Abstract
Proteomics, the study of the proteome (the collection of all the proteins expressed from the genome in all isoforms, polymorphisms and post-translational modifications), is a rapidly developing field in which there are numerous new and often expensive technologies, making it imperative to use the most appropriate technology for the biological system and hypothesis being addressed. This review provides some guidelines on approaching a broad-based proteomics project, including strategies on refining hypotheses, choosing models and proteomic approaches with an emphasis on aspects of sample complexity (including abundance and protein characteristics), and separation technologies and their respective strengths and weaknesses. Finally, issues related to quantification, mass spectrometry and informatics strategies are discussed. The goal of this review is therefore twofold: the first section provides a brief outline of proteomic technologies, specifically with respect to their applications to broad-based proteomic approaches, and the second part provides more details about the application of these technologies in typical scenarios dealing with physiological and pathological processes. Proteomics at its best is the integration of carefully planned research and complementary techniques with the advantages of powerful discovery technologies that has the potential to make substantial contributions to the understanding of disease and disease processes.
Physiology, and scientific research in general, has tended towards a reductionist approach in understanding diseases and the disease processes. Traditionally this strategy has been necessary due to the limitations of most technologies, the complex nature of physiological and pathological systems and the successes related to drilling deeply into a focused area eliminating confounding variables. However, new proteomic technologies have created a renaissance for the classical approaches that deal with understanding complex systems and diseases at a global level, thus allowing an expanded experimental view. Proteomic approaches were initially explored with the advent of two-dimensional gel electrophoresis (O'Farrell, 1975) and application of chromatography techniques (Neverova & Van Eyk 2002); however, the difficulty in identifying proteins suppressed the broad application of proteomics. This technological hurdle has for the most part been overcome through the application of mass spectrometry (MS) used in combination with developing genomics databases. The goal of this review is twofold: the first section provides a brief outline of proteomic technologies, specifically with respect to their applications to broad-based proteomic approaches, and the second part provides more details about how to apply these technologies in typical scenarios dealing with physiological and pathological processes.
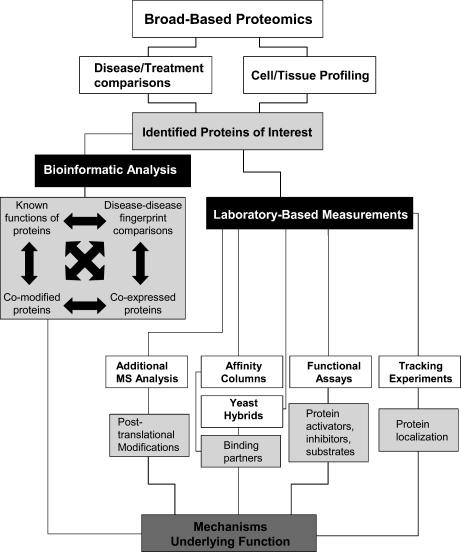
Proteomics defined in its most broad terms is the study of the proteome: the collection of all the proteins expressed from the genome in all isoforms, polymorphisms and post-translational modifications (PTMs). Broad-based proteomics is a strategy wherein disease or physiological models are analysed with the most well-suited proteomics technologies to reveal changes or differences in protein make-up between the experimental condition and controls. This approach allows for the optimal discovery of the changes which define the model system. Screening for the maximum number of protein changes limits experimental bias by not excluding groups of proteins, therefore making it imperative to use the most appropriate technology for the biological system and hypothesis being addressed. The initial proteomic discovery is only the beginning of such a study and must be complemented using a variety of laboratory-based techniques to put the protein changes into their cellular context, as outlined in Fig. 1. This multitiered approach allows us to ask global questions arising from an overview of the disease or physiological condition, and is particularly powerful when applied to studies of protein function by knockout, knock-in or transgenic expression in animals like the mouse. For example, identification of the primary protein target and the composition of related downstream targets; whether the changes involve PTM or expression differences and how these two parameters may be regulated; the timeline for the response (do the protein changes occur in parallel or sequentially?); and the relationships between protein synthesis and degradation of the target – to name a few. Thus, initial data obtained by broad-based proteomics are the framework upon which downstream experimental decisions are based. In the second tier, the initial proteomic screen will pinpoint which subproteome or approach to use in the next level of analysis, or whether to look at multiple different disease models to determine the converging responses. Occasionally, very direct conclusions can be drawn by the results that reveal the changes that are unique from one condition. For instance, in cardiac disease a condition like hypoxia may be differentiated from another condition like ischaemia or in general terms a diseased tissue may be differentiated from a control tissue.
Figure 1. Proteomic process for disease-related protein discovery followed by bioinformatic modelling or laboratory-based biochemical validation.
Technologies and considerations
Create a general hypothesis specific to the study system
The key to increasing the chances for success in any proteomics experiment is to have a thorough understanding of the physiological model or disease process being studied, from which a hypothesis is formed that will drive the choice and selection of the particular proteomic/analytical approach. For example, the proteomic approach will differ for studies in which the pathological perturbation changes the high copy number (high abundance) proteins which dominate a cellular proteome (cytoskeletal proteins, many housekeeping proteins, etc.), versus those that are confined to low copy number – low abundance proteins (e.g. chaperones, signal transduction molecules, etc.). Understanding the experimental system will greatly improve the chances of success on proceeding to the next phase of a proteomics project, or at least determine if proteomics is an appropriate experimental design for the goals.
Choice of model
Proteomics is built upon the foundations of genomics. Therefore, a lack of genomic information on a particular species can substantially limit the success in a proteomics project by increasing the difficulty of successful protein identification. Because of this, the first question a physiologist must address is the choice of model system in which to attempt to apply proteomics, and whether adequate genome coverage exists, while balancing the appropriateness of the model for the specific biological question. Although genome projects involving many of the traditional species used as classical physiological models are now under way (e.g. rabbit, dog and sheep), the current lack of a genome sequence for a particular species, especially animals classically used in many in vivo studies, can be a tremendous stumbling block for a proteomics project, since even having proteins with 80% sequence identity is often insufficient for protein identification through homology by peptide mass fingerprinting (PMF) (Wilkins & Williams 1997) or MS (it is only with de novo sequencing that protein or genome databases are not relied upon for homology; Steen & Mann 2004). Therefore, until the genome of the model species of interest is fully sequenced, the first recommendation is to work with a model where a full genome is in place, or to choose a collaborator that has experience managing projects involving these species. Animal models, however, are of extreme utility in proteomics studies as they eliminate the heterogeneity often observed when using human tissue and allow careful control of temporal and perturbation studies.
Choice of proteomic approach
With broad-based proteomic approaches one is generally interested in maximizing coverage of the proteome or subproteome (a subfractionated or enriched subset of the proteome) under study. Often termed protein expression profiling, the relative changes between proteomes (or subproteomes) are examined either quantitatively (protein quantity) or qualitatively (determination of PTMs and isoforms present). It is important to realize that not all proteomics technologies provide both qualitative and quantitative data – thus the choice of technologies will be dictated by the importance of these two kinds of data in the system under study (Table 1). For instance, in acute injury the immediate cellular response will primarily be changes in the PTM status. General mining (brute-force identification of proteins) to identify all the proteins in a particular sample, and interaction mapping are more specialized applications that require either a high-throughput proteomics laboratory or a facility utilizing yeast genetics as a starting point for identifying potential interaction partners, although most laboratories are capable of using this approach in a limited manner with affinity purification strategies. Today, there is a general lack of PTM characterization. However, there are several approaches including affinity approaches, and the use of new and specific dyes that enrich the detection of specific PTMs (Peng et al. 2003; Ge et al. 2004). Certainly, in the future development of robust methods for detection of specific PTMs and the identification of the exact amino acid residue(s) that are modified will result in an explosion of information.
Table 1.
Common separation technologies and their applications
| Technology | Quantitative | PTMs | Separation | Ease of sample preparation | Experience level | Separation protein (PR)/ peptides (PEP) | Protein abundance (copy number) |
|---|---|---|---|---|---|---|---|
| Gel Based | |||||||
| One-dimensional electrophoresis | NoA | NoB | Low | Simple | Novice | PR/PEP | Med–high |
| Two-dimensional electrophoresis | Relative | YesB | Medium/high | Moderate | Advanced | PR | Med–high |
| Relative quantification | |||||||
| 2DE-DIGE | Relative | Yes | N/A | Simple | Novice | PR | Med–high |
| Chromatography based | |||||||
| rp-HPLC | NoA | NoB | Medium | Simple | Intermediate | PR/PEP | Low–high |
| Cation-exchange | NoA | No | Medium | Simple | Advanced | PR | Med–high |
| Affinity | NoA | No | High | Simple | Novice | PR/PEP | Low–high |
| Capillary electrophoresis | NoA | NoC | High | Simple | Advanced | PEP | Low–high |
| Two-dimensional chromatography | NoA | YesB | High | Simple | Advanced | PR/PEP | Low–highD |
| Combination Approaches | |||||||
| Ion exchange/rp-HPLC (MUDPIT) | NoA | YesC | High | Moderate | Advanced | PR/PEP | Low–HighD |
| Relative quantification | |||||||
| ICAT/iTRAQ/IDBEST/H2018 | RelativeE | Yes | N/A | Moderate | Intermediate/advanced | PEP | Low–HighD |
Must be a spot/band/peak representing a single protein to quantify;
isoelectric shift dependent for 2DE, hydrophobic and isoelectric dependent for 2DLC, some hydrophobic for HPLC, some mass shifts (phosphorylation/glycosylation) for 1DE;
can resolve differences in PTMs between peptide fragments;
low limit detection if coupled to MS for detection;
absolute quantitation of specific peptides can be obtained with iTRAQ if a known quantity is used as a standard.
Sample complexity
Prior to the initiation of any proteomic project, some basic attributes of the sample being analysed must be considered. Broadly these are protein concentration, dynamic range (highest to lowest abundant proteins), degree of protein solubility and the copy number of the protein classes in which one is attempting to assess changes. Despite the femtomolar sensitivity of modern mass spectrometers, issues involved in separating complex protein mixtures (protein interactions, etc.) and obtaining good quality MS data (residual contaminants, etc.) often raise the sample requirements for biological samples, both from an aspect of protein separation and MS identification. A rule of thumb is to expect reasonable success in identifying your protein of interest if the protein can be visualized by coomassie blue staining (> 300 ng), and with developed skill and good instrumentation, success can be obtained in identifying high abundant proteins that are visible by silver staining (> 5 ng) (Shevchenko et al. 1996). Combining samples for multiple preparations or from preparative analysis will increase the likelihood of protein identification.
Applications to disease screening
Choice of sample
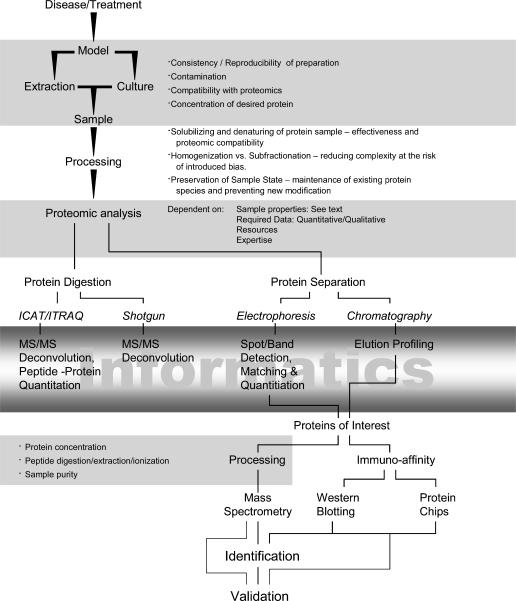
Figure 2 illustrates the decision schema for assessing the feasibility of a proteomic project. The first consideration as outlined above is identifying the complexity of the sample, and the source of the sample. Typical targets for sample acquisition from humans are blood (serum/plasma/cells), cerebrospinal fluid (CSF), urine, mucosal secretions and tissue (biopsy/postmortem), where ease of collection is generally the driving motive, however, quantities may be limited. Ideally, samples from humans are obtained from individuals over time and cohorts have to be carefully controlled to minimize any variability within sample groups. Animal models (depending on size and cost) generally allow researchers to obtain more abundant samples and specifically control where a sample is obtained. In some cases, an appropriate cell culture model exists, where the amount of material can be expanded to suit the large quantity of protein needed for a project. It is necessary to guard against the frustrating position in which the quantity of material is only sufficient to identify the number of proteins changing, and it is impossible to obtain more of the same sample for follow-up experiments such as the actual protein identification, characterization of isoform/PTM, or functional analysis. To avoid this, it is best to have abundant sample (preferably an expendable control sample), where multiple experiments optimizing conditions can be performed, prior to the use of precious samples. As mentioned above, the complexity of the sample must be addressed, as illustrated by the triangle in Fig. 2 in which the interplay between complexity, extraction conditions and reproducibility is shown. Basically, the simplest most reproducible approach should be utilized to obtain the proteome of interest, and if necessary, the same rule should apply to obtaining the subproteome of interest. Generally, one should target the source organ or tissue directly, sampling diseased tissue with an internal control of non-diseased tissue if possible, or at least an exactly age and condition matched control.
Figure 2. An overview of the potential steps of proteomic analysis processes with highlighted considerations and concerns.
This schema is intended to guide the researcher to the most beneficial (most accurate, appropriately scoped, least-biased) analysis of the sample. The gradient informatics box highlights the majority of the proteomic data analysis and often is the bottleneck in proteomic analysis.
Separation technologies
The choice of protein separation tool used is critical as the protein/peptide separation technology must match the experimental hypothesis. Table 1 illustrates some of the potential tools at the disposal of a typical investigator. Protein separation technologies exploit the intrinsic properties of proteins (molecular mass, isoelectric point (charge), hydrophobicity and biospecificity (or immuno-specificity)).
System familiarity
Due to the complexity of any proteomic study the investigator needs to be familiar with the technical aspects even when done in collaboration with a core laboratory. Care must be taken when using technologies and bioinformatics that an investigator does not have direct control over or is not able to directly participate in the process as there are many places where judgement is required. Proteomics is not yet (and most likely will never be) a cook-book approach. There are still many issues that confound results, from protein solubility issues and issues in chromatography (elution profiling) to deconvolution of large data sets, MS coverage, automated analysis settings and bioinformatics analysis choice. Unless an investigator has direct access to the raw data for analysis, and a level of understanding where they can manipulate the raw data, it is unwise to present the data as a discovery; rather it is best to confirm the results by classical biochemical technique. The bottom-line ‘keep it simple’ strategy is most often the best choice for most investigators in regard to proteomics unless they are either making a commitment to proteomics or in collaboration with proteomic investigators.
Protein abundance/protein characteristics/sample complexity
The choice of separating proteins or their peptide complements (obtained from a globally digestion of the proteome) and the separation technology must be based foremost on the abundance and the character of the proteins suspected to change in the experiment. For instance, if PTMs are thought to dominate the proteomic changes, then the separation of intact proteins is often needed. Additionally, care must be taken if the goal is the detection of low copy number proteins as they will be under-represented by most techniques. Digestion of the proteome and analysis of the resulting peptide fragments by liquid chromatography (Table 1) can often enhance the detection of lower abundance proteins, but this can be at the cost of losing information about the nature of the proteins (isoform and PTM status). Specific enhancement of low abundant proteins can be also be used with multiple different techniques. This approach involves breaking the proteome into subproteomes – a group of proteins linked by either cellular location (organelle or specific protein complex, e.g. proteins that bind a specific kinase), or by sharing a chemical propriety (binding ATP or having isoelectric point (pI) values above 9.0). Stasyk & Huber (2004) provide a recent review on fractionation strategies. In terms of broad based analysis, multiple subproteomes may need to be analysed and then the global cellular picture can be put together as the information is integrated.
Qualitative versus quantitative
The next most important choice is whether direct quantification (protein concentration or ratio of protein with respect to one or more condition) of changes is required versus simply analysing the changes qualitatively (all or none or prominent), and whether a goal is to identify post-translational modifications or protein isoform differences. Table 1 illustrates which technologies are appropriate for these choices; however, each has its limitations (see the table and its footnotes). For instance, two-dimensional electrophoresis (2DE) is an excellent choice for relative quantification (by two dimensional difference gel electrophoresis (DIGE) or image analysis software using various dyes), and separation of PTMs (for a review of 2DE, see Gorg et al. 2004); however, the software required for analysis, and scanner equipment needed to take advantage of the robust fluorescent dyes available are very expensive. The length of time required for data analysis and proper spot identification can also be extensive. Liquid chromatography combining chromatographic focusing and hydrophobicity (reversed phase) increases the number of PTMs observed in intact proteins, which can be lost by other methods. It must be recognized that chasing down a PTM, determining the type (phosphorylation versus oxidation or glycosylation) and the exact amino acid residues that are modified, is difficult and often requires additional validation using traditional biochemical methods. Therefore, PTMs are often overlooked and only differences in protein quantification are reported. This is a huge loss to the field; for instance, a method like isotope coded affinity tags (ICAT) with multidimensional liquid chromatography (LC) separation may identify changes in relative abundance of a protein, but may overlook a post-translational modification that is critical in relation to the disease or treatment condition. The future development of more robust methods to track down and monitor PTM is therefore critical. However, the emphasis on developing and expanding the capabilities of each protein separation technology is ongoing and the future is bright. For example see Cantin & Yates (2004) for a recent review on shotgun approaches to PTM analysis.
Relative quantification
Certainly, one of the major breakthroughs in proteomics came with the development of reagents that allow for differential labelling of multiple samples, and then running the samples together in the separation phase (multiplexing). To date, the most successful and widely used examples of this technology is DIGE during 2DE and ICAT using combined chromatography methods. ICAT, which uses different molecular weight mass tags, and affinity purification to attempt to quantify the relative abundance of peptide fragments were the only option available for high-throughput chromatography based applications (Yan et al. 2004; Sethuraman et al. 2004). However, the recent development of iTRAQ, which uses much smaller mass tags of the same molecular weight, but with different fragmentation patterns during tandem MS identification (MS/MS analysis), to determine relative abundance is now another option where the same peptide from different samples is quantified at the same scan in the MS (a limitation of ICAT). Newer reagents are also available including bromine-based mass tags that promise wider applications and ease of use. Table 1 illustrates some of the currently available reagents and their potential applications (for review see Leitner & Lindner 2004).
Mass spectrometry/informatics
Another factor to consider is the degree of difficulty not only in preparation of the sample for separation, but also in the preparation of the fractionated sample for subsequent identification by MS. This can range from simple with methods like reversed phase–HPLC analysis (separating proteins/peptides based on hydrophobicity), since fractions can be easily lyophilized and digested and spotted for either matrix associated laser desorption ionization (MALDI)–time of flight (TOF) or for electrospray ionization (ESI)–MS, to difficult with methods like 2DE, which involves spot cutting, digestion, extraction of peptides and preparation for spotting or MS/MS analysis. Table 1 provides a simplistic summary of the relative difficulty of each method (see Kussmann & Roepstorff (2000) for a review of sample preparation strategies for MALDI).
The application of bioinformatics to a proteomics study is unavoidable. The ominous grey box in Fig. 1 illustrates that bioinformatics must be an integrated portion of any proteomics project and will bear different weights at different stages, depending on which separation strategy is utilized. For instance, methods like ICAT require the tracking of information at all stages, and deconvolution of data to reconstruct which peptides match to which proteins, whereas 2DE may require substantial data analysis for absolute and relative quantification. Regardless, all proteomics project will eventually require substantial data management and organization when they reach the protein identification stage. Before starting a proteomics project, one must consider the type of data generated, and how it will be stored, or the risk of becoming overwhelmed with data organization becomes a very real possibility.
MS dominates protein identification, even though immuno-based identification can be used to answer specific questions or be used for validation of ambiguous MS-based identifications. MS identification, regardless of instrumentation can be divided into two strategies, PMF in which purified proteins are identified by the masses of peptide fragments after digestion with different enzymes, or MS/MS analysis. MS/MS utilizes data obtained by instruments with the ability to isolate one peptide and then induce its fragmentation through either its collision with an inert gas (traditional MS/MS analysis), or by molecular energy transfer during laser desorption with MALDI (known as post-source decay or PSD). Immuno-based identification utilizes traditional blotting techniques to confirm MS identifications that are of low statistical confidence. Immuno-chip-based technologies can by-pass the entire need for separation strategies and MS analysis, for the analysis of a known subset of proteins, or as a validation strategy for proteomic data. Briefly, these techniques generally require the adsorption of an antibody (or bait) to a solid phase that is then incubated with a sample and developed similar to an ELISA-based assay (see Table 2 and Espina et al. (2004) for review). Arrays can be used to screen large number of proteins, and holds promise for even broader analysis as number of proteins capable of being observed quantitatively increase into the hundreds.
Table 2.
Chip-Based technologies and their applications
| Technology | Reproducibility | Quantification | Multiplex | Automation | Capture |
|---|---|---|---|---|---|
| SELDI | Varied | Limited | No | Semi | Yes |
| A2 MicroArray System (Beckman Coulter) | Excellent | Excellent | Yes (= 13) | Yes | Yes |
| Chip-Based Protein Arrays (Sigma, Bio-Rad, etc.) | Varied | Relative | Yes (Chip Area) | Yes | No |
| Chip-Based Multiplex Elisa (FASTQuant – Schleicher and Schuell) | Excellent | Excellent | Yes (Chip Area) | Yes | No |
| Solution Based Arrays (Luminex) | Excellent (CV < 10%) | Excellent | Yes (= 100) | Yes | No |
| Chip-Solution Micro-Array (3D HydroArray, Biocept) | Excellent (CV < 10%) | Excellent | Yes (Currently 45) | Yes | No |
*Capture can be done in different types of conditions (solution based versus solid phase, etc.).
MS analysis is often the stage at which most proteomics projects fall apart – when a great deal of effort has been applied to separate and isolate proteins, and there is a failure to obtain good MS data. Good quality MS data are required to provide unambiguous protein identification and, if possible, information about modifications to the protein. Unfortunately, MS can be very unforgiving with regard to sample preparation requirements (keratin contamination, protein abundance issues, etc.). Also, as mentioned previously, the question of species (and hence matching to the existing protein/genome databases), separation method (amount of protein or peptides present will impact on the resolution of the separation), and sample preparation method will drive the choice of the type of instrumentation that can be used. Table 3A outlines the ionization sources and detectors available in current mass spectrometers, and some of their strengths and weaknesses. The choice of MS instrumentation available will often influence the project design; for instance, if one is interested in obtaining PTM information on a sequence, the ability to achieve good MS/MS data is essential, although use of this type of instrument generally limits the sample throughput that can be achieved. The MS that most beginners will be exposed to is MALDI-TOF. MALDI-TOF instruments are very user friendly and robust, and result in very accurate masses that can then be used for identification by PMF. However, MALDI is typically limited to complex mixtures of only a few proteins, whereas liquid chromatography (LC) front ends, due to the additional separation capabilities, can often handle even more complex mixtures. Newer instruments combine the benefits of MALDI with the power of obtaining good MS/MS data by using more sophisticated mass analysers.
Table 3A.
Typical Ionization sources for mass spectrometers
| Ionization source | Advantages | Disadvantages | Manufacturers |
|---|---|---|---|
| Matrix associated desorption Ionization (MALDI) | Simple sample application and acquisition, high-throughput capable, low down-time | Variable sample preparation for different samples | 3, 4, 5, 7, 8, 10, 12, 14, 15, 16, 17 |
| Electrospray Ionization (ESI) | Consistent sample ionization efficiency, capable of ionizing complex mixtures | Rigid sample preparation requirements, Can have high downtime | 1–5, 9–13, 15 and 18 |
| SELDI (surface enhanced laser desorption ionization) | Affinity capture | Sensitivity, resolution, separation | 6 |
A large degree of effort is expended at the last stage of a proteomics project where identifications must be made from the peak lists obtained from a mass spectrometer. Fenyo (2000) provides a detailed overview of the most common search tools available. Briefly, peak lists are generated by carefully analysing the data generated from a mass spectrometer. For PMF experiments, complex algorithms are used to determine if a peak is real versus electronic noise (peak width, threshold over background, relative ratio of isotopes, mass differences between peaks, etc. are used to make this determination), and then a list is generated for searching. Generally the degree of success on getting a match from the database depends on several factors, including the coverage of the protein digested (actual/theoretical number of fragments), the mass accuracy of the measurement, and the purity of the sample (homogeneity). Recently, guidelines for publishing protein identification data were extablished (Carr et al. 2004). Briefly these include the degree of coverage (number of fragments matched versus theoretical peptides for PMF, and number of fragmentation ions obtained by MS/MS analysis matching the theoretical sequence), the MS analysis software settings used to select peaks for searching, and the parameters used for searching the peak lists against databases.
There is still much advancement required in this area – for example algorithms must be generated to help identify peptide fragments that are modified, databases that include information on small nucleotide polymorphisms, and MS/MS databases that can robustly identify masses corresponding to complex PTMs.
Collaboration or going it alone
The last consideration a researcher new to proteomics should consider relates to whether the investigator should choose a collaborator or go it alone with or without the use of a core facility. These choices are typically situation specific, but some general guidelines to consider are the degree of support offered by a core facility, and whether the staff allow participation in a project, or if they will just accept a sample and run it. Often core facilities do not have the time to invest in a substantial degree of optimization of an experimental system, or to necessarily invest the time involved in the extensive data analysis required downstream (at a reasonable cost). This is detrimental to the field of proteomics. Although less common than core facilities, laboratories specializing in proteomics are often a good choice at minimum in an advisory capacity, and often produce very fruitful collaborations. Regardless, the questions for both scenarios are exactly those outlined above, and each stage of the project should be discussed in depth so that reasonable expectations can be realized. Proteomics often requires the juggling of several technologies, so some facilities and proteomics labs may be more experienced at one than another. Additionally, classical biochemistry techniques can be used effectively to complement proteomic discoveries. Often a protein chemistry laboratory that specializes in separation techniques is an ideal initial collaborator, and then the choice remains whether to collaborate or go it alone with protein identification. The last factor is cost and availability of MS instrumentation. MALDI instruments of varying quality are widely available, and more MS/MS instruments are being brought into core facilities, although the sample throughput capabilities of these instruments are much lower. If a project requires an instrument like a Fourier transform ion cyclotron resonance mass spectrometer (FT-ICR MS), there are few groups in the world in academic settings that have these instruments so time on such instruments is limited.
The future
It is fun, although perhaps dangerous, to speculate on the future in this rapidly changing field, but ideally the future will involve a much higher degree of integration of different experimental approaches, so that minimal experimental findings on a protein's properties (pI, hydrophobicity or molecular mass), plus very limited MS information (one unique fragment) may be used to identify proteins with ease. This will require the development of real-time informatics, to choose the targets that are of high value for mass spectrometry. Another required breakthrough which will assist proteomic experiments is the development of techniques similar to cluster analysis in genomics to determine which classes of proteins are related, so that relative changes between control groups and experimental groups can be differentiated with more ease. Also, as mentioned above, advancements also need to be made with databases to include information on SNPs and PTMs. Finally, as technologies improve to increase the depth of proteome coverage (including instrumentation that can obtain 100% coverage of proteins and interaction mapping projects are completed) interactions between proteins will become more apparent, so that changes in one or two proteins that are visible by current techniques may illuminate changes in other interacting partners that can be chased down using traditional biochemical techniques.
Summary
Proteomics at its best is the integration of carefully planned research with the advantages of powerful discovery and analytical technologies. If planned properly, proteomic projects can be extremely fruitful, and lay the groundwork for years of further research and investigation. Although it is not high-throughput with respect to the amount of time invested, it certainly is with regard to the breadth of what can be observed. Used appropriately in the context of physiological models of diseases and protein function studies using knockout, knock-in and transgenic expression in animals, several new therapeutic targets and biomarkers can be discovered, and the potential contribution towards the understanding of disease and disease processes is unparalleled. That is the eventual promise of proteomics; the technology is the tool to carry us toward this goal, but it has to be the science that drives the process.
Table 3B.
Typical mass analysers for mass spectrometers
| Mass analyser | Applications: | Advantages | Disadvantages | Manufacturers | Availability |
|---|---|---|---|---|---|
| Time of flight (TOF) | High-throughput PMF | High throughput, ease of use | Most unable to do MS/MS (PSD) | 3–8, 12–17 | Common |
| TOF-reflectron/linear or curved | High-throughput PMF, postsource decay (PSD) | High throughput, High mass accuracy | PSD sometimes difficult to ID and hard to obtain | 16 | Common |
| Quadropole (Q)–TOF | MS, MS/MS | Good MS/MS, good mass accuracy, quantification possible | Not high-throughput, Cost | 4 and 5 | Less common |
| Quadropole–ion trap | MS, MS/MS (CID), MSn | Good MS/MS, ability to do multiple rounds of fragmentation, sensitive | Not high-throughput | 18 | Less common |
| Triple quadropole | MS, MS/MS (CID) | Excellent MS/MS with CID | Cost | 4 and 18 | Less common/rare |
| Quadropole/hexapole/TOF | MS, MS/MS (CID), DeNovo Sequencing* | Excellent MS, MS/MS and mass accuracy | Cost | 15 *(Q-TOF Ultima) | Less Common |
| Ion trap | MS, MS/MS (CID), MSn | Good MS/MS, ability to do multiple rounds of fragmentation | Mass range | 1, 5, 9 | Less Common |
| Magnetic Sector | MS, MS/MS (CID) | Wide range of sample concentrations | Mass range (unless high-end) | 2 and 11 | Rare |
| FTMS | MS, MS/MS (CID), MSn | Exquisite mass accuracy, wide mass range, fragmentation | Very costly, Difficult to operate | 10 | Very rare |
| Cryo-cooled detector | MS | Extremely high mass detection | Ionization efficiency, cost | 7 | Very rare |
Manufacturers: (1) Agilent Technologies, (2) AMD Intectra GmbH, (3) Amersham Pharmacia Biotech, (4) Applied Biosystems, (5) Bruker Daltonics Inc., (6) Ciphergen Biosystems Inc., (7) Comet, (8) GSG Mess-und Analysengeräte GmbH, (9) Hitachi Instruments Inc., (10) IonSpec Corp., (11) JEOL USA Inc., (12) Kratos Analytical, (13) LECO Corp., (14) Mass Technologies, (15) Micromass-Waters, (16) Scientific (SAI) Ltd, (17) Stanford Research Systems, (18) Thermo Finigan. (Manufacturers and corresponding mass analysers based upon information from The Scientist (2001), vol. 15, p. 31.)
Acknowledgments
The authors would like to acknowledge the following funding sources that supported this review: NHLBI (contract N0-HV-28180), NIH (R01 HL56370 and R01 HL61638), and a grant from the Donald W. Reynolds Foundation. A special thanks to Brian Stanley for critical review of the manuscript.
References
- Carr S, Aebersold R, Baldwin M, Burlingame A, Clauser K, Nesvizhskii A. The need for guidelines in publication of peptide and protein identification data. Working Group on Publication Guidelines for Peptide and Protein Identification Data. Mol Cell Proteomics. 2004;3:531–533. doi: 10.1074/mcp.T400006-MCP200. Epub 2004 April. 09. [DOI] [PubMed] [Google Scholar]
- Cantin GT, Yates JR., 3rd Strategies for shotgun identification of post-translational modifications by mass spectrometry. J Chromatogr A. 2004;1053:7–14. doi: 10.1016/j.chroma.2004.06.046. [DOI] [PubMed] [Google Scholar]
- Espina V, Woodhouse EC, Wulfkuhle J, Asmussen HD, Petricoin EF, Liotta LA., 3rd Protein microarray detection strategies: focus on direct detection technologies. J Immunol Meth. 2004;290:121–133. doi: 10.1016/j.jim.2004.04.013. 10.1016/j.jim.2004.04.013. [DOI] [PubMed] [Google Scholar]
- Fenyo D. Identifying the proteome: software tools. Curr Opin Biotechnol. 2000;11:391–395. doi: 10.1016/s0958-1669(00)00115-4. 10.1016/S0958-1669(00)00115-4. [DOI] [PubMed] [Google Scholar]
- Ge Y, Rajkumar L, Guzman RC, Nandi S, Patton WF, Agnew BJ. Multiplexed fluorescence detection of phosphorylation, glycosylation, and total protein in the proteomic analysis of breast cancer refractoriness. Proteomics. 2004;4:3464–3467. doi: 10.1002/pmic.200400957. 10.1002/pmic.200400957. [DOI] [PubMed] [Google Scholar]
- Gorg A, Weiss W, Dunn MJ. Current two-dimensional electrophoresis technology for proteomics. Proteomics. 2004;15:15. doi: 10.1002/pmic.200401031. [DOI] [PubMed] [Google Scholar]
- Kussmann M, Roepstorff P. Sample preparation techniques for peptides and proteins analyzed by MALDI-MS. Meth Mol Biol. 2000;146:405–424. doi: 10.1385/1-59259-045-4:405. [DOI] [PubMed] [Google Scholar]
- Leitner A, Lindner W. Current chemical tagging strategies for proteome analysis by mass spectrometry. J Chromatogr B Anal Technol Biomed Life Sci. 2004;813:1–26. doi: 10.1016/j.jchromb.2004.09.057. [DOI] [PubMed] [Google Scholar]
- Neverova I, Van Eyk JE. Application of reversed phase high performance liquid chromatography for subproteomic analysis of cardiac muscle. Proteomics. 2002;2:22–31. 10.1002/1615-9861(200201)2:1<22::AID-PROT22>3.3.CO;2-C. [PubMed] [Google Scholar]
- O'Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Sethuraman M, McComb ME, Heibeck T, Costello CE, Cohen RA. Isotope-coded affinity tag approach to identify and quantify oxidant-sensitive protein thiols. Mol Cell Proteomics. 2004;3:273–278. doi: 10.1074/mcp.T300011-MCP200. 10.1074/mcp.T300011-MCP200. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Stasyk T, Huber LA. Zooming in: Fractionation strategies in proteomics. Proteomics. 2004;12:3704–3716. doi: 10.1002/pmic.200401048. 10.1002/pmic.200401048. [DOI] [PubMed] [Google Scholar]
- Steen H, Mann M. The ABC's (and XYZ's) of peptide sequencing. Nat Rev Mol Cell Biol. 2004;5:699–711. doi: 10.1038/nrm1468. 10.1038/nrm1468. [DOI] [PubMed] [Google Scholar]
- Wilkins MR, Williams KL. Cross-species protein identification using amino acid composition, peptide mass fingerprinting, isoelectric point and molecular mass: a theoretical evaluation. J Theor Biol. 1997;186:7–15. doi: 10.1006/jtbi.1996.0346. 10.1006/jtbi.1996.0346. [DOI] [PubMed] [Google Scholar]
- Yan W, Lee H, Deutsch EW, Lazaro CA, Tang W, Chen E, Fausto N, Katze MG, Aebersold R. A dataset of human liver proteins identified by protein profiling via isotope-coded affinity tag (ICAT) and tandem mass spectrometry. Mol Cell Proteomics. 2004;3:1039–1041. doi: 10.1074/mcp.D400001-MCP200. 10.1074/mcp.D400001-MCP200. [DOI] [PubMed] [Google Scholar]