Abstract
The Jerusalem artichoke (Helianthus tuberosus) xenobiotic inducible cytochrome P450, CYP76B1, catalyzes rapid oxidative dealkylation of various phenylurea herbicides to yield nonphytotoxic metabolites. We have found that increased herbicide metabolism and tolerance can be achieved by ectopic constitutive expression of CYP76B1 in tobacco (Nicotiana tabacum) and Arabidopsis. Transformation with CYP76B1 conferred on tobacco and Arabidopsis a 20-fold increase in tolerance to linuron, a compound detoxified by a single dealkylation, and a 10-fold increase in tolerance to isoproturon or chlortoluron, which need successive catalytic steps for detoxification. Two constructs for expression of translational fusions of CYP76B1 with P450 reductase were prepared to test if they would yield even greater herbicide tolerance. Plants expressing these constructs had lower herbicide tolerance than CYP76B1 alone, which is apparently a consequence of reduced stability of the fusion proteins. In all cases, increased herbicide tolerance results from more extensive metabolism, as demonstrated with exogenously fed phenylurea. Beside increased herbicide tolerance, expression of CYP76B1 has no other visible phenotype in the transgenic plants. Our data indicate that CYP76B1 can function as a selectable marker for plant transformation, allowing efficient selection in vitro and in soil-grown plants. Plants expressing CYP76B1 may also be a potential tool for phytoremediation of contaminated sites.
Engineering of herbicide tolerance in higher plants can be achieved in many ways: via introduction of an altered target protein that is insensitive to the herbicide, overexpression of a wild-type target, or modification of herbicide transport, compartmentation, or metabolism. Increasing metabolism may be the best strategy because the phytotoxic compound is chemically altered and there is no interference with primary metabolism and no residual herbicide remains in the plant. So far, most crops genetically modified for herbicide metabolism have been transformed with genes isolated from microorganisms (Duke, 1996); however, plants themselves offer a wide choice of herbicide-detoxifying enzymes. The introduction of different plant genes or appropriate alterations in expression levels in crop plants could be considered as an accelerated adjunct to classical breeding techniques to engender gene transfer between plants.
The genes for herbicide-detoxifying enzymes in higher plants are just starting to be characterized. Efforts are focused on multigene families like those of glutathione S-transferases (McGonigle et al., 2000) or glycosyl transferases (Ross et al., 2001; Brazier et al., 2002), and cytochrome P450 monooxygenases (Werck-Reichhart et al., 2000). The latter, which is by far the largest family of enzymatic proteins in higher plants (272 P450 genes are found in the diminutive genome of Arabidopsis), offers the widest resource in terms of diversity and possible substrate specificity (http://drnelson.utmem.edu/cytochromep450.html; http://www.biobase.dk/P450/p450.shtml; Schuler, 1996; Chapple, 1998; Kahn and Durst, 2000; Werck-Reichhart and Feyerei-sen, 2000; Werck-Reichhart et al., 2002).
In vivo and in vitro experimentation has largely demonstrated the involvement of P450s in the metabolism of all major classes of herbicides and their contribution to herbicide selectivity and weed resistance (Werck-Reichhart et al., 2000). Numerous reports confirm that the xenobiotic metabolizing P450s are induced by chemicals such as drugs, metals, ethanol, herbicide safeners, or herbicides themselves (Barrett, 1995; Batard et al., 1995; Frear, 1995; Moreland et al., 1995; Persans and Schuler, 1995; Potter et al., 1995). The latter observation led us to isolate and characterize cDNAs of P450s expressed upon plant treatment with xenobiotics (Batard et al., 1998; Cabello-Hurtado et al., 1998), among them CYP76B1, which shows high transcriptional activation after treatment with MnCl2 or drugs such as aminopyrine or phenobarbital in Jerusalem artichoke (Helianthus tuberosus).
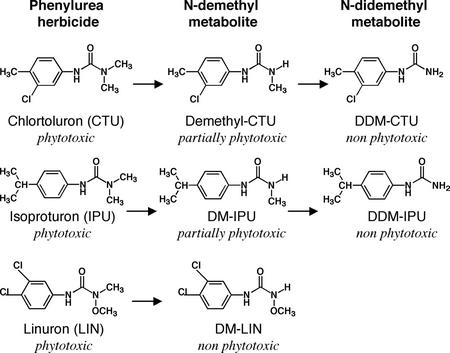
Expression of CYP76B1 in yeast (Saccharomyces cerevisiae) revealed its ability to metabolize a broad range of exogenous molecules with a catalytic efficiency often comparable with that observed for P450-dependent metabolism of endogenous compounds (Robineau et al., 1998). Among the xenobiotics tested, CYP76B1 was particularly effective at one or more N-demethylation reactions with phenylurea herbicides (Fig. 1). The plant detoxification of these compounds is well understood (Cole, 1983), and early investigators have established that the demethyl derivative of compounds like diuron and monuron are less phytotoxic than the parent, and that the di-demethyl derivative is essentially nonphytotoxic (Geissbühler et al., 1963; Swanson and Swanson, 1968). Similar results have been obtained with a variety of related compounds, including chlortoluron (Ryan et al., 1981; Ryan and Owen, 1982) and isoproturon (Cabanne et al., 1987; Glässgen et al., 1999). The removal of the single N-methyl substituent of linuron results in the nearly complete elimination of phytotoxicity (Kuratle et al., 1969; Nashed and Ilnicki, 1970), similar to the effect of two-step detoxification with most other herbicides in this class.
Figure 1.
CYP76B1 metabolism of phenylurea.
Therefore, CYP76B1 is a good candidate gene to use for engineering herbicide resistance in sensitive crops or for increasing the bioremediation potential of some plants. We have tested the effects of its ectopic constitutive expression on phenylurea tolerance of tobacco (Nicotiana tabacum) and Arabidopsis. To optimize the activity of the recombinant P450 protein and to circumvent possible limiting input of reducing equivalents for CYP76B1, translational fusions of the CYP76B1 cDNA with that of a P450 reductase previously isolated from the same species were constructed. In the present paper, we compare the increased tolerance obtained in the two plant species with the different constructs. A very significant increase in tolerance was observed for several phenylureas, allowing in vitro selection of the transformants and also conferring resistance to formulated herbicide applied by foliar treatment to plants grown in soil.
RESULTS
Construction of cDNAs Encoding CYP76B1-P450 Reductase Fusion Proteins
Recent data have suggested that proteins catalyzing the electron transfer to P450 enzymes may become rate limiting in planta. For example, high expression of a cytochrome b5 gene is needed for full activity of the flavonoid 3′,5′-hydroxylase activity in the petunia (Petunia hybrida) flower (De Vetten et al., 1999). Furthermore, plants contain different P450 reductases, and a coordinate activation of a cinnamate 4-hydroxylase and a specific P450 reductase gene upon elicitor treatment, infection, or UV light irradiation has been observed in parsley (Petroselinum crispum; Koopmann and Hahlbrock, 1997). The availability of an electron donor is particularly likely to be a rate-limiting factor in the case of recombinant overexpression of a P450 terminal oxidase, and the electron drain caused by the recombinant enzyme may interfere with the activity of endogenous proteins with essential functions. For this reason, we constructed chimeric cDNAs coding for fusion proteins between CYP76B1 and a P450 reductase. Natural forms of such fusion proteins were found in bacteria (Narhi and Fulco, 1986) and eukaryotes (McMillan et al., 1992; Nakayama et al., 1996), and artificial chimera were shown to be catalytically active in yeast and Escherichia coli (Murakami et al., 1987; Hotze et al., 1995; Fisher et al., 1996). Fusions of yeast P450 reductase and mammalian P450 enzymes have been used to engineer herbicide resistance in tobacco and potato (Solanum tuberosum; Shiota et al., 1994; Inui et al., 1999).
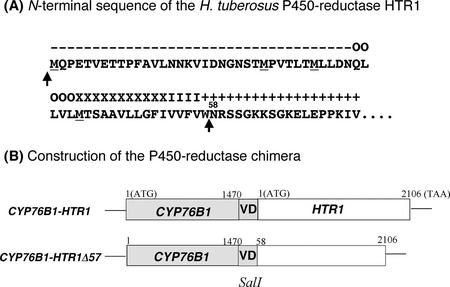
The source of the coexpressed reductase influences the stability of recombinant P450s and the efficiency of coupling of the electron transport to substrate oxidation (Louerat-Oriou et al., 1998; Robineau et al., 1998). Thus, we chose to fuse the coding sequence of CYP76B1 to that of a P450 reductase previously isolated from the same plant (Jerusalem artichoke), HTR1 (Hasenfratz, 1992; GenBank accession no. Z26250). Two types of translational fusions were constructed (Fig. 2). One included the full-length reductase sequence attached to the 3′ end of CYP76B1 (76B1-HTR1). This fusion comprises the longest open reading frame of HTR1, including the first of several ATGs, codons for a 35-amino acid hydrophilic stretch predicted to form a loop, and those for a hydrophobic helical segment, which is predicted to direct membrane insertion. In the second fusion (76B1-HTR1Δ57), the first 57 codons for the first loop and the membrane-insertion segment were removed to obtain a direct fusion of the flavoprotein domain of the reductase to the P450 hemoprotein (Shibata et al., 1990). In both cases, the fusion was achieved by PCR modification of the coding sequences so as to introduce a SalI site 3′ and 5′ of the P450 and reductase sequences, respectively.
Figure 2.
Construction of the translational fusions between CYP76B1 and the Jerusalem artichoke P450 reductase HTR1. Both sequences were modified by PCR with a proof-reading polymerase using the primers given in text. BamHI and SalI sites were added just at the 5′ and 3′ ends of the CYP76B1 sequence, respectively, the stop codon being removed. SalI and EcoRI sites were inserted 5′ and 3′ of HTR1 so as to generate two different translational fusions. A, The HTR1 sequence has four ATG codons (underlined) that may constitute possible translation starts. Arrows indicate the connecting points between the P450 and the reductase. Predicted structures: +, inside loop; −, outside loop; O, outside helix cap; X, central transmembrane helix segment; and I, inside helix cap. B, Our first construct contains the largest reading frame. In the second construct, all ATGs, the first loop, and the transmembrane helix were removed, the two globular domains of the proteins being joined by just a short loop structure. In both cases, the SalI site created a Val-Asp linker predicted to maintain a loop structure between the two proteins.
Expression and Activity of the Fusion Proteins in Yeast
The reductase-P450 fusion proteins were first tested in yeast for expression and activity. The different constructs were inserted in a multicopy plasmid under the control of the Gal-inducible promoter GAL10-CYC1. This plasmid was transformed into W303-1B and WHT1, a yeast strain where the endogenous P450 reductase has been replaced with the Jerusalem artichoke P450 reductase HTR1 under the control of GAL10-CYC1.
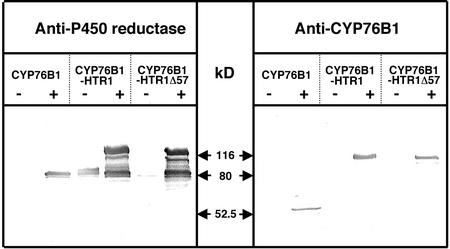
Immunoblot analysis of the microsomal fractions of Gal-induced recombinant yeast with anti-CYP76B1 and anti-Jerusalem artichoke P450 reductase polyclonal antibodies (Fig. 3) indicated that the fusion proteins were expressed at levels at least equivalent to the wild-type CYP76B1. However, multiple lower Mr bands were evident from both of them, suggesting that they are degradation products and that the fusion proteins are more labile in vivo than the CYP76B1 alone. Staining with anti-P450 reductase antibodies also revealed some reductase protein, not fused to P450, produced in the absence and presence of Gal from the full fusion (76B1-HTR1). This was particularly evident in the W303–1B blots, where there was no crosshybridization with the endogenous reductase (CYP76B1 lanes), yet there was strong crosshybridization in the 76-HTR lanes. This was unexpected and could be due to the presence of a cryptic promoter in the coding sequence of CYP76B1 or in the 5′ segment of HTR1.
Figure 3.
Expression in yeast (WHT1) of CYP76B1 and its translational fusions with HTR1. The immunoblots (20 μg of protein lane−1) were revealed with rabbit polyclonal antibodies directed against His-tagged recombinant CYP76B1 (Robineau et al., 1998) or purified Jerusalem artichoke P450 reductases (Benveniste et al., 1989). −, Microsomes isolated from yeast grown on Glc; +, microsomes from yeast induced for 16 h on Gal; M, Mr markers.
To verify if functional enzymes were produced, P450 absorbance, reductase, and monooxygenase activities were also measured. The results of this analysis (Table I) confirmed the expression of reductase in the absence of induction from the 76B1-HTR1 construct, and, to a much lesser extent, from the 76B1-HTR1Δ57 (ATG-less) chimera. MROD, easy to monitor by fluorometry, was chosen as reporter monooxygenase activity (Robineau et al., 1998). The full-length fusion 76B1-HTR1 showed no MROD activity. This was probably the result of impaired folding or of the instability of the P450 domain because no CO-binding hemoprotein was detected in the recombinant yeast microsomes, whereas reductase was active for cytochrome c reduction. In a converse manner, the truncated fusion 76B1-HTR1Δ57 was functional in yeast microsomes. Calculation of specific activities indicates that its turnover is similar or is slightly increased compared with that of CYP76B1 separately coexpressed with HTR1 in yeast microsomes.
Table I.
Expression and activity in yeast of CYP76B1, single or fused to the HTR1 reductase
| Strain Construct | Growth Medium | MROD Activity | MROD Turnover Rate | P450 Content | Cyt c Reductase |
|---|---|---|---|---|---|
| pmol s−1 mg−1 protein | min−1 | nmol mg−1 protein | nmol s−1 mg−1 protein | ||
| W303-1B | |||||
| CYP76 | Glc | n.d. | – | n.d. | 1.39 |
| CYP76 | Gal | 4.1 | 1.7 | 141 | 1.04 |
| 76-HTR1 | Gal | 0.46 | – | n.d. | 14.12 |
| 76-HTR1Δ57 | Gal | 14.5 | 33.4 | 26 | 40.8 |
| WHT1 | |||||
| CYP76 | Glc | n.d. | – | n.d. | 0.05 |
| CYP76 | Gal | 100 | 92 | 65 | 4.44 |
| 76-HTR1 | Glc | n.d. | – | n.d. | 2.82 |
| 76-HTR1 | Gal | n.d. | – | n.d. | 8.46 |
| 76-HTR1Δ57 | Glc | n.d. | – | n.d. | 0.17 |
| 76-HTR1Δ57 | Gal | 208 | 106 | 117 | 7.77 |
CYP76B1 alone, and the 76-HTR1 and 76-HTR1Δ57 constructs were expressed in W303-1B and WHT1 where the yeast reductase gene was replaced with the coding sequence of the Jerusalem artichoke reductase HTR1 under the control of the Gal-inducible promoter GAL10-CYC1. MROD, 7-Methoxyresorufin O-deethylase; n.d., not detectable.
Herbicide Tolerance of Transformed Tobacco
For expression in tobacco, all constructs were cloned into a T-DNA binary vector under the control of a strong constitutive duplicated 35S promoter. T1 (primary) transformants were selected on kanamycin, and the number of integration loci in the host genome was determined by Southern blot. Plants with single insertions were self-pollinated, and subsequent generations were selected on kanamycin and diuron.
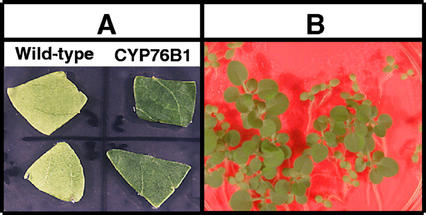
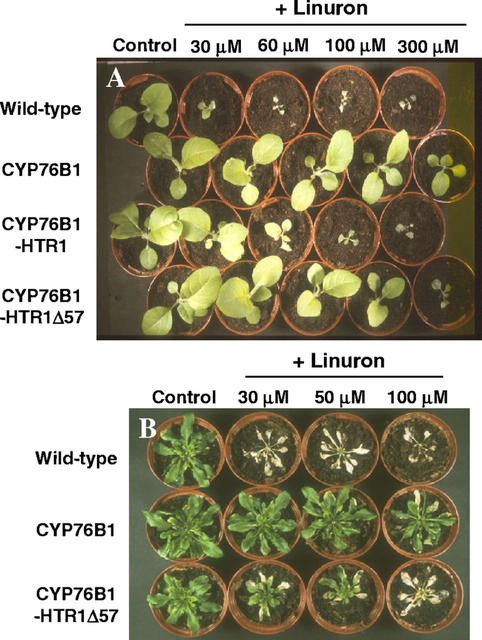
The herbicide tolerance of the transformants was assayed at all stages of the process using different methods. Phenylurea herbicides inhibit photosynthesis by blocking electron transport in the photosystem II complex (Oettmeier, 1999). Susceptible plant treatment leads to leaf chlorosis followed by complete necrosis, resulting in a dramatic dose-dependent reduction of growth. The tolerance of the T1 transformants was tested by aging leaf pieces on agar plates containing different concentrations of herbicide under strong light. Leaves of the transformants appeared resistant to bleaching by 0.5 mm linuron (Fig. 4A).
Figure 4.
Increased herbicide tolerance of T1 and T2 CYP76B1 tobacco transformants. A, Leaf pieces were washed with sterile water and cut under sterile conditions before being aged under strong light for 10 d on Murashige and Skoog medium containing 0.5 mm linuron. B, Selection of the T2 on Murashige and Skoog medium containing 0.9 μm diuron. Growth of wild-type plants is strongly inhibited.
Selection of the T2 and T3 generations was performed, and the segregation of the T3 progeny was checked by growing the seeds on sterile plates containing 0.9 μm diuron (Fig. 4B). Selection of the transformants on diuron or kanamycin was equally effective.
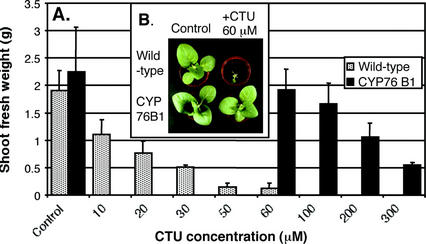
T3 homozygous plants were then tested for herbicide tolerance after leaf treatment of soil-grown plants with commercial formulations of the phenylurea herbicides linuron, chlortoluron, and isoproturon. A dramatic reduction of plant growth was observed following treatment with herbicide concentrations around 10 μm (Figs. 5A and 6). The increase in phenylurea resistance of transgenic plants was evaluated by comparing concentrations leading to 50% (I50) inhibition of shoot growth (Table II). The highest increase in herbicide tolerance (around 20-fold) was obtained with linuron for the plants expressing CYP76B1 alone, not fused to any reductase (Fig. 5). The CYP76B1-HTR1Δ57 fusion also conferred a significant tolerance (around 10-fold). CYP76B1-HTR1 had a reduced efficiency. The same general trend was observed when tolerance was assayed with chlortoluron or isoproturon, although the overall resistance factor was not as high as with linuron. Linuron was completely detoxified by a single demethylation, whereas chlortoluron and isoproturon needed to be doubly dealkylated to yield a nonphytotoxic metabolite (Fig. 1). We propose that the need for two catalytic steps to achieve detoxification with chlortoluron and isoproturon results in the overall lower resistance factor with these compounds.
Figure 5.
Comparison of the increase in phenylurea tolerance obtained with the different constructs. Tests were performed with 1-month-old homozygous T3 plants, leaves being treated twice at a 4-d interval with dilutions of commercial formulations of linuron. A, Tobacco plants 12 d after the first treatment. B, Arabidopsis plants 10 d after first treatment
Table II.
Compared growth inhibition by phenylureas of wild-type and transgenic tobacco and Arabidopsis
| Herbicide Treatment | Tobacco
|
|||
|---|---|---|---|---|
| Wild type | CYP76B1 | 76B1-HTR1 | 76B1-HTR1Δ | |
| I50 (μm) | ||||
| CTU | 14 | 180 (12.9) | 36 (2.6) | 55 (3.9) |
| IPU | 28 | 90 (3.2) | 49 (1.8) | 64 (2.3) |
| LIN | 10 | 220 (22) | 50 (5) | 108 (10.8) |
| Arabidopsis | ||||
| Wild type | CYP76B1 | 76B1-HTR1 | 76B1-HTR1Δ | |
| IPU | 7 | 44 (6.3) | – | 23 (3.3) |
| LIN | 7 | 80 (11.4) | – | 21 (3) |
I50 for chlortoluron (CTU), isoproturon (IPU), and linuron (LIN) were determined from the dose-response curves based on three independent measurements of shoot fresh weight 9 d after herbicide treatment. Resistance factors (I50R/I50S) are shown in brackets. R, Resistant to phenylurea herbicides; S, susceptible to phenylurea herbicides.
Transgene Expression and Herbicide Metabolism in Transformed Tobacco
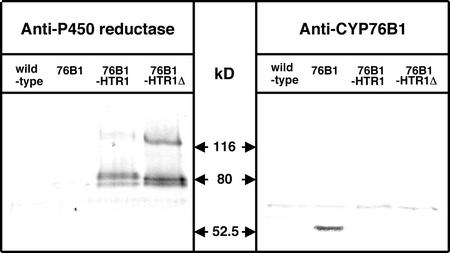
Data obtained with yeast microsomes indicated that the CYP76B1-HTR1Δ57 fusion had the highest monooxygenase activity when expressed in microsomes also containing wild-type (not fused) plant P450 reductase. This fusion did not provide the highest level of herbicide tolerance in tobacco, therefore, this discrepancy was further investigated. A microsomal fraction was prepared from leaves of the different transformants, and the CYP76B1 and reductase contents were estimated by immunoblot (Fig. 7). The staining with antireductase antibodies revealed a much higher expression of the CYP76B1-HTR1Δ57 than of the CYP76B1-HTR1 construct in plant tissues. As in the case of recombinant yeast microsomes, expression of nonfused reductase was also observed. When the immunoblots were stained with anti-CYP76B1 antibodies, only CYP76B1 expressed alone was detected. As the fusion proteins were readily detected in yeast microsomes (Fig. 3), a selective epitope masking in plant membranes, although possible, seems rather unlikely. Therefore, it suggests that the levels of expression or stability of the fusions in plants is much lower than that of the P450 expressed alone. The P450 segment of the protein seems particularly prone to misfolding or degradation by proteases.
Figure 7.
Immunoblot analysis of the transgenic tobacco microsomes. Immunoblots were revealed with the same polyclonal sera as in Figure 3. Twenty-five micrograms of microsomal protein was loaded in each lane.
No reliable measurement of cytochrome c reductase activity was possible in microsomes from light- or dark-grown plants, in vitro or in soil. In the same way, P450 content and CYP76B1-associated demethylase activity could not be measured in plant microsomes, probably due to contamination by chloroplast pigments and redox components, or to plant secondary metabolites such as phenols and alkaloids.
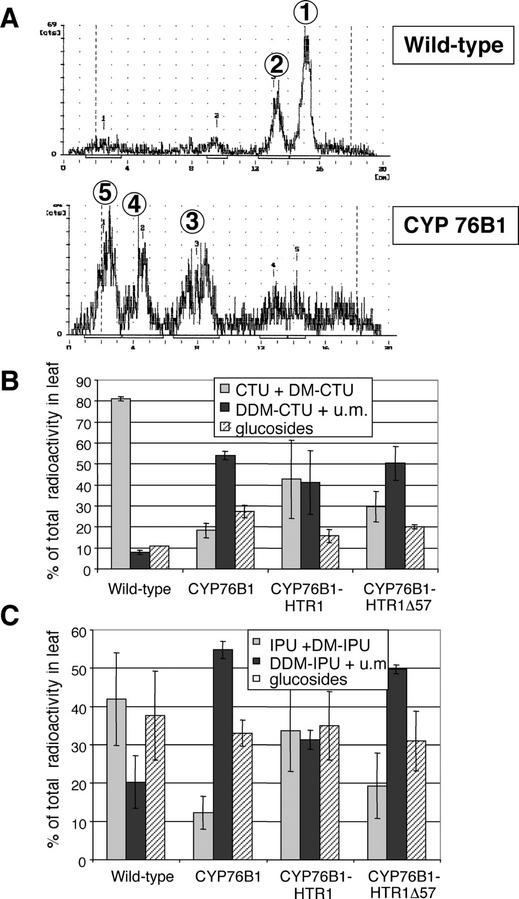
To directly evaluate the metabolism of phenylurea herbicides in the transgenic plants, individual leaves of T3 homozygotes were excised and fed with radiolabeled chlortoluron or isoproturon. All transgenic lines showed a higher capacity to metabolize the herbicide than wild-type tobacco (Fig. 8). The capacity to produce nonphytotoxic metabolites paralleled herbicide tolerance of the transformants: CYP76B1 > CYP76B1-HTR1Δ57 > CYP76B1-HTR1 > wild type. The products of CYP76B1-dependent metabolism of phenylurea were further converted by other tobacco enzymes into other more polar metabolites (e.g. glucosyl conjugates; see Fig. 8A). Wild-type plants more actively metabolized isoproturon than chlortoluron (Fig. 8, B and C). Therefore, the increase in herbicide metabolism of the recombinant plants was smaller for isoproturon than for chlortoluron.
Figure 8.
Chlortoluron and isoproturon metabolism in wild-type and recombinant tobacco leaves. Excised leaves petioles were fed with 0.84 kBq [phenyl-U-14C]chlortoluron or 1.55 kBq [phenyl-U-14C]isoproturon. After complete absorption, the herbicide solution was replaced with water and leaves were maintained under continuous illumination for 12 h. 14C-labeled parent herbicide and its metabolites were extracted in methanol and were quantified by thin-layer chromatography (TLC). Glc conjugates were identified after treatment of extracts with β-glucosidase. A, TLC profiles of the chlortoluron metabolites: (1) CTU, (2) N-demethyl CTU (DM-CTU), (3) N-didemethyl CTU (DDM-CTU) + unknown metabolites, (4) unknown metabolites, and (5) glucosides. B, Proportion of residual glucosides and phytotoxic (CTU +DM-CTU) and nonphytotoxic (DDM-CTU + unknown metabolites) aglycones recovered after chlortoluron metabolism. C, Proportion of residual glucosides and phytotoxic (IPU + DM-PU) and nonphytotoxic (DDM-IPU + unknown metabolites) aglycones recovered after isoproturon metabolism.
Herbicide Tolerance and Transgene Expression in Arabidopsis
Immunoblot of the microsomes from transgenic Arabidopsis gave essentially the same results as those obtained with tobacco (data not shown).
DISCUSSION
Factors Influencing Herbicide Tolerance
To our knowledge, the present study constitutes the first systematic investigation on the level of herbicide tolerance in transgenic plants by the overexpression of a plant P450 enzyme. There are few reports on the engineering of herbicide metabolism with plant genes (Jepson et al., 1996; Siminszky et al., 1999), but the rather scarce data available do not indicate how large increases in herbicide tolerance have been achieved. CYP76B1, a gene inducible by chemical stress isolated from Jerusalem artichoke, was previously shown to metabolize herbicides of the class of phenylurea with high turnover rates (Robineau et al., 1998). We show here that its 35S promoter-driven expression confers a 3- to 20-fold increase in resistance to phenylurea on tobacco and Arabidopsis. The level of resistance that is obtained varies with the active compound used for treatment in each plant. In tobacco, for example, a 2- to 3-fold higher increase in tolerance is achieved against linuron compared with chlortoluron, and the increase in chlortoluron resistance is, in turn, 2- to 4-fold higher than the increase in tolerance to isoproturon. This reflects, in part, the affinity and turnover rates for each herbicide, but also depends on the number of catalytic steps needed for complete detoxification and on the initial level of tolerance of the transformed plant. CYP76B1 metabolizes phenylurea exclusively via N-dealkylation. Compounds with two N-methyl substituents (e.g. isoproturon, diuron, and chlortoluron) require two dealkylation steps to become nonphytotoxic. Compounds with N-methoxy and N-methyl substituents, such as linuron and related molecules, lose phytotoxicity with a single demethylation.
In addition, their affinity for CYP76B1 was shown to be higher than that of related structures with two methyl substituents (turnover rates are not reported so far; Robineau et al., 1998). This readily explains why CYP76-expressing plants are more tolerant to linuron than to chlortoluron and isoproturon. Wild-type tobacco plants are more tolerant to isoproturon than to chlortoluron (Table II) and, in good agreement with this observation, metabolize isoproturon much faster than chlortoluron (Fig. 8). Catalytic efficiency of chlortoluron metabolism by recombinant CYP76B1 is one order of magnitude higher than that of isoproturon (Robineau et al., 1998). Thus, it is not surprising if transformed tobacco shows a higher increase in tolerance to chlortoluron than isoproturon. Thus, the increase in herbicide tolerance that will be obtained by ectopic expression of CYP76B1 is expected to vary with the phenylurea molecule.
Impact of the P450 Reductase
The highest increase in herbicide resistance was obtained via plant transformation with just CYP76B1. Attempts to further optimize CYP76B1 efficiency in planta, using translational fusions with a homologous P450 reductase to overcome possible rate-limiting electron transfer, proved disappointing. The plant P450 reductase, HTR1, as many other plant P450 reductases, differs from animal and yeast enzymes in that it presents a longer hydrophilic segment with a predicted loop structure at its N terminus. We tested if the presence of this structure as a linker between the two intact proteins results in a functional chimera, with both domains anchored and sufficiently mobile in the membrane. We compared the efficiency of this fusion with another construct deleted of the N terminus of the reductase so as to remove the potentially labile linker between the two globular proteins. The complete fusion, CYP76B1-HTR1, was barely effective. The best fusion protein, CYP76B1-HTR1Δ57, was two to four times less efficient than CYP76B1 alone. There are two possible explanations for the lower activity of the fusion proteins: a low expression or stability of the fusions in planta or a poor intramolecular electron transfer compared with that between single CYP76B1 and the P450 reductases from the host plant.
The second alternative is very difficult to verify with plant microsomes, but western-blot analysis of plant membranes and measurements of activities in recombinant yeast largely support the first hypothesis. As in previous reports on yeast or E. coli expression of mammalian or yeast fusion proteins (Shibata et al., 1990; Helvig and Capdevilla, 2000), the deletion of the membrane-anchoring segment of the P450 reductase was required to obtain chimeric protein expressed at significant levels in yeast and plants. Almost no fusion protein was detected in plants transformed with CYP76B1-HTR1 (Fig. 7), in agreement with the low metabolism (Fig. 8) and increased tolerance (Table II) observed in the recombinant plants. In a converse manner, the higher levels of fusion protein detected in plants transformed with CYP76B1-HTR1Δ57 are also in agreement with the higher levels of metabolism and herbicide tolerance achieved in the transformants. Thus, fusion proteins seem to be prone to protease degradation, and only systematic optimization of the connecting segment between the two proteins could possibly lead to really stable and catalytically effective chimera (Govindaraj and Poulos, 1995).
A large proportion of the reductase was recovered as single, nonfused protein in yeast and plant. This may indicate activity of a cryptic promoter upstream of the reductase sequence, but also misfolding and fast degradation of the P450 segment of the fused protein. Expression of single reductase in yeast transformed with the fusion constructs in the absence of Gal induction strongly supports the first hypothesis. In support of the second hypothesis, the fusion proteins, which are readily detected by anti-CYP76B1 antibodies in yeast microsomes (Fig. 3), are not detected in microsomal fractions isolated from recombinant plants (Fig. 7), which suggests a strong alteration in P450 conformation. Instability of the hemoprotein domain compared with the reductase is also apparent upon expression in yeast (Table I). Thus, it is possible that combined effects of a cryptic promoter and P450 degradation contribute to the accumulation of isolated reductase protein. An interesting consequence of this high reductase accumulation is that, a least in the case of the CYP76B1-HTR1 transformants, some increased herbicide metabolism might result from reductase-mediated activation of endogenous enzymes. Such a possibility will be further investigated by construction of reductase-overexpressing plants.
Potential Applications
Ectopic expression of the CYP76B1 protein has no visible impact on plant growth and development. This indicates that its overexpression does not directly or indirectly lead to major perturbation of an essential pathway or to accumulation of a phytotoxic compound. The P450 reductase isoforms and their constitutive expression in the plants seem to ensure efficient, if not optimal, reduction of the recombinant P450 oxidase. Thus, it seems possible to use the CYP76B1 gene as a positive marker of plant selection. Our experiments indicate that selection is easily achieved in vitro by including herbicide in the growth medium or via leaf treatment of soil-grown plants with commercial phenylurea formulations. The only selection system using a P450 gene described so far is a negative selection system using a bacterial P450 gene that is also effective in vitro and in soil (O'Keefe et al., 1994; Koprek et al., 1999). CYP76B1 has the advantage over other positive selection systems in that it is a plant gene conferring resistance to a whole class of generic herbicides and also leading to their detoxification. It is interesting to note that phenylurea dealkylation by CYP76B1 in planta allows further metabolism, not only conjugation, by the plant enzymes. Thus, CYP76B1 is also a potential tool for phytoremediation of contaminated soils and wastewater, or containment of herbicide leaching into groundwater (Dietz and Schnoor, 2001).
MATERIALS AND METHODS
Chemicals
Umbelliferone was purchased from Sigma (St. Quentin-Fallavier, France) and 7-methoxyresorufin was purchased from Molecular Probes (Leiden, The Netherlands). Linuron, chlortoluron, and isoproturon were purchased from Promochem (Molsheim, France). [phenyl-U-14C]Chlortoluron (64 GBq mol−1), [phenyl-U-14C]isoproturon (344 GBq mol−1), and reference metabolites were generous gifts from Syngenta (Basel). The commercial formulations of herbicides used for plant treatments in soil were Afalon 50L (active ingredient: linuron, distributed by Bayer Crop Science, Lyon, France), and Chlorto-Stef GT and Iso-Stef GT (active ingredients: chlortoluron and isoproturon, respectively, distributed by Stefes, Senlis, France).
Construction of the WHT1 Yeast Strain
The WHT1 yeast strain was constructed by replacing the gene of the endogenous P450 reductase of W303–1B with the coding sequence of a P450 reductase from Jerusalem artichoke (Helianthus tuberosus), HTR1, under the control of the Gal-inducible promoter GAL10-CYC1. BamHI and EcoRI sites were generated 5′ and 3′ of the HTR1 coding sequence, respectively, using the PCR primers N-terminal (5′-cgggatccATGCAACCGGAAACCGTCG-3′) and C-terminal (5′-ccggaattcTCACCAAACATCACGGAGGTATC-3′). The Pfu DNA polymerase-amplified fragment was inserted in the integrative plasmid pYeDP110 (Pompon et al., 1996) using these restriction sites. The plasmid was linearized with NotI for integration of HTR1 and GAL10-CYC1 at the locus of the yeast (Saccharomyces cerevisiae) reductase by homologous recombination.
Construction of the Plant Expression Vectors
Three vectors were constructed for plant transformation: one with the CYP76B1 (accession no. Y0992) coding sequence alone, and the two others designed to direct expression of translational fusions of CYP76B1 and of the Jerusalem artichoke P450 reductase HTR1 (accession no. Z26250). To optimize the fusion proteins, predictions of secondary structures and trans-membrane segments were performed using the online PSIpred prediction server (http://bioinf.cs.ucl.ac.uk/psipred/). Junctions between P450 and reductase were chosen so that the reductase was predicted to start with a loop structure (Fig. 2). The first construct, CYP76B1-HTR1, included a predicted membrane-spanning segment near the reductase N terminus. In the second construct, CYP76B1-HTR1Δ57, this segment and the upstream loop were deleted. For the construction of the fusions, the coding region of CYP76B1 was PCR amplified so as to create BamHI (5′) and SalI (3′) sites with the primers CYP76-2 (5′-cgcggatccATGGATTTTCTTATAATAGTGAGTAC-3′) and CYP76-3 (5′-cgcgtcgacGTTCAATGGTATTGGAACAACACAC-3′). CYP76B1 was also inserted alone in the BamHI and SacI sites of the expression vector using CYP76-2 in conjunction with CYP76-4 (5′-cgcgagctcCTAGTTCAATGGTATTGGAACAACAC-3′). The primers used for amplification of HTR1 were designed to create SalI and SmaI sites, respectively, at the 5′ and 3′ ends of the PCR products: HTR1-1 (5′-cgcgtcgacATGCAACCGGAAACCGTCGAAACG-3′), HTR1-2 (5′-cgccccgggTCACCAAACATCACGGAGGTATC-3′), HTR1-3 (5′-cgcgtcgacAATAGATCGTCCGGTAAGAAGTCCG-3′). HTR1-1 with HTR1-2 amplified the complete coding region, and HTR1-2 with HTR1-3 produced the truncated form lacking the sequence coding for amino acids 1 through 57.
The PCR mixtures contained 500 ng of template, 20 pm of both primers, and 0.2 mm dNTPs in a total volume of 50 μL. They were preheated for 2 min at 94°C before the addition of 5 units of Pfu DNA polymerase (Stratagene, La Jolla, CA). After 3 min of additional heating at 94°C, 30 cycles of amplification were carried out as follows: 1 min of denaturation at 94°C, 2 min of annealing at 55°C, and a 2-min extension at 72°C. The reaction was completed by a 10-min extension at 72°C, and an additional 20-min PCR extension step was carried on with Taq DNA polymerase (Roche Molecular Diagnostics, Basel) to add an extra 3′-terminal adenosine to both ends. PCR products were then cloned in pGEM-T plasmid (Promega, Madison, WI).
To construct the fusions, after digestion with BamHI and SalI, the CYP76B1 fragment was subcloned into the plasmid pLacZi (Clontech, Palo Alto, CA). The resulting plasmid was digested with SalI and SmaI, and the HTR1 fragments, excised from pGEM by SalI and SmaI digestion, were inserted 3′ from CYP76B1. The resulting fusions were digested with BamHI and SmaI and were cloned into pBDX. The single CYP76B1 sequence terminated by a stop codon, excised from pGEM with BamHI and SacI, was also directly subcloned into the corresponding sites of pBDX. The pBDX binary vector, derived from pBD515.3 (Husselstein et al., 1999), contains between the left and right T-DNA borders a kanamycin-resistance gene and a modified multiple cloning site flanked in 5′ by a duplicated 35S promoter, and by a nopaline synthase terminator at the 3′ end (H. Schaller and P. Crevenat, unpublished data).
Yeast Expression
The fusions constructed in pBDX were excised by BamHI and EcoRI digestion, and were ligated in pYeDP60 (Pompon et al., 1996) previously digested with the same restriction enzymes. The CYP76B1 sequence previously cloned in pGEM-T was transferred into pYeDP60 using the BamHI and SacI restriction sites. The different constructs were used to transform the W303–1B (wild-type) and WHT1 yeast strains. Yeast transformation was performed essentially according to Schiestl and Gietz (1989). Yeast growth, induction, and preparation of yeast microsomes were carried out according to Pompon et al. (1996).
Plant Transformation and Characterization of Transgenic Plants
PBDX and the constructs pBDX-CYP76B1, pBDX-CYP76B1-HTR1, and pBDX-CYP76-HTR1Δ57 were transferred from Escherichia coli XL1 Blue into Agrobacterium tumefaciens LB4404 (Hoekema et al., 1983) by triparental mating with the helper plasmid pRK2013 (Bevan, 1984).
Five-week-old roots from Arabidopsis C24 were infected with the resulting A. tumefaciens LBA4404 strains, and T1 plants were regenerated as described by Valvekens et al. (1988). Culture room conditions were 23°C with a 16-h light/8-h dark cycle.
Tobacco (Nicotiana tabacum var. Xanthi) leaves were transformed via A. tumefaciens as in Schaller et al. (1998). In vitro cultures were grown under a 16-h light/8-h dark cycle at 23°C.
Transformed T1 Arabidopsis and tobacco plants were selected in vitro for kanamycin resistance (on 50 and 100 mg L−1 kanamycin, respectively). The T2 segregation was analyzed in vitro on a selective medium containing kanamycin or 0.9 μm diuron. T3 homozygous transgenic plants were selected among the self-progeny of kanamycin- and diuron-resistant T2 plants on the same media. The segregation of the T3 self-progeny was checked by growing the seeds on a sterile medium containing 0.9 μm diuron.
Biochemical Characterization of the Transformed Plants
Microsomes of recombinant plant leaves were prepared essentially according to Halkier and Møller (1989), except that the ratio of plant material to polyvinylpolypyrrolidone was 1:1 (w/w). Quantification of microsomal protein was carried out using a protein assay (Bio-Rad, Hercules, CA). SDS-PAGE, electroblotting onto nitrocellulose (Hybond C; Amersham Biosciences, Piscataway, NJ), and immunostaining with rabbit polyclonal antibodies raised against recombinant CYP76B1 (Robineau et al., 1998) or purified Jerusalem artichoke reductases (Benveniste et al., 1989) were performed as in Werck-Reichhart et al. (1993). Herbicide metabolism and P450 reductase assays have been described in Cabello-Hurtado et al. (1998) and in Benveniste et al. (1986), respectively.
Herbicide Treatments and Phytotoxicity Evaluation
Foliar treatments of soil-grown plants were performed at the three- to four-leaf stage in the case of tobacco and the six- to eight-rosette leaf stage for Arabidopsis. Plants were sprayed twice with 10 mL of water dilutions of commercial herbicides at 4-d intervals. Plant tolerance was assessed 9 to 12 d after the first treatment by measuring shoots wet weight. Data are means ± se of triplicate experiments.
In Vivo Herbicide Metabolism
In vivo metabolism was assayed with excised leaves from the wild-type and transgenic tobacco plants fed with 0.84 kBq [phenyl-U-14C] chlortoluron or 1.55 kBq [phenyl-U-14C] isoproturon dissolved in water, as described by Siminszky et al. (1999). Herbicide and metabolites were quantified by TLC analysis of methanol extracts. Metabolites were identified by comparison with reference compounds using the solvent system, hexane:chloroform:acetone:ethanol (8:8:4:1, v/v). Metabolites that did not display mobility upon TLC analysis were submitted to hydrolysis with β-glucosidase, as described by Shiota et al. (1996), for quantification of Glc conjugates.
Figure 6.
Shoot growth inhibition by chlortoluron of CYP76B1-transformed and wild-type plants. A, Dose-response: shoot fresh weight (average ± se of three measurements) was determined 9 d after foliar herbicide treatment. B, Appearance of the control and 60 μm-treated plants.
ACKNOWLEDGMENTS
We thank Denis Pompon and Philippe Urban for the providing the pYeDP60 and pYeDP110 vectors and WAT11 yeast strain, and Marie Paule Hasenfratz, Agnēs Lesot, and I. Benveniste for the HTR1 cDNA. The technical help of Monique Schmitz and Marta Ramel is greatly appreciated.
Footnotes
This work was supported by E.I. DuPont de Nemours and Company (grant to L.D.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.005801.
LITERATURE CITED
- Barrett M. Metabolism of herbicides by cytochrome P450 in corn. Drug Metab Drug Interact. 1995;12:299–315. doi: 10.1515/dmdi.1995.12.3-4.299. [DOI] [PubMed] [Google Scholar]
- Batard Y, LeRet M, Schalk M, Robineau T, Durst F, Werck-Reichhart D. Molecular cloning and functional expression in yeast of CYP76B1, a xenobiotic-inducible 7-ethoxycoumarin O-deethylase from Helianthus tuberosus. Plant J. 1998;14:111–120. doi: 10.1046/j.1365-313x.1998.00099.x. [DOI] [PubMed] [Google Scholar]
- Batard Y, Zimmerlin A, Le Ret M, Durst F, Werck-Reichhart D. Multiple xenobiotic-inducible P450s are involved in alkoxycoumarins and alkoxyresorufins metabolism in higher plants. Plant Cell Environ. 1995;18:523–533. [Google Scholar]
- Benveniste I, Gabriac B, Durst F. Purification and characterisation of the NADPH-cytochrome P450 (cytochrome c) reductase from higher plant microsomal fraction. Biochem J. 1986;235:365–373. doi: 10.1042/bj2350365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste I, Lesot A, Hasenfratz MP, Durst F. Immunochemical characterization of NADPH-cytochrome P450 reductase from Jerusalem artichoke and other higher plants. Biochem J. 1989;259:847–853. doi: 10.1042/bj2590847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 1984;12:8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazier M, Cole DJ, Edwards R. O-Glucosyltransferase activities toward phenolic natural products and xenobiotics in wheat and herbicide-resistant and herbicide susceptible black-grass (Alopecurus myosuroides) Phytochemistry. 2002;59:149–156. doi: 10.1016/s0031-9422(01)00458-7. [DOI] [PubMed] [Google Scholar]
- Cabanne F, Huby D, Gaillardon P, Scalla R, Durst F. Effect of the cytochrome P450 inhibitor 1-aminobenzotriazole on the metabolism of chlortoluron and isoproturon in wheat. Pestic Biochem Physiol. 1987;28:371–380. [Google Scholar]
- Cabello-Hurtado F, Batard Y, Salaün JP, Durst F, Pinot F, Werck-Reichhart D. Cloning, expression in yeast and functional characterization of CYP81B1, a plant P450 which catalyzes in-chain hydroxylation of fatty acids. J Biol Chem. 1998;273:7260–7267. doi: 10.1074/jbc.273.13.7260. [DOI] [PubMed] [Google Scholar]
- Chapple C. Molecular genetics analysis of plant cytochrome P450-dependent monooxygenases. Annu Rev Plant Physiol Mol Biol. 1998;49:311–343. doi: 10.1146/annurev.arplant.49.1.311. [DOI] [PubMed] [Google Scholar]
- Cole D. Oxidation of xenobiotics in plants. In: Hutson DH, Roberts TR, editors. Progress in Pesticide Biochemistry and Toxicology. Vol. 3. New York: John Wiley & Sons; 1983. pp. 199–254. [Google Scholar]
- De Vetten N, Ter Horst J, Van Schaik HP, De Boer A, Mol J, Koes R. A cytochrome b5 is required for full activity of flavonoid 3′,5′-hydroxylase, a cytochrome P450 involved in the formation of blue flower colors. Proc Natl Acad Sci USA. 1999;96:778–783. doi: 10.1073/pnas.96.2.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz AC, Schnoor JL. Advances in phytoremediation. Environ Health Perspect. 2001;109:163–168. doi: 10.1289/ehp.01109s1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duke SO. Herbicide-Resistant Crops: Agricultural, Environmental, Economic, Regulatory, and Technical Aspects. Boca Raton, FL: CRC Lewis Publishers; 1996. [Google Scholar]
- Fisher CW, Shet MS, Estabrook RW. Construction of plasmids and expression in Escherichia coli of enzymatically active fusion proteins containing the heme-domain of a P450 linked to the NADPH-P450 reductase. Methods Enzymol. 1996;272:15–25. doi: 10.1016/s0076-6879(96)72004-9. [DOI] [PubMed] [Google Scholar]
- Frear DS. Wheat microsomal cytochrome P450 monooxygenases: characterization and importance in the metabolic detoxification and selectivity of wheat herbicides. Drug Metab Drug Interact. 1995;12:329–357. doi: 10.1515/dmdi.1995.12.3-4.329. [DOI] [PubMed] [Google Scholar]
- Geissbühler H, Haselbach H, Aebi H, Ebner L. The fate of N′-(4-chlorophenoxy)-phenyl-N,N-dimethylurea (C1983) in soils and plants: breakdown in soils and plants. Weed Res. 1963;3:277–297. [Google Scholar]
- Glässgen WE, Komossa D, Bohnenkämper O, Haas M, Hertkorn N, May RG, Szymczak W, Sandermann H. Metabolism of the herbicide isoproturon in wheat and soybean cell suspension cultures. Pestic Biochem Physiol. 1999;63:97–113. [Google Scholar]
- Govindaraj S, Poulos TL. Role of the linker region connecting the reductase and heme domains in cytochrome P450BM-3. Biochemistry. 1995;34:11221–11226. doi: 10.1021/bi00035a031. [DOI] [PubMed] [Google Scholar]
- Halkier BA, Møller BL. Biosynthesis of the cyanogenic glucoside dhurrin in seedlings of Sorghum bicolor (L.) Moench and partial purification of the enzyme system involved. Plant Physiol. 1989;90:1552–1559. doi: 10.1104/pp.90.4.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasenfratz MP. Clonage de la NADPH-cytochrome P450 reductase et d'une protéine calnexine-like chez Helianthus tuberosus. PhD thesis. Strasbourg, France: Université Louis Pasteur; 1992. [Google Scholar]
- Helvig C, Capdevilla JH. Biochemical characterization of rat P450 2C11 fused to rat or bacterial NADPH-P450 reductase domains. Biochemistry. 2000;39:5196–5205. doi: 10.1021/bi992578v. [DOI] [PubMed] [Google Scholar]
- Hoekema A, Hirsch PR, Hooykas PJJ, Schilperoort RA. A binary plant vector strategy based on separation of vir-and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature. 1983;303:179–180. [Google Scholar]
- Hotze M, Schroeder G, Schroeder J. Cinnamate 4-hydroxylase from Catharanthus roseus, and a strategy for the functional expression of plant cytochrome P450 proteins as translational fusions with P450 reductase in Escherichia coli. FEBS Lett. 1995;374:345–350. doi: 10.1016/0014-5793(95)01141-z. [DOI] [PubMed] [Google Scholar]
- Husselstein T, Schaller H, Gachotte D, Benveniste P. Δ7-Sterol-C5-desaturase: molecular characterization and functional expression of wild-type and mutant alleles. Plant Mol Biol. 1999;39:891–906. doi: 10.1023/a:1006113919172. [DOI] [PubMed] [Google Scholar]
- Inui H, Ueyama Y, Shiota N, Ohkawa Y, Ohkawa H. Herbicide metabolism and cross-tolerance in transgenic potato plants expressing human CYP1A1. Pestic Biochem Physiol. 1999;64:33–46. [Google Scholar]
- Jepson I, Holt DC, Roussel V, Wright SY, Greenland AJ. Transgenic plant analysis as a tool for the study of maize glutathione S-transferases. In: Hatzios KK, editor. Regulation of Enzymatic Systems Detoxifying Xenobiotics in Plants: NATO ASI Series: High Technology. Vol. 37. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1996. pp. 313–323. [Google Scholar]
- Kahn R, Durst F. Function and evolution of plant cytochrome P450. In: Romeo JT, Ibrahim R, Varin L, DeLuca V, editors. Recent Advances in Phytochemistry. Vol. 34. Oxford: Elsevier Science; 2000. pp. 151–189. [Google Scholar]
- Koopmann E, Hahlbrock K. Differentially regulated NADPH:cytochrome P450 oxidoreductases in parsley. Proc Natl Acad Sci USA. 1997;94:14954–14959. doi: 10.1073/pnas.94.26.14954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprek T, McElroy D, Louwerse J, Williams-Carrier R, Lemaux PG. Negative selection systems for transgenic barley (Hordeum vulgare L.): comparison of bacterial codA- and cytochrome P450 gene-mediated selection. Plant J. 1999;19:719–726. doi: 10.1046/j.1365-313x.1999.00557.x. [DOI] [PubMed] [Google Scholar]
- Kuratle H, Rahn EM, Woodmansee CW. Basis for selectivity of linuron on carrot and common ragweed. Weed Sci. 1969;17:216–219. [Google Scholar]
- Louerat-Oriou B, Perret A, Pompon D. Differential redox and electron-transfer properties of purified yeast, plant and human NADPH-cytochrome P450 reductases highly modulate cytochrome P450 activities. Eur J Biochem. 1998;258:1040–1049. doi: 10.1046/j.1432-1327.1998.2581040.x. [DOI] [PubMed] [Google Scholar]
- McGonigle B, Keeler SJ, Lau S-MC, Koeppe MK, O'Keefe DP. A genomic approach to the comprehensive analysis of the glutathione S-transferase gene family in soybean and maize. Plant Physiol. 2000;124:1105–1120. doi: 10.1104/pp.124.3.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan K, Bredt DS, Hirsch DJ, Snyder SH, Clarck JE, Master BSI. Cloned, expressed rat cerebellar nitric oxide synthase contains stoichiometric amounts of heme, which binds carbon monoxide. Proc Natl Acad Sci USA. 1992;89:11141–11145. doi: 10.1073/pnas.89.23.11141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreland DE, Corbin FT, Fleischmann TJ, McFarland JE. Partial characterization of microsomes isolated from mung bean cotyledons. Pestic Biochem Physiol. 1995;52:98–108. [Google Scholar]
- Murakami H, Yabusaki T, Sakaki M, Ohkawa H. A genetically engineered P450 monooxygenase: construction of the functional fused enzyme between rat cytochrome P450c and NADPH-cytochrome P450 reductase. DNA. 1987;6:189–197. doi: 10.1089/dna.1987.6.189. [DOI] [PubMed] [Google Scholar]
- Nakayama N, Takamae A, Shoun H. Cytochrome P450foxy, a catalytically self-sufficient fatty acid hydroxylase of the fungus Fusarium oxysporum. J Biochem. 1996;119:435–440. doi: 10.1093/oxfordjournals.jbchem.a021260. [DOI] [PubMed] [Google Scholar]
- Narhi LO, Fulco AJ. Characterization of a catalytically self-sufficient 119,000-dalton cytochrome P-450 monooxygenase induced by barbiturates in Bacillus megaterium. J Biol Chem. 1986;261:7160–7169. [PubMed] [Google Scholar]
- Nashed RB, Ilnicki RD. Absorption, distribution, and metabolism of linuron in corn, soybean, and crabgrass. Weed Sci. 1970;18:25–28. [Google Scholar]
- Oettmeier W. Herbicide resistance and supersensitivity in photosystem II. Cell Mol Life Sci. 1999;15:1255–1277. doi: 10.1007/s000180050370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkawa H, Imaishi H, Shiota N, Inui H, Ohkawa Y. Molecular mechanisms of herbicide resistance. Rev Toxicol. 1998;2:245–252. [Google Scholar]
- O'Keefe DP, Tepperman JM, Dean C, Leto KJ, Erbes DL, Odell JT. Plant expression of a bacterial cytochrome P450 that catalyzes activation of a sulfonylurea pro-herbicide. Plant Physiol. 1994;105:473–482. doi: 10.1104/pp.105.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persans MW, Schuler MA. Differential induction of cytochrome P450-mediated triasulfuron metabolism by naphthalic anhydride and triasulfuron. Plant Physiol. 1995;109:1483–1490. doi: 10.1104/pp.109.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompon D, Louerat B, Bronine A, Urban P. Yeast expression of animal and plant P450s in optimized redox environment. Methods Enzymol. 1996;272:51–64. doi: 10.1016/s0076-6879(96)72008-6. [DOI] [PubMed] [Google Scholar]
- Potter S, Moreland DE, Kreuz K, Ward E. Induction of cytochrome P450 genes by ethanol in maize. Drug Metab Drug Interact. 1995;12:317–327. doi: 10.1515/dmdi.1995.12.3-4.317. [DOI] [PubMed] [Google Scholar]
- Robineau T, Batard Y, Nedelkina S, Cabello-Hurtado F, LeRet M, Sorokine O, Didierjean L, Werck-Reichhart D. The chemically inducible plant cytochrome P450 CYP76B1 actively metabolizes phenylureas and other xenobiotics. Plant Physiol. 1998;118:1049–1056. doi: 10.1104/pp.118.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JK, Li Y, Lim E, Bowles DJ. Higher plant glycosyltransferases. Gen Biol. 2001;2:3004.1–3004.6. doi: 10.1186/gb-2001-2-2-reviews3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan PJ, Gross D, Owen WJ, Laanio TL. The metabolism of chlortoluron, diuron, and CGA 43 057 in tolerant and susceptible plants. Pestic Biochem Physiol. 1981;16:213–221. [Google Scholar]
- Ryan PJ, Owen WJ. The mechanism of selectivity of chlortoluron between cereals and grassweeds. Proc Br Crop Prot Conf Weeds. 1982;i:317–324. [Google Scholar]
- Schaller H, Bouvier-Navé P, Benveniste P. Overexpression of an Arabidopsis thaliana (L.) Heynh. cDNA encoding a sterol-C241-methyltransferase in Nicotiana tabacum L. modifies the ratio of 24-methyl cholesterol to sitosterol and is associated with growth reduction. Plant Physiol. 1998;118:461–469. doi: 10.1104/pp.118.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl RH, Gietz RD. High efficiency transformation of intact cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Schuler MA. Plant cytochrome P450 monooxygenases. Crit Rev Plant Sci. 1996;15:235–284. [Google Scholar]
- Shibata M, Sakaki T, Yabusaki Y, Murakami H, Ohkawa H. Genetically engineered monooxygenases: construction of bovine P45c17/yeast reductase fused enzymes. DNA Cell Biol. 1990;9:27–36. doi: 10.1089/dna.1990.9.27. [DOI] [PubMed] [Google Scholar]
- Shiota N, Inui H, Ohkawa I. Metabolism of the herbicide chlortoluron in transgenic tobacco plants expressing the fused enzyme between rat cytochrome P4501A1 and yeast NADPH-cytochrome P450 reductase. Pestic Biochem Physiol. 1996;54:190–198. [Google Scholar]
- Shiota N, Nagasawa A, Sakaki T, Yabusaki Y, Ohkawa H. Herbicide-resistant tobacco plants expressing the fused enzyme between rat cytochrome P4501A1 (CYP1A1) and yeast NADPF-cytochrome P450 oxidoreductase. Plant Physiol. 1994;106:17–23. doi: 10.1104/pp.106.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siminszky B, Corbin FT, Ward ER, Fleischmann TJ, Dewey RE. Expression of a soybean cytochrome P450 monooxygenase cDNA in yeast and tobacco enhances the metabolism of phenylurea herbicides. Proc Natl Acad Sci USA. 1999;96:1750–1755. doi: 10.1073/pnas.96.4.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson CR, Swanson HR. Metabolic fate of monuron and diuron in isolated leaf discs. Weed Sci. 1968;16:137–143. [Google Scholar]
- Valvekens D, Van Montagu M, Van Lijsebettens M. Agrobacterium tumefaciens-mediated transformation of Arabidopsis root explants using kanamycin selection. Proc Natl Acad Sci USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werck-Reichhart D, Bak S, Paquette S. Cytochromes P450. In: Somerville CR, Meyerowitz EM, editors. The Arabidopsis Book. Rockville, MD: American Society of Plant Biologists; 2002. http://www.aspb.org/publications/arabidopsis , doi/10.1199/tab.0028. http://www.aspb.org/publications/arabidopsis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werck-Reichhart D, Batard Y, Kochs G, Lesot A, Durst F. Monospecific polyclonal antibodies directed against purified cinnamate 4-hydroxylase from Helianthus tuberosus: immunopurification, immunoquantitation, and interspecies cross-reactivity. Plant Physiol. 1993;102:1291–1298. doi: 10.1104/pp.102.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werck-Reichhart D, Feyereisen R. Cytochrome P450: a success story. Gen Biol. 2000;1:3003.1–3003.0. doi: 10.1186/gb-2000-1-6-reviews3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werck-Reichhart D, Hehn H, Didierjean L. Cytochromes P450 for engineering herbicide tolerance. Trends Plant Sci. 2000;5:116–123. doi: 10.1016/s1360-1385(00)01567-3. [DOI] [PubMed] [Google Scholar]