Abstract
To further our understanding of how plants defend against the harmful effects of ultraviolet (UV) light, we characterized an Arabidopsis mutant hypersensitive to UV-B. This mutant, UV resistance locus 8-1 (uvr8-1), contains a single recessive mutation at the bottom of chromosome 5. Fine-scale mapping localized uvr8-1 to a 21-kb locus containing five predicted open reading frames. Sequencing of this entire region revealed that the uvr8-1 allele contains a 15-nucleotide deletion in a gene similar to the human guanine nucleotide exchange factor regulator of chromatin condensation 1. This mutation reduces the UV-B-mediated induction of flavonoids and blocks chalcone synthase mRNA and protein induction. In contrast, uvr8-1 has enhanced induction of PR1 and PR5 proteins in response to UV-B, an indication of increased UV-B injury. These results suggest that UVR8 acts in a UV-B signal transduction pathway leading to induction of flavonoid biosynthesis.
Plants must resist the deleterious effects of UV light because they are dependent on sunlight for photosynthesis and cannot avoid UV light exposure. Although UV is defined as the region of the spectrum from 200 to 400 nm, only the levels of UV-B (280–320 nm) reaching the earth's surface are increased by the thinning of the stratospheric ozone layer (Caldwell et al., 1989; Frederick et al., 1989; Stolarski et al., 1992; Kerr and McElroy, 1993). Thus, studies have focused on UV-B tolerance mechanisms because plants are directly affected by changes in terrestrial UV-B fluence.
UV-B is known to cause DNA damage predominantly through cyclobutyl pyrimidine dimer formation and, to a lesser extent, pyrimidine-pyrimidinone (6, 4) photoproducts, both of which form by covalent bonding of adjacent pyrimidines (for review, see Britt, 1995; Landry et al., 1997; Nakajima et al., 1998). Failure to repair these DNA lesions interferes with DNA synthesis and transcription, and can result in heritable mutations (for review, see Britt, 1995). Studies of Arabidopsis have identified a variety of UV-B-hypersensitive mutants deficient in DNA repair (uvr1 [Britt et al., 1993], uvr2 [Jiang et al., 1997; Landry et al., 1997], uvr3 [Jiang et al., 1997; Nakajima et al., 1998], and uvh1 [Harlow et al., 1994]). For example, photolyases, enzymes that use blue light energy to repair pyrimidine dimers (Todo et al., 1993; Sancar, 1994), are critical for plant survival under UV-B in the laboratory (Ahmad et al., 1997; Landry et al., 1997; Nakajima et al., 1998). Other light-independent DNA repair mechanisms in plants are currently under study.
In addition to directly causing DNA damage, UV-B generates oxidative stress through the formation of reactive oxygen species (ROS; Strid, 1992; Krizek et al., 1993 ; Doke et al., 1994; Foyer et al., 1994b), which in turn causes enhanced lipid and protein oxidation (Kramer et al., 1991; Landry et al., 1995). Plants counteract this increased ROS by increasing antioxidant enzymes (Foyer et al., 1994a; Kangasjarvi et al., 1994). For example, exposure to UV-B induces guaiacol-peroxidases, ascorbate peroxidases, cytosolic Cu/Zn-superoxide dismutase (SOD), and coniferyl alcohol peroxidases (Rao et al., 1996; Kliebenstein et al., 1998; Mazza et al., 1999). In addition, a role for ROS in UV-B-mediated plant damage is further evidenced by mutants deficient in ascorbic acid synthesis that are sensitive to UV-B irradiation (Conklin et al., 1999).
Although reactionary defense mechanisms abate the secondary effects of ROS generated by UV-B, plants utilize UV-absorptive secondary metabolites from the phenylpropanoid biosynthetic pathway as sunscreens to avoid UV-B. These compounds, especially the colorless flavonoids (Chappell and Hahlbrock, 1984; Day, 1993; Day et al., 1993) and hydroxycinnamic acids (Li et al., 1993; Landry et al., 1995; Liu et al., 1995; Ormrod et al., 1995), accumulate in plants in response to UV. Several studies using Arabidopsis mutants deficient in flavonoids and hydroxycinnamic acids underscore the importance of chemical sunscreens in protecting against UV radiation (tt4 and tt5 [Li et al., 1993], uvs [Lois and Buchanan, 1994], fah1 [Landry et al., 1995], and uvt1 [Bieza and Lois, 2001]). Field studies in soybean (Glycine max) showed that UV-B was specifically required for sunscreen compound induction and this induction leads to a decrease in UV-B-mediated DNA (Mazza et al., 2000).
Here, we report the isolation and characterization of the UV-B-sensitive mutant, UV resistance locus 8-1 (uvr8-1), which defines a new class of UV-resistance gene. Unlike previously reported mutants that are defective in DNA damage repair or sunscreen biosynthetic enzymes, uvr8-1 has altered UV-B signal transduction as shown by a lack of UV-induced accumulation of flavonoids and chalcone synthase (CHS) mRNA and protein. Map-based cloning of uvr8-1 identified a gene with extensive sequence similarity to the human guanine nucleotide exchange factor regulator of chromatin condensation 1 (RCC1). In other eukaryotes, RCC1 functions as a nucleotide exchange factor for the Ran G-protein to regulate diverse biological processes, including RNA processing and nucleocytoplasmic transport (Renault et al., 1998). These results suggest that UVR8 plays a role in UV-B-mediated induction of flavonoid biosynthesis and plant defense against UV-B.
RESULTS
UV-B Hypersensitivity of uvr8-1
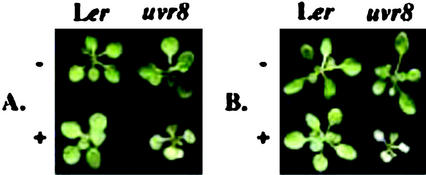
The uvr8-1 mutant was identified as having increased UV-B sensitivity compared with the progenitor Landsberg erecta (Ler) tt5 chalcone isomerase-deficient mutant line based on increased leaf injury and stunted growth under continuous cool-white fluorescent light chronically supplemented with 0.2 kJ UV-BBE m−2 h−1 UV-B for 10 d (see “Materials and Methods”). It is interesting to note that the parental chalcone isomerase-deficient tt5 line is already quite UV-B hypersensitive under growth chamber conditions due to decreased accumulation of flavonoids and sinapate esters (Li et al., 1993; Landry et al., 1995). uvr8-1 was subsequently outcrossed four consecutive times to the wild-type Ler TT5 to generate homozygous Ler uvr8-1 TT5 lines. Homozygous uvr8-1 TT5 lines are indistinguishable from wild-type Ler UVR8 TT5 in the absence of UV-B, as shown in Figure 1A. However, uvr8-1 shows enhanced UV-B sensitivity in comparison with the wild-type Ler after subjecting 10-d-old plants to 3 d of constant 0.2 kJ UV-BBE m−2 h−1 (Fig. 1A). This sensitivity is displayed as necrosis of the first true leaves and cotyledons, as well as folding of the youngest leaves (Fig. 1A). In addition, leaf necrosis progressively worsens during 3 d of recovery under white light minus UV-B (Fig. 1B).
Figure 1.
uvr8-1 is hypersensitive to UV-B in comparison with wild-type Ler. Wild-type Ler and uvr8-1 plants were grown in the absence of UV-B for 10 d. They were then treated with 72 h of 0.2 kJ UV-BBE m−2 h−1 (+) and were allowed a 72-h recovery period in white light without UV-B. Identically aged control plants were grown in the absence of UV-B throughout the experiment (−). Plants were photographed immediately after the UV-B treatment or following a 72-h recovery period. A, Plants photographed immediately after a 72-h UV-B treatment. B, Plants photographed after a 72-h recovery period following a 72-h UV-B treatment.
uvr8-1 Alters Phenylpropanoid Metabolism
The accumulation of phenylpropanoid-derived metabolites, flavonoids, tannins, and anthocyanins is controlled by developmental (for example, in the seed coat) and environmental (e.g. under high-intensity white light) signals in Arabidopsis and other plants. UV-B also induces anthocyanin pigment accumulation in the hypocotyl of wild-type Arabidopsis seedlings, and this response is nearly abolished in uvr8-1 (J.E. Lim and D.J. Kliebenstein, unpublished data). However, the seeds from these plants have the wild-type brown seed coat coloration, suggesting that uvr8-1 is defective in environmental but not developmental regulation of anthocyanin accumulation.
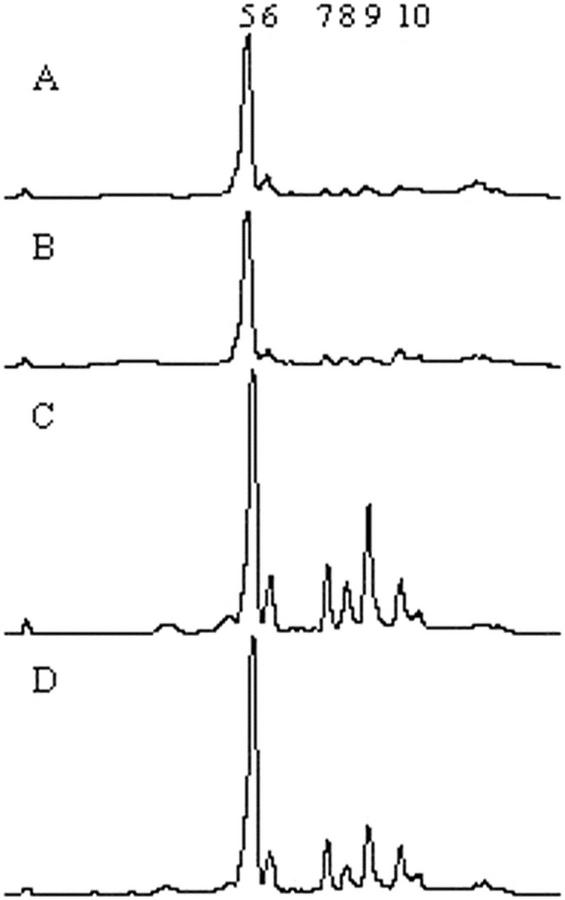
We next tested the hypothesis that uvr8-1 is altered in UV-B-mediated regulation of flavonoid or sinapate ester concentrations. As shown in Figure 2, HPLC analysis revealed that untreated uvr8-1 plants have normal sunscreen accumulation (Fig. 2, A and B). As expected, irradiating wild-type Ler TT5 with UV-B leads to increased sinapate esters (peaks 5 and 6) and flavonoids (peaks 7–10; Fig. 2, A versus C). In contrast, uvr8-1 accumulates approximately 50% less total flavonoids than wild type following UV-B exposure (Fig. 2, B versus D). It is surprising that sinapate ester induction is normal in uvr8-1. Thus, uvr8-1 seems to have altered UV-B regulation of flavonoid and anthocyanin metabolism.
Figure 2.
uvr8-1 reduces the UV-B-mediated induction of flavonoid accumulation. Plants were grown in the absence of UV-B for 14 d and were treated with 0.4 kJ UV-BBE m−2 h−1 for 3 d. Control plants were grown under white light in the absence of UV-B. Leaf tissue was harvested and methanol extracts were fractionated by reverse-phase HPLC. Numbers indicate the sinapate esters (peaks 5 and 6) and flavonoids (peaks 7–10), as previously identified by Li et al. (1993). A, Chromatogram of extract from Ler untreated control. B, Chromatogram of extract from uvr8-1 untreated control. C, Chromatogram of extract from Ler treated with UV-B for 3 d. D, Chromatogram of extract from uvr8-1 treated with UV-B for 3 d.
uvr8-1 Blocks Induction of CHS mRNA and Protein
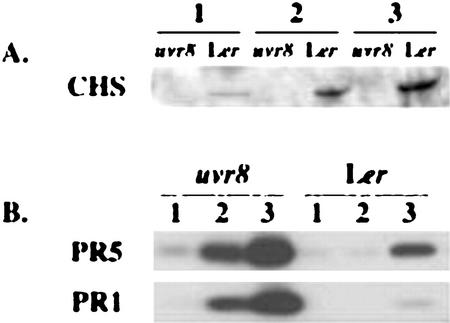
CHS is the committing enzyme for flavonoid and anthocyanin biosynthesis (for review, see Bharti and Khurana, 1997 and Jackson et al., 1995), and it is positively regulated by UV-B (Chappell and Hahlbrock, 1984; Christie and Jenkins, 1996; Fuglevand et al., 1996). To ask whether the uvr8-1-decreased flavonoid induction is controlled at the level of CHS protein accumulation, we compared CHS protein accumulation in uvr8-1 and wild-type Ler following UV-B treatment. As shown in Figure 3A, CHS protein continually increased over three consecutive days of UV-B treatment in wild-type Ler. In comparison, uvr8-1 completely blocked the UV-B-mediated induction of CHS protein. The inhibition of CHS induction in uvr8-1 is not caused by a global loss of stress responsive gene expression, as PR-1 and PR-5 proteins are induced more rapidly and to a higher level in uvr8-1 than in wild-type Ler (Fig. 3B). This suggests that UVR8 is required for transduction of a UV-B response signal.
Figure 3.
Induction of CHS, PR1, and PR5 proteins by UV-B. Plants were grown in the absence of UV-B for 14 d and were then treated with 0.4 kJ UV-BBE m−2 h−1 for 1, 2, or 3 d. Control plants were grown to the same age in the absence of UV-B. A, Immunoblot with CHS antisera. Control plants showed no detectable CHS protein. B, Immunoblot with PR1- and PR5-specific antisera.
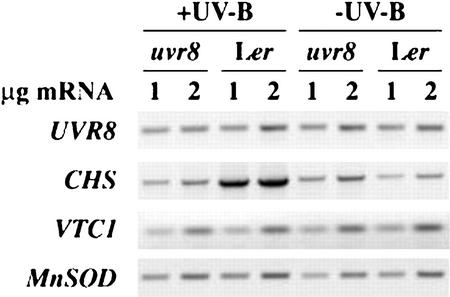
To test whether the decreased induction of CHS protein in uvr8-1 is controlled at the mRNA level, we compared CHS mRNA accumulation in uvr8-1 and wild-type Ler following UV-B treatment. As shown in Figure 4, uvr8-1 and wild-type plants grown in white light without supplementary UV-B had comparable CHS mRNA. However, UV-B-mediated induction of CHS mRNA is nearly blocked in uvr8-1 as compared with wild type. The expression of VTC1 (vitamin C deficient; an ascorbic acid biosynthetic enzyme) and manganese SOD 1 (MSD1) were assayed to examine if uvr8-1 is impaired in antioxidant defense capacity (Kliebenstein et al., 1998; Conklin et al., 1999). These genes are expressed at similar levels in uvr8-1 and wild-type Ler before and after UV-B treatment, suggesting that uvr8-1 is not deficient in antioxidant defense (Fig. 4). MSD1 was previously shown to not respond to UV-B treatment and, therefore, functions as a control showing the use of equal cDNA amounts in the different reactions (D.J. Kliebenstein, unpublished data). These results further support the hypothesis that UVR8 transduces a UV-B-specific signal.
Figure 4.
Analysis of CHS mRNA induction following UV-B treatment. Reverse transcriptase (RT)-PCR analysis of CHS mRNA induction by UV-B. Fourteen-day-old plants were grown in the absence of UV-B and were treated with 0.4 kJ UV-BBE m−2 h−1 for 24 h or were left under −UV-B conditions for an additional 24 h. Tissue was then harvested for RNA extraction. Ethidium bromide-stained products are shown from quantitative RT-PCR. For each sample, 1 and 2 μg of total RNA were used for cDNA synthesis prior to PCR amplification. The results are representative of three independent experiments.
Map-Based Cloning of UVR8
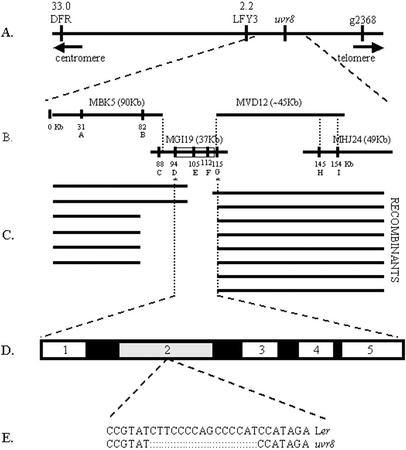
To prepare for map-based cloning of UVR8, we initiated genetic analysis of uvr8-1. The mutant was genetically characterized by testing segregation of UV-B sensitivity in the F2 generation of a cross between homozygous uvr8-1 TT5 in the Ler genetic background and wild-type UVR8 TT5 Ler. Analysis of >540 segregating F2 progeny showed a 3:1 ratio of UV-B-resistant:UV-B-sensitive plants (χ2 = 0.241), suggesting that uvr8-1 is a monogenic recessive mutation. An F2 mapping population was created by crossing homozygous uvr8-1 TT5 in the Ler genetic background to Colombia-0 (Col-0). The F2 progeny were scored for UV-B sensitivity and were genotyped (see http://www.Arabidopsis.org for information about available markers). As shown in Figure 5A, uvr8-1 is on chromosome 5, centromere distal to LFY3 (Konieczny and Ausubel, 1993; Bell and Ecker, 1994).
Figure 5.
Chromosome walk to uvr8-1 locus. A, Markers used for localizing uvr8-1 to chromosome 5. Numbers at top represent genetic distance to uvr8-1 in centromeres. B, The four P1 clones (MBK5, MGI19, MVD12, and MHJ24) covering the uvr8-1 locus, with their sizes indicated in parentheses. The physical positions of markers MBK5-1 (A), MBK5C3 (B), MGI19-1 (C), MGI19C7 (D), MGI19C9 (E), MGI19C8 (F), MGI19C6 (G), MHJ24C1 (H), and MHJ24-2 (I) are indicated. C, Map of approximate locations of recombination breakpoints used for fine mapping. D, Smallest region genetically identified to contain uvr8-1 mutation. The five ORFs are putative amino acid transporter (1), RCC1 homolog (2), Ser/Thr protein phosphatase (3), hypothetical protein (4), and histidinol dehydrogenase (5). E, DNA sequence of the uvr8-1 15-bp deletion.
Fine-scale genetic mapping of the mutation required the identification of polymorphic markers tightly linked to UVR8. Using publicly available wild-type Col-0 genomic sequences of MBK5, MGI19, and MHJ24 P1 clones (Fig. 5B and http://mips.gsf.de/proj/thal/db/index.html), we developed nine new markers. These simple sequence length polymorphisms (SSLPs) (MBK5-1, MGI19-1, and MHJ24-2), cleaved-amplified polymorphic sequences (CAPS; MBK5C3, MGI19C7, MGI19C9, and MHJ24C1), and single nucleotide polymorphic (SNP) markers (MGI19C8 and MGI19C6) are documented in Table I (Fig. 5B). Genotyping 1,254 UV-B-sensitive F2 individuals with the flanking markers MBK5-1 (Fig. 5B, marker A) and MHJ24-2 (Fig. 5B, marker I) identified 14 recombinants (Fig. 5C). The 14 recombinants were tested in the F3 progeny to verify the UV-B sensitivity, and the genotype was tested for the other seven markers. The location of the recombination breakpoints indicates that UVR8 is between markers MGI19C7 and MGI19C6 (Fig. 5C). This 21-kb interval is predicted to contain five open reading frames (ORFs; Fig. 5D; http://mips.gsf.de/cgi-bin/proj/thal/ bac_cosmid?MGI19).
Table I.
SSLP, CAPS, and SNP markers created in this study
| Marker | Class | Enzyme | Lera | Cola |
|---|---|---|---|---|
| MBK5-1 | SSLP | – | ∼180 | 207 |
| MBK5C3 | CAPS | XbaI | ∼326; ∼530 | 856 |
| MGI19-1 | SSLP | – | NPb | 205 |
| MGI19C7 | CAPS | BsmAI | 2,069 | ∼1969, ∼100 |
| MGI19C9 | CAPS | AflIIIb | 1,303, 468, 142, 75 | 1,013, 468, 290, 142, 75 |
| MGI19C8 | SNP | – | Tc | Ac |
| MGI19C6 | SNP | – | Td | Cd |
| MHJ24C1 | CAPS | BsmAI, MluI | ∼950; ∼650; ∼400 | ∼950; ∼600; ∼400; ∼50 |
| MHJ24-2 | SSLP | – | ∼142 | 145 |
Size of products produced in bp or sequence difference.
No product produce, dominant Col-0 marker.
Base no. 30296 in MGI19.
Base no. 34324 in MGI19.
uvr8-1 Contains a 15-bp Deletion in an RCC1 Homolog
To identify the molecular lesion responsible for the UV-B sensitivity, the entire 21-kb region containing UVR8 was sequenced from uvr8-1 and wild-type Ler to detect uvr8-1-specific polymorphisms (the Ler genomic sequence is GenBank accession no. AF130442, and the experimentally verified cDNA is GenBank accession no. AF130441). The only difference between the uvr8-1 and the wild-type Ler sequence in the entire 21-kb region was a 15-bp deletion in ORF2, renamed UVR8 from here in this manuscript (Fig. 5, D and E). The predicted UVR8 protein has sequence similarity to the RCC1 family of proteins (35% identity and 50% similarity), which are nuclear-localized guanine nucleotide exchange factors for the small G-protein Ran. In mammals and fungi, RCC1 functions with the Ran G-protein to regulate diverse biological processes, nucleocytoplasmic transport, and the cell cycle (for review, see Renault et al., 1998). Genomic sequencing has identified Ran homologs in Arabidopsis, suggesting that the Ran G-protein regulatory mechanism may also function in plants.
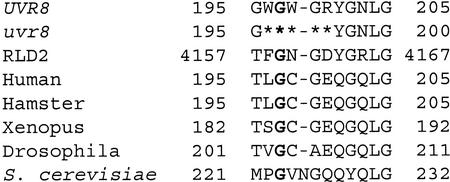
The RCC1 secondary structure contains seven β sheet blades whose structural integrity is dependent on four absolutely conserved Gly and one invariant cis-Pro (Renault et al., 1998). All of these conserved Gly and Pro residues are present in the wild-type UVR8 protein, except one Pro to Asn change. In addition, UVR8 contains the eight amino acids considered essential for RCC1 activity (Azuma et al., 1996; Renault et al., 1998). In contrast, the five-amino acid deletion in uvr8-1 removes one of these absolutely conserved Gly and changes the spacing between two others (Fig. 6, conserved Gly are in bold print).
Figure 6.
Deletion in uvr8-1 removes a Gly conserved among RCC1 homologs. An alignment of the 15 amino acids surrounding the uvr8-1 deletion. The sequences are UVR8, uvr8-1, RCC1 from human (Homo sapiens; U50078), and homologs from hamster (P23800), Xenopus (D00646), fruit fly (Drosophila melanogaster; S15028), and yeast (Saccharomyces cerevisiae; P21827). RLD2 is a human protein that contains a domain similar to the entire RCC1 protein (Rosa et al., 1996). Asterisks mark the uvr8-1 deletion. The numbers designate the distance from the carboxyl terminus. Gly described in the text are in bold.
Complementation of uvr8-1
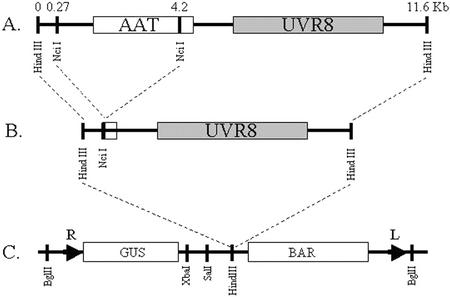
To confirm that the 15-bp deletion in uvr8-1 is the basis of the UV-B sensitivity, we attempted to rescue the uvr8-1 UV-B phenotype through transformation with plasmid pGPTV-UVR8, which contains a UVR8 genomic fragment in the binary transformation vector pGPTV, as shown in Figure 7. Wild-type Ler and mutant uvr8-1 were transformed with pGPTV-UVR8 or empty pGPTV, and the transgenic progeny were tested for UV-B tolerance. All 16 uvr8-1 lines containing pGPTV-UVR8 were UV-B tolerant, indicating that UVR8 complements the uvr8-1 mutation and confirming that the 15-bp deletion in uvr8-1 leads to increased UV-B sensitivity. In addition, all six uvr8-1-transgenic lines containing pGPTV were UV-B sensitive, and all Ler transgenics, with pGPTV (five lines) or pGPTV-UVR8 (12 lines), were UV-B resistant, suggesting that transformation did not affect UV-B tolerance (Table II). UV-B-irradiated wild-type Ler transformants containing extra copies of UVR8 in the form of pGPTV-UVR8 also displayed an enhanced purple coloration in the leaves and anthocyanin pigment accumulation in the hypocotyl in comparison with transgenics containing the empty pGPTV vector alone. Thus, UVR8 may be a positive regulator for anthocyanin pigment accumulation. In summary, the deletion in uvr8-1 is the basis of the UV-B sensitivity, possibly through the removal of a protein required for the induction of UV-B defense mechanisms.
Figure 7.
Generation of the UVR8 complementation construct. Boxes represent ORFs with gene labels contained within each box. Numbers at the top represent location of restriction enzyme digestion sites in kilobases. A, HindIII fragment containing the putative amino acid transporter and UVR8. B, Final HindIII cassette containing UVR8 obtained after removing the NcII fragment containing the amino acid transporter. C, Restriction map of pGPTV-BAR binary vector. L, Left T-DNA border; R, right T-DNA border.
Table II.
Primers used in this study
| Primer | Dira | Sequence (5′–3′) |
|---|---|---|
| 221EF1 | F | CTACTGGTGGTGAGAAAATGTC |
| R1013 | R | TGTTGTCCACTTGCTCCATCG |
| MBK5C3 | F | CCAATGGGGGTGCTCTAATG |
| R | TTCACCTTTGTCTTACGATTCTAC | |
| MBK5-1 | F | ATGACTGTTGTTTACCATTA |
| R | GAGCATTTCACAGAGACG | |
| MGI19C4 | F | GAAGATGGCGAGTGAAGAGATAAG |
| R | CTGCCCAGCGACGGTTTT | |
| MGI19C6 | F | GAAGATGGCGAGTGAAGAGATAAG |
| R | CTGCCCAGCGACGGTTTT | |
| MGI19C7 | F | GTCACGTAACTACCTAACTCTT |
| R | AGCTGGCCGTGGAACATC | |
| MGI19C8 | F | TACACTATCGCCCACAACTACAAG |
| R | CCGGCGACACTCAAACTCAA | |
| MGI19C9 | F | AGGCGCGATGGAGATTTG |
| R | GCACTTTGAAGCATTTATTG | |
| MGI19-1 | F | TACTTGAGATGCCCCGTGACAG |
| R | CATGACCTTCTTTTCCTATTGA | |
| MGI19-3 | F | GTCCGGCATCAAAACCTAAACA |
| R | TGCCGGATACAAAACATACAC | |
| MGI19-5 | F | CCGTGAAAAGCGAAAGGAC |
| R | GTAGTTGTGGGCGATAGTGTAGAT | |
| MGI19-7 | F | ACCGCCATTATCCGTTAGT |
| R | TTTTTCTCCACCACCTTCTT | |
| MGI19-8 | F | ATAAGGGCCGAGGTTTC |
| R | CACAGCTGCTAAGATGATTC | |
| MGI19-9 | F | GATGGAGTATGAAGTGGGTTTGTC |
| R | TATCGATCCATCTTCAGGTATTTA | |
| MGI19-11 | F | TCTCTGCAGGCGACACC |
| R | TGCAGGGAATACGAATCAAAATGG | |
| MGI19-13 | F | GAGTCTTCTAGCTTACACCAGT |
| R | CGCGCCTTTATTACCATCTACCG | |
| MGI19-14 | F | TTCGCGAAGAGCACAGCCTACTA |
| R | GCGCCTTACACCTGTGAATGATG | |
| MGI19-15 | F | AGAGAAGCGAAAGACGAGACAG |
| R | CAGCAGCAACAAGGACAGAAC | |
| MHJ24C1 | F | TTCCCTCACCAGTAAAGC |
| R | TTGCAGGAGAAAAGAAGG | |
| MHJ24-2 | F | AACAGAATTAGCCGGAGTAGAT |
| R | ATTTAAAAGTATTGCGAACGAT |
F, Forward primer; R, reverse primer.
DISCUSSION
UVR8 Positively Regulates UV-B Induction of Phenylpropanoid Metabolism
Our results suggest that UVR8 is a positive regulator involved in a UV-B signal transduction pathway. First, the uvr8-1 mutation blocks the UV-B-mediated induction of CHS mRNA and protein, as well as reduces flavonoid and anthocyanin pigment accumulation (Figs. 2–4). Second, the presence of transgenic copies of UVR8 in uvr8-1 rescues anthocyanin production in response to UV-B and enhances the up-regulation of anthocyanin accumulation in wild-type Ler (J.E. Lim and D.J. Kliebenstein, unpublished data). Considering these results, UVR8 appears to be a positive regulator at least in the UV-B signal transduction pathway for CHS. The apparent complete block in CHS mRNA and protein induction only leads to a 50% reduction in flavonoid accumulation. This could result from a UV-B-mediated increase in metabolite flow through the phenylpropanoid pathway that acts to push the production of flavonoids in the absence of increased CHS levels.
In contrast to the positive action of UVR8, AtMYB4, a previously identified transcription factor involved in regulating phenylpropanoid metabolism in response to UV-B, is a negative regulator. AtMYB4 represses cinnamate 4-hydroxylase expression, whereas it has minimal impact upon CHS (Jin et al., 2000). Upon exposure to UV-B, AtMYB4 transcript levels decrease, allowing increased cinnamate 4-hydroxylase expression and increased sinapate ester accumulation (Jin et al., 2000). Furthermore, an AtMYB4 knockout mutation leads to elevated sinapate ester and cinnamate 4-hydroxylase accumulation, but does not alter CHS or flavonoid levels (Jin et al., 2000). This suggests that the different phenylpropanoid biosynthetic pathway components are controlled by different regulatory mechanisms involving positive and negative control elements.
Specificity of UV-B-Mediated CHS Regulation
A vast amount of work has shown that CHS mRNA accumulation is up-regulated following exposure to UV-B (for review, see Jenkins et al., 2001). This UV-B-mediated induction can be attenuated by red light through phyB and can be amplified by blue light independent of cry1 (Wade et al., 2001). Thus, UV-B is detected and the signal is transduced through an independent pathway not dependent on the known photoreceptors (Jenkins et al., 2001). Other work has shown that UV-B-dependent signals can be transduced through signal transduction pathways using nitric oxide, salicylic acid, jasmonic acid, ethylene, and/or ROS (Mackerness, 2000). Of all of these signals, only nitric oxide plays a role in UV-B-mediated induction of CHS mRNA or protein (Mackerness et al., 2001). Thus, UVR8 is functioning in a very specific UV-B signal transduction pathway that may use nitric oxide. Further evidence for this specificity comes from the observation that uvr8 does not alter tannin accumulation in seeds or anthocyanin accumulation in response to methyl jasmonate treatment (J.E. Lim and D.J. Kliebenstein, unpublished data). Identification of additional UVR8 signal transduction components should provide unique insight into how UV-B regulates CHS. In addition, analysis of CHS regulation in uvr8 under a diverse array of conditions will enable analysis of the UV-B specificity of this pathway.
uvr8-1 Is UV-B Sensitive Due to a Deletion in an RCC1 Homolog
Complementation and mapping experiments show that a 15-bp deletion in a gene similar to RCC1 causes uvr8-1's UV-B sensitivity. The similarity between UVR8 and RCC1 suggests that UVR8 may have guanine nucleotide exchange activity (Aebi et al., 1990; Klebe et al., 1995). It is interesting that RCC1 mutations in Saccharomyces cerevisiae alter a wide variety of processes, including pre-mRNA processing and transport (Aebi, 1990; Kadowaki et al., 1993), mating behavior (Clark and Sprague, 1989), initiation of mitosis (Matsumoto and Beach, 1991), and chromatin decondensation (Sazer and Nurse, 1994). Although RCC1 mutations in fungi and other species are lethal or highly pleiotropic, the uvr8-1 mutation has no discernible effect on growth/development of growth chamber grown plants except in the presence of UV-B. This lack of pleiotropy could be an effect of this specific uvr8-1 mutation or an indication that UVR8 is not an RCC1 ortholog. Evidence for the latter comes from the observation that UVR8 is significantly smaller than RCC1 and does not contain the nuclear localization sequences conserved among the animal and fungal RCC1 proteins. The functional significance of the similarity between UVR8 and RCC1 remains to be determined.
uvr8-1 Appears to Be Deficient in Multiple UV-B Defense Mechanisms
uvr8-1 appears to impair UV-B tolerance mechanisms in addition to decreased CHS and flavonoid induction. This conclusion comes from the observation that uvr8-1 was identified as UV-B sensitive in a tt5 background that blocks flavonoid accumulation (Shirley et al., 1992). Additional support for this hypothesis stems from the observation that the phenotypes associated with uvr8-1 UV-B sensitivity (necrosis and leaf cupping) continue to worsen up to 72 h after UV-B removal. In comparison, UV-B sensitivity in the tt5 mutant does not show a continual progression of UV-B damage after removal of the UV-B (Li et al., 1993). Thus, uvr8-1 may be defective in the induction of mechanisms that detoxify UV-B-mediated damage. However, these mechanisms do not block the accumulation of PR1 and PR5 stress-responsive proteins or alter the regulation of VTC1 and MnSOD antioxidant mRNAs (Figs. 3 and 4). Additional work is necessary to fully understand how the impaired UV-B signal transduction in uvr8-1 relates to the displayed UV-B sensitivity.
Future Work and Implications
Further studies into the biological role of UVR8 should enhance our understanding of UV-B signal transduction and sensitivity. Identifying the specific mechanism by which uvr8-1 generates increased UV-B sensitivity and isolation of additional uvr8-1 mutations and new mutations that alter different components of the UVR8 UV-B tolerance mechanisms will boost our understanding of how plants resist UV-B. Because G proteins have not been shown to regulate CHS or phenylpropanoid metabolism, biochemical testing of the hypothesis that UVR8 is a guanine nucleotide exchange factor and identification of its substrate will help to elucidate new components in the regulation of secondary metabolism. In addition, investigating where UVR8 is in the UV-B signal transduction pathway and how uvr8-1 affects the synergy between UV-B, UV-A, and white light in regulating CHS will clarify how these signal transduction pathways are coordinated.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Wild-type Arabidopsis accessions Ler and Col-0 were used in this study, as were the mutant lines Ler tt5 (Shirley et al., 1992). All plants were grown under constant light (60–100 μE m−2 s−1 photosynthetically active radiation) using CW1500 cool-white fluorescent lamps (General Electric, Fairfield, CT) with filters to remove all UV-B, before and after UV-B treatment. Plants used for UV-B induction studies were grown on nutrient agar plates. Plants for UV-B sensitivity assays were grown in Cornell soil-less mix (Landry et al., 1995). All experiments were independently replicated at least twice.
UV-B Treatment
UV-B treatments were conducted, and UV-B fluences are expressed as previously described (Caldwell, 1971; Landry et al., 1995, 1997). White light (100 μEi m−2 s−1) was supplemented with UV-B from F40 UV-B fluorescent lamps (Phillips, Somerset, NJ) or T12F40 UV-B lamps (UV Resources International, Cleveland). Light was filtered through 3-mm-thick Pyrex glass plates to remove UV-C, wavelengths <280 nm, and to attenuate UV-B levels; control plants were additionally shielded with 0.13-mm-thick Mylar (AIN Plastics, Mt. Vernon, NY) to remove UV-B, wavelengths <310 nm. For plants grown on soil, 0.13-mm-thick cellulose acetate was used instead of Pyrex glass to remove UV-C. UV-A levels were identical between control (−UV-B) and UV-B treatments. For mutant selection, 4-d-old seedlings were irradiated with 0.2 kJ UV-BBE m−2 h−1 (0.7 kJ m−2 h−1 unweighted UV-B) for 10 d. For analysis of uvr8-1, plants were grown for 10 d before being irradiated with 0.2 kJ UV-BBE m−2 h−1 (or 1.4 kJ m−2 h−1 unweighted UV-B) for 3 d.
Quantitative RT-PCR Analysis
RNA was isolated from approximately 100 μg of leaf tissue using Trizol reagent (Invitrogen, Grand Island, NY). cDNA synthesis was per the manufacturer's instructions (SuperScript II; Invitrogen). PCR reactions were as described for SSLP reactions using primers 221EF1 and R1013 for the UVR8 cDNA (Table II), primers p3-GMP and 3′GMP for VTC1 (Conklin et al., 1999), primers MnSOD1F and 1R for MSD1 (Kliebenstein et al., 1998), and primers CHS-1F and CHS-1R for CHS (Shirley et al., 1995). Titration studies showed the optimal range for each primer pair is 1 or 2 μg of mRNA for cDNA synthesis followed by 3 μg of the diluted cDNA mix in a 25-cycle PCR reaction.
Immunoblot Analysis
Protein sample preparation, quantitation, electrophoresis, and immunoblotting were as described by Zhao and Last (1995). Twenty micrograms of total leaf protein was loaded per lane, except for 60 μg of total leaf protein per lane for CHS. Proteins were detected using published antiserum concentrations and chemiluminescent detection (Cain et al., 1997; Kliebenstein et al., 1998).
uvr8-1 Isolation and Genetic Characterization
Ler tt5 seeds were gamma irradiated with 50 krad, grown on soil, and M1 plants were allowed to generate M2 populations by self-pollination. One M2 plant was identified as more UV-B sensitive than the tt5 parent under continuous UV-B and was allowed to self-fertilize for two generations to obtain M4 seeds. A UV-B-sensitive M4 progeny was outcrossed to Ler TT5 to generate F2 for segregation analysis. UV-B-sensitive F2 progeny with a wild-type testa phenotype were allowed to self-pollinate. Homozygous uvr8-1/uvr8-1; TT5/TT5 lines were selected from the F3 and were outcrossed to Ler three times. Progeny from the fourth Ler TT5outcross (OC4) were used for all biochemical analyses unless otherwise noted.
To generate a mapping population, uvr8-1/uvr8-1; TT5/TT5 plants were crossed to wild-type Col-0. F2 seeds were grown on plates without UV-B for 14 d and were then treated with UV-B for 3 d to identify homozygous uvr8-1 plants. All UV-B-sensitive plants were left to recover on plates for 2 weeks before being transferred to soil and allowed to self-cross to generate F3 seed.
DNA Extraction, PCR, Restriction Enzyme Digestion, and Gel Electrophoresis Conditions
DNA was extracted as described (Conklin et al., 1999). All PCR was done in a DNA Thermal Cycler 480 or a GeneAmp PCR System 9600 (PerkinElmer Instruments, Norwalk, CT). SSLP markers were amplified using a standard PCR mixture and program (Bell and Ecker, 1994), except that 1.2 mm MgCl2 was used with MHJ24-2. SSLP products were separated on 4% (w/v) agarose and were visualized with ethidium bromide using an EagleEye II (Stratagene, La Jolla, CA). Standard PCR conditions were used for CAPS and SNP markers (Konieczny and Ausubel, 1993). CAPS markers were digested with the respective enzyme (Table I) and were separated on 1.5 or 2.5% (w/v) agarose gels.
Mapping Analysis, Sequencing, and Sequence Analysis
SSLP markers were identified by searching published Col-0 genomic sequences for di- or trinucleotide repeats longer than six units. PCR primers were designed to amplify products smaller than 200 bp and were synthesized by the Great American Gene Company (http://www.geneco.com) or by the Cornell BioResource Center (Cornell University, Ithaca, NY; Table II). These primers were used to screen for polymorphisms between Col-0 and Ler as previously described, and the polymorphic markers are listed in Table I. SNP and CAPS makers were identified by designing primers to amplify approximately 2 kb of presumed noncoding regions (Table II). These primers were used to amplify Ler genomic DNA, and the resulting PCR products were purified using the QIAquick PCR purification kit (Qiagen, Santa Clarita, CA) and sequenced by the Cornell BioResource Center. The sequence was compiled and compared with the published Col-0 genomic sequence using Sequencer 3.1 (Gene Codes, Ann Arbor, MI) and DNAstar (Madison, WI).
Complementation Analysis
The construct scheme is shown in Figure 7. Escherichia coli containing the P1 clone MGI19 (Arabidopsis Resource Center, Ohio State University, Columbus) was grown overnight, and the P1 clone was isolated using a plasmid maxi kit (Qiagen). MGI19 was digested with HindIII and the fragment, including UVR8, and an amino acid transporter gene (AAT) was subcloned into pBluescript II SK+ phagemid (Fig. 7, A and B). This plasmid was digested with NcII, and the digested plasmid minus the 4-kb AAT fragment was relegated to produce a HindIII cassette containing only the complete UVR8 gene. This final cassette was then inserted into HindIII-digested pGPTV-BAR (Fig. 7C; Becker et al., 1992) to generate pGPTV-UVR8. pGPTV-UVR8 was transformed into uvr8-1 and Ler using Agrobacterium tumefaciens pMP90 strain GB3101 (Bent and Clough, 1998; Conklin et al., 1999). uvr8-1 and Ler were also transformed with the empty pGPTV-BAR as controls. T1 progeny were screened for BASTA resistance and the BAR gene using PCR (Conklin et al., 1999). In addition, primers 221EF1 and R1013 were used to screen uvr8-1 transformed with pGPTV-UVR8 to for wild-type UVR8. BASTA-resistant T1 and T2 progeny were tested for UV-B sensitivity as described above.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
ACKNOWLEDGMENTS
We thank Dr. Patricia L. Conklin (State University of New York, Cortland) for help with complementation experiments and VTC1 primers, Dr. Ann Stapleton (University of North Carolina, Wilmington) for immunoassay of cyclobutyl pyrimidine dimer and 6,4-photoproduct repair, Dr. Brenda Shirley (Virginia Polytechnic Institute and University, Blacksburg) for CHS antibody, the Arabidopsis Biological Resource Center for EST and P1 clones, and Dr. Anne Britt (University of California, Davis) for reviewing the paper.
Footnotes
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.005041.
LITERATURE CITED
- Aebi T, Clark MW, Vijayraghavan U, Abelson J. A yeast mutant, PRP20, altered in mRNA metabolism and maintenance of the nuclear structure, is defective in a gene homologous to the human gene RCC1 which is involved in the control of chromosome condensation. J Mol Gen Genet. 1990;224:72–80. doi: 10.1007/BF00259453. [DOI] [PubMed] [Google Scholar]
- Ahmad M, Jarillo JA, Klimczak LJ, Landry LG, Peng T, Last RL, Cashmore AR. An enzyme similar to animal type II photolyases mediates photoreactivation in Arabidopsis. Plant Cell. 1997;9:199–207. doi: 10.1105/tpc.9.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma Y, Seino H, Seki T, Uzawa S, Klebe C, Ohba T, Wittinghofer A, Hayashi N, Nishimoto T. Conserved histidine residues of RCC1 are essential for nucleotide exchange on Ran. J Biochem. 1996;120:82–91. doi: 10.1093/oxfordjournals.jbchem.a021397. [DOI] [PubMed] [Google Scholar]
- Becker D, Kemper E, Schell J, Masterson R. New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol. 1992;20:1195–1197. doi: 10.1007/BF00028908. [DOI] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Bent AF, Clough SJ. Agrobacterium germ-line transformation: transformation of Arabidopsis without tissue culture. In: Gelvin SB, Schilperoort RA, editors. Plant Molecular Biology Manual. Ed 2. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 1–14. [Google Scholar]
- Bharti AK, Khurana JP. Mutants of Arabidopsis as tools to understand the regulation of phenylpropanoid pathway and UVB protection mechanisms. Photochem Photobiol. 1997;65:765–776. doi: 10.1111/j.1751-1097.1997.tb01923.x. [DOI] [PubMed] [Google Scholar]
- Bieza K, Lois R. An Arabidopsis mutant tolerant to lethal ultraviolet-B levels shows constitutively elevated accumulation of flavonoids and other phenolics. Plant Physiol. 2001;126:1105–1115. doi: 10.1104/pp.126.3.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britt AB. Repair of DNA damage induced by ultraviolet radiation. Plant Physiol. 1995;108:891–896. doi: 10.1104/pp.108.3.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britt AB, Chen J-J, Wykoff D, Mitchell D. A UV-sensitive mutant of Arabidopsis defective in the repair of pyrimidine-pyrimidinone(6-4) dimers. Science. 1993;261:1571–1574. doi: 10.1126/science.8372351. [DOI] [PubMed] [Google Scholar]
- Cain CC, Saslowsky DE, Walker RA, Shirley BW. Expression of chalcone synthase and chalcone isomerase proteins in Arabidopsis seedlings. Plant Mol Biol. 1997;35:377–381. doi: 10.1023/a:1005846620791. [DOI] [PubMed] [Google Scholar]
- Caldwell MM. Solar UV irradiation and the growth and development of higher plants. In: Giese AC, editor. Photophysiology. New York: Academic Press; 1971. pp. 131–177. [Google Scholar]
- Caldwell MM, Teramura AH, Tevini M. The changing solar ultraviolet climate and the ecological consequences for higher plants. Trends Ecol Evol. 1989;4:363–367. doi: 10.1016/0169-5347(89)90100-6. [DOI] [PubMed] [Google Scholar]
- Chappell J, Hahlbrock K. Transcription of plant defense genes in response to UV light or fungal elicitor. Nature. 1984;311:76–78. [Google Scholar]
- Christie JM, Jenkins GI. Distinct UV-B and UV-A/blue light signal transduction pathways induce chalcone synthase gene expression in Arabidopsis cells. Plant Cell. 1996;8:1555–1567. doi: 10.1105/tpc.8.9.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KI, Sprague GF., Jr Yeast pheromone response pathway: characterization of a suppressor that restores mating to receptorless mutants. Mol Cell Biol. 1989;9:2682–2694. doi: 10.1128/mcb.9.6.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin PL, Norris SR, Wheeler GL, Smirnoff N, Williams EH, Last RL. Genetic evidence for the role of GDP-mannose in plant vitamin C biosynthesis. Proc Natl Acad Sci USA. 1999;96:4198–4203. doi: 10.1073/pnas.96.7.4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day TA. Relating UV-B radiation screening effectiveness of foliage to absorbing-compound concentration and anatomical characteristics in a diverse group of plants. Oceologia. 1993;95:542–550. doi: 10.1007/BF00317439. [DOI] [PubMed] [Google Scholar]
- Day TA, Martin G, Vogelmann TC. Penetration of UV-B radiation in foliage: evidence that the epidermis behaves as a non-uniform filter. Plant Cell Environ. 1993;16:735–741. [Google Scholar]
- Doke N, Miura Y, Sanchez LM, Kawakita K. Involvement of superoxide in signal transduction: responses to attack by pathogens, physical and chemical shocks, and UV irradiation. In: Foyer CH, Mullineaux PM, editors. Causes of Photooxidative Stress and Amelioration of Defense Systems in Plants. Boca Raton, FL: CRC Press; 1994. pp. 177–198. [Google Scholar]
- Foyer CH, Descourvieres P, Kunert KJ. Protection against oxygen radicals: an important defense mechanism studied in transgenic plants. Plant Cell Environ. 1994a;17:507–523. [Google Scholar]
- Foyer CH, Lelandais M, Kunert KJ. Photooxidative stress in plants. Physiol Plant. 1994b;92:696–717. [Google Scholar]
- Frederick JE, Snell HE, Haywood EK. Solar ultraviolet radiation at the earth's surface. Photochem Photobiol. 1989;50:443–450. [Google Scholar]
- Fuglevand G, Jackson JA, Jenkins GI. UV-B, UV-A, and blue light signal transduction pathways interact synergistically to regulate chalcone synthase gene expression in Arabidopsis. Plant Cell. 1996;8:2347–2357. doi: 10.1105/tpc.8.12.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow GR, Jenkins ME, Pittalwala TS, Mount DW. Isolation of uvh1, an Arabidopsis mutant hypersensitive to ultraviolet light and ionizing radiation. Plant Cell. 1994;6:227–235. doi: 10.1105/tpc.6.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JA, Fuglevand G, Brown BA, Shaw MJ, Jenkins GI. Isolation of Arabidopsis mutants altered in the light-regulation of chalcone synthase gene expression using a transgenic screening approach. Plant J. 1995;8:369–380. doi: 10.1046/j.1365-313x.1995.08030369.x. [DOI] [PubMed] [Google Scholar]
- Jenkins GI, Long JC, Wade HK, Shenton MR, Bibikova TN. UV and blue light signaling: pathways regulating chalcone synthase gene expression in Arabidopsis. New Phytol. 2001;151:121–131. doi: 10.1046/j.1469-8137.2001.00151.x. [DOI] [PubMed] [Google Scholar]
- Jiang C-Z, Yee J, Mitchell DL, Britt AB. Photorepair mutants of Arabidopsis. Proc Natl Acad Sci USA. 1997;94:7441–7445. doi: 10.1073/pnas.94.14.7441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 2000;19:6150–6161. doi: 10.1093/emboj/19.22.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T, Goldfarb D, Spitz LM, Tartakoff AM, Ohno M. Regulation of RNA processing and transport by a nuclear guanine nucleotide release protein and members of the Ras superfamily. EMBO J. 1993;12:2929–2937. doi: 10.1002/j.1460-2075.1993.tb05955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kangasjarvi J, Talvinen J, Utriainen M, Karjalainen K. Plant defense system induced by ozone. Plant Cell Environ. 1994;17:783–794. [Google Scholar]
- Kerr JB, McElroy CT. Evidence for large upward trends of ultraviolet-B radiation linked to ozone depletion. Science. 1993;262:1032–1034. doi: 10.1126/science.262.5136.1032. [DOI] [PubMed] [Google Scholar]
- Klebe C, Prinz H, Wittinghofer A, Goody RS. The kinetic mechanism of Ran-nucleotide exchange catalyzed by RCC1. Biochemistry. 1995;34:12543–12552. doi: 10.1021/bi00039a008. [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Monde R-A, Last RL. Superoxide dismutase in Arabidopsis: an eclectic enzyme family with disparate regulation and protein localization. Plant Physiol. 1998;118:637–650. doi: 10.1104/pp.118.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutants using codominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Kramer GF, Norman HA, Krizek DT, Mirecki RM. Influence of UV-B radiation on polyamines, lipid peroxidation and membrane lipids in cucumber. Phytochemistry. 1991;30:2101–2108. [Google Scholar]
- Krizek DT, Kramer GF, Upadhyaya A, Mirecki RM. UV-B response of cucumber seedlings grown under metal halide and high pressure sodium/deluxe lamps. Physiol Plant. 1993;88:350–358. [Google Scholar]
- Landry LG, Chapple CCS, Last RL. Arabidopsis mutants lacking phenolic sunscreens exhibit enhanced ultraviolet-B injury and oxidative damage. Plant Physiol. 1995;109:1159–1166. doi: 10.1104/pp.109.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry LG, Stapleton AE, Lim J, Hoffman P, Jays BJ, Walbot V, Last RL. An Arabidopsis photolyase mutant is hypersensitive to ultraviolet-B radiation. Proc Natl Acad Sci USA. 1997;94:328–332. doi: 10.1073/pnas.94.1.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ou-Lee T-M, Raba R, Amundson RG, Last RL. Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. Plant Cell. 1993;5:171–179. doi: 10.1105/tpc.5.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Gitz DC, McClure JW. Effects of UV-B on Flavonoids, ferulic acid, growth and photosynthesis in barley primary leaves. Physiol Plant. 1995;93:725–733. [Google Scholar]
- Lois R, Buchanan BB. Severe sensitivity to ultraviolet radiation in an Arabidopsis mutant deficient in flavonoid accumulation. Planta. 1994;194:504–509. [Google Scholar]
- Mackerness SAH. Plant responses to ultraviolet-B (UV-B:280–320 nm) stress: What are the key regulators? Plant Growth Regul. 2000;32:27–39. [Google Scholar]
- Mackerness SAH, John CF, Jordan B, Thomas B. Early signaling components in ultraviolet-B responses: distinct roles for different reactive oxygen species and nitric oxide. FEBS Lett. 2001;489:237–242. doi: 10.1016/s0014-5793(01)02103-2. [DOI] [PubMed] [Google Scholar]
- Matsumoto T, Beach D. Premature initiation of mitosis in yeast lacking RCC1 or an interacting GTPase. Cell. 1991;66:347–360. doi: 10.1016/0092-8674(91)90624-8. [DOI] [PubMed] [Google Scholar]
- Mazza CA, Battista D, Zima AM, Szwarcberg-Bracchitta M, Giordano CV, Acevedo A, Scopel AL, Ballare CL. The effects of solar UV-B radiation on the growth and yield of barley are accompanied by increased DNA damage and antioxidant responses. Plant Cell Environ. 1999;22:61–70. [Google Scholar]
- Mazza CA, Boccalandro HE, Giordano CV, Battista D, Scopel AL, Ballare CL. Functional significance and induction by solar radiation of ultraviolet-absorbing sunscreens in field-grown soybean crops. Plant Physiol. 2000;122:117–125. doi: 10.1104/pp.122.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima S, Sugiyama M, Iwai S, Hitomi K, Otoshi E, Kim S-T, Jiang C-Z, Todo T, Britt AB, Yamamoto K. Cloning and characterization of a gene (UVR3) required for photorepair of 6-4 photoproducts in Arabidopsis thaliana. Nucleic Acids Res. 1998;26:638–644. doi: 10.1093/nar/26.2.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormrod DP, Landry LG, Conklin PL. Short-term UV-B radiation and ozone exposure effects on aromatic secondary metabolite accumulation and shoot growth of flavonoid-deficient Arabidopsis mutants. Physiol Plant. 1995;93:602–610. [Google Scholar]
- Rao MV, Paliyath G, Ormrod DP. Ultraviolet-B- and ozone-induced biochemical changes in antioxidant enzymes of Arabidopsis thaliana. Plant Physiol. 1996;110:125–136. doi: 10.1104/pp.110.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault L, Nassar N, Vetter I, Becker J, Klebe C, Roth M, Wittinghofer A. The 1.7 Å crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-blade propeller. Nature. 1998;392:97–101. doi: 10.1038/32204. [DOI] [PubMed] [Google Scholar]
- Rosa JL, Casaroli-Marano RP, Bucker AJ, Vilaro S, Barbacid M. p619, a giant protein related to the chromosome condensation regulator RCC1, stimulates guanine nucleotide exchange on ARF1 and Rab proteins. EMBO J. 1996;15:4262–4273. [PMC free article] [PubMed] [Google Scholar]
- Sancar A. Structure and function of DNA photolyase. Biochemistry. 1994;33:2–9. doi: 10.1021/bi00167a001. [DOI] [PubMed] [Google Scholar]
- Sazer S, Nurse P. A fission yeast RCC1-related protein is required for the mitosis to interphase transition. EMBO J. 1994;13:606–615. doi: 10.1002/j.1460-2075.1994.tb06298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley BW, Hanley S, Goodman HM. Effects of ionizing radiation on a plant genome: analysis of two Arabidopsis transparent testa mutations. Plant Cell. 1992;4:333–347. doi: 10.1105/tpc.4.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM. Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 1995;8:659–671. doi: 10.1046/j.1365-313x.1995.08050659.x. [DOI] [PubMed] [Google Scholar]
- Stolarski R, Bojkov R, Bishop L, Zerefos C, Staehelin J, Zawodny J. Measured trends in stratospheric ozone. Science. 1992;256:342–349. doi: 10.1126/science.256.5055.342. [DOI] [PubMed] [Google Scholar]
- Strid Å, Porra RJ. Alterations in pigment content in leaves of Pisum sativum after exposure to supplementary UV-B. Plant Cell Physiol. 1992;33:1015–1023. [Google Scholar]
- Todo T, Takemori H, Ryo H, Ihara M, Matsunaga T, Nikaido O, Sato K, Nomura T. A new photoreactivating enzyme that specifically repairs ultraviolet light-induced (6-4) photoproducts. Nature. 1993;361:371–374. doi: 10.1038/361371a0. [DOI] [PubMed] [Google Scholar]
- Wade HK, Bibikova TN, Valentine WJ, Jenkins GI. Interactions within a network of phytochrome, cryptochrome and UV-B phototransduction pathways regulate chalcone synthase gene expression in Arabidopsis leaf tissue. Plant J. 2001;25:675–685. doi: 10.1046/j.1365-313x.2001.01001.x. [DOI] [PubMed] [Google Scholar]
- Zhao J, Last RL. Immunological characterization and chloroplast localization of the tryptophan biosynthetic enzymes of the flowering plant Arabidopsis thaliana. J Biol Chem. 1995;270:6081–6087. doi: 10.1074/jbc.270.11.6081. [DOI] [PubMed] [Google Scholar]