Abstract
Ribosome-inactivating proteins are N-glycosidases that remove a specific adenine from the sarcin/ricin loop of the large rRNA, thus arresting protein synthesis at the translocation step. In the present study, a novel type I ribosome-inactivating protein, termed PAP-H, was purified from Agrobacterium rhizogenes-transformed hairy roots of pokeweed (Phytolacca americana). The protein was purified by anion- and cation-exchange chromatography. PAP-H has a molecular mass of 29.5 kD as detected by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and its isoelectric point was determined to be 7.8. Yeast (Saccharomyces cerevisiae) ribosomes incubated with PAP-H released the 360-nucleotide diagnostic fragment from the 26S rRNA upon aniline treatment, an indication of its ribosome-inactivating activity. Using immunofluorescence microscopy, PAP-H was found to be located in the cell walls of hairy roots and root border cells. PAP-H was determined to be constitutively secreted as part of the root exudates, with its secretion enhanced by a mechanism mediated by ethylene induction. Purified PAP-H did not show in vitro antifungal activity against soil-borne fungi. In contrast, root exudates containing PAP-H as well as additional chitinase, β-1,3-glucanase, and protease activities did inhibit the growth of soil-borne fungi. We found that PAP-H depurinates fungal ribosomes in vitro and in vivo, suggesting an additive mechanism that enables PAP-H to penetrate fungal cells.
Ribosome-inactivating proteins (RIPs) are widely distributed plant enzymes that inhibit protein synthesis by virtue of their N-glycosidic activity, selectively cleaving an adenine residue from a highly conserved and surface-exposed stem loop structure in the 28S rRNA (Endo and Tsurugi, 1987). This cleavage prevents the binding of the EF-2/GTP complex, with the subsequent arrest of protein synthesis leading to autonomous cell death (Osborn and Hartley, 1990). RIPs are either enzymatically active single polypeptides (type I) or heterodimers (type II). A type II RIP consists of an A chain, functionally equivalent to a type I RIP, which is attached to a sugar-binding B chain (for review, see Mehta and Boston, 1998; Tumer et al., 1999; Nielsen and Boston, 2001). Besides RNA N-glycosidase activity, some RIPs have ribonuclease, DNase, DNA glycosylase, and apurinic/apyrimidic lyase activities (Li et al., 1991; Roncuzzi and Gasperi-Campani, 1996; Nicolas et al., 1997, 1998, 2000; Hudak et al., 2000). In addition, RIPs from Trichosanthes kirilowii cell cultures have been demonstrated to possess chitinase activity (Remi Shih et al., 1997). Certain type I RIPs display a variety of antimicrobial activities, including antifungal, antibacterial (Vivanco et al., 1999), and broad-spectrum antiviral effects against different plant and animal viruses (Ussery et al., 1977; Chen et al., 1991), including a human immunodeficiency virus (Zarling et al., 1990). In addition, pokeweed (Phytolacca americana) antiviral protein (PAP) showed the inhibition of tumor cell growth (Stirpe et al., 1992), and the RIPs from Ricinus communis (ricin) and Saponaria officinalis (saporin) have been shown to possess insecticidal properties against Coleopteran species (Gatehouse et al., 1990). Despite extensive enzymatic and antimicrobial characterization, the significance of RIPs for plant biology remains largely unknown.
Pokeweed produces a suite of constitutive and induced RIPs in its leaves and seeds. For instance, PAP is a 29-kD constitutive RIP found in pokeweed leaves and localized in the cell wall matrix of leaf mesophyll cells (Irvin, 1975; Irvin et al., 1980; Ready et al., 1986; Lin et al., 1991). PAP II is a seasonal 30-kD RIP found in pokeweed leaves harvested in late summer (Irvin et al., 1980), and PAP-S (29.8 kD) is expressed in seeds (Barbieri et al., 1982). Amino acid comparisons show 80% homology of PAP with PAP-S, and 33% homology of PAP with PAP II. Accordingly, PAP-S cross-reacts with PAP antibodies, but PAP II does not react with PAP antibodies (Barbieri et al., 1982). PAP is thought to play a defense role because it depurinates ribosomes from all organisms tested, and because its expression in transgenic tobacco (Nicotiana tabacum cv Samsan and Nicotiana benthamiana) plants leads to resistance against viral and fungal infection (Lodge et al., 1993; Zoubenko et al., 1997). However, a clear understanding of the role of PAP in pokeweed has not been achieved.
To better understand the functional significance of RIPs in pokeweed, we developed a hairy root system for the expression and manipulation of constitutive RIPs. Hairy roots show stable expression of root-specific biosynthetic pathways, and thus have been used as an experimental system to study the biology and biochemistry of underground organs (Flores and Curtis, 1992; Flores et al., 1999). Plant roots synthesize and store various macromolecules, including storage and defense-related proteins such as chitinase and β-1,3-glucanase, to cope with pathogenic challenge (Mauch et al., 1988; Linthorst, 1991; Savary and Flores, 1994; Savary et al., 1997). Here, we report the identification of a novel RIP, termed PAP-H, located in the cell walls of hairy roots and root border cells of pokeweed. PAP-H was also found to be constitutively secreted as part of the root exudates, and its secretion was enhanced by elicitation with ethylene. Hairy root exudates of pokeweed containing PAP-H and other defense-related proteins showed strong antifungal activity against fungi causing root rot. We have previously reported that in vitro root secretions of secondary metabolites and proteins compare with root secretions under natural settings (Flores et al., 1999), suggesting that pokeweed may secrete RIPs into the soil. This paper reveals a new mechanism by which roots are able to secrete RIPs into the rhizosphere to prevent pathogen infection.
RESULTS
Development of Pokeweed Hairy Roots
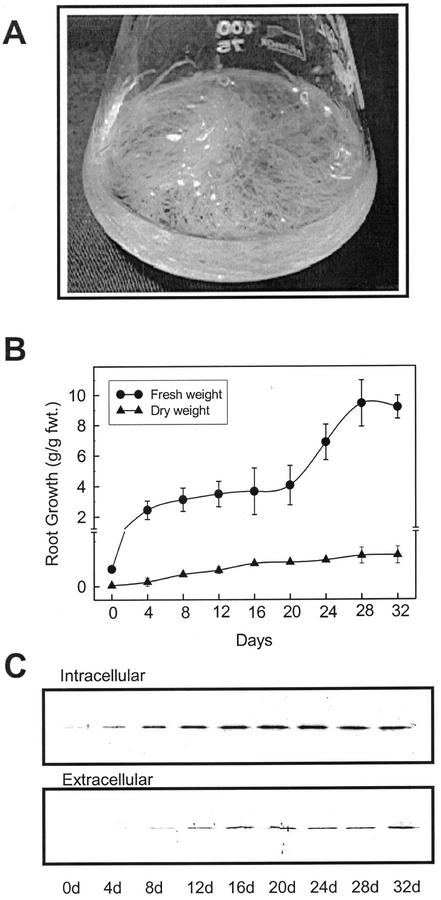
The transformation of pokeweed with Agrobacterium rhizogenes American Type Culture Collection (ATCC) no. 15834 was accomplished. Several hairy root clones were established and selected based on growth and stability, and root cultures were established in 125-mL flasks as indicated in “Materials and Methods” (Fig. 1A). Pokeweed root cultures showed stable growth and phenotype, producing a substantial biomass yield. As shown in Figure 1B, pokeweed hairy roots showed a biphasic root growth until d 30, which contained two periods of exponential growth. Maximum tissue accumulation under these conditions was approximately 180 g fresh weight L−1 medium, representing about a 900-fold increase in biomass starting from a single root tip inoculum. Root intracellular (in organ) and extracellular (secreted) proteins that accumulated in the culture medium during the time course were also examined by SDS-PAGE followed by western blotting using a PAP-specific antibody (Fig. 1C). Cross-reactivities with the PAP antibody were found in both the intracellular and the extracellular protein fractions. In the intracellular protein fraction, PAP antibody cross-reactivity increased during very early stages of growth, and maximum protein accumulation occurred before the end of exponential root growth phase at approximately 20 d. PAP cross-reactivity developed in culture media (extracellular proteins) after d 8, and increased through the time course.
Figure 1.
Establishment of pokeweed hairy roots, and time course of root growth and PAP-H accumulation. A, Developed hairy roots of pokeweed as described in “Materials and Methods.” B, Growth curve of fresh and dry weight accumulation over 32 d. C, Western-blot analysis of intra- and extracellular PAP-H accumulation over 32 d. The same amount (0.2 g fresh weight) of hairy roots from each sample was harvested to extract total proteins, and 1 mL of media from each sample was collected to concentrate the root secreted proteins followed by trichloroacetic acid (TCA) precipitation as described in “Materials and Methods.”
Identification and Purification of RIP from Hairy Roots of Pokeweed
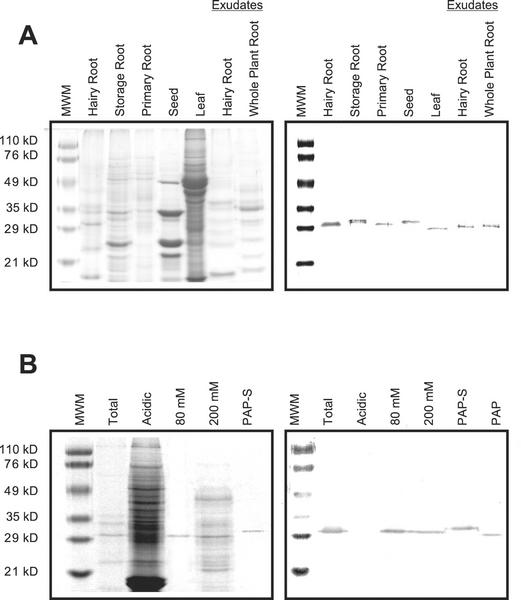
To ascertain whether PAP expressed in established hairy roots is similar to PAP isoforms produced in leaves, seeds, and roots of pokeweed, the protein profiles of these different organs were probed with a PAP antibody by western blotting. Total protein was extracted from 40-d-old hairy root cultures of pokeweed, and from pokeweed leaves, seeds, and roots (Fig. 2A). Western-blot analysis indicated slight differences in Mr among immunodetected proteins from different extracts. The immunoreactive band observed in hairy roots by western blotting was slightly larger in size than that of PAP (from leaves) but smaller than that of PAP-S (from seeds). It also differed in size from the immunoreactive bands of RIPs detected in storage roots, but was of a similar size to that of an RIP from primary roots. Based on these results, we concluded that RIP is produced in the transformed hairy roots of the pokeweed plant. Interestingly, RIP production was also detected as part of the root exudates secreted both in vitro from hairy root cultures and from whole-plant cultures.
Figure 2.
Identification and purification of PAP-H. A, Profiling of SDS-PAGE and western blot. Total proteins prepared from different organs of the pokeweed plant, and media where the hairy roots and whole pokeweed plant were grown were concentrated and run through 12.5% (w/v) SDS-PAGE. The antibody raised against PAP was used for western blotting. Approximately 25 μg of protein was loaded per lane. B, Cation-exchange chromatography of the total hairy root proteins from pokeweed. Total protein and samples of each fraction were run in 12.5% (w/v) SDS-PAGE, and western blot was performed using the PAP antibody. Approximately 5 μg of total, basic fractions, PAP-S and PAP, and 15 μg of acidic fraction were loaded per lane.
To biochemically purify the RIP from hairy roots of pokeweed, total proteins were subjected to ion-exchange chromatography as outlined in “Materials and Methods.” Because RIPs are mostly basic proteins, the unretained protein solution from the anion-exchange column was applied to a cation-exchange column (Sep-Pack, Waters, Milford, MA), and basic proteins were eluted with NaCl step gradients from 40 to 600 mm. A 29.5-kD protein corresponding to the RIP detected in hairy roots was resolved in the 80 mm fraction and analyzed by western blotting (Fig. 2B). The RIP was further resolved using UNO S1 cation-exchange column chromatography (Bio-Rad, Hercules, CA). The RIP was determined to have a molecular mass of 29.5 kD as determined by SDS-PAGE.
Characterization and Enzymatic Activity of PAP-H
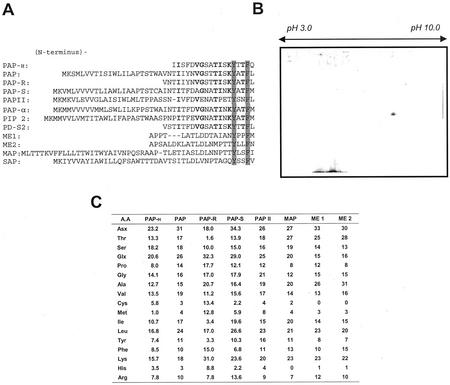
The N-terminal region of the RIP purified from hairy roots of pokeweed was sequenced and compared with those from PAP isoforms and other RIPs (Fig. 3A). The data showed that the N-terminal amino acid sequence of the hairy root RIP differed from those of all known PAP isoforms and other RIPs. The N-terminal regions of the hairy root RIP had 61% homology with PAP, and 56% homology with PAP-S. The N-terminal region of the hairy root RIP also showed a significant homology with PAP-α (Lin et al., 1991), as well as PIP 2 from P. insularis (Song et al., 2000) and PD-S2 from P. dioica (Del Vecchio Blanco et al., 1997). Importantly, the data indicated that highly conserved hydrophobic residues reported in the N-terminal region of all other RIPs, such as a Tyr-14 and Phe-17 (Funatsu et al., 1991) were found in the N-terminal region of the hairy root RIP. Based on these results, we concluded that the RIP purified from transformed hairy roots of pokeweed is a novel type of PAP, and named it PAP-H. PAP-H was determined to be, unexpectedly, a neutral protein with a pI of 7.8 by isoelectric focusing (IEF)-PAGE (Fig. 3B), and amino acid composition analysis showed that the amino acid distribution of PAP-H was similar to that of other RIPs (Fig. 3C).
Figure 3.
Characterization of PAP-H. A, Comparison of the N-terminal sequences of PAP-H. PAP (GenBank accession no. X55383); PAP-R (Bolognesi et al., 1990); PAP-S (X98079); PAP II (X78628); PAP-α, antiviral protein precursor from pokeweed (D10600); PIP 2, RIP 2 from Phytolacca insularis (AF141331); PD-S2, protein synthesis inhibitor from Phytolacca dioica (P34967); ME1 and 2, Mirabilis expansa protein 1 and 2 (Vivanco et al., 1999); MAP, Mirabilis jalapa antiviral protein (D10227); SAP, saporin from S. officinalis (CAA41948). Shaded boxes represent two amino acids that are absolutely conserved in all RIPs (Funatsu et al., 1991). Bold characters indicate homology regions among RIPs purified from pokeweed plants. B, Determination of pI. SDS-PAGE of the purified PAP-H was performed as described in “Materials and Methods.” pH range of the first dimension gel was pH 3 to 10 using Bio-Lyte ampholytes (Bio-Rad). C, Amino acid composition of PAP-H, PAP (Irvin et al., 1980), PAP-R (Bolognesi et al., 1990), PAP-S (Barbieri et al., 1982), PAP II (Irvin et al., 1980), MAP (Wong et al., 1992), and ME 1 and ME 2 (Vivanco et al., 1999).
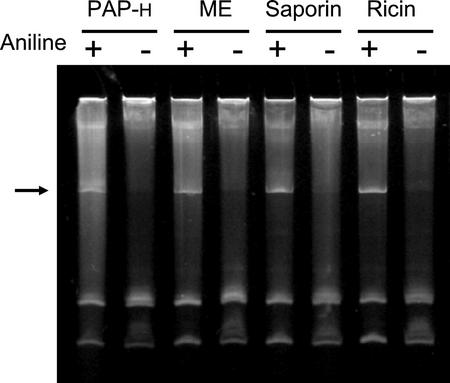
The RNA N-glycosidase activity of PAP-H was tested using yeast (Saccharomyces cerevisiae) ribosomes as a substrate. When depurinated rRNAs are treated with aniline, cleavage occurs at the depurinated site and a small nucleotide fragment is released from the 26S rRNA (Stirpe and Barbieri, 1986). Yeast ribosomes were incubated with PAP-H and with RIPs from M. expansa (ME), S. officinalis (saporin), and R. communis (ricin). As shown in Figure 4, PAP-H dupurinated the rRNAs and released the 367-nucleotide fragment upon treatment with aniline (Stirpe and Barbieri, 1986). These results demonstrate the enzymatic activity of PAP-H as an RIP.
Figure 4.
Enzymatic activity of PAP-H in vitro. Ribosomes were isolated from yeast and incubated with PAP-H, ME, saporin, and ricin as described in “Materials and Methods.” rRNAs were extracted, treated with aniline, separated on a 4.5% (w/v) urea-polyacrylamide gel, and stained with ethidium bromide. The presence (+) or absence (−) of aniline is denoted. The arrow shows the presence of the diagnostic 367-nucleotide cleavage product of rRNA.
PAP-H Is Localized at the Cell Wall Matrix in the Hairy Roots and Root Border Cells of Pokeweed
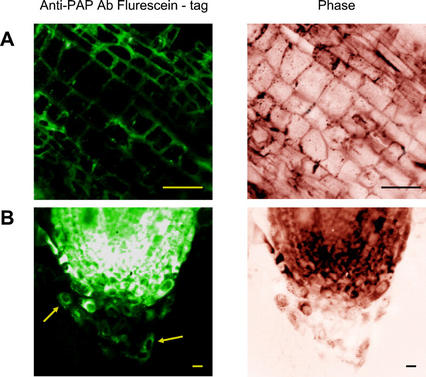
We examined the location of PAP-H in hairy roots of pokeweed using fluorescent microscopy. As shown in Figure 5A, thin longitudinal-sectioned hairy roots were treated with an anti-PAP antibody and a fluorescein-labeled secondary antibody as described in “Materials and Methods.” Green fluorescence, which indicates PAP-H presence as measured by the fluorescent tag-antibody interaction, was observed in the walls of every cell in the hairy roots. According to the broad density of fluorescence imaging along the entire circumference of the cell wall, PAP-H is most likely embedded in the cell wall matrix rather than bound to the cell wall (Fig. 5A). Interestingly, we found that PAP-H was also located in the cell walls of root border cells released from the root cap as the root grows (Fig. 5B). This observation suggested a possible secretion mechanism of PAP-H from the roots into the rhizosphere.
Figure 5.
Localization of PAP-H in hairy roots of pokeweed. The tips of hairy root tissues were cut into thin sections and fixed as described in “Materials and Methods.” Sections of cells were then stained with PAP antibody, followed by fluorescein-labeled secondary antibody reaction. Green fluorescence as observed in a fluorescent microscope indicates the presence of PAP-H in the cells. A, Longitudinal section of hairy roots. B, Longitudinal section of hairy roots showing root border cells (the arrows) near the root cap. Phase images were taken by light microscopy. Control root images not incubated with PAP antibody and fluorescein-labeled secondary antibody did not show fluorescent signal, indicating no root endogenous fluorescence (data not shown). Bars = 20 μm.
cDNA Cloning of PAP-H
Total RNA was isolated from hairy roots of pokeweed using the RNeasy Plant Mini Kit (Qiagen USA, Valencia, CA; see “Materials and Methods”). To clone the cDNA corresponding to the PAP-H protein, two gene-specific primers (GSPs) were designed for RACE. The sequences of these primers were formulated based upon the partial amino acid sequence of the N terminus region of PAP-H, as well as the codon preferences of several previously cloned PAP-related genes from pokeweed. One outer primer covered amino acids 6 through 14, including the first absolutely conserved hydrophobic residue, Tyr-14 (Y), whereas a second primer was designed from amino acids 9 through 18, overlapping the outer primer and covering the two absolutely conserved hydrophobic residues, Tyr-14 (Y) and Phe-17 (F). A 968-bp fragment obtained by 3′-RACE was cloned into the pCR4Blunt-TOPO and pCR4TA-TOPO (Invitrogen, Carlsbad, CA), and its nucleotide sequence was determined as addressed in “Materials and Methods.” A 3′-untranslated region of 51 bp with a polyadenylation site was found downstream from the stop codon. To obtain a 5′ sequence of PAP-H, 5′-RACE was performed using two GSPs designed from the fragments produced by 3′-RACE. These primers were designed so that a full-length cDNA clone sequence could be assembled from sequences of overlapping 5′- and 3′-RACE amplification products. A GSP of 27 bp was designed; it corresponded to the DNA sequence located approximately 100 bp downstream from the GSPs used for 3′-RACE. A second GSP of 29 bp was designed, corresponding to the DNA sequence located approximately 200 bp downstream of the 3′-RACE GSPs. 5′-RACE was performed and amplification products were cloned and sequenced as described for the 3′-RACE procedures.
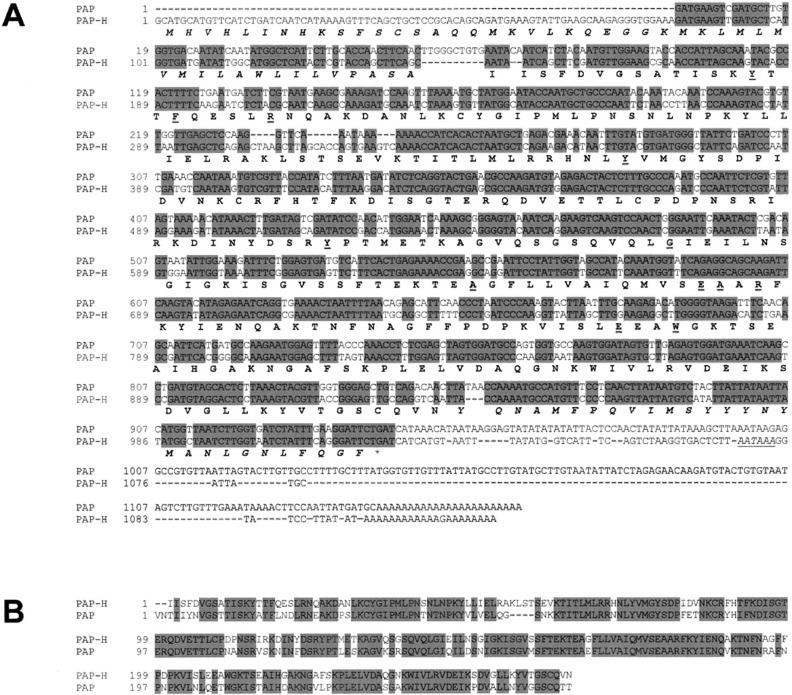
After DNA sequencing of the multiple clones for each amplification product produced with the various GSPs, the PAP-H cDNA sequence was assembled using the Jellyfish 1.5 Gene Analysis software package (http://www.biowire.com) and found to be 1,125 bp in length with an open reading frame of 1,074 bp (Fig. 6A; GenBank accession no. AY071928). The deduced translated amino acid sequence indicated that PAP-H cDNA has a putative signal peptide sequence of 47 amino acids, and a coding sequence of 292 amino acid residues. The first 18 amino acid residues from the deduced mature PAP-H polypeptide were found to be identical to those obtained by N-terminal sequencing of the purified PAP-H protein (Fig. 3C). A polyadenylation signal (AATAAA) was found between 10 and 30 bases upstream of the polyadenylation site, as found in most plant and animal mRNA (Joshi, 1987). Using the gene analysis software package mentioned above, the coding region of the PAP-H cDNA was found to be slightly longer than that of PAP (GenBank accession no. X55383), especially on the 5′-upstream region. When compared with the previously cloned PAP cDNA, PAP-H was found to contain an additional 25 amino acids upstream of the N-terminal sequence. Approximately 50% amino acid sequence homology was found to exist between PAP-H and PAP in the N-terminal extension region. Analysis also indicated that the C-terminal extension region of PAP-H might comprise 28 instead of 29 amino acids as found in PAP, with six amino acids differing between the two translated sequences. The cDNA sequence of the open reading frame region from PAP-H has 59.1% homology with PAP, 69.7% with PAP-S, 64.7% with PAP-α, and 52.1% with PAP II (GenBank accession nos. X55383, X98079, D10600, and X78628). The analysis also shows that the predicted mature protein from PAP-H cDNA has 66.8% identity with PAP, 64.4% with PAP-S, 58.4% with PAP-α, and 36.0% with PAP II. Highly conserved hydrophobic residues reported by Funatsu et al. (1991) were found in the deduced amino acid sequences of the PAP-H cDNA (Tyr-14, Phe-17, Arg-22, Tyr-74, Tyr-125, Gly-143, Ala-166, Glu-178, Ala-179, Arg-181, Glu-207, and Trp-210), and the relatively well-conserved region (172AIQMVSEAARFKYI186) thought to be the active site of enzymatic activity of RIPs (Frankel et al., 1989; Lin et al., 1991) was also found in PAP-H (Fig. 6B).
Figure 6.
cDNA and deduced amino acid sequences of PAP-H. A, The amino acid sequence of the mature protein (PAP-H) is in dark bold and the absolutely conserved amino acid residues among RIPs (Funatsu et al., 1991) are in dark bold and underlined. The dark bold and italic characters represent amino- and carboxyl-terminal extensions. The polyadenylation sites are indicated in italics and underlined. The cDNA sequence (X55358) of PAP is aligned along with PAP-H. B, Comparison of the mature proteins between PAP-H and PAP (X55358).
Ethylene Enhances the Secretion of PAP-H into the Rhizosphere
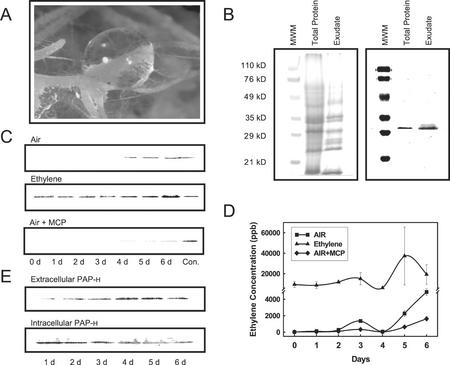
Based on PAP-H cell wall localization and on the observation that PAP-H was secreted as part of the root exudates (Fig. 2A), we decided to study the exudation mechanism of PAP-H. Upon closer examination, we observed exudates resembling water drops that were secreted from the root hairs of pokeweed root cultures growing in solid culture media (Fig. 7A). The exudate was collected using a pipette, filtered to avoid collecting root border cells, and run on an SDS-PAGE gel (Fig. 7B). The exudate showed slight differences in protein patterns compared with the proteins expressed in hairy roots. To determine if PAP-H was also secreted as a part of the root exudates, the intracellular (in organ) and extracellular (in exudate) proteins of pokeweed hairy roots were probed with the PAP antibody (Fig. 7B). The antibody strongly cross-reacted with both intra- and extracellular proteins and showed the same band, PAP-H (29.5 kD), which indicated that PAP-H is secreted as part of the root exudates as well as with the root border cells. In accordance, in vitro-grown pokeweed plants growing in liquid media also released PAP-H, presumably through roots (Fig. 2A).
Figure 7.
Identification and ethylene induction of extracellular PAP-H. A, Water drop-shaped exudates secreted from hairy roots of pokeweed grown in solid culture media. B, SDS-PAGE and western blotting of total protein and exudates from hairy roots of pokeweed. The PAP antibody strongly cross-reacted with intra- and extracellular proteins and showed the same band, PAP-H (29.5 kD), indicating that PAP-H was secreted as part of the root exudates. Approximately 20 μg of protein was loaded per lane. C, Western-blot analysis of elicitation experimentation during one time course study. Hairy roots of pokeweed grown for 30 d were transferred into 50 mL of fresh Murashige and Skoog media, and treated with air (top), ethylene (middle), and air + methylcyclopropane (MCP; bottom) through 6 d. The medium from which the hairy roots were grown for 30 d was used as control. One milliliter of media from each sample was collected to concentrate the root secreted proteins followed by TCA precipitation as described in “Materials and Methods,” and subsequently the concentrated protein was loaded in each lane. D, Ethylene concentration in the headspace of flasks during the course of each treatment. E, Western-blot analysis of intra- and extracellular PAP-H elicited by ethylene in the time course. One milliliter of media from each sample was collected, concentrated, and loaded as described above.
To analyze the biological significance of PAP-H exudation, hairy root cultures of pokeweed were challenged with the stress-related chemical ethylene. As shown in Figure 7C, an enhanced induction of PAP-H was observed in the root exudates when the hairy roots were treated with ethylene. The secretion of PAP-H was induced shortly after beginning the ethylene treatment, and constantly increased through d 6 of ethylene treatment. On d 6, the secretion of PAP-H was increased up to 8-fold compared with a control, which consisted of roots secreting PAP-H for 30 d without any treatment. Air treatment showed no induction of PAP-H until 3 d after culture transfer, showing PAP-H secretion on d 4. In contrast, treatment with an ethylene inhibitor (MCP) suppressed the natural secretion of PAP-H. This result suggested that MCP blocked the binding of the endogenous ethylene and thus PAP-H secretion. The concentration of ethylene in the headspace of the flask was measured in replicates of all three treatments. Endogenous ethylene produced in the hairy roots treated with air began to increase in d 4, and reached 5 μL L−1 at d 6, a level similar to the concentration supplied in the ethylene treatment (Fig. 7D). The increase of endogenous ethylene concentration correlates to the increase of PAP-H production starting on d 4, as detected by western blotting. The result shown in Figure 7D indicated that MCP also blocked the production of endogenous ethylene. Thus, our results clearly show that PAP-H is secreted under natural conditions by a mechanism mediated by ethylene induction. Interestingly, the western-blot analysis of intracellular PAP-H indicates that its concentration inside the hairy roots slightly decreased upon ethylene treatment, which correlates to increasing PAP-H accumulation in the media (Fig. 7E). These results suggest that ethylene induces secretion of PAP-H rather than the novo production of this RIP. Taken together, these results suggest that ethylene induces secretion of PAP-H into the rhizosphere but does not induce production of PAP-H.
Antifungal Activity of PAP-H
To explore the biological significance of PAP-H, we examined the N-glycosidase activity of PAP-H against fungal ribosomes. The depurination experiment described for PAP-H against yeast ribosomes was repeated using ribosomes isolated from Trichoderma reesei. After aniline treatment, a small fragment of about 360 nucleotides was detected by urea-acrylamide gel electrophoresis (Fig. 8A). This result indicates that PAP-H may be involved in inhibiting fungal growth by actively depurinating the fungal ribosomes. Based on this result, we tested the antifungal activity of PAP-H against an array of fungi (see “Materials and Methods”). However, PAP-H did not show fungal inhibitory activity in plate assays. Subsequently, we tested the total secreted proteins (exudates) from pokeweed hairy roots for fungal inhibition to check any possible antifungal effect with other pathogenesis-related (PR) proteins that may allow PAP-H entrance onto fungal cells. As shown in Figure 8B, 50 μg of total secreted proteins produced a significant inhibition of the growth of R. solani as well as of T. reesei (data not shown). These data indicate that PAP-H may participate in an active defense mechanism of roots against rhizosphere microbes. Thus, we studied whether fungal inhibition was due to the direct effect of PAP-H on fungal ribosomes. We cocultured R. solani with hairy roots of pokeweed that were grown for 2 weeks before coculture in Murashige and Skoog basal media (Fig. 8C, a). Fungal hyphae growing in the proximity of the roots were collected after a week of coculture and washed briefly with water to cleanse of medium (solids) attached to hyphae, and ribosomes were then isolated from collected fungal mycelium tissues. Interestingly, a small fragment appeared when the rRNA derived from these ribosomes was run on urea PAGE, indicating rRNA cleavage due to RIP depurination after RNA extraction and aniline treatment (Fig. 8C, b–d). This result demonstrated that PAP-H entered into the cytosolic region of fungal cells, and depurinated fungal ribosomes.
Figure 8.
Antifungal activity of pokeweed hairy root exudates. A, Depurination of T. reesei ribosomes in vitro. Ribosomes were isolated and incubated with PAP-H, ME, saporin, and ricin. The presence (+) or absence (−) of aniline is denoted. The arrow shows the presence of the diagnostic nucleotide cleavage product of rRNA. B, Radial growth inhibition assay of pokeweed hairy root exudates against Rhizoctonia solani. Twenty-five millimolar NaPO4 buffer, pH 7.5 (a), and filter-sterilized 50 μg of total root exudates (b) were applied to the discs, and tested for antifungal activity. C, Enzymatic activity of exudated PAP-H from hairy roots against R. solani. R. solani was cocultured with hairy roots of pokeweed that were grown for 2 weeks before the coculture in Murashige and Skoog basal media (a), and ribosomes of fungal hyphae growing in the proximity of the roots were isolated after a week of coculture. Then, rRNA was isolated from both ribosomes followed by treatment with aniline (b), and compared with ribosomes isolated from normally grown R. solani, incubated with PAP-H and then treated with aniline (c). Fungal hyphae of R. solani growing alone in a petri dish were collected, and their ribosomes were isolated and examined for no natural occurrence of rRNA depurination in fungal cells without prior incubation with PAP-H and/or the root exudates (d). The presence (+) or absence (−) of aniline is denoted. The arrow shows the presence of the diagnostic nucleotide cleavage product of rRNA. D, Enzymatic activities of chitinase and β-1,3-glucanase in root exudates. For each activity, triplicate samples were assayed at two different dilutions with each assay run in triplicate. Error bars indicate ±sd. E, Determination of proteolytic activity of root exudates. Zymogram gel electrophoresis containing gelatin was performed as described in “Materials and Methods.” The gel was then stained with 0.5% (w/v) Coomassie Brilliant Blue R-250. The arrow indicates protease activity in root exudates.
Enzymatic Activity Assay of PR Proteins in Root Exudates of Pokeweed
Based on previous results, we hypothesized that root exudates of pokeweed contained other PR proteins that could facilitate the entry of PAP-H into fungal cells by lysing fungal cell walls. Chitinase, β-1,3-glucanase, and protease enzymatic activities were tested as possible candidates as described in “Materials and Methods.” As shown in Figure 8, D and E, the root exudates demonstrated chitinase and β-1,3-glucanase (10.18 and 16.82 units mg−1 total protein) activities, as well as protease activities detected by zymogram gel analysis. Correlating the concentration of chitinase and β-1,3-glucanase in pokeweed root exudates with previously reported enzymatic activity for these enzymes in other plants (approximately 14 units mg−1 for chitinase and 18.5 units mg−1 for β-1,3-glucanase; Kombrink et al., 1988; Vögeli et al., 1988; Kragh et al., 1990; Wyatt et al., 1991; Qiu et al., 1997), we conclude that the concentration of these PR proteins in the root exudates is sufficient to damage fungal cell walls. These results suggest that these PR proteins in root exudates may generate an additive effect to facilitate entrance of PAP-H into fungal cells.
DISCUSSION
We established transformed root clones of pokeweed with A. rhizogenes ATCC number 15834. These transformed roots (“hairy roots”) displayed similar morphological characteristics to primary roots, including a well-developed cortex with new tissues developing from an apical meristem. The hairy roots of pokeweed showed stable and fast growth, and produced PAP-H steadily and constitutively. Hairy roots of pokeweed grew rapidly after d 20, and reached the stationary stage in approximately 30 d. The production of intracellular PAP-H increased during the very early stage, and remained constitutive after 15 d. In contrast, extracellular PAP-H accumulated to detectable levels in approximately 8 d, and continuously increased with the growth of roots. These results indicate that extracellular PAP-H accumulation occurs over time along with root growth, whereas the expression of intracellular PAP-H is constant.
In this communication, we report the isolation of PAP-H, a new constitutively produced RIP, from A. rhizogenes-transformed hairy roots of pokeweed. The pI of PAP-H was determined to be 7.8 by IEF-PAGE, suggesting that PAP-H is the first neutral and active protein among known RIPs (Fig. 3B). As shown in Figure 3C, the lower levels of basic amino acids such as Lys and Arg in PAP-H compared with the levels in PAP and other isoforms may contribute to its lower pI. PAP-H showed N-terminal amino acid sequence similarity with other RIPs found in pokeweed, such as PAP and PAP-S, and to a lesser extent with PAP-II (Irvin et al., 1980; Barbieri et al., 1982). PAP-H cross-reacts with PAP antibodies, but does not react with the PAP-II antibody (data not shown). Amino acid comparisons show 75.8% homology between PAP and PAP-S, and 33% homology between PAP and PAP II. In accordance, PAP-S cross-reacts with PAP antibodies, but PAP II does not react with PAP antibodies (Barbieri et al., 1982). We suggest that because PAP, PAP-S, and PAP-H are constitutively produced, they share more sequence and antigenic specificity, in contrast to PAP II, in which expression is environmentally regulated.
Comparing PAP isoforms detected among different types of roots (Fig. 2A), PAP-H showed size differences with RIPs found in storage roots. However, PAP-H has an Mr similar to the protein expressed in primary roots, which cross-reacted with the PAP antibody. A. rhizogenes-transformed hairy roots, in general, exhibit morphological and genetic similarity with fibrous primary roots (for review, see Vivanco and Flores, 2000; Bais et al., 2001). Savary and Flores (1994) have shown that secondary growth induction leading to storage root formation shifts the protein production and accumulation patterns of RIPs. Therefore, PAP-H may be considered a biochemically identical isoform to the protein found in the primary roots. Our results also showed that storage roots have two immunoreactive bands that indicate a molecular mass (30 kD) close to that of PAP-S isolated from seeds (Barbieri et al., 1982). Furthermore, hairy root and whole-plant root exudates cross-reacted with the PAP antibody, and the immunoreactive band produced by these exudates showed the same Mr as PAP-H, suggesting that PAP-H is released from roots to the rhizosphere as a part of the root exudates (Figs. 2A and 7B).
The amino acid sequence of the PAP-H cDNA was deduced and found to be identical to the protein sequence corresponding to the N terminus of the purified PAP-H protein (Figs. 3C and 6, A and B). Mature PAP-H contains 264 amino acid residues, which is similar to the 262 and 261 amino acid residues found in PAP and PAP-S (Monzingo et al., 1993). Although the numbers of amino acid residues from mature proteins are similar to each other, molecular weights of PAP-H, PAP, and PAP-S clearly showed differences (Fig. 2A), suggesting that posttranslational modification may occur in pokeweed RIPs.
Recently, a number of mutagenesis studies using PAP have revealed certain specific sites important for its enzymatic activities. Transgenic plants expressing nontoxic mutated PAP forms such as PAPx (active site mutant), PAPn (N-terminal mutant), and PAPc (C-terminal mutant) have shown that Glu-176, Gly-75, and 25 C-terminal amino acids are critical amino acid residues involved in ribosome depurination (Smirnov et al., 1997; Tumer et al., 1997; Zoubenko et al., 1997, 2000; Hudak et al., 2000). Comparison of cDNA sequences of PAP-H and PAP shows that PAP-H also contains those amino acid residues involved in ribosome depurination. The deduced amino acid sequences of PAP-H from its cDNA show Glu-178 and Gly-77 corresponding to Glu-176 and Gly-75 from PAP; the latter amino acids are responsible for the N-glycosidase activity of PAP. PAP-H also has high sequence homology with PAP in 25 C terminus amino acid residues. Furthermore, the PAP active site residues (Tyr-72, Tyr-123, and Arg-179) directly participated in the catalytic deadenylation of RNA, and Trp-208 involved in the stabilization of ribosome binding (Rajamohan et al., 2000) may correspond to Tyr-74, Tyr-125, Arg-181, and Trp-210 from PAP-H. These sequence analyses indicate that PAP-H shares with PAP amino acid residues involved in cytotoxicity, so that the N-glycosidic activity of PAP-H may be similar to that of PAP.
Interestingly, the cDNA analysis demonstrates that PAP-H contains 75 amino acid residues corresponding to the N- and C-terminal extensions (47 from N terminus and 28 from C terminus). The sequence comparison of both amino and carboxy extension regions from PAP-H and PAP suggest that these regions from PAP-H may function as cell wall-targeting sequences in hairy roots as was previously reported for PAP (Ready et al., 1986; Monzingo et al., 1993). However, the N-terminal extension of PAP-H contains an additional 25 amino acids compared with PAP and PAP-S, and the sequences of these two proteins differ from each other. Therefore, the N terminus sequence of PAP-H may include additional functions such as directing secretion of PAP-H to the rhizosphere under ethylene regulation.
We found that PAP-H is located in the cell wall matrix of hairy roots (Fig. 5A), similar to PAP being localized in the cell wall matrix of leaf mesophyll cells (Ready et al., 1986). Because PAP is an exported protein, it may have an endoplasmic reticulum (ER) signal sequence at the N terminus to enter the secretory pathway (for review, see Lodish et al., 2000). As described above, PAP-H contains potentially functional N-terminal extension residues, and shows localization similar to that of PAP. In accordance, PAP-H may share the same processing mechanisms as PAP. Interestingly, PAP-H was also found to be localized in the cell walls of root border cells and released from the root tip as the root grew (Fig. 5B). Root border cells are considered to be one of the key factors in root-microbe communication because they are programmed to be detached from the root and enter the rhizosphere (for review, see Hawes et al., 2000). Border cells are involved in triggering various responses by producing secondary metabolites, chemo-attractants, repellents, and signals that can lead to agglutination against infesting microbes and root parasites. However, no specific enzyme has been isolated from root border cells. Recently, Hawes and coworkers have suggested the production of a low-pH galactosidase in the root border cells of the pea (Pisum sativum; Hawes et al., 2000). Our data showed that pokeweed root border cells produce and store RIPs. As mentioned above, we also found that hairy roots of pokeweed constitutively secreted water drop-shaped exudates while growing in solid media (Fig. 7A). Western-blot analysis (Fig. 7B) of filtered exudates collected from hairy roots indicated that PAP-H was released as part of the root exudates. To our knowledge, PAP-H is the first RIP shown to be secreted in the root exudates, as well as the first shown to be compartmentalized in secreted root border cells. These root-specific functions may act as potential plant defense mechanism against pathogen infection.
PAP is regarded as a defense-related protein because it can deadenylate ribosomes from all organisms, and its expression in transgenic plants leads to resistance to viral and fungal infection (Lodge et al., 1993; Zoubenko et al., 1997). Although many elicitors and signals that accompany pathogen recognition and defense responses of plants have been found to induce PR proteins, only limited information has been determined about the induction of RIPs. Some studies have addressed the induction of RIPs caused by stress and stress-related compounds such as jasmonate (Reinbothe et al., 1994), osmotic stress or heat shock (Stirpe et al., 1996), and salt shock (Rippmann et al., 1997). Reinbothe et al. (1997) reported that methyl jasmonate also rapidly induces and accumulates RIP in leaf tissues of barley (Hordeum vulgare) at the transcriptional level. Ethylene is induced in response to environmental stress, including infection by pathogens (for review, see Ohme-Takagi et al., 2000). Ethylene has also been shown to be an important factor in plant cell culture systems, affecting the stimulation of secondary metabolites from various tissue and cell cultures (Cho et al., 1988; Phisalaphong and Linden, 1999). Thus, ethylene is biologically active at a very low concentration—less than 1 μL L−1—and its production is temporarily increased severalfold within 25 to 30 min when tissues are wounded or mechanically perturbed (Taiz and Zeiger, 1991). Exogenous application of ethylene has been shown to induce the transcription of several genes encoding basic-type PR proteins such as class I basic chitinases and class I β-1,3-glucanase (for review, see Ohme-Takagi et al., 2000). Chen et al. (2001) have reported recently that the ethylene receptor-like protein ETR1 is located in the ER membrane in Arabidopsis. Ethylene receptors such as ETR1 are two-component signaling systems containing a receptor with His kinase activity and a response regulator (Schaller et al., 1995, 2000; Hall et al., 2000). In response to ethylene, the receptor is autophosphorylated and a phosphate is then transferred to the response regulator, which mediates downstream responses (Schaller et al., 2000). Because PAP-H showed strong secretory induction by ethylene treatment and is assumed to be secreted through the ER, the ethylene receptor located in the ER membrane may be involved in activating the secretory mechanism of PAP-H. In conclusion, we feel that ethylene induced by biological stresses, such as pathogen attack, binds to the receptor protein at the ER membrane, initiating a downstream signaling cascade to activate the PAP-H secretory pathway as a defense mechanism.
As part of a secretory root defense mechanism, PAP-H creates an antifungal scenario with the aid of other PR proteins secreted from hairy roots of pokeweed. As shown in Figure 8A, PAP-H has in vitro N-glycosidase activity against fungal ribosomes, which indicates that PAP-H can recognize and depurinate fungal ribosomes; however, purified PAP-H did not show in vitro antifungal activity against R. solani. In contrast, total root exudates of pokeweed showed inhibitory activity against fungi (Fig. 8B). Upon close examination, the ribosomes isolated from R. solani grown in the presence of hairy roots of pokeweed (Fig. 8C, a) showed depurination traces (Fig. 8C, c). We hypothesize that PAP-H depurinates fungal rRNA and inhibits fungal growth by disrupting protein synthesis in fungal cells. The penetration of PAP-H into the fungal cells may be facilitated by PR proteins such as chitinase, β-1,3-glucanase, and proteases (Fig. 8, D and E). Both chitinase and β-1,3-glucanase are widely distributed enzymes in higher plants, and have been hypothesized to function synergistically in plant defense against fungal pathogens (Abeles et al., 1971; for review, see Leubner-Metzger and Meins, 1999; Neuhaus, 1999).
We suggest that PAP-H is constitutively produced as a host defense protein, and secreted into the rhizosphere as a barrier against soil-borne microbe infection. The secretion of PAP-H is regulated in response to endo- and exogenous stresses. Our results indicate that PAP-H may prevent the multiplication of pathogens in the soil, acting in an additive effect with other PR proteins. Another hypothesis is that PAP-H is constitutively secreted to produce organic matter in the soil by depurinating the ribosomes of soil microorganisms. These and other hypotheses are currently being tested to shed light on the function of RIPs in the rhizosphere.
MATERIALS AND METHODS
Plant Material
Seeds of pokeweed (Phytolacca americana) were collected in New Brunswick (NJ). Seeds were washed five times with sterile water and germinated on filter papers on a petri dish. The seeds were then transferred to pots and placed in the greenhouse.
Establishment of Agrobacterium rhizogenes-Transformed Hairy Roots of Pokeweed
Shoots from pokeweed were collected from greenhouse-grown plants and surface sterilized with 10% (v/v) commercial bleach for 15 min and then washed four times with sterile water. Shoot cultures were placed separately in Magenta GA-7 vessels containing Murashige and Skoog basal medium (Murashige and Skoog, 1962) solidified with 0.3% (w/v) Phytagel (Sigma, St. Louis). Cultures were kept in a light chamber maintained at 24°C with a light intensity of 100 μmol m−2 s−1 PAR. To produce hairy root cultures, 1-month-old in vitro plants were infected with a 3-d-old culture of A. rhizogenes (ATCC no. 15834) grown in TY medium (0.8% [w/v] tryptone, 0.5% [w/v] yeast extract, and 0.25% [w/v] NaCl) at 30°C (Verveliet et al., 1975). In brief, stems of pokeweed were punctured in several places with A. rhizogenes and then placed in a light chamber. Roots that developed at the infection sites were transferred to petri dishes containing solid Murashige and Skoog medium supplemented with 250 μg mL−1 Claforan (Hoechst-Roussel Pharmaceuticals, Somerville, NJ) and kept in the dark chamber at 24°C. After 14 d, 1-cm root tips were subcultured twice to eradicate excess bacteria before transferring them to fresh medium in the absence of antibiotic. Clonal root lines established after serial transfers of root tips to fresh Murashige and Skoog medium were subcultured into 125-mL Erlenmeyer flasks containing 50 mL of liquid Murashige and Skoog medium and placed on a gyratory shaker set at 90 rpm in a dark chamber.
Protein Extraction from Hairy Roots
Hairy roots of pokeweed were immersed in liquid N2 and ground to a powder using a mortar and pestle. Ground root tissue was dissolved into two volumes of extraction buffer (25 mm NaPO4 [pH 7.0] with 250 mm NaCl, 10 mm EDTA, 5 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and 1.5% [w/v] polyvinylpolypyrrolidone), homogenized, and centrifuged for 30 min at 10,000g. The supernatant was brought to 20% (w/v) ammonium sulfate, and centrifuged again for 20 min at 10,000g. The supernatant was dialyzed against 20 mm NaPO4 buffer (pH 7.0) until it was free from sulfate ion. All extraction procedures were conducted at 4°C, and the crude extract was stored at 4°C until use.
Electrophoresis and Western-Blot Analysis
SDS-PAGE gel electrophoresis was performed with 12.5% (w/v) acrylamide discontinuous gels (Laemmli, 1970) using an electrophoresis cell (Mini-Protein 3 Cell, Bio-Rad) according to manufacturer's instructions. Low-molecular mass protein markers (21.1–110 kD, Bio-Rad) were run simultaneously for each electrophoresis gel. The gel was stained with Coomassie Brilliant Blue R-250 (EM Science, Gibbstown, NJ). Proteins were electroblotted to Immun-Blot polyvinylidene difluoride membranes (Bio-Rad) using a Mini Trans-Blot Electrophoretic Transfer Cell (Bio-Rad). The blot was then probed with protein A-purified polyclonal rabbit anti-PAP antibody obtained from Dr. Nilgun Tumer (Rutgers University, New Brunswick, NJ), and the membranes were developed using an Opti-4CN Detection Kit (Bio-Rad), following the manufacturer's instructions. An antiserum titer of 1:1,000 (w/v) was used for all experiments.
Chromatography and IEF
Ion-exchange separation was performed using Sep-Pack Plus Cartridges (Waters). Waters Accell Plus QMAs were equilibrated with 20 mm NaPO4 buffer (pH 7.0), and the flow-through solution containing unretained (basic) proteins was collected. Basic proteins were applied to Waters Accell Plus CMs equilibrated with 20 mm NaPO4 buffer (pH 7.0) and unbound (acidic) proteins were collected. Basic proteins were eluted with step gradients from 40 to 600 mm of NaCl. The target protein was resolved in the 80 mm fraction. The 80 mm fraction was dialyzed against 20 mm NaPO4 buffer (pH 7.0) and subsequently separated by cation-exchange chromatography using a UNO S1 column (Bio-Rad). Diluted protein solutions and fractions were concentrated by ultrafiltration using a Stirred Ultrafiltration Cell 8050 (Millipore, Bedford, MA). Protein purity and peak size were confirmed by SDS-PAGE stained with Coomassie Brilliant Blue R-250 (EM Science) and Silver Stain Plus (Bio-Rad). Protein concentration was determined by the Bradford (1976) method using a protein assay kit (Bio-Rad).
The pI of purified PAP-H was estimated by IEF using a Mini-Protean II 2-D Cell (Bio-Rad) with Bio-Lyte ampholytes (pH range 3–10; Bio-Rad) following manufacturer's instructions. Second dimension was performed in an SDS-PAGE gel with 12.5% (w/v) acrylamide gel using a Mini-Protein 3 Cell (Bio-Rad), and stained with Silver Stain Plus (Bio-Rad).
N-Terminal Sequencing and Amino Acid Analysis
The purified protein was N-terminally sequenced on a Precise Protein Sequencer System (Applied Biosystems, Foster City, CA) at the Macromolecular Resources Facility (Department of Biochemistry, Colorado State University). Amino acid analysis and composition were obtained by the Protein Structure Core Facility at the University of Nebraska (Lincoln).
Isolation of Ribosomes and rRNA Depurination Assay
Yeast (Saccharomyces cerevisiae) strain YPH500 (Sikorski and Hieter, 1989) was grown in YPD medium (1% [w/v] yeast extract, 2% [w/v] peptone, and 2% [w/v] Glc), and fungi, Trichoderma reesei, and Rhizoctonia solani were grown in potato dextrose media. Yeast was then pelleted by centrifugation and mycelial tissue of T. reesei and R. solani were collected by vacuum filtration. To isolate ribosomes, 10 g of pelleted yeast and fungal hyphae was ground in a mortar with liquid N2, and dissolved in 100 mL of extraction buffer (200 mm KCl, 25 mm MgCl2, 25 mm EGTA, 200 mm Suc, and 25 mm β-mercaptoethanol in 200 mm Tris-HCl [pH 9.0]). The supernatant, collected by centrifugation at 10,000g for 20 min at 4°C, was pipetted onto a Suc cushion (1 m Suc, 20 mm KCL, and 5 mm MgCl2 in 25 mm Tris-HCl [pH 7.6]) in 70 Ti tubes (Beckman Instruments, Fullerton, CA), and centrifuged at 55,000 rpm for 4 h at 4°C (L-70 Ultracentrifuge, Beckman Instruments). The pellets were resuspended in 25 mm Tris-HCl buffer (pH 7.6) with 25 mm KCl and 5 mm MgCl2, and stored at −80°C.
The depurination assay was conducted according to Tumer et al. (1997). In brief, ribosomes were resuspended in RIP buffer (167 mm KCl, 100 mm MgCl2, and 100 mm Tris-HCl [pH 7.2]) and incubated with RIPs at 30°C for 30 min in a total volume of 100 μL. RNA incubated in the absence of RIPs served as a negative control. After incubation, the RIPs were removed from the mixture by phenol:chloroform extraction and the RNA was divided in half. One-half of the extracted RNA was incubated on ice for 30 min with 1 m aniline acetate (pH 4.5) and precipitated with ethanol. Both aniline-treated and untreated RNAs were subjected to electrophoresis in a 7 m urea/6% (w/v) polyacrylamide gel and stained with ethidium bromide.
cDNA Cloning of PAP-H
Total RNA was isolated from hairy roots of pokeweed using the RNeasy Plant Mini Kit (Qiagen USA) according to manufacturer's instructions. cDNA was cloned by the RACE-PCR method using the SMART RACE cDNA Amplification Kit following the manufacturer's instructions (CLONTECH, Palo Alto, CA). To amplify the 3′ end of the PAP-H cDNA(s), two GSPs (5′-CCT TCG ATG TTG GAA GTG CAA CCA TTA GC and 5′-GGA AGT GCA ACC ATT AGC AAG TAT ACC ACC) were designed based on the N-terminal amino acid sequence of the purified PAP-H protein and considering codon preferences in previously cloned PAP-related genes from pokeweed. PCR reactions were performed in a GeneAmp System 2400 (Applied Biosystems) as follows: 94°C for 1 min; 30 cycles of 94°C for 5 s, 68°C for 10 s, and 72°C for 3 min. The nucleotide sequences of the GSPs used for 5′-RACE (5′-TGG CAA CCA ATA GGA ATC CTG CCT CGG and 5′-TGG CAA CCA ATA GGA ATC CTG CCT CGG) were designed based on the nucleotide sequences derived from the 3′-RACE product. The second PCR was performed as follows: 32 cycles of 94°C for 5 s, 60°C for 10 s, and 72°C for 3 min. Each PCR product was purified using Quantum Prep Gel Slice Kit (Bio-Rad), and cloned into pCR4Blunt-TOPO or pCR4TA-TOPO (Invitrogen) for sequencing. The DNA sequencing was performed using an ABI Prism 377 DNA Sequencer (Applied Biosystems) at the Macromolecular Resources Facility (Department of Biochemistry, Colorado State University).
Microscopy
The tips of hairy root tissues were cut into 2-mm segments and fixed for 1 h at 4°C with 1.5% (w/v) formaldehyde in Sorenson's phosphate buffer (SPB; 0.03 m sodium phosphate monobasic and 0.12 m sodium phosphate dibasic [pH 7.5]). Samples were placed into gelatin capsules containing 15% (w/v) gelatin solution, and overlaid with one or two drops of the gelatin solution by gentle pipetting. The gelatin capsules containing samples were incubated for 24 h at 4°C to be polymerized. After the resins were completely polymerized, gelatin blocks were removed from the capsules, and sections were cut 30 to 50 μm thick in prechilled SPB using a Vibratome (Sorvall MT2-B, Kendro, Newtown, CT). The sections were placed on slides and allowed to adhere for a few seconds. A drop of 50 mm Gly was added and sections were incubated for approximately 10 min. After washing twice with SPB, the sections were incubated with labeling-blocking buffer (LBB; 10% [v/v] goat serum and 1% [v/v] Triton X) for 30 min at 4°C, and incubated overnight with diluted PAP primary antibody (1:1,000 [w/v]) in LBB at 4°C in dark. Negative controls were incubated with only LBB under the same conditions. After 17 h, the sections were rinsed with LBB four times for 10 min at 4°C, and incubated with fluorescein goat anti-rabbit IgG (1:1,000 [v/v] dilution in LBB; Molecular Probes, Eugene, OR) for 4 h in dark at 4°C. The sections were washed with LBB again and SPB was used as a final wash. The sections were then mounted with antifade reagent (SlowFade Antifade Kit; Molecular Probes) in glycerol/PBS and sealed, and fluorescent images were taken using a microscope (2000 EXII, JEOL, Tokyo). PAP antibodies were obtained from Dr. Nilgun Tumer (Rutgers University).
Preparation of Proteins from Liquid Media
Root cultures grown in liquid media were vacuum filtered with a 0.8-μm cellulose nitrate membrane filter (Whatman, Maidstone, UK), and the media were supplementary filtered to avoid debris using a 0.22-μm Millex-GP syringe-driven filter (Millipore). The samples were precipitated with TCA according to the method of Peterson (1977). In brief, to each 1 mL of sample containing approximately 5 to 100 mg of protein, 100 μL of Na-deoxycholate was added and incubated for 10 min at room temperature. Then, 100 μL of 72% (w/v) TCA was dispensed, mixed, and incubated on ice for 15 min and centrifuged for 10 min. The supernatants were immediately removed, and the pellets were washed three times with ice-cold acetone. Pellets were then dissolved in SDS-PAGE sample buffer (Laemmli, 1970).
Elicitation Experiments and Ethylene Measurement
Hairy roots of pokeweed grown for 30 d were transferred into 50 mL of fresh Murashige and Skoog media, and treated with air, ethylene, and air + MCP (Biotechnologies for Horticulture, Burr Ridge, IL). Air treatment was performed using an air-permeable silicon cap, and ethylene treatment was conducted by feeding 10 μL L−1 ethylene gas into a flask connected to a gas-mixing apparatus (Mirjalili and Linden, 1995) at a total flow rate of 15 mL min−1; air was used as the balance gas. These flasks were connected by means of a second tubing to remove effluent gas outside of the incubator. MCP was added as powder at a final concentration of 20 μL L−1. Each treatment was incubated on a gyratory shaker in a dark chamber for a week and 2 mL of each culture media was collected daily for 6 d. Collected samples were stored at 4°C and concentrated using TCA precipitation (Peterson, 1977).
Ethylene was measured on a model 5840A gas chromatography system (Hewlett-Packard, Palo Alto, CA) equipped with a flame ionization detector. A Porapak N (6 ft, 0.2-mm i.d., stainless steel, Alltech, Nicholasville, KY) column was used at 75°C. The injection port and detector temperatures were 90°C and 180°C, respectively; the mobile gas was helium at 20 mL min−1; sample size was 0.1 mL.
Antifungal Assay
Antifungal activity of purified and exudated proteins was determined by a radial growth inhibition assay adapted from the method of Schlumbaum et al. (1986). Various fungal plugs were placed in the center of potato dextrose agar plates, and sterile paper discs were placed next to the fungal plugs. Fifty micrograms of each protein, which was sterilized using Ultrafree-MC Sterile (0.22-μm GV Durapore, Millipore), was pipetted onto the discs. The plates were then incubated in the dark at room temperature. Antifungal activity was observed as a crescent-shaped zone of inhibition at the mycelial front. The effect on fungal growth was expressed qualitatively, according to the procedure of Schlumbaum et al. (1986).
Enzyme Activity Determinations
A colorimetric assay for chitinase and β-1,3-glucanase activities in root exudates, with chitinase azure and laminarin azure as substrates, was performed as described by Qiu et al. (1997). The reaction mixture contained 550 μL of water, 200 μL of substrate (4 mg mL−1 in 0.2 m sodium acetate buffer [pH 5.0]), and 50 μL of root exudates (1 mg mL−1). After incubating the mixtures at 37°C for 5 min, 200 μL of 1 n HCl was added, placed on ice for 10 min, and centrifuged for 5 min at 12,000g. The resulting supernatants (900 μL) were measured spectrophotometrically at 550 nm. All assays were performed in triplicate, and blanks were prepared without the addition of root exudates during incubation. Enzymatic activities were calculated according to Wirth and Wolf (1992) and expressed as international units per milligram protein. One international unit is defined as the amount of enzyme required to catalyze the formation of 1 nmol of product per minute.
Zymogram Ready Gel was used for proteolytic activity. A 10% (v/v) SDS-PAGE gel containing gelatin (Bio-Rad) was used and electrophoresis was performed according to the manufacture's instructions. After electrophoretic separation, the gel was incubated at room temperature for 30 min in 100 mL of 2.5% (v/v) Triton X-100 with agitation, and incubated overnight at 37°C in 100 mL of development buffer (50 mm Tris-base, 200 mm NaCl, 5 mm CaCl2 anhydrous, and 0.02% [v/v] Brij-35 [pH 7.5]). The gel was then stained with 0.5% (w/v) Coomassie Brilliant Blue R-250 (EM Science).
ACKNOWLEDGMENTS
The authors thank Dr. Ramarao Vepachedu and Dr. Kevin Morey for technical assistance on cDNA cloning, and Dr. Stephen Wallner for editing suggestions in the preparation of the manuscript.
Footnotes
This work was supported by the National Science Foundation (CAREER award no. MCB–0093014 to J.M.V.) and by the Colorado State University Agricultural Experiment Station (to J.M.V.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.000794.
LITERATURE CITED
- Abeles FB, Bosshart RP, Forrence LE, Habig WH. Preparation and purification of glucanase and chitinase from bean leaves. Plant Physiol. 1971;47:129–134. doi: 10.1104/pp.47.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bais HP, Loyola-Vargas VM, Flores HE, Vivanco JM. Root-specific metabolism: the biology and biochemistry of underground organs. In Vitro Cell Dev Plant. 2001;37:730–741. [Google Scholar]
- Barbieri I, Aron GM, Irvin JD, Stirpe F. Purification and partial characterization of another form of the antiviral protein from the seeds of Phytolacca americanaL. (pokeweed) Biochem J. 1982;203:55–59. doi: 10.1042/bj2030055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolognesi A, Barbieri L, Abbondanza A, Ida Falasca A, Carnicelli D, Giulia Battelli M, Stirpe F. Purification and properties of new ribosome-inactivating proteins with RNA N-glycosidase activity. Biochim Biophys Acta. 1990;1087:293–302. doi: 10.1016/0167-4781(90)90002-j. [DOI] [PubMed] [Google Scholar]
- Chen Y-F, Randlett MD, Findell JL, Schaller GE. Proceeding of Plant Biology 2001, Providence, RI, Poster No. 959. Rockville, MD: American Society of Plant Biology; 2001. Ethylene receptor ETR1 localizes to the ER of Arabidopsis. p. 190. [Google Scholar]
- Chen ZC, White RF, Antoniw JF, Lin Q. Effect of pokeweed antiviral protein (PAP) on the infection of viruses. Plant Pathology. 1991;40:416–620. [Google Scholar]
- Cho GH, Kim DI, Pedersen H, Chin CK. Ethaphon enhancement of secondary metabolite synthesis in plant cell cultures. Biotechnol Prog. 1988;4:184–188. [Google Scholar]
- Del Vecchio Blanco F, Bolognesi A, Malorni A, Sande MJW, Savino G, Parente A. Complete amino acid sequence of PD-S2, a new ribosome-inactivating protein from seeds of Phytolacca dioicaL. Biochim Biophys Acta. 1997;1338:137–144. doi: 10.1016/s0167-4838(96)00182-3. [DOI] [PubMed] [Google Scholar]
- Endo Y, Tsurugi K. RNA N-glycosidase activity of ricin A-chain: mechanism of action of the toxic lectin ricin on eukaryotic ribosomes. J Biol Chem. 1987;263:8735–8739. [PubMed] [Google Scholar]
- Flores HE, Curtis WR. Approaches to understanding and manipulating the biosynthetic potential of plant roots. Ann NY Acad Sci. 1992;665:188–209. doi: 10.1111/j.1749-6632.1992.tb42584.x. [DOI] [PubMed] [Google Scholar]
- Flores HE, Vivanco JM, Loyola-Vargas V. “Radicle” biochemistry: the biology of root-specific metabolism. Trends Plant Sci. 1999;4:220–226. doi: 10.1016/s1360-1385(99)01411-9. [DOI] [PubMed] [Google Scholar]
- Frankel A, Schlossman D, Welsh P, Hertler A, Withers D, Johnston S. Selection and characterization of Ricin toxin A-chain mutation in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:415–420. doi: 10.1128/mcb.9.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funatsu G, Islam MR, Minami Y, Sung-Sil K, Kimura M. Conserved amino acid residues in ribosome-inactivating proteins from plants. Biochimie. 1991;73:1157–1161. doi: 10.1016/0300-9084(91)90160-3. [DOI] [PubMed] [Google Scholar]
- Gatehouse AMR, Barbieri L, Stirpe F, Croy RRD. Effects of ribosome inactivating proteins on insect development: differences between Lepidoptera and Coleoptera. Entomol Exp Appl. 1990;54:43–51. [Google Scholar]
- Hall AE, Findell JL, Schaller GE, Sisler EC, Bleecker AB. Ethylene perception by the ERS1 protein in Arabidopsis. Plant Physiol. 2000;123:1449–1458. doi: 10.1104/pp.123.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawes MC, Cunawardena U, Miyasaka S, Zhao X. The role of root border cells in plant defense. Trends Plant Sci. 2000;5:128–133. doi: 10.1016/s1360-1385(00)01556-9. [DOI] [PubMed] [Google Scholar]
- Hudak KA, Wang P, Tumer NE. A novel mechanism for inhibition of translation by pokeweed antiviral protein: depurination of the capped RNA template. RNA. 2000;6:369–380. doi: 10.1017/s1355838200991337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvin JD. Purification and partial characterization of the antiviral protein from Phytolacca americanawhich inhibits eukaryotic protein synthesis. Arch Biochem Biophys. 1975;169:522–528. doi: 10.1016/0003-9861(75)90195-2. [DOI] [PubMed] [Google Scholar]
- Irvin JD, Kelly T, Robertus JD. Purification and properties of a second antiviral protein from Phytolacca americanawhich inactivates eukaryotic ribosomes. Arch Biochem Biophys. 1980;200:418–425. doi: 10.1016/0003-9861(80)90372-0. [DOI] [PubMed] [Google Scholar]
- Joshi CP. Putative polyadenylation signals in nuclear genes of higher plants: a compilation and analysis. Nucleic Acids Res. 1987;15:9627–9640. doi: 10.1093/nar/15.23.9627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kombrink E, Schroder M, Hanbrock K. Several “pathogenesis-related” proteins in potato are 1,3-β-glucanase and chitinases. Proc Natl Acad Sci USA. 1988;85:782–786. doi: 10.1073/pnas.85.3.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragh KM, Jacobsen S, Mikkelsen JD. Induction, purification and characterization of barley leaf chitinase. Plant Sci. 1990;71:55–68. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leubner-Metzger G, Meins F., Jr . Functions and regulation of plant β-1,3-glucanase (PR-2) In: Datta SK, Muthukrisknan S, editors. Pathogenesis-Related Proteins in Plants. Boca Raton, FL: CRC Press; 1999. pp. 49–76. [Google Scholar]
- Li MX, Yeung HW, Pan LP, Chan SI. Trichosanthin, a potent HIV-1 inhibitor, can cleave supercoiled DNA in vitro. Nucleic Acids Res. 1991;19:6309–6312. doi: 10.1093/nar/19.22.6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Chen ZC, Antoniw JF, White RF. Isolation and characterization of a cDNA clone encoding the anti-viral protein from Phytolacca americana. Plant Mol Biol. 1991;17:609–614. doi: 10.1007/BF00037047. [DOI] [PubMed] [Google Scholar]
- Linthorst HJM. Pathogenesis-related proteins of plants. CRC Crit Rev Plant Sci. 1991;10:305–308. [Google Scholar]
- Lodge JK, Kaniewski WK, Tumer NE. Broad-spectrum virus resistance in transgenic plants expressing pokeweed antiviral protein. Proc Natl Acad Sci USA. 1993;90:7089–7093. doi: 10.1073/pnas.90.15.7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell JE. Protein sorting: organelle biogenesis and protein secretion. In: Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell JE, editors. Molecular Cell Biology. Ed 4. New York: WH Freeman and Company; 2000. pp. 675–750. [Google Scholar]
- Mauch F, Hadwiger LA, Boller T. Antifungal hydrolases in pea tissue: I. Purification and characterization of two chitinases and two β-1,3-glucanases differentially regulated during development in response to fungal infection. Plant Physiol. 1988;87:325–333. doi: 10.1104/pp.87.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta AD, Boston RS. Ribosome-inactivating protein. In: Bailey-Serres J, Gallie DR, editors. A Look beyond Transcription: Mechanisms Determining mRNA Stability and Translation in Plants. Rockville, MD: American Society of Plant Physiologists; 1998. pp. 145–152. [Google Scholar]
- Mirjalili N, Linden JC. Gas phase composition effects on suspension cultures of Taxus cuspidata. Biotechnol Bioeng. 1995;48:123–132. doi: 10.1002/bit.260480206. [DOI] [PubMed] [Google Scholar]
- Monzingo AF, Collins EJ, Ernst SR, Irvin JD, Robertus JD. The 2·5 Å structure of pokeweed antiviral protein. J Mol Biol. 1993;233:705–715. doi: 10.1006/jmbi.1993.1547. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Neuhaus J-M. Plant chitinases. In: Datta SK, Muthukrisknan S, editors. Pathogenesis-Related Proteins in Plants. Boca Raton, FL: CRC Press; 1999. pp. 77–105. [Google Scholar]
- Nicolas E, Beggs JM, Haltiwanger BM, Taraschi TF. Direct evidence for the deoxyribonuclease activity of the plant ribosome-inactivating protein gelonin. FEBS Lett. 1997;406:162–164. doi: 10.1016/s0014-5793(97)00267-6. [DOI] [PubMed] [Google Scholar]
- Nicolas E, Beggs JM, Haltiwanger BM, Taraschi TF. A new class of DNA glycosylase/apurinic/apyrimidinic lyases that act on specific adenines in single-stranded DNA. J Biol Chem. 1998;273:17216–17220. doi: 10.1074/jbc.273.27.17216. [DOI] [PubMed] [Google Scholar]
- Nicolas E, Beggs JM, Taraschi TF. Gelonin is an unusual DNA glycosylase that removes adenine from single-stranded DNA, normal base pairs and mismatches. J Biol Chem. 2000;275:31399–31406. doi: 10.1074/jbc.M004505200. [DOI] [PubMed] [Google Scholar]
- Nielsen K, Boston RS. Ribosome-inactivating proteins: a plant perspective. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:785–816. doi: 10.1146/annurev.arplant.52.1.785. [DOI] [PubMed] [Google Scholar]
- Ohme-Takagi M, Suzuki K, Shinshi H. Regulation of ethylene-induced transcription of defense genes. Plant Cell Physiol. 2000;41:1187–1192. doi: 10.1093/pcp/pcd057. [DOI] [PubMed] [Google Scholar]
- Osborn RW, Hartley MR. Dual effects of the ricin A chain on protein synthesis in rabbit reticulocyte lysate. Inhibition of initiation and translocation. Eur J Biochem. 1990;193:401–407. doi: 10.1111/j.1432-1033.1990.tb19353.x. [DOI] [PubMed] [Google Scholar]
- Peterson GL. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal Biochem. 1977;83:346–356. doi: 10.1016/0003-2697(77)90043-4. [DOI] [PubMed] [Google Scholar]
- Phisalaphong M, Linden JC. Kinetic studies of paclitaxel production by Taxus canadensiscultures in batch and semicontinuous with total cell recycle. Biotechnol Prog. 1999;15:1072–1077. doi: 10.1021/bp990098p. [DOI] [PubMed] [Google Scholar]
- Qiu J, Hallmann J, Kokalis-Burelle N, Weaver DB, Rodriguez-Kabana R, Tuzun S. Activity and differential induction of chitinase isozymes in soybean cultivars resistant or susceptible to root-knot nematodes. J Nematol. 1997;29:523–530. [PMC free article] [PubMed] [Google Scholar]
- Rajamohan F, Pugmire MJ, Kurinow IV, Uckun FM. Modeling and alanine scanning mutagenesis studies of recombinant pokeweed antiviral protein. J Biol Chem. 2000;275:3382–3390. doi: 10.1074/jbc.275.5.3382. [DOI] [PubMed] [Google Scholar]
- Ready MP, Brown DT, Robertus JD. Extracellular localization of pokeweed protein. Proc Natl Acad Sci USA. 1986;84:5053–5056. doi: 10.1073/pnas.83.14.5053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe C, Parthier B, Reinbothe S. Temporal pattern of jasmonate-induced alterations in gene expression of barley leaves. Planta. 1997;201:281–287. doi: 10.1007/s004250050067. [DOI] [PubMed] [Google Scholar]
- Reinbothe S, Reinbothe C, Lehman J, Becker W, Apel K, Parthier B. JIP60, a methyl jasmonate-induced ribosome-inactivating protein involved in plant stress reactions. Proc Natl Acad Sci USA. 1994;91:7012–7016. doi: 10.1073/pnas.91.15.7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remi Shih NR, McDonald KA, Jackman AP, Girbes T, Lglesias R. Bifunctional plant defense enzymes with chitinase and ribosome inactivating activities from Trichosanthes kirilowiicell cultures. Plant Sci. 1997;130:145–150. [Google Scholar]
- Rippmann JF, Michalowski CB, Nelson DE, Bohnert HJ. Induction of a ribosome-inactivating protein upon environmental stress. Plant Mol Biol. 1997;35:701–709. doi: 10.1023/a:1005871023944. [DOI] [PubMed] [Google Scholar]
- Roncuzzi L, Gasperi-Campani A. DNA-nuclease activity of the single-chain ribosome-inactivating proteins dianthin 30, saporin 6 and gelonin. FEBS Lett. 1996;392:16–20. doi: 10.1016/0014-5793(96)00776-4. [DOI] [PubMed] [Google Scholar]
- Polito L. Activities associated with the presence of ribosome-inactivating proteins increase in senescent and stressed leaves. FEBS Lett. 1996;382:309–312. doi: 10.1016/0014-5793(96)00188-3. [DOI] [PubMed] [Google Scholar]
- Savary BJ, Flores HE. Biosynthesis of defense-related proteins in transformed root cultures of Trichosanthes kirilowii Maxim. var japonicum(Kitam.) Plant Physiol. 1994;106:1195–1204. doi: 10.1104/pp.106.3.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savary BJ, Flores HE, Hill JJ. Isolation of a class III chitinase produced in root cultures of Trichosanthes kirilowiiand assessment of accumulation patterns and antifungal activity. Plant Physiol Biochem. 1997;35:543–551. [Google Scholar]
- Schaller GE, Gamble RL, Randlett M, Zhao X, Qu X. Ethylene receptors and the two-component paradigm. In: Walker J, Randall D, editors. Current Topics in Plant Biochemistry, Physiology and Molecular Biology. Vol. 18. Columbia: University of Missouri; 2000. pp. 68–69. [Google Scholar]
- Schaller GE, Ladd AN, Lanahan MB, Spanbauer JM, Bleecker AB. The ethylene response mediator ETR1 Arabidopsisforms a disulfide-linked dimer. J Biol Chem. 1995;270:12526–12530. doi: 10.1074/jbc.270.21.12526. [DOI] [PubMed] [Google Scholar]
- Schlumbaum A, Mauch F, Vogeli U, Boller T. Plant chitinases are potent inhibitors of fungal growth. Nature. 1986;324:36–367. [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:12–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnov S, Shualev V, Tumer NE. Expression of pokeweed antiviral protein in transgenic plants induces virus resistance in grafted wild-type plants independently of salicylic acid accumulation and pathogenesis-related protein synthesis. Plant Physiol. 1997;114:1113–1121. doi: 10.1104/pp.114.3.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S-K, Choi Y, Moon YH, Kim S-G, Choi YD, Lee JS. Systemic induction of a Phytolacca incularisantiviral protein gene by mechanical wounding, jasmonic acid, and abscisic acid. Plant Mol Biol. 2000;43:439–450. doi: 10.1023/a:1006444322626. [DOI] [PubMed] [Google Scholar]
- Stirpe F, Barbieri L. Ribosome-inactivating proteins up to date. FEBS Lett. 1986;195:1–8. doi: 10.1016/0014-5793(86)80118-1. [DOI] [PubMed] [Google Scholar]
- Stirpe F, Barbieri L, Batelli MG, Soria M, Lappi DA. Ribosome inactivating proteins from plants: present status and future prospects. Biotechnology. 1992;10:405–412. doi: 10.1038/nbt0492-405. [DOI] [PubMed] [Google Scholar]
- Stirpe F, Barbieri L, Gorini P, Valbonesi P, Bologenesi A, Polito L. Activities associated with the presence of ribosome-inactivating proteins increase in senescent and stressed leaves. FEBS Lett. 1996;382:309–312. doi: 10.1016/0014-5793(96)00188-3. [DOI] [PubMed] [Google Scholar]
- Taiz L, Zeiger E. Ethylene and abscisic acid. In: Taiz L, Zeiger E, editors. Plant Physiology. Redwood City, CA: The Benjamin/Cummings Publishing Company; 1991. pp. 473–489. [Google Scholar]
- Tumer NE, Hudak K, Di R, Coetser C, Wang R, Zoubenko O. Pokeweed antiviral protein and its applications. Curr Top Microbiol Immunol. 1999;240:139–158. doi: 10.1007/978-3-642-60234-4_7. [DOI] [PubMed] [Google Scholar]
- Tumer NE, Hwang D-J, Bonness M. C-terminal deletion mutant of pokeweed antiviral protein inhibits viral infection but does not depurinate ribosomes. Proc Natl Acad Sci USA. 1997;94:3866–3871. doi: 10.1073/pnas.94.8.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ussery MA, Irvin JD, Hardesty B. Inhibition of poliovirus replication by a plant antiviral peptide. Ann NY Acad Sci. 1977;284:431–440. doi: 10.1111/j.1749-6632.1977.tb21979.x. [DOI] [PubMed] [Google Scholar]
- Verveliet G, Holsters M, Teuchy H, Van Montagu M, Schell J. Characteristics of different plaque forming and defective temperate phages in Agrobacteriumstrains. J Gel Virol. 1975;26:33–48. doi: 10.1099/0022-1317-26-1-33. [DOI] [PubMed] [Google Scholar]
- Vivanco JM, Flores HE. Control of root formation by plant growth regulators. In: Basra AS, editor. Plant Growth Regulators in Agriculture and Horticulture: Their Role and Commercial Uses. Food Products Press, New York: an Imprint of The Haworth Press; 2000. pp. 1–25. [Google Scholar]
- Vivanco JM, Savary BJ, Flores HE. Characterization of two novel type I ribosome-inactivating proteins from the storage roots of the Andean crop Milabilis expansa. Plant Physiol. 1999;119:1447–1456. doi: 10.1104/pp.119.4.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vögeli U, Meins F, Jr, Boller B. Co-ordinated regulation of chitinase and β-1,3-glucanase in bean leaves. Planta. 1988;174:364–372. doi: 10.1007/BF00959522. [DOI] [PubMed] [Google Scholar]
- Wirth SJ, Wolf GA. Micro-plate colourimetric assay for endo-acting cellulase, xylanase, chitinase, 1.3-β-glucanase and amylase extracted from forest soil horizons. Soil Biol Biochem. 1992;24:511–519. [Google Scholar]
- Wong R, Ng TB, Chan SH, Dong TX, Xeung HW. Characterization of Mirabilis jalapa antiviral protein: a ribosome inactivating protein from Mirabilis jalapaL. Biochem Int. 1992;28:585–593. [PubMed] [Google Scholar]
- Wyatt SE, Pan SQ, Kuc J. β-1,3-Glucanase, chitinase, and peroxidase activities in tobacco tissues resistant and susceptible to blue mould as related to flowing, age and sucker development. Physiol Mol Plant Pathol. 1991;39:433–440. [Google Scholar]
- Zarling JM, Moran RA, Haffar O, Sias J, Richmann DD, Spina CA, Myers DE, Kuelbeck V, Ledbetter JA, Uckun FM. Inhibition of HIV replication by pokeweed antiviral protein targeted to CD4+cells by monoclonal antibodies. Nature. 1990;347:92–95. doi: 10.1038/347092a0. [DOI] [PubMed] [Google Scholar]
- Zoubenko O, Hudak K, Tumer NE. A non-toxic pokeweed antiviral protein mutant inhibits pathogen infection via a novel salicylic acid-independent pathway. Plant Mol Biol. 2000;44:219–229. doi: 10.1023/a:1006443626864. [DOI] [PubMed] [Google Scholar]
- Zoubenko O, Uckun F, Hur Y, Chet I, Tumer NE. Plant resistance to fungal infection induced by nontoxic pokeweed antiviral protein mutants. Nat/Biotechnol. 1997;15:992–996. doi: 10.1038/nbt1097-992. [DOI] [PubMed] [Google Scholar]