Abstract
T cell receptor (TCR) β variable region genes are assembled in progenitor T cells from germ-line Vβ, Dβ, and Jβ segments via an ordered two-step process in which Dβ to Jβ rearrangements occur on both alleles before appendage of a Vβ to a preexisting DJβ complex. Direct joining of Vβ segments to nonrearranged Dβ or Jβ segments, while compatible with known restrictions on the V(D)J recombination mechanism, are infrequent within the endogenous TCRβ locus. We have analyzed mechanisms that mediate ordered Vβ, Dβ, and Jβ assembly via an approach in which TCRβ minilocus recombination substrates were introduced into embryonic stem cells and then analyzed for rearrangement in normal thymocytes by recombinase-activating gene 2-deficient blastocyst complementation. These analyses demonstrated that Vβ segments are preferentially targeted for rearrangement to Dβ as opposed to Jβ segments. In addition, we further demonstrated that Vβ segments can be appended to nonrearranged endogenous Dβ segments in which we have eliminated the ability of Dβ segments to join to Jβ segments. Our findings are discussed in the context of the mechanisms that regulate the ordered assembly and utilization of V, D, and J segments.
T cell receptor (TCR) and immunoglobulin (Ig) variable region genes are assembled during progenitor lymphocyte development from variable (V), diversity (D), and joining (J) segments by V(D)J recombination (1, 2). V(D)J recombination is initiated via introduction of DNA double-strand breaks between two participating variable gene segments and their associated recombination signal sequences (RSSs) (3). This reaction is carried out by the lymphocyte-specific recombinase-activating gene 1 (RAG-1) and RAG-2 proteins, which are specifically targeted by the RSSs (4, 5). Subsequently, the cleaved gene segments are joined by a set of generally expressed DNA repair enzymes, which catalyze a nonhomologous end-joining reaction (6). Thus, the specificity of the reaction is provided by the recognition of RSSs by the RAG proteins.
RSSs consist of conserved heptamer and nonamer sequences that flank nonconserved spacers of 12 or 23 bp (hereafter referred to as 12-RSS and 23-RSS, respectively) (1). V(D)J recombination occurs only between gene segments flanked by a 12-RSS and a 23-RSS, a phenomenon referred to as the 12/23 rule (1). This restriction in V(D)J joining is mediated at the level of RAG recognition and cleavage (7, 8). The assembly of all of the different families and types of TCR and Ig variable region gene segments is mediated by the RAG and nonhomologous end-joining proteins, a property based on the relative conservation of the RSSs among the different gene segments. However, despite the relatively generic nature of the basic reaction, V(D)J recombination is tightly regulated in the context of lineage specificity (e.g., TCR variable region genes are assembled in T but not B cells), the context of developmental stage (e.g., TCRβ variable region genes are assembled before TCRα genes), and in the context of allelic exclusion (9). These observations led to the notion that V(D)J recombination is directed by modulating the accessibility of the various classes of variable region gene segments to the common V(D)J recombinase (9).
TCRβ variable region genes are assembled from Vβ, Dβ, and Jβ gene segments in progenitor thymocytes by an ordered process in which Dβ to Jβ rearrangement generally occurs on both alleles before appendage of a Vβ to a preexisting DJβ complex. In addition, expression of a TCRβ protein from a productive VβDJβ rearrangement prevents Vβ to DJβ rearrangement on the second allele and ensures allelic exclusion (10, 11). The mechanisms responsible for ordered assembly of V, D, and J segments or for allelic exclusion of V region gene assembly are not known; however, various considerations suggests that they may be mechanistically linked (12). In this context, the V, D, and J segments of the TCRβ and IgH loci, which are both assembled in an ordered fashion, also are regulated in the context of allelic exclusion. In contrast, TCRδ locus V, D, and J segment assembly is not ordered (13), and this locus does not exhibit allelic exclusion (14).
The murine TCRβ locus is composed of ≈35 Vβ segments and two Dβ-Jβ-Cβ clusters. Vβ segments are flanked by 3′ 23-RSSs and Jβ segments are flanked by 5′ 12-RSSs whereas Dβ segments are flanked by 5′ 12-RSSs and 3′ 23-RSSs (Fig. 1 A and B). Thus, whereas assembly of complete VβDJβ rearrangements occurs in the context of the 12/23 rule, direct Vβ to Jβ joining is also permissible by the 12/23 rule. Yet, most TCRβ rearrangements include Dβ nucleotides, suggesting that mechanisms exist to ensure both ordered rearrangement and Dβ segment utilization. The complex organization of the TCRβ locus presents a challenging obstacle to analyses of mechanisms that direct rearrangement within the locus. To address this issue, we have developed a transgenic TCRβ minilocus that rearranges in developing T cells and recapitulates many features of the endogenous TCRβ gene assembly process (15, 16). For example, TCRβ minilocus rearrangement is ordered, with Dβ to Jβ rearrangement occurring before Vβ to Dβ rearrangement. Moreover, direct Vβ to Jβ rearrangements within the construct appear infrequent, despite the relatively close proximity of these segments in the minilocus (15). Thus, this TCRβ minilocus should provide an ideal model system to study cis-acting elements that regulate ordered TCRβ gene segment rearrangement.
Figure 1.
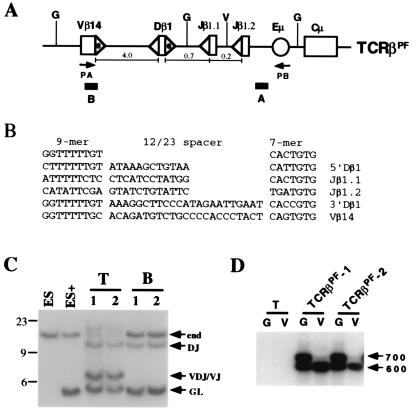
Rearrangement of TCRβPF in T and B cells. (A) TCRβPF contains germ-line Vβ14, Dβ1, and Jβ1.1/Jβ1.2 gene segments linked to the IgH intronic enhancer (Eμ) and constant region gene (Cμ). Shown are the locations of probes A and B, BglII (G) and EcoRV (V) sites, and the PA and PB oligonucleotide primers. 23-RSSs (dotted ▵) and 12-RSSs (▵) are indicated. The distances (kb) between gene segments are noted. The minilocus is not drawn to scale. (B) The sequences of the Vβ14, Dβ1, and Jβ1.1 and Jβ1.2 RSSs are diagrammed below the consensus heptamer and nonamer sequences (29). (C) BglII-digested genomic DNA subjected to Southern blot analysis by using probe A. DNA was isolated from nontransfected ES cells (ES); ES cells transfected with TCRβPF (ES+); thymocytes (T) and B cells (B) from two independently derived mice, TCRβPF-1 (1) and -2 (2), containing the TCRβPF minilocus. Shown are the expected size bands from the nonrearranged endogenous TCRβ locus (end) and TCRβPF minilocus (GL). Also shown are the expected size bands from DJ and VDJ/VJ rearrangements of TCRβPF. The 23-, 9-, and 6-kb markers are indicated. (D) PCR analysis was carried out on thymocyte DNA isolated from a non-minilocus-containing wild-type mouse (T) and the TCRβPF-1 and -2 mice. Before PCR, genomic DNA was digested with either BglII (G) or EcoRV (V). EcoRV digestion diminishes the amount of VDJβ1.1 template. Indicated are the 700- and 600-bp products expected for Vβ rearrangements to Jβ1.1/DJβ1.1 and Jβ1.2, respectively.
Here we describe two experimental approaches to investigate ordered assembly of TCRβ variable region gene segments during T lymphocyte development in vivo. First, we assay TCRβ minilocus rearrangements in lymphocytes isolated from chimeric mice generated via RAG-2-deficient blastocyst complementation (RDBC) by using embryonic stem (ES) cells into which the minilocus was introduced by transfection (17). This approach permits more rapid in vivo analyses than the conventional transgenic method and facilitates analysis of modified constructs. In addition, we have extended such analyses by introducing one of the minilocus alterations into the endogenous chromosomal locus of an ES cell line with a modified endogenous TCRβ locus that contains only one D-Jβ cluster (DJβ1) (18). We have used these approaches to elucidate potential mechanisms that contribute to the ordered rearrangement of Vβ, Dβ, and Jβ segments.
Materials and Methods
TCRβ Minilocus and Targeting Constructs.
The TCRβPE minilocus was described (ref. 15; referred to as no. 9 transgene). The TCRβPE BamHI/KpnI fragment containing the germ-line Dβ1, Jβ1.1, and Jβ1.2 gene segments was independently subcloned, thereby generating pDJβGL. The M2 mutation was introduced by ligating the annealed oligonucleotides DBAA1GTCCTTTTTTGTATAAAGCTGTAACATTGTGGGGACAGGGGGCATTTTAAATTC and DBAA2-AATTGAATTTAAAATGCCCCCTGTCCCCACAATGTTACAGCTTTATACAAAAAAG and the Eco0109/EcoRI-digested 150-bp PCR product generated with primers 5′-CGAATTCTATGGGAAGCCTTTAC-3′ and 5′-CCTCTCTCAAGGTCCATCAA-3′ into Eco0109-digested pDJβGL to generate pDJβM2. The BamHI/KpnI fragment from pDJβM2 was subcloned into BamHI/KpnI-digested TCRβPF to generate TCRβM2.
The DJβRPF minilocus was constructed by replacing the BamHI/KpnI fragment of TCRβPF with a fragment containing a DJβ1.1 rearrangement (no join diversity) and a germ-line Jβ1.2 gene segment. To construct DJβRC, the BamHI/KpnI fragment from the DJβRPF was independently subcloned, thereby generating p5-1. p5-1 was digested with EcoRV and StuI and religated generating p5-1A. PCR product C1 was generated with primers 5′-CGAATTCCAGACTCACAGTTGTAG-3′ and 5′-TATCAGGACCTACACGGAGGACATGCTTT-3′ and product C2 with primers 5′-ACCAAGGTCCCATATTCGAGTATCTGTATT-3′ and 5′-TGAATTCCCACACCCAAAGACCC-3′. PCR products C1 and C2 were digested with EcoRI and Eco0109 and subcloned into Eco0109-cut p5-1A to generate p5-1B followed by digestion with EcoRV and StuI and religation to generate p5-1C. The BamHI/KpnI fragment from p5-1C was subcloned into BamHI/KpnI-digested DJβRPF to generate DJβRC. To construct the DJβRD, minilocus fragment D1 was generated by PCR amplification of the oligonucleotide 5′-GATCAAGTTCGGGATCCGGGTCCTTTTTTGTATAAAGCTGTAACATTGTGCAAACTCCGACTACACCTTCGGCTCAGGGACCAGGCTTTTGGTAATAGGTAAGATATCCCACAG-3′ and fragment D2 was generated by PCR amplification of oligonucleotide 5′-GTAGGTAAGATATCTTTCAGGTAAATTTCCAGGTCTCTCTTCGGACAAAGCATGTCCTCCGTGTCCATATTCGAGTATCTGTATTCTGATGTGGGGACAGGGGGCAAACACAGAAGTCTTCTTTGGTAAAGGAACCAGACTCACAGTTGTAGGTAAGGCCTCGAGCCGG-3′. Fragment D1 was digested with Eco0109 and EcoRV and fragment D2 with EcoRV and StuI; these two fragments were subcloned into Eco0109- and StuI-digested p5-1 to generate p5-1D. The BamHI/KpnI fragment from p5-1D was subcloned into DJβRPF to generate the DJβRD minilocus.
pM2KI was constructed as described for pM4KI except that the 5′ homology region was generated by subcloning the Eco0109 fragment from pDJβM2 into a 2-kb NotI/BglII TCRβ genomic DNA fragment (18).
Chimeric mice were generated by RDBC and directly analyzed or bred for germ-line transmission of mutations as described (17, 18).
Southern and Northern Blot Analyses.
Southern and Northern analyses were carried out as described (19). Probe A is a 300-bp EcoRV fragment that spans Jβ1.2 and the downstream region. Probe B is a 600-bp AccI Vβ14 fragment. Probe C is a 0.8-kb DrdI/DrdI fragment. Probe D is a 0.4-kb BglII/AccI fragment. Probe E is a 0.7-kb AflII/HaeII fragment. Probe 1 is a 1-kb EcoRI fragment. Probe 2 is a 2-kb HindIII fragment. The Jβ1 probe is a 2-kb PstI fragment. Glyceraldehyde-3-phosphate dehydrogenase and CD3ɛ cDNA probes have been described (19).
PCR Analyses.
The oligonucleotide primers used were PA-GGCAAGCAAGCTGGTGTGT, PB-GCATCTCCCTCAAATGAGCC, P1-CCTCTCTCAAGGTCCATCAA, PR1-GCAGAAGAGGATTTCCCTGC, and Vβ14-GGCAAGCAAGCTGGTGTGT. The PV Vβ primer set has been described (20). PCR conditions were as described with the following cycle conditions: 92°C for 90 s; 60°C for 150 s; 72°C for 60 s for 30 cycles (20).
Flow Cytometry.
Single-cell suspensions were prepared from thymus and spleen as described (19). Cells were stained with FITC-conjugated anti-CD8 and phycoerythrin-conjugated anti-CD4 antibodies (PharMingen) and analyzed by a FACScan (Becton Dickinson).
Results
Efficient Lineage-Specific Recombination of the TCRβ Minilocus in Chimeric Mice Generated by RDBC.
We have described a transgenic TCRβ minilocus (hereafter referred to as TCRβPF) that contains germ-line Vβ14, Dβ1, Jβ1.1, and Jβ1.2 gene segments linked to the IgH μ constant region (Cμ) gene (Fig. 1A) (15, 16). In transgenic mice, this minilocus undergoes enhancer-dependent Dβ to Jβ rearrangement in B and T cells, with Vβ to DJβ rearrangement occurring only in T cells (Fig. 1C) (15, 16). Furthermore, minilocus Dβ to Jβ rearrangement generally precedes Vβ to Dβ rearrangement (Fig. 2B) (15). To facilitate this approach, we used cotransfection with a PGK-Neor-selectable marker gene to generate ES cell lines with low copy numbers of TCRβPF (Fig. 1C; compare ES to ES+ lanes). These transfected ES cells were used to generate chimeric mice by RDBC; in these mice, the vast majority of thymocytes and all peripheral T cells derive from the transfected ES cells (17).
Figure 2.
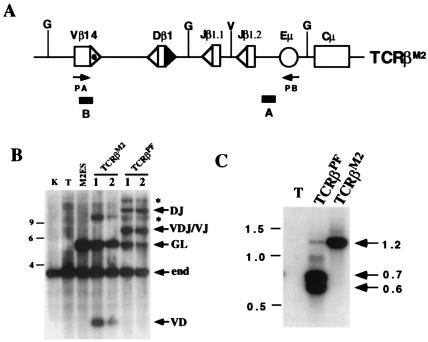
Analysis of TCRβM2 rearrangement in T cells. (A) TCRβM2 is similar to TCRβPF (Fig. 1A) except for mutation of the 3′ Dβ1 23-RSS heptamer (▴) as described in the text. The remaining 23-RSS (dotted ▵) and 12-RSSs (▵) are indicated. (B) BglII-digested genomic DNA isolated from kidney (K), thymocytes (T), ES cells transfected with TCRβ M2 (M2ES), or thymocytes isolated from mice containing TCRβM2 or TCRβPF miniloci was subjected to Southern blot analysis by using probe B. Shown are the expected size bands from the nonrearranged endogenous TCRβ locus (end), and the TCRβM2 and TCRβPF miniloci in the nonrearranged (GL), DJ, V(D)J/VJ, and VD rearrangement configurations. Also indicated (*) are the bands representing pseudonormal joins between tandemly integrated copies of the miniloci which have been described (15). The 9-, 6-, and 4-kb markers are shown. (C) PCR was carried out with primers PA and PB on thymocyte DNA from a wild-type (T), TCRβPF, and TCRβM2 mice. Indicated are the 1.2-, 0.7-, and 0.6-kb products expected for Vβ rearrangements to Dβ1, (D)Jβ1.1, and (D)Jβ1.2, respectively. The 1.5-, 1.0-, and 0.5-kb markers are shown.
Southern blot analyses of genomic DNA isolated from thymocytes and purified peripheral B cells revealed TCRβPF Dβ to Jβ rearrangement in B and T cells, with Vβ to DJβ rearrangements occurring only in T cells (Fig. 1C). PCR analyses of thymocyte DNA demonstrated that efficient Vβ(D)Jβ rearrangement occurred to both Jβ1.1 and Jβ1.2 gene segments (Fig. 1D). In addition, Vβ to Dβ rearrangement within TCRβPF was not detectable by Southern blotting, although low levels were detectable by PCR (Fig. 2 B and C). Together, these data demonstrate that the TCRβPF minilocus undergoes efficient lineage-specific recombination that recapitulates many aspects of endogenous TCRβ rearrangement when transfected into ES cells and assayed for rearrangement in T lineage cells generated by RDBC. In addition, these studies show that germ-line passage of the TCRβPF minilocus is not required to set up normal regulatory constraints and that such constraints are not markedly influenced by the cotransfected PGK-Neor cassette.
DJβ Assembly Is Not Required for Vβ Rearrangement.
To determine whether Vβ segments must rearrange to an assembled DJβ, a version of TCRβPF (TCRβM2) was generated in which the 3′Dβ1 RSS heptamer (CACAGTG) was replaced with an irrelevant sequence (ATTTTAA) and assayed by RDBC (Fig. 2A). As predicted, the TCRβM2 minilocus failed to undergo Dβ to Jβ rearrangement (Fig. 2B). However, Vβ to Dβ rearrangement within TCRβM2, in contrast to TCRβPF, was readily detectable by Southern blotting, as well as by PCR (Fig. 2 B and C). Together, these data demonstrate that DJβ rearrangement per se is not required for Vβ to Dβ rearrangement within the minilocus. Furthermore, direct Vβ to Jβ rearrangements within TCRβM2 were not observed, despite 12/23 compatibility of the Vβ 23-RSS and the Jβ 12-RSSs (Fig. 2B).
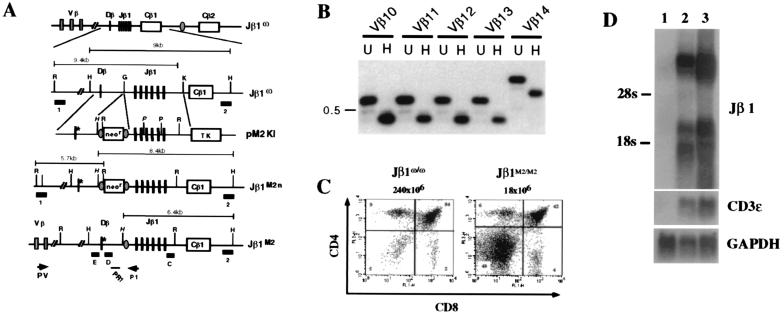
To investigate whether the rearrangement patterns observed in the TCRβ minilocus accurately reflected those of the endogenous TCRβ locus, gene targeting was used to introduce the M2 mutation into a single TCRβ allele (Jβ1M2) of the Jβ1ω/ω ES cell line (Fig. 3A). The DJβ2 gene cluster was deleted by gene targeting and replaced by single loxP sites on both TCRβ alleles of the Jβ1ω/ω ES line (Fig. 3A) (18). Jβ1ω/ω mice exhibit normal T cell development with TCRβ rearrangements limited to the DJβ1 gene cluster (18). Jβ1M2/ω ES cells were used to generate chimeric mice by RDBC that were analyzed directly or bred for germ-line transmission of the mutant Jβ1M2 allele. Flow cytometric analyses revealed that T cell development proceeded normally in Jβ1M2/ω mice (data not shown). Southern blot analysis of Jβ1M2/ω αβ T cell hybridomas showed that all had Jβ1 rearrangements on the Jβ1ω allele, whereas only one had undergone a Jβ1 rearrangement on the Jβ1M2 allele (Table 1). Of these hybridomas, 22 (21%) had undergone a Vβ to Dβ1 rearrangement on the Jβ1M2 allele (Table 1). Furthermore, these Vβ to Dβ1 rearrangements used a diverse Vβ repertoire (Fig. 3B). Together, these data demonstrate that Dβ to Jβ rearrangement of either the minilocus or the endogenous TCRβ locus is not required for rearrangement of a diverse set of Vβ segments to a previously nonrearranged Dβ segment. In addition, these findings further confirm that the TCRβ minilocus accurately recapitulates rearrangement patterns of the endogenous TCRβ locus.
Figure 3.
Generation and analysis of mice with M2 mutation in the endogenous TCRβ locus. (A) Schematic of the Jβ1ω allele showing some of the Vβ gene segments, the DJβ1 gene cluster, the loxP site (shaded oval) that replaces the DJβ2 gene cluster, and the Cβ1 and Cβ2 genes (not to scale). Jβ1ω/ω ES cells were transfected with the pM2KI targeting vector to generate the Jβ1M2 n allele. Cre-mediated deletion of the loxP-flanked neor gene generates the Jβ1M2 allele which differs from the Jβ1ω allele by the presence of the M2 mutant heptamer (*), an introduced HindIII site (H), and a single loxP site that is not contiguous with the mutation. Shown are EcoRI (R), HindIII (H), and KpnI (K) sites. The position of probes 1, 2, C, D, and E are shown as filled rectangles. Probes 1 and 2 were used to identify correctly targeted alleles (data not shown). Also shown is the position of oligonucleotide primer P1 and probe PR1. PV is a described (20) set of Vβ gene segment primers. (B) PCR analyses of Jβ1M2/ω thymocyte DNA by using the PV panel of Vβ primers and the P1 primer. Shown are PCR reactions by using Vβ10- to Vβ14-specific primers with the P1 primer. Southern blot analysis of undigested (U) or HindIII-digested (H) PCR products was carried out by using the PR1 oligonucleotide probe. PCR products from Vβ to Dβ rearrangements on the Jβ1M2 allele are reduced in size on HindIII digestion. The 0.5-kb marker is indicated. (C) Flow cytometric analysis of thymocytes from 3- to 4-week-old Jβ1ω/ω and Jβ1M2/M2 mice by using CD4-PE and CD8-FITC. Shown is a representative analysis of the eight Jβ1ω/ω and Jβ1M2/M2 mice analyzed. (D) Whole-cell RNA was isolated from RAG-2−/− spleen (lane 1), RAG-2−/− thymus (lane 2), and Jβ1M2/M2:RAG-2−/− thymus (lane 3) and subjected to Northern blot analysis by using a probe that spans the Jβ1 gene segments (Jβ1) and the CD3ɛ and glyceraldehyde-3-phosphate dehydrogenase probes as controls.
Table 1.
Analysis of TCRβ rearrangements in Jβ1ω/ω and Jβ1M2/ω αβ T cell hybridomas
| Hybridoma | Total no. | Allele | No. with rearrangements
|
|||
|---|---|---|---|---|---|---|
| GL | DJ | VD | V(D)J | |||
| Jβ1ω/ω | 50 | ω | 0 | 34 | 0 | 16 |
| ω | 0 | 0 | 0 | 50 | ||
| Jβ1M2/ω | 106 | ω | 0 | 1 | 0 | 105 |
| M2 | 83 | 0 | 22 | 1 | ||
TCRβ allele configuration in T cell hybridomas was determined by Southern blot analysis of HindIII- or EcoRI-digested genomic DNA by using probes C, D, and E (Fig. 3A). GL, nonrearranged.
Despite 12/23 compatibility between Vβ 23-RSSs and Jβ 12-RSSs, only one of the 106 Jβ1M2/ω splenic αβ T cell hybridomas had undergone a Vβ to Jβ1 rearrangement on the Jβ1M2 allele (Table 1). Consistent with this, Jβ1M2/M2 mice exhibited a significant block in thymocyte development at the CD4−/CD8− (double negative) stage (Fig. 3C) and reduced numbers of mature splenic T cells (data not shown). Failure of Vβ to Jβ joining on the Jβ1M2 allele was not caused by diminished germ-line Jβ transcripts as evidenced by the equivalent levels of these transcripts in double-negative thymocytes isolated from RAG-2−/− and RAG-2−/−:Jβ1M2/M2 mice (Fig. 3D). Together, these data demonstrate that there is a significant bias for Vβ to Dβ versus Vβ to Jβ rearrangement despite the 12/23 compatibility of both steps.
Efficient TCRβ Minilocus Vβ Rearrangement to a Preassembled DJβ Complex.
To elucidate cis-acting elements that promote preferential rearrangement of Vβ segments to Dβ versus Jβ segments, we conducted a systematic mutational analysis of the TCRβPF minilocus. First, we constructed a modified TCRβPF construct (DJβRPF) in which the germ-line Dβ1 and Jβ1.1 gene segments were replaced with a DJβ1.1 rearrangement (Fig. 4A). As assayed by RDBC and Southern blot analysis of thymocyte and B cell genomic DNA, DJβRPF showed efficient Vβ to DJβ rearrangement in T but not B lymphocytes (Fig. 4B). PCR analyses of DJβRPF revealed that Vβ rearrangement occurred exclusively to DJβ1.1 (Fig. 4C), despite the observation that the Jβ1.2 gene segment is competent for rearrangement as indicated by efficient TCRβPF Dβ to Jβ1.2 rearrangement (Fig. 1D) (15, 16, 21, 22). Consequently, the DJβRPF minilocus, like the endogenous TCRβ locus, exhibits a significant bias for Vβ to Dβ versus Vβ to Jβ rearrangement.
Figure 4.
Analysis of DJβRPF, DJβRC, and DJβRD rearrangement in T cells. (A) Schematic of DJβRPF, DJβRC, and DJβRD miniloci. In DJβRPF, the germ-line Dβ1/Jβ1.1 gene segments and intervening sequence of TCRβPF has been replaced by a preassembled DJβ1.1 rearrangement. In DJβRC, the position of DJβ1.1 and Jβ1.2 and their flanking RSSs have been exchanged. In DJβRD, the position of the coding region of DJβ1.1 and Jβ1.2 has been exchanged with the RSSs remaining in their native positions. The Jβ1.2 12-RSS (shaded triangle), Dβ1 12-RSS (open triangle), and Vβ14 23-RSS (dotted open triangle) are indicated. (B) BglII-digested genomic DNA isolated from kidney (K) and ES cells transfected with DJβRPF or thymocytes (T) and B cells (B) from a mouse with the DJβRPF minilocus was subjected to Southern blot analysis by using probe A (Fig. 1). Shown are the expected size bands from the nonrearranged endogenous TCRβ locus (end) and nonrearranged (GL) or VDJ/VJ-rearranged DJβRPF minilocus. The 9- and 6-kb markers are indicated. (C) The PA and PB primers were used to PCR thymocyte DNA from wild-type non-miniloci-containing mice (T) or mice that have the TCRβPF (−1), DJβRPF (−8, −19), DJβRC (−24, −35), or DJβRD (−11, −13) miniloci. Analyses shown are from mice generated from independently derived ES cells. Genomic DNA was digested with BglII before PCR; PCR products were subjected to Southern blot analysis by using probe A. Vβ rearrangement to the Jβ1.2 RSS (shaded triangle) yields a 700-bp PCR product from DJβRC and 600-bp PCR products from DJβRPF and DJβRD.
TCRβ Vβ to DJβ Rearrangement Bias Is Not Positional or Coding Sequence Dependent.
The bias for Vβ to Dβ versus Vβ to Jβ rearrangement could be enforced by the relative position of the Dβ and Jβ gene segments, with Vβ rearrangement targeted to the most proximal 12-RSS. To test this notion, we constructed a modified DJβRPF minilocus, DJβRC, in which the position of the DJβ1.1 and Jβ1.2 coding sequences and their associated 12-RSSs were exchanged (Fig. 4A). Southern blot analysis revealed that DJβRC undergoes efficient rearrangement in thymocytes (data not shown). Moreover, PCR analyses of thymocyte DNA revealed significant DJβRC Vβ to DJβ1.1 rearrangement with no readily detectable Vβ to Jβ1.2 rearrangement (Fig. 4C). Therefore, the bias for Vβ to DJβ rearrangement is not specified to the most proximal 5′12-RSS, but is rather targeted specifically to the Dβ versus the Jβ elements.
Recombination substrate studies have shown that V(D)J recombination efficiency can be influenced by coding sequences (23–27). To address this possibility, we constructed a modified DJβRPF minilocus, DJβRD, in which the DJβ1.1 and Jβ1.2 coding sequences were exchanged, but the native positions of their associated 5′ RSSs were maintained (Fig. 4A). After RDBC, Southern blot analysis of thymocyte DNA again confirmed that DJβRD undergoes efficient rearrangement in thymocytes (data not shown). Significantly, however, PCR analyses of DJβRD revealed Vβ to Jβ1.2 rearrangement with no detectable Vβ to DJβ1.1 rearrangement (Fig. 4C). This finding shows that the bias in Vβ to DJβ1.1 rearrangement is not determined by Dβ coding sequences, and therefore, most likely is specified by the Dβ versus Jβ 5′12-RSS sequences.
Discussion
Restraints Beyond 12/23 Compatibility Restrict Joining of Vβ to Dβ Versus Jβ Segments.
Direct Vβ to Jβ rearrangement satisfies the 12/23 rule. However, our analyses of the M2 mutation clearly revealed a substantial bias for Vβ to Dβ versus Vβ to Jβ rearrangement, despite 12/23 compatibility of the Vβ and Jβ RSSs. To further elucidate elements responsible for this bias, we analyzed a modified version of the TCRβPF minilocus with a preassembled DJβ1.1 and germ-line Jβ1.2 gene segments (DJβRPF). These studies showed preferential Vβ to Dβ rearrangement when the DJβ1.1 complex is positioned downstream of Jβ1.2, demonstrating that the bias is not imparted by relative proximity of Dβ and Jβ segments to Vβ. In addition, we observed specific Vβ targeting to the Jβ1.2 coding sequence when this coding sequence was adjacent to the 5′Dβ 12-RSS (DJβRC), whereas rearrangement was not observed to the Dβ coding sequence when it was flanked by the Jβ1.2 12-RSS (DJβRD). Taken together, these findings reveal that unanticipated regulatory constraints, independent of simple positional effects or the 12/23 rule, target Vβ rearrangement to Dβ segments and prevent efficient Vβ to Jβ rearrangement.
Our mutational analyses strongly suggest that this novel constraint in the V(D)J recombination reaction is determined by distinct features of the 5′Dβ1 and Jβ1 12-RSSs beyond simply enforcing the 12/23 rule. In parallel studies, we tested whether these conclusions apply to endogenous loci by generating and analyzing mice with appropriate mutations in the Jβ1ω TCRβ allele (18). These studies clearly confirmed that the 5′ Dβ1 12-RSS, and not the Jβ 12-RSSs, specifically and precisely targets Vβ rearrangement (18). Whereas the mechanism responsible for this phenomenon remains to be fully elucidated, the observed rearrangement bias clearly results from RSS sequence constraints beyond 12/23 compatibility. Such constraints may have important implications for Vβ repertoire development and potentially for enforcing allelic exclusion at the Vβ to Dβ step (18). Finally, we note that our studies show that direct Vβ to Jβ joining can occur at low frequency in Jβ1M2/M2 mice and lead to the generation of T cells. Given that the productive TCRβ rearrangements generated from direct VβJβ joins use a large repertoire of different Vβ gene segments, most or all Vβ segments appear to retain a similarly inefficient ability to be joined directly to a Jβ segment.
Factors That Effect Ordered Rearrangement of TCRβ V, D, and J Segments.
Endogenous TCRβ V region gene assembly is ordered with Dβ to Jβ rearrangements occurring on both alleles before Vβ appendage to a DJβ complex. Yet, Vβ to Dβ rearrangement occurred quite readily in the TCRβM2 minilocus that lacked a functional 3′ Dβ 23-RSS (M2 mutation) and which is, thus, incapable of Dβ to Jβ rearrangement. It seemed possible that this unanticipated observation may not apply to the endogenous locus, in which much greater distances and other sequences separate the rearranging elements. To address this issue, we introduced the M2 mutation into an ES cell line with a modified TCRβ locus (Jβ1ω/ω) that contains only the D-Jβ1 gene cluster (18). Consistent with the TCRβM2 minilocus results, significant levels of Vβ to Dβ rearrangements, but few Vβ to Jβ rearrangements, occurred in the absence of Dβ to Jβ rearrangement within the endogenous TCRβ locus.
Ordered rearrangement was first described in the IgH locus (DH to JH before VH appendage to DJH) and proposed to be important for regulation of IgH V region gene assembly in the context of allelic exclusion (12). Similar conclusions were reached regarding the TCRβ V region gene locus (10). In the IgH locus, the VH and JH segments have 23-RSSs, whereas the DH segments have 5′ and 3′ 12-RSSs. Therefore, direct joining of VH to JH segments would be prohibited by the 12/23 rule. However, whereas the RSS structure of the IgH locus prescribes DH to JH and VH to DH joining, it does not prescribe an order for these events. The novel constraints imposed by RSSs that we observed in the TCRβ minilocus also enforce Dβ to Jβ and Vβ to Dβ joining, but in a fashion that goes beyond the 12/23 rule. However, as was the case for the IgH locus, these constraints again would not prescribe a precise order.
Our studies of the M2 mutation provide additional insight into ordered rearrangement by showing that Dβ to Jβ rearrangement per se is not a prerequisite for Vβ appendage to a Dβ. In addition, this process does not require deletion of sequences between the 3′ Dβ 23-RSS and Jβ or formation of a DJβ complex. Therefore, our finding that direct Vβ to Dβ joins are readily detected in normal thymocytes that harbor the M2 allele suggests other testable possibilities. One would be that Dβ to Jβ rearrangement would enhance Vβ rearrangement to the 5′Dβ 12-RSS by removal of the 3′Dβ 23-RSS that could theoretically be a higher affinity site for RAG interaction. Another, based on prior observations, would be that Dβ to Jβ rearrangement occurs at an earlier developmental stage than Vβ rearrangements, but that progenitor T cells can transit to the Vβ-rearranging stage (perhaps at reduced frequency) without generating a DJβ rearrangement (28). In either case, our overall findings, including the 12/23 restriction, raise the intriguing possibility that productive TCRβ rearrangement could mediate allelic exclusion through the generation of a signal that modulates recombinase access to the 5′ Dβ 12-RSSs.
Acknowledgments
This work was supported in part by the National Institutes of Health Grants AI01297 (B.P.S.) and AI20047 (F.W.A.). B.P.S. is a recipient of a Career Development Award from the Burroughs Wellcome Fund. C.H.B. was a fellow of the Irvington Institute for Immunological Research. F.W.A. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
- TCR
T cell receptor
- RAG
recombinase-activating gene
- V
variable
- D
diversity
- J
joining
- RDBC
RAG-2-deficient blastocyst complementation
- RSS
recombination signal sequence, ES, embryonic stem
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.130190597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.130190597
References
- 1.Tonegawa S. Nature (London) 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 2.Blackwell T K, Alt F W. Annu Rev Genet. 1989;23:605–636. doi: 10.1146/annurev.ge.23.120189.003133. [DOI] [PubMed] [Google Scholar]
- 3.Gellert M. Adv Immunol. 1997;64:39–64. doi: 10.1016/s0065-2776(08)60886-x. [DOI] [PubMed] [Google Scholar]
- 4.Schatz D G, Oettinger M A, Schlissel M S. Annu Rev Immunol. 1992;10:359–383. doi: 10.1146/annurev.iy.10.040192.002043. [DOI] [PubMed] [Google Scholar]
- 5.McBlane J F, van Gent D C, Ramsden D A, Romeo C, Cuomo C A, Gellert M, Oettinger M A. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 6.Weaver D T. Adv Immunol. 1995;58:29–85. doi: 10.1016/s0065-2776(08)60619-7. [DOI] [PubMed] [Google Scholar]
- 7.Eastman Q M, Leu T M, Schatz D G. Nature (London) 1996;380:85–88. doi: 10.1038/380085a0. [DOI] [PubMed] [Google Scholar]
- 8.van Gent D C, McBlane J F, Ramsden D A, Sadofsky M J, Hesse J E, Gellert M. Cell. 1995;81:925–934. doi: 10.1016/0092-8674(95)90012-8. [DOI] [PubMed] [Google Scholar]
- 9.Sleckman B P, Gorman J R, Alt F W. Annu Rev Immunol. 1996;14:459–481. doi: 10.1146/annurev.immunol.14.1.459. [DOI] [PubMed] [Google Scholar]
- 10.Uematsu Y, Ryser S, Dembic Z, Borgulya P, Krimpenfort P, Berns A, von Boehmer H, Steinmetz M. Cell. 1988;52:831–841. doi: 10.1016/0092-8674(88)90425-4. [DOI] [PubMed] [Google Scholar]
- 11.Malissen M, Trucy J, Jouvin-Marche E, Cazenave P A, Scollay R, Malissen B. Immunol Today. 1992;13:315–322. doi: 10.1016/0167-5699(92)90044-8. [DOI] [PubMed] [Google Scholar]
- 12.Alt F W, Yancopoulos G D, Blackwell T K, Wood C, Thomas E, Boss M, Coffman R, Rosenberg N, Tonegawa S, Baltimore D. EMBO J. 1984;3:1209–1219. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krangel M S, Hernandez-Munain C, Lauzurica P, McMurry M, Roberts J L, Zhong X-P. Immunol Rev. 1998;165:131–147. doi: 10.1111/j.1600-065x.1998.tb01236.x. [DOI] [PubMed] [Google Scholar]
- 14.Sleckman B P, Khor B, Monroe R, Alt F W. J Exp Med. 1998;188:1465–1471. doi: 10.1084/jem.188.8.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferrier P, Krippl B, Blackwell T K, Furley A J, Suh H, Winoto A, Cook W D, Hood L, Costantini F, Alt F W. EMBO J. 1990;9:117–125. doi: 10.1002/j.1460-2075.1990.tb08087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okada A, Mendelsohn M, Alt F W. J Exp Med. 1994;180:261–272. doi: 10.1084/jem.180.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J, Lansford R, Stewart V, Young F, Alt F W. Proc Natl Acad Sci USA. 1993;90:4528–4532. doi: 10.1073/pnas.90.10.4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bassing C H, Alt F W, Hughes M H, D'Auteuil M, Wehrly T, Woodman B B, Gartner F, White M, Davidson L, Sleckman B P. Nature (London) 2000;405:583–586. doi: 10.1038/35014635. [DOI] [PubMed] [Google Scholar]
- 19.Sleckman B P, Bardon C G, Ferrini R, Davidson L, Alt F W. Immunity. 1997;7:505–515. doi: 10.1016/s1074-7613(00)80372-6. [DOI] [PubMed] [Google Scholar]
- 20.Gartner F, Alt F W, Monroe R, Chu M, Sleckman B P, Davidson L, Swat W. Immunity. 1999;10:537–546. doi: 10.1016/s1074-7613(00)80053-9. [DOI] [PubMed] [Google Scholar]
- 21.Ferrier P, Covey L R, Suh H, Winoto A, Hood L, Alt F W. Int Immunol. 1989;1:66–74. doi: 10.1093/intimm/1.1.66. [DOI] [PubMed] [Google Scholar]
- 22.Demengeot J, Oltz E M, Alt F W. Int Immunol. 1995;7:1995–2003. doi: 10.1093/intimm/7.12.1995. [DOI] [PubMed] [Google Scholar]
- 23.Boubnov N V, Wills Z P, Weaver D T. Nucleic Acids Res. 1995;23:1060–1067. doi: 10.1093/nar/23.6.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerstein R M, Lieber M R. Genes Dev. 1993;7:1459–1469. doi: 10.1101/gad.7.7b.1459. [DOI] [PubMed] [Google Scholar]
- 25.Nadel B, Feeney A J. J Immunol. 1995;155:4322–4329. [PubMed] [Google Scholar]
- 26.Ezekiel U R, Engler P, Stern D, Storb U. Immunity. 1995;2:381–389. doi: 10.1016/1074-7613(95)90146-9. [DOI] [PubMed] [Google Scholar]
- 27.VanDyk L F, Wise T W, Moore B B, Meek K. J Immunol. 1996;157:4005–4015. [PubMed] [Google Scholar]
- 28.Livak F, Tourigny M, Schatz D G, Petrie H T. J Immunol. 1999;162:2575–2580. [PubMed] [Google Scholar]
- 29.Hesse J E, Lieber M R, Mizuuchi K, Gellert M. Genes Dev. 1989;3:1053–1061. doi: 10.1101/gad.3.7.1053. [DOI] [PubMed] [Google Scholar]