Abstract
The Arabidopsis Salt Overly Sensitive 2 (SOS2) gene encodes a serine/threonine (Thr) protein kinase that has been shown to be a critical component of the salt stress signaling pathway. SOS2 contains a sucrose-non-fermenting protein kinase 1/AMP-activated protein kinase-like N-terminal catalytic domain with an activation loop and a unique C-terminal regulatory domain with an FISL motif that binds to the calcium sensor Salt Overly Sensitive 3. In this study, we examined some of the biochemical properties of the SOS2 in vitro. To determine its biochemical properties, we expressed and isolated a number of active and inactive SOS2 mutants as glutathione S-transferase fusion proteins in Escherichia coli. Three constitutively active mutants, SOS2T168D, SOS2T168DΔF, and SOS2T168DΔ308, were obtained previously, which contain either the Thr-168 to aspartic acid (Asp) mutation in the activation loop or combine the activation loop mutation with removal of the FISL motif or the entire regulatory domain. These active mutants exhibited a preference for Mn2+ relative to Mg2+ and could not use GTP as phosphate donor for either substrate phosphorylation or autophosphorylation. The three enzymes had similar peptide substrate specificity and catalytic efficiency. Salt overly sensitive 3 had little effect on the activity of the activation loop mutant SOS2T168D, either in the presence or absence of calcium. The active mutant SOS2T168DΔ308 could not transphosphorylate an inactive protein (SOS2K40N), which indicates an intramolecular reaction mechanism of SOS2 autophosphorylation. Interestingly, SOS2 could be activated not only by the Thr-168 to Asp mutation but also by a serine-156 or tyrosine-175 to Asp mutation within the activation loop. Our results provide insights into the regulation and biochemical properties of SOS2 and the SOS2 subfamily of protein kinases.
During growth and development of many multicellular organisms, protein kinases function in a variety of signaling pathways critical for cell division, metabolism and response to hormonal, developmental, and environmental signals. The activity of protein kinases can either be stimulatory or inhibitory to downstream targets (Simon, 1994; Perrimon, 1995). Knowledge of how the relevant protein kinases are regulated, therefore, is one key to understanding basic cellular processes involved in growth and development. The SNF1/AMPKs are highly conserved Ser/Thr protein kinases identified in fungi, fruitfly (Drosophila melanogaster), Caenorhabditis elegans, mammals, and plants (McCartney and Schmidt, 2001). Many SNF1-related protein kinase genes (SnRKs) have been isolated in plants, and these SnRK kinases have been classified into three subgroups (SnRK1, SnRK2, and SnRK3) based on sequence similarity (Halford and Hardie, 1998).
The Arabidopsis Salt Overly Sensitive 2 (SOS2) and Salt Overly Sensitive 3 (SOS3) genes were isolated through positional cloning and were shown to be required for sodium and potassium ion homeostasis and salt tolerance (Liu and Zhu, 1997, 1998). SOS2 encodes a 446-amino acid Ser/Thr protein kinase with an N-terminal kinase catalytic domain similar to SNF1/AMPK and a novel C-terminal regulatory domain (Liu et al., 2000). SOS2 can be classified as a member of the SnRK3 subgroup of SNF1-related protein kinases. SOS3 encodes a myristoylated EF-hand calcium-binding protein (Liu and Zhu, 1998; Ishitani et al., 2000) that may sense the calcium signal elicited by salt stress (Knight et al., 1997). SOS3 physically interacts with SOS2 in the yeast (Saccharomyces cerevisiae) two-hybrid system and in vitro (Halfter et al., 2000). Both the catalytic and regulatory domains are essential for SOS2 function in salt tolerance (Liu et al., 2000). A 21-amino acid sequence in the regulatory domain of SOS2, the FISL motif, has been determined to be necessary and sufficient to bind SOS3 (Guo et al., 2001). Salt stress up-regulation of the Salt Overly Sensitive 1 (SOS1) gene encoding a Na+/H+ antiporter is partially under control of the SOS2-SOS3 regulatory pathway (Shi et al., 2000). SOS2 and SOS3 are more importantly both required for the posttranslational activation of SOS1 Na+/H+ exchange activity (Qiu et al., 2002).
Recently, we have characterized the functional domains in SOS2 kinase (Guo et al., 2001). Recombinant SOS2 protein produced in bacteria exhibits no substrate phosphorylation activity in the absence of SOS3, although it has autophosphorylation activity (Halfter et al., 2000). In the presence of calcium, SOS3 activates the substrate phosphorylation activity of SOS2 (Halfter et al., 2000). The substrate phosphorylation activity of SOS2 could also be activated by a Thr-168 to Asp mutation within the activation loop or by removal of the autoinhibitory FISL motif (Guo et al., 2001; Qiu et al., 2002). In this study, we used the constitutively activated SOS2 mutants to characterize the biochemical properties of SOS2. These properties include divalent cation preference, phosphate donor specificity, steady-state substrate kinetics, and the reaction mechanism of autophosphorylation. We also discovered that the substitution of Ser-156 or Tyr-175 within the activation loop with Asp could also activate SOS2. These results help understand the biochemical characteristics and the regulation of SOS2 protein kinase.
RESULTS
Expression, Purification, and Kinase Activities of Recombinant SOS2 Mutants
Bacterially expressed SOS2 recombinant protein is inactive by itself in peptide substrate phosphorylation and becomes active in the presence of SOS3 that binds to the autoinhibitory FISL motif of SOS2 (Halfter et al., 2000; Guo et al., 2001). Three SOS3-independent, constitutively active mutants SOS2T168D, SOS2T168DΔF, and SOS2T168DΔ308 were produced, which contain the activation loop Thr-168 to Asp mutation or combine the activation loop mutation with removal of the FISL motif or regulatory domain (Guo et al., 2001; Qiu et al., 2002). These were expressed here as glutathione S-transferase (GST)-tagged fusion proteins in E. coli and affinity-purified on glutathione-Sepharose. The eluting proteins were analyzed for purity by SDS-PAGE (data not shown). Each protein migrated as predicted from its molecular mass. The purity of these preparations was estimated to be above 95%, and their identities were confirmed by western analysis (data not shown). These purified kinase samples were used for the remainder of this study.
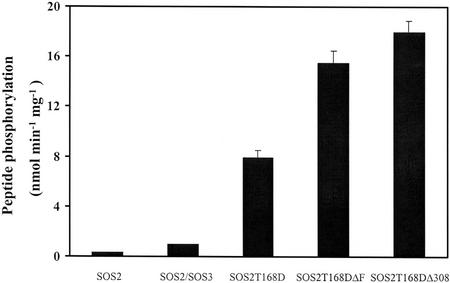
An exogenous peptide, p3 (ALARAASAAALARRR), derived from the recognition sequences of protein kinase C and SNF1/AMPK, was earlier shown to be phosphorylated by SOS2 in the presence of SOS3 (Halfter et al., 2000). Kinase activity of the purified recombinant proteins was evaluated by measuring phosphorylation activity toward this peptide substrate, without addition of SOS3. A standard kinase assay with 5 mm Mg2+ showed that these SOS2 mutants displayed much higher phosphorylation of the peptide substrate p3 (Fig. 1) and autophosphorylation (data not shown) than either SOS2 alone or SOS2 in the presence of SOS3 (designated SOS2/SOS3) did. These active SOS2 mutants were, thus, chosen for further biochemical characterization.
Figure 1.
Kinase activities of the purified recombinant SOS2T168D, SOS2T168DΔF, and SOS2T168DΔ308 fusion proteins. The SOS2, SOS3, and SOS2 mutant cDNAs were expressed as GST-tagged fusion proteins in E. coli and purified by glutathione-Sepharose affinity chromatography. Peptide phosphorylation activities of SOS2 and SOS2 mutants were measured using p3 as a peptide substrate in the presence of 5 mm Mg2+. Error bars indicate ±sd (n = 3).
Phosphorylation of the peptide substrate by all SOS2 mutants was linear during 30 min (data not shown). The autophosphorylation activity of these proteins was detectable in 5 min, the first time point assayed, and completed after 30 min (data not shown). All subsequent kinase assays, therefore, were routinely terminated at 30-min incubations to obtain a good estimate of the initial rate. In addition, we have found that the purified enzyme is highly stable when maintained in a concentrated solution, even at room temperature but rapidly loses activity upon dilution. Enzyme inactivation after dilution could result from alterations in either the tertiary structure of the enzyme or its aggregation state. Our observations are similar to previous reports demonstrating that the aggregation state of a type 1 receptor Tyr kinase catalytic domain significantly affects the rate of catalysis (Mohammadi et al., 1993; Gregoriou et al., 1995).
Divalent Cation Preference
Kinases, like other phosphotransferases, require a divalent cation to coordinate the phosphate groups of the nucleotide triphosphate substrate. These enzymes can also be activated or inactivated by binding of a cation to an additional site of interaction (Armstrong et al., 1979; Sun and Budde, 1997). To determine the divalent cation preferences in vitro of these mutants, we measured peptide substrate phosphorylation in the presence of various concentrations of either Mg2+ or Mn2+ (Fig. 2, A and B; SOS2T168D data not shown). With all kinases, there was no substrate phosphorylation in the absence of added divalent cation, and all of them showed higher rates with Mn2+ than with Mg2+. Minimal concentrations for any activity were 0.25 mm for Mn2+ and 0.5 mm for Mg2+, and optimal concentrations were 2.5 and 5 mm, respectively. Above 2.5 mm, Mn2+ was inhibiting for all kinases. ATP was held constant at 10 μm in the experiment, and so any concentration of cation above 10 μm is essentially free from bound nucleotide. The difference must, therefore, reflect different affinity of Mn2+ and Mg2+ for binding to a cation site on the enzyme. The apparent Km and Kcat values for ATP with either Mg2+ or Mn2+ were determined at a constant concentration of peptide substrate p3. These experiments were performed at the optimal concentration of free Mg2+ or Mn2+ as seen in Figure 2. The titrations of ATP with Mg2+ or Mn2+ were conducted in parallel, and the reactions were initiated with the same diluted enzyme mixture to ensure that the results are directly comparable. The results are summarized in Table I. For all enzymes, the Kcat/Km was 4- to 5-fold higher for ATP with Mn2+ than for ATP with Mg2+.
Figure 2.
Divalent cation dependence of peptide phosphorylation. Peptide phosphorylation reactions by the kinases were performed using peptide substrate p3 at various concentrations of Mn2+ (as MnCl2) or Mg2+ (as MgCl2) as indicated. Initial rates were measured and plotted against the Mn2+ or Mg2+ concentrations. A, SOS2T168DΔ308. B, SOS2T168DΔF.
Table I.
Kinetic parameters for ATP metal
| Enzyme | ATP-Mg2+
|
ATP-Mn2+
|
||||||
|---|---|---|---|---|---|---|---|---|
| Mg2+ | Kcat | Km | Kcat/Km | Mn2+ | Kcat | Km | Kcat/Km | |
| mm | s−1 | μm | m−1 s−1 | mm | s−1 | μm | m−1 s−1 | |
| SOS2T168D | 5.0 | 0.79 | 3.0 | 2.60 × 105 | 2.5 | 1.54 | 1.3 | 11.9 × 105 |
| SOS2T168DΔF | 5.0 | 0.82 | 3.4 | 2.41 × 105 | 2.5 | 1.85 | 1.5 | 12.3 × 105 |
| SOS2T168DΔ308 | 5.0 | 0.86 | 4.0 | 2.15 × 105 | 2.5 | 1.89 | 2.0 | 9.5 × 105 |
Peptide substrate phosphorylation reactions were performed using indicated enzymes and peptide p3 (150 μm). Mn2+ (as MnCl2) and Mg2+ (as MgCl2) were added at the optimum concentration as indicated. Three experiments were performed for each enzyme substrate combination using concentration of ATP metal described in Fig. 2. The sd values of the parameter estimates were all less than 10% of the value shown.
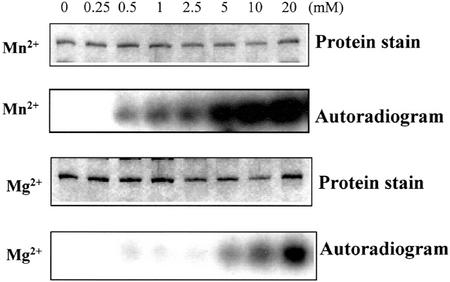
To determine the metal cation requirement of the SOS2 mutants for autophosphorylation, we used the same concentration series of the two divalent cations in the assay system. Mn2+ provided significant activation by 1 mm with all enzymes (Fig. 3 for SOS2T168DΔ308; SOS2T168D and SOS2T168DΔF data not shown). A similar level of activation required 10 mm of Mg2+. It is apparent that autophosphorylation prefers Mn2+ over Mg2+, but there are very different optimal levels than those seen in peptide phosphorylation. In subsequent studies, we used the concentration of Mn2+ (2.5 mm) optimal for substrate phosphorylation and adequate for autophosphorylation.
Figure 3.
Dependence of autophosphorylation of SOS2T168DΔ308 on divalent cations. Autophosphorylation of SOS2T168DΔ308 in the presence of various concentrations of Mn2+ (as MnCl2) or Mg2+ (as MgCl2), as indicated, was presented as the density of autoradiographic bands. Three independent experiments were performed, and a typical result is shown here.
A small number of protein kinases use GTP and ATP as a phosphate donor. These include a receptor-like kinase from Madagascar periwinkle (Schulze-Muth et al., 1996), a human STE20-like Ser/Thr protein kinase (Schinkmann and Blenis, 1997), and an Arabidopsis Ser/Thr protein kinase CK2 (Sugano et al., 1998). To test for GTP use with our enzymes, increasing concentrations of unlabeled GTP were used to compete with [γ-32P]ATP in the substrate phosphorylation and autophosphorylation assays. Cold GTP did not compete with ATP for all kinases in substrate phosphorylation of p3 or in autophosphorylation (data not shown). To further confirm the specificity of ATP as a phosphate donor, kinase assays were performed using either [γ-32P]ATP or [γ-32P]GTP at identical specific activities. None of them could use GTP as a phosphate donor for both autophosphorylation and peptide phosphorylation (data not shown).
Steady-State Peptide Substrate Kinetics
Both substrate phosphorylation and autophosphorylation of the three kinases had pH optima between 7.0 and 7.5, and activity was optimal at 30°C for all kinases (data not shown). In addition to p3, SOS2 could phosphorylate two other synthetic peptides: p1 (LRRASLG) and p2 (VRKRTLRRL), derived from the recognition sequences of protein kinase C or SNF1/AMPK (Halfter et al., 2000). To evaluate peptide substrate preference of these kinases, we analyzed the steady-state kinetic parameters toward the three peptides. Apparent Km and Kcat values for p1, p2, and p3 of the three kinases were determined from Eadie-Hofstee plots of V0 versus V0/[S] (data not shown). The Kcat to Km ratios show clearly that all kinases prefer p3 as a substrate to either p1 or p2 (Table II). Although p3 is not based on a physiological substrate for SOS2, all these Km values are within the range of those found for plant SNF1-related kinases with peptide substrates that do reflect true physiological substrates. For one example, cauliflower (Brassica oleracea) 3-hydroxy-3-methylglutaryl-CoA reductase kinasehad a Km of 95 μm for the SAMS peptide based on conserved residues of known physiological substrates (Weekes et al., 1993).
Table II.
Peptide substrate steady-state kinetic parameters with three peptide substrates
| SOS2 Mutant | Peptide Substrate | Kcat | Km | Kcat/Km |
|---|---|---|---|---|
| s−1 | μm | m−1 s−1 | ||
| SOS2T168D | p1 | 1.89 | 211 ± 3.9 | 0.89 × 104 |
| p2 | 2.30 | 119 ± 2.8 | 1.93 × 104 | |
| p3 | 2.76 | 99 ± 1.1 | 2.79 × 104 | |
| SOS2T168DΔF | p1 | 2.01 | 201 ± 3.1 | 1.00 × 104 |
| p2 | 2.26 | 111 ± 1.8 | 2.04 × 104 | |
| p3 | 2.62 | 95 ± 1.5 | 2.76 × 104 | |
| SOS2T168DΔ308 | p1 | 1.90 | 198 ± 5.1 | 0.96 × 104 |
| p2 | 2.32 | 145 ± 4.1 | 1.60 × 104 | |
| p3 | 2.51 | 113 ± 4.8 | 2.22 × 104 |
Phosphorylation of the peptide substrate p1, p2, or p3 by SOS2T168D, SOS2T168DΔF, or SOS2T168DΔ308 was measured at optimal concentration of Mn2+ (2.5 mm). The kinetic parameters were determined by varying [p1], [p2], and [p3] while holding ATP at 10 μm. Kcat/Km is presented here as a measure of overall enzyme efficiency for each peptide substrate. Values are the means ± sd from three separate experiments.
Effect of SOS3 on Kinase Activity of SOS2T168D
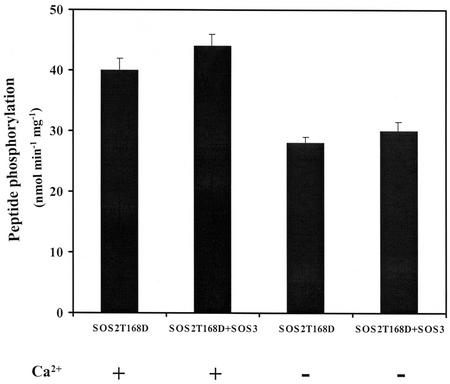
The regulatory protein SOS3 has been shown to activate SOS2 in a Ca2+-dependent manner (Halfter et al., 2000) by binding to the autoinhibitory FISL motif in the C-terminal domain of SOS2 (Guo et al., 2001). To test whether SOS3 still could enhance the activity of the activation loop Thr-168 to Asp mutant SOS2T168D (with no deletions), we compared the phosphorylation of p3 by SOS2T168D with or without SOS3, either in the presence or absence of 0.5 mm calcium. SOS3 had no significant effect on p3 phosphorylation by SOS2T168D either in the presence or absence of calcium (Fig. 4). SOS3 also exhibited little effect on autophosphorylation activity, either with or without calcium (data not shown). These observations suggest that the Thr-168 to Asp mutation within the activation loop could release (at least partially) the autoinhibitory effect of the FISL motif on SOS2 kinase activity, thus, making SOS2 independent of the regulatory protein SOS3. In addition, calcium (0.5 mm) was not required for kinase activity of SOS2T168D, although it seemed to slightly activate SOS2T168D either in the presence or absence of SOS3 (Fig. 4). At the present time, the significance and potential mechanism of this slight calcium enhancement of SOS2T168D activity is unclear.
Figure 4.
Effect of SOS3 on kinase activity of the activation loop mutant SOS2T168D. Substrate phosphorylation of SOS2T168D using peptide substrate p3 was measured with SOS3 or without SOS3 in the presence or absence of 0.5 mm Ca2+ (as CaCl2) in the kinase buffer. Error bars indicate ±sd (n = 3).
Autophosphorylation Mechanism
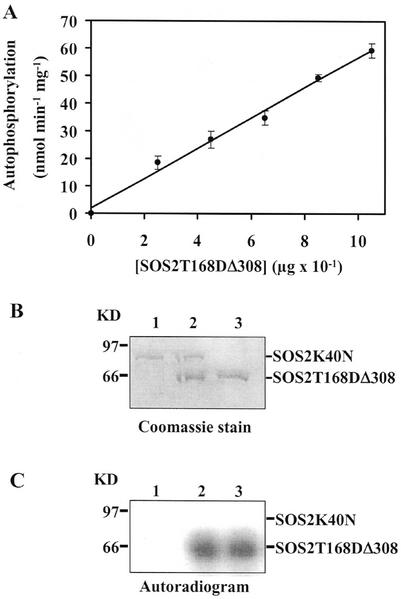
In many cases autophosphorylation of a protein kinase has been shown to proceed by an intermolecular mechanism (Johnson et al., 1996) in which the catalytic domains and phosphorylation sites reside on separate molecules. To test by which mechanism SOS2 kinase could autophosphorylate, we determined the dependence of autophosphorylation activity on protein concentration. The autophosphorylation reactions of all these kinases showed first order kinetics (a linear increase in rate with increasing kinase protein) rather than second order (rate increases with the square of kinase concentration; Fig. 5A for SOS2T168DΔ308; data not shown), which suggests that SOS2 may autophosphorylate by an intramolecular reaction (Horn and Walker, 1994).
Figure 5.
Intramolecular autophosphorylation mechanism of SOS2. A, A plot of SOS2T168DΔ308 autophosphorylation versus its protein concentration. Actual amounts of SOS2T168DΔ308 varied from 0 to 1.05 μg per 30-μL reaction. A 30-μL reaction mixture contained various concentrations of the purified SOS2T168DΔ308 protein as indicated, 5 μCi of [γ-32P]ATP, and 2.5 mm Mn2+ (as MnCl2) in kinase assay buffer. Reactions were incubated for 30 min at 30°C. After separation on 10% (w/v) SDS-PAGE, the resulting gels were autoradiographed using a phosphor imager. The first order kinetics suggests an intramolecular autophosphorylation mechanism. B, Protein stain. Lanes 1, 2, and 3 represent Coomassie Blue-stained gel corresponding to lanes 1, 2, and 3 in C. C, Autoradiographs of phosphorylation assays of SOS2K40N and SOS2T168DΔ308 proteins. Eight hundred nanograms of either protein as shown in B was incubated alone or together in the presence of 5 μCi of [γ-32P]ATP in kinase assay buffer as described above, fractionated by 10% (w/v) SDS-PAGE, and exposed to x-ray film. Lane 1, SOS2K40N; lane 2, SOS2K40N and SOS2T168DΔ308; and lane 3, SOS2T168DΔ308. Lack of a labeled band of the same size in lane 2 of B shows that SOS2T168DΔ308 (62 kD) cannot transphosphorylate SOS2K40N (80 kD), again suggesting an intramolecular autophosphorylation mechanism.
To provide additional evidence, we tested whether the truncated active protein SOS2T168DΔ308 was able to transphosphorylate the kinase-dead mutant SOS2K40N. This mutant has the residue Lys-40, a conserved amino acid in the catalytic site required for phosphotransfer activity in all protein kinases (Hanks et al., 1988; Knighton et al., 1991), changed to Asn through site-directed mutagenesis. The active kinase SOS2T168DΔ308 could be clearly resolved from the full-length protein SOS2K40N by SDS-PAGE (Fig. 5B). This inactive protein was then co-incubated with SOS2T168DΔ308 in the kinase assay. As expected, SOS2K40N failed to autophosphorylate (Fig. 5C, lane 1), and SOS2T168DΔ308 had high autophosphorylation activity (Fig. 5C, lanes 2 and 3). The SOS2K40N was not trans-phosphorylated in the presence of SOS2T168DΔ308 (Fig. 5C, lane 2), and no autophosphorylation of the inactive SOS2 mutant was detectable even after very long exposures (data not shown). These results are strong evidence for an intramolecular rather than intermolecular reaction mechanism of SOS2 autophosphorylation. Thus, unlike some other Ser/Thr protein kinases (Horn and Walker, 1994; Oh et al., 2000; Shah et al., 2001), there is no need to postulate oligomerization of the protein as part of the mechanism.
Activation by Substitution of Either Ser or Tyr with Asp within the Activation Loop
Many protein kinases are activated by phosphorylation of one or more residues within an activation loop. The introduction of a phosphate results in ionic interactions that are critical to kinase activity (Johnson et al., 1996). In some protein kinases, such as phosphorylase b kinase and phosphoenolpyruvate carboxylase kinase (Hartwell et al., 1999), the phosphorylation site within the activation loop is replaced by a negatively charged residue. These kinases are indeed constitutively active, and do not require phosphorylation.
SOS2 protein is not constitutively active in substrate phosphorylation. In the SOS2 kinase subfamily, the activation loop is located between the conserved DFG and APE residues in the kinase catalytic domain (Guo et al., 2001). A comparison of the activation loops of 23 members of the kinase subfamily showed that in addition to a Thr residue, either a Ser or Tyr residue is completely conserved in all members of this subfamily (Fig. 6; data not shown). By mutating the conserved Thr to Asp in its activation loop, we earlier created a constitutively active SOS2 kinase (Guo et al., 2001). Here, we wanted to see if changing either the conserved Ser or Tyr to Asp could also make these kinases constitutively active. We constructed two activation loop single mutants, designated SOS2S156D and SOS2Y175D by mutating Ser-156 to Asp and Tyr-175 to Asp, respectively via site-directed mutagenesis.
Figure 6.
Sequence alignment of the SOS2 activation loop compared with the corresponding region of the sequence of two SOS2-like kinases PKS11 and PKS18. A, Shown is a schematic diagram of the domain structure for SOS2. FISL, FISL motif. B, An alignment of SOS2, PKS11, and PKS18 activation loop. PKS11 and PKS18 are identical to gene products with the following GenBank accession numbers: T09903 and BAB09310, respectively. Amino acids are numbered on the left. Identical residues and conservative replacements are shown with black and gray shading, respectively. The three conserved residues in all members of SOS2 subfamily kinase are marked with asterisks (Ser and Tyr) and a dot (Thr), respectively. Dashed lines represent spaces that were introduced to maximize alignment.
We expressed these mutants, and also the wild-type protein as GST fusion proteins in E. coli, and purified them on glutathione-Sepharose (data not shown). SDS-PAGE analysis shows that the expression level of the recombinant mutant proteins was similar to that of the wild type (data not shown). Kinase assays showed that SOS2S156D and SOS2Y175D exhibited 19- and 23-fold higher activity, respectively, in p3 phosphorylation than the wild-type kinase (data not shown). Both activation loop mutant kinases also had higher autophosphorylation activity compared with the wild type (data not shown).
DISCUSSION
SOS2 Exhibits Unusual But Not Unique ATP-Metal Substrate Preference
The biochemical characteristics of SOS2 and mechanisms of the regulation of its kinase activity are not fully understood. The current evaluation of the biochemical and kinetic properties of the SOS2 has been greatly facilitated by the availability of recombinant SOS2 mutants that show robust kinase activities. All of these three SOS2 mutants exhibited a strong preference for Mn2+ over Mg2+ for peptide substrate phosphorylation (Fig. 2) and autophosphorylation (Fig. 3).
We conducted ATP kinetic studies in the presence of the optimal divalent metal concentration. These studies show that all of these proteins have a higher affinity and catalytic efficiency toward ATP with Mn2+ than with Mg2+ (Table I). Activity of a number of other kinases also works better with Mn2+ than with Mg2+ (Horn and Walker, 1994; Muranaka et al., 1994; Su et al., 1996; Schinkmann and Blenis, 1997; Stocchetto et al., 1997). This divalent metal preference has been thought to reflect involvement of the kinase in a complex for full activation (Guy et al., 1992; Su et al., 1996).
The intracellular concentration of Mn2+ is in the micromolar range, whereas the Mg2+ concentration is in the millimolar range (Schinkmann and Blenis, 1997). The minimal and optimal activation concentrations of Mn2+ for SOS2 mutants in vitro (Figs. 2 and 3) suggest that Mn2+ may not play a role in SOS2 activity regulation under physiological conditions.
Peptide Substrate Selectivity
SOS2 can be classified as a member of the SnRK3 family of SNF1-related protein kinases (Hardie, 2000). The physiological substrate (s) of SOS2 kinase is still unknown. Testing the three different peptide substrates with these kinases, the Kcat to Km ratios show clearly a preference order of p3 > p2 > p1 (Table II). The apparent Km values toward the three peptides for these kinases (Table II) are similar to those for SNF1/AMPK from yeast, mammalian, and higher plants with Ser-containing peptides as substrates (Barker et al., 1996; Hardie, 1999). The preferred peptide substrate p3 (ALARAASAAALARRR) contains within it the following sequence: hydrophobic-X-basic-X (2)-Ser-X (3)-hydrophobic residue. The same motif has been previously established as a minimal recognition motif (Dale et al., 1995b) for the cauliflower AMPK/SNF1 homolog (Ball et al., 1995). An alignment of sequences around the phosphorylation sites on 3-hydroxy-3-methylglutaryl-CoA reductases, nitrate reductases, and Suc phosphate synthases from different plant species identified a consensus recognition motif for SnRK1 protein kinases (Sugden et al., 1999b), and this is shared by p3. Therefore, the presence of the hydrophobic and basic residues may be a determinant for the substrate specificity of the SOS2. Comparison of the kinetic data of these SOS2 mutants shows that removing the regulatory domain of SOS2 does not seem to significantly affect cofactor preference, peptide substrate specificity, and catalytic efficiency toward peptide substrates.
We have previously reported that salt stress up-regulation of a Na+/H+ antiporter SOS1 gene is controlled by the SOS3-SOS2 regulatory pathway (Shi et al., 2000). We have recently shown that SOS3-SOS2 also directly activates SOS1 sodium-proton exchange activity (Qiu et al., 2002). The C-terminal part of the SOS1 protein contains some putative consensus recognition motifs found in the preferred peptide substrate p3 of the SOS2 mutants. An SOS1-His tagged protein purified from yeast membranes has been recently observed to be phosphorylated in vitro by the SOS2T168DΔ308 (Quintero et al., 2002). Therefore, SOS1 is likely one physiological substrate of SOS2.
Activation of SOS2 by Multisite Phosphorylation within the Activation Loop
Regulation of protein kinases is achieved through many different mechanisms, including protein phosphorylation by other kinase(s) (Elion, 1998), autophosphorylation (Cooper and MacAuley, 1988; Sato et al., 1996), or control by regulatory domains or subunits. A key feature for regulation in many protein kinases is thought to be the phosphorylation of one or more residues within the activation loop of the catalytic subunit (Vertommen et al., 2000; McCartney and Schmidt, 2001). An unphosphorylated activation loop can block access of substrates to the active site, whereas phosphorylation can cause an outward rotation of the activation loop, making substrate accessible to the active site residues for catalysis (Jeffrey et al., 1995; Sicheri and Kuriyan, 1997; Xu et al., 1999).
Three residues—Ser-156, Thr-168, and Tyr-175—within the SOS2 activation loop are completely conserved among all members of the SOS2 kinase subfamily (Fig. 6; data not shown). In addition to activation by converting the conserved Thr-168 to Asp (Guo et al., 2001), we have demonstrated here activation by changing the conserved Ser or Tyr to Asp (data not shown). These results are similar to findings with a number of other protein kinases in both plants (Torruella et al., 1986; Ali et al., 1994; Iglesias et al., 1998; Sugden et al., 1999a) and animals (Waldron et al., 2001). It has been thought that phosphorylation of the activation loop shifts the equilibrium toward a conformation that accommodates protein substrate binding, and some data in the literature support this concept (Hubbard, 1997; Hubbard et al., 1998; Johnson et al., 1998; Shah et al., 2001).
Finally, recombinant SOS2 mutants have been useful because of their relative abundance compared with native SOS2 protein, the purification of which from Arabidopsis has not been possible because of its extreme low abundance (J.-K. Zhu, unpublished data). The recombinant SOS2 mutants are easily purified, and being catalytically active, have permitted biochemical analysis. Although these studies demonstrate the catalytic potential of the proteins, the biologically relevant form of the SOS2 kinase is most likely in the complex it makes with SOS3 and perhaps additional proteins; it will be of interest to see whether there are any significant differences in activity between the complex and the mutated active forms of SOS2.
MATERIALS AND METHODS
Site-Directed Mutagenesis
A cDNA containing the complete open reading frame of SOS2 was obtained by reverse transcription-PCR as described by Liu et al. (2000). Substitution of either Ser or Tyr with Asp within the activation loop of SOS2 was introduced using oligonucleotide-directed in vitro mutagenesis. The sequences of mutagenic oligonucleotide primers (MWG-Biotech, High Point, NC) were as follows: 5′-TTTCGGATTTCGGACTCGACGCATTGCCTCAGGAAGGAG-3′ (SOS2S156D, forward), 5′-TCCTTCCTGAGGCAATGCGTCGAGTCCGAAATCCGAAACC-3′ (SOS2S156D, reverse), 5′-ACATGTGGAACTCCGAACGACGTAGCTCCAGAGGTACTTAG-3′ (SOS2Y175D, forward), and5′-AAGTACCTCTGGAGCTACGTCGTTCGGAGTTCCACATGTGG-3′ (SOS2Y175D, reverse). In vitro mutagenesis reactions were performed on the plasmid DNA with a 1:1 (v/v) enzyme mix of LA Tag (TaKaRa Shuzo, Ltd., Kyoto) and Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA) using the following PCR cycle: 95°C for 30 s, followed by 16 cycles of 95°C for 30 s, 60°C for 1.0 min, and 72°C for 7 min. The PCR products were gel-purified and treated with DpnI to digest the parental double-stranded DNA. The digested PCR products were then transformed into DH5α-competent cells. The sequences of mutation and the fidelity of the rest of the DNA in all constructs were confirmed by DNA sequencing. GST-SOS2K40N and GST-SOS3 constructs were produced as described by Liu et al. (2000) and Halfter et al. (2000), respectively.
Expression of Kinase Fusions in E. coli and Protein Purification
All constructs were expressed in bacteria as a C-terminal fusion protein with the bacterial GST under control of the isopropyl β-d-thiogalactopyranoside-inducible tac promoter. All mutant and wild-type GST fusion constructs were transformed into E. coli BL21 (codon plus) cells (Stratagene). Freshly transformed single colonies were grown overnight at 37°C, transferred to fresh 1,000 mL of Luria-Bertani media, and further cultured until the A600 reached approximately 0.8. Recombinant protein expression was induced by 0.6 mm isopropyl β-d-thiogalactopyranoside for 4 h. The cells were harvested by centrifugation (4,000g, 25 min, 4°C), and the pellets were resuspended in a ice-cold bacterial lysis buffer containing 140 mm NaCl, 2.7 mm KCl, 10.1 mm Na2HPO4, 1.8 mm KH2PO4, pH 7.5, 10% (v/v) glycerol, 5 mm dithiothreitol, 2 μg aprotinin mL−1, 2 μg leupeptin mL−1, and 2 mm phenylmethanesulfonyl fluoride. Lysozyme (1 mg mL−1) and Triton X-100 (1%, v/v) were added to the suspension and incubated on ice with gentle shaking for 30 min before sonication. The sonicate was clarified by centrifugation at 15,000g for 30 min at 4°C, and the supernatant was recovered. Recombinant proteins were then purified from the bacterial lysates by glutathione-Sepharose (Amersham Pharmacia Biotech) affinity chromatography. Glutathione-Sepharose 4B beads were added to the supernatant, and the mixture was incubated with gentle agitation for 60 min on ice. The Sepharose beads were then sedimented, and the pellets were carefully washed six times with the cell lysis buffer and resuspended in kinase assay buffer. SDS-PAGE (10%, w/v) analysis was used to evaluate the protein composition of each preparation. Gels were stained with Coomassie Brilliant Blue.
Kinase Activity Assays
In vitro phosphorylation assays were performed as described previously (Halfter et al., 2000) with some modification. Peptide phosphorylation was measured as the incorporation of radioactivity from [γ-32P]ATP (Perkin Elmer Life Sciences, Boston) into the peptide substrate. Forty microliters of the reaction mixture contained 20 mm Tris (pH 7.2), 2.5 mm MnCl2 or 5 mm MgCl2, 0.5 mm CaCl2, 10 μm ATP, 5 μCi [γ-32P]ATP, 150 μm peptide substrate, and 2 mm dithiothreitol. Three peptide substrates used were p1 (LRRASLG; Kemptide, St. Louis), p2 (VRKRTLRRL; Sigma, St. Louis), and p3 (ALARAASAAALARRR, Research Genetics, Huntsville, AL). Enzymatic reactions were initiated by adding 5 μCi of [γ-32P]ATP, and reaction mixtures were immediately incubated at 30°C with gentle shaking. Reactions were terminated after 30 min by adding 1 μL of 0.5 m EDTA, and the GST fusion proteins bound to glutathione-Sepharose beads were pelleted. Fifteen microliters of the supernatant was applied onto P-81 phosphocellulose paper (Whatman, Clifton, NJ) for peptide phosphorylation analysis. The P-81 paper was then washed three times with 1% (v/v) phosphoric acid, and 32P incorporation into the peptide was quantified by phosphor imaging on a STORM 860 PhosphorImager (Molecular Dynamics, Sunnyvale, CA). For autophosphorylation assays, the remaining 25-μL reaction mixture was added with 5 μL of 6× Laemmli sample buffer (Laemmli, 1970) and then separated by a 10% (w/v) SDS-PAGE gel and autoradiographed. For the analysis of divalent cation requirements, kinase assays were performed in the kinase assay buffers containing 0 to 20 mm of MnCl2 or MgCl2, using 150 μm p3 and 10 μm ATP. For ATP substrate kinetics analysis, 0 to 25 μm of ATP was used while keeping p3 constant (150 μm). Peptide substrate kinetic parameters were determined by varying the concentrations of the peptides (0–250 μm) at a fixed ATP concentration (10 μm).
Data Analysis
Initial rates were determined by measuring the amount of phosphorylated peptide formed in 30 min, because this time period produced adequate amounts of product for both enzymes and was within the linear portion of the reaction progress curve. The kinetic parameters were determined by nonlinear least squares analysis of the averaged initial velocity data fitting to the Henri-Michaelis-Menten equation (Eq. 1).
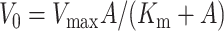 |
1 |
In this equation, V0 is the measured initial velocity; Vmax is the maximum velocity; A is the concentration of ATP-metal; and Km is the apparent Km. The Kcat values were calculated by dividing Vmax by the total enzyme concentration. Three experiments were performed for all kinetic studies, and the average data were fit to the equation.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes.
Footnotes
This work was supported by the National Institutes of Health (grant no. R01GM59138 to J.-K.Z.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.004507.
LITERATURE CITED
- Ali N, Halfter U, Chua N-H. Cloning and biochemical characterization of a plant protein kinase that phosphorylates serine, threonine, and tyrosine. J Biol Chem. 1994;269:31626–31629. [PubMed] [Google Scholar]
- Armstrong RN, Kondo H, Granot J, Kaiser ET, Mildvan AS. Magnetic resonance and kinetic studies of the manganese (II) ion and substrate complexes of the catalytic subunit of adenosine 3′, 5′-monophosphate dependent protein kinase from bovine heart. Biochemistry. 1979;18:1230–1238. doi: 10.1021/bi00574a018. [DOI] [PubMed] [Google Scholar]
- Ball KL, Barker J, Halford NG, Hardie DG. Immunological evidence that HMG-CoA reductase kinase-A is the cauliflower homologue of the RKIN1 subfamily of plant protein kinases. FEBS Lett. 1995;377:189–192. doi: 10.1016/0014-5793(95)01343-1. [DOI] [PubMed] [Google Scholar]
- Barker JH, Slocombe SP, Ball KL, Hardie DG, Shewry PR, Halford NG. Evidence that barley HMG-CoA reductase kinase is a member of the SNF1-related protein kinase family. Plant Physiol. 1996;112:1141–1149. doi: 10.1104/pp.112.3.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JA, MacAuley A. Potential positive and negative autoregulation of p60c-src by intermolecular autophosphorylation. Proc Natl Acad Sci USA. 1988;85:4232–4236. doi: 10.1073/pnas.85.12.4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale S, Wilson WA, Edelman AM, Hardie DG. Similar substrate recognition motifs for mammalian AMP-activated protein kinase, higher plant HMG-CoA reductase kinase-A, yeast SNF1, and mammalian calmodulin-dependent protein kinase I. FEBS Lett. 1995b;361:191–195. doi: 10.1016/0014-5793(95)00172-6. [DOI] [PubMed] [Google Scholar]
- Elion EA. Routing MAP kinase cascades. Science. 1998;281:1625–1626. doi: 10.1126/science.281.5383.1625. [DOI] [PubMed] [Google Scholar]
- Gregoriou M, Jones PF, Timms JF, Yang JJ, Radford SE, Rees AR. Physicochemical characterization of the cytoplasmic domain of the epidermal growth factor receptor and evidence for conformational changes associated with its activation by ammonium sulphate. Biochem J. 1995;306:667–678. doi: 10.1042/bj3060667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Halfer U, Ishitani M, Zhu J-K. Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell. 2001;13:1383–1399. doi: 10.1105/tpc.13.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy PM, Carraway KL, Cerione RA. Biochemical comparisons of the normal and oncogenic forms of insect cell-expressed new tyrosine kinases. J Biol Chem. 1992;267:13851–13856. [PubMed] [Google Scholar]
- Halford NG, Hardie DG. SNF1-related protein kinases: global regulators of carbon metabolism in plants? Plant Mol Biol. 1998;37:735–748. doi: 10.1023/a:1006024231305. [DOI] [PubMed] [Google Scholar]
- Halfter U, Ishitani M, Zhu J-K. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci USA. 2000;97:3735–3740. doi: 10.1073/pnas.040577697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- Hardie DG. Plant protein serine/threonine kinases: classification and functions. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:97–131. doi: 10.1146/annurev.arplant.50.1.97. [DOI] [PubMed] [Google Scholar]
- Hardie DG. Plant protein-serine/threonine kinases: classification into subfamilies and overview of function. In: Kreis M, Walker JC, editors. Plant Protein Kinases. San Diego: Academic Press; 2000. pp. 1–44. [Google Scholar]
- Hartwell J, Gill A, Nimmo GA, Wilkinsa MB, Jenkins GI, Nimmo HG. Phosphoenolpyruvate carboxylase kinase is a novel protein kinase regulated at the level of expression. Plant J. 1999;20:333–342. doi: 10.1046/j.1365-313x.1999.t01-1-00609.x. [DOI] [PubMed] [Google Scholar]
- Horn MA, Walker JC. Biochemical properties of the autophosphorylation of RLK5, receptor-like protein kinase from Arabidopsis thaliana. Biochim Biophys Acta. 1994;1208:65–74. doi: 10.1016/0167-4838(94)90160-0. [DOI] [PubMed] [Google Scholar]
- Hubbard SR. Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. EMBO J. 1997;16:5572–5581. doi: 10.1093/emboj/16.18.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard SR, Mohammadi M, Schlessinger J. Autoregulatory mechanisms in protein-tyrosine kinases. J Biol Chem. 1998;273:11987–11990. doi: 10.1074/jbc.273.20.11987. [DOI] [PubMed] [Google Scholar]
- Iglesias T, Waldron RT, Rozengurt E. Identification of in vivo phosphorylation sites required for protein kinase D activation. J Biol Chem. 1998;273:27662–27667. doi: 10.1074/jbc.273.42.27662. [DOI] [PubMed] [Google Scholar]
- Ishitani M, Liu J, Halfter U, Kim C-S, Wei M, Zhu J-K. SOS3 function in plant salt tolerance requires myristoylation and calcium binding. Plant Cell. 2000;12:1667–1677. doi: 10.1105/tpc.12.9.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich NP. Mechanism of CDK activation revealed by the structure of a cyclin A-CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- Johnson LN, Lowe ED, Noble ME, Owen DJ. The eleventh datta lecture: the structural basis for substrate recognition and control by protein kinases. FEBS Lett. 1998;430:1–11. doi: 10.1016/s0014-5793(98)00606-1. [DOI] [PubMed] [Google Scholar]
- Johnson LN, Noble ME, Owen DJ. Active and inactive protein kinases: structural basis for regulation. Cell. 1996;85:149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- Knight H, Trewavas AJ, Knight MR. Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant J. 1997;12:1067–1078. doi: 10.1105/tpc.8.3.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knighton DR, Zheng J, Ten EL, Ashford VA, Xuong NH, Taylor SS, Sowadski JM. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:407–414. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
- Laemmli U. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Liu J, Ishitani M, Halfter U, Kim C-S, Zhu J-K. The Arabidopsis thaliana SOS2gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci USA. 2000;97:3730–3734. doi: 10.1073/pnas.060034197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhu J-K. An Arabidopsis mutant that requires increased calcium for potassium nutrition and salt tolerance. Proc Natl Acad Sci USA. 1997;94:14960–14964. doi: 10.1073/pnas.94.26.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhu J-K. A calcium sensor homolog required for plant salt tolerance. Science. 1998;280:1943–1945. doi: 10.1126/science.280.5371.1943. [DOI] [PubMed] [Google Scholar]
- McCartney RR, Schmidt MC. Regulation of Snf1 kinase: activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J Biol Chem. 2001;276:36460–36466. doi: 10.1074/jbc.M104418200. [DOI] [PubMed] [Google Scholar]
- Mohammadi M, Honegger A, Sorokin A, Ullrich A, Schlessinger J, Hurwitz DR. Aggregation-induced activation of the epidermal growth factor receptor protein tyrosine kinase. Biochemistry. 1993;32:8742–8748. doi: 10.1021/bi00085a004. [DOI] [PubMed] [Google Scholar]
- Muranaka T, Banno H, Machida Y. Characterization of tobacco protein kinase NPK5, a homolog of Saccharomyces cerevisiae SNF1 that constitutively activates expression of the glucose-repressive SUC2 gene for a secreted invertase of S. cerevisiae. Mol Cell Biol. 1994;14:2958–2965. doi: 10.1128/mcb.14.5.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh M-H, Ray WK, Huber SC, Asara JM, Gage DA, Clouse SD. Recombinant brassinosteroid insensitive 1 receptor-like kinase autophosphorylates on serine and threonine residues and phosphorylates a conserved peptide motif in vitro. Plant Physiol. 2000;124:751–766. doi: 10.1104/pp.124.2.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrimon N. Hedgehog and beyond. Cell. 1995;80:517–520. doi: 10.1016/0092-8674(95)90503-0. [DOI] [PubMed] [Google Scholar]
- Qiu Q-S, Guo Y, Dietrich MA, Schumaker KS, Zhu J-K. Regulation of SOS1, a plasma membrane Na+/H+exchanger in Arabidopsis by SOS2 and SOS3. Proc Natl Acad Sci USA. 2002;99:8436–8441. doi: 10.1073/pnas.122224699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero FJ, Ohta M, Shi H, Zhu J-K, Pardo JM. Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+homeostasis. Proc Natl Acad Sci USA. 2002;99:9061–9066. doi: 10.1073/pnas.132092099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Aoto M, Mori K, Akasofu S, Tokmakov AA, Sahara S, Fukami Y. Purification and characterization of a Src-related p57 protein-tyrosine kinase from Xenopusoocytes. J Biol Chem. 1996;271:13250–13257. doi: 10.1074/jbc.271.22.13250. [DOI] [PubMed] [Google Scholar]
- Schinkmann K, Blenis J. Cloning and characterization of a human STE20-like protein kinase with unusual cofactor requirements. J Biol Chem. 1997;272:28695–28703. doi: 10.1074/jbc.272.45.28695. [DOI] [PubMed] [Google Scholar]
- Schulze-Muth P, Irmler S, Schroder G, Schroder J. Novel type of receptor-like protein kinase from a higher plant (Catharanthus roseus): cDNA, gene, intramolecular autophosphorylation, and identification of a threonine important for auto- and substrate phosphorylation. J Biol Chem. 1996;271:26684–26689. doi: 10.1074/jbc.271.43.26684. [DOI] [PubMed] [Google Scholar]
- Shah K, Vervoort J, Vries SC. Role of threonines in the Arabidopsis thalianasomatic embryogenesis receptor kinase 1 activation loop in phosphorylation. J Biol Chem. 2001;276:41263–41269. doi: 10.1074/jbc.M102381200. [DOI] [PubMed] [Google Scholar]
- Shi H, Ishitani M, Kim C-S, Zhu J-K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+antiporter. Proc Natl Acad Sci USA. 2000;97:6896–6901. doi: 10.1073/pnas.120170197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicheri F, Kuriyan J. Structures of Src-family tyrosine kinases. Curr Opin Struct Biol. 1997;7:777–785. doi: 10.1016/s0959-440x(97)80146-7. [DOI] [PubMed] [Google Scholar]
- Simon MA. Signal transduction during the development of the DrosophilaR7 photoreceptor. Dev Biol. 1994;166:431–442. doi: 10.1006/dbio.1994.1327. [DOI] [PubMed] [Google Scholar]
- Stocchetto S, Marin O, Carignani G, Pinna LA. Biochemical evidence that Saccharomyces cerevisiae YGR262cgene, required for normal growth, encodes a novel Ser/Thr-specific protein kinase. FEBS Lett. 1997;414:171–175. doi: 10.1016/s0014-5793(97)00980-0. [DOI] [PubMed] [Google Scholar]
- Su JY, Eriksob E, Maller JL. Cloning and characterization of a novel serine/threonine protein kinase expressed in early Xenopusembryos. J Biol Chem. 1996;271:14430–14437. doi: 10.1074/jbc.271.24.14430. [DOI] [PubMed] [Google Scholar]
- Sugano S, Andronis C, Green RM, Wang Z-Y, Tobin EM. Protein kinase CK2 interacts with and phosphorylates the Arabidopsis circadian clock-associated 1 protein. Proc Natl Acad Sci USA. 1998;95:11020–11025. doi: 10.1073/pnas.95.18.11020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden C, Crawford RM, Halford NG, Hardie DG. Regulation of spinach SNF1-related (SnRK1) kinases by protein kinases and phosphatases is associated with phosphorylation of the T loop and is regulated by 5-AMP. Plant J. 1999a;19:433–439. doi: 10.1046/j.1365-313x.1999.00532.x. [DOI] [PubMed] [Google Scholar]
- Sugden C, Donaghy PG, Halford NG, Hardie DG. Two SNF1-related protein kinases from spinach leaf phosphorylate and inactivate 3-hydroxy-3-methylglutaryl-coenzyme A reductase, nitrate reductase, and sucrose phosphate synthase in vitro. Plant Physiol. 1999b;120:257–274. doi: 10.1104/pp.120.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Budde RJ. Requirement for an additional divalent metal cation to activate protein tyrosine kinases. Biochem. 1997;36:2139–2146. doi: 10.1021/bi962291n. [DOI] [PubMed] [Google Scholar]
- Torruella M, Casano LM, Vallejos RH. Evidence of the activity of tyrosine kinase(s) and of the presence of phosphotyrosine proteins in pea plantlets. J Biol Chem. 1986;261:6651–6653. [PubMed] [Google Scholar]
- Vertommen D, Rider M, Ni Y, Waelkens E, Merlevede W, Vandenheede JR, Lint JV. Regulation of protein kinase D by multisite phosphorylation. J Biol Chem. 2000;275:19567–19576. doi: 10.1074/jbc.M001357200. [DOI] [PubMed] [Google Scholar]
- Waldron RT, Rey O, Iglesias T, Tugal T, Cantrell D, Rozengurt E. Activation loop Ser744 and Ser748in protein kinase D are transphosphorylated in vivo. J Biol Chem. 2001;276:32606–32615. doi: 10.1074/jbc.M101648200. [DOI] [PubMed] [Google Scholar]
- Weekes J, Ball KL, Caudwell FB, Hardie DG. Specificity determinants for the AMP-activated protein kinase and its plant homologue analyzed using synthetic peptides. FEBS Lett. 1993;334:335–339. doi: 10.1016/0014-5793(93)80706-z. [DOI] [PubMed] [Google Scholar]
- Xu W, Doshi A, Lei M, Eck MJ, Harrison SC. Crystal structures of c-Src reveal features of its autoinhibitory mechanism. Mol Cell. 1999;3:629–638. doi: 10.1016/s1097-2765(00)80356-1. [DOI] [PubMed] [Google Scholar]