Abstract
The first committed step in the conversion of cycloartenol into Δ5 C24-alkyl sterols in plants is catalyzed by an S-adenosyl-methionine-dependent sterol-C24-methyltransferase type 1 (SMT1). We report the consequences of overexpressing SMT1 in tobacco (Nicotiana tabacum), under control of either the constitutive carnation etched ring virus promoter or the seed-specific Brassica napus acyl-carrier protein promoter, on sterol biosynthesis in seed tissue. Overexpression of SMT1 with either promoter increased the amount of total sterols in seed tissue by up to 44%. The sterol composition was also perturbed with levels of sitosterol increased by up to 50% and levels of isofucosterol and campesterol increased by up to 80%, whereas levels of cycloartenol and cholesterol were decreased by up to 53% and 34%, respectively. Concomitant with the enhanced SMT1 activity was an increase in endogenous 3-hydroxy-3-methylglutaryl coenzyme A reductase activity, from which one can speculate that reduced levels of cycloartenol feed back to up-regulate 3-hydroxy-3-methylglutaryl coenzyme A reductase activity and thereby control the carbon flux into sterol biosynthesis. This potential regulatory role of SMT1 in seed sterol biosynthesis is discussed.
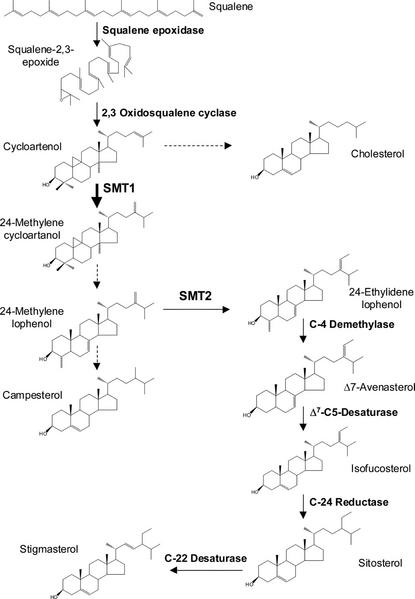
Sterols are ubiquitous in plant cells, where they serve crucial functions to control the fluidity and permeability of membranes and as precursors to steroid growth regulators such as brassinosteroids (Ikekawa, 1990; Bach and Benveniste, 1997). The sterol pathway is divided into two distinct parts (Nes, 2000). First, acetate is converted into squalene epoxide via several steps in the mevalonate pathway. Second, in the first committed step of sterol biosynthesis, squalene epoxide is cyclized to give cycloartenol, which is transformed into end product sterols in a series of enzyme catalyzed methylations, demethylations, and desaturations (Fig. 1). In contrast to animals, higher plants contain mixtures of 24-alkyl-Δ5-sterols, of which campesterol is the principal 24-methyl sterol, and sitosterol and stigmasterol are the predominant 24-ethyl sterols (Schaller et al., 1998). It has been proposed that the C-24 methylation of cycloartenol is a major site of regulation in the sterol biosynthetic pathway (Nes et al., 1991; Chappell et al., 1995; Nes, 2000). Important evidence supporting this view is that transgenic tobacco (Nicotiana tabacum) leaf-overexpressing 3-hydroxy-3-methylglutaryl CoA reductase (HMGR) accumulates high levels of cycloartenol, but only relatively moderate levels of 24-alkyl-Δ5-sterols (Chappell et al., 1995; Schaller et al., 1995).
Figure 1.
Schematic representation of the sterol pathway post-squalene. Solid lines indicate that one enzymatic step is involved in the conversion and dashed lines indicate the involvement of more than one enzyme.
The conversion of cycloartenol into 24-methylene cycloartenol is principally catalyzed by an S-adenosyl-l-Met-dependent sterol C-24 methyltransferase type 1 (SMT1). Genes encoding SMT1 enzyme have been cloned from yeast (Saccharomyces cerevisiae) and a range of plants such as Arabidopsis, tobacco, Ricinus communis, soybean (Glycine max), and Oryza sativa (Schaeffer et al., 2000). SMT1 preferentially catalyzes the methyl addition to cycloartenol, but it can also catalyze the second methylation step, namely the conversion of 24-methylene lophenol to 24-ethylidene lophenol, albeit at a much lower rate (Husselstein et al., 1996; Bouvier-Navé et al., 1998). Conversely, sterol-C24 methyltransferase 2 (SMT2) is mainly responsible for the second methyl addition, but it can also catalyze the first sterol methylation (Schaeffer et al., 2001). Therefore, it was not a surprise that an Arabidopsis SMT1 deletion mutant was still able to accumulate alkylated sterols, but at altered levels (Diener et al., 2000). The perturbed leaf sterol composition of the SMT1 mutant was associated with several phenotypical abnormalities such as poor growth and fertility, a loss of proper embryo morphogenesis, and sensitivity of the root to calcium (Diener et al., 2000). This emphasizes that perturbed sterol composition in vegetative tissue can have severe consequences for plant development.
To date, all effort has been devoted to elucidate the function of SMT1 in vegetative plant tissues in relation to sterol composition and plant development (Deiner at al., 2000; Schaeffer et al., 2000; Sitbon and Jonsson, 2001). However, little is known of the role of SMT1 in sterol biosynthesis in seed tissue. An effective way to study the function of genes in plants is to generate transgenic plants, which overexpress the gene under investigation. In this report, we investigate the function of SMT1 in sterol biosynthesis in tobacco seed by overexpressing SMT1 under control of either a constitutive or a seed-specific promoter. Herein, we present the metabolic effects that overexpression of SMT1 in tobacco has on SMT enzyme activity, sterol composition, and total sterol content in seed tissue. Furthermore, we also show that the activity of one key enzyme in sterol biosynthesis, HMGR, is up-regulated in seed tissue of plants overexpressing SMT1.
RESULTS AND DISCUSSION
Overexpression of SMT1 in Tobacco
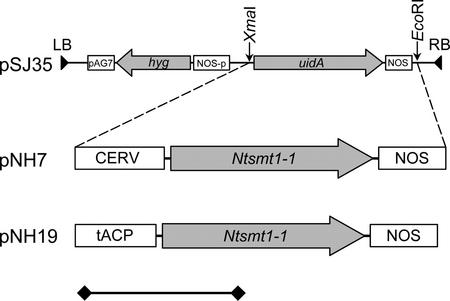
The Ntsmt1-1 gene, encoding SMT1, was cloned from a pool of tobacco cDNAs as described in “Materials and Methods.” Plant expression vectors were constructed by placing Ntsmt1-1 under control of either the constitutive carnation etched ring virus (CERV; pNH7) or the seed-specific truncated acyl-carrier protein (ACP) promoter (pNH19; Fig. 2). The CERV promoter has similar strength and developmental regulation pattern as the more commonly used enhanced cauliflower mosaic virus 35S promoter (M. Harker, N. Holmberg, J.C. Clayton, C.L. Gibbard, A.D. Wallace, S. Rawlins, S.A. Hellyer, A. Lanot, and R. Safford, unpublished data). The ACP promoter is active during the second and third stages of rape seed development, when sterol biosynthesis occurs (DeSilva et al., 1990; M. Harker and A. Lanot, unpublished data).
Figure 2.
Binary plant transformation vector pNH7 and pNH19 holding the CERV-Ntsmt1-1-nopaline synthase (NOS) and ACP-Ntsmt1-1-NOS expression cassettes, respectively. LB, Left border; RB, right border. Arrows indicate the orientation of the Ntsmt1-1 and the hygromycine resistance genes. The size bar indicates 1,000 bp.
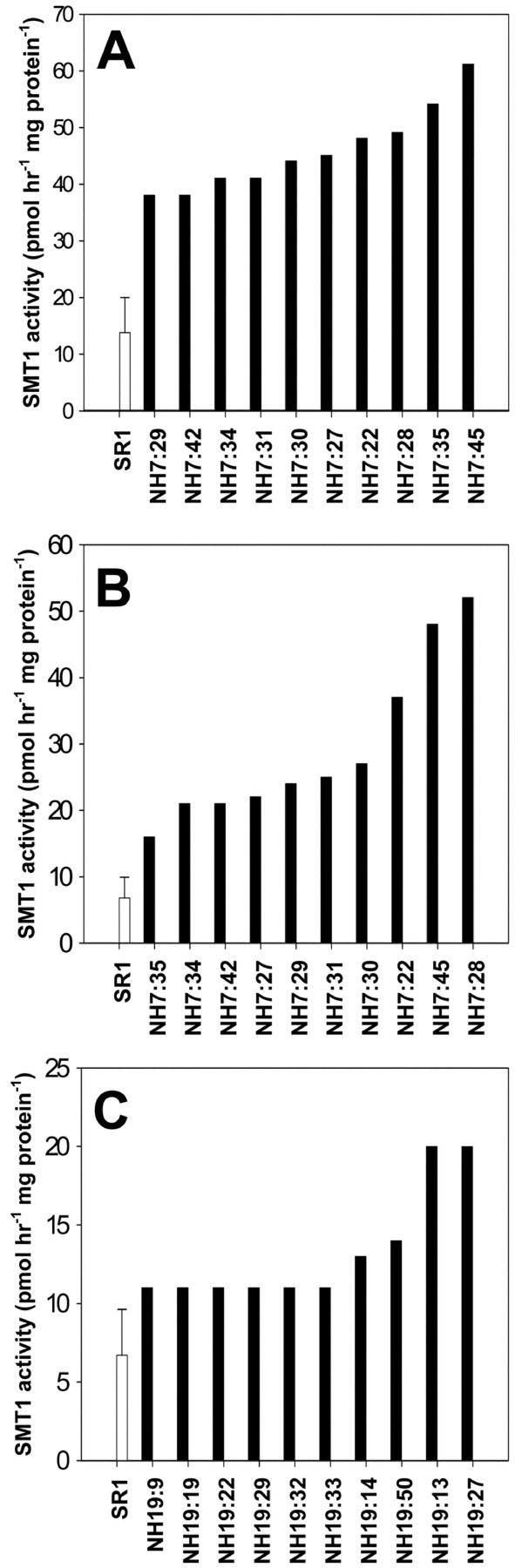
Binary vectors pNH7 (CERV-Ntsmt1-1-NOS) and pNH19 (ACPp-Ntsmt1-1-NOS), and the empty vector control, pSJ35, were transformed into tobacco. Thirty individually transformed NH7 and NH19 lines were generated and screened for SMT1 activity in leaf and seed tissue. As demonstrated in Figure 3, A and B, transgenic NH7 lines exhibited up to 4.3-fold more SMT1 activity in leaf and up to 7.4-fold more activity in seed tissue than the average of control plants. Although the overall trend of increased activity in the population of leaf samples was mirrored in the seed samples, there was only a loose correlation between the SMT1 activity in leaf and seed tissue. The transgenic NH19 lines also demonstrated significant increases in SMT1 activity in seed tissue (Fig. 3C). Plants NH19:13 and NH19:27 exhibited up to 3.5-fold more SMT1 activity than the average of control plants. Seeds of selected high-expressing plants were germinated on hygromycin and grown on to the subsequent generation (T1).
Figure 3.
The SMT1 activity of T0 transgenic and control lines. Transgenic lines are indicated in black staples and control in white staples. The average activities of four independent tobacco SR1 control lines are given and the sds are indicated with error bars. A, The SMT1 activity in expanding leaves transformed with pNH7 (CERV-Ntsmt1-1-nos). B, The SMT1 activity in seeds collected 17 d after anthesis of tobacco transformed with pNH7. C, The SMT1 activity in seeds collected 17 d after anthesis of tobacco transformed with pNH19 (ACP-Ntsmt1-1-nos).
Genetic Characterization of SMT1-Expressing Lines
Transgenic NH7 and NH19 lines in the T0 generation, which exhibited high SMT1 activity, were subjected to Southern-blot analysis to deduce the number of transgene integration loci because previous reports have indicated that multiple copies may lead to cosuppression (Hobbs et al., 1990). As shown in Table I, the copy number varied in the transgenic lines. Surprisingly, there was not a clear correlation between the insertion of the transgene into a single locus and high SMT1 activity. Lines exhibiting high SMT activity had the transgene inserted into one, two, or four genomic loci (Table I; Fig. 3).
Table I.
No. of transgene insertion loci in SMT1-expressing tobacco lines
| Line | No. of Insertion Locia |
|---|---|
| NH7:2 | 2 |
| NH7:13 | 1 |
| NH7:21 | 2 |
| NH7:27 | 1 |
| NH7:28 | 2 |
| NH7:33 | 1 |
| NH7:45 | 1 |
| NH19:10 | 3 |
| NH19:13 | 4 |
| NH19:15 | 1 |
| NH19:27 | 4 |
The no. of transgene insertion loci was determined by Southern-blot analysis using a digoxigenin (DIG)-labeled probe complementary to a fragment of the hygromycin resistance marker.
Segregation of the transgene in T1 lines NH7:27 and NH19:27 was assessed by germinating 25 seeds on hygromycin-containing media. NH7:27 and NH19:27 exhibited Mendelian segregation ratios of 3:1 (χ2 = 0.26, P > 0.9) and 4:0 (χ2 = 0.04, P > 0.99), respectively (data not shown). This observation correlates well with the Southern-blot data, which demonstrated that NH7:27 contains a single transgene insertion locus and that NH19:27 contains several insertion loci (Table I).
Enhanced Carbon Flux into End Product Sterols in Seed But Not in Leaf Tissue
The sterol level and composition of leaf tissue from five individual T1 NH7 lines was analyzed (Table II). In agreement with Schaeffer et al. (2000) and Sitbon and Jonsson (2001), we did not observe any significant changes in the amounts of total sterols in leaf tissue of SMT1-overexpressing plants. Moreover, overexpression of SMT1 in tobacco did not lead to an altered visual phenotype, despite a perturbed sterol composition, which is also in accord with both Schaeffer et al. (2000) and Sitbon and Jonsson (2001). In contrast, transgenic plants overexpressing SMT2 were shown to have an altered campesterol to sitosterol ratio, which resulted in a stunted growth phenotype (Schaller et al., 1998; Schaeffer et al., 2001).
Table II.
Sterol composition of leaf from T1 tobacco transformed with CERV-Ntsmt1-1 (μg g dry wt−1)
| Compound | Controla (stdev) | NH7:27 | NH7:29 | NH7:34 | NH7:42 | NH7:45 |
|---|---|---|---|---|---|---|
| Cycloartenol | 53 (13) | 42 | n.d.b | n.d. | n.d. | n.d. |
| Cholesterol | 189 (22) | 35 | 57 | 55 | 58 | 45 |
| 24-Methylene cycloartanol | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| 24-Methylene lophenol | 29 (25) | 74 | 65 | 48 | 66 | n.d. |
| 24-Ethylidene lophenol | 18 (31) | n.d. | n.d. | n.d. | n.d. | n.d. |
| Isofucosterol | 216 (26) | 199 | 162 | 150 | 179 | 160 |
| Campesterol | 1,118 (81) | 1,221 | 1,188 | 1,233 | 1,135 | 1,083 |
| Sitosterol | 462 (61) | 718 | 471 | 395 | 450 | 465 |
| Stigmasterol | 1,047 (62) | 1,145 | 1,124 | 1,027 | 1,084 | 1,008 |
| Total sterolsc | 3,080 (12.7) | 3,420 | 3,070 | 2,910 | 3,070 | 2,760 |
The average values were obtained by collecting mature leaves from two individual empty vector transformed plants and one SR1 wild-type plant.
Nondetectable levels (<2 μg g dry wt−1).
The total sterol levels were calculated by adding the above shown intermediates and end product sterols.
The sterol composition of SMT1-overexpressing leaf tissue, however, was altered, with cycloartenol and cholesterol levels dramatically reduced (Table II). The level of cycloartenol was reduced from approximately 50 μg g−1 dry weight to non-detectable levels (<2 μg g−1 dry weight) in four of five lines and the level of cholesterol was reduced by up to 5.4-fold as compared with wild-type lines. These decreases in cholesterol and cycloartenol levels are more striking than those observed by Schaeffer et al. (2000) when expressing the tobacco SMT1 in tobacco, and by Sitbon and Jonsson (2001) when expressing soybean SMT1 in tobacco. A possible explanation for the more pronounced decreases in cycloartenol and cholesterol in the present study is that higher levels of SMT1 activity were achieved. The increases in SMT1 activity achieved by Sitbon and Jonsson (2001) and Schaeffer et al. (2000) were up to 54% and 63% over wild-type levels, respectively. These are moderate effects when compared with the 5-fold increases in SMT1 activity observed in the NH7 lines (Fig. 3A). Although it should be pointed out that the SMT1 assays were not performed in an identical fashion, Schaeffer et al. (2000), for instance, used the true SMT1 substrate, cycloartenol, when assaying the SMT1 activity, whereas lanosterol was used in the current study and by Sitbon and Jonsson (2001). However, it is unlikely that the use of lanosterol as substrate would give rise to higher SMT1 activity.
In contrast to leaf, seed of transgenic tobacco overexpressing SMT1, under control of either the CERV or ACP promoter, exhibited elevated total sterol levels (Tables III and IV). The total sterol level was 27% and 44% higher in the NH7:27 and NH19:27 T1 lines, respectively. Interestingly, the relative decrease of cycloartenol (percent of wild type level) was less dramatic in the seed tissue than in the leaf tissue of the NH7 lines (Tables II and III). However, it should be pointed out that the absolute amount of cycloartenol was higher in wild-type seed than leaf (approximately 300 versus approximately 50 μg g−1 dry weight) and that the absolute decrease in cycloartenol in the NH7 lines was greater in seed than in leaf (approximately 200 versus approximately 50 μg). Moreover, the cholesterol and cycloartenol levels were reduced in NH7:27 by 25% and 53%, respectively, as compared with wild-type values (Table III). Similar decreases in cholesterol and cycloartenol were observed in the seed of NH19:27 (34% and 26%; Table IV). The absolute decrease in cycloartenol levels was greater in line NH7:27 than in line NH19:27 (184 versus 73 μg; Tables III and IV). In addition, the levels of 24-methylene and 24-ethylidene sterols, downstream of cycloartenol, were significantly increased. Another observation was that the increase in 24-methylene lophenol was greater than the increase 24-ethylidene lophenol in the seed of NH7:27 and NH19:27. It has been shown previously that SMT1 can convert 24-methylene lophenol into 24-ethylidene lophenol, albeit at a very low rate (Husselstein et al., 1996). Therefore, one would predict that SMT1 would alter the balance in favor of 24-ethylidene lophenol, but this was shown not to be the case. The slight differences in sterol profile observed between NH7:27 and NH19:27 might be explained by minor differences in temporal regulation afforded by the CERV and the ACP promoters.
Table III.
Sterol composition of seed from T1 tobacco transformed with CERV-Ntsmt1-1 (pNH7; μg g dry wt−1)
| Compound | SR1a | NH7:27b | Ratio (NH7:27/SR1) |
|---|---|---|---|
| Cycloartenol | 350 (48)c | 166 (39) | 0.47 |
| Cholesterol | 237 (49) | 179 (33) | 0.79 |
| 24-Methylene cycloartanol | 19 (1) | 43 (7) | 2.3 |
| 24-Methylene lophenol | 81 (3) | 161 (34) | 2.0 |
| 24-Ethylidene lophenol | 346 (16) | 654 (60) | 1.9 |
| Δ7-Avenasterol | 54 (9) | 74 (8) | 1.4 |
| Isofucosterol | 698 (33) | 960 (97) | 1.4 |
| Campesterol | 508 (25) | 692 (83) | 1.4 |
| Sitosterol | 1,522 (82) | 2,026 (191) | 1.3 |
| Stigmasterol | 365 (52) | 341 (18) | 0.93 |
| Total sterolsd | 4,179 (7) | 5,295 (380) | 1.3 |
The average values were obtained by collecting mature seeds from two individual SR1 plants.
The average values were obtained by collecting mature seeds from four individual NH7:27 T1 plants.
The sd is given in parentheses.
The total sterol levels were calculated by adding the above shown intermediates and end product sterols.
Table IV.
Sterol composition of seed from T1 tobacco transformed with ACP-Ntsmt1-1 (pNH19; μg/g dry wt−1)
| Compound | SR1a | NH19:27b | Ratio (NH19:27/SR1) |
|---|---|---|---|
| Cycloartenol | 277 (67)c | 204 (27) | 0.74 |
| Cholesterol | 200 (26) | 132 (9) | 0.66 |
| 24-Methylene cycloartanol | 29 (8) | 63 (9) | 2.2 |
| 24-Methylene lophenol | 69 (29) | 130 (13) | 1.9 |
| 24-Ethylidene lophenol | 362 (47) | 600 (7.1) | 1.7 |
| Δ7-Avenasterol | 33 (3) | 24 (5) | 0.7 |
| Isofucosterol | 614 (94) | 1,091 (88) | 1.8 |
| Campesterol | 463 (57) | 818 (68) | 1.8 |
| Sitosterol | 1,473 (78) | 2,152 (156) | 1.5 |
| Stigmasterol | 345 (24) | 352 (33) | 1.0 |
| Total sterolsd | 3,930 (395) | 5,650 (372) | 1.4 |
The average values were obtained by collecting mature seeds from four individual SR1 plants.
The average values were obtained by collecting mature seeds from five individual NH19:27 T1 plants.
The sd is given in parentheses.
The total sterol levels were calculated by adding the above shown intermediates and end product sterols.
The end product sterols, isofucosterol, campesterol, and sitosterol, were all significantly increased in the seeds of NH7:27 and NH19:27 tobacco (Tables III and IV), but levels of stigmasterol were unaltered in seed as well as leaf tissue (Tables II–IV). The latter finding is easy to explain in leaf tissue because the level of the immediate precursor of stigmasterol, sitosterol, is unaltered. However, the sitosterol level in seed tissue was increased by 30% to 50%, without impacting on the level of stigmasterol, suggesting that the C-22 desaturase that catalyzes the conversion of sitosterol into stigmasterol is a critical slow step in seed tissue (Tables III and IV; M. Harker, N. Holmberg, J.C. Clayton, C.L. Gibbard, A.D. Wallace, S. Rawlins, S.A. Hellyer, A. Lanot, and R. Safford, unpublished data).
The increased levels of end product sterols cannot simply be explained by the conversion of cycloartenol into the concomitant alkylated sterols because cycloartenol is only a minor intermediate (approximately 8% of total sterols in seed). An alternative explanation is that the decreased levels of cycloartenol and/or cholesterol levels feed back to enzymes upstream of SMT1, up-regulating enzyme activities and enhancing carbon flux into the sterol biosynthetic pathway. One potential target enzyme is HMGR, which has been shown to control the flux of carbon into sterol biosynthesis (Chappell et al., 1995; Schaller et al., 1995). Therefore, we measured the HMGR activity in the seed of SMT1-overexpressing plants as described below.
Increased Endogenous HMGR Activity in Seed with Reduced Levels of Cycloartenol
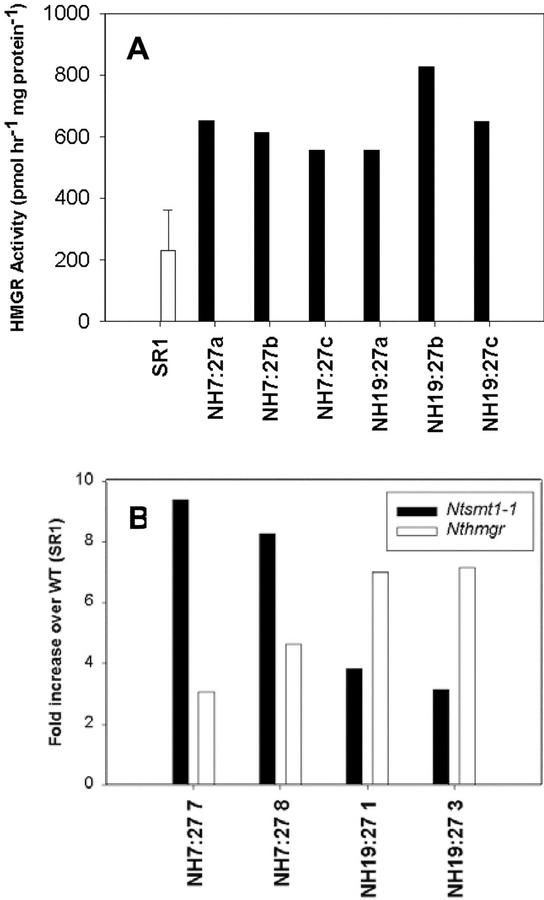
HMGR activity and transcript levels were measured in seed tissue of two high T1 sterol lines (NH7:27 and NH19:27). As shown in Figure 4A, lines NH7:27 and NH19:27 have elevated HMGR activity, up to 2.7- and 3.6-fold, respectively. Moreover, these lines exhibit not only elevated Ntsmt1-1 transcript levels, which are due to the transgene expression, but also elevated levels of endogenous hmgr transcripts (Fig. 4B). The relative differences in the increases of hmgr and Ntsmt1-1 transcript levels between the NH7:27 and NH19:27 lines are most likely due to differences in the promoter strength and temporal expression patterns of the CERV and ACP promoters (Fig. 4B).
Figure 4.
A, Endogenous HMGR activity in the seed of T1 transgenic tobacco lines NH7:27 (CERV-Ntsmt1-1-nos) and NH19:27 (ACP-Ntsmt1-1-nos) collected 14 d after anthesis. The average HMGR activity of SR1 tobacco was calculated from the activities of seed samples of four individual plants. The error bar corresponds to the sd. B, Real-time PCR to detect the Ntsmt1-1 and the Nthmgr transcript levels in transgenic seed of NH7:27 and NH19:27.
The most obvious conclusion from these experiments is that the overexpression of SMT1 reduces the level of cycloartenol, which in turn feeds back to up-regulate the activity of HMGR. The elevated transcript levels suggest that up-regulation of HMGR activity occurs via transcriptional control, but it cannot be excluded that HMGR is regulated on other levels as well.
Concluding Remarks
It has been shown in several studies that the activity of HMGR is important in regulating the carbon flux from acetate to sterols (e.g. Chappell et al., 1995; Schaller et al., 1995). Deregulation of HMGR activity in tobacco via gene overexpression demonstrated that the intermediate cycloartenol overaccumulated (85-fold), whereas the end product sterols (e.g. sitosterol) increased to a much lesser extent (2-fold; Schaller et al., 1995). Therefore, it was proposed that alkylation of cycloartenol is another key regulatory point in sterol biosynthesis (Guo et al., 1995). Yet, when SMT1 or SMT2 were overexpressed in tobacco a concomitant increase of total sterol could not be detected in leaf tissue (Schaeffer et al., 2000; Sitbon and Jonsson, 2001). However, it has been shown that an Arabidopsis smt1 deletion mutant accumulated elevated levels of cholesterol in leaf tissue, which indicates that SMT1 plays an important regulatory role (Diener et al., 2000). In accord with Diener et al. (2000), we demonstrated that overexpression of SMT1 reduces the level of cholesterol to undetectable levels in leaf and by about 35% in seed. This raises the interesting possibility to produce vegetables and plant oils with reduced levels of cholesterol.
We have demonstrated for the first time, to our knowledge, that overexpression of SMT1 leads to an increase in sterol accumulation in seed, while also confirming the lack of effect on sterol levels in leaf tissue. Hence, we conclude that the carbon fluxes into the sterol pathways in leaf and seed tissue are regulated via different mechanisms. Although overexpressing SMT1 in seed only gave rise to modest absolute decreases in cycloartenol levels, greater significant increases in total sterol levels were observed (Tables III and IV). Furthermore, we also detected increases in endogenous HMGR activity in the SMT1-overexpressing tobacco (Fig. 4). Therefore, we hypothesize that lowering the level of the key intermediate, cycloartenol, feeds back and up-regulates HMGR activity to increase the carbon flux to cycloartenol. However, it cannot be ruled out that lowering the cholesterol level also affects HMGR activity. Furthermore, the elevated hmgr transcript levels suggest that transcriptional regulation is involved.
The question of why sterol biosynthesis is regulated via different mechanisms in seed and leaf tissues remains unanswered. However, it has been shown that germinating seed lacks an active de novo sterol pathway, but important changes in sterol composition still occur during germination and seedling development (Fang and Baisted, 1975; Guo et al., 1995). Therefore, it is plausible that the reduced cycloartenol level, imposed by SMT1 overexpression, enhances the carbon flux as a compensatory mechanism to ensure that the seed has sufficient amounts of sterols for efficient germination and seedling development to occur. However, more research is clearly required to further elucidate the precise mechanism by which HMGR activity is up-regulated when SMT1 is overexpressed.
MATERIALS AND METHODS
Strains, Plasmids, Media, and Culture Conditions
Escherichia coli strain DH5α (Invitrogen, Carlsbad, CA) was used as the host strain in all cloning procedures. Binary vector pSJ35 was created by filling in the BamHI restriction site of the pGPTV-HYG with the Klenow enzyme (Becker et al., 1992). Plasmid pTZ5BS holding the Brassica napus ACP promoter and the ACP5 gene was previously described by DeSilva et al. (1990). Predigested vector pGEM-T Easy containing poly-T overhangs was obtained from Promega (Madison, WI) and used according to the supplier's recommendations. Bacteria were cultivated in Luria-Bertani medium (10 g L−1 tryptone, 5 g L−1 yeast [Saccharomyces cerevisiae] extract, and 5 g L−1 NaCl) supplemented with ampicillin (100 μg mL−1) on a rotary shaker (210 rpm) at 37°C.
Plant Material
Tobacco (Nicotiana tabacum) cv SR1 Petite Havana was grown in either Murashige and Skoog medium or a compost:perlite mixture (2:1 [v/v]; Murashige and Skoog, 1962). The temperature in the growth rooms was kept at 22°C, and a day/night cycle of 16 h/8 h was used. The light intensity was 40 μmol m−2 s−1.
Oligonucleotide Synthesis
All oligonucleotides were synthesized by MWG (Milton Keynes, UK) and they are compiled in Table V.
Table V.
Oligonucleotide primers (given in 5′ to 3′ direction)
| Primer | Sequence |
|---|---|
| ACP5a | cca tcg atc tga ttg gta aga tat gg |
| ACP3b | gtt gcc atg gtc gat att cac gag aga gat c |
| CERV1 | atc ctc aac ttc caa tca ga |
| CERV2 | ttc ttg aga gat agc ttg a |
| CERV1S | gtc tgt cta aag taa agt aga tgc g |
| Clsmt5c | gga agg acA TGT CAA AAC AAG GG |
| Clsmt3d | CGG AAT TCC ATA TTA CTG AGA |
| NosAs | ccg gca aca gga ttc aat ctt |
| Ntsmt13 | CAA CGC CCT CCA AAA ACT GTA |
| Ntsmt15 | tgg gaa aga aAT GTC AAA A |
| PCR628e | gat ccc atg gcc cgc tag cca att gga gct |
| PCR629e | cca att ggc tag cgg gcc atg g |
| PCR645 | gat cca cct cga gtg taa acc ATG gcc cg |
| PCR646 | cta gcg ggc cat ggt tta cac tcg agg tg |
| pUC/M13 forward | ttt ccc agt cac gac gtt gt |
| pUC/M13 reverse | gta aaa cga cgg cca gt |
| RoRidT17 | aag gat ccg tcg aca tcg ata ata cga ctc act ata ggg att ttt ttt ttt ttt ttt ttt |
| Sal1f | aat tcg ctg gtg tcg acc tta ctt |
| Sal2f | cta gaa gta agg tcg aca cca gcg |
| Seqsmt3 | agc tac tga gcg aga agt g |
| Seqsmt5 | gca ccc agg tgg aaa gg |
| TGN1 | cgg acc cag ctt ctc agt g |
| TGN2 | tct ccc cag ccg tat tcg tag |
| TaqA1 | tgc tga gcg ttt ccg ttg |
| TaqA2 | ccg gca gct tcc att cc |
| TaqN1 | caa cag gat caa ggc cga aa |
| TaqN2 | agg cac tgt gtt gtc aat cta acc |
| TaqNtH1 | gac ggg atg acg agg gct |
| TaqNtH2 | aac aat tcc gac gcc cta |
| Xma1g | agc tta ctc ttc ccg gga ttg tta t |
| Xma2g | cga taa caa tcc cgg gaa gag ta |
| 181 | gga aac agc tat gac cat gat tac |
The primers hold the following restriction enzyme sites (underlined): a, ClaI; b, NcoI; c, AflIII; d, EcoRI; e, NcoI/NheI/MunI; f, SalI; and g, XmaI. Translation initiation sites (ATG) are indicated in bold letters.
The primer holds the ClaI restriction enzyme site.
The primer holds the NcoI restriction enzyme site.
The primer holds the AflIII restriction enzyme site.
The primer holds the EcoRI restriction enzyme site.
The primer holds the NcoI/NheI/MunI restriction enzyme site.
The primer holds the SalI restriction enzyme site.
The primer holds the XmaI restriction enzyme site.
Cloning of Ntsmt1-1
Two-week-old seedlings of tobacco cv SR1 Petit Havana were ground up in a mortar and the total RNA was isolated using the Purescript RNA isolation kit from Flowgen (Shenstone, UK). Total RNA (5 μg) was mixed with primer RoRidT17 (10 pmol) in 11.34 μL of diethyl pyrocarbonate-treated water. The mixture was incubated at 68°C for 4 min and thereafter placed in wet ice for 2 min. First strand buffer (1×), dithiothreitol (0.1 μmol), RNAsin (22 units), dNTP (20 nmol), and Superscript (200 units) were added to give a final volume of 20 μL. The mix was incubated at 37°C for 60 min. The Ntsmt1-1 cDNA (accession no. U81312) was amplified by gene-specific primers Ntsmt15 and Ntsmt13 using 35 thermal cycles (30 s at 94°C, 30 s at 53°C, and 90 s at 72°C) and a mix of Taq and Pyrococcus furiosus (10% [v/v]) DNA polymerase. The amplification products were separated on a 1.2% (w/v) agarose gel and the fragment corresponding to the full-length Ntsmt1-1 cDNA was excised and ligated into pGEM-T. Plasmid pGEM-T plasmid harboring the Ntsmt1-1 gene was sequenced using an automatic 373 sequencer (Perkin-Elmer Applied Biosystems, Foster City, CA) and the following primers: pUC/M13 forward, pUC/M13 reverse, Seqsmt5, and Seqsmt3. Sequencing demonstrated that the cloned gene contained two base substitutions (T553 to G and G555 to T) as compared with the published Ntsmt1-1 sequence (Bouvier-Navé et al., 1998). It is difficult to assess whether these nucleotide substitutions have arisen from infidelity during the reverse transcription or PCR amplification, or if the discrepancies found are due to natural variation among tobacco varieties. Bouvier-Navé et al. (1998) cloned Ntsmt1-1 from a cDNA library of 5-week-old tobacco var Xanthi SH6 calli, derived from leaf protoplasts, and we cloned Ntsmt1-1 from a pool of cDNA derived from 2-week-old SR1 (Petit Havana) seedlings. However, because the identical nucleotide substitutions appeared in five individual clones from two different amplification reactions, we concluded that the most likely explanation is that the discrepancies are due to varietal differences. In addition, the two mutations led to the conservative Leu-167 to Val amino acid substitution. The side chains of these amino acids are nonpolar and very similar, and they only differ by one methyl group. Furthermore, alignment of SMT sequences from tobacco, Arabidopsis (accession no. AAG28462), Triticum aestivum (accession no. AAB49338), yeast (accession no. NP013706), Neurospora crassa (accession no. CAB97289), and Pneumocystis carinii (accession no. AAK54439) using ClustalW revealed that the amino acids at that position, which corresponds to 167 in NtSMT1, are not conserved. Therefore, it is highly unlikely that that the Leu-167 to Val substitution will result in altered substrate specificity or binding.
Construction of Shuttle Vectors
The 308-bp HindIII/BamHI-digested CERV promoter fragment from pSJ103 and the SacI/EcoRI-digested 275-bp NOS from pSJ35 was inserted into pUC19, giving pUCN. A synthetic DNA linker, assembled by annealing oligonucleotides PCR628 and PCR629, was inserted into the BamHI/SacI-digested pUCV, yielding pUCV2. This linker introduces the following restriction enzyme sites (5′ to 3′): NcoI, NheI, and MunI. Another DNA linker was assembled thereafter by annealing PCR645 and PCR646. The sequence of this linker was designed to mimic the consensus Kozak sequence from plants (taa acc ATG G), which has been shown to render efficient translation (Joshi et al., 1997). This linker was inserted into BamHI/NheI-digested pUCV2, yielding shuttle vector pNH1. A synthetic DNA linker, holding a SalI site, was assembled by annealing SalI and SalII and inserted between the EcoRI and XbaI sites of pNH1, yielding pNH2. Another synthetic DNA linker holding an XmaI site, assembled by annealing oligonucleotides XbaI and XmaII, was inserted between the HindIII and ClaI site of pNH2, giving pNH3.
Cloning of Promoters
The CERV promoter was isolated by PCR from infected Dianthus barbatus leaf material using the primers CERV1 and CERV2 under standard conditions. The approximately 380-bp fragment (corresponding to nucleotides 6,737–7,118 of Hull et al., 1986) was cloned into the TA cloning vector pT7Blue (Invitrogen) with the promoter in the same orientation as the T7 promoter yielding pSJ103. Sequence analysis showed that the isolated CERV promoter differed from the published sequence at several positions (T6790 to C, C6826 to T, A6872 to G, and T6729 to A). The most significant change was a small deletion in a poly(A+) tract in the 5′-untranslated leader (four As versus nine As in the published sequence). These changes probably represent differences in virus isolates rather than PCR errors, but the latter cannot be ruled out.
A truncated 289-bp portion of the ACP promoter and 69 bp of the 5′-untranslated region of the ACP5 gene were amplified from vector pTZ5BS (DeSilva et al., 1990) using primers ACP5 and ACP3. ACP5 and ACP3 were designed with 5′ overhangs holding restriction enzyme sites for ClaI and NcoI, respectively. The amplified promoter fragment was digested with ClaI and NcoI and inserted into ClaI/NcoI-digested pNH3, yielding pNH12.
Construction of Ntsmt1-1 Expression Vectors
The 1,040-bp Ntsmt1-1 gene was amplified from plasmid pGEM-T, harboring Ntsmt1-1, by PCR using standard conditions. To facilitate cloning, the primers used, clsmt5 and clsmt3, were designed with 5′ overhangs containing AflIII and EcoRI restriction sites, respectively. A mix of Taq and Pyrococcus furiosus (10% [v/v]) DNA polymerase was used to ensure high amplification fidelity during the amplification. The amplification products were separated on a 1.2% (w/v) agarose gel and the 1,040-bp Ntsmt1-1 gene was excised, purified, digested by AflIII and EcoRI, and inserted into vector pNH3 downstream of the CERV promoter and upstream of the NOS terminator giving pNH6. Plasmid pNH6 was sequenced with primers CERV1S, clsmt5, seqsmt5, seqsmt3, clsmt3, and NosAs to confirm its authenticity. The CERV-Ntsmt1-1-NOS cassette was excised from pNH6 using XmaI and EcoRI and subsequently inserted into XmaI/EcoRI-digested pSJ35, giving pNH7 (Fig. 1). Vector pNH7 was sequenced using primers CERV1S and NosAs to confirm that the junction between promoter, Ntsmt1-1, and terminator were intact.
The AflIII/EcoRI-digested Ntsmt1-1 amplification product was inserted into NcoI/MunI-digested pNH12, rendering pNH14. Plasmid pNH14 was sequenced with primers 181, clsmt5, seqsmt5, seqsmt3, and NosAs to confirm that the PCR amplification had not introduced any mutations. Vector pNH14 was digested by XmaI and EcoRI to excise the ACPp-Ntsmt1-1-NOS expression cassette. This cassette was subsequently inserted into XmaI/EcoRI-digested pSJ35, resulting in binary expression vector pNH19 (Fig. 1). Vector pNH19 was sequenced with primers ACP5, Ntsmt15, and NosAs to confirm that the junction regions between promoter, gene, and terminator were intact.
Plant Transformation and Growth Conditions
Binary vectors pNH7, pNH19, and pSJ35 were transformed into Agrobacterium tumefaciens LBA4404 using electroporation as described by Shen and Forde (1989). A. tumefaciens-mediated transformation of tobacco SR1 was carried out using the leaf disc method as described previously by Horsch et al. (1985). Hygromycin (25 mg L−1)-resistant seedlings were screened by PCR to identify transformants using primer pairs TGN1/TGN2 (NH7) and ACP5/TGN2 (NH19) (Edwards et al., 1991). Antisense primer TGN2 annealed to the coding sequence of Ntsmt1-1, and sense primers TGN1 and ACP5 annealed to the CERV and ACP promoter, respectively. PCR positive transformants were transferred from tissue culture to growth chambers and potted up into a compost:perlite mix (2:1 [v/v]). Pods were tagged and seeds were collected for SMT and HMGR enzyme activity measurements 14 and 17 d after anthesis. Mature seeds were collected after about 12 weeks.
Southern-Blot Analysis
Genomic DNA was isolated from leaf tissue of 4-week-old soil-grown primary transgenics using the cetyl-trimethyl-ammonium bromide method as described by Rogers and Bendich (1985). Approximately 10 μg of genomic DNA was digested with EcoRI and separated on a 0.7% (w/v) agarose gel. The DNA fragments were transferred onto a Hybond N+ nylon membrane using capillary transfer as described by Sambrook et al. (1989). Southern-blot analysis was performed using a DIG-labeled probe directed against the hygromycin resistance marker gene according to the method described in the DIG System Users Guide (Roche Molecular Biochemicals, Mannheim, Germany).
SMT1 Activity Assay
All steps were carried out at 4°C unless otherwise stated. Approximately 150 mg of tagged seeds (collected 17 d after anthesis), which had been stored previously at −80°C, were homogenized in the ratio of 1:10 (w/v) seed:buffer using an Ultra-turrax at maximum speed. The homogenization buffer consisted of: 0.2 m potassium phosphate, pH 7.5; 0.4 m Suc; 10 mm EDTA; MgCl2; 5 mm glutathione; and 40g L−1 polyvinylpolypyrrolidone. Aliquots of homogenate (400 μL) were centrifuged at 1,200g for 5 min at 4°C. The supernatant was removed using a syringe and needle and discarded. The lipid and pellet fractions were extracted together with 400 μL of homogenization buffer containing 10 mm CHAPS to extract SMT activity. The tobacco samples were vortexed and left on ice for 20 min. The samples were again centrifuged at 1,200g for 5 min at 4°C and the supernatant removed. This process was then repeated. The two detergent extracts were then mixed 1:1 (v/v) and assayed immediately for SMT enzyme activity.
Crude extracts of young developing tobacco leaves were homogenized 1:10 in homogenization buffer (same as above) using an Ultra-turrax at maximum speed. After centrifugation for 5 min at 1,200g, the resulting supernatant was immediately assayed for SMT1 activity.
The standard assay system consisted of 50 μm S-adenosyl-l-[methyl-14C] Met (57 mCi mmol−1), 125 μm lanosterol emulsified in Tween-80 (final concentration 0.1% [v/v]), and 80 μL of enzyme extract in a final volume of 100 μL. Control assays contained no enzyme extract. The assays were incubated for 1 h at 30°C and terminated by the addition of 100 μL of 12% (w/v) KOH in ethanol. Lanosterol and cholesterol, 15 μg each, were added as carriers. The neutral lipids were extracted with hexane (2 × 600 μL) and the combined eluant was evaporated to dryness under nitrogen. The lipid residue was resuspended in 15 μL of toluene. The toluene containing extract (10 μL) was applied to precoated silica thin-layer chromatography (TLC) plates (silica gel 60 F254, Merck, Rahway, NJ). The TLC plates were developed in dichloromethane. The 4-desmethyl and 4,4-dimethyl sterols were visualized with iodine vapor. The band corresponding to lanosterol (4,4-dimethyl sterol) was scraped off into scintillation vials. Liquid scintillation cocktail (Readysafe, Beckman Instruments, Fullerton, CA) was added and radioactivity was measured with a Beckman LS650 scintillation counter. It should be noted that endogenous substrate, in particular cycloartenol, may also be methylated by SMT1 and hence contribute to the measured SMT1 activity.
HMGR Activity Assay
All steps were carried out at 4°C unless otherwise stated. Homogenization buffer was 0.2 m potassium phosphate, pH 7.5, containing 0.4 m Suc, 10 mm EDTA, 5 mm MgCl2, 5 mm glutathione, and 4 g/100 μL insoluble polyvinylpyrrolidone. Developing seeds from tagged pods (approximately 150 mg), collected 14 d after anthesis, were homogenized in the ratio 1:10 (w/v) seed:buffer using an Ultra-turrax at maximum speed. After centrifugation for 5 min at 1,200g, the lipid and pellet fractions were extracted together with 400 μL of homogenization buffer containing 2% (w/v) Brij. The samples were vortexed and incubated on ice for 20 min. The samples were again centrifuged at 1,200g for 5 min at 4°C. This process was immediately repeated. The two detergent extracts were mixed 1:1 (v/v) and assayed for HMGR enzyme activity. The assay system consisted of 100 mm potassium phosphate, pH 7.5; 3 mm NADPH; 20 μm [14C] HMG CoA (30 nCi); and 20 μL of enzyme extract in a final volume of 26 μL. In control assays, the enzyme extract was omitted. After incubation for 15 min at 30°C, reactions were terminated by the addition of 5 μL of 6 m HCl. To this mixture, 5 μL of mevalonate lactone (1 mg μL−1) was added to act as the carrier. The samples were left to lactonize for 15 min at room temperature and clarified by centrifugation at 13,000 rpm for 2 min at room temperature. Aliquots of supernatant (10 μL) were applied to precoated silica TLC plates (silica gel 60 F254, Merck). Unreacted substrate was separated from product by developing plates in diethyl ether:acetone (3:1 [v/v]). Mevalonate lactone was visualized with iodine vapor and scraped off into scintillation vials. Radioactivity was measured as described above.
Real-Time PCR
Total RNA was isolated from developing tobacco seed (14 d after anthesis) of NH7:27, NH19:27, and SR1 plants using the RNaqueous kit according to the supplier's instructions (Ambion, Austin, TX). The total RNA was treated with DNase to remove any contamination of genomic DNA and subsequently converted into cDNA using the 3′ RACE System from Invitrogen. Taqman primer pairs directed against Ntsmt1-1 (TaqN1 and TaqN2), tobacco tac9 actin (TaqA1 and TaqA2), and tobacco hmgr (TaqNtH1 and TaqNtH2) genes were designed using the Primer Express software (PE-Applied Biosystems). These primer pairs were used together with Sybr Green (PE-Applied Biosystems, Foster City, CA) in Taqman PCR reactions to detect transcript levels of Ntsmt1-1, hmgr, and tac9 in transgenic and control samples. The Ntsmt1-1 and hmgr transcript levels in the transgenic tobacco were calculated in relation to the transcript levels in SR1 tobacco according to the manual supplied by PE-Applied Biosystems.
Sterol Analysis
Mature seed and leaf tissue was collected and freeze dried. Freeze-dried tissue (approximately 50 mg) was ground with a pestle and mortar. Dihydrocholesterol (250 μL, 0.2% [w/v]) in chloroform was added to act as the internal standard. Sterols were extracted in 5 mL of 2:1 (v/v) chloroform:methanol for 30 min at 80°C. The mixture was allowed to cool and then filtered. The extract was reduced to dryness under nitrogen. Transmethylation of the extract was performed by resuspending the extracts in 1 mL of toluene and 2 mL of sodium methoxide (0.5 m). The mixture was maintained at 80°C for 30 min and allowed to cool. Boron trifluoride solution (2 mL, 14% [w/v]) was added and the mixture was heated for a further 10 min at 80°C. Diethyl ether (3 mL) was added to the mixture when cool, followed by deionized water (5 mL) with vigorous shaking. The ether layer was removed and a second extraction with ether performed. The ether layers were combined and backwashed with 5 mL of water. The ether phase was dried over anhydrous sodium sulfate overnight.
Extracts were resuspended in 300 μL of toluene and the free sterols silylated by the addition of 150 μL of 95:5 (v/v) N,O-bis(trimethylsilyl)acetamide:trimethylchlorosilane followed by incubation at 50°C for 10 min. Sterol derivatives were analyzed by gas chromatography using a Perkin-Elmer Autosystem XL GC with flame ionization detection and a BPX5 capillary column (25-m × 0.32-mm i.d., 0.25-μm film thickness, and helium 8 pounds per square inch), ex SGE. The temperature program used a fast rise from 80°C to 230°C (45°C min−1), a slow rise from 230°C to 280°C (4°C min−1), and 280°C to 355°C (20°C min−1), and finally 5 min at 355°C. Peak areas were calculated automatically using Turbochrom software (PerkinElmer, San Jose, CA). Sterol structures were identified by mass spectrometry.
ACKNOWLEDGMENTS
We thank Ann Scarborough, Rachel Payne, and Bernadette Marsh for skillful sequencing assistance.
Footnotes
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.004226.
LITERATURE CITED
- Bach TJ, Benveniste P. Cloning of cDNAs or genes encoding enzymes of sterol biosynthesis from plants and other eukaryotes: heterologous expression and complementation analysis of mutations for functional characterisation. Prog Lipid Res. 1997;36:197–226. doi: 10.1016/s0163-7827(97)00009-x. [DOI] [PubMed] [Google Scholar]
- Becker D, Kemper E, Schell J, Masterson R. New plant binary vectors with selectable markers located proximal to the left t-DNA border. Plant Mol Biol. 1992;20:1195–1197. doi: 10.1007/BF00028908. [DOI] [PubMed] [Google Scholar]
- Bouvier-Navé P, Husselstein T, Benveniste P. Two families of sterol methyltransferases are involved in the first and second methylation steps of plant sterol biosynthesis. Eur J Biochem. 1998;256:88–96. doi: 10.1046/j.1432-1327.1998.2560088.x. [DOI] [PubMed] [Google Scholar]
- Chappell J, Wolf F, Prouix J, Cueller R, Saunders C. Is the reaction catalyzed by 3-hydroxy-3-methylglutaryl coenzyme A reductase a rate limiting step for isoprenoid biosynthesis in plants? Plant Physiol. 1995;109:1337–1343. doi: 10.1104/pp.109.4.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSilva J, Loader NM, Jarman C, Windust JHC, Hughes SG, Safford R. The isolation and sequence-analysis of 2 seed-expressed acyl carrier protein genes from Brassica napus. Plant Mol Biol. 1990;14:537–548. doi: 10.1007/BF00027499. [DOI] [PubMed] [Google Scholar]
- Diener AC, Li H, Zhou W-X, Whoriskey WJ, Nes WD, Fink GR. Sterol methyltransferase 1 controls the level of cholesterol in plants. Plant Cell. 2000;12:853–870. doi: 10.1105/tpc.12.6.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C. A simple and rapid method for the preparation of genomic DNA for PCR analysis. Nucleic Acids Res. 1991;16:1349. doi: 10.1093/nar/19.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang T-Y, Baisted DJ. 2,3-Oxidosqualene cyclase and cycloartenol-S-adenosylmethionine methyltransferase activities in vivoin cotyledon and axis tissues of germinating pea seeds. Biochem J. 1975;150:323–328. doi: 10.1042/bj1500323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Venkatramesh M, Nes WD. Developmental regulation of sterol biosynthesis in Zea mays. Lipids. 1995;30:203–219. doi: 10.1007/BF02537823. [DOI] [PubMed] [Google Scholar]
- Hobbs SLA, Kpodar P, Dlong CMO. The effect of T-DNA copy number, position and methylation on reporter gene-expression on tobacco transformants. Plant Mol Biol. 1990;16:851–864. doi: 10.1007/BF00039425. [DOI] [PubMed] [Google Scholar]
- Horsch RD, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT. A simple and general method for transferring genes into plants. Science. 1985;227:1229–1231. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- Hull R, Sadler J, Longstaff M. The sequence of carnation etched ring virus-DNA-comparison with cauliflower mosaic-virus and retroviruses. EMBO J. 1986;5:3083–3090. doi: 10.1002/j.1460-2075.1986.tb04614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husselstein T, Gachotte D, Desprez T, Bard M, Benveniste P. Transformation of Saccharomyces cerevisiae with a cDNA encoding a sterol C-methyltransferase from Arabidopsis thalianaresults in the synthesis of 24-ethyl sterols. FEBS Lett. 1996;381:87–92. doi: 10.1016/0014-5793(96)00089-0. [DOI] [PubMed] [Google Scholar]
- Ikekawa N. The analysis of brassinosteroids: plant growth promoting substances. Trends Anal Chem. 1990;9:337–342. [Google Scholar]
- Joshi CP, Zhou H, Huang X, Chiang VL. Context sequences of translation initiation codon in plants. Plant Mol Biol. 1997;3:993–1001. doi: 10.1023/a:1005816823636. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Nes WD. Sterol methyl transferase: enzymology and inhibition. Biochim Biophys Acta. 2000;1529:63–88. doi: 10.1016/s1388-1981(00)00138-4. [DOI] [PubMed] [Google Scholar]
- Nes WD, Janssen GG, Norton RA, Kalinowska M, Crumely FG, Tal B, Bergentsrahle A, Jonsson L. Regulation of sterol biosynthesis in sunflower by 24(R,S),25-epiminolanosterol, a novel C-24 methyl transferase inhibitor. Biochem Biophys Res Commun. 1991;177:566–574. doi: 10.1016/0006-291x(91)92021-b. [DOI] [PubMed] [Google Scholar]
- Rogers SO, Bendich AJ. Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol Biol. 1985;5:69–74. doi: 10.1007/BF00020088. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Frisch EF, Maniatis FE. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schaeffer A, Bouvier-Navé P, Benveniste P, Schaller H. Plant sterol-C24-methyl transferases: different profiles of tobacco transformed with SMT1 or SMT2. Lipids. 2000;35:263–269. doi: 10.1007/s11745-000-0522-1. [DOI] [PubMed] [Google Scholar]
- Schaeffer A, Bronner R, Benveniste P, Schaller H. The ratio of campesterol to sitosterol that modulates growth in Arabidopsis is controlled by sterol methyltransferase 2,1. Plant J. 2001;25:605–615. doi: 10.1046/j.1365-313x.2001.00994.x. [DOI] [PubMed] [Google Scholar]
- Schaller H, Bouvier-Navé P, Benvenist P. Overexpression of an Arabidopsis cDNA encoding a sterol-C241-methyltransferase in tobacco modifies the ratio of 24-methyl cholesterol to sitosterol and is associated with growth reduction. Plant Physiol. 1998;118:461–469. doi: 10.1104/pp.118.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller H, Grausem B, Benveniste P, Chye M-L, Tan C-T, Song Y-H, Chua N-H. Expression of the Hevea brasiliensis (H.B.K.) Müll. Arg. 3-hydroxy-3-methylglutaryl-coenzyme A reductase 1 in tobacco results in sterol overproduction. Plant Physiol. 1995;109:761–770. doi: 10.1104/pp.109.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitbon F, Jonsson L. Sterol composition and growth of transgenic tobacco plants expressing type-1 and type-2 sterol methyltransferases. Planta. 2001;212:568–572. doi: 10.1007/s004250000417. [DOI] [PubMed] [Google Scholar]
- Shen W-J, Forde BF. Efficient transformation of Agrobacterium spp. by high voltage electroporation. Nucleic Acids Res. 1989;17:8385. doi: 10.1093/nar/17.20.8385. [DOI] [PMC free article] [PubMed] [Google Scholar]