wesselius JC. Response of light-grown wild type and au-mutant tomato plants to end-of-day far-red light. J Photochem Photobiol B. 1990;4:391–405. [Google Scholar]
- Mathews S, Sharrock RA. The phytochrome gene family in grasses (Poaceae): a phylogeny and evidence that grasses have a subset of the loci found in dicot angiosperms. Mol Biol Evol. 1996;13:1141–1150. doi: 10.1093/oxfordjournals.molbev.a025677. [DOI] [PubMed] [Google Scholar]
- Mockler TC, Guo H, Yang H, Duong H, Lin C. Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development. 1999;126:2073–2082. doi: 10.1242/dev.126.10.2073. [DOI] [PubMed] [Google Scholar]
- Muramoto T, Kohchi T, Yokota A, Hwang I, Goodman HM. The Arabidopsis photomorphogenic mutant hy1 is deficient in phytochrome chromophore biosynthesis as a result of a mutation in a plastid heme oxygenase. Plant Cell. 1999;11:335–348. doi: 10.1105/tpc.11.3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BM, Shanklin J, Koorneef, Kendrick RE, Quail PH. Immunochemically detectable phytochrome is present at normal levels but is photochemically nonfunctional in the hy1 and hy2 long hypocotyl mutants of Arabidopsis. Plant Mol Biol. 1989;12:425–437. doi: 10.1007/BF00017582. [DOI] [PubMed] [Google Scholar]
- Quail PH. Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol. 2002;3:85–93. doi: 10.1038/nrm728. [DOI] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J. Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell. 1993;5:147–157. doi: 10.1105/tpc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson PR, McCormac AC, Irvine AS, Smith H. Genetic engineering of harvest index in tobacco through overexpression of a phytochrome gene. Nat Biotechnol. 1996;14:995–998. doi: 10.1038/nbt0896-995. [DOI] [PubMed] [Google Scholar]
- Roth R, Hall LN, Brutnell T, Langdale JA. bundle sheath defective2, a mutation that disrupts the coordinated development of bundle sheath and mesophyll cells in the maize leaf. Plant Cell. 1996;8:915–927. doi: 10.1105/tpc.8.5.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt J. Is photomorphogenic shade avoidance adaptive? Perspectives from population biology. Plant Cell Environ. 1997;20:826–830. [Google Scholar]
- Schneider-Poetsch HAW, Kolukisaoglu U, Clapham DH, Hughes J, Lamparter T. Non-angiosperm phytochromes and the evolution of vascular plants. Physiol Plant. 1998;102:612–622. [Google Scholar]
- Shlumukov LR, Barro F, Barcelo P, Lazzeri P, Smith H. Establishment of far-red high irradiance responses in wheat through transgenic expression of an oat phytochrome A gene. Plant Cell Environ. 2001;24:703–712. [Google Scholar]
- Smith H. Phytochromes and light signal perception by plants: an emerging synthesis. Nature. 2000;407:585–591. doi: 10.1038/35036500. [DOI] [PubMed] [Google Scholar]
- Stern DB, Higgs DC, Yang J. Transcription and translation in chloroplasts. Trends Plant Sci. 1997;2:308–315. [Google Scholar]
- Takano M, Kanegae H, Shinomura T, Miyao A, Hirochika H, Furuya M. Isolation and characterization of rice phytochrome A mutants. Plant Cell. 2001;13:521–534. doi: 10.1105/tpc.13.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry MJ. Phytochrome chromophore-deficient mutants. Plant Cell Environ. 1997;20:740–745. [Google Scholar]
- Terry MJ. Biosynthesis and analysis of bilins. In: Smith AG, Witty M, editors. Heme, Chlorophyll, and Bilins: Methods and Protocols. Totowa, NJ: Humana Press Inc; 2001. pp. 273–292. [Google Scholar]
- Terry MJ, Kendrick RE. The aurea and yellow-green-2 mutants of tomato are deficient in phytochrome chromophore synthesis. J Biol Chem. 1996;271:21681–21686. doi: 10.1074/jbc.271.35.21681. [DOI] [PubMed] [Google Scholar]
- Terry MJ, Kendrick RE. Feedback inhibition of chlorophyll synthesis in the phytochrome chromophore-deficient aurea and yellow-green-2 mutants of tomato. Plant Physiol. 1999;119:143–152. doi: 10.1104/pp.119.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry MJ, McDowell MT, Lagarias JC. (3Z)- and (3E)-phytochromobilin are intermediates in the biosynthesis of the phytochrome chromophore. J Biol Chem. 1995;270:11111–11118. doi: 10.1074/jbc.270.19.11111. [DOI] [PubMed] [Google Scholar]
- Terry MJ, Ryberg M, Raitt CE, Page AM. Altered etioplast development in phytochrome chromophore-deficient mutants. Planta. 2001;214:314–325. doi: 10.1007/s004250100624. [DOI] [PubMed] [Google Scholar]
- Thomas B, Vince-Prue D. Photoperiodism in Plants. New York: Academic Press; 1997. [Google Scholar]
- Tiller K, Eisermann A, Link G. The chloroplast transcription apparatus from mustard (Sinapis alba L.). Evidence for three different transcription factors which resemble bacterial sigma factors. Eur J Biochem. 1991;198:93–99. doi: 10.1111/j.1432-1033.1991.tb15990.x. [DOI] [PubMed] [Google Scholar]
- Tobin EM, Silverthorne J. Light regulation of gene expression in higher plants. Annu Rev Plant Physiol. 1986;36:569–593. [Google Scholar]
- Van Tunen AJ, Koes RE, Spelt CE, van Der Krol AR, Stuitje AR, Mol JNM. Cloning of two chalcone flavanone isomerase genes from Petunia hybrida: coordinate, light regulated and differential expression of flavonoid genes. EMBO J. 1988;7:1257–1263. doi: 10.1002/j.1460-2075.1988.tb02939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Terry MJ, Rameau C, Reid JB, Kendrick RE. The phytochrome-deficient pcd1 mutant of pea is unable to convert heme to biliverdin IXα. Plant Cell. 1996;8:55–67. doi: 10.1105/tpc.8.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Terry MJ, Reid JB, Kendrick RE. The phytochrome-deficient pcd2 mutant of pea is unable to convert biliverdin IXα to 3Z-phytochromobilin. Plant J. 1997;11:1177–1186. doi: 10.1105/tpc.8.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam GC, Patel S, Devlin PF. Phytochromes and photomorphogenesis in Arabidopsis. Philos Trans R Soc Lond B Biol Sci. 1998;353:1445–1453. doi: 10.1098/rstb.1998.0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoo T, Okuno K. Genetic analysis of earliness mutations induced in the rice cultivar Norin 8. Japan J Breed. 1993;43:1–11. [Google Scholar]
- Young DA, Allen RL, Harvey AJ, Lonsdale DM. Characterization of a gene encoding a single-subunit bacteriophage-type RNA polymerase from maize which is alternatively spliced. Mol Gen Genet. 1998;260:30–37. doi: 10.1007/s004380050867. [DOI] [PubMed] [Google Scholar]