Abstract
Reasons for the variable amylose content of endosperm starch from waxy cultivars of barley (Hordeum vulgare) were investigated. The mature grains of most such cultivars contain some amylose, although amounts are much lower than in wild-type cultivars. In these low-amylose cultivars, amylose synthesis starts relatively late in grain development. Starch granules in the outer cell layers of the endosperm contain more amylose than those in the center. This distribution corresponds to that of granule-bound starch synthase I (GBSSI), which is more severely reduced in amount in the center of the endosperm than in the outer cell layers, relative to wild-type cultivars. A second GBSSI in the barley plant, GBSSIb, is not detectable in the endosperm and cannot account for amylose synthesis in the low-amylose cultivars. The change in the expression of GBSSI in the endosperm of the low-amylose cultivars appears to be due to a 413-bp deletion of part of the promoter and 5′-untranslated region of the gene. Although these cultivars are of diverse geographical origin, all carry this same deletion, suggesting that the low-amylose cultivars have a common waxy ancestor. Records suggest a probable source in China, first recorded in the 16th century. Two further families of waxy cultivars have no detectable amylose in the endosperm starch. These amylose-free cultivars were selected in the 20th century from chemically mutagenized populations of wild-type barley. In both cases, 1-bp alterations in the GBSSI gene completely eliminate GBSSI activity.
The aim of this work was to investigate the reported variation in the amylose content of starch from the endosperm of waxy mutants of barley (Hordeum vulgare). Amylose is synthesized by granule-bound starch synthase I (GBSSI), an isoform of starch synthase of approximately 60 kD. GBSSI is encoded at the Waxy loci in cereals. In most cereal species, waxy mutants lack any detectable amylose in the starch of the endosperm. The major exception is barley, in which waxy mutant cultivars are reported to have between 0% and 13% amylose in their starch. For most such low-amylose cultivars, endosperm starch is reported to contain between 0.4% and 9% amylose (Banks et al., 1970; Morrison et al., 1986; McDonald et al., 1991; Song and Jane, 2000), but starch from a few cultivars has undetectable amylose (barley cv Yon M Kei, Ishikawa et al., 1995; barley cv CDC Alamo [line SB94794], Bhatty and Rossnagel, 1997). In this paper, the waxy mutants with detectable amylose will be referred to as low-amylose cultivars and those with undetectable amylose will be referred to as amylose-free.
In one low-amylose barley line (SW7142-92), the residual amylose has been shown to be concentrated in the outer layer of cells of the endosperm. Starch in the cells in this subaleurone layer stained blue-black with iodine solution, whereas that in the remainder of the endosperm stained red. The amylose contents of starch from tissues dissected from the outer and innermost parts of the grains of this cultivar were 8.6% and 2.2%, respectively (Oscarsson et al., 1997; Andersson et al., 1999).
Two possible explanations for the wide variation in amylose content of the starch of barley waxy mutants are suggested by recent studies of GBSSI. First, it has been shown that wheat (Triticum aestivum) possesses two isoforms of GBSSI with different spatial distributions in the plant. In developing wheat grains, one GBSSI isoform accounts for amylose synthesis in the endosperm and a second accounts for much of the amylose synthesis in the pericarp, aleurone, and embryo (Fujita and Taira, 1998; Nakamura et al., 1998; Vrinten and Nakamura, 2000). The pea (Pisum sativum) plant also has two, differently expressed isoforms of GBSSI (Denyer et al., 1997). It is likely that in barley, as in wheat, two isoforms of GBSSI are present and expressed in different tissues. However, if in barley the isoform expressed primarily in other parts of the plant was also expressed in the endosperm of barley in addition to the endosperm-specific isoform of GBSSI, then loss of either GBSSI could result in a low-amylose content. Loss of both forms of GBSSI from the endosperm would result in amylose-free starch.
Second, several independently derived waxy cultivars of barley have been shown to possess identical deletions in a GBSSI gene expressed in the endosperm—the only GBSSI gene thus far identified in barley. The deletion overlaps a TATA box, and reverse transcriptase (RT)-PCR failed to reveal any mRNA for GBSSI in endosperm of the mutant cultivars (Drescher et al., 2000). However, the data do not rule out the possibility that the deletion drastically reduces but does not eliminate expression of the gene. Thus, the presence of amylose in the endosperms of the low-amylose cultivars of barley might be explained by the widespread occurrence of a mutation that reduces but does not entirely eliminate expression of the gene encoding endosperm GBSSI.
To discover which, if either, of these explanations is correct, we have examined the occurrence and distribution of amylose and GBSSI protein(s), and the nature and expression of genes encoding GBSSI in the developing grains of low-amylose and amylose-free mutants of barley.
RESULTS
The Distribution of Amylose in the Endosperm
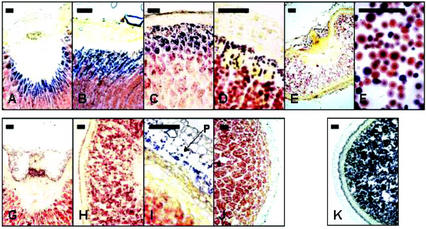
Iodine-stained sections of developing grain revealed great variation between waxy cultivars in the proportion of endosperm cells that contained significant amounts of amylose. In the amylose-free barley cv Yon M Kei and cv CDC Alamo, we observed no blue-staining granules at any stage of development (Fig. 1, G–I). Barley cv Arizona Hulless Waxy, a parent of CDC Alamo, also had no blue-staining granules (Fig. 1J); therefore, we consider it to be an amylose-free cultivar. In the low-amylose barley cv Iyatomi Mochi, cv Waxy Oderbrucker, cv Waxy Hector, and line SB85750 (the other parent of CDC Alamo; Bhatty and Rossnagel, 1997), no blue-staining granules were present in young endosperm (from grain up to about 20 mg fresh weight), but blue-staining granules appeared in outer cells of the endosperm during the later part of development (Fig. 1, A–F). In some of the low-amylose cultivars, blue-staining granules in the endosperm were largely confined to cells immediately adjacent to the groove (inside the basal endosperm transfer cell layer; Olsen et al., 1999), whereas in others, blue-staining starch granules were present in cells at the outer edge of the endosperm all around the grain (Fig. 1, A–D). In most cases, blue-staining granules were not confined to a single layer of cells. There was a gradation from the outer edge of the endosperm of cells with blue staining, through cells in which the peripheral region of the granule stained red and the core stained blue, to cells in which the entire granule stained red. In barley cv Waxy Hector, a low-amylose cultivar reported to have up to 8% amylose in its starch (Morrison et al., 1986), granules containing some amylose were present from early in development (in grains of less than 20 mg fresh weight). In more mature endosperms of barley cv Waxy Hector, most of the granules stained either completely or partly blue with iodine solution (Fig. 1F).
Figure 1.
A through K, Developing endosperm and starch from waxy barley mutants. For endosperm sections, whole grains were fixed in formaldehyde, embedded in wax, sectioned, and stained with iodine solution. All samples were taken from grain of 50 to 70 mg fresh weight (starting to turn yellow) except for those in E and G, which were from grain of approximately 20 mg fresh weight. Bars represent a distance of 50 μm. A through F, Low-amylose cultivars. G through H, Amylose-free cultivars. K, Wild-type cultivar. P, Pericarp. A, Barley cv Iyatomi Mochi. B, Barley cv Iyatomi Mochi. C, Barley cv Waxy Oderbrucker. D, Barley cv SB85750. E, Barley cv Iyatomi Mochi, young grain. F, Starch extracted from barley cv Waxy Hector. G, Barley cv Yon M Kei. H, Barley cv CDC Alamo. I, Barley cv CDC Alamo, young grain. J, Barley cv Arizona Hulless Waxy. K, Barley cv Shikoku Hadaka.
In all of the cultivars, including those with no amylose in the endosperm, starch in the pericarp (Fig. 1I) and in the embryo (not shown) stained blue with iodine at all of the developmental stages at which it was present.
The Presence of GBSSI Protein in the Endosperm
In all of the low-amylose cultivars, the starch contained a protein of approximately 60 kD, immunologically related to the GBSSI present in wild-type barley but present in very much lower concentrations than in wild-type starch (Fig. 2A). In the amylose-free waxy barley cv Yon M Kei, as reported previously (Ishikawa et al., 1995), no 60-kD protein was detectable (Fig. 2A). However, in the amylose-free barley cv CDC Alamo, the amount of the 60-kD protein was very similar to that in representative wild-type cultivars. Measurements of starch synthase activity associated with granules in developing endosperm of barley cv CDC Alamo suggested that most or all of this GBSSI protein was inactive. The activity was comparable with or lower than that of other waxy cultivars, and only about 10% of that of wild-type cultivars (data not shown). Much or all of this residual activity is likely to be due to isoforms of starch synthase other than GBSSI (Hylton et al., 1995).
Figure 2.
A and B, Presence of GBSSI-like proteins in starch from the developing endosperm of barley. After gelatinization by boiling in SDS-containing buffer, samples of starch from developing endosperms (from grains of approximately 50 mg fresh weight) were subjected to electrophoresis on 7.5% (w/v) SDS-polyacrylamide gels. Gels were either stained with Coomassie Brilliant Blue (right) or blotted onto nitrocellulose (left). Blots were developed with serum containing antibodies against GBSSI of pea embryos at a dilution of 1:2,000 (v/v; A) or 1:750 (v/v; B). A, Gel and blot of starch granule-bound proteins from whole endosperms. The phenotypes of the cultivars with respect to their amylose contents are indicated. WT, Wild type. LA, Low amylose. AF, amylose free. B, Gel and blot of starch from dissected outer layers and inner part of the endosperm of the low-amylose barley cv Iyatomi Mochi, and from whole endosperm of the wild-type barley cv Hector.
To study the distribution of the GBSSI-like 60-kD protein in the endosperms of a low-amylose cultivar, the outer layers were dissected away from the inner part of the maturing endosperm of barley cv Iyatomi Mochi. Starch extracted from the outer layers contained considerably more of the 60-kD protein than starch from the inner part (Fig. 2B).
Comparison of GBSSI Isoforms
To discover whether in barley, as in wheat, there is a second form of GBSSI, we searched for a barley expressed sequence tag (EST) similar to the nonendosperm form of GBSSI in wheat (GBSSII; Vrinten and Nakamura, 2000). A barley EST (accession no. AL508718) was identified and used to clone a cDNA of a second form of barley GBSSI, which we called GBSSIb (submitted to GenBank; accession no. AF486521). The predicted mature GBSSIb protein shares 96.4% identity with wheat GBSSII and 65.3% identity with barley GBSSI. Thus, in barley, as in wheat, there are two forms of GBSSI that are similar in amino acid sequence but differ particularly at the N termini of the mature proteins (Table I). To investigate whether GBSSIb is expressed in the outer cell layers of the endosperm of barley, we compared the predicted protein sequences of the two isoforms of GBSSI with protein sequences obtained experimentally from starch from wild-type and waxy barley endosperms.
Table I.
Comparison of the N-terminal sequences of GBSSI from wheat and barley
| Cultivar | N-Terminal Sequence | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Waxy Oderbrucker | A | T | G | S/ | — | M | N | L | V | F | (V) | G | |
| (N) | |||||||||||||
| Shikoku Hadaka | A | T | G | S | G | M | N | L | V | F | V | G | |
| Vogelsanger Gold | A | T | G | S | G | M | N | L | V | F | V | G | |
| Wheat GBSSI | A | T | G | S | G | G | M | N | L | V | F | V | G |
| Wheat GBSSII | S | T | G | M | P | I | I | F | V | A | |||
| Barley GBSSIb | S | T | G | M | P | I | I | F | V | A | |||
Sequences for Waxy Oderbrucker (low amylose) and Shikoku Hadaka (wild type) were obtained experimentally from GBSSI proteins purified from starch granules extracted from developing barley endosperm. The N-terminal sequence of the mature barley GBSSIb was predicted from the cDNA sequence (AF486521) using TargetP (Emanuelsson et al., 2000). The other sequences were reported previously (barley GBSSI, Vogelsanger Gold, Rohde et al., 1988; wheat GBSSI, Ainsworth et al., 1993; and wheat GBSSII, Nakamura et al., 1998). –, Identification was not possible. Parentheses indicate uncertainty.
GBSSI proteins in wild-type and low-amylose cultivars were compared by matrix-assisted laser-desorption ionization (MALDI)-time of flight (TOF) mass spectrometry (MS) and by N-terminal sequencing. MALDI-TOF MS was performed on tryptic digests of GBSSI purified from starch from the wild-type barley cv Hector and the low-amylose barley cv Waxy Hector and Iyatomi Mochi. For barley cv Hector, 13 peptides were identified that accounted for 29% of the amino acids in the mature GBSSI protein. For barley cv Waxy Hector, 16 peptides were identified that accounted for 32% of the amino acids in GBSSI. For barley cv Iyatomi Mochi, 14 peptides were identified that accounted for 32% of the amino acids in GBSSI. For all three samples, the best match of peptide masses obtained was to the amino acid sequence predicted from the barley GBSSI cDNA sequence (accession no. X07932; Rohde et al., 1988). These results are consistent with the idea that the GBSSI-like protein in the low-amylose barley cv Iyatomi Mochi and cv Waxy Hector is the product of the same gene that encodes the endosperm GBSSI in wild-type barley.
Protein sequencing revealed that the N-terminal 12 amino acids of the GBSSI protein from starch from the outer part of the endosperm of the low-amylose barley cv Waxy Oderbrucker matched the sequence of the GBSSI protein from the endosperm of the wild-type barley cv Shikoku Hadaka and cv Vogelsanger Gold (Table I). The sequences of these proteins were also very similar to that of the GBSSI expressed in wheat endosperm (Ainsworth et al., 1993; Taira et al., 1995). They differed considerably from the N-terminal sequences of the nonendosperm form of GBSSI in wheat (GBSSII) and barley (GBSSIb). These data again suggest strongly that the GBSSI in the endosperm of the low-amylose waxy cultivars of barley is the same protein as that in the endosperm of wild-type barley, rather than a different isoform expressed primarily in other parts of the plant.
Mutations in the GBSSI Gene of Waxy Barleys
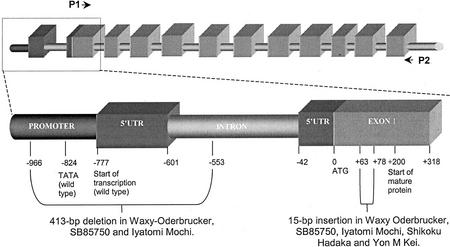
To provide further evidence about the identity of the GBSSI in the endosperm of waxy barley cultivars, we cloned and sequenced the cDNA encoding GBSSI and 1 kb of the promoter region of the GBSSI gene from a wild-type cultivar, barley cv Oderbrucker, and from several low-amylose (Waxy Oderbrucker, Iyatomi Mochi, and SB85750) and amylose-free (Yon M Kei and CDC Alamo) lines and cultivars. The sequence of GBSSI obtained from barley cv Oderbrucker (accession no. AF486514) was almost identical to that of the wild-type barley cv Vogelsanger Gold, published earlier (accession no. X07931). The 5′-untranslated region (UTR) of these wild-type alleles includes intron 1 (Fig. 3), and the region upstream of the 5′-UTR contains a predicted transcription complex-binding site (TATA box) 43 bp upstream of the transcription start site (Rohde et al., 1988).
Figure 3.
Diagrammatic representations of the gene structure of GBSSI. Upper diagram, The entire GBSSI gene. Blocks represent the 5′-UTR and the exons. Lower diagram, the 5′-UTR and exon 1 expanded to show the 413-bp deletion and 15-bp insertion. The sequences for the promoter and 5′-UTRs of GBSSI have been submitted to GenBank (accession nos.: barley cv Oderbrucker, AF486508; barley cv Waxy Oderbrucker, AF486509; barley cv Iatoma Mochi, AF486510; SB85750, AF486511; barley cv CDC Alamo, AF486512; and barley cv Yon M Kei, AF486513).
The GBSSI sequences for all of the low-amylose cultivars were very similar to one another and different from the sequences from wild-type barley in two main respects. First, in the GBSSI alleles from the low-amylose cultivars, there was a 413-bp deletion in the promoter and 5′-UTR including the TATA box, the start of transcription, and part of intron 1 (Fig. 3). Second, there was also a 15-bp insertion in exon 1 that does not cause a frame shift but results in the addition of five extra amino acids to the transit peptide of the protein. To discover more about the distribution of this 15-bp insertion among barley cultivars, we sequenced the same region from barley cv Shikoku Hadaka, from which the waxy barley cv Yon M Kei was derived. The 15-bp insertion was present in the GBSSI allele in this cultivar (Table II, column 1). Thus, the insert represents allelic variation that has little or no impact upon amylose content: It cannot be responsible for the low-amylose phenotype. We conclude that the reduction in amylose content in low-amylose cultivars is probably due to the 413-bp deletion that is common to all of the cultivars of this type that we have examined.
Table II.
Comparisons of the cDNA and predicted protein sequences of GBSSI from barley
| Group 1 | 56 78 | 100 121 | 178 186 | 205 213 | 562 570 | 856 864 |
| Vogelsanger Gold | ACCGACAGA . . . . . . TTCCGGCGTCCAGGT | AACCCGGCGGATGCGGCGCTT | GGGAGCCGG | GTGAGCGCC | CAGCAGCGC | ATTGACGGC |
| (X07932) | T D R . . . . F R R P G | N P A D A A L | G S R | V S A | Q Q R | I D G |
| CDC Alamo | ACCGACAGA . . . . . . TTCCGGCGTCCAGGT | AACCCGGCGGATGCGGCGCTT | GGGAGCCGG | GTGAGCGCC | CAGCAGCGC | ATTGTCGGC |
| (AF486519) | T D R . . . . F R R P G | N P A D A A L | G S R | V S A | Q Q R | I V G |
| Oderbrucker | ACCGACAGA . . . . . . TTCCGGCGTCCAGGT | AACCCGGCGGATGCGGCGCTT | GGGAGCCGG | GTGAGCGCC | CAGCAGCGC | ATTGACGGC |
| (AF486514) | T D R . . . . F R R P G | N P A D A A L | G S R | V S A | Q Q R | I D G |
| Group 2 | 56 93 | 115 136 | 193 101 | 220 228 | 577 585 | 871 879 |
| Iyatomi Mochi | ACCGACAGGTCGGCGCCGTCCATGTTCCGGCATGCTGGT | AAGCCCGCAGATGGGACGTTT | GGGAACCGG | GTGCGTGCC | CAGCAGCGC | ATTGACGGC |
| (AF486517) | T D R S A P S M F R E A G | K P A D G T F | G N R | V R A | Q Q R | I D G |
| Waxy Oderbrucker | ACCGACAGGTCGGCGCCGTCCATGTTCCGGCATGCTGGT | AAGCCCGCAGATGGGACGTTT | GGGAACCGG | GTGCGTGCC | CAGCAGCGC | ATTGACGGC |
| (AF486515) | T D R S A P S M F R H A G | K P A D G T F | G N R | V R A | Q Q R | I D G |
| SB85750 | ACCGACAGGTCGGCGCCGTCCATGTTCCGGCATGCTGGT | AAGCCCGCAGATGGGACGTTT | GGGAACCGG | GTGCGTGCC | CAGCAGCGC | ATTGACGGC |
| (AF486518) | T D R S A P S M F R H A G | K P A D G T F | G N R | V R A | Q Q R | I D G |
| Shikoku Hadaka | ACCGACAGGTCGGCGCCGTCCATGTTCCGGCATGCTGGT | AAGCCCGCAGATGGGACGTTT | GGGAACCGG | GTGCGTGCC | CAGCAGCGC | ATTGACGGC |
| (AF486520) | T D R S A P S M F R H A G | K P A D G T F | G N R | V R A | Q Q R | I D G |
| Yon M Kei | ACCGACAGGTCGGCGCCGTCCATGTTCCGGCATGCTGGT | AAGCCCGCAGATGGGACGTTT | GGGAACCGG | GTGCGTGCC | CAGTAGCGC | ATTGACGGC |
| (AF486516) | T D R S A P S M F R E A G | K P A D G T F | G N R | V R A | Q * R | I D G |
Sequences of six regions of GBSSI cDNA from different barley cultivars are compared in the six columns. The GenBank accession nos. for the cDNA sequences are given in parentheses. The positions of the bases relative to the start of translation are indicated. Bases in the cDNA sequences that vary are underlined. Amino acids in the protein sequences that vary are shown in bold. The stop codon in the Yon M Kei protein sequence is indicated by an asterisk.
The GBSSI sequence from the amylose-free barley cv CDC Alamo was identical to that of the wild-type barley cv Vogelsanger Gold except for one base substitution (T instead of A) at position 860 in barley cv CDC Alamo. This is predicted to result in the substitution of the aliphatic amino acid (Val) for the acidic amino acid (Asp). Asp is conserved in this position in all of the other GBSSI alleles of barley (Table II) and in GBSSI from a wide range of other species (data not shown). This single base change in barley cv CDC Alamo is likely to be the cause of the observed production of an inactive GBSSI protein in this cultivar (see above). Thus, it defines an Asp that is essential for GBSSI activity. The sequence of GBSSI from the amylose-free barley cv Yon M Kei also contained a single base substitution compared with wild-type sequences, at position 580 (T instead of C). This is predicted to create a stop codon, and thus is likely to be responsible for the complete lack of GBSSI protein that was observed in this mutant.
In addition to the 15-bp insertion in the GBSSI gene already mentioned (Fig. 3), there are other sequence differences between the GBSSI alleles for which we have complete sequences. These are summarized in Table II. These minor differences are not correlated with amylose content and therefore are unlikely to contribute to the waxy phenotype. However, they represent at least some of the allelic variation in GBSSI that exists within cultivated barleys. On the basis of cDNA sequence comparisons, the GBSSI alleles can be divided into two groups representing two haplotypes. Group 1 contains GBSSI alleles from the wild-type barley cv Vogelsanger Gold and cv Oderbrucker and the amylose-free barley cv CDC Alamo. All of the members of this group lack the 15-bp insertion and vary from the group 2 alleles in 10 other positions (Table II, columns 1–4). Group 2 contains GBSSI alleles from the wild-type barley cv Shikoku Hadaka, the low-amylose lines/cultivars, Iyatomi Mochi, Waxy Oderbrucker, and SB85750, and the amylose-free barley cv Yon M Kei. All of the members of this group have the 15-bp insertion.
Comparison of GBSSI Transcripts
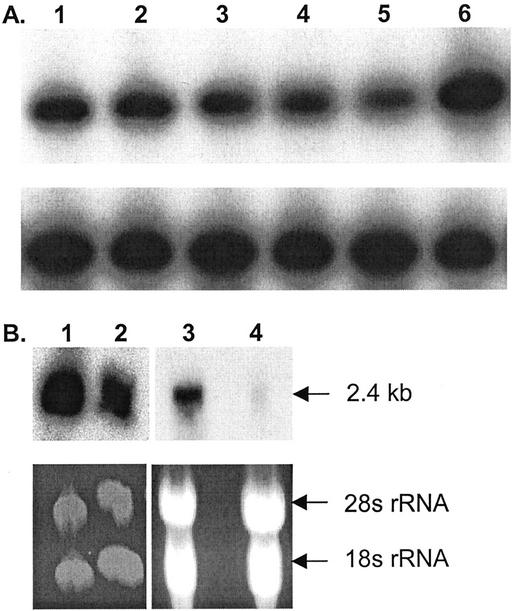
The relative amounts of GBSSI transcripts in developing endosperms of wild-type and waxy barleys were compared in two ways. First, semiquantitative RT-PCR (Fig. 4A) showed that the low-amylose line/cultivars Waxy Oderbrucker, Iyatomi Mochi, and SB85750 all had normal or only slightly reduced levels of GBSSI transcript in endosperms from grains of 30 to 45 mg fresh weight The amylose-free barley cv Yon M Kei (containing a GBSSI allele with an introduced stop codon in the coding region) also had a normal level of transcript. However, the low-amylose barley cv CDC Alamo, which had a normal amount of an inactive form of GBSSI protein, had elevated levels of transcript. Second, the transcript levels in barley cv Oderbrucker and cv Waxy Oderbrucker at two developmental stages were compared by northern analysis (Fig. 4B). In the older endosperms (from grains of 30–45 mg fresh weight), the transcripts in these cultivars were of the expected size and were of similar abundance. The GBSSI transcript was also abundant in young endosperms (from grains of 12–16 mg fresh weight) of barley cv Oderbrucker. However, in young endosperms of barley cv Waxy Oderbrucker, there was very little, if any, transcript.
Figure 4.
A and B. Comparison of GBSSI transcripts. A, Semiquantitative RT-PCR. RNA was extracted from endosperms of grains of 30 to 45 mg fresh weight Tracks are: 1, barley cv Oderbrucker; 2, barley cv Waxy Oderbrucker; 3, barley cv Yon M Kei; 4, barley cv Iyatomi Mochi; 5, SB85750; and 6, barley cv CDC Alamo. Upper, Product generated using primers designed to amplify GBSSI. Lower, Product generated using primers designed to amplify Mub-1 (ubiquitin). B, Northern blots. Tracks are: 1 and 3, barley cv Oderbrucker; and 2 and 4, barley cv Waxy Oderbrucker. RNA was extracted from endosperms of grains of 30 to 45 mg fresh weight was in tracks 1 and 2 and RNA from grains of 14 to 16 mg fresh weight was in tracks 3 and 4. Upper, Products generated using primers designed to amplify GBSSI. The approximate size of the GBSSI transcript is indicated. Lower, Ethidium bromide-stained gels used to prepare the blots shown in the upper panels.
Attempts to map the transcription start site for GBSSI in the low-amylose mutants and to search for alternative transcription start sites in the 5′-upstream sequences have been unsuccessful so far. We assume that there is an alternative transcription start site either in the remaining part of intron 1 or further upstream. This results in a longer pre-RNA that is spliced to give a mature RNA of similar size to the mature RNA of the wild type.
DISCUSSION
Our results suggest that there is only one GBSSI gene expressed in barley endosperm and that this is responsible for the amylose synthesized in low-amylose waxy cultivars as well as that in wild-type cultivars. MALDI-TOF analysis and amino acid sequence from the GBSSI protein in the endosperm of these waxy cultivars show that this protein is indistinguishable from the GBSSI of wild-type endosperms. The protein is different from the predicted product of a second GBSSI gene, GBSSIb. GBSSIb, like the homologous gene in wheat (GBSSII, Vrinten and Nakamura, 2000), is probably expressed in parts of the plant other than the endosperm. It is likely to be responsible for the synthesis of the amylose in the pericarp, including that observed in pericarps of the amylose-free cultivars.
The GBSSI alleles of all of the low-amylose waxy barleys we examined carry a 413-bp deletion in the promoter and 5′-UTR. This discovery strongly suggests that the alleles in all of these cultivars are derived from a single origin. The cultivars we examined are extremely diverse in phenotype and geographical origin. They include cultivars bred in Japan, Canada, Europe, and the United States, and a wide range of awn, row, pigment, hull, and growth habit characteristics. Nonetheless, records on the origins of the cultivars are consistent with the idea that the waxy characteristic in most or all of these barleys is derived from waxy barleys native to Asia (Takahashi, 1955). The Asian waxy barleys are probably all descended from a glutinous (waxy) form of barley recorded in China in the 16th century. Glutinous barleys are believed to have been introduced into Japan from Korea at some time before the 17th century (Takahashi, 1955). The European and North American waxy barleys stem from a U.S. Department of Agriculture-Agricultural Research Service (USDA-ARS) breeding program in Aberdeen, Idaho (1940–1950) in which a Japanese cultivar, barley cv Murasaki mochi, was crossed to a European cultivar, barley cv Oderbrucker (Harold Bockelman, USDA-ARS, personal communication). The resulting barley cv Waxy Oderbrucker was then used in a breeding program in the United States and Canada. We can trace many of the modern waxy cultivars back to barley cv Waxy Oderbrucker. For example, it was a parent of barley cv Waxy Betzes (Fox, 1981), which was then used as a parent in the breeding program in Saskatchewan from which the line SB85750 was derived. Barley cv Waxy Hector was also derived from either cv Waxy Betzes or cv Waxy Oderbrucker.
Our analysis indicates that in the low-amylose barley mutants, the 413-bp deletion alters the spatial and/or temporal expression of GBSSI in the endosperm. The deletion removes the normal transcription complex-binding site (TATA box) and transcription start site. We assume that an alternative, upstream promoter region and transcription start site are used to produce the GBSSI transcript in the mutants. The mutant transcript is produced only late in endosperm development, consistent with the appearance of amylose only later in endosperm development. The fact that amylose and GBSSI protein are found mainly or exclusively in the outer cells of the endosperm indicates that the alternative promoter specifies a different spatial and/or temporal pattern of expression from the normal promoter.
Effects on amylose content of mutations in the promoter or 5′-UTR of GBSSI have also been reported in other species. In rice, variations in amylose content between cultivars were shown to be due to differences in the efficiency with which intron 1 in the 5′-UTR was removed from the GBSSI pre-mRNA (Wang et al., 1995; Bligh et al., 1998). Low-amylose (6.7%–16.0% amylose) cultivars of rice had lower levels of mature GBSSI mRNA than high-amylose (20.0%–27.8% amylose) cultivars as well as incompletely processed GBSSI mRNA. In these cultivars, the inefficient processing of the pre-mRNA was due to a single base mutation at the 5′ splice site of intron 1. The reduced efficiency of GBSSI pre-mRNA processing also resulted in alternate splicing at multiple sites, some of which had non-consensus splice site sequences. Growth temperature can also affect the efficiency of pre-mRNA processing in low-amylose cultivars of rice. Plants grown at 18°C had higher steady-state levels of mature GBSSI mRNA than plants grown at 25 or 32°C (Larkin and Park, 1999). At lower temperatures, when splicing was more efficient, the activity of GBSSI in the low-amylose cultivars was higher (Umemoto et al., 1995) and more amylose was synthesized (Hirano and Sano, 1998).
In potato (Solanum tuberosum), GBSSI activity and amylose content are affected by the presence or absence of a 140-bp fragment at a site in the promoter region approximately 0.5 kb upstream of the ATG start codon (van de Wal et al., 2001). Alleles of the gene that contain the 140-bp fragment result in lower GBSSI activity and amylose content than alleles without this fragment. The basis for this effect is not known because the variations in GBSSI activity could not be attributed to large differences in the amounts of either GBSSI RNA or protein.
The highly variable amylose content of starch granules from low-amylose cultivars of barley may be an important consideration in assessing the physicochemical properties of starch. Individual endosperms contain granules that stain blue with iodine that probably have near-normal levels of amylose, granules with blue-staining cores that are likely to have a severely reduced amylose content, and granules that stain almost completely red and probably have near-zero amylose contents. This contrasts with the situation in low-amylose starches from other species. For example, in the low-amylose lines of potato created by expression of antisense GBSSI constructs (Kuipers et al., 1994; Tatge et al., 1999), there was less granule-to-granule variation in amylose content than in the low-amylose barley cultivars. Whether the physicochemical properties of low-amylose starches of comparable bulk amylose content are influenced by the extent of heterogeneity of amylose contents between individual granules remains to be determined.
Two of the waxy cultivars with no amylose in the endosperm, barley cv Arizona Hulless Waxy and cv Yon M Kei, were produced by mutagenesis of wild-type cultivars rather than by breeding from low-amylose cultivars traceable to Japan (see below). Barley cv Yon M Kei was generated by mutagenesis of the wild-type barley cv Shikoku Hadaka with sodium azide (Ishikawa et al., 1995). Barley cv Arizona Hulless Waxy was generated by mutagenesis of a wild-type line, 76-19-7, with diethyl sulfate (PI 560053; USDA-ARS, National Genetic Resources Program, 1991). The third amylose-free cultivar, barley cv CDC Alamo, was derived by breeding from barley cv Arizona Hulless Waxy, and probably carries the same GBSSI allele as this parent (Bhatty and Rossnagel, 1997). Barley cv Yon M Kei and cv CDC Alamo do not have the 413-bp deletion seen in the low-amylose cultivars. Instead, they have different mutations in the GBSSI gene that account for the complete absence of GBSSI activity. The endosperm of barley cv Yon M Kei contains no detectable GBSSI protein. There is a single base substitution in this gene that creates a stop codon. This prevents the production of a full-length GBSSI protein. Presumably, the incomplete GBSSI protein is either unstable and is rapidly degraded or it is not capable of binding to the starch granules and, therefore, would not have been detected in our experiments. Barley cv CDC Alamo has wild-type levels of GBSSI protein. The mutant protein is able to bind tightly to starch granules like normal GBSSI, but it is unable to synthesize amylose due to a single base substitution that results in a conserved Asp residue (Asp-217) being replaced by a Val. It is known that enzymes in the glycosyltransferase family of which GBSSI is a member have Asp residues at the catalytic center that participate in the enzymatic reaction (Tarbouriech et al., 2001). Mutational analysis of several glycosyltransferases, including an isoform of starch synthase, has shown an absolute requirement for certain conserved Asp residues (for example, cellulose synthase, Saxena and Brown, 1997; chitin synthase 2, Nagahashi et al., 1995; and starch synthase IIb, Nichols et al., 2000). Substitution of these—even conservative substitution with similar amino acids—results in inactive enzymes. However, whether the conserved Asp-217 is at the catalytic center of GBSSI remains to be discovered.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Grains of barley (Hordeum vulgare) cultivars were obtained from the John Innes Centre and Crop Development Centre Germplasm collections, from Dr. Tom Blake (Montana State University, Bozeman, barley cv Nubet), and from Dr. Naoyuki Ishikawa (Tochigi Agricultural Experiment Station, Tochigi, Japan, barley cv Shikoku Hadaka 84 and cv Yon M Kei 286). Barley plants were grown in a greenhouse in individual pots at a minimum temperature of 12°C, with supplementary lighting in winter.
Extraction of Starch
Endosperms, free of pericarp and embryos, were dissected from developing grains of 50 mg fresh weight. For starch extraction from the inner and outer layers of the endosperm, whole endosperms were frozen on dry ice, then “peeled” with a fine razor blade to remove tissue to a depth of very approximately 0.5 mm from the surface. Starch was extracted as described by Hylton et al. (1995) and stored at −20°C.
SDS-PAGE and Immunoblotting
Starch samples were washed twice by suspension and centrifugation in 2% (w/v) aqueous SDS, at 30 mg starch mL−1. Washed starch was suspended in gel sample buffer (Hylton et al., 1995) at 50 mg starch mL−1, boiled for 5 min, and allowed to cool. Unlike the wild-type starches, centrifugation of the starches from the waxy lines did not result in a supernatant. Therefore, none of the samples were centrifuged after boiling. Instead, approximately 30 μL of the resulting paste was loaded directly into wells on 7.5% (w/v) SDS-polyacrylamide gels (10 cm long, 1 mm thick). Due to the starch in the samples, which remained in the wells, there was some unavoidable distortion of the protein bands on the gel (see Fig. 2). After electrophoresis, gels were stained with Coomassie Brilliant Blue R or electroblotted onto nitrocellulose. Blots were developed with serum containing antibodies against GBSSI of pea (Pisum sativum) embryos (Smith, 1990) at a dilution of 1:2,000 (v/v), as described by Denyer et al. (1997).
Identification of GBSSI Proteins by MALDI-TOF and by Sequencing
Starch granule-bound proteins were subjected to SDS-PAGE and the separated proteins were stained with Coomassie Brilliant Blue R-250. The major, approximately 60-kD, protein bands were excised and subjected to tryptic digestion according to Speicher (2000) followed by analysis by MALDI-TOF MS. Mass fragment sizes were used to query the National Center for Biotechnology Information database using the MASCOT search tool (http://www.matrixscience.com). All matched sequences showed mass errors of <75 μL L−1.
Protein was blotted from an SDS polyacrylamide gel onto a polyvinylidene difluoride membrane (Immobilon P, Millipore, Bedford, MA) and stained with Coomassie Brilliant Blue R250. The excised band was sequenced directly from the membrane by Edman degradation on a model 494 Procise protein sequencer (PE-Applied Biosystems, Foster City, CA) using the pulsed-liquid mode.
Light Microscopy
Freshly harvested tissue was fixed in a formaldehyde solution and dehydrated through a graded ethanol series according to Johnson et al. (1994). After transfer to Histoclear (Agar Scientific, Stansted, Essex, UK), tissue was embedded in Paramat wax (BDH, Poole, UK), and sectioned. Wax was removed with Histoclear and sections were transferred through an ethanol series into water and photographed through a light microscope after staining with iodine solution (dilutions of 2- or 5-fold of Lugol's solution; Sigma, Poole, UK). To improve contrast, red/brown and blue colors in photographs were enhanced using Adobe Photoshop software (Adobe Systems, Mountain View, CA).
DNA Extraction and the Cloning and Sequencing of Alleles of GBSSI
DNA was extracted from 0.1-g samples of young barley leaves with the DNeasy plant mini-kit (Qiagen, Hilden, Germany). The promoters were amplified using Pfu-Turbo DNA polymerase (Stratagene, La Jolla, CA) with primers designed to GBSSI from barley cv Vogelsanger Gold (X07931) forward (5′-TATATGACGCACTCCACACCCACACACACA-3′) and reverse (5′-CTGTTCCTGAAATCTAAGATCGTTTGCAGA-3′). The blunt-ended PCR products had adenine residues added by incubation with deoxyadenylate triphosphate and Taq polymerase at 72°C. The products were then ligated into pGEM-T-easy vector (Promega, Madison, WI) for sequencing. Overlapping sequencing primers were designed at 400-bp intervals.
RNA Extraction and DNA Synthesis
RNA was extracted from 0.5 g of endosperms from grains of 30 to 45 mg each or from 21 whole grains of 12 to 16 mg each using Concert RNA reagent (Invitrogen Ltd., Paisley, UK). RNA was treated with DNaseI (Roche, Basel) and cleaned again with phenol:chloroform. cDNA was synthesized at 58°C from a reverse primer designed to Vogelsager Gold cDNA (X07932; P2, 5′-TGCTCCATGCACCAGAATGT-3′) using Thermoscript RT (Invitrogen Ltd.). The enzyme was denatured at 85°C and RNA removed from the duplex by incubation at 37°C with RNase H (Roche).
RT-PCR
RT-PCR with the cDNA synthesis primer (P2) and a forward primer (P1, 5′-TGCTCTCTCACTGCAGGTAG-3′) was done using Pfx polymerase or Platinum Taq DNA polymerase and PCRx reaction buffer (Invitrogen Ltd.). Semiquantitative RT-PCR was done in tandem with primers to ubiquitin (mub1-M60175; 5′-CGGACACCATCGACAACGTCCAG-3′ and 5′-GCCA-GTTCTAAGCCTTCTGGTTGTAG-3′). PCR cycles were paused after 15 cycles and 5-μL aliquots removed. Products were separated on 1% (w/v) agarose gel and transferred to Duralon nylon membrane (Stratagene) by capillary transfer. Membranes were hybridized in phosphate buffer (pH 7.4), 10 mm EDTA, and 7% (w/v) SDS with 25 ng of 32P[α-dCTP]-labeled GBSSI-cDNA or ubiquitin-cDNA amplified from Oderbrucker, for 4 h at 65°C and washed with 0.1× SSC containing 0.1% (w/v) SDS.
Cloning and Sequencing of GBSSIb
RNA was extracted from the entire seeds of barley cv Nubet 3 DPA. Ten micrograms of total RNA was used to synthesize cDNA using the Generacer Kit (Invitrogen Ltd.). RACE products amplified with primers designed to EST AL508718 (5′-TCCTACAACTGGAACAGACTTCCGAGATAA-3′ and 5′-ACGGTTCTGCTTTTGTGCTTGCTGCATT-3′) were cloned into the TOPO-10 vector (Invitrogen Ltd.) and sequenced.
Northern Blotting
Ten micrograms of total RNA was separated on a 1% (w/v) agarose denaturing gel with size standards (Promega). RNA was visualized with ethidium bromide under UV light to ensure equal loading. RNA was transferred to Duralon nylon membrane (Stratagene) by capillary transfer. Hybridization conditions were identical to those used for semiquantitative RT-PCR.
ACKNOWLEDGMENTS
The authors are very grateful to Dr. Naoyuki Ishikawa (Tochigi Agricultural Experiment Station) and Dr. Tom Blake (Montana State University) for the gift of grains; to Dr. Harold Bockelman (USDA-ARS, Aberdeen, ID), Dr. Walter Newman (Montana State University), Dr. Allan Simons (Arizona Crop Improvement Association, Tucson), and Dr. Dale Clark (Western Plant Breeders, Bozeman, MT) for helpful discussions; and to Dr. David Laurie (John Innes Centre, Norwich, UK) for constructive criticism of the manuscript.
Footnotes
This work was supported by the Biotechnology and Biological Sciences research Council (UK; competitive strategic grant to the John Innes Centre).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.005454.
LITERATURE CITED
- Ainsworth C, Clark J, Balsdon J. Expression, organisation and structure of the genes encoding the waxy protein (granule-bound starch synthase) in wheat. Plant Mol Biol. 1993;22:67–82. doi: 10.1007/BF00038996. [DOI] [PubMed] [Google Scholar]
- Andersson L, Fredriksson H, Bergh MO, Andersson R, Åman P. Characterisation of starch from inner and peripheral parts of normal and waxy barley kernels. J Cereal Sci. 1999;30:165–171. [Google Scholar]
- Banks W, Greenwood CT, Walker JT. Studies on the starches of barley genotypes: the waxy starch. Stärke/Starch. 1970;22:149–180. [Google Scholar]
- Bhatty RS, Rossnagel BG. Zero amylose lines of hulless barley. Cereal Chem. 1997;74:190–191. [Google Scholar]
- Bligh HFJ, Larkin PD, Roach PS, Jones CA, Fu H, Park WD. Use of alternate splice sites in granule-bound starch synthase mRNA from low-amylose rice varieties. Plant Mol Biol. 1998;38:407–415. doi: 10.1023/a:1006021807799. [DOI] [PubMed] [Google Scholar]
- Denyer K, Barber LM, Edwards EA, Smith AM, Wang TL. Two isoforms of the GBSSI class of granule-bound starch synthase are differentially expressed in the pea plant. Plant Cell Environ. 1997;20:1566–1572. [Google Scholar]
- Drescher A, Schreiber H, Habekuss A. Mutations at the waxy locus of different barleys and their influence on starch synthesis. Barley Genetics VIII. 2000;3:142–144. [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Fox GJ. The effect of the waxy endosperm, short awn, and hulless seed genes upon biochemical and physiological seed characteristics important in barley (Hordeum vulgare L.). PhD thesis. Bozeman: Montana State University; 1981. [Google Scholar]
- Fujita N, Taira T. A 56-kD protein is a novel granule-bound starch synthase existing in the pericarps, aleurone layers, and embryos of immature seed in diploid wheat (Triticum monococcum L.) Planta. 1998;207:125–132. doi: 10.1007/s004250050464. [DOI] [PubMed] [Google Scholar]
- Hirano H-Y, Sano Y. Enhancement of Wx gene expression and the accumulation of amylose in response to cool temperatures during seed development in rice. Plant Cell Physiol. 1998;39:807–812. [Google Scholar]
- Hylton CM, Denyer K, Keeling PL, Chang M-T, Smith AM. The effect of waxy mutations on the granule-bound starch synthases of barley and maize endosperms. Planta. 1995;198:230–237. [Google Scholar]
- Ishikawa N, Ishihara J, Itoh M. Artificial induction and characterization of amylose-free mutants of barley. Barley Genet Newslett. 1995;24:49–53. [Google Scholar]
- Johnson S, Liu CM, Hedley CL, Wang TL. An analysis of seed development in Pisum sativum: XVIII. The isolation of mutants defective in embryo development. J Exp Bot. 1994;45:1503–1511. [Google Scholar]
- Kuipers AGJ, Jacobsen E, Visser RGF. Formation and deposition of amylose in the potato tuber are affected by the reduction of granule-bound starch synthase gene expression. Plant Cell. 1994;6:43–52. doi: 10.1105/tpc.6.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin PD, Park WD. Transcript accumulation and utilization of alternate and non-consensus splice sites in rice granule-bound starch synthases are temperature-sensitive and controlled by a single-nucleotide polymorphism. Plant Mol Biol. 1999;40:719–727. doi: 10.1023/a:1006298608408. [DOI] [PubMed] [Google Scholar]
- McDonald AML, Stark JR, Morrison WR, Ellis RP. The composition of starch granules from developing barley genotypes. J Cereal Sci. 1991;13:93–112. [Google Scholar]
- Morrison WR, Scott DC, Karkalas J. Variation in the composition and physical properties of barley starches. Stärke/Starch. 1986;38:374–379. [Google Scholar]
- Nagahashi S, Sudoh M, Ono N, Sawada R, Yamaguchi E, Uchida Y, Mio T, Takagi M, Arisawa M, Yamada-Okabe H. Characterization of chitin synthase 2 of Saccharomyces cerevisiae. Implication of two highly conserved domains as possible catalytic sites. J Biol Chem. 1995;270:13961–13967. doi: 10.1074/jbc.270.23.13961. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Vrinten P, Hayakawa K, Ikeda J. Characterization of a granule-bound starch synthase isoform found in the pericarp of wheat. Plant Physiol. 1998;118:451–459. doi: 10.1104/pp.118.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols DJ, Keeling PL, Spalding M, Guan H. Involvement of conserved aspartate and glutamate residues in the catalysis and substrate binding of maize starch synthase. Biochem. 2000;39:7820–7825. doi: 10.1021/bi000407g. [DOI] [PubMed] [Google Scholar]
- Olsen O-A, Linnestad C, Nichols SE. Developmental biology of the cereal endosperm. Trends Plant Sci. 1999;4:253–257. doi: 10.1016/s1360-1385(99)01431-4. [DOI] [PubMed] [Google Scholar]
- Oscarsson M, Parkkonen T, Autio K, Åman P. Composition and microstructure of waxy, normal and high amylose barley samples. J Cereal Sci. 1997;26:259–264. [Google Scholar]
- Rohde W, Becker D, Salamini F. Structural analysis of the waxy locus from Hordeum vulgare. Nucleic Acids Res. 1988;16:7185–7186. doi: 10.1093/nar/16.14.7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena IM, Brown RM. Identification of cellulose synthase(s) in higher plants: sequence analysis of processive β-glycosyltransferases with the common motif “D,D,D35Q(R,Q)XRW.”. Cellulose. 1997;4:33–49. [Google Scholar]
- Smith AM. Evidence that the “waxy” protein of pea is not the major starch-granule-bound starch synthase. Planta. 1990;182:599–604. doi: 10.1007/BF02341037. [DOI] [PubMed] [Google Scholar]
- Song Y, Jane J. Characterization of barley starches of waxy, normal and high amylose varieties. Carbohydr Polymers. 2000;41:365–377. [Google Scholar]
- Speicher KD. Systematic analysis of peptide recoveries from in-gel digestions for protein identifications in proteome studies. J Biomol Technol. 2000;11:74–86. [PMC free article] [PubMed] [Google Scholar]
- Taira T, Fujita N, Takaoka K, Uematsu M, Wadano A, Kozaki S, Okabe S. Variation in the primary structure of waxy proteins (granule-bound starch synthase) in diploid cereals. Biochem Genetics. 1995;33:269–281. [PubMed] [Google Scholar]
- Takahashi R. The origin and evolution of cultivated barley. Adv Genetics. 1955;7:227–266. [Google Scholar]
- Tarbouriech N, Charnock SJ, Davies GJ. Three-dimensional structures of the Mn and MgdTDP complexes of the family GT-2 glycosyltransferase SpsA: a comparison with related NDP-sugar glycosyltransferases. J Mol Biol. 2001;314:655–661. doi: 10.1006/jmbi.2001.5159. [DOI] [PubMed] [Google Scholar]
- Tatge H, Marshall J, Martin C, Edwards EA, Smith AM. Evidence that amylose synthesis occurs within the matrix of the starch granule in potato tubers. Plant Cell Environ. 1999;22:543–550. [Google Scholar]
- Umemoto T, Nakamura Y, Ishikura N. Activity of starch synthase and the amylose content in rice endosperm. Phytochemistry. 1995;40:1613–1616. [Google Scholar]
- USDA-ARS, National Genetic Resources Program. PI 560053. Germplasm Resources Information Network (GRIN)-Online database www.ars-grin.gov. Beltsville, MD: National Germplasm Resources Laboratory; 1991. [Google Scholar]
- van de Wal MHBJ, Jacobsen E, Visser RGF. Multiple allelism as a control mechanism in metabolic pathways: GBSSI allelic composition affects the activity of granule-bound starch synthase I and starch composition in potato. Mol Genet Genomics. 2001;265:1011–1021. doi: 10.1007/s004380100496. [DOI] [PubMed] [Google Scholar]
- Vrinten P, Nakamura T. Wheat granule-bound starch synthase I and II are encoded by separate genes that are expressed in different tissues. Plant Physiol. 2000;122:255–263. doi: 10.1104/pp.122.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z-Y, Zheng F-Q, Shen G-Z, Goa J-P, Snustad DP, Li M-G, Zhang J-L, Hong M-M. The amylose content in rice endosperm is related to the post-transcriptional regulation of the waxy gene. Plant J. 1995;7:613–622. doi: 10.1046/j.1365-313x.1995.7040613.x. [DOI] [PubMed] [Google Scholar]