Abstract
Representational Oligonucleotide Microarray Analysis (ROMA) detects genomic amplifications and deletions with boundaries defined at a resolution of ∼50 kb. We have used this technique to examine 243 breast tumors from two separate studies for which detailed clinical data were available. The very high resolution of this technology has enabled us to identify three characteristic patterns of genomic copy number variation in diploid tumors and to measure correlations with patient survival. One of these patterns is characterized by multiple closely spaced amplicons, or “firestorms,” limited to single chromosome arms. These multiple amplifications are highly correlated with aggressive disease and poor survival even when the rest of the genome is relatively quiet. Analysis of a selected subset of clinical material suggests that a simple genomic calculation, based on the number and proximity of genomic alterations, correlates with life-table estimates of the probability of overall survival in patients with primary breast cancer. Based on this sample, we generate the working hypothesis that copy number profiling might provide information useful in making clinical decisions, especially regarding the use or not of systemic therapies (hormonal therapy, chemotherapy), in the management of operable primary breast cancer with ostensibly good prognosis, for example, small, node-negative, hormone-receptor-positive diploid cases.
As cancers evolve, their genomes undergo many alterations, including point mutations, rearrangements, deletions, and amplifications, which presumably alter the ability of the cancer cell to proliferate, survive, and spread in the host (Balmain et al. 2003; DePinho and Polyak 2004). An understanding of these changes will allow the design of more rational therapies and, by providing precise diagnostic criteria, allow fitting the correct therapy to each patient according to need. Primary breast cancers in particular exhibit a wide range of outcomes and degrees of benefit from systemic therapies, which are incompletely predicted by conventional clinical and clinico-pathological features. This is especially apparent in the case of small primaries without axillary lymph node involvement, which usually have a good prognosis but are sometimes associated with eventual metastatic dissemination and death.
Breast tumors have long been known to suffer multiple genomic rearrangements during their development, and thus it is reasonable to hypothesize that clinical heterogeneity may be caused by the existence of genetically distinct subgroups. One common approach to the molecular characterization of breast cancer has been “expression profiling,” measuring the entire transcriptome by microarray hybridization. Expression profiling has been very effective at revealing phenotypic subtypes of breast cancer and clinically useful diagnostic patterns of gene expression in tumors (Perou et al. 2000; Sorlie et al. 2001; Ahr et al. 2002; van’t Veer et al. 2002; Sotiriou 2003; Paik et al. 2004). Expression profiling does not look directly at underlying genetic changes, and its dependence on RNA, a fragile molecule, creates some problems in standardization and cross-validation of microarray platforms. Moreover, variation in the physiological context of the cancer within the host, such as the proportion of normal stroma and the degree of inflammatory response, or the degree of hypoxia, as well as methods used for extraction and preservation of sample, are all potentially useful but confounding factors (Edén et al. 2004).
Direct analysis of the tumor genome provides an alternative and perhaps, complementary, means of comparing breast tumors by revealing the genetic events accumulated during tumor progression. We have begun a long-term genomic study of clinically well-defined sets of breast cancer patients with a high-resolution microarray technique called Representational Oligonucleotide Microarray Analysis (ROMA) (Lucito et al. 2003). ROMA is based on the principle that noise in microarray hybridization can be significantly reduced by reducing the complexity of the labeled DNA target in the hybridization mix. In its present configuration, ROMA uses a “representation” of the genome created by PCR amplification of the smallest fragments of a BglII restriction digest. The representation contains <3% of the complexity of the normal human genome and is specifically matched with a unique microarray containing >83,000 oligonucleotide probes designed to pair with the amplified fragments. Coupled with an efficient edge-detection or segmentation algorithm, ROMA yields highly precise profiles of even closely spaced amplicons and deletions. At present, ROMA is capable of detecting the breakpoints of chromosomal events at a resolution of 50 kb. This study is intended to explore whether high resolution of the genetic events in tumors can form an additional basis for the clinical assessment of breast cancer.
The first global studies capable of resolving deletions and amplifications combined comparative genomic hybridization (CGH) and cytogenetics (Kallioniemi et al. 1992a, b, c), and this approach has been applied to breast tumors (Kallioniemi et al. 1994; Ried et al. 1997; Tirkkonen et al. 1998). Subsequently, microarray methods using CGH have increased resolution and reproducibility and improved throughput (Ried et al. 1995; Pollack et al. 2002; Albertson 2003; Lage et al. 2003). These published microarray studies have largely validated the results of cytogenetics CGH, but have not had sufficient resolution to significantly improve our knowledge of the role of genetic events in the etiology of disease, nor assist in the treatment of the patient. On the other hand, knowledge of specific genetic events, like amplification of ERBB2, as studied by fluorescence in situ hybridization (FISH) or Q-PCR, has been clinically useful (van de Vijver et al. 1987; Slamon et al. 1989; Menard et al. 2001). ROMA provides an extra measure of resolution in genomic analysis that might be useful in clinical evaluation, as well as delineating loci important in disease evolution.
We sought to determine whether there were features in the genomes of tumor cells that correlated with clinical outcome in a uniform population of women with “diploid” breast cancers. We chose this population because a significant number of cases culminate in death despite their clinical and histo-pathological parameters that would predict a favorable outcome. Our population of 99 diploid cancers drew from a bank at the Karolinska Institute (KI), and was comprised of long-term and short-term survivors who were similar for node status, grade, and size. For part of our analysis, we draw on additional studies in progress, one using 41 aneuploid (defined as >2n DNA content) (see Methods) cancers from KI, and the other using an additional 103 cancers from the Oslo Micrometastasis Study, Oslo, Norway (OMS). The latter set was not scored for ploidy and has only an average of 8 yr follow up and is included in this study only for comparison of overall frequency of events. The individual genome profiles from the KI data set but not the OMS data set are in the Supplemental material and at (http://roma.cshl.edu). The OMS data set will be posted as part of a second paper specifically dealing with that group. The makeup of these sample sets with respect to clinical parameters is summarized in Table 1.
Table 1.
Distribution of patients and clinical parameters in the Swedish and Norwegian data sets
Numbers will not add up exactly because of partial information on certain individual cases.
aProgesterone (PR) and estrogen (ER) receptors measured by ligand binding; (pos) ≥0.5 fg/μg protein.
bERBB2 amplification scored by ROMA as segmented ratio >0.1 above baseline.
Our studies demonstrate a striking similarity of genome profiles from two different study populations, as well as the commonality of affected loci in aneuploid and diploid cancers. Significantly, we observe a different genome profile between diploid tumors with good and poor outcome. The complexity and the number of events, captured in a mathematical measure, has led to our working hypothesis that genomic profiling may be useful for the molecular staging of breast cancer, and, when validated by further studies, may have implications for clinical practice.
Results
The clinical makeup of the sample sets included in this study is summarized in Table 1. The KI tumor data set was assembled from a collection of >10,000 fresh frozen surgical tumor samples with detailed pathology profiles and long-term follow up. The patients in this study underwent surgery between 1987 and 1992 yielding follow-up data for survival of 15–18 yr. The sample set was assembled with the goal of studying a statistically significant population of otherwise rare outcomes, particularly diploid tumors that led to death within 7 yr, and aneuploid tumors with long-term survival (described in Methods). At the same time, the sample was balanced with respect to tumor size, grade, node involvement, and hormone receptor status. Treatment information is also available in the clinical table available in the Supplemental material; however, the sample set was not stratified according to treatment because the treatment groups are too fragmented to be significant. The Norwegian tumor set was selected from a trial previously described by Wiedswang et al. (2003) designed to identify markers associated with micrometastasis at the time of diagnosis (i.e., disseminating tumor cells in blood and bone marrow). The patients included in the study were recruited between 1995 and 1998, and fresh frozen tumors were available for a subset that was not selected for particular characteristics.
Processing individual cancer genomes
We examined all breast cancer genomes with ROMA, an array-based hybridization method that uses genomic complexity reduction based on representations. In the present case, we performed comparative hybridization using BglII representations, and arrays of 85,000 oligonucleotide (50-mer) probes with a Poisson distribution throughout the genome and a mean interprobe distance of 35 kb (Lucito et al. 2003). In all cases, we compared tumor DNA from a patient to a standard unrelated male human genome. We performed hybridizations in duplicate with color-reversal, and data were rendered as normalized ratios of probe hybridization intensity of tumor to normal.
The normalized ratios are influenced by many factors, including the signal-to-noise characteristics that differ for each probe, sequence polymorphisms in the genomes that affect the BglII representation, DNA degradation of the sample, and other variation in reagents and protocols during the hybridization and scan. Statistical processing called “segmentation” identifies the most likely state for each block of probes, thus reducing the noise in the graphical presentation of the profile.
Within each raw ROMA profile, segmentation places consecutive probe intensity ratios into a series of distinct distributions, reflecting the alterations that occur when blocks of the genome are amplified, duplicated, or deleted. Several methods for segmentation have been published by us and others (Daruwala et al. 2004; Olshen et al. 2004), but in the present case, and in the interest of having very solid findings, we have used a simplified method that recognizes distinct distributions of ratio based on minimization of variance and a Kolmogorov-Smirnov test with P-values set at 10−5 (see Methods). All methods converge on roughly the same segmentation pattern, especially at the boundaries, or edges, of events, but the simplified method used herein does not consider short segments (sets of probes less than six). On average, the resolution of the edges of a gene copy number alteration event is ∼50 kb under our present conditions. We report each probe ratio as the mean of the medians of the ratios within the segment to which that probe belongs, producing a “segmented profile” of each cancer. Both raw ratios and segmented ratios are posted on our Web site. Events less than six probes in length are, of course, visible in the unsegmented data and can be segmented by other methods, such as Hidden Markov Models (HMM); however, these very narrow events do not affect the conclusions of this report and are excluded from the statistical analysis for simplicity.
Single nucleotide polymorphisms (SNPs), found in all profiles, are present in our methods that use restriction endonuclease-based representations. These are most often the result of sequence differences between sample and reference that alter the restriction sites used in the representation process. For purposes of this report, they merely contribute to noise and do not significantly affect segmentation. However, both rare copy number variants (CNVs) and more prevalent copy number polymorphisms (CNPs) (Sebat et al. 2004) will be present in any high-resolution copy number scan, regardless of method, when comparing one person to another. All of our tumor profiles are obtained by comparison to an unrelated standard normal male. If these CNPs and CNVs are not masked, analysis could mistake either for a cancer lesion. We have compiled a list of common CNPs and rare CNVs by profiling healthy cells from 482 individuals, and we used these to mask the “normal” CNPs in our tumor profiles as described in Methods, yielding a “masked segmented profile.” We post the masked segmented profiles in the Supplemental material. The collection of CNPs used for masking includes but is not limited to Scandinavian individuals and represents at most a few hundred probes being removed from consideration for segmentation in any sample. A CNP falling under a larger (cancer-related) event does not affect the segmentation of that event. Both the Kolmogorov-Smirnov segmentation software and the CNP masking algorithms are posted at http://roma.cshl.edu in the forms of scripts interpretable by R or S+ statistical analysis software.
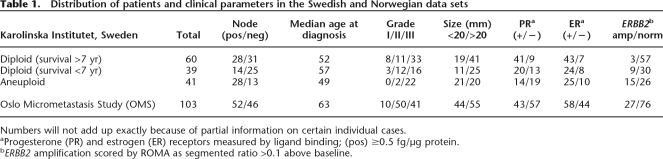
The mean ratios within segments are not directly proportional to true copy number. The unknown proportion of “normal” stroma in the surgical biopsies, the potential for clonal variation, and nonspecific hybridization background signal all contribute to a measured segment ratio below the actual copy number. Although ratios do not directly measure copy number, differences between the median ratios of segments do reflect differences in gene copy within a given experiment. This has been extensively validated by interphase FISH (see e.g., Fig. 3A, Bbelow).
Figure 3.
Validation of peaks and valleys in ROMA profiles by interphase FISH. (A) Expanded ROMA profile of a firestorm on chromosome 8 in the diploid tumor WZ11. The graph shows the normalized raw data (gray) and segmented profile (red) along with the genes for which the probes shown in the FISH images were constructed. Several distinct conditions are exemplified in the images. First, the ROMA profile indicates that the 8p arm is deleted distal to the 8p12 cytoband yielding a single copy of DBC1 (green), but >10 tightly clustered copies of BAG4, which is located in the frequently amplified 8p12 locus (Garcia et al. 2005). Tight clusters of multiple copies corresponding to ROMA peaks are also shown in the FISH images for CKS1A, MYC, TPD52, and the uncharacterized ORF AK096200. Note that the FISH signals corresponding to distinct loci cluster together irrespective of their distance on the same arm (CKS1A/MYC) or across the centromere (BAG4/AK096200). Finally, the spaces between ROMA peaks on 8q, exemplified by NBN (formerly known as NBS1), uniformly show two copies as indicated by the ROMA profile. (B) Expanded view of the centromere and 11q arm from diploid tumor WZ17 showing correspondence of the copy number as measured by FISH with the copy number predicted by the ROMA profile. The y-axis represents the segmented ratios of sample versus control. Chromosome position on the x-axis is in megabases according to Freeze 15 (April 2003) on the UCSC Genome Browser (Karolchik et al. 2003). FISH probes were amplified from primers identified from specific loci using PROBER software (Methods).The insert outlined in black is magnified to show specific details. Comparative data for the probes shown in black are not shown but are available on our Web site. In the boxed region, note that in the nonamplified regions the ROMA profile predicts two copies of the arm proximal to the leftmost amplification. Consistent with the profile, the FISH image shows two copies of probe 11Q3, with one of the spots located in the cluster along with the amplified copies. The amplicon to the right yields four copies by FISH (probe 11Q4). The ROMA profile for the amplicon represented by probe 11Q6 suggests that it is in a region in which the surrounding nonamplified portion of the arm is deleted. This arrangement is commonly observed in firestorms and is confirmed by the FISH image showing one pair of the loci 11q5 and 11Q6 together, representing the intact arm, and no copy of probe 11Q5 in the amplified cluster of spots for 11Q6. (C) Profile of tumor WZ19 in which two firestorms are observed on chromosomes 11q and 17q. In contrast to the overlapping clusters shown in A, amplifications on unrelated arms visualized using FISH probes for CCND1 and ERBB2 cluster independently in the nucleus.
Event frequency plots in breast cancer and their correlation with outcome
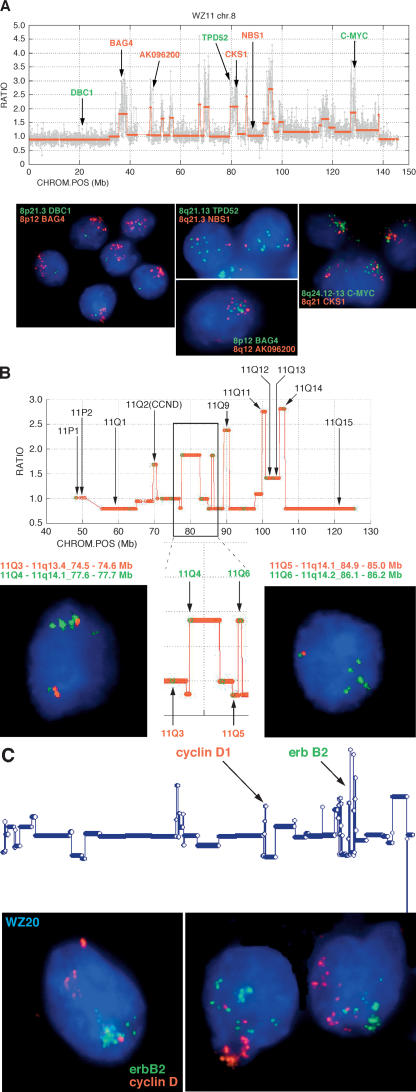
Once all the individual profiles are accumulated, they can be examined and compared as subpopulations. A straightforward, albeit simplistic, view of genome alterations is the frequency plot, a measure at each probe of the frequency with which the probe is amplified or deleted above a threshold in the genome profiles of a set of cancers. To obtain an overview of breast cancer lesions, we show plots from the Swedish group, the Norwegian group, and for the combined set, plotting amplification frequencies as above the line and deletions below (Fig. 1A). Even at this crude view, it is evident that amplifications and deletions do not occur at random throughout the genome, and regions that are amplified tend not to be deleted, and vice versa. Many of the well-known loci known to be deleted or amplified, such as TP53, CDKN2A, MYC, CCND1, and ERBB2, are at or near the centers of frequently altered regions. Additionally, there are frequent “peaks” and “valleys” where none of the familiar suspects are found. The data are posted at our Web site, for detailed inspection by the interested reader.
Figure 1.
Comparative frequency plots of amplification (up) and deletion (down) in various data sets. Frequency calculated on normalized, segmented ROMA profiles using a minimum of six consecutive probes identifying a segment with a minimum mean of 0.1 above (amplification) or below (deletion) baseline. Frequencies are plotted only for chromosomes 1–22. (A) Total Swedish data set (red) versus total Norwegian data set (blue). (B) Swedish diploid subset (blue) versus total Swedish aneuploid subset (red). (C) Swedish diploid 7-yr survivors (red) versus Swedish diploid 7-yr nonsurvivors (blue).
The Swedish (combined aneuploid and diploid) and Norwegian breast cancers display similar frequency profiles, with slightly higher frequencies in the Norwegian set. This discrepancy is most likely explained by the high proportion of diploid cancers in the Swedish set. While the Norwegian set is sequential and unselected, the Swedish set is >70% pseudo-diploid, selected according to our working hypothesis that diploids would provide the most information about tumor development. When we compare the diploid to aneuploid Swedish cancers (Fig. 1B), we again observe similar profiles along with a similar difference in overall frequencies. This difference is not apparent when Swedish aneuploids are compared to the Norwegian group (data not shown). Thus the two cancer types, diploid and aneuploid, share the same loci of amplification and deletion.
The decreased frequency observed in the diploid set relative to the aneuploid set can be attributed to the presence of long-term survivors in the former group. Frequency plots comparing 7-yr (long-lived) survivors to those who do not survive as long (short-lived) is shown in Figure 1C. Clearly, designating a patient as a “survivor” or “nonsurvivor” at a specific time is not accurate in terms of the real progression of the disease. However, it is useful for understanding the relationship of disease progression to molecular events. We used 7 yr as a demarcation because it reflects the point at which the rate of death from cancer in the worst prognosis group drops to near zero. For the studies described in this paper, demarcation values between 7 yr and 10 yr can be used without changing the basic conclusions. It is quite apparent that there are fewer overall events, both amplifications and deletions, in the diploid survivors. Using 25 events as a divider, we obtain the most significant association of the long-lived versus the short-lived cancer patients, with a P-value of 4.2 × 10−4 by Fisher’s exact test.
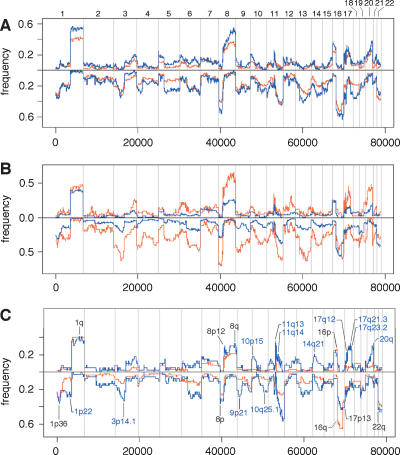
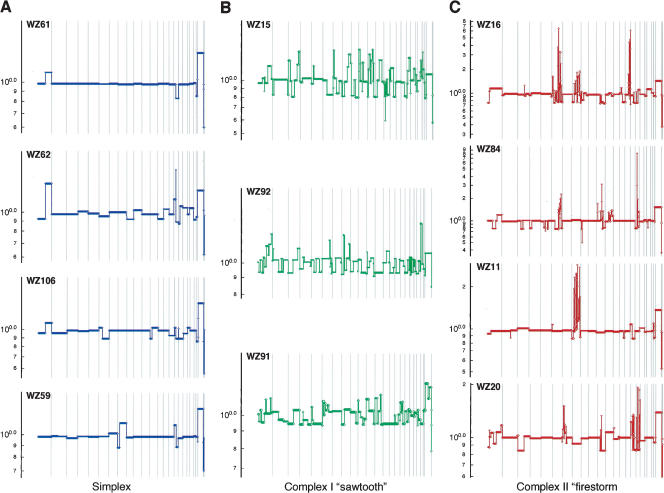
Patterns of genome profiles
Visual inspection of segmented profiles suggests that they come in three basic patterns (Fig. 2), which we present as qualitative heuristic tools for distinguishing apparently distinct processes of genomic rearrangement. The first profile pattern (Fig. 2A), which we call “simplex,” has broad segments of duplication and deletion, usually comprising entire chromosomes or chromosome arms, with occasional isolated narrow peaks of amplification. Simplex tumors make up ∼60% of the diploid data set, while the rest fall into two distinct categories of “complex” patterns. One of these complex patterns is the “sawtooth” (Fig. 2B), characterized by many narrow segments of duplication and deletion, often alternating, more or less affecting all the chromosomes. Little of the genome remains at normal copy number, yet the events typically do not involve high copy number amplification. Note that the scale of the y-axis in Figure 2B is identical to that in Figure 2A. It should be further noted that the X-chromosome peak is often low in sawtooth profiles (e.g., WZ15 in Fig. 2B), indicating that the X chromosome is not exempt from frequent loss in these tumors.
Figure 2.
Major types of tumor genomic profiles. Segmentation profiles for individual tumors representing each category: (A) simplex; (B) complex type I or sawtooth; (C) complex type II or firestorm. Scored events consist of a minimum of six consecutive probes in the same state. The y-axis displays the geometric mean value of two experiments on a log scale. Note that the scale of the amplifications in C is compressed relative to A and B owing to the high levels of amplification in firestorms. Chromosomes 1–22 plus X and Y are displayed in order from left to right according to probe position.
The third pattern (Fig. 2C) resembles the simplex type except that the cancers contain at least one localized region of clustered, relatively narrow peaks of amplification, with each cluster confined to a single chromosome arm. We denote these clusters by the descriptive term “firestorms” because we believe that the clustering of multiple amplicons on single chromosome arms reflects a concerted mechanism of repeated recombination on that arm rather than a series of independent amplification events. The high copy number of these amplicons is reflected in the scale of the y-axis in Figure 2C.
The two complex patterns, firestorm (25%) and sawtooth (5%), make up ∼30% of the diploid tumors in this data set. We cannot perfectly classify all profiles with this system, but the patterns appear to represent genomic lesions resulting from distinctly different mechanisms, and more than one mechanism may be operant to varying degrees within any given tumor.
A fourth type is the “flat” profile, in which we observe no clear amplifications or deletions other than copy number polymorphisms and single probe events, as discussed above, and the expected difference in the sex chromosomes. These examples are few in number (14/140) and are not presented graphically here. Some may result from the analysis of biopsies comprised mostly of stroma, or some may comprise a clinically relevant set of cancers with no detectable amplifications or deletions. Performing the analyses described in this paper with or without these flat profiles does not alter our conclusions; hence, we include them in the analyses presented here.
Firestorms
We used interphase FISH to validate that segmentation is not an artifact of ROMA or statistical processing of ROMA data. Either BAC clones or probes created by primer amplification were labeled and hybridized to preparations of the same frozen tumor specimens profiled by ROMA (Methods). Probes were selected from 33 loci representing both peaks and valleys in the ROMA profile. In each case, the segmentation values were confirmed by FISH. We show here representative instances of these data for the complex pattern of amplification we call “firestorms.”
Firestorms are represented in ROMA profiles as clustered narrow peaks of elevated copy number. The pattern is limited to one or a few chromosome arms in each tumor, with the remainder of the genome remaining more or less quiet, often indistinguishable from the simplex pattern. The individual amplicons in these firestorms are separated by segments that are not amplified, and are, in fact, often deleted, yielding a pattern of interdigitated amplification and LOH as shown for chromosome 8 (WZ11) in Figure 3A and chromosome 11q (WZ17) in Figure 3B. We infer from this that the phenomenon is a result of sequential replication and recombination events or breakage and rejoining events that occur on a particular chromosome arm rather than a general tendency toward amplification throughout the genome.
One might imagine that the individual peaks in a cluster arise from clonal subpopulations within the tumor. They do not. The FISH images of Figure 3 clearly indicate that amplifications at neighboring peaks of a cluster occur in the same cell. Moreover, they colocalize in the nucleus. In those cases in which a cell harbors two firestorms, each on different chromosomes, these too occur in the same cell, but individually segregate within the nucleus by chromosome arm, as shown in Figure 3C for CCND1 (cyclin D1) on chromosome 11q and ERBB2 (HER-2/neu) on 17q. A total of 18 BAC probes representing amplicons and intervening spaces were used in verifying the structure of chromosome 8 in WZ11 and 15 primer amplified probes were used for chromosome 11 in WZ17. Summary data for all probes are available in the Supplemental material.
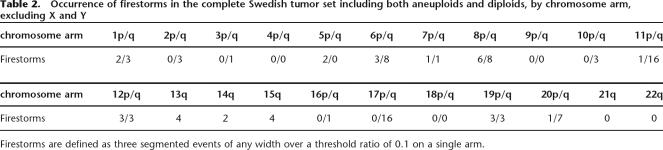
Firestorms have been observed at least once on most chromosomes in the tumors we have analyzed, but certain arms clearly undergo this process more frequently (see Table 2). In particular, chromosomes 6, 8, 11, 17, and 20 are often affected, with 11q and 17q being the most frequently subject to these dramatic rearrangements. Within the latter, the loci containing CCND1 on 11q and ERBB2 on 17q are most frequently amplified and may “drive” the selection of the events. Chromosomes 6, 8, and 20 have a comparable frequency of firestorms, but the “drivers” for these events are less obvious. However, these potential “driver” genes are likely not to be the sole reason for the complex amplification patterns seen in firestorms. The other peaks in the firestorms are not randomly distributed. Each chromosome appears to undergo selective pressure to gain or lose specific regions as exemplified by the frequency plot of chromosome 17 shown in Figure 4. The histogram of amplification (blue) or deletion (red) for 27 Grade II and Grade III tumors exhibiting firestorms on chromosome 17 from both Scandinavian data sets shows distinct peaks and valleys when compared to the equivalent histogram for a set of tumors of equivalent grade but without chromosome 17 firestorms (black and gray histograms). As shown in Figure 4, there is a strong tendency for deletion of the distal p arm including TP53 and for deletion of 17q21 including BRCA1. Conversely, there are at least four distinct peaks of high-frequency amplification on the long arm of 17 in addition to the peak containing ERBB2. As noted in the figure, several genes of interest for breast cancer are located near the epicenters of these peaks, including TOB1 (transducer of ERBB2) and BCAS3 (breast carcinoma amplified sequence). Furthermore, in contrast to accepted dogma (Jarvinen and Liu 2003), a fraction of the firestorms on 17q (5%–10%) do not include amplification of ERBB2, giving weight to the notion that other loci in the region may contribute to oncogenesis. In contrast, broad duplications and deletions are detectable in the non-firestorm subset, but they do not form clear peaks.
Table 2.
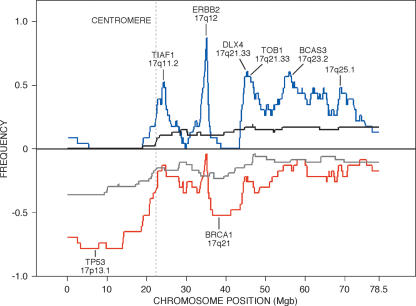
Occurrence of firestorms in the complete Swedish tumor set including both aneuploids and diploids, by chromosome arm, excluding X and Y
Firestorms are defined as three segmented events of any width over a threshold ratio of 0.1 on a single arm.
Figure 4.
Frequency plots of amplification and deletions in tumors containing clustered amplifications (firestorms) on chromosome 17. Lines represent histograms of the number of events for each probe in segmented ROMA profiles over threshold as in Figure 1 for two subsets extracted from the combined Scandinavian data set. Blue and red lines represent amplifications and deletions, respectively, in the subset of 23 tumors containing firestorms on chromosome 17, each showing clear peaks (valleys) of activity. Black and gray lines represent equivalent events in a set of 53 tumors in which firestorms are not observed on chromosome 17.
Frequently amplified and deleted loci
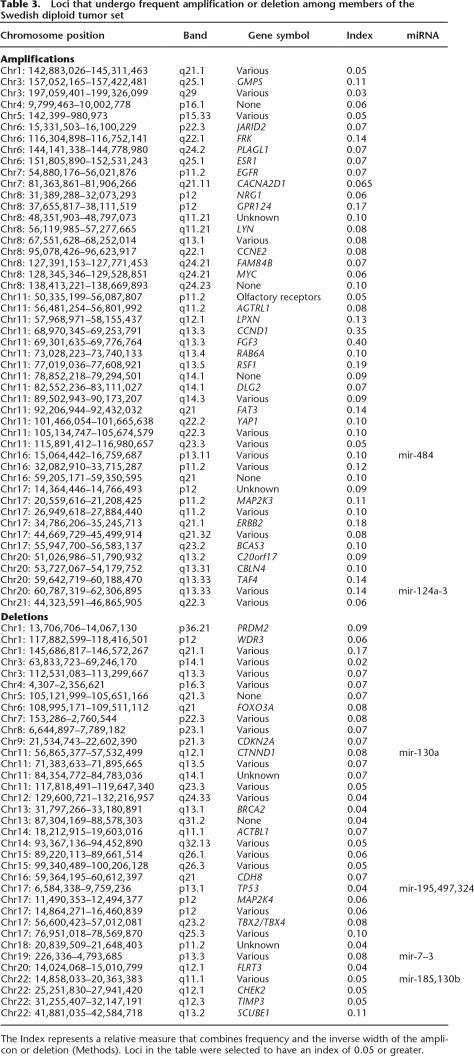
It is of interest to note the regions that are most frequently amplified or deleted in a large data set such as the one presented here. There is no single accepted algorithm for deciding which regions are of most interest, and the parameters used will depend on the goals of the individual researcher. In Table 3 we present the results of one such algorithm (see “Frequently Amplified and Deleted Loci” in Methods) that reflects a component of frequency at any locus plus a factor that gives weight to the inverse of the width of any given event. The latter is based on the rationale that narrow events centered on a given locus should carry more weight than a broad event that happens to encompass that locus. In the table, the relative value for each locus is shown in the Index column. Representative genes that have some potential relation to breast cancer are included for reference purposes, but we do not presume knowledge of the direct involvement of specific genes in tumorigenesis based on this analysis. While several specific amplicons have been reported previously for specific chromosomes, such as 11q (Ormandy et al. 2003) and the ERBB2 region of 17q (Jarvinen and Liu 2003), we know of no other report cataloging a data set of comparable size and resolution permitting this level of detailed analysis. For example, Ormandy et al. (2003) report three narrow (<2 Mb) “core” amplicons in the 11q13 bands along with an independent 17-Mb amplicon spanning the other three. Our analysis yields roughly equivalent peaks of high significance (index value) at 11q13.3 and 13.4 in agreement with their data, along with at least 11 additional distinct peaks where repeated amplification events have occurred on that arm. A graphical version of this analysis is available in the Supplemental material.
Table 3.
Loci that undergo frequent amplification or deletion among members of the Swedish diploid tumor set
Rearrangements in Grade I tumors
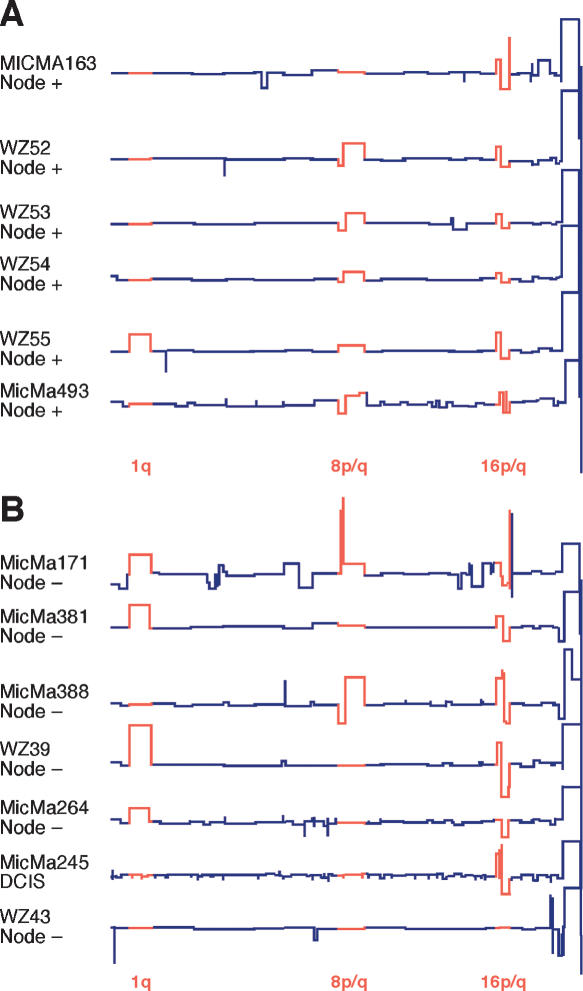
Tumors in which the cells maintain their differentiation as shown by histological examination are generally considered to be less aggressive and to have a good prognosis irrespective of migration to the lymph nodes. Ten examples of these so-called Grade I tumors were available from the Swedish samples and 13 from the Norwegian collection, including eight in which one or more nodes were affected. A single noninvasive DCIS (ductal carcinoma in situ) sample (MicMa245) was also present in the Norwegian set. All of the Swedish samples were medium to large tumors between 20 and 30 mm in size, while the Norwegian samples ranged from 0.5 to 25 mm.
Although the number of samples is small, the similarity in ROMA profiles among the 13 representative samples depicted in Figure 5 is dramatic and may provide insight into some of the earliest events leading to invasive breast cancer. Four of the 23 Grade I samples yielded no detectable events (data not shown). Eighteen of the 19 tumors with any detectable events showed a characteristic rearrangement in chromosome 16 in which one copy of 16q appears to be deleted (assuming diploidy) and 16p is concomitantly duplicated. This rearrangement was also present in the DCIS sample (MicMa245 in Fig. 5B). The rearrangement of chromosome 16 is often coupled with either a converse rearrangement of the arms of chromosome 8 (8p deleted and 8q duplicated) or a duplication of the q arm of chromosome 1. All three of these events are seen in more highly rearranged breast cancer genomes such as those in Figure 2C and, in fact, are among the most common events by frequency in all samples (see Fig. 1B).
Figure 5.
Comparison of Grade I and DCIS tumors by ROMA. Segmented ROMA profiles of six node-positive (Fig. 5A) and seven node-negative (Fig. 5B) Grade I or DCIS tumors, representing a total of 24 examples from the combined Swedish and Norwegian collections. Most frequent rearrangements are depicted in red.
Grade I tumors generally display relatively few genomic events but rarely show more complex patterns of advanced simplex tumors (see MicMa171 in Fig. 5B), indicating that despite a strong correspondence, there is not a strict relation between genomic state and histological grade. MicMa171 has progressed to the point of achieving the common amplicons at 8p12 (Garcia et al. 2005) and 17q11.2, both of which are noted in Table 3. The sole Grade I tumor not showing rearrangement of 16p/q (WZ43 in Fig. 5B) exhibits a different pattern with rearrangements of chromosome 20q and deletion of 22q, indicating that the 16p/q rearrangement is not the only pathway to tumorigenesis. Although certain of these rearrangements contain obvious candidate driver genes such as the duplication of MYC on 8q24 or the loss of the cadherin (CDH) complex on 16q, the actual target genes remain the target of further study.
Relation of patterns to clinical outcome
On first inspection, the highly rearranged “sawtooth” and “firestorm” patterns appeared to correlate with shorter survival in the diploid tumors, presumably because selection of novel genetic combinations afforded the cancer cells the opportunity for accelerated recombination. We sought to confirm this observation by rigorous mathematical and statistical analysis. Using the total number of segments, or events, as a measure does not clearly distinguish a sample with a single firestorm from the simplex pattern with a similar number of events, but the effects of the firestorm on survival are clear. We chose a mathematical measure that would separate the sawtooth and firestorm patterns from the flat and simplex patterns by scoring the close-packed spacing of the firestorm events, while at the same time incorporating the total number of events. The sum of the reciprocals of the mean of lengths of all adjacent segment pairs accomplishes this goal:
 |
where i enumerates all the discontinuities with a magnitude above a numerical threshold of 0.1 in the segmented profile, and where liR (liL) denotes the number of probes in the closest neighboring discontinuity on the right (left), or to a chromosome boundary, whichever is closer. We call this the “inverse adjacent segment length measure.” This calculation is performed after masking for CNPs, and does not include the X or Y chromosomes. The measure works equally well if absolute position in the genome is substituted for probe number. Using this algorithm, the sawtooth patterns achieve a high F because of the sheer number of distributed events, while the firestorm patterns achieve high F-values even if only a single arm is affected because of the contribution of proximity (see WZ11 in Fig. 2C).
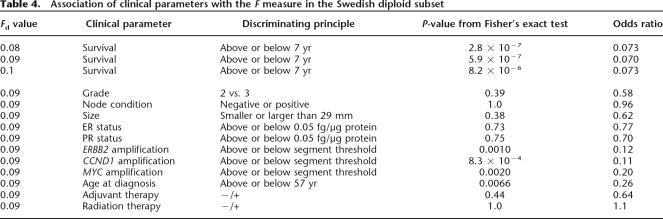
F is a robust measure separating the diploid cancers into two populations that have different survival rates. F ranges in value from zero to a maximum of ∼0.86 for the Swedish diploid group. For a range of values of F, from 0.08 to 0.1 we find both a significant and strong association between the discriminant value and survival beyond 7 yr. The optimum value for F separating by survival does not change appreciably when calculated for survival at 10 yr. As shown in Table 4, 0.08 and 0.09 yield the lowest P-values (2.8 × 10−7 and 5.9 × 10−7 by Fisher’s exact test), with 0.09 showing the strongest association with the long-lived versus the short-lived cancer patients, with an odds ratio of 0.07. Analysis was performed using the fisher.test function in the R data analysis software, which computes an estimate of the odds ratio for a 2×2 contingency table using the conditional maximum likelihood estimate. In contrast, the divider based solely on the number of events without regard to size or proximity has a lower significance, with a P-value of 4.2 × 10−4.
Table 4.
Association of clinical parameters with the F measure in the Swedish diploid subset
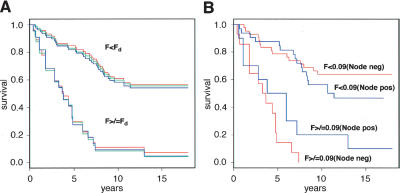
A strong association between F and survival is also found using an alternative statistical procedure that makes no explicit reference either to a particular discriminant value of F or to a particular survival time threshold: We divide the Swedish diploid set into quartiles with respect to F, then apply a log-rank test for differences in survival in these four groups. The four groups are found to have different survival properties, with a P-value of 10−7. In Figure 6A, we display the Kaplan-Meier plots of survival for all Swedish diploids, with a range of discriminant values for F from 0.08 to 0.1. These plots show dramatically different rates of survival for tumors above or below the F-discriminant (Fd). The discriminatory power of F with respect to survival is even more dramatic when node-positive and node-negative cases are plotted separately as in Figure 6B, using F = 0.09.
Figure 6.
Kaplan-Meier plots of the Swedish diploid subset grouped according to the Firestorm Index (F). (A) Complete Swedish diploid data set grouped according to three different discriminator settings (Fd) of F: Fd = 0.08 (red); Fd = 0.09 (blue); Fd = 0.1 (green). (B) Swedish diploid data set separated into node-negative (red) and node-positive (blue) subsets with Fd set to 0.09.
While we find association between F and survival, we find no significant association between F and either tumor size, lymph node status, grade, or expression of the estrogen (ER) and progesterone (PR) receptors (see Table 4; Methods). In other words, F is an independent clinical parameter. This result does not imply that these other parameters do not predict disease recurrence, or that in a random accrual F would not associate with them. Rather, it reflects that our two groups of diploids, short-term and long-term survivors, were picked to be balanced for lymph node status, tumor size, and so forth, and that F has predictive value independent of these traditional clinical measures. We do find significant association between F, on the one hand, and age at diagnosis and amplifications of the CCND1, MYC, and ERBB2 loci, on the other hand. However, as we show in the following, F retains its predictive value for survival after adjustment for the effects of these four factors.
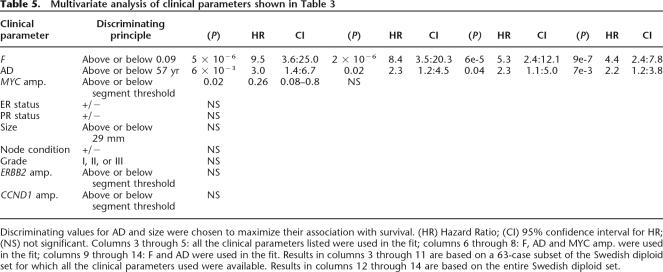
To further study the effect of F on survival, we fit our data to a Cox proportional hazards model, starting with a 63-case subset of the Swedish diploid data set for which we have complete information on all the clinical parameters listed in Table 4. A clinical parameter is considered significant for survival if the corresponding P-value is below 0.05. As shown in Table 5, we perform several rounds of analysis, each time removing from consideration clinical parameters not found significant in the previous round. This reduction in the number of parameters, in turn, allows us to increase the data set for which the information on the remaining parameters is complete. As a result, we find that F and the age at diagnosis are the only covariates that remain statistically significant through all the rounds of analysis. A fit to the entire Swedish diploid data set gives 4.4 as a hazard ratio for F, adjusted for the age at diagnosis.
Table 5.
Multivariate analysis of clinical parameters shown in Table 3
Discriminating values for AD and size were chosen to maximize their association with survival. (HR) Hazard Ratio; (CI) 95% confidence interval for HR; (NS) not significant. Columns 3 through 5: all the clinical parameters listed were used in the fit; columns 6 through 8: F, AD and MYC amp. were used in the fit; columns 9 through 14: F and AD were used in the fit. Results in columns 3 through 11 are based on a 63-case subset of the Swedish diploid set for which all the clinical parameters used were available. Results in columns 12 through 14 are based on the entire Swedish diploid set.
Discussion
To the best of our knowledge, this study represents the first large sample set of primary breast tumors profiled for copy number at a resolution of <50 kb, and using a set of probes designed specifically to cover the genome evenly without regard to gene position. Coupled with a segmentation algorithm that accurately reflects event boundaries, this design has allowed us to examine genome rearrangements in tumors at an unprecedented level of detail. At this resolution, narrow and closely spaced amplifications and deletions, some as narrow as 100 kb, are clearly distinguished, and can be validated as discrete events by interphase FISH.
Cataloging the events observed in these tumor sets has allowed us to create a high-resolution map of the regions most frequently affected in this collection of tumors as compiled in Table 3. Furthermore, examination of the ROMA patterns has led us to discern three distinct profile types, described as simplex, sawtooth, and firestorm, that provide insights into the natural history of tumor development and, moreover, provide prognostic and predictive information that may be of use in clinical practice.
ROMA profiles
Each of the three characteristic profiles shown by example in Figure 2 provides a different insight into the biology of primary breast tumors. Simplex profiles are characterized by multiple duplications and deletions of whole chromosomes or chromosome arms. Moreover, certain specific chromosome arm gains and losses are highly favored, and at least a subset appears in nearly all simplex tumors, even those low-grade tumors with less than three total events (Fig. 5). These lesions, all of which have been reported elsewhere by various methods (Kallioniemi et al. 1994; Ried et al. 1995; Tirkkonen et al. 1998; Pollack et al. 2002; Nessling et al. 2005), are duplication of 1q, 8q, and 16p, and deletion of 8p, 16q, and 22q. Each of these shows high frequency in the set of diploid tumors (Fig. 1B). Not all of the events occur together in the same tumor, and there is not enough data as yet to test whether there is any intrinsic order to the timing of their appearance. We do note, however, that the frequency of these specific changes remains constant when we compare tumors from surviving patients (or those with few events) with subsets of tumors that have poor survival (and many more total events) (Fig. 1B). One interpretation of these results is that in the early stages of tumor development, cells undergo a subset of these specific gain or loss events as they give rise to proliferating clones. Subsequently, as these clones become less differentiated and gain potential to spread in the host, additional events accumulate. Thus it is reasonable to speculate that there are early and late genomic events that can be separated according to the degree of progression exhibited by the cancer.
Comparing Figure 2, A and C, it is apparent that the complex firestorm profiles display a spectrum of whole arm events reminiscent of the simplex profiles, but with the notable difference that certain chromosomes are covered almost completely with high copy number, closely spaced amplicons. We call these features firestorms because they must be the result of violent disruptions of at least one homolog, probably involving multiple rounds of breakage, copying, and rejoining to form chains of many copies (up to 30 copies in some cases, as measured by FISH). The copies apparently remain contiguous since in all cases tested, FISH results indicate that the copies fall in tight clusters within the nucleus.
Firestorms might arise through one or more previously characterized genetic mechanisms that have been previously characterized in cultured cells, such as breaks at fragile sites (Coquelle et al. 1997; Hellman et al. 2002) or recombination at pre-existing palindromic sites (Tanaka et al. 2005), perhaps by shortened telomeres. Initial joining of chromatids or chromosomes can lead to breakage-fusion-bridge (BFB) processes first described by McClintock (1938, 1941). The process of chromatid fusion and bridge formation is often seen in tumor cells (Gisselsson et al. 2000; Shuster et al. 2000) and has the potential to result in repeated rounds of segmental amplification while remaining limited to a single arm as we have documented for firestorm events. This in itself might be a mechanism for genetic instability that augurs poor outcome, for example, by enabling the cancer cell to “search” locally for combinations of genes that by amplification or deletion promote resistance to natural controls on cell growth, invasion, or metastasis.
Finally, the alternative complex pattern, which we call sawtooth, demonstrates the operation of a path to complex genomic alteration distinct from that leading to firestorms. In contrast to firestorms, the sawtooth pattern consists of up to 30 duplication or deletion events, mostly involving chromosomal segments significantly broader than firestorm amplicons and distributed nearly evenly across the genome. Sawtooth profiles seldom show high copy number amplification as noted by the difference in the y-axis scale between Figure 2, A and B, versus Figure 2C. Sawtooth profiles, like firestorms, are associated with a poor prognosis, but their relatively high F index comes from the sheer number of events rather than the close spacing of the amplicons in firestorms. Taken together, these differences indicate that a genome-wide instability has been established in these tumors, perhaps distinguishing a distinct ontogeny and pathway toward metastasis.
The “Firestorm Index”
The high resolution of the ROMA technique along with our segmentation algorithm has enabled us to visualize narrow and closely spaced chromosomal rearrangements, in particular, those that make up the complex firestorm patterns. The validity of the amplicon assignments, and hence of the Kolmogorov-Smirnov methodology, has been validated by FISH in all cases tested. Coupled with the long-term survival and ploidy data available for the Swedish data set, we derived a working hypothesis consistent with previously reported work (Al-Kuraya et al. 2004; Loo et al. 2004) that complexity of rearrangement is a negative prognostic factor, but with the novel addition that the closely spaced events in firestorms make a disproportionately large contribution to that prognosis.
We have, therefore, derived a molecular signature, F, that correlates with survival in a subset of tumors, namely, pseudo-diploid tumors of patients from Scandinavia. The signature is a simply defined mathematical measure that incorporates two features of the genome copy number profile, namely, the number of distinguishable amplification and deletion segments, and the close packing of these segments. It is easy to imagine that the number of distinguishable events can serve as a marker for malignant “progression.” A large number of events might reflect either an unstable genome, a cancer that has been growing for a longer time within the patient and hence has had more opportunity to metastasize, or a cancer that has undergone more selective events than a cancer with fewer “scars” in its genome. It is worth noting that even a single case of the clustered amplifications that we call firestorms appears to be a prognostic indicator of poor outcome.
Our preliminary analyses of this selected sample set indicate that prognoses in primary breast cancer, measured by the probability of overall survival, are correlated with the morphology of the gene copy number signature. Within the balanced group of our samples, the magnitude of the signature is independent of such established clinical markers as node status, histologic grade, and primary tumor size. Hence, it is reasonable to expect that the signature will contribute to the prediction of outcome, perhaps—as suggested by our data—in combination with other known factors. A particularly valuable role for the signature may be in the estimation of survival for patients with ostensibly good prognosis, node-negative breast cancer, a group that may or may not benefit from systemic therapy. A clear potential application of such a measure is in the determination of prognosis, with a focus on the identification of patients with such excellent prognoses that systemic therapy is not required or, conversely, such poor prognoses—in spite of clinical measurements that might be misleading in this regard—that systemic treatment is absolutely indicated. For example, a patient with a small, estrogen-receptor-positive, node-negative primary breast cancer—all factors that usually indicate a good prognosis—might have an especially poor prognosis as predicted by our method. Further work with unselected sample sets will, of course, be required to extend these findings beyond the working hypothesis stage.
Event mapping
We expect further gains in outcome prediction that uses knowledge of which individual loci are amplified or deleted in a specific cancer. Indeed, there are clearly loci, such as 1q, 8p and 8q, 16p and 16q, and 22q that are present in both outcome groups with almost equal frequency, and others, such as 1p12–13, 11q12 and 11q13, 9p, 10q, 17q, and 20q that are present predominantly in the cancers from patients with poor outcomes. We can improve the separation of the two groups in our own data set by adding rules that proscribe amplification or deletion at specific loci or combinations of loci. However, despite exhaustive attempts, we could not convince ourselves that additional improvement in outcome prediction based on knowledge of specific loci was more than one would expect by chance, given overall event frequencies. The literature does contain many reports that specific amplifications or deletions correlate with poor prognosis (Berns et al. 1995; Jarvinen and Liu 2003; Al Kuraya et al. 2004; Chunder et al. 2004; Knoop et al. 2005; Madjd et al. 2005). While these reports may, indeed, be correct, they may also be a consequence of the larger picture, namely, that there are more lesions in “progressed” cancers. The copy numbers of specific genes may also be useful in clinical decision-making, following the clear demonstration that ERBB2 amplification—now determined by FISH—conveys both prognostic and therapeutic information. For example, patients with amplified ERBB2, as determined by FISH, are now treated with Herceptin. This determination can be made as well by ROMA or other methods for genome profiling, and such profiling may be more informative about which patients have amplifications and which benefit from such treatment. Other events in the genome can also indicate different choices of therapy. For example, two of the patients in our study exhibit amplification at the EGFR locus rather than ERBB2, and such patients might benefit from treatment with drugs targeted to that oncogene such as Tarceva. There are other such examples in the data set. More data than we now have will be needed to fully test a better outcome predictor model based on specific loci.
Scandinavian tumor sets
In the course of this study, and to gain a perspective, we have compared ROMA profiles from two independent sets of tumors from Sweden and Norway, and shown a basic similarity in the profiles independent of source or collection method. It is noteworthy that the diploid tumors with poor outcome show a very similar overall profile to the aneuploid tumors. Thus, whether or not the two classes of tumors, diploid and aneuploid, have different mechanisms for malignant genome evolution, a subset of loci recurred in amplifications and deletions in both types.
It is perhaps not surprising that the tumors from Swedish and Norwegian populations selected for this study have very similar frequency profiles, given the ethnic and environmental homogeneity in Scandinavia. It is unclear to us at the moment whether these populations will show similarity to other breast tumor sample sets. In any event, the ability to profile cancers from populations of restricted ethnicity and environment adds a new tool for those who wish to study the effects of genetics and environment on cancer. It will be of great interest to assess genome profiles of other geographically defined groups, with particular attention to the possibility of inherited patterns of disease susceptibility or gene–environment interactions.
Future directions
In this study, we have focused on a restricted question, the relationship between complex genomic rearrangements and tumor progression as determined by eventual outcome in breast cancer. There are many other interesting questions that we do not address in the present paper. We do not examine the related question of genomic and molecular markers for survival among aneuploid cancers. We have not analyzed what the collective profiles teach us about the location of candidate oncogenes and tumor suppressors. The latter is a deceptively complex problem that we will address subsequently. In the meantime, we post our genome profiles and associated data on our Web site (http://roma.cshl.edu) for others to explore. It is evident from even superficial inspection that many recurrent events encompass known oncogenes (such as ERBB2, CCND1, MYC) and tumor suppressors (such as CDKN2A and TP53), but many do not, such as a commonly amplified and very narrow region at 8p12, for which the driver gene has not been definitively identified (marked with a probe for BAG4 in Fig. 3A; Garcia et al. 2005). We are also currently analyzing the important question of whether certain lesions show covariance.
Finally, it is becoming clear through the identification of gene copy number alterations in tumors in numerous CGH studies, that there is likely to be a genetic pathway, albeit a complex one, at work in the evolution of tumors. As the collection of tumor genomic profiles increases and can be compared with treatment regimes as well as patient outcomes, that prognostic information regarding clinical outcome will likely become apparent. Thus existence of some systematic organization to the genomic events in these tumors raises the intriguing possibility that we may soon be able to dissect the pathways that determine the bridge from noninvasive to invasive to metastatic cancer.
Methods
Patient samples
A total of 140 frozen tumor specimens was selected from the archives at the Cancer Center of the Karolinska Institute, Stockholm, Sweden. Samples in this particular data set were selected to represent several distinct diagnostic categories in order to populate groups for comparison by FISH and ROMA. From a total of 5782 cases, analyzed for ploidy at the Division for Cellular and Molecular Pathology at the Karolinska Hospital at the time of primary diagnosis (1987–1991), 1601 pseudo-diploids were available with complete clinical information including ploidy, grade, node status, and clinical follow up for 14 to 18 yr. Of these, 4.0% or 64 cases were node-negative nonsurvivors at 7 yr, and 8.0% or 127 cases were node-positive nonsurvivors. Of these, 47 cases were locally available as frozen tissue and made up the group of node-negative and node-positive nonsurvivors. The diploid survivor group was selected from the remainder of the samples in order to match tumor size and grade.
From the Oslo Micrometastasis study (OMS) (Wiedswang et al. 2003), fresh frozen samples from the primary tumor from 103 cases were available for analyses by ROMA.
Clinical parameters
Status of the estrogen and progesterone receptors (ER, PR) was determined by ligand binding with a threshold value of >0.05 fg/μg DNA for classification as receptor positive for the Swedish samples. For the Norwegian samples, automatic immunostaining was performed using mouse monoclonal antibodies against ER and PgR (clones 6F11 and 1A6, respectively; Novocastra). Immunopositivity was recorded if ≥10% of the tumor cell nuclei were immunostained. Amplification of the ERBB2 gene was assessed by FISH on tissue microarray sections using the PathVysion HER-2 DNA Probe kit (Vysis Inc.).
ROMA DNA microarray analysis
ROMA was performed on a high-density oligonucleotide array containing ∼85,000 features, manufactured by Nimblegen. Hybridization conditions and statistical analysis have been described previously (Lucito et al. 2003).
Sample preparation, microarray hybridization, and image analysis
The preparation of genomic representations, labeling, and hybridization were performed as described previously (Lucito et al. 2003). Briefly, the complexity of the samples was reduced by making BglII genomic representations, consisting of small (200–1200 bp) fragments amplified by adaptor-mediated PCR of genomic DNA (Sebat et al. 2004). For each experiment, two different samples were prepared in parallel. DNA samples (10 μg) were then labeled differentially with Cy5-dCTP or Cy3-dCTP using the Amersham-Pharmacia Megaprime labeling Kit, and hybridized in comparison to each other. Each experiment was hybridized in duplicate, where in one replicate, the Cy5 and Cy3 dyes were swapped (i.e., “color reversal”). Hybridizations consisted of 25 μL of hybridization solution (50% formamide, 5× SSC, and 0.1% SDS) and 10 μL of labeled DNA. Samples were denatured in an MJ Research Tetrad for 5 min at 95°C, and then pre-annealed for 30 min at 37°C. This solution was then applied to the microarray and hybridized under a coverslip for 14–16 h at 42°C. After hybridization, slides were washed for 1 min in 0.2% SDS/0.2× SSC, 30 sec in 0.2× SSC, and 30 sec in 0.05× SSC. Slides were dried by centrifugation and scanned immediately. An Axon GenePix 4000B scanner was used setting the pixel size to 5 μm. GenePix Pro 4.0 software was used for quantitation of intensity for the arrays.
Data processing
Array data were imported into S-PLUS for further analysis. Measured intensities without background subtraction were used to calculate ratios. Data were normalized using an intensity-based lowess curve fitting algorithm. Log ratio values obtained from color reversal experiments were averaged and displayed as presented in the figures.
Statistics and segmentation algorithm
Segmentation views the probe ratio distribution as an ordered series of probe log ratios, placed in genome order, and breaks it into intervals each with a mean and a standard deviation. At the end of this process, the probe data, in genome order, is divided into segments (long and certain intervals), each segment and feature with its own mean and standard deviation, and each feature associated with a likelihood that the feature is not the result of chance clustering of probes with deviant ratios.
The ratio data are processed in three phases. In the first phase, we iteratively segment the log ratio data by minimizing variance, then test the segment boundaries by setting a very stringent Kolmogorov-Smirnov (K-S) P-value statistic for each segment relative to its neighboring segment (P = 10−5). No segment smaller than six probes in length is considered. In the second phase, we compute the “residual string” of segmented log ratio data, adjusting the mean and standard deviation of each segment so that the residual string has a mean of 0 and a standard deviation of 1. “Outliers” are defined based on deviance within the population, and features are defined as clusters of outliers (at least two). In the third phase, the features are assigned likelihood. We determine a “deviance measure” for each feature that reflects its deviance from the remainder of the data string. We then, in effect, either randomize or model randomization of the residual string (i.e., look at the residual data in a randomized order) many times, and collect deviance measures of all features generated by purely random processes. After binning the features by their length and their deviance measure, we can determine the likelihood that a given feature with a given length and deviance measure would have been generated by random processes if the probe data were noise.
Statistical analysis of segmented data was performed using R and S+ statistical languages. In particular, the R Survival package was used for survival analysis.
Masking of frequent CNPs
A large fraction of our collection of genome profiles are of a self–nonself type, that is, a cancer genome and a reference genome originate in different individuals. As a result, not all of the relative copy number variation in the cancer genome is due to cancer: Some of it reflects copy number polymorphisms (CNPs) present in the healthy genome of the affected individual. This noncancerous signal can potentially contaminate subsequent analysis and must be filtered out. To this end, we examine our collection of ROMA profiles derived from cancer-free genomes (∼500 cases in our most recent study). From that collection we determine the contiguous regions (here to be understood as series of consecutive ROMA probes) in the genome where CNP frequencies satisfy two conditions: (1) These frequencies are higher than certain fe everywhere in the region; (2) these frequencies are higher than certain fs ≥ fe somewhere in the region. This determination is done separately for the amplification and for the deletion CNPs. With our present cancer-free collection, the optimal values are fe = 0.006, fs = 0.03. Once the mask, that is, the set of CNP-prone regions of the genome, is known, it is used for masking likely noncancerous CNPs in cancer genome profiles. Here we describe the masking algorithm for amplifications; the algorithm for deletions is completely analogous. If an amplified segment in a cancer genome profile falls entirely within a mask, a point (a probe) is selected at random in the segment, and the neighboring segments on the right and on the left are extended to that point. If one of the segment’s endpoints is at a chromosome boundary, the neighboring segment is extended from the other endpoint to the boundary. In effect, the CNPs are excised from the profile in a minimally intrusive fashion.
Frequently amplified and deleted loci
For the purpose of compiling a list of frequently amplified loci, amplification events are defined as follows. First, the logarithm of the relative copy number is computed for every segment in the genome (the segmentation method is described earlier in this section). Denote the resulting piecewise constant function L(x), where x is the genome position. Next, (1) the values of L(x) below a threshold t are replaced by 0. Then (2) we identify event blocks, that is, contiguous intervals of the genome such that L(x) > 0 everywhere within the interval. For every block, (3) an event extending over the entire block is added to the list of events. Next (4) a minimal nonzero value of L(x) is found in each block, and that value is subtracted form L(x) within that block. The steps (1) through (4) are iterated as long as L(x) > 0 anywhere in the genome. The event counting rule for deletions is completely analogous, with obvious sign changes made throughout the description. We used a value of 0.1 for t in the present study. Once the events have been identified, we compute for every position in the genome an event density measure, defined as the sum of inverse lengths of all the events containing that position. We then identify positions with the highest event density in every chromosome arm.
Fluorescence in situ hybridization
FISH analysis was performed using interphase cells, and probes were prepared either from BACs or amplified from specific genomic regions by PCR. Based on the human genome sequence, primers (1–2 kb in length) were designed from the repeat-masked sequence of each CNP interval, and limited to an interval no larger than 100 kb. For each probe, a total of 20–25 different fragments were amplified, then pooled, and purified by ethanol precipitation. Probe DNA was then labeled by nick translation with SpectrumOrange or SpectrumGreen (Vysis Inc.). Denaturation of probe and target DNA was performed for 5 min at 90°C, followed by hybridization in a humidity chamber overnight at 47°C. The cover glasses were then removed, and the slides were washed in 2× SSC for 10 min at 72°C, and slides were dehydrated in graded alcohol. The slides were mounted with antifade mounting medium containing DAPI (4′,6-diamino-2-phenylindole; Vectashield) as a counterstain for the nuclei. Evaluation of signals was carried out in an epifluorescence microscope. Selected cells were photographed in a Zeiss Axioplan 2 microscope equipped with an Axio Cam MRM CCD camera and Axio Vision software.
Probe design for FISH
Hybridization probes for FISH were constructed in one of two methods. For the interdigitation analysis, probes were created from bacterial artificial chromosomes (BAC) selected using the UCSD Genome Browser. For the determination of copy number in the deletions and amplifications of the aneuploid tumors, probes were made with PCR amplification of primers identified through the PROBER algorithm designed in this laboratory (Navin et al. 2006). Genomic sequences of 100 kb containing target amplifications were tiled with 50 probes (800–1400 bp).
Oligonucleotide primers were ordered in 96-well plates from Sigma Genosys and resuspended to 25 μM. Probes were amplified with the PCR Mastermix kit from Eppendorf (Cat. 0,032,002.447) from EBV immortalized cell line DNA (Chp-Skn-1) DNA (100 ng) with 55°C annealing, 72°C extension, 2 min extension time, and 23 cycles. Probes were purified with Qiagen PCR purification columns (Cat. 28,104) and combined into a single probe cocktail (10–25 μg total probes) for dye labeling and Metaphase/Interphase FISH.
Measurement of DNA content
The ploidy of each tumor was determined by measurement of DNA content using Feulgen photocytometry (Forsslund and Zetterberg 1990; Forsslund et al. 1996) The optical densities of the nuclei in a sample are measured and a DNA index is calculated and displayed as a histogram (Kronenwett et al. 2004) Normal cells and diploid tumors display a major peak at 2c DNA content with a smaller peak of G2-phase replicating cells that corresponds to the mitotic index. Highly aneuploid tumors display broad peaks that often center on 4c copy number but may include cells from 2c to 6c or above.
Patient consent
KI samples were collected from patients undergoing radical mastectomy at the Karolinska Insitutet between 1984 and 1991. This project was approved by the Ethical Committee of the Karolinska Institute, Stockholm, Sweden (772003). Samples in the OMS set were collected during 1995–1998 after informed written consent and analysis protocols approved by the Regional Committee for Research Ethics, Health Region II, Oslo, Norway (approval S97103).
Acknowledgments
This work was supported by grants to M.W. from the National Institutes of Health 5R01-CA078544-07; Department of the Army W81XWH04-1-0477; W81XWH-05-1-0068; W81XWH-04-0905; The Simons Foundation; Miracle Foundation; Breast Cancer Research Foundation; Long Islanders Against Breast Cancer; West Islip Breast Cancer Foundation; Long Island Breast Cancer (1 in 9); Elizabeth McFarland Breast Cancer Research Grant; and Breast Cancer Help Inc. M.W. is an American Cancer Society Research Professor. This work was supported by grants to A.Z. from the Swedish Cancer Society (grant number 0046-B05-39XBC), from the Stockholm Cancer Society (grant number 03:17), and from the Swedish Research Council (grant number K2006-31X-20081-01-3). The OMS study has been supported by the Norwegian Cancer Society. We are also grateful for critical review by Knut Liestøl, Institute for Informatics, University of Oslo and for useful comments by Xiaoyue Zhao, Cold Spring Harbor Laboratory.
Footnotes
[Supplemental material is available online at www.genome.org and at http://roma.cshl.edu.]
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.5460106
References
- Ahr A., Karn T., Solbach C., Seiter T., Strebhardt K., Holtrich U., Kaufmann M., Karn T., Solbach C., Seiter T., Strebhardt K., Holtrich U., Kaufmann M., Solbach C., Seiter T., Strebhardt K., Holtrich U., Kaufmann M., Seiter T., Strebhardt K., Holtrich U., Kaufmann M., Strebhardt K., Holtrich U., Kaufmann M., Holtrich U., Kaufmann M., Kaufmann M. Identification of high risk breast-cancer patients by gene expression profiling. Lancet. 2002;359:131–132. doi: 10.1016/S0140-6736(02)07337-3. [DOI] [PubMed] [Google Scholar]
- Albertson D.G. Profiling breast cancer by array CGH. Breast Cancer Res. Treat. 2003;78:289–298. doi: 10.1023/a:1023025506386. [DOI] [PubMed] [Google Scholar]
- Al Kuraya K., Schraml P., Torhorst J., Tapia C., Zaharieva B., Novotny H., Spichtin H., Maurer R., Mirlacher M., Kochli O., Schraml P., Torhorst J., Tapia C., Zaharieva B., Novotny H., Spichtin H., Maurer R., Mirlacher M., Kochli O., Torhorst J., Tapia C., Zaharieva B., Novotny H., Spichtin H., Maurer R., Mirlacher M., Kochli O., Tapia C., Zaharieva B., Novotny H., Spichtin H., Maurer R., Mirlacher M., Kochli O., Zaharieva B., Novotny H., Spichtin H., Maurer R., Mirlacher M., Kochli O., Novotny H., Spichtin H., Maurer R., Mirlacher M., Kochli O., Spichtin H., Maurer R., Mirlacher M., Kochli O., Maurer R., Mirlacher M., Kochli O., Mirlacher M., Kochli O., Kochli O., et al. Prognostic relevance of gene amplifications and coamplifications in breast cancer. Cancer Res. 2004;64:8534–8540. doi: 10.1158/0008-5472.CAN-04-1945. [DOI] [PubMed] [Google Scholar]
- Balmain A., Gray J., Ponder B., Gray J., Ponder B., Ponder B. The genetics and genomics of cancer. Nat. Genet. 2003;33:238–244. doi: 10.1038/ng1107. [DOI] [PubMed] [Google Scholar]
- Berns E.M., de Klein A., van Putten W.L., van Staveren I.L., Bootsma A., Klijn J.G., Foekens J.A., de Klein A., van Putten W.L., van Staveren I.L., Bootsma A., Klijn J.G., Foekens J.A., van Putten W.L., van Staveren I.L., Bootsma A., Klijn J.G., Foekens J.A., van Staveren I.L., Bootsma A., Klijn J.G., Foekens J.A., Bootsma A., Klijn J.G., Foekens J.A., Klijn J.G., Foekens J.A., Foekens J.A. Association between RB-1 gene alterations and factors of favourable prognosis in human breast cancer, without effect on survival. Int. J. Cancer. 1995;64:140–145. doi: 10.1002/ijc.2910640212. [DOI] [PubMed] [Google Scholar]
- Chunder N., Mandal S., Roy A., Roychoudhury S., Panda C.K., Mandal S., Roy A., Roychoudhury S., Panda C.K., Roy A., Roychoudhury S., Panda C.K., Roychoudhury S., Panda C.K., Panda C.K. Analysis of different deleted regions in chromosome 11 and their interrelations in early- and late-onset breast tumors: Association with cyclin D1 amplification and survival. Diagn. Mol. Pathol. 2004;13:172–182. doi: 10.1097/01.pas.0000124337.49401.0b. [DOI] [PubMed] [Google Scholar]
- Coquelle A., Pipiras E., Toledo F., Buttin G., Debatisse M., Pipiras E., Toledo F., Buttin G., Debatisse M., Toledo F., Buttin G., Debatisse M., Buttin G., Debatisse M., Debatisse M. Expression of fragile sites triggers intrachromosomal mammalian gene amplification and sets boundaries to early amplicons. Cell. 1997;89:215–225. doi: 10.1016/s0092-8674(00)80201-9. [DOI] [PubMed] [Google Scholar]
- Daruwala R.S., Rudra A., Ostrer H., Lucito R., Wigler M., Mishra B., Rudra A., Ostrer H., Lucito R., Wigler M., Mishra B., Ostrer H., Lucito R., Wigler M., Mishra B., Lucito R., Wigler M., Mishra B., Wigler M., Mishra B., Mishra B. A versatile statistical analysis algorithm to detect genome copy number variation. Proc. Natl. Acad. Sci. 2004;101:16292–16297. doi: 10.1073/pnas.0407247101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePinho R.A., Polyak K., Polyak K. Cancer chromosomes in crisis. Nat. Genet. 2004;36:932–934. doi: 10.1038/ng0904-932. [DOI] [PubMed] [Google Scholar]
- Edén P., Ritz C., Rose C., Ferno M., Peterson C., Ritz C., Rose C., Ferno M., Peterson C., Rose C., Ferno M., Peterson C., Ferno M., Peterson C., Peterson C. “Good Old” clinical markers have similar power in breast cancer prognosis as microarray gene expression profilers. Eur. J. Cancer. 2004;40:1837–1841. doi: 10.1016/j.ejca.2004.02.025. [DOI] [PubMed] [Google Scholar]
- Forsslund G., Zetterberg A., Zetterberg A. Ploidy level determinations in high-grade and low-grade malignant variants of prostatic carcinoma. Cancer Res. 1990;50:4281–4285. [PubMed] [Google Scholar]
- Forsslund G., Nilsson B., Zetterberg A., Nilsson B., Zetterberg A., Zetterberg A. Near tetraploid prostate carcinoma. Methodologic and prognostic aspects. Cancer. 1996;78:1748–1755. [PubMed] [Google Scholar]
- Garcia M.J., Pole J.C., Chin S.F., Teschendorff A., Naderi A., Ozdag H., Vias M., Kranjac T., Subkhankulova T., Paish C., Pole J.C., Chin S.F., Teschendorff A., Naderi A., Ozdag H., Vias M., Kranjac T., Subkhankulova T., Paish C., Chin S.F., Teschendorff A., Naderi A., Ozdag H., Vias M., Kranjac T., Subkhankulova T., Paish C., Teschendorff A., Naderi A., Ozdag H., Vias M., Kranjac T., Subkhankulova T., Paish C., Naderi A., Ozdag H., Vias M., Kranjac T., Subkhankulova T., Paish C., Ozdag H., Vias M., Kranjac T., Subkhankulova T., Paish C., Vias M., Kranjac T., Subkhankulova T., Paish C., Kranjac T., Subkhankulova T., Paish C., Subkhankulova T., Paish C., Paish C., et al. A 1 Mb minimal amplicon at 8p11-12 in breast cancer identifies new candidate oncogenes. Oncogene. 2005;24:5235–5245. doi: 10.1038/sj.onc.1208741. [DOI] [PubMed] [Google Scholar]
- Gisselsson D., Pettersson L., Hoglund M., Heidenblad M., Gorunova L., Wiegant J., Mertens F., Dal Cin P., Mitelman F., Mandahl N., Pettersson L., Hoglund M., Heidenblad M., Gorunova L., Wiegant J., Mertens F., Dal Cin P., Mitelman F., Mandahl N., Hoglund M., Heidenblad M., Gorunova L., Wiegant J., Mertens F., Dal Cin P., Mitelman F., Mandahl N., Heidenblad M., Gorunova L., Wiegant J., Mertens F., Dal Cin P., Mitelman F., Mandahl N., Gorunova L., Wiegant J., Mertens F., Dal Cin P., Mitelman F., Mandahl N., Wiegant J., Mertens F., Dal Cin P., Mitelman F., Mandahl N., Mertens F., Dal Cin P., Mitelman F., Mandahl N., Dal Cin P., Mitelman F., Mandahl N., Mitelman F., Mandahl N., Mandahl N. Chromosomal breakage-fusion-bridge events cause genetic intratumor heterogeneity. Proc. Natl. Acad. Sci. 2000;97:5357–5362. doi: 10.1073/pnas.090013497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman A., Zlotorynski E., Scherer S.W., Cheung J., Vincent J.B., Smith D.I., Trakhtenbrot L., Kerem B., Zlotorynski E., Scherer S.W., Cheung J., Vincent J.B., Smith D.I., Trakhtenbrot L., Kerem B., Scherer S.W., Cheung J., Vincent J.B., Smith D.I., Trakhtenbrot L., Kerem B., Cheung J., Vincent J.B., Smith D.I., Trakhtenbrot L., Kerem B., Vincent J.B., Smith D.I., Trakhtenbrot L., Kerem B., Smith D.I., Trakhtenbrot L., Kerem B., Trakhtenbrot L., Kerem B., Kerem B. A role for common fragile site induction in amplification of human oncogenes. Cancer Cell. 2002;1:89–97. doi: 10.1016/s1535-6108(02)00017-x. [DOI] [PubMed] [Google Scholar]
- Jarvinen T.A., Liu E.T., Liu E.T. HER-2/neu and topoisomerase IIα in breast cancer. Breast Cancer Res. Treat. 2003;78:299–311. doi: 10.1023/a:1023077507295. [DOI] [PubMed] [Google Scholar]
- Kallioniemi A., Kallioniemi O.P., Sudar D., Rutovitz D., Gray J.W., Waldman F., Pinkel D., Kallioniemi O.P., Sudar D., Rutovitz D., Gray J.W., Waldman F., Pinkel D., Sudar D., Rutovitz D., Gray J.W., Waldman F., Pinkel D., Rutovitz D., Gray J.W., Waldman F., Pinkel D., Gray J.W., Waldman F., Pinkel D., Waldman F., Pinkel D., Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992a;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- Kallioniemi A., Kallioniemi O.P., Waldman F.M., Chen L.C., Yu L.C., Fung Y.K., Smith H.S., Pinkel D., Gray J.W., Kallioniemi O.P., Waldman F.M., Chen L.C., Yu L.C., Fung Y.K., Smith H.S., Pinkel D., Gray J.W., Waldman F.M., Chen L.C., Yu L.C., Fung Y.K., Smith H.S., Pinkel D., Gray J.W., Chen L.C., Yu L.C., Fung Y.K., Smith H.S., Pinkel D., Gray J.W., Yu L.C., Fung Y.K., Smith H.S., Pinkel D., Gray J.W., Fung Y.K., Smith H.S., Pinkel D., Gray J.W., Smith H.S., Pinkel D., Gray J.W., Pinkel D., Gray J.W., Gray J.W. Detection of retinoblastoma gene copy number in metaphase chromosomes and interphase nuclei by fluorescence in situ hybridization. Cytogenet. Cell Genet. 1992b;60:190–193. doi: 10.1159/000133333. [DOI] [PubMed] [Google Scholar]
- Kallioniemi O.P., Kallioniemi A., Kurisu W., Thor A., Chen L.C., Smith H.S., Waldman F.M., Pinkel D., Gray J.W., Kallioniemi A., Kurisu W., Thor A., Chen L.C., Smith H.S., Waldman F.M., Pinkel D., Gray J.W., Kurisu W., Thor A., Chen L.C., Smith H.S., Waldman F.M., Pinkel D., Gray J.W., Thor A., Chen L.C., Smith H.S., Waldman F.M., Pinkel D., Gray J.W., Chen L.C., Smith H.S., Waldman F.M., Pinkel D., Gray J.W., Smith H.S., Waldman F.M., Pinkel D., Gray J.W., Waldman F.M., Pinkel D., Gray J.W., Pinkel D., Gray J.W., Gray J.W. ERBB2 amplification in breast cancer analyzed by fluorescence in situ hybridization. Proc. Natl. Acad. Sci. 1992c;89:5321–5325. doi: 10.1073/pnas.89.12.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallioniemi A., Kallioniemi O.P., Piper J., Tanner M., Stokke T., Chen L., Smith H.S., Pinkel D., Gray J.W., Waldman F.M., Kallioniemi O.P., Piper J., Tanner M., Stokke T., Chen L., Smith H.S., Pinkel D., Gray J.W., Waldman F.M., Piper J., Tanner M., Stokke T., Chen L., Smith H.S., Pinkel D., Gray J.W., Waldman F.M., Tanner M., Stokke T., Chen L., Smith H.S., Pinkel D., Gray J.W., Waldman F.M., Stokke T., Chen L., Smith H.S., Pinkel D., Gray J.W., Waldman F.M., Chen L., Smith H.S., Pinkel D., Gray J.W., Waldman F.M., Smith H.S., Pinkel D., Gray J.W., Waldman F.M., Pinkel D., Gray J.W., Waldman F.M., Gray J.W., Waldman F.M., Waldman F.M. Detection and mapping of amplified DNA sequences in breast cancer by comparative genomic hybridization. Proc. Natl. Acad. Sci. 1994;91:2156–2160. doi: 10.1073/pnas.91.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoop A.S., Knudsen H., Balslev E., Rasmussen B.B., Overgaard J., Nielsen K.V., Schonau A., Gunnarsdottir K., Olsen K.E., Mouridsen H., Knudsen H., Balslev E., Rasmussen B.B., Overgaard J., Nielsen K.V., Schonau A., Gunnarsdottir K., Olsen K.E., Mouridsen H., Balslev E., Rasmussen B.B., Overgaard J., Nielsen K.V., Schonau A., Gunnarsdottir K., Olsen K.E., Mouridsen H., Rasmussen B.B., Overgaard J., Nielsen K.V., Schonau A., Gunnarsdottir K., Olsen K.E., Mouridsen H., Overgaard J., Nielsen K.V., Schonau A., Gunnarsdottir K., Olsen K.E., Mouridsen H., Nielsen K.V., Schonau A., Gunnarsdottir K., Olsen K.E., Mouridsen H., Schonau A., Gunnarsdottir K., Olsen K.E., Mouridsen H., Gunnarsdottir K., Olsen K.E., Mouridsen H., Olsen K.E., Mouridsen H., Mouridsen H., et al. Retrospective analysis of topoisomerase IIa amplifications and deletions as predictive markers in primary breast cancer patients randomly assigned to cyclophosphamide, methotrexate, and fluorouracil or cyclophosphamide, epirubicin, and fluorouracil: Danish Breast Cancer Cooperative Group. J. Clin. Oncol. 2005;23:7483–7490. doi: 10.1200/JCO.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Kronenwett U., Huwendiek S., Ostring C., Portwood N., Roblick U.J., Pawitan Y., Alaiya A., Sennerstam R., Zetterberg A., Auer G., Huwendiek S., Ostring C., Portwood N., Roblick U.J., Pawitan Y., Alaiya A., Sennerstam R., Zetterberg A., Auer G., Ostring C., Portwood N., Roblick U.J., Pawitan Y., Alaiya A., Sennerstam R., Zetterberg A., Auer G., Portwood N., Roblick U.J., Pawitan Y., Alaiya A., Sennerstam R., Zetterberg A., Auer G., Roblick U.J., Pawitan Y., Alaiya A., Sennerstam R., Zetterberg A., Auer G., Pawitan Y., Alaiya A., Sennerstam R., Zetterberg A., Auer G., Alaiya A., Sennerstam R., Zetterberg A., Auer G., Sennerstam R., Zetterberg A., Auer G., Zetterberg A., Auer G., Auer G. Improved grading of breast adenocarcinomas based on genomic instability. Cancer Res. 2004;64:904–909. doi: 10.1158/0008-5472.can-03-2451. [DOI] [PubMed] [Google Scholar]
- Lage J.M., Leamon J.H., Pejovic T., Hamann S., Lacey M., Dillon D., Segraves R., Vossbrinck B., Gonzalez A., Pinkel D., Leamon J.H., Pejovic T., Hamann S., Lacey M., Dillon D., Segraves R., Vossbrinck B., Gonzalez A., Pinkel D., Pejovic T., Hamann S., Lacey M., Dillon D., Segraves R., Vossbrinck B., Gonzalez A., Pinkel D., Hamann S., Lacey M., Dillon D., Segraves R., Vossbrinck B., Gonzalez A., Pinkel D., Lacey M., Dillon D., Segraves R., Vossbrinck B., Gonzalez A., Pinkel D., Dillon D., Segraves R., Vossbrinck B., Gonzalez A., Pinkel D., Segraves R., Vossbrinck B., Gonzalez A., Pinkel D., Vossbrinck B., Gonzalez A., Pinkel D., Gonzalez A., Pinkel D., Pinkel D., et al. Whole genome analysis of genetic alterations in small DNA samples using hyperbranched strand displacement amplification and array-CGH. Genome Res. 2003;13:294–307. doi: 10.1101/gr.377203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo L.W., Grove D.I., Williams E.M., Neal C.L., Cousens L.A., Schubert E.L., Holcomb I.N., Massa H.F., Glogovac J., Li C.I., Grove D.I., Williams E.M., Neal C.L., Cousens L.A., Schubert E.L., Holcomb I.N., Massa H.F., Glogovac J., Li C.I., Williams E.M., Neal C.L., Cousens L.A., Schubert E.L., Holcomb I.N., Massa H.F., Glogovac J., Li C.I., Neal C.L., Cousens L.A., Schubert E.L., Holcomb I.N., Massa H.F., Glogovac J., Li C.I., Cousens L.A., Schubert E.L., Holcomb I.N., Massa H.F., Glogovac J., Li C.I., Schubert E.L., Holcomb I.N., Massa H.F., Glogovac J., Li C.I., Holcomb I.N., Massa H.F., Glogovac J., Li C.I., Massa H.F., Glogovac J., Li C.I., Glogovac J., Li C.I., Li C.I., et al. Array comparative genomic hybridization analysis of genomic alterations in breast cancer subtypes. Cancer Res. 2004;64:8541–8549. doi: 10.1158/0008-5472.CAN-04-1992. [DOI] [PubMed] [Google Scholar]
- Lucito R., Healy J., Alexander J., Reiner A., Esposito D., Chi M., Rodgers L., Brady A., Sebat J., Troge J., Healy J., Alexander J., Reiner A., Esposito D., Chi M., Rodgers L., Brady A., Sebat J., Troge J., Alexander J., Reiner A., Esposito D., Chi M., Rodgers L., Brady A., Sebat J., Troge J., Reiner A., Esposito D., Chi M., Rodgers L., Brady A., Sebat J., Troge J., Esposito D., Chi M., Rodgers L., Brady A., Sebat J., Troge J., Chi M., Rodgers L., Brady A., Sebat J., Troge J., Rodgers L., Brady A., Sebat J., Troge J., Brady A., Sebat J., Troge J., Sebat J., Troge J., Troge J., et al. Representational oligonucleotide microarray analysis: A high-resolution method to detect genome copy number variation. Genome Res. 2003;13:2291–2305. doi: 10.1101/gr.1349003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madjd Z., Spendlove I., Pinder S.E., Ellis I.O., Durrant L.G., Spendlove I., Pinder S.E., Ellis I.O., Durrant L.G., Pinder S.E., Ellis I.O., Durrant L.G., Ellis I.O., Durrant L.G., Durrant L.G. Total loss of MHC class I is an independent indicator of good prognosis in breast cancer. Int. J. Cancer. 2005;117:248–255. doi: 10.1002/ijc.21163. [DOI] [PubMed] [Google Scholar]
- McClintock B. The production of homozygous deficient tissues with mutant characteristics by means of the aberrant mitotic behavior of ring-shaped chromosomes. Genetics. 1938;23:315–376. doi: 10.1093/genetics/23.4.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The stability of broken ends of chromosomes in Zea mays. Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menard S., Fortis S., Castiglioni F., Agresti R., Balsari A., Fortis S., Castiglioni F., Agresti R., Balsari A., Castiglioni F., Agresti R., Balsari A., Agresti R., Balsari A., Balsari A. HER2 as a prognostic factor in breast cancer. Oncology. 2001;61:67–72. doi: 10.1159/000055404. [DOI] [PubMed] [Google Scholar]
- Navin N., Grubor V., Hicks J., Leibu E., Thomas E., Troge J., Riggs M., Lundin P., Maner S., Sebat J., Grubor V., Hicks J., Leibu E., Thomas E., Troge J., Riggs M., Lundin P., Maner S., Sebat J., Hicks J., Leibu E., Thomas E., Troge J., Riggs M., Lundin P., Maner S., Sebat J., Leibu E., Thomas E., Troge J., Riggs M., Lundin P., Maner S., Sebat J., Thomas E., Troge J., Riggs M., Lundin P., Maner S., Sebat J., Troge J., Riggs M., Lundin P., Maner S., Sebat J., Riggs M., Lundin P., Maner S., Sebat J., Lundin P., Maner S., Sebat J., Maner S., Sebat J., Sebat J., et al. PROBER: Oligonucleotide FISH probe design software. Bioinformatics. 2006;22:2437–2438. doi: 10.1093/bioinformatics/btl273. [DOI] [PubMed] [Google Scholar]
- Nessling M., Richter K., Schwaenen C., Roerig P., Wrobel G., Wessendorf S., Fritz B., Bentz M., Sinn H.-P., Radwimmer B., Richter K., Schwaenen C., Roerig P., Wrobel G., Wessendorf S., Fritz B., Bentz M., Sinn H.-P., Radwimmer B., Schwaenen C., Roerig P., Wrobel G., Wessendorf S., Fritz B., Bentz M., Sinn H.-P., Radwimmer B., Roerig P., Wrobel G., Wessendorf S., Fritz B., Bentz M., Sinn H.-P., Radwimmer B., Wrobel G., Wessendorf S., Fritz B., Bentz M., Sinn H.-P., Radwimmer B., Wessendorf S., Fritz B., Bentz M., Sinn H.-P., Radwimmer B., Fritz B., Bentz M., Sinn H.-P., Radwimmer B., Bentz M., Sinn H.-P., Radwimmer B., Sinn H.-P., Radwimmer B., Radwimmer B., et al. Candidate genes in breast cancer revealed by microarray-based comparative genomic hybridization of archived tissue. Cancer Res. 2005;65:439–447. [PubMed] [Google Scholar]
- Olshen A.B., Venkatraman E.S., Lucito R., Wigler M., Venkatraman E.S., Lucito R., Wigler M., Lucito R., Wigler M., Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- Ormandy C.J., Musgrove E.A., Hui R., Daly R.J., Sutherland R.L., Musgrove E.A., Hui R., Daly R.J., Sutherland R.L., Hui R., Daly R.J., Sutherland R.L., Daly R.J., Sutherland R.L., Sutherland R.L. Cyclin D1, EMS1 and 11q13 amplification in breast cancer. Breast Cancer Res. Treat. 2003;78:323–335. doi: 10.1023/a:1023033708204. [DOI] [PubMed] [Google Scholar]
- Paik S., Shak S., Tang G., Kim C., Baker J., Cronin M., Baehner F.L., Walker M.G., Watson D., Park T., Shak S., Tang G., Kim C., Baker J., Cronin M., Baehner F.L., Walker M.G., Watson D., Park T., Tang G., Kim C., Baker J., Cronin M., Baehner F.L., Walker M.G., Watson D., Park T., Kim C., Baker J., Cronin M., Baehner F.L., Walker M.G., Watson D., Park T., Baker J., Cronin M., Baehner F.L., Walker M.G., Watson D., Park T., Cronin M., Baehner F.L., Walker M.G., Watson D., Park T., Baehner F.L., Walker M.G., Watson D., Park T., Walker M.G., Watson D., Park T., Watson D., Park T., Park T., et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- Perou C.M., Sorlie T., Eisen M.B., de van Rijn M., Jeffrey S.S., Rees C.A., Pollack J.R., Ross D.T., Johnsen H., Aksien L.A., Sorlie T., Eisen M.B., de van Rijn M., Jeffrey S.S., Rees C.A., Pollack J.R., Ross D.T., Johnsen H., Aksien L.A., Eisen M.B., de van Rijn M., Jeffrey S.S., Rees C.A., Pollack J.R., Ross D.T., Johnsen H., Aksien L.A., de van Rijn M., Jeffrey S.S., Rees C.A., Pollack J.R., Ross D.T., Johnsen H., Aksien L.A., Jeffrey S.S., Rees C.A., Pollack J.R., Ross D.T., Johnsen H., Aksien L.A., Rees C.A., Pollack J.R., Ross D.T., Johnsen H., Aksien L.A., Pollack J.R., Ross D.T., Johnsen H., Aksien L.A., Ross D.T., Johnsen H., Aksien L.A., Johnsen H., Aksien L.A., Aksien L.A., et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Pollack J.R., Sorlie T., Perou C.M., Rees C.A., Jeffrey S.S., Lonning P.E., Tibshirani R., Botstein D., Borresen-Dale A.L., Brown P.O., Sorlie T., Perou C.M., Rees C.A., Jeffrey S.S., Lonning P.E., Tibshirani R., Botstein D., Borresen-Dale A.L., Brown P.O., Perou C.M., Rees C.A., Jeffrey S.S., Lonning P.E., Tibshirani R., Botstein D., Borresen-Dale A.L., Brown P.O., Rees C.A., Jeffrey S.S., Lonning P.E., Tibshirani R., Botstein D., Borresen-Dale A.L., Brown P.O., Jeffrey S.S., Lonning P.E., Tibshirani R., Botstein D., Borresen-Dale A.L., Brown P.O., Lonning P.E., Tibshirani R., Botstein D., Borresen-Dale A.L., Brown P.O., Tibshirani R., Botstein D., Borresen-Dale A.L., Brown P.O., Botstein D., Borresen-Dale A.L., Brown P.O., Borresen-Dale A.L., Brown P.O., Brown P.O. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc. Natl. Acad. Sci. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ried T., Just K.E., Holgreve-Grez H., Du Manoir S., Speicher M.R., Schröck E., Latham C., Blegen H., Zetterberg A., Cremer T., Just K.E., Holgreve-Grez H., Du Manoir S., Speicher M.R., Schröck E., Latham C., Blegen H., Zetterberg A., Cremer T., Holgreve-Grez H., Du Manoir S., Speicher M.R., Schröck E., Latham C., Blegen H., Zetterberg A., Cremer T., Du Manoir S., Speicher M.R., Schröck E., Latham C., Blegen H., Zetterberg A., Cremer T., Speicher M.R., Schröck E., Latham C., Blegen H., Zetterberg A., Cremer T., Schröck E., Latham C., Blegen H., Zetterberg A., Cremer T., Latham C., Blegen H., Zetterberg A., Cremer T., Blegen H., Zetterberg A., Cremer T., Zetterberg A., Cremer T., Cremer T., et al. Comparative genomic hybridization of formalin-fixed, paraffin-embedded breast tumors reveals different patterns of chromosomal gains and losses in fibroadenomas and diploid and aneuploid carcinomas. Cancer Res. 1995;5:5415–5423. [PubMed] [Google Scholar]
- Ried T., Liyanage M., Du Manoir S., Heselmeyer K., Auer G., Macville M., Schrock E., Liyanage M., Du Manoir S., Heselmeyer K., Auer G., Macville M., Schrock E., Du Manoir S., Heselmeyer K., Auer G., Macville M., Schrock E., Heselmeyer K., Auer G., Macville M., Schrock E., Auer G., Macville M., Schrock E., Macville M., Schrock E., Schrock E. Tumor cytogenetics revisited: Comparative genomic hybridization and spectral karyotyping. J. Mol. Med. 1997;75:801–814. doi: 10.1007/s001090050169. [DOI] [PubMed] [Google Scholar]
- Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Lundin P., Maner S., Massa H., Walker M., Chi M., Maner S., Massa H., Walker M., Chi M., Massa H., Walker M., Chi M., Walker M., Chi M., Chi M., et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Shuster M.I., Han L., Le Beau M.M., Davis E., Sawicki M., Lese C.M., Park N.H., Colicelli J., Gollin S.M., Han L., Le Beau M.M., Davis E., Sawicki M., Lese C.M., Park N.H., Colicelli J., Gollin S.M., Le Beau M.M., Davis E., Sawicki M., Lese C.M., Park N.H., Colicelli J., Gollin S.M., Davis E., Sawicki M., Lese C.M., Park N.H., Colicelli J., Gollin S.M., Sawicki M., Lese C.M., Park N.H., Colicelli J., Gollin S.M., Lese C.M., Park N.H., Colicelli J., Gollin S.M., Park N.H., Colicelli J., Gollin S.M., Colicelli J., Gollin S.M., Gollin S.M. A consistent pattern of RIN1 rearrangements in oral squamous cell carcinoma cell lines supports a breakage-fusion-bridge cycle model for 11q13 amplification. Genes Chromosomes Cancer. 2000;28:153–163. [PubMed] [Google Scholar]
- Slamon D.J., Godolphin W., Jones L.A., Holt J.A., Wong S.G., Keith D.E., Levin W.J., Stuart S.G., Udove J., Ullrich A., Godolphin W., Jones L.A., Holt J.A., Wong S.G., Keith D.E., Levin W.J., Stuart S.G., Udove J., Ullrich A., Jones L.A., Holt J.A., Wong S.G., Keith D.E., Levin W.J., Stuart S.G., Udove J., Ullrich A., Holt J.A., Wong S.G., Keith D.E., Levin W.J., Stuart S.G., Udove J., Ullrich A., Wong S.G., Keith D.E., Levin W.J., Stuart S.G., Udove J., Ullrich A., Keith D.E., Levin W.J., Stuart S.G., Udove J., Ullrich A., Levin W.J., Stuart S.G., Udove J., Ullrich A., Stuart S.G., Udove J., Ullrich A., Udove J., Ullrich A., Ullrich A. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- Sorlie T., Perou C.M., Tibshirani R., Aas T., Geisler S., Johnsen H., Hastie T., Eisen M.D., de van Rijn M., Jeffrey S.S., Perou C.M., Tibshirani R., Aas T., Geisler S., Johnsen H., Hastie T., Eisen M.D., de van Rijn M., Jeffrey S.S., Tibshirani R., Aas T., Geisler S., Johnsen H., Hastie T., Eisen M.D., de van Rijn M., Jeffrey S.S., Aas T., Geisler S., Johnsen H., Hastie T., Eisen M.D., de van Rijn M., Jeffrey S.S., Geisler S., Johnsen H., Hastie T., Eisen M.D., de van Rijn M., Jeffrey S.S., Johnsen H., Hastie T., Eisen M.D., de van Rijn M., Jeffrey S.S., Hastie T., Eisen M.D., de van Rijn M., Jeffrey S.S., Eisen M.D., de van Rijn M., Jeffrey S.S., de van Rijn M., Jeffrey S.S., Jeffrey S.S., et al. Gene expression patterns of carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiriou C. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc. Natl. Acad. Sci. 2003;100:10393–10398. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H., Bergstrom D.A., Yao M.C., Tapscott S.J., Bergstrom D.A., Yao M.C., Tapscott S.J., Yao M.C., Tapscott S.J., Tapscott S.J. Widespread and nonrandom distribution of DNA palindromes in cancer cells provides a structural platform for subsequent gene amplification. Nat. Genet. 2005;37:320–327. doi: 10.1038/ng1515. [DOI] [PubMed] [Google Scholar]
- Tirkkonen M., Tanner M., Karhu R., Kallioniemi A., Isola J., Kallioniemi O.P., Tanner M., Karhu R., Kallioniemi A., Isola J., Kallioniemi O.P., Karhu R., Kallioniemi A., Isola J., Kallioniemi O.P., Kallioniemi A., Isola J., Kallioniemi O.P., Isola J., Kallioniemi O.P., Kallioniemi O.P. Molecular cytogenetics of primary breast cancer by CGH. Genes Chromosomes Cancer. 1998;21:177–184. [PubMed] [Google Scholar]
- de van Vijver M., de van Bersselaar R., Devilee P., Cornelisse C., Peterse J., Nusse R., de van Bersselaar R., Devilee P., Cornelisse C., Peterse J., Nusse R., Devilee P., Cornelisse C., Peterse J., Nusse R., Cornelisse C., Peterse J., Nusse R., Peterse J., Nusse R., Nusse R. Amplification of the neu (c-erbB-2) oncogene in human mammmary tumors is relatively frequent and is often accompanied by amplification of the linked c-erbA oncogene. Mol. Cell. Biol. 1987;7:2019–2023. doi: 10.1128/mcb.7.5.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van’t Veer L.J., Dai H., de van Vijver M.J., He Y.D., Hart A.A., Mao M., Peterse H.L., van der Kooy K., Marton M.J., Witteveen A.T., Dai H., de van Vijver M.J., He Y.D., Hart A.A., Mao M., Peterse H.L., van der Kooy K., Marton M.J., Witteveen A.T., de van Vijver M.J., He Y.D., Hart A.A., Mao M., Peterse H.L., van der Kooy K., Marton M.J., Witteveen A.T., He Y.D., Hart A.A., Mao M., Peterse H.L., van der Kooy K., Marton M.J., Witteveen A.T., Hart A.A., Mao M., Peterse H.L., van der Kooy K., Marton M.J., Witteveen A.T., Mao M., Peterse H.L., van der Kooy K., Marton M.J., Witteveen A.T., Peterse H.L., van der Kooy K., Marton M.J., Witteveen A.T., van der Kooy K., Marton M.J., Witteveen A.T., Marton M.J., Witteveen A.T., Witteveen A.T., et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Wiedswang G., Borgen E., Karesen R., Kvalheim G., Nesland J.M., Qvist H., Schlichting E., Sauer T., Janbu J., Harbitz T., Borgen E., Karesen R., Kvalheim G., Nesland J.M., Qvist H., Schlichting E., Sauer T., Janbu J., Harbitz T., Karesen R., Kvalheim G., Nesland J.M., Qvist H., Schlichting E., Sauer T., Janbu J., Harbitz T., Kvalheim G., Nesland J.M., Qvist H., Schlichting E., Sauer T., Janbu J., Harbitz T., Nesland J.M., Qvist H., Schlichting E., Sauer T., Janbu J., Harbitz T., Qvist H., Schlichting E., Sauer T., Janbu J., Harbitz T., Schlichting E., Sauer T., Janbu J., Harbitz T., Sauer T., Janbu J., Harbitz T., Janbu J., Harbitz T., Harbitz T., et al. Detection of isolated tumor cells in bone marrow is an independent prognostic factor in breast cancer. J. Clin. Oncol. 2003;21:3469–3478. doi: 10.1200/JCO.2003.02.009. [DOI] [PubMed] [Google Scholar]