Abstract
Nucleosome positions within the chromatin landscape are known to serve as a major determinant of DNA accessibility to transcription factors and other interacting components. To delineate nucleosomal patterns in a model genetic organism, Caenorhabditis elegans, we have carried out a genome-wide analysis in which DNA fragments corresponding to nucleosome cores were liberated using an enzyme (micrococcal nuclease) with a strong preference for cleavage in non-nucleosomal regions. Sequence analysis of 284,091 putative nucleosome cores obtained in this manner from a mixed-stage population of C. elegans reveals a combined picture of flexibility and constraint in nucleosome positioning. As has previously been observed in studies of individual loci in diverse biological systems, we observe areas in the genome where nucleosomes can adopt a wide variety of positions in a given region, areas with little or no nucleosome coverage, and areas where nucleosomes reproducibly adopt a specific positional pattern. In addition to illuminating numerous aspects of chromatin structure for C. elegans, this analysis provides a reference from which to begin an investigation of relationships between the nucleosomal pattern, chromosomal architecture, and lineage-based gene activity on a genome-wide scale.
As a major subunit of eukaryotic chromatin, nucleosome cores consist of 146 bp of DNA wrapped 1.65 times around a histone protein octamer containing two each of the four core histones, H2A, H2B, H3, and H4 (Luger et al. 1997; Davey et al. 2002). The length of DNA in the core is conserved, while the “linker” DNA between nucleosomes can vary between species, chromatin regions, and cells. In many situations, an additional set of histones (H1 or H5) associate with linker DNA immediately flanking the core (Muyldermans and Travers 1994; Buttinelli et al. 1999).
Numerous cellular systems that interact with DNA have been shown to respond dramatically to the presence of nucleosomes in a region. Some critical processes are categorically excluded or facilitated on nucleosome-bound regions, while the efficiency of other processes is influenced by the precise setting of the DNA on the nucleosomal surface. Hence, the dynamic localization properties for nucleosomes and their flanking regions play key roles (permissive and regulatory) in nuclear function. At the extremes of possibility, nucleosomes could be locked in a single position or completely free to (rapidly) diffuse along the DNA (Kornberg 1974). In some formalisms, two types of interdependent DNA features that determine nucleosome positioning have been described: “translational signal” and “rotational setting.” A translational signal determines placement of, and a rotational setting determines orientation of DNA in relation to the histone octamer (Lu et al. 1994). Numerous studies of nucleosomal positioning have been carried out for individual genomic regions, indicating that positioning can be both flexible and constrained (e.g., Lu et al. 1994; Kornberg and Lorch 1999; Sekinger et al. 2005). Recently, these studies have been extended genome-wide through the analysis of populations of nucleosomes using high-density microarrays (for the yeast Sacchromyces cerevisiae) (Yuan et al. 2005) and using the Chromatin Array technique (for human cell lines) (Weil et al. 2004). These studies have shed considerable light on DNA sequence and other factors influencing nucleosome occupancy; certainly the results serve as a strong argument for pursuing a genome-wide analysis of nucleosome positioning and dynamics at an increasingly detailed level (Satchwell et al. 1986; Dong et al. 1990; Lu et al. 1994; Travers and Muyldermans 1996; Flaus and Richmond 1998; Crawford et al. 2006).
Caenorhabditis elegans provides an intriguing system in which to study global aspects of nucleosomal positioning. The C. elegans genome is well characterized on both a structural level (sequence) and in terms of genetic function (Riddle et al. 1997; C. elegans Sequencing Consortium 1998). In addition, the C. elegans genome can be readily modified by transgenesis (introduction of foreign DNA in a variety of contexts) or by mutation. These manipulations, combined with an unusually high incidence of putative nucleosomal positioning cues in a fraction of the genome (e.g., VanWye et al. 1991; Widom 1996; Fukushima et al. 2002; Cohanim et al. 2006; Fire et al. 2006; Kumar et al. 2006; Moreno-Herrero et al. 2006) should eventually facilitate the experimental analysis of connections between DNA sequence, nucleosome dynamics, and genetic function.
Methods to understand nucleosome positioning can be broadly classified into reconstitution and “natural position” approaches. Reconstitution experiments generally begin with naked DNA and purified histones that are assembled in vitro into a chromatin-like structure whose relationship to DNA sequence can then be analyzed (Dong et al. 1990; Flaus et al. 1996; Flaus and Richmond 1998; Brower-Toland et al. 2002). While these studies have greatly increased our fundamental understanding of many of the translational and rotational constraints conferred by DNA sequences, they all suffer from the simplicity of the assay, in particular the reconstitution of nucleosomes away from their native nuclear context. Complementary to these are in situ experiments in which chromatin is isolated from cells and then nucleosome positioning and structure are analyzed (Satchwell et al. 1986; Muyldermans and Travers 1994; Travers and Muyldermans 1996). Accurate retention and representation of nucleosome positioning has been a long-standing issue in the field, and numerous debates have addressed the questions of whether different isolation and analysis protocols might themselves influence the distributions of observed nucleosome positions. In particular, one notes a tradeoff between conditions that produce a highly pure and uniform population of nucleosome cores (often involving a high-salt step that strips most nonhistone proteins off of the DNA and therefore potential reassortment of nucleosome positions) and treatments that operate on more native chromatin templates that are consequently much more complex than a simple DNA::histone core complex. Despite the uncertainties and debates inherent in methodologies used to determine nucleosome position, each of the different approaches has provided a valuable contribution to understanding this critical protein::nucleic acid interaction.
In this work, we used a high-throughput single-molecule approach to analyze nucleosome core positions in C. elegans chromatin. This analysis yielded an extensive picture of chromatin structure for the C. elegans genome, while providing examples of both constrained and flexible nucleosome positioning.
Results
Isolation, optimization, and sizing of mononucleosome core DNAs
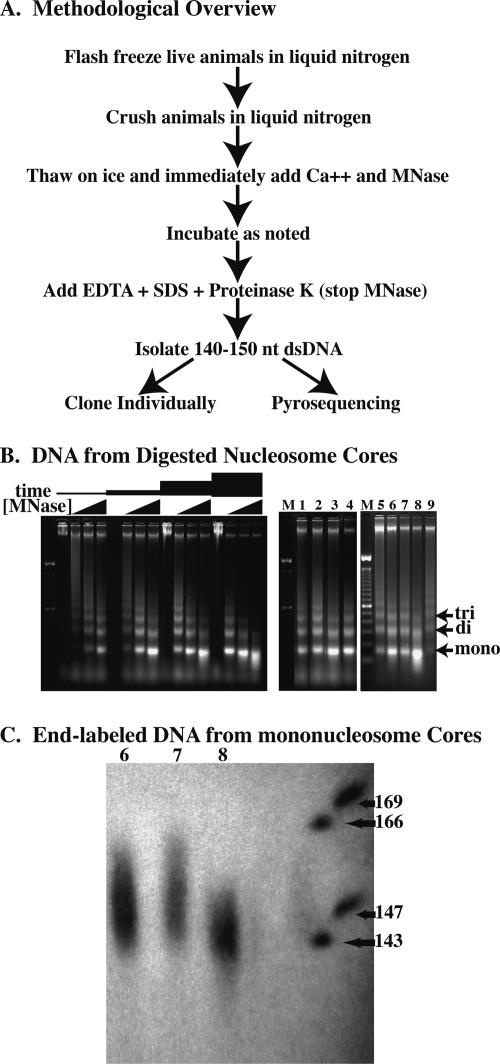
We first sought a method for isolating DNA fragments from nucleosomal cores that would rely on physiological conditions that minimally disrupt the association of nonhistone proteins while providing a relatively uniform DNA preparation. One goal, in particular, was to avoid protracted isolation protocols or stripping of nonhistone proteins, since these might be expected to allow relocalization of nucleosome cores on DNA. Our strategy was thus to digest chromatin to nucleosome cores as soon as possible after cell lysis using physiological salt conditions with moderate to low temperatures and short digestion times to minimize nucleosome movement. Briefly, we isolated mononucleosome core DNAs by grinding flash-frozen wild-type (N2) worms in liquid nitrogen with subsequent thawing, micrococcal nuclease (MNase) digestion, proteinase K treatment, agarose gel isolation, and purification as described below (Methods) (Fig. 1A). Animals were alive when flash frozen, remained frozen during the grinding steps, and were immediately added to a concentrated solution of micrococcal nuclease upon thawing. micrococcal nuclease digestion was stopped within a short time, followed by isolation of DNA fragments and analysis. We obtained varying degrees of digestion depending on the amount of micrococcal nuclease used, the temperature at which the digestions occurred, and the length of time of the digestion. As expected, these digestions produced ladders consisting of mono-, di-, tri-, and higher order nucleosome DNA fragments on agarose gels. Following electrophoresis, we excised the bands representing the mononucleosome cores for further analysis (Fig. 1B).
Figure 1.
Digestion of C. elegans chromatin in situ to generate core fragments. (A) Overview of the methodology used to isolate nucleosome core DNAs. (B) Electorphoretic gel analysis of micrococcal nuclease-digested chromatin showing the descending-ladder effect with increased micrococcal nuclease digestion. Gel at left shows digestions performed with increasing concentrations of micrococcal nuclease (0, 0.2, 0.8, and 3.2 U/μL) for various amounts of time (2, 6, 18, and 54 min) at 25°C. Lanes 1–9 of the right gel correspond to DNA isolated from the various digestion conditions (digests #1–#9) as described in Table 1. (C) Sequencing gel showing end-labeled mononucleosome core DNA from digestion conditions #6, #7, and #8 in lanes 6, 7, and 8, respectively.
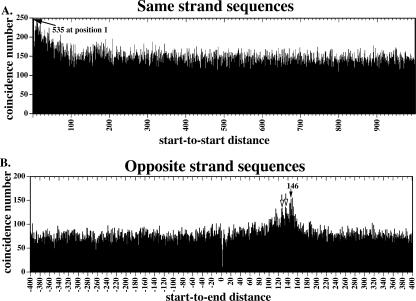
To determine the extent to which the mononucleosome cores were digested, we cloned cores isolated from various conditions and sequenced a total of 346, with a number of clones from each condition to determine a rough size distribution (Table 1; Supplemental Fig. 1). Further confirmation of size distribution was obtained following end labeling of bulk samples (with polynucleotide kinase) followed by electrophoresis on DNA-sequencing gels (Fig. 1C). Ideally, all cores would be the same size of 146 bp, but in practice we isolated a range of fragments from each of the samples. Size distribution depended on micrococcal nuclease concentration, digestion temperature, and duration. We chose conditions with the tightest distribution for further analysis. These conditions (100 μL of newly thawed C. elegans powder treated with 1280 units of micrococcal nuclease at 16°C for 12 min) resulted in a substantial band corresponding to DNA digested to mononucleosomes as assayed by both agarose and acrylamide gel electorphoresis (Fig. 1B,C lane 8). From the 37 fragments from this DNA that were cloned and sequenced, we obtained a size distribution with a median of 147 and standard deviation of 8.1 bp (Supplemental Fig. 1; Table 1).
Table 1.
Summary of manually cloned mononucleosome core DNAs
aMicrococcal nuclease (MNase) concentrations in units/μL final concentration.
bEnds were either unprocessed (none), processed with T4 DNA polymerase (Filled), or processed with T4 polynucleotide kinase (PNK) and Filled (see Methods).
Accuracy and precision of nucleosome footprinting by micrococcal nuclease digestion
Nucleosomes engage a segment of 146 bp of DNA in a tight complex. Micrococcal nuclease is a rather efficient tool for footprinting pure DNA-core complexes (Noll 1974b; Axel 1975), but we note that preparation of such complexes has generally involved high salt or other treatments removing non-nucleosomal proteins in chromatin, and thus, the substantial likelihood of nucleosome repositioning (McGhee and Felsenfeld 1980). Avoiding such treatments serves to allow greater preservation of nucleosome position; at the same time, the complex mixture of proteins bound to linker DNA in intact chromatin will provide some protection of the linker sequence to micrococcal nuclease digestion. A tradeoff thus exists, in that greater amounts of nuclease used to more completely digest the linker will also result in some cleavage within the nucleosome. The deviations we observe for in situ digested nucleosome core size are substantially greater than that observed in chromatin that has been stripped of non-nucleosomal protein or with nucleosome cores reconstituted from pure histones mixed with DNA. Since our conditions yield fragments that are both larger and smaller than 146 bp, one can assume that the fragment population includes instances in which the micrococcal nuclease digestion extended partially into the structural core (yielding a shorter fragment) and instances in which the nuclease digestion has left a DNA tail attached to the nucleosome core (yielding a longer fragment). Given the size ranges observed from electrophoresis and cloning, it appears that under- and overdigestion extents are most often in the range of a few base pairs.
Genomic positions of cores
In our initial sample of 346 clones, cores were found throughout the genome with examples from exons, introns, intergenic, repeated elements, exon/intron junctions, 5′UTRs, 3′UTRs, etc. Based on this small sample, it appears that nucleosomes can be found in every category of DNA in the genome.
Using our optimized isolation procedure, we produced microgram quantities of nonamplified mononucleosome DNA cores for bulk pyrosequencing. We obtained 312,492 individual end sequences, using the pyrosequencing-based “454” platform described in Margulies et al. (2005). In general, the pyrosequencing reads yielded 50–200 bases of sequence. This is certainly sufficient information to unambiguously place the sequenced end in any unique region of the C. elegans genome.
A sequence alignment for each of the pyrosequencing reads against the C. elegans genome (WS154) was carried out with a local installation of BLAT (ver. 32x1) using default parameters (Kent 2002). Of the 312,492 sequence reads, 283,745 (90.8%) showed significant matches to C. elegans genome assembly WS154. Additionally, there were 1919 sequences (0.6%) matching the Escherichia coli K12 genome, most likely representing contamination of the preparation with DNA from the bacterial lawn used as a food source for C. elegans (E. coli strain OP50), and 24 sequences matching fragments of the C. elegans mitochondrial genome. Of the 28,747 runs that did not show significant matches to the C. elegans genome using the BLAT algorithm, an arbitrary sample of 200 pyrosequencing runs were subjected to Batch BLAST analysis using public resources from the Netherlands Bioinformatics Center (http://www.nbic.nl/bioassist/servicetools/BatchBlast/). Of these 200 runs, 128 (64.0%) matched the C. elegans genome, 15 (7.5%) matched bacterial genomes (E. coli or Shigella), 34 (17.0%) matched other miscellaneous organisms, and 23 (11.5%) had no match. Extrapolation from this limited BLAST/curation analysis would suggest that an additional ∼18,400 of the unidentified pyrosequencing reads are C. elegans sequence, while as many as 10,000 DNA fragments may have come from other sources (most prominently from the E. coli).
In order to have positional data that is directly comparable to other resources, we filtered the sequence data to focus on individual sequences that could unambiguously be placed on the C. elegans physical map (WS120) as represented by the UCSC Genome Web Browser (http://www.nbic.nl/bioassist/servicetools/BatchBlast/). A certain fraction of sequences are excluded by this analysis, including putative nucleosome cores in repeated genomic regions, a number of sequences with ambiguous termini, and sequences corresponding to the small fraction of the C. elegans genome for which no sequence is available. Approximately 60% of the raw sequencing reads (187,863 total) were sufficiently accurate, unique, and unambiguous to be definitively placed in the C. elegans genome. We refer to this data set as the Unique Unambiguous Pyro-core (UUPc) data set. As observed with our 346 manually cloned cores, these 187,863 putative pyrosequencing cores (pyro-cores) were found on every chromosome and were well distributed throughout the genome (Supplemental Fig. 2) with 28,230 hits on chromosome I, 30,310 hits on chromosome II, 26,111 hits on chromosome III, 30,177 hits on chromosome IV, 39,547 hits on chromosome V, and 33,488 hits on the X chromosome. Densities of unique sequence pyro-cores recovered and assigned using this analysis were remarkably constant over the genome, with enrichment (approximately twofold) observed in a small number of regions. The enriched signals might represent regions of the genome with increased nucleosome density, increased accessibility and recovery in the sequencing protocol, or increased copy number (e.g., through tandem duplications not annotated in the draft genome sequence) in the population of wild-type animals used for this analysis. The latter seems a particularly intriguing possibility, given that C. elegans appears to duplicate genomic regions of tens to hundreds of kilobases relatively frequently (in evolutionary time) (Thomas 2006).
Our initial data set is sufficient to obtain a pyro-core map for the C. elegans unique genome at a representation of one nucleosome per 300–400-bp segment. We have taken two approaches toward analyzing characteristics of the pyro-core distribution on a finer scale. (1) Focusing on unique sequences in the genome, we could make certain inferences on relative position based on cases where two pyro-core sequences fall (by chance) into a limited DNA sequence interval. (2) Focusing on repetitive sequences in the genome, we could derive a small number of much higher density consensus nucleosome maps. We note that these two approaches operate on different data sets (unique DNA versus repetitive regions).
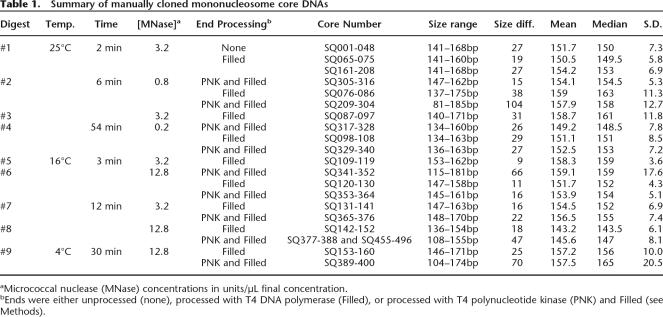
As a first analysis of positioning in unique DNA, we used the UUPc data set to analyze nucleotide and dinucleotide composition at every position relative to core fragment termini. One feature illustrated by this analysis is modest sequence bias at the sites of micrococcal nuclease cleavage. Previous studies of micrococcal nuclease specificity have suggested preferential cleavage at A/T-rich target sequences (Wingert and Von Hippel 1968). We observe evidence for this, as G/C residues are considerably underrepresented on both residues immediately flanking the site of cleavage (Supplemental Fig. 3A). In contrast, we observe an enrichment of G/C sequences at positions just downstream from the micrococcal nuclease cleavage site (e.g., bases 3 and 4). This suggests a situation where biases in terminal nucleotide distribution represent only slightly imprecise cleavage of nucleosomes with terminal G/C residues. Under such conditions, nucleosomes with G/C termini would most frequently show cleavage at more susceptible A/T residues located a few nucleotides into the linker. An alternative explanation, that the micrococcal nuclease cleavage specificity actively shifts the nucleosome position, is not suggested since there is no corresponding anomaly in the A/T versus G/C ratios at position 146 in the sequence profiles (Supplemental Fig. 3B). A situation in which the micrococcal nuclease cleavage at either end could reposition the nucleosome would generate a symmetry of the nucleosome-dependence plot for both ends of the nucleosome. The observed unidirectional dependence is most consistent with an influence of sequence on cleavage, but not nucleosome position. As will be discussed below, the nonrandom character of micrococcal nuclease digestion has rather significant implications for certain analyses (e.g., rotational setting), while having much less drastic effects on other analyses such as larger scale translational positioning.
Analysis of dinucleotide distributions revealed a strong periodicity of AA and TT dinucleotides with an interval of ∼10 nt extending across the entire core, but becoming less pronounced in the sequence surrounding the putative dyad of the nucleosome (base pair 73) (Fig. 2). Periodicity is seen to a lesser extent extending upstream from the start of the core. This periodicity was completely absent when “random cores” were generated (described in Methods) from the C. elegans genome and subjected to the same analysis. This AA/TT dinucleotide periodicity is similar to AA/TT dinucleotide periodicity observed by others when looking at sequences of a much smaller set of experimentally derived nucleosome cores (Ioshikhes et al. 1996). The sequence data sets also indicate a potential for additional sequence-dependent biases in translational positioning of nucleosomes. This is evident from the relative enrichment of AT and TA dinucleotides in linker DNA and enrichment of GC and CG dinucleotides in core DNA. Micrococcal nuclease terminus specificities, although a concern in this analysis, are unlikely to represent the major source of the observed periodicities. Most significantly, the peaks of AA and TT dinucleotides are retained when examining a subset of pyro-cores in which AA or TT are either required to be at position −1/1 (Supplemental Fig. 4, middle) or are excluded from positions −2/−1, −1/1, and 1/2 (Supplemental Fig. 4, bottom). These filtered spectra are inconsistent with terminal cleavage specificity being the source of the AA/TT dinucleotide periodicity.
Figure 2.
Positional dependence of dinucleotide composition in core and flanking sequences. Graphs show incidence of each dinucleotide at every position in the set of 163,651 unique pyro-core sites. Positions 1–146 show incidence in the presumed core nucleosome, while flanking sequences show the 60 nt preceding and following the pyro-core (positions −60 to −1 and 147–206, respectively). RND TT is a parallel analysis performed on a simulated set of random core segments constructed in silico from C. elegans’ DNA. In each graph, the scale of the ordinate is the same and shows a 5000 core range.
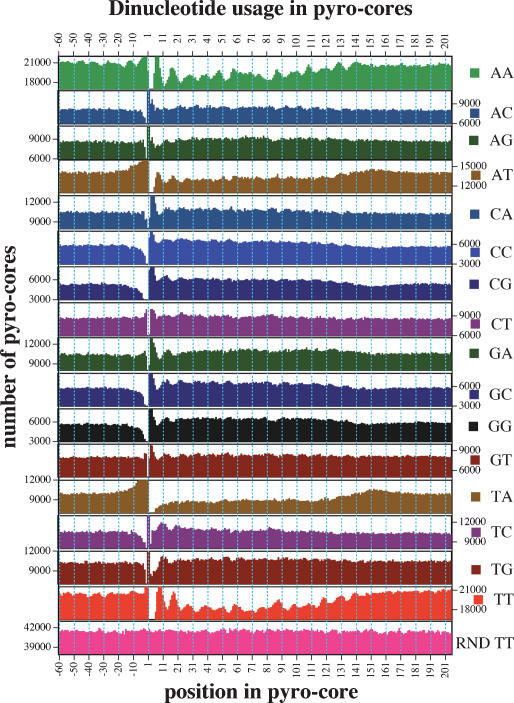
A key feature of the chromatin dynamics for a given chromosomal region is the variability of nucleosome position in a population of cells. As an initial test of this, we examined the distribution of Start-to-Start distances between pairs of unambiguously located pyro-core starts within 1000 bases of each other in the genome. To avoid any assumptions about pyro-core length, we limited this analysis to cores that were sequenced on the same strand. From the resulting plot (Fig. 3A), a large number of instances in which pyro-cores are closely positioned in the genome can be observed. These can be divided into two groups: cases in which two different reads have identical sequence and those where there is a finite but subnucleosomal separation (e.g., 1–150). Cases in which two samples give identical sequence almost certainly include many spurious repeat events, in which two separate beads are included in the same emulsion droplet in emulsion-PCR (during preparation of samples for 454 sequencing). Even with these repeats ignored, there is still evidence for preferential positioning of core positions in the data set, since cases of a single-base differential between adjacent pyro-cores are still more common (535) than any other separation (<250 for any other separation). Conversely, even if all of the same-base matches in the data set were considered, the majority of potentially overlapping pairs of pyro-core sequences (within 150 nt) are spread between 10 and 150 bp apart and not present at the close range of 0–10 bp. This and the relative constancy of the separation distribution illustrates a substantial degree of local plasticity in nucleosome position.
Figure 3.
Positional relationships between neighboring cores. (A) Relative positions of unique/unambiguous pyro-core start (Start-to-Start distances for cores sequenced on the same strand). Histogram shows total pairs of unique/unambiguous nucleosomes with a Start-to-Start distance corresponding to the value on the x-axis. A total of 535 pyro-core starts are found at position 1 and are noted on the graph by an arrow. (B) Start-to-End distances for cores sequenced on opposite strands. Histogram shows total pairs of unique/unambiguous nucleosomes with a Start-to-End distance corresponding to the value on the x-axis. A filled arrow notes a peak at 146 nt and open arrows note peaks ∼10 and 20 bp shorter, which suggest possible rotational setting preferences.
A second global view of relative positioning comes from comparing distances between starts of pyro-core sequences on different strands (Start-to-End distances). In this case, there is no ambiguity of spurious duplication of sequencing runs from a single initial molecule (since the strands are kept together during preparation for 454 sequencing). The Start-to-End analysis, shown in Figure 3B, demonstrates a clear peak (146 nt) consistent with some degree of nucleosome constraint. Smaller peaks at positions ∼10 and ∼20 bp shorter than the main peak are certainly intriguing. These are consistent with some degree of rotational phasing for nucleosomes, but could also reflect nuclease preferences. Confirming the independent Start-to-Start analysis, a major conclusion from the Start-To-End profile is the substantial heterogeneity in core positions for the diverse cell populations used for these studies. This is evidenced, for example, by the relatively constant incidence of Start-to-End distances over the range of the graph.
High-resolution nucleosomal maps of the 5S and 18/26S rDNA clusters
The pyro-cores that map to tandemly repeated rDNA loci in C. elegans provide an independent data set from the Unique/Unambiguous Pyro-core data set analyzed above, since the repeated character of rDNA sequences caused these pyro-cores to be excluded in the above analysis. As in other systems (Baer and Kornberg 1979; Dong et al. 1990; Dammann et al. 1995; Langst et al. 1997), the repetitive nature of rDNA genes makes them ideal loci in which to look for nonrandom positioning of nucleosomes. In order to make use of these gene clusters we performed a default BLAT analysis of the 312,492 pyrosequencing reads versus the 5S rDNA (Ce5SSL1) (chromosome V: ∼110 copes, 982 bp) (Nelson and Honda 1985; Krause and Hirsh 1987) and 18/26S rDNA genes (chromosome I: ∼55 copies, 7203 bp) (Files and Hirsh 1981; Ellis et al. 1986). A total of 786 of 312,492 total sequencing runs corresponded to the Ce5SSL1 repeat (0.25%). This is slightly greater than the 0.11% that would be expected for a locus of 982 bp repeated 110 times in a genome of 108 bp. The 18/26S rDNA gene cluster may likewise be overrepresented, with 6346 (2.0%) of the pyrosequencing runs instead of the expected 0.40% for a locus of 7203 bp repeated 55 times in the C. elegans genome. We have not further investigated the apparent overrepresentation, which could represent higher nucleosome density, greater nuclease accessibility due to sequence or structure, preferential recovery of fragments, differences in sequencing quality, or differences in true higher copy numbers of the two loci in our wild-type C. elegans stock. We note, however, that overrepresentation of rDNA-derived sequences in C. elegans libraries was frequently observed in genome-wide library construction prior to genome sequencing (Coulson et al. 1986).
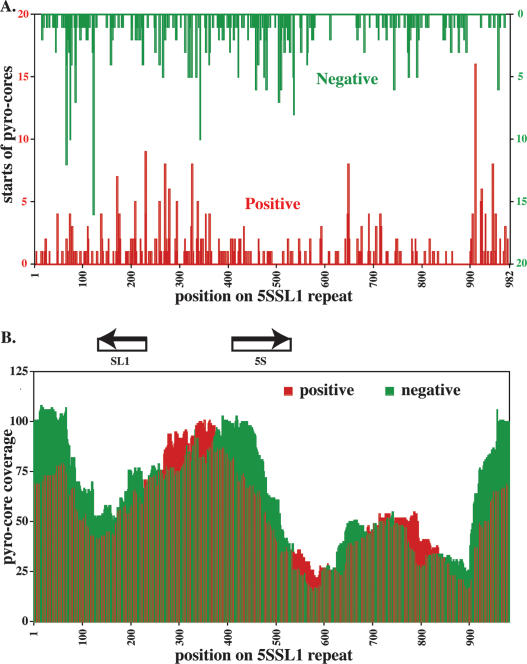
For the Ce5SSL1 cluster, we observe individual termini broadly distributed through the sequence (Fig. 4A). We also assessed coverage as a function of position within the repeat, as represented by the number of putative nucleosomes covering each site in the repeat, assuming a total length of 146 for each core (Fig. 4B). Although both termini and coverage show nonrandom character, the biases are modest, with the most striking preference representing two putative nucleosome positions just downstream of the SL1 encoding transcript. Despite this and other apparent preferences, we note that all positions within the 982-bp segment exhibit apparent nucleosome occupancies within two- to threefold of the calculated mean.
Figure 4.
Core positioning and coverage on C. elegans 5S/SL1 DNA. (A) Positive (red) and negative (green) starts of pyro-core sequences represented on a single element of the tandemly repeated Ce5SSL1 gene cluster. (B) Coverage plot of positive (red) and negative (green) pyro-core data for the Ce5SSL1 locus based on an assumption that each sequenced core represents a 146-bp single nucleosome segment.
Figure 5 shows an analysis of positioning of sequence reads on the larger Ce18/26SrDNA locus. Much of this locus displays a pattern of modest variation in nucleosome densities similar to that observed with the Ce5SSL1 locus. A considerable amount of the variation may be due to modest experimental biases or noise, as is evidenced by the difference observed between profiles for the two DNA strands. In contrast, we observe one very distinct feature at the 3′ end of the 18/26S transcript, which is a region of 100–120 bp, and which is substantially underrepresented in the putative nucleosome cores. This is also the site at which nucleosome-depleted regions have been observed in rDNAs of other organisms (e.g., yeast and mouse) (Dammann et al. 1995; Langst et al. 1997).
Figure 5.
Core coverage on C. elegans 18/26S rDNA. (A) Positive (red) and negative (green) starts of pyro-core sequences on a single 7203-bp copy of the 18/26S rDNA gene repeat. (B) Coverage plot of positive (red) and negative (green) pyro-core data for the 18/26S rDNA gene. Between the two coverage plots is a map of the position of the rRNA-coding elements in the rDNA repeat.
Positioned nucleosomes in psx1 repeat locus
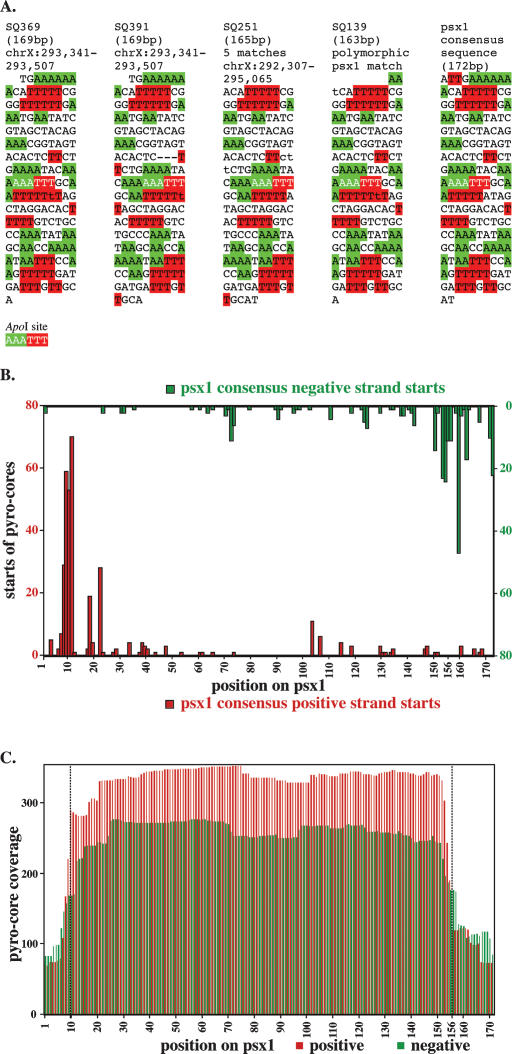
In our initial cloning of 346 cores we were surprised to isolate four independent nucleosome cores covering a genomic element, representing over 1% of our total clones (Fig. 6A). These cores corresponded most closely to a region of the left end of the X chromosome annotated to have 19.2 copies of a 172- or 175-bp repeat (3331 bp total). Interestingly, the four core clones of this element had very similar termini. We have named this repeat the positioning sequence on X (psx1). This sequence is a component of a previously recognized repeat, CE#000122 (Bao and Eddy 2002). An analysis using the UCSC Genome Browser revealed imperfect hits on all five of the other chromosomes in addition to the closer (in some cases perfect) hits on the X chromosome. In total, there are ∼40 copies of the psx1 repeat in version WS120 of the C. elegans genome with 34 of those repeats contained in two clusters on X and one cluster on V. Given the >100-fold discrepancy between the number of times psx1 was cloned and expected in our initial libraries, we performed a Southern blot analysis of the psx1 repeat element with two distinct strains of C. elegans that had been cultured separately for over 30 yr [wild-type strain (N2) and unc-93(e1500)] (Greenwald and Horvitz 1980). Using either restriction enzyme Sau96I or HaeIII, both of which fail to cut inside of the psx1 repeat, we observed not the expected 3.7-kb or 4.0-kb bands of psx1 hybridizing material, but instead, in each case, an intense band migrating at the exclusion limit of the gel (>15 kb; data not shown). The large size of this band indicates a much higher copy number for the tandem psx1 repeat than is seen in current drafts of the genome sequence.
Figure 6.
A reiterated genetic element with constrained core positioning. (A) Sequences of four closely matching cores from the original (manually isolated) series of 346 clones. These four clones, isolated from three different chromatin digestion conditions and four separate ligations, correspond to variants of a repeated sequence, designated psx1, present in the C. elegans genome (Bao and Eddy 2002). Individual polymorphisms relative to a consensus version (right) of psx1 are printed in lowercase. Runs of two or more consecutive A’s or T’s are highlighted in green and red, respectively and the sequences are displayed in 10-nt sections. (B) Starts of pyro-core sequences mapping to psx1 copies. Red and green are data from positive and negative starts, respectively. (C) Coverage plot of positive (red) and negative (green) pyro-core data for psx1.
The frequency and uniformity of the initial clones indicated that psx1 might serve as a valuable example of a highly repeated sequence with well-positioned nucleosomes. The ability to detect strongly nonrandom positioning in a population of nucleosome-length DNA is critical to the utility of any high-throughput chromatin analysis. We thus investigated the character of this repeated sequence further using a variety of techniques: analysis of the high-throughput sequence data, Southern blotting, primer extension, and PCR-based cloning of termini.
Of our 312,492 pyrosequencing reads, 635 reads, or ∼0.2%, correspond to psx1 (∼20 reads would be expected based on current genome annotation). Consistent with the initial analysis of the four manually cloned psx1 copies, the majority of pyro-cores corresponding to psx1 evidenced a unique position (Fig. 6B,C). Positive strand reads (i.e., reads in the annotated “sense” orientation) most frequently started within a few nucleotides of a single site (ACATTT), while negative-strand reads most frequently began ∼146 bp downstream. The start positions on the positive-strand pyro-cores clustered in a range of about 4 nt and were more uniform than those of the negative strands (which were clustered within a stretch of ∼14 nt). This difference in precision of start site could be explained to some extent by variations in the size of psx1 repeats (either 172 or 175 bp) as well as by slightly variable cleavage (as discussed above). Pyro-cores initiating within two bases of the main peak (around nucleotide 10) account for 59.8% of psx1 positive-strand sequences recovered. Additional pyro-cores were distributed through the psx1 sequence, generally at a much lower density, but with a possible secondary peak ∼8–12 nt downstream of the major terminus.
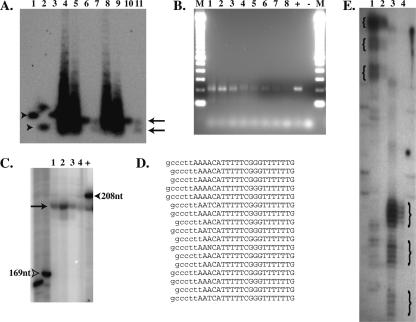
Southern blot analysis of positioning made use of a unique ApoI restriction site roughly in the center of each copy of psx1 (Fig. 7A). Digestion of C. elegans DNA to ∼146 bp nucleosome cores generates, as expected, a psx1 hybridizing band of comparable size. This band is cleaved roughly in half by ApoI, generating a single novel band roughly consistent with the expected 77/72-bp fragments (Fig. 7A). A nonpositioned population of nucleosomes on psx1 would have been expected to produce a smear running from 0 to 146 bp, while the observed result is consistent with considerable positioning.
Figure 7.
Evidence for Nucleosome Positioning on psx1. (A) Southern blot probed for psx1. Arrowheads indicate uncut (lanes 1,3) and ApoI cut (lane 2) cloned control psx1 DNA, while arrows indicate positions of uncut (lanes 6,10) and ApoI cut (lanes 7,11) mononucleosome core bands. Lanes 4, 5, 8, and 9 contain nucleosome ladders from micrococcal nuclease-digested chromatin. (B) Agarose gel electrophoresis of unique PCR product bands indicating positioning of micrococcal nuclease termini on psx1. Lanes 1–8 correspond to PCR products from reactions using ligated cores isolated with conditions 1, 3, 4, 5, 6, 7, 8, and 9, respectively (Table 1). “M”, “+”, and “−” indicate lanes containing 100-bp ladder markers (Invitrogen), positive control PCR reaction, and negative control (no template) PCR reactions, respectively. (C) Polyacrylamide gel electrophoresis with lanes 1–4 and + containing end-labeled PCR products from corresponding reactions in lanes 1–4 and + in B. Arrow indicates uniform size of psx1 core products, while open arrowhead indicates 169-nt marker and filled arrowhead indicates control reaction product of 208 nt (∼6 nt larger than the psx1 core products). (D) Sequences of vector-core junctions of psx1 nucleosome cores cloned from PCR reactions shown in lanes 1 and 2 of B. Sequence from the cloning vector is indicated in lowercase, and sequence from psx1 cores is indicated in uppercase letters, respectively. (E) Polyacrylamide gel electrophoresis of psx1 primer extension products from primer AF-SJ-6 with 10 rounds (lane 1) or one round (lane 2) of extension, and from primer AF-SJ-7 with 10 rounds (lane 3) or one round (lane 4) of extension. Curly braces demark positions of three different size ranges of products seen in both sets of primers with the largest size products in each sample being the most abundant as observed in the lanes in which samples underwent only a single round of extension, 2 and 4.
As a third approach to measure position, we ligated isolated mononucleosome cores to a DNA vector of known sequence and amplified the resulting product by PCR with one primer in psx1 and one in the vector. This analysis was carried out using cores derived from a variety of micrococcal digestion conditions (see Fig. 7 legend), with the resulting products comigrating as a single band on a 2% agarose gel (Fig. 7B). Following gel isolation and end labeling, samples were resolved on sequencing polyacrylamide gels, with migration closely corresponding to the expected length of 202 bp (Fig. 7C). Fine-scale analysis of PCR ligation products was carried out by cloning and sequencing psx1-derived PCR products; of 18 ligation products characterized, 17 exhibited termini within three nucleotides of the expected junction (Fig. 7D; the 18th product started 32 nt further upstream).
Primer extension analysis provides an additional tool for mapping termini in a complex DNA sample (Wilson et al. 1980). Using nonligated mononucleosome cores we performed primer extension reaction with primers that hybridize to psx1 at two different positions. The extended products were analyzed on a sequencing polyacrylamide gel. As expected, the two different primers each gave products within 3–5 nt of the expected size range (Fig. 7E). Fainter sets of bands from products ∼10 and ∼20 nt shorter than the major bands may correspond to minor termini.
Discussion
As a model system for animal development and physiology, C. elegans provides a remarkable combination of well-characterized structures and powerful tools. Given the paramount roles of chromatin and epigenetics in development and maintenance of species, C. elegans would certainly provide an ideal platform in which to explore the connections between chromatin structures and epigenetic functions. In this study, we have presented an initial analysis of plasticity and constraint in nucleosome positions in mixed-stage populations of C. elegans hermaphrodites grown under conditions of ample food. We envision the data resulting from this analysis to be of use in several contexts. First, it is hoped that this data will contribute toward understanding genomic function in C. elegans. Second, this analysis provides an unusually large data set of nucleosome positions for structural analyses of nucleosome::DNA interactions.
Before delving into the details of the observed localization, it is important to understand both the power and limitations of the primary method we used to define nucleosome cores. First, we stress a tradeoff between complexity in the assay and physiological relevance of the inferred position. Pure nucleosomes can readily be assembled on pure DNA, examined by a wide variety of physical and biochemical techniques, and used thereby to illuminate an intrinsic component of the nucleosomal energy landscape. Although positions obtained from analysis with purified components are likely to diverge dramatically from those in vivo, they certainly provide an important window into the biochemistry and energetics of nucleosome::DNA interactions. As a somewhat more physiologically related view of nucleosomal pattern, a number of procedures have been developed that strip nonhistone proteins from in vivo chromatin (in many cases with high salt) to yield a relatively manipulable population of nucleosome::DNA complexes. Certainly such treatment would be expected to modify positions of some nucleosomes; nonetheless, the data obtained from such analysis has unquestionably been of value. Attempts to determine nucleosome position in chromatin that has not been vigorously stripped are a means to get closer to natural pattern; at the same time, these methods present some challenges in that the distinction in accessibility between nucleosome and non-nucleosome DNA can be compromised by tightly binding non-nucleosomal components.
Our approach to this question in C. elegans has been to develop a method that relies on natural chromatin from cells that have been frozen while alive, disrupted while frozen, and thawed directly into a mix containing a high concentration of a reagent with the capacity to distinguish nucleosomal from non-nucleosomal DNA. Numerous probes with different degrees of specificity have been used in bulk chromatin analysis. At one extreme of this spectrum are some chemical probes, such as hydroxyl radical (Tullius and Dombroski 1986) and MPE (Cartwright et al. 1983), that show a modest degree of preferential cleavage relative to the nucleosome position, but which have little or no bias in terms of the underlying sequence. At an opposite end of the specificity spectrum are enzymatic probes, such as micrococcal nuclease (Wingert and Von Hippel 1968; Horz and Altenburger 1981) and the apoptotic nuclease DFF40 (Widlak et al. 2000), which have some sequence bias, but which show a much stronger respect for nucleosome boundaries in their initial attack on a chromatin template. Some studies have also used pancreatic DNaseI, which is selective for different regions of nucleosomes (Noll 1974a) and has been used particularly successfully to delineate open regions and sites in chromatin (Lutter 1979; Crawford et al. 2006). We have begun with micrococcal nuclease because this appears to be the probe that most definitively identifies a set of fragments as nucleosomal in character. While we consider the resulting set of “putative nucleosome cores” as an appropriate data set, we stress two major biases of micrococcal nuclease that are likely to skew the precise distribution of cloned cores. First, there are sequence-related differences in the precise extent of micrococcal nuclease trimming activities around individual nucleosome cores (see Supplemental Fig. 3 and text of Results). A second bias relates to the ability of micrococcal nuclease to digest certain nucleosome-bound sequences more efficiently than others, with a considerable rotational preference in this digestion ability (McGhee and Felsenfeld 1983). Although we cannot rule out specific internal nuclease cleavage events, the rather precise sizing of core fragments generated during our micrococcal nuclease treatment is suggestive of at most a limited loss of nucleosome-length fragments due to internal digestion.
One final set of “known” biases that we point out involves the amplification and pyrosequencing technique used for the analysis. We can only observe nucleosome core ends in this assay if the resulting fragments can be amplified and unambiguously identified by pyrosequencing. It should be stressed that amplification by emulsion PCR occurs only after the DNAs have been diluted to concentrations of ≪1 molecule per emulsion droplet. Thus, competitive aspects of PCR will not affect the relative representation in the final data set. Nonetheless, any fragment that was recalcitrant to either the initiation or amplification phases of PCR would certainly be underrepresented in the final data set. Pyrosequencing, likewise, appears remarkably efficient, but by no means perfect as a means to assign fragment identities. We note that the 42 rounds of pyrosequencing used in this analysis generates between 39 and 303 bp of moderately high-quality sequence (with a mode of 108 bp, a mean of 113.1 bp, and standard deviation of 15.4); this is more than sufficient to place any nonrepetitive segment uniquely in the C. elegans genome, even with small numbers of pyrosequencing-related errors. Nevertheless, we found a number of sequences for which ambiguities arose from sequencing-related issues such as the precise placement of the 5′ end. Given the unique characteristics of error generation during pyrosequencing (in particular, confounding effects at and downstream of long homopolymer runs) (Ronaghi et al. 1999), there is certainly potential for a modest pyrosequencing-related bias.
Despite the various potential biases at various stages of the analysis, we expect the data resulting from high-throughput sequencing of nucleosome core preparations to have substantial value. In particular, these data generate a positional profile of chromatin at a level of detail and precision that should allow physiological rearrangements at a locus to be readily detected. The analysis of psx1 and of the Ce5SSL1 ribosomal RNA loci, in particular, demonstrate that we can indeed distinguish regions with highly positioned nucleosomes (e.g., psx1) from those with relatively arbitrary positioning (e.g., Ce5SSL1), with the basic aspects of both positioning (in the case of psx1) and flexibility (in the case of Ce5SSL1) being relatively insensitive to the modality of analysis used to characterize the putative nucleosome cores. Although all of the techniques used to address positioning (Southern blots, primer extension, PCR, and high-throughput sequencing) have significant strengths and weaknesses, only the sequencing approach has a true potential for genome-wide analysis.
In addition to providing a valuable example of nucleosome constraint, the psx1 element provides a rather unexpected enigma. Our data (both representation in libraries and Southern blots) indicate that repeats of this segment make up a much larger fraction of the genome than expected from the available genome assembly. One presumes that the difficulties in assembling tandemly arranged segments with similar sequences accounts for this discrepancy. Although the function and origin of psx1 are not clear, the strong nucleosome positioning character suggests potential roles in chromosome structure and function. In the course of this analysis, we observed a number of other repeated loci that may be underrepresented in the annotated genome (data not shown). Functions for these elements may also be an intriguing area of investigation.
Finally, we note that the basic chromatin characterization and high-throughput sequencing analysis described herein provide a bulk characterization of chromatin that should be useful as a reference for studies of specific loci and/or specific developmental and physiological conditions. As a first component of this, the nucleosome ladders seen by agarose gel electrophoresis of micrococcal nuclease-digested chromatin allowed us to view the bulk size distributions for di-, tri-, tetra-, and pentanucleosomes from C. elegans. The discrete bands produced by micrococcal nuclease digestion seen on our agarose gels confirmed results of Dixon et al. (1990), indicating that a fraction of C. elegans chromatin is compacted into regularly spaced nucleosomes. From our agarose gels, an estimate of bulk linker DNA size for this portion of the genome was 16 ± 4 bp, similar to the internucleosomal separation of ∼14 bp reported by Dixon et al. (1990) for higher order polynucleosomes. In addition to the higher order internucleosomal repeat, Dixon et al. (1990) reported an anomalously long mononucleosome band (186 bp); we have not observed this under the conditions of our assay.
Two types of constraint in nucleosomal position can be envisioned. Constraint in relative position would occur where neighboring nucleosomes were frequently separated from each other by a specific linker length but not necessarily fixed to specific spots on the DNA. From the bulk agarose gel analysis described above, it is clear that nucleosomes are arranged relative to each other in an orderly (end-to-end) fashion over a significant fraction of cells and positions within the genome. Constraint in absolute position would occur in regions where nucleosomes reproducibly prefer specific positions relative to the underlying DNA sequence. The lack of a strong dinucleosome peak in the Start-to-Start analysis of the sequencing data (Fig. 3A), combined with the modest character of the ∼146-nt peak in the Start-to-End analysis from sequencing data (Fig. 3B) indicate that fixed constraints on absolute nucleosome positions are relatively rare within the mixed-stage population of C. elegans cells that we analyzed. Whether the variability in absolute positions reflects distinct patterns in diverse cell types in the mixed-stage population of C. elegans or flexible patterning within each individual cell type will be an intriguing question for further investigation.
While the sequence data supports the existence of considerable positional flexibility for C. elegans nucleosomes, there is also ample evidence for positional constraint. This is indicated by the 146-nt peak in the plot of Start-to-End incidences in Figure 3B. Additional indication of such constraint can be derived from a more sensitive analysis based on a Fourier transform of the Start-to-Start data in Figure 3A (see Supplemental Fig. 5). The Fourier analysis shows two strong periodicities. A broad peak ranging from 157 to 171 bp with a maximal signal at ∼167 bp (Supplemental Fig. 5A) suggests a fraction of genome in which nucleosomes are reproducibly packed with an average linker size of ∼21 bp. A second periodicity, at ∼9.9 bp (Supplemental Fig. 5B) is consistent with two possible explanations, the first of which is operational (a bias in preparation or amplification/sequencing of nucleosome cores that might recapitulate the strong underlying 10.n periodicity in the C. elegans genome) (McGhee and Felsenfeld 1983) and the second of which is fundamental (significant rotational constraint of nucleosomes on the DNA template). As the parameters for rotational and translational constraint in positioning of nucleosomes become clear, it will be of particular interest to compare these with the large number of potential nucleosome positioning guide sequences that have been detected bioinformatically in the C. elegans genome (VanWye et al. 1991; Widom 1996; Fukushima et al. 2002; Cohanim et al. 2006; Fire et al. 2006; Kumar et al. 2006).
Methods
Isolation of core DNA fragments
To isolate the DNA from nucleosome cores we first isolated mixed stage, wild-type (N2) C. elegans cultured on OP50 bacteria and flash froze them into pellets in 0.34 M sucrose/Buffer A (15 mM Tris-HCl at pH 7.4, 15 mM NaCl, 1 mM DTT, 60 mM KCl, 0.5 mM spermidine, 0.15 mM spermine, 25 mM bisulfite) using liquid nitrogen. Next, the worm pellets were ground to a fine powder in liquid nitrogen using a mortar and pestle. The powder was allowed to thaw to 4°C on ice. CaCl2 was then added to a final concentration of 1 mM and micrococcal nuclease (Roche) resuspended at 30 or 300 U/μL in NEBuffer 1 and was added to final concentrations ranging from 0 to 12.8 U/μL and incubated at 4, 16, or 25°C for varying amounts of time as described in the Results and Table 1. The micrococcal nuclease digestion was stopped by addition of an equal volume of worm lysis buffer (0.1 M Tris-HCl at pH 8.5, 0.1 M NaCl, 50 mM EDTA, 1% SDS) and one-tenth volume of proteinase K (20 mg/mL in TE at pH 7.4) and incubated at 65°C for 45 min with frequent vortexing throughout the incubation to remove and digest the histone and other proteins. The nucleosome core DNA was isolated by phenol, phenol/chloroform, and chloroform extractions followed by ethanol precipitation in the presence of one-tenth volume of saturated ammonium acetate and RNase treatment, followed by phenol/chloroform and chloroform extractions and ethanol precipitation. The DNA was resuspended in TE (pH 7.4).
Micrococcal nuclease-digested DNA was separated into mononucleosome, dinucleosome, trinucleosome, etc., DNAs by agarose gel electrophoresis running the DNA samples on a 4% NuSieve GTG Agarose (Cambrex) gel at 100 V for 4 h. Mono-, di-, and trinucleosome bands were visualized, photographed, and excised from the gel, and the DNA was extracted by melting the band in a 65°C water-bath and subsequent phenol/chloroform and chloroform extractions and ethanol precipitation. The DNA was resuspended in TE (pH 7.4) for cloning or 5 mM Tris (pH 7.9) for pyrosequencing (454 Life Sciences).
Preparation of core DNA ends for cloning
Micrococcal nuclease leaves a 3′-phosphate and a 5′-hydroxyl when it cuts double-stranded DNA, leaving ends that may have 5′ or 3′ single-stranded overhangs. In order to produce uniform blunt ends on all of the isolated nucleosome cores we removed the 3′-phosphates by treating the isolated cores with T4 Polynucleotide Kinase (NEB) in 100 mM MES (∼pH 6.0), 10 mM MgCl2, 1 mM DTT, and 20 mg/mL BSA for 1 h at 37°C followed by 10 min heat inactivation at 70°C and phenol/chloroform and chloroform extraction and resuspension in TE. Next, any single-stranded overhangs were filled by treating the cores with T4 DNA polymerase (NEB) in 1× NEBuffer 3 (50 mM Tris-HCL, 10 mM MgCl2, 100 mM NaCl, 1 mM dithiothreitol at pH 7.9) (NEB) supplemented with 200 μM dNTPs and 1 μg/μL BSA for 15 min at room temperature, followed by phenol/chloroform and chloroform extraction and resuspension in TE. Treating the cores this way increased our cloning efficiency by over an order of magnitude. Nucleosome cores were ligated to pCR4Blunt-TOPO and transformed into chemically competent TOP10 cells (Invitrogen). Cores were either sequenced by isolating and sequencing plasmids or by PCR amplification of bacterial colonies harboring core-containing plasmids.
Virtual generation of “random cores”
A simulated population of randomized core sequences was generated for comparison purposes. This database was derived by random sampling of sites from C. elegans genome build WS154. A total of 328,094 random sites from the genome were chosen, given arbitrary directions (sense or antisense), and used as initiation points for simulated pyrosequencing runs.
Sequence alignment
Pyro-core sequences were aligned against either the WS120 build or the WS154 build of the C. elegans genome with a local installation of BLAT v. 32x1 using default parameters. The quality of each alignment was judged by three criteria: the alignment score (>80% of the maximum score), the first sequenced base from the pyro-core sequence that matches (must be the first sequenced base), and the difference in scores between the “best match” and the “second best match” (>10 arbitrary score units). Alignment scores are calculated by adding the number of nucleotides that match, subtracting the number of nucleotides that mismatch, and subtracting the number of gap positions (score = matches − mismatches − gaps). No gap opening penalties were included due to the types of sequencing errors experienced with pyrosequencing. Perl scripts are available upon request.
Southern blotting
Isolated mononucleosome core DNA and a control DNA fragment containing a single copy of psx1 were either digested with the restriction enzyme ApoI or left undigested at 50°C for 3.0 h followed by phenol/chloroform and chloroform extractions. The samples were separated by electrophoresis on a 1.2% agarose gel run in 1× TAE at 80 V for 6 h and then blotted onto Hybond-N+ membrane (Amersham Biosciences) by capillary transfer overnight using alkaline transfer followed by UV cross-linking (Stratalinker 2400 [Stratagene] 1200 ujoules ×100), baking at 80°C for 2 h. Probing was performed using a random-primed 32P-labeled probe for psx1. Following prehybridization at 64°C in 50 mL of phosphate-SDS buffer [0.5 M sodium phosphate at pH 7.2, 1 mM EDTA at pH 8.0, 7% (w/v) SDS, 1% (w/v) bovine serum albumin], the probe was added and then hybridized overnight, and washed twice with 100 mL [40 mM sodium phosphate buffer at pH 7.2, 1 mM EDTA at pH 8.0, 5% (w/v) SDS, 0.5% (w/v) bovine serum albumin] and four times with 100 mL [40 mM sodium phosphate buffer at pH 7.2, 1 mM EDTA at pH 8.0, 1% (w/v) SDS].
PCR-based local analysis of core positions
The PCR experiment to determine unique positioning of nucleosomes on psx1 was performed in a total reaction volume of 50 μL containing both M13 Reverse (5′-CAGGAAACAGCTATGAC-3′) and AF-SJ-1 (5′-GCAGACAAAAAGTGTCC-3′) primers each at 5 ng/μL final concentration (f.c.), dNTPs (400 μM f.c.), PCR Thermo Buffer (1X f.c.), and either the experimental samples of 0.5 μL of mononucleosome cores ligated into pCR4Blunt-TOPO vector (as described above) or control reactions with 2.0 μL). The 1:1000 dilution of the positive control plasmid pSJ391 containing the psx1 nucleosome core DNA fragment (∼40 pg/μl). The samples were heated at 95°C for 10 min and then cycled for 35 cycles at 95°C for 30 sec, 46°C for 30 sec, and 72°C for 1 min, followed by an additional extension at 72°C for 10 min. For each sample, 10 μL were run on a 2% agarose gel in 1XTAE containing 30 ng/mL EtBr for 2 h at 80 V.
To remove primer dimers and unincorporated nucleotides and primers, 10 μL of each PCR reaction were run at 70 V for 2 h on a 1% low-melt agarose gel and wide bands were excised for each sample starting just above the primer dimer band and extending an equal distance above the PCR product band. The DNA was extracted from each gel slice by adding 400 μL of 1 M NH4OAc, 10 mM EDTA, 0.2% SDS, and 50 ng/μL glycogen, and heating at 68°C until the gel slice was melted, then extracting the DNA by phenol, phenol/chloroform and chloroform extraction, and ethanol precipitation. The cleaned up PCR products were then either cloned into the pCR4Blunt-TOPO vector as above (without end modification) or radioactively end labeled with [γ-32P]ATP using T4 Polynucleotide Kinase (NEB) and run on a Polyacrylamide/UREA sequencing gel.
Primer extension analysis of core positions
Two reactions with 40 pmols of either primer AF-SJ-6 (5′-AGGT GCCACGAATTTGCTCCTTTATTTGGGCAGACAAAAAGTGTCC-3′) or AF-SJ-7 (5′-AGGTGCCACGAATTTGCTCCTTTTCAGAA GAGTGTACTACCGTTTC-3′) in a total reaction volume of 20 μL containing 1X ThermoPol Reaction Buffer [20 mM tris-HCl at pH 8.8, 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4, 0.1% Triton x-100], dCTP, dGTP, dTTP (all 200 μM f.c.), and [α-32P]dATP (5 μM f.c.) were prepared. A total of 1 μL of Taq DNA polymerase was added to each reaction and then the tubes were heated sequentially at 94°C for 2 min, 50°C for 2 min, and 72°C for 2 min. The reactions were then split in two, with one set of reactions being stopped by the addition of 2 μL of 100 mM EDTA and the other set of reactions cycled nine more times at 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min. After stopping the second set of reactions with EDTA, 30 pmols of the 3′-biotinylated primer AF-SJ-5 (5′-AAGGAGCAAATTCGTGGCACCTAA-Bio-3′) were added to each tube, followed by 5 min of boiling and cooling to room temperature to hybridize the psx1 extension products to the biotinylated oligo complementary to the 5′ portions of the extended primers. Streptavidin-coated magenetosphere beads and a magnet (Tong and Smith 1992) were used to isolate the primer extension products: Each reaction was brought up to 50 μL of volume and added to 500 μg of Dynabeads M-270 Streptavidin (Dynal Biotech) with final elution of the products from the beads in 5 μL of T.E. Each eluate had 2 μL of Formamide-loading Buffer [80% (w/v) deionized formamide, 10 mM EDTA at pH 8.0, 1 mg/mL xylene cyanol FF, 1 mg/mL bromophenol blue] added, was boiled for 5 min, cooled on ice, and then run on an 8% polyacrylamide TBE Urea sequencing gel at 50 W for 1.5 h.
Acknowledgments
We thank Lia Gracey, Roger Kornberg, Mike Cherry, Bob Kingston, Geeta Narlikar, Chaya Krishna, Poornima Parameswaran, Ky Sha, Julia Pak, Rayka Yokoo, Jonathan Gent, Jamie Fleenor, Len Lutter, Aaron Straight, Craig Kaplan, Mei Hsu, Blake Hill, Arend Sidow, Zhi-Ying Chen, Mark Kay, and John Leamon for their help and suggestions over the course of this work and acknowledge National Institutes of Health (Grant NIGMS R01-GM37706 to A.Z.F.), (Stanford Genome Training Program HG00044; H.L.M.), (Grant T32GM07231; F.J.T.), American Cancer Society (postdoctoral fellowship PF-05-121-01-DDC to S.M.J.), and Stanford University (Stanford Graduate Fellowship; D.P.R.) for financial support.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5560806
References
- Axel R. Cleavage of DNA in nuclei and chromatin with staphylococcal nuclease. Biochemistry. 1975;14:2921–2925. doi: 10.1021/bi00684a020. [DOI] [PubMed] [Google Scholar]
- Baer B.W., Kornberg R.D., Kornberg R.D. Random location of nucleosomes on genes for 5 S rRNA. J. Biol. Chem. 1979;254:9678–9681. [PubMed] [Google Scholar]
- Bao Z., Eddy S.R., Eddy S.R. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 2002;12:1269–1276. doi: 10.1101/gr.88502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower-Toland B.D., Smith C.L., Yeh R.C., Lis J.T., Peterson C.L., Wang M.D., Smith C.L., Yeh R.C., Lis J.T., Peterson C.L., Wang M.D., Yeh R.C., Lis J.T., Peterson C.L., Wang M.D., Lis J.T., Peterson C.L., Wang M.D., Peterson C.L., Wang M.D., Wang M.D. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc. Natl. Acad. Sci. 2002;99:1960–1965. doi: 10.1073/pnas.022638399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttinelli M., Panetta G., Rhodes D., Travers A., Panetta G., Rhodes D., Travers A., Rhodes D., Travers A., Travers A. The role of histone H1 in chromatin condensation and transcriptional repression. Genetica. 1999;106:117–124. doi: 10.1023/a:1003745315540. [DOI] [PubMed] [Google Scholar]
- Cartwright I.L., Hertzberg R.P., Dervan P.B., Elgin S.C., Hertzberg R.P., Dervan P.B., Elgin S.C., Dervan P.B., Elgin S.C., Elgin S.C. Cleavage of chromatin with methidiumpropyl-EDTA. iron(II) Proc. Natl. Acad. Sci. 1983;80:3213–3217. doi: 10.1073/pnas.80.11.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Cohanim A.B., Kashi Y., Trifonov E.N., Kashi Y., Trifonov E.N., Trifonov E.N. Three sequence rules for chromatin. J. Biomol. Struct. Dyn. 2006;23:559–566. doi: 10.1080/07391102.2006.10507081. [DOI] [PubMed] [Google Scholar]
- Coulson A., Sulston J., Brenner S., Karn J., Sulston J., Brenner S., Karn J., Brenner S., Karn J., Karn J. Toward a physical map of the genome of the nematode Caenorhabditis elegans. Proc. Natl. Acad. Sci. 1986;83:7821–7825. doi: 10.1073/pnas.83.20.7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford G.E., Holt I.E., Whittle J., Webb B.D., Tai D., Davis S., Margulies E.H., Chen Y., Bernat J.A., Ginsburg D., Holt I.E., Whittle J., Webb B.D., Tai D., Davis S., Margulies E.H., Chen Y., Bernat J.A., Ginsburg D., Whittle J., Webb B.D., Tai D., Davis S., Margulies E.H., Chen Y., Bernat J.A., Ginsburg D., Webb B.D., Tai D., Davis S., Margulies E.H., Chen Y., Bernat J.A., Ginsburg D., Tai D., Davis S., Margulies E.H., Chen Y., Bernat J.A., Ginsburg D., Davis S., Margulies E.H., Chen Y., Bernat J.A., Ginsburg D., Margulies E.H., Chen Y., Bernat J.A., Ginsburg D., Chen Y., Bernat J.A., Ginsburg D., Bernat J.A., Ginsburg D., Ginsburg D., et al. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS) Genome Res. 2006;16:123–131. doi: 10.1101/gr.4074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann R., Lucchini R., Koller T., Sogo J.M., Lucchini R., Koller T., Sogo J.M., Koller T., Sogo J.M., Sogo J.M. Transcription in the yeast rRNA gene locus: Distribution of the active gene copies and chromatin structure of their flanking regulatory sequences. Mol. Cell. Biol. 1995;15:5294–5303. doi: 10.1128/mcb.15.10.5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey C.A., Sargent D.F., Luger K., Maeder A.W., Richmond T.J., Sargent D.F., Luger K., Maeder A.W., Richmond T.J., Luger K., Maeder A.W., Richmond T.J., Maeder A.W., Richmond T.J., Richmond T.J. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 & resolution. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- Dixon D.K., Jones D., Candido E.P., Jones D., Candido E.P., Candido E.P. The differentially expressed 16-kD heat shock genes of Caenorhabditis elegans exhibit differential changes in chromatin structure during heat shock. DNA Cell Biol. 1990;9:177–191. doi: 10.1089/dna.1990.9.177. [DOI] [PubMed] [Google Scholar]
- Dong F., Hansen J.C., van Holde K.E., Hansen J.C., van Holde K.E., van Holde K.E. DNA and protein determinants of nucleosome positioning on sea urchin 5S rRNA gene sequences in vitro. Proc. Natl. Acad. Sci. 1990;87:5724–5728. doi: 10.1073/pnas.87.15.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R.E., Sulston J.E., Coulson A.R., Sulston J.E., Coulson A.R., Coulson A.R. The rDNA of C. elegans: Sequence and structure. Nucleic Acids Res. 1986;14:2345–2364. doi: 10.1093/nar/14.5.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Files J.G., Hirsh D., Hirsh D. Ribosomal DNA of Caenorhabditis elegans. J. Mol. Biol. 1981;149:223–240. doi: 10.1016/0022-2836(81)90299-0. [DOI] [PubMed] [Google Scholar]
- Fire A., Alcazar R., Tan F., Alcazar R., Tan F., Tan F. Unusual DNA structures associated with germline genetic activity in Caenorhabditis elegans. Genetics. 2006;173:1259–1273. doi: 10.1534/genetics.106.057364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaus A., Richmond T.J., Richmond T.J. Positioning and stability of nucleosomes on MMTV 3′LTR sequences. J. Mol. Biol. 1998;275:427–441. doi: 10.1006/jmbi.1997.1464. [DOI] [PubMed] [Google Scholar]
- Flaus A., Luger K., Tan S., Richmond T.J., Luger K., Tan S., Richmond T.J., Tan S., Richmond T.J., Richmond T.J. Mapping nucleosome position at single base-pair resolution by using site-directed hydroxyl radicals. Proc. Natl. Acad. Sci. 1996;93:1370–1375. doi: 10.1073/pnas.93.4.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima A., Ikemura T., Kinouchi M., Oshima T., Kudo Y., Mori H., Kanaya S., Ikemura T., Kinouchi M., Oshima T., Kudo Y., Mori H., Kanaya S., Kinouchi M., Oshima T., Kudo Y., Mori H., Kanaya S., Oshima T., Kudo Y., Mori H., Kanaya S., Kudo Y., Mori H., Kanaya S., Mori H., Kanaya S., Kanaya S. Periodicity in prokaryotic and eukaryotic genomes identified by power spectrum analysis. Gene. 2002;300:203–211. doi: 10.1016/s0378-1119(02)00850-8. [DOI] [PubMed] [Google Scholar]
- Greenwald I.S., Horvitz H.R., Horvitz H.R. unc-93(e1500): A behavioral mutant of Caenorhabditis elegans that defines a gene with a wild-type null phenotype. Genetics. 1980;96:147–164. doi: 10.1093/genetics/96.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horz W., Altenburger W., Altenburger W. Sequence specific cleavage of DNA by micrococcal nuclease. Nucleic Acids Res. 1981;9:2643–2658. doi: 10.1093/nar/9.12.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioshikhes I., Bolshoy A., Derenshteyn K., Borodovsky M., Trifonov E.N., Bolshoy A., Derenshteyn K., Borodovsky M., Trifonov E.N., Derenshteyn K., Borodovsky M., Trifonov E.N., Borodovsky M., Trifonov E.N., Trifonov E.N. Nucleosome DNA sequence pattern revealed by multiple alignment of experimentally mapped sequences. J. Mol. Biol. 1996;262:129–139. doi: 10.1006/jmbi.1996.0503. [DOI] [PubMed] [Google Scholar]
- Kent W.J. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R.D. Chromatin structure: A repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Kornberg R.D., Lorch Y., Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- Krause M., Hirsh D., Hirsh D. A trans-spliced leader sequence on actin mRNA in C. elegans. Cell. 1987;49:753–761. doi: 10.1016/0092-8674(87)90613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar L., Futschik M., Herzel H., Futschik M., Herzel H., Herzel H. DNA motifs and sequence periodicities. In Silico Biol. 2006;6:0008. [PubMed] [Google Scholar]
- Langst G., Blank T.A., Becker P.B., Grummt I., Blank T.A., Becker P.B., Grummt I., Becker P.B., Grummt I., Grummt I. RNA polymerase I transcription on nucleosomal templates: The transcription termination factor TTF-I induces chromatin remodeling and relieves transcriptional repression. EMBO J. 1997;16:760–768. doi: 10.1093/emboj/16.4.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Wallrath L.L., Elgin S.C., Wallrath L.L., Elgin S.C., Elgin S.C. Nucleosome positioning and gene regulation. J. Cell. Biochem. 1994;55:83–92. doi: 10.1002/jcb.240550110. [DOI] [PubMed] [Google Scholar]
- Luger K., Mader A.W., Richmond R.K., Sargent D.F., Richmond T.J., Mader A.W., Richmond R.K., Sargent D.F., Richmond T.J., Richmond R.K., Sargent D.F., Richmond T.J., Sargent D.F., Richmond T.J., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Lutter L.C. Precise location of DNaseI cutting sites in the nucleosome core determined by high resolution gel electrophoresis. Nucleic Acids Res. 1979;6:41–56. doi: 10.1093/nar/6.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Berka J., Braverman M.S., Chen Y.J., Chen Z., Braverman M.S., Chen Y.J., Chen Z., Chen Y.J., Chen Z., Chen Z., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J.D., Felsenfeld G., Felsenfeld G. Nucleosome structure. Annu. Rev. Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- McGhee J.D., Felsenfeld G., Felsenfeld G. Another potential artifact in the study of nucleosome phasing by chromatin digestion with micrococcal nuclease. Cell. 1983;32:1205–1215. doi: 10.1016/0092-8674(83)90303-3. [DOI] [PubMed] [Google Scholar]
- Moreno-Herrero F., Seidel R., Johnson S.M., Fire A., Dekker N.H., Seidel R., Johnson S.M., Fire A., Dekker N.H., Johnson S.M., Fire A., Dekker N.H., Fire A., Dekker N.H., Dekker N.H. Structural analysis of hyperperiodic DNA from C. elegans. Nucleic Acids Res. 2006;34:3057–3066. doi: 10.1093/nar/gkl397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyldermans S., Travers A.A., Travers A.A. DNA sequence organization in chromatosomes. J. Mol. Biol. 1994;235:855–870. doi: 10.1006/jmbi.1994.1044. [DOI] [PubMed] [Google Scholar]
- Nelson D.W., Honda B.M., Honda B.M. Genes coding for 5S ribosomal RNA of the nematode Caenorhabditis elegans. Gene. 1985;38:245–251. doi: 10.1016/0378-1119(85)90224-0. [DOI] [PubMed] [Google Scholar]
- Noll M. Internal structure of the chromatin subunit. Nucleic Acids Res. 1974a;1:1573–1578. doi: 10.1093/nar/1.11.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M. Subunit structure of chromatin. Nature. 1974b;251:249–251. doi: 10.1038/251249a0. [DOI] [PubMed] [Google Scholar]
- Riddle D., Blumenthal T., Meyer B., Priess J., Blumenthal T., Meyer B., Priess J., Meyer B., Priess J., Priess J. C. ELEGANS II. Cold Spring Harbor Laboratory Press; Plainview, NY: 1997. [Google Scholar]
- Ronaghi M., Nygren M., Lundeberg J., Nyren P., Nygren M., Lundeberg J., Nyren P., Lundeberg J., Nyren P., Nyren P. Analyses of secondary structures in DNA by pyrosequencing. Anal. Biochem. 1999;267:65–71. doi: 10.1006/abio.1998.2978. [DOI] [PubMed] [Google Scholar]
- Satchwell S.C., Drew H.R., Travers A.A., Drew H.R., Travers A.A., Travers A.A. Sequence periodicities in chicken nucleosome core DNA. J. Mol. Biol. 1986;191:659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- Sekinger E.A., Moqtaderi Z., Struhl K., Moqtaderi Z., Struhl K., Struhl K. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol. Cell. 2005;18:735–748. doi: 10.1016/j.molcel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Thomas J.H. Analysis of homologous gene clusters in Caenorhabditis elegans reveals striking regional cluster domains. Genetics. 2006;172:127–143. doi: 10.1534/genetics.104.040030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X., Smith L.M., Smith L.M. Solid-phase method for the purification of DNA sequencing reactions. Anal. Chem. 1992;64:2672–2677. doi: 10.1021/ac00046a004. [DOI] [PubMed] [Google Scholar]
- Travers A.A., Muyldermans S.V., Muyldermans S.V. A DNA sequence for positioning chromatosomes. J. Mol. Biol. 1996;257:486–491. doi: 10.1006/jmbi.1996.0178. [DOI] [PubMed] [Google Scholar]
- Tullius T.D., Dombroski B.A., Dombroski B.A. Hydroxyl radical “footprinting”: High-resolution information about DNA-protein contacts and application to λ repressor and Cro protein. Proc. Natl. Acad. Sci. 1986;83:5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanWye J.D., Bronson E.C., Anderson J.N., Bronson E.C., Anderson J.N., Anderson J.N. Species-specific patterns of DNA bending and sequence. Nucleic Acids Res. 1991;19:5253–5261. doi: 10.1093/nar/19.19.5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil M.R., Widlak P., Minna J.D., Garner H.R., Widlak P., Minna J.D., Garner H.R., Minna J.D., Garner H.R., Garner H.R. Global survey of chromatin accessibility using DNA microarrays. Genome Res. 2004;14:1374–1381. doi: 10.1101/gr.1396104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widlak P., Li P., Wang X., Garrard W.T., Li P., Wang X., Garrard W.T., Wang X., Garrard W.T., Garrard W.T. Cleavage preferences of the apoptotic endonuclease DFF40 (caspase-activated DNase or nuclease) on naked DNA and chromatin substrates. J. Biol. Chem. 2000;275:8226–8232. doi: 10.1074/jbc.275.11.8226. [DOI] [PubMed] [Google Scholar]
- Widom J. Short-range order in two eukaryotic genomes: Relation to chromosome structure. J. Mol. Biol. 1996;259:579–588. doi: 10.1006/jmbi.1996.0341. [DOI] [PubMed] [Google Scholar]
- Wilson J.T., Wilson L.B., Reddy V.B., Cavallesco C., Ghosh P.K., deRiel J.K., Forget B.G., Weissman S.M., Wilson L.B., Reddy V.B., Cavallesco C., Ghosh P.K., deRiel J.K., Forget B.G., Weissman S.M., Reddy V.B., Cavallesco C., Ghosh P.K., deRiel J.K., Forget B.G., Weissman S.M., Cavallesco C., Ghosh P.K., deRiel J.K., Forget B.G., Weissman S.M., Ghosh P.K., deRiel J.K., Forget B.G., Weissman S.M., deRiel J.K., Forget B.G., Weissman S.M., Forget B.G., Weissman S.M., Weissman S.M. Nucleotide sequence of the coding portion of human α globin messenger RNA. J. Biol. Chem. 1980;255:2807–2815. [PubMed] [Google Scholar]
- Wingert L., Von Hippel P.H., Von Hippel P.H. The conformation dependent hydrolysis of DNA by micrococcal nuclease. Biochim. Biophys. Acta. 1968;157:114–126. doi: 10.1016/0005-2787(68)90270-0. [DOI] [PubMed] [Google Scholar]
- Yuan G.C., Liu Y.J., Dion M.F., Slack M.D., Wu L.F., Altschuler S.J., Rando O.J., Liu Y.J., Dion M.F., Slack M.D., Wu L.F., Altschuler S.J., Rando O.J., Dion M.F., Slack M.D., Wu L.F., Altschuler S.J., Rando O.J., Slack M.D., Wu L.F., Altschuler S.J., Rando O.J., Wu L.F., Altschuler S.J., Rando O.J., Altschuler S.J., Rando O.J., Rando O.J. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]