Abstract
In screens for regulators of root morphogenesis in Arabidopsis we isolated six new recessive mutants with irregular cell expansion. Complementation analyses placed the mutations in two loci, PLEIADE (PLE) and HYADE (HYA). Phenotypic analyses revealed multinucleated cells, cell wall stubs, and synchronized cell divisions in incompletely separated cells that are all characteristics of defective cytokinesis. These defects were pronounced in roots and undetectable in aerial organs. In addition, fertility and germination were not affected by the mutations. Thus, the alleles that we have isolated of PLE and HYA suggest that the genes may encode organ-specific components needed primarily during root development. Analysis of microtubule arrays during cell cycle in ple and hya roots indicates that the presence of several synchronized nuclei influences the position of preprophase band, mitotic spindles, and phragmoplasts. The enhanced and synergistic phenotype of PLE/ple.hya/hya seedlings and double mutants point to a role of PLE and HYA in the same process. These mutants provide tools to elucidate the regulation of nuclear cytoskeletal interactions during cell division and cytokinesis.
The Arabidopsis root has a well-defined and simple structure with a radial pattern of single layers of epidermis, cortex, endodermis, and pericycle tissues, which are produced by rings of meristematic initials located at the tip of the root (Dolan et al., 1993). In the meristematic zone, cells divide transversely giving rise to organized files along the root axis. Root meristems vary in size depending on environmental conditions, and, thus, the number of mitotic cells actively dividing is flexible (Baskin, 2000; Hauser and Bauer, 2000).
In plants conventional somatic cell divisions and in particular cytokinesis differ in some aspects from other organisms (Glotzer, 1997). One major difference is that in contrast to the contractile ring, which is used for partitioning the cytoplasm of most animal cells (Field et al., 1999), plant cells divide the cytoplasm by constructing a new wall inside the cell. Furthermore, plants have developed several different mechanisms to execute cytokinesis. Before prophase of conventional somatic cell divisions, plants develop a special microtubule structure, the preprophase band (PPB) at the plasma membrane. The cortical position of the PPB correlates with the location where the cell plate will fuse with the parental cell walls at the end of cytokinesis. In telophase of conventional cell divisions, another special microtubule structure (the phragmoplast, which has similarity to the midbody of animal cells) forms from spindle remnants, microfilaments, and Golgi-derived vesicles between separated chromosomes (Samuels et al., 1995). In plants, the Golgi-derived vesicles, which contain cell wall material, fuse to form the cell plate. The cell plate expands outward and finally reaches the parental cell wall at points previously marked by the PPB. However, not all plant cells need the formation of a PPB to divide correctly. For example, no PPB has been detected at the onset of the first mitotic division during pollen-grain development. The mechanism by which the cell plate is guided to the parental cell wall is unknown in divisions with or without the PPB.
In Arabidopsis, “nonconventional” syncytial cytokinesis occurs during nuclear endosperm development (Otegui and Staehelin, 2000a; Otegui et al., 2001), during male and female meiosis (Otegui and Staehelin, 2000b), and during female gametophyte cellularization (Webb and Gunning, 1990, 1994; Christensen et al., 1998). In these cases, nuclear divisions and cytokinesis are uncoupled. This leads to the formation of multinucleated/syncytial cells. Syncytial cytokinesis does not involve the formation of PPBs, and the position of the new cell walls is determined by interacting arrays of microtubules that radiate from the nuclear envelope surfaces. The onset of cellularization occurs simultaneously. In endosperm, the MTs form groups of miniphragmoplasts between a pair of nuclei leading to a patchwork of syncytial-type cell plates (Otegui and Staehelin, 2000a).
From recently isolated mutants and their affected gene products, a picture has begun to emerge of the mechanism and spatial regulation of plant cytokinesis (Nacry et al., 2000). Characteristics of all cytokinesis mutants are the presence of incomplete cell walls in enlarged cells. Furthermore, the affected cells are multinucleated and/or contain enlarged polyploid nuclei. The mutants can be categorized according to their cell biological defects and their affected genes according to their similarity to biochemically characterized gene products. For example, the pilz mutant embryos and endosperms lack both interphase and mitotic microtubule arrays, indicating that the PILZ gene products are involved in microtubule array formation in conventional and syncytial-type cytokinesis (Mayer et al., 1999). Related phenotypes were observed in titan mutants (Liu and Meinke, 1998). The molecular analysis revealed that TTN1 encodes a regulatory protein known as tubulin-folding cofactor D and that TTN5 encodes a small GTP-binding protein-related ADP ribosylation factor-like protein (ARL2; McElver et al., 2000; Tzafrir et al., 2002). Recent experiments showed that ARL2 interacts with tubulin-folding cofactor D in fission yeast (Schizosaccharomyces pombe) and humans (Bhamidipati et al., 2000; Radcliffe et al., 2000).
Another group of cytokinesis mutants is defective in vesicle trafficking and cell plate formation (Assaad et al., 1996; Lukowitz et al., 1996; Nacry et al., 2000). A cytokinesis-specific syntaxin (KNOLLE) and a SEC1 homolog (KEULE) have been isolated and shown to bind to each other (Waizenegger et al., 2000; Assaad et al., 2001). Furthermore, some cytokinesis-defective mutants can be grouped based on their severe deficiencies in the formation of the primary cell wall. For example the CYT1 gene has been cloned, and it encodes a Man-1-phosphate guanylyltransferase (Lukowitz et al., 2001). The cyt1 mutants exhibit a 5-fold decrease in cellulose content and hyper-accumulate callose. The deficiency in N-glycosylation in cyt1 indicates that N-glycosylation is required for cellulose biosynthesis, and improper cell wall synthesis leads to cytokinesis defects. A relationship between improper cell wall synthesis leading to cytokinesis defects fits the phenotype of mutations in the KORRIGAN (KOR) gene, which encodes a putative membrane-bound endo-1,4-β-glucanase. Whereas weak kor/rsw2 mutants have reduced cellulose and increased pectin content (His et al., 2001; Lane et al., 2001; Sato et al., 2001), a strong allele exhibits characteristic cytokinesis defects such as aberrant cell plates, incomplete cell walls, and multinucleated cells (Zuo et al., 2000).
Most cytokinesis-related genes were identified from embryo- or endosperm-defective mutants, indicating that some components are shared between the different modes of plant cytokinesis. However, there is evidence that some components of conventional somatic cytokinesis are developmentally regulated. For example, TSO1 is a gene that appears to be involved in cytokinesis primarily during flower development (Liu et al., 1997; Hauser et al., 1998a; 2000; Song et al., 2000). In addition, although no cytokinesis occurs from the first zygotic division onward of double mutants between KNOLLE and KEULE, the haploid gametophytes are functional (Waizenegger et al., 2000). Moreover, several genes have been identified because of their abnormal male gametophytic meiosis and mitosis (Chen and McCormick, 1996; Hülskamp et al., 1997; Spielman et al., 1997; Park et al., 1998; Park and Twell, 2001), but the molecular basis of these genes is still unknown.
In this report, we present the genetic identification of two new Arabidopsis loci, PLE and HYA. Mutations in PLE and HYA result in irregularly expanded root cells. At the cellular level, the ple and hya alleles contain partially formed transverse cell walls and multiple nuclei, characteristics of cytokinesis-defective mutants. During cell division, these multinucleated cells divide synchronously and influence the position of microtubule arrays including the PPBs, the mitotic spindle, and the phragmoplasts. The strong root phenotypes of the ple and hya alleles indicate that the genes may encode components required for organ-specific cytokinesis. We discuss the possible mode of action of the PLE and HYA gene products during cytokinesis.
RESULTS
Isolation and Genetic Classification
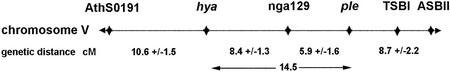
In genetic screens for root morphogenesis mutants of Arabidopsis, six lines were isolated with irregular root expansion phenotypes (Table I). All six mutants segregated as single nuclear recessive mutations (Tables II and III). Reciprocal complementation analyses revealed that they belonged to two different genetic loci, PLE and HYA. The PLE locus included two ethyl methanesulfonate (EMS) and one T-DNA mutagenized allele, and the HYA locus comprised three T-DNA mutagenized alleles. The chromosomal locations of the two loci were determined by the use of molecular markers. Both loci are located on the bottom of chromosome V and linked with a map distance of 14.5 centiMorgans (cM) to each other. HYA maps between the simple sequence length polymorphism (SSLP) markers AthS0191 and nga129 and PLE between nga129 and the cleaved amplified polymorphic sequences (CAPS) marker ASBII (Fig. 1).
Table I.
Overview of the ple and hya alleles
| Mutant
|
Mutagen | Genetic Status | Map Positiona (Chromosome) | Root Lengthb
|
Meristem Sizeb
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Allele | Ecotype | 4 DAG | 8 DAG | Ep | Co | En | |||
| mm | |||||||||
| ple-1 | Col | EMS | Recessive | 109 (V) | 1.3 ± 0.4 | 2.0 ± 0.4 | 7.7 ± 1.6 | 7.9 ± 1.5 | 7.8 ± 1.6 |
| ple-2 | Col | EMS | Recessive | 3.4 ± 1.0 | 5.9 ± 1.5 | 10 ± 0.9 | 10.4 ± 1.3 | 9.9 ± 1.6 | |
| ple-3 | Ws | T-DNA | Recessive | 1.5 ± 0.7 | 2.2 ± 1.3 | 9.5 ± 1.0 | 9.7 ± 1.3 | 9.5 ± 1.3 | |
| hya-1 | Ws | T-DNA | Recessive | 95 (V) | 3.3 ± 0.9 | 6.1 ± 1.0 | 6.9 ± 1.1 | 7.8 ± 1.3 | 7.6 ± 0.9 |
| hya-2 | Ws | T-DNA | Recessive | 5.5 ± 1.6 | 10.3 ± 3.6 | n.d. | |||
| hya-3 | Ws | T-DNA | Recessive | 5.2 ± 1.0 | 15.6 ± 3.0 | n.d. | |||
| wt | Col | 9.9 ± 2.0 | 26.9 ± 3.0 | 8.6 ± 1.6 | 8.8 ± 1.4 | 8.6 ± 1.4 | |||
Approximate map positions determined by mapping with CAPS and microsatellite markers (see “Materials and Methods”). The positions correspond to the RI-map of Lister and Dean (1993) (http://www.arabidopsis.org/servlets/mapper).
±sd.
Table II.
Complementation tests of the cytokinesis mutants
| Parental Genotype | Generation | Total | Seedling Phenotype
|
|
|---|---|---|---|---|
| Wild type | Mutant | |||
| ple-1 × ple-3 | F1 | 10 | 0 | 10 |
| ple-2 × ple-1 | F1 | 6 | 0 | 6 |
| ple-2 × ple-3 | F1 | 3 | 0 | 3 |
| hya-2 × hya-3 | F1 | 9 | 0 | 9 |
| hya-3 × hya-1 | F1 | 32 | 0 | 32 |
| ple-1 × hya-1 | F1 | 33 | 33 | 0 |
| ple-1 × hya-2 | F1 | 4 | 4 | 0 |
| ple-1 × hya-3 | F1 | 6 | 6 | 0 |
| ple-2 × hya-1 | F1 | 40 | 40 | 0 |
| ple-3 × hya-1 | F1 | 33 | 33 | 0 |
Table III.
Segregation analysis of ple and hya alleles
| Cross | Generation | Total | Seedling Phenotype
|
χ2 | |
|---|---|---|---|---|---|
| Wild type Obs./Exp. | Mutant Obs./Exp. | ||||
| ple-1 × Ler | F2 | 530 | 394 /397.5 | 136 /132.5 | 0.12 |
| ple-2 × Ler | F2 | 228 | 179 /171 | 49 /57 | 1.49 |
| ple-3 × Ws | F2 | 205 | 162 /153.75 | 43 /51.25 | 1.77 |
| hya-1 × Ws | F2 | 178 | 123 /133.5 | 55 /44.5 | 3.31 |
| Ler × hya-2 | F2 | 250 | 194 /187.5 | 56 /62.5 | 0.91 |
| Ler × hya-3 | F2 | 167 | 129 /125.25 | 38 /41.75 | 0.45 |
χ2 calculation is based on expected ratio of three wild-type to one mutant phenotypes. The χ2 values indicate no significant deviation (P > 0.05) from the expected ratio. See Table IV for definition of abbreviations.
Figure 1.
Genetic map of the PLE and HYA loci. Around 300 F2 mutants were used for mapping of each locus with the SSLP markers nga129, AthS0191, and TSBI and the CAPS marker ASBII. The genetic distance in cM was calculated from the recombination frequency after Kosambi (1944). Both loci map to the bottom of chromosome V. They are linked with a calculated distance of 14.5 cM.
PLE and HYA Are Involved in Cytokinesis
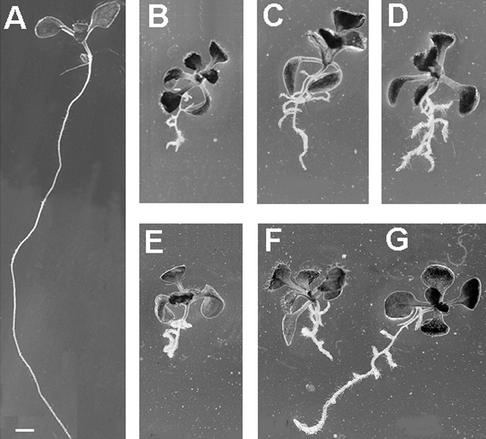
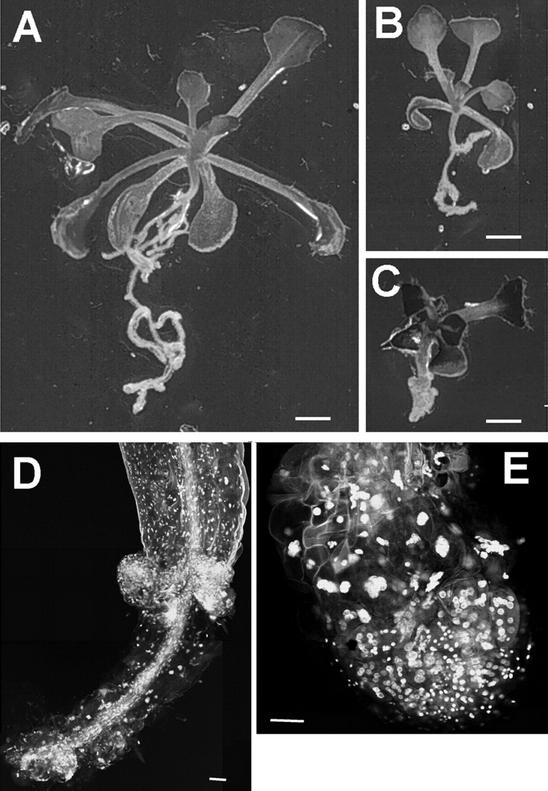
Typical of these new mutants is an irregularly expanded root. The phenotype ranges from very thick and extremely short roots to elongated roots with a wavy growth pattern and enhanced lateral root initiation (Fig. 2). In ple-2 and ple-3, development of aerial organs appears normal as evidenced by microscopic analyses of shoot and floral meristems (see Fig. 4), which produce normal leaves and flowers. Furthermore, no problems with fertility were observed in all ple and hya alleles. Ple-1 plants exhibited a smaller rosette and shorter inflorescence. This suggests that the affected genes may be active predominately in roots.
Figure 2.
Phenotype of wild-type (WT) and cytokinesis-defective seedlings on vertical nutrient agar medium. Mutant and WT seedlings were photographed 12 and 8 d after germination (DAG), respectively. A, WT Col; B, hya-1; C, hya-2; D, hya-3; E, ple-1; F, ple-3; and G, ple-2. The primary roots of the cytokinesis mutants are significantly shorter, irregularly expanded, exhibit a wavy growth pattern and develop more lateral roots. The shoots were indistinguishable from WT under these conditions. Bars in A and B through G = 1 mm.
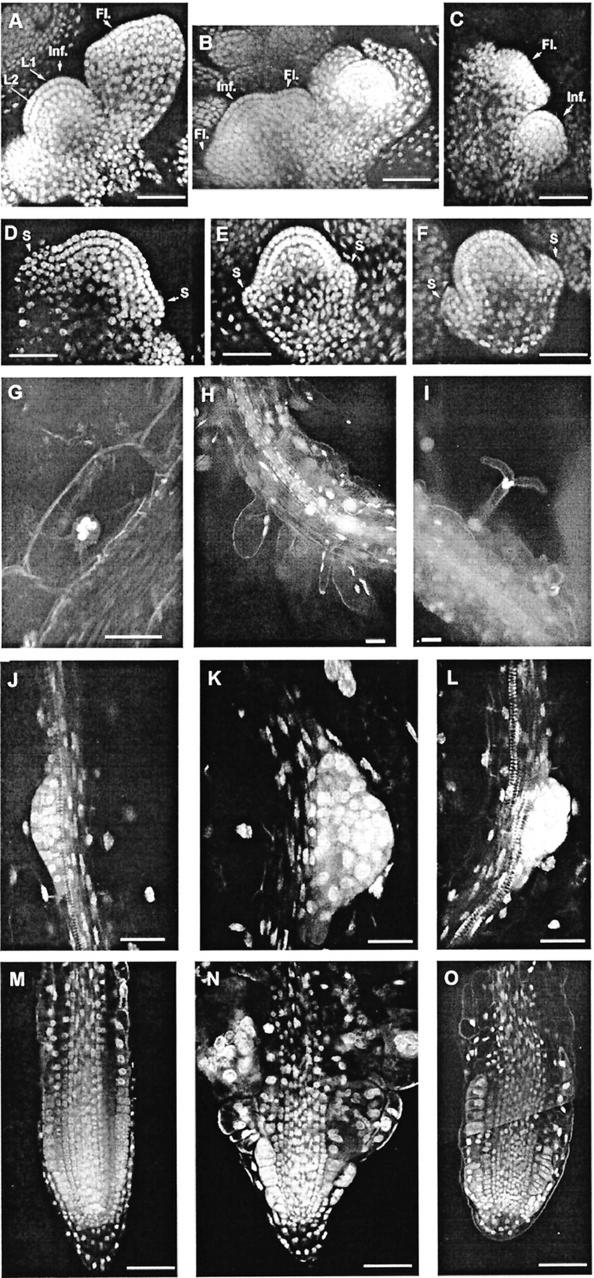
Figure 4.
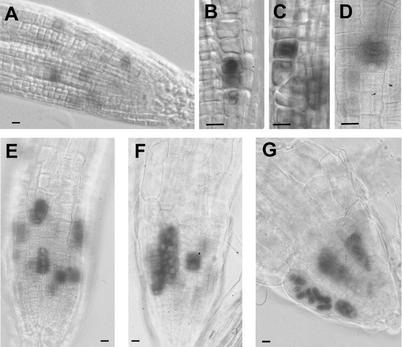
Visualization of nuclei in different organs of wild-type (WT) and cytokinesis-defective mutants. The nuclei were either stained with YO-PRO (A–F) or with DAPI (H and I) and visualized with confocal laser scanning microscopy (CLSM) or a fluorescence microscope, respectively. Optical longitudinal sections of inflorescence and floral meristems of wild type (A and D), ple-1 (B and E), and hya-2 (C and F). The two single-layered L1 and L2 cell files are clearly defined. A through C, Inflorescence meristems are flanked by floral meristems in different stages. D through F, Floral meristems at the stage of early sepal development. The primary and lateral root meristems of ple (K and N) and hya (L and O) accumulate multiple nuclei. Root hairs of ple-1 emerge with altered morphology as bifurcation (I) or as large bulges (H). G, Multiple nuclei phenotype in cortex cells of roots of flowering ple-1 mutants grown in soil. Fewer cells with less morphological defects are detectable in differentiated parts of roots of these plants. S, Sepal; Inf, inflorescence meristem; Fl, floral meristem. Bars = 50 μm in A through F, H, I, M, and O and 25 μm in G and J through L.
Wavy root growth patterns have been described for agravitropic mutants. To test whether the newly isolated mutants respond to gravity, seedlings were exposed to an altered gravitropic stimulus by rotating the nutrient plates by 90°. All mutant roots grew toward the gravitropic stimulus. Therefore, we conclude that the wavy growth behavior is caused primarily by the irregularly expanded root cells.
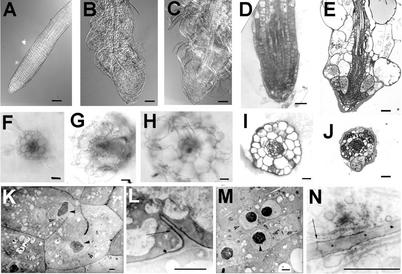
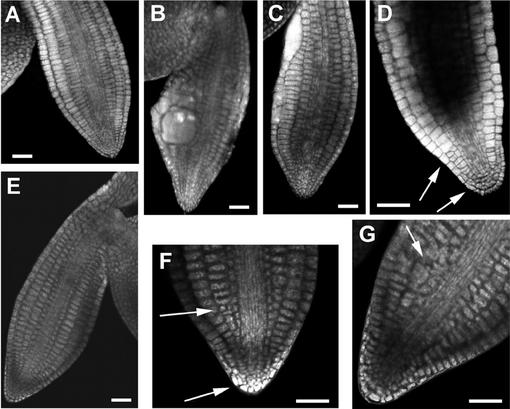
Transverse sections through the differentiation zone of roots revealed that some of the epidermis and cortex cells are grossly expanded (Fig. 3). These spherical cells are visible in whole-mount preparations (Fig. 3). Although root cells are malformed and the radial organization of different root tissues is more variable, mutant roots have clearly distinguishable cell layers (Fig. 3). Sections revealed that cell wall stubs and multiple nuclei are often present—a characteristic feature of cytokinesis-defective mutants (Fig. 3). All root tissues are affected to different degrees with the epidermis and cortex forming larger cells than the endodermis or stele tissues. Cells containing wall stubs appear to differentiate as witnessed by the presence of plasmodesmata and of suberin in endodermal cells (Fig. 3). Thus, the cytokinesis defect does not appear to change the differentiated characteristics of the affected cells.
Figure 3.
Root morphology of wild type (WT), ple, and hya. Whole-mount preparations of cleared WT (A), hya-3 (B), and ple-2 (C) root tips grown on nutrient agar medium using differential interference contrast microscopy. Median longitudinal and transverse sections of primary root tips of hya-3 (D and I) and ple-1 (E and J) resin embedded and stained with basic fuchsine/toluidine. In E and J, every tissue has aberrant cell wall stubs and multiple nuclei. Fresh transverse sections through the differentiation zone of a primary WT (F), hya-3 (G), and ple-2 (H) roots. The surface areas of hya roots are about 3 times and of ple-1 roots 3.5 to 4 times larger than WT. The three tissue layers epidermis, cortex and endodermis are clearly outlined but radially enlarged. H, In ple, the diarch symmetry of the vascular tissues is disrupted. K through M, TEM of ple-1 root cells. Note the darkly stained uneven nucleoli (arrowheads), the nuclear membrane (open arrows), and the incomplete cell walls. Longitudinal sections through cortex (K and N), endodermis (L), and vascular (M) cells. Although the newly developed cell wall is incomplete, suberin lamellas of the casparian stripe and plasmodesmata are incorporated (L and N, arrows). Bars = 50 μm in A through E, 25 μm in F through J, and 1 μm in K through N.
Another feature of cytokinesis-defective mutants is their multinucleated phenotype. Visualization of the nuclei in mutants by YO-PRO and 4,6-diamidino-2-phenylindole (DAPI) staining revealed that the giant root cells contain up to 32 nuclei, indicating that these cells undergo karyokinesis without cytokinesis (Fig. 4). This multinucleated phenotype is the reason for naming the genes after the stellar constellations, the PLEIADES (PLE) and HYADES (HYA).
Longitudinal sections demonstrate that apical cells in the root meristems are less affected. Quiescent center and root cap cells rarely contain more than one nucleus. In addition, there is a phenotypic gradient from the outside (epidermis) with the most severely affected cells toward the inside (stele) with only a few multinucleated cells.
Although the polarity of giant multinucleated cells is affected, not all of the multinucleated cells have lost their anisotropic expansion (Fig. 4). For example, the epidermal cells are still able to differentiate into root hair cells. But most of the multinucleated root hairs exhibit abnormal bulging, produce several tips, or become bifurcated (Fig. 4).
Previous analyses of cell expansion mutants indicated that growth conditions can modulate the expansion phenotype (Hauser et al., 1995). By decreasing the Suc concentration in the nutrient plates and by isolating roots of adult plants from the soil, we found that the multinucleated phenotype was attenuated (Fig. 4).
The number of nuclei in one cell indicates how many divisions were aberrant. Thus, to accumulate 32 nuclei, five rounds of cell divisions are necessary. This suggests that most of the defective cell divisions may occur after germination. Morphometric analyses revealed that the size of mature embryonic root meristem is similar between wild type and the cytokinesis mutants (Table I). However, we also identified multinucleated cells in mature embryos of two alleles, ple-1 and ple-3, of which 20.9% (n = 67) and 48.8% (n = 41) contained multinucleated cells in the embryonic roots, in hypocotyls, or in both (Fig. 5). As in adult roots some nuclei seem to be larger, suggesting that DNA endoreduplication or nuclear fusion might occur during mitosis. Because the nuclei of multinucleated cells cluster, we were not able to address these hypotheses by measuring their DNA content.
Figure 5.
Embryonic phenotype of ple mutants. Optical sections through mature embryos of ple-3 (A–D) and ple-1 (F–G) mutants stained with YO-PRO. The overall architecture of the embryos is normal with only a few cells in ple-3 being enlarged and containing multiple or giant nuclei (B and C). In some embryos only minor deviations of the architecture could be detected (arrows in D, F, and G). Bar = 25 μm.
To analyze the phenotype during cell division, we crossed the mutants with the cycB1;1:CDB:β-glucuronidase (GUS) marker line, which labels cells in late G2 and mitosis. We have shown that the subcellular localization of the histochemical GUS staining changes during mitosis and discriminates different mitotic phases (Hauser and Bauer, 2000). In prophase and telophase, the dye precipitates around the nucleus, whereas in meta- and anaphase, it is distributed in the whole cell. In mid-late anaphase the dye is denser on both sides of the metaphase plate and may point to a spindle localization. After completion of cytokinesis, the dye disappears gradually (Fig. 6). The histochemical GUS staining in mutant background revealed that mitosis is synchronized in incompletely separated cells. Their shared cytoplasm probably accounts for this synchronization. Furthermore, the subcellular localization of the dye in the prophase and telophase is similar to wild type. Only in ple alleles was the anaphase-specific spindle localization more frequently detectable than in wild-type roots. This might indicate that cell cycle is slowed down in these mutants.
Figure 6.
Histochemical staining of mitotic cells of WT, ple, and hya roots. The CYCB1;1:CDB:GUS marker line stains cells from late G2 to the onset of G1 and exhibits also a subcellular localization during mitosis of WT (A–D), hya-2 (E), and ple-3 (F and G) roots. In pro- and telophase, the staining accumulates around the nucleus (A and B), whereas in meta- and anaphase, it is distributed in the whole cell (B through D) with a slightly denser accumulation on both sides of the metaphase plate in mid-late anaphase (C, D, and G). After completion of cytokinesis, the dye disappears gradually. E through G, Cells with multiple nuclei divide synchronously. Bar = 10 μm.
Our phenotypic analyses suggest that PLE and HYA may be primarily active in roots. Because the analyzed alleles may not be null mutants, we are aware that some caution needs to be invoked in the interpretation of their spatial-temporal specificity.
Microtubule Organization during the Cell Cycle
During the cell cycle, plant microtubules (MT) undergo a series of conformational changes. In interphase of rapidly expanding cells, cortical microtubules are perpendicular to the axis of cell elongation (Fig. 7A). In late G2 before cell division, the PPB is formed (Fig. 7, B and C), and its localization correlates with the position and fusion site of the phragmoplast, cell plate, and new cell wall (Vantard et al., 2000). To characterize further the role of PLE and HYA at the cellular level, we studied MT organization in wild-type and mutant plants.
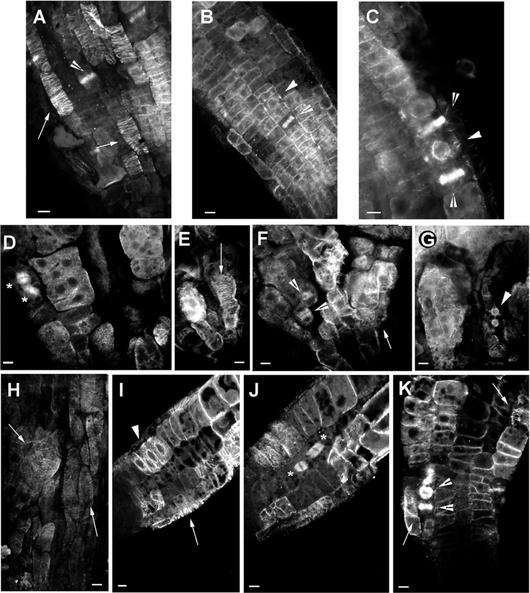
Figure 7.
Immunolocalization of microtubules in wild-type, ple-1, and hya-3 roots. A through C, Optical sections through the epidermis of wild-type roots showing cortical MTs (A; black arrow), phragmoplasts (B and C; white arrow), and the MTs of the PPB (arrowhead). The PPB is always accompanied with strong perinuclear MTs (C). D through G, Optical sections through the epidermis of ple-1 roots showing mitotic spindles (stars), dense accumulation of cytoplasmic MTs (D), cortical (E and F), and the perinuclear MTs of the PPB (G). Note the dark gray nuclei in multinucleated cells (D) and the cell wall stubs where MTs seems to nucleate (G). H through K, Optical sections through the epidermis and cortex of hya-3 roots showing slightly misoriented cortical (H, I, and K), perinuclear MTs and PPBs (I), mitotic spindles (J) of synchronized two nuclei containing cortical cells. K, Shows the misaligned phragmoplasts of a multinucleated epidermis cell. Bar = 10 μm.
In ple and hya, the helical arrays of cortical MTs are present but misplaced in giant cells with multiple nuclei (Fig. 7, E, H, and J). Furthermore, in ple, short, diffuse cytoplasmic MTs were detectable in interphase cells (Fig. 7, D and G). During late G2, the perinuclear MTs stained strongly in both mutants but were not symmetrically distributed around the nucleus in the ple mutants. Although the PPB is present in cells with single nuclei, it is displaced in hya (Fig. 7I) and is not discernible in multinucleated ple cells (Fig. 7G). Thus, cells with numerous nuclei do not exhibit an equatorial arrangement of the PPB. During mitosis, the position of spindles and phragmoplasts is also misplaced in multinucleated cells of both mutants (Fig. 7, D, F, J, and K).
Thus, MT aggregates in multinucleated G2 cells could be interpreted as an attempt to produce PPBs, but signals from several nuclei interfere with correct positioning. These interfering signals could also be the reason why spindles and phragmoplasts are misplaced in mitotic cells. These phenotypes support the hypothesis that signals from nuclei together with signals from the cortex are responsible for positioning the diverse MT arrays in plant cells.
Cell Plate Formation in ple and hya
The MT analysis revealed that the phragmoplast is formed in both mutants. One of its proposed functions is to direct Golgi-derived vesicles to the division plane where the cell wall material-containing vesicles fuse to form the cell plate. The cell plate is a transient membrane-bound compartment, which undergoes complex transformations while expanding from the middle out to the division site of the parental cell walls. Callose was identified as the predominant lumenal component of forming cell plates. During cell plate maturation, the callose is replaced by cellulose, xyloglycans, and pectin (Samuels et al., 1995; Otegui and Staehelin, 2000b).
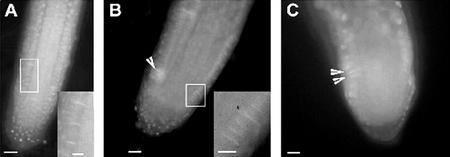
To gain insight into the aberrant cell wall formation of ple and hya, we studied callose deposition into the cell plate by histochemical staining with aniline blue. For better orientation, the nuclei were counter-stained with DAPI. Callose-containing cell plates were observed between sister nuclei in root meristems by focusing through the whole cell, because fluorescence of aniline blue and DAPI could rarely be focused in one optical plane (Fig. 8A). In multinucleated cells of ple and hya, cell plates develop synchronously and with a slight angle to each other (Fig. 8, B and C). This displacement is comparable with misplaced phragmoplastic MTs in multinucleated cells of fairly normal morphology. In multinucleated root cells with a stronger cell morphological defect, we rarely spotted cell plates in a focal plane. Callose was further observed at the end of cell wall stubs in already expanded multinucleated cells of ple (data not shown). Such plugs of callose can be induced by stress and might be a secondary effect. In summary, cell plates are formed in ple and hya but misplaced, indicating that the phragmoplast is functional.
Figure 8.
Callose staining of cell plates in root meristems of wild type ple and hya. A, An epidermal cell file of wild type with callose in the cell plates. B and C, ple-2 and hya-1, respectively, show callose deposition in the cell plate of a synchronously dividing cells with multiple nuclei. Bars = 25 μm in A through C.
Double Mutant Analysis
To determine possible genetic interactions between PLE and HYA, double mutants were generated by crossing homozygous hya-1 and hya-2 with ple-1 and ple-2 mutants. F1 progeny of these crosses were all phenotypically wild type. The F2 seedlings were grouped into five classes: wild type, very weak, weak, ple, and double mutant phenotypes (Table IV). Among F2 progeny, double mutants were classified as those with extremely short roots (n = 36, root length 1.31 ± 0.5 mm; Fig. 9C). CLSM analyses revealed that most of the root and more hypocotyl cells of double mutants were multinucleated, indicative of a synergistic enhancement of the cytokinesis defects (Fig. 9, D and E).
Table IV.
Double mutant and segregation analysis of the progeny of selfed ple/PLE.hya/HYA plants
| Phenotypic Classification | Wild Type | Very Weak | Weak | ple | Strong Dbl. |
|---|---|---|---|---|---|
| PLE/PLE.HYA/HYA | hya/hya | PLE/ple.hya/hya | ple/ple | ple/ple.hya/hya | |
| PLE/PLE.HYA/hya | ple/ple.HYA/hya | ||||
| PLE/ple.HYA/HYA | |||||
| PLE/ple.HYA/hya |
| Total | Exp./Obs. | Exp./Obs. | Exp./Obs. | Exp./Obs. | Exp./Obs. |
|---|---|---|---|---|---|
| Unlinked | 9 : | 1 : | 2 : | 3 : | 1 |
| Linkeda | 0.084 × 0.059 × N | ||||
| ple-2 × hya-1 | |||||
| 280 | 157.5 /179 | 17.5 /13 | 35 /33 | 52.5 /54 | 1.4 /1 |
| hya-1 × ple-1 | |||||
| 240 | 135 /122 | 15 /15 | 30 /46 | 45 /55 | 1.2 /2 |
| ple-1 × hya-2 | |||||
| 142 | 80.1 /64 | 8.9 /7 | 17.8 /32 | 26.7 /37 | 0.7 /2 |
Dbl., Double mutants; Exp., expected; Obs., observed.
Because PLE and HYA are linked, the expected number of double mutants was calculated by multiplying the expected frequencies of recombination between hya and nga129 (0.084) and nga129 and ple (0.059) with the total number of F2 plants analyzed (N).
Figure 9.
Double mutant analysis of ple and hya. For double mutant analyses hya-1 (A) and ple-1 (B) were crossed and the genotype of different phenotypic classes were confirmed with molecular markers. C, The enhanced phenotype of the hya-1 ple-1 double mutant is exemplified. Optical sections through the root and the hypocotyl of hya-1 ple-1 double mutants show that more cells in both organs contain multiple nuclei. Bars = 1 mm (A–C) and 25 μm (D and E).
The genotype of members of these phenotypic classes was confirmed using closely linked markers (Fig. 1) and segregation analyses of the F2 progeny. The homozygous hya seedlings exhibited the weakest phenotypes, whereas those grouped as weak phenotype were PLE/ple.hya/hya. In contrast to the single mutants, the double mutants were not viable on soil and were not fertile. Furthermore, in the progeny of selfed PLE/ple.hya/hya and ple/ple.HYA/hya plant, a significant number of seeds did not germinate, indicating that they are embryonic lethal (Table V). Together with the synergistic phenotype of the PLE/ple.hya/hya seedlings, this suggests that PLE and HYA genetically interact and might be involved in the same process.
Table V.
Segregation analysis of the progeny of selfed ple/ple.HYA/hya and PLE.ple/hya.hya plants
| F2 Genotype | F3 Line | Total | Phenotype Strong Dbl.a | Not Germinated | Genotype Confirmed |
|---|---|---|---|---|---|
| ple-2.HYA/hya-1 | A | 109 | 12 | 10 | 8 × ple/ple.hya/hya |
| E | 141 | 25 | 11 | 25 × ple/ple.hya/hya | |
| PLE/ple-2.hya-1 | B | 49 | 6 | 11 | 5 × ple/ple.hya/hya |
| C | 100 | 24 | 5 | 19 × ple/ple.hya/hya | |
| 5 × PLE/ple/hya/hya |
The genotype of the double mutants (Dbl.) was confirmed with molecular markers.
These plants did not survive on soil.
DISCUSSION
Spatio-Temporal Specificity of PLE and HYA Function
We have identified two new loci, PLE and HYA, whose mutant alleles exhibit typical features of cytokinesis-defective mutants. Whereas in ple and hya all root tissues are affected by the mutations, the aerial portions of these mutants have a wild-type appearance and no detectable fertility or germination defects. Homozygous plants of five alleles survive on soil and are indistinguishable from wild type. Only the strongest allele of ple-1 forms a smaller plant. Moreover, ple hya double mutants show synergistic cytokinesis defects and are not viable on soil. Our interpretation is that the root functions of weak alleles are still able to support normal aboveground development, whereas the cytokinesis defects of roots in severe alleles cannot sustain normal plant growth. Plants cannot survive if, in addition to the root, aerial parts are affected, as the hypocotyl in double mutants. Moreover, the synergistic phenotype of double mutants points to a potential genetic interaction between PLE and HYA.
The segregation data of homozygous and heterozygous ple and hya alleles indicate that PLE and HYA are not required for gametophytic development. The phenotype of ple and hya alleles is predominantly detectable in primary and secondary roots of seedlings grown on nutrient agar plates. Although most root tissues display cytokinesis defects, no dramatic radial pattern changes are detectable in the root or embryo. The spatial arrangement of initials is the result of early cell divisions, and at the early torpedo stage of embryogenesis, the tissues of both root and hypocotyl are produced (Scheres et al., 1994). The finding that the maximal number of nuclei within root cells was 32 is in agreement with the approximately four cell divisions that each cell undergoes in postembryonic root meristems of Arabidopsis (Fujie et al., 1993). This provides support for the hypothesis that most of the defective cell divisions occur after germination and points to a developmental stage-specific function of the PLE and HYA gene products.
However, multinucleated cells are detectable at low frequency in mature embryos of strong ple alleles. Our PLE and HYA alleles, alternatively, may not be null mutants, and the phenotypic analyses may lead to an overestimate of their spatio-temporal specificities. Another possibility is that the PLE and HYA genes belong to a gene family with partially redundant functions. Other members of this gene family may act during embryogenesis, in mitotic divisions of somatic cells, in shoot and floral meristems, and during gametophytic cell divisions.
Asymmetric Cell Plate Formation in ple and hya
We observed cell wall stubs in the mutants, indicating that the growing cell plate reaches the parental cell wall at late stages of cytokinesis but only on one side of the cell. This implies that as soon as one side of the cell plate reaches the parental wall, it is stabilized, whereas the other side of the plate is degraded. Thus, PLE and HYA gene products may be involved in the spatial-temporal coordination of cell plate growth and/or stabilization during cell wall maturation.
Asymmetric cell wall stubs are characteristic for cytokinesis defects either induced by mutations or upon caffeine inhibition of cytokinesis. The possibility that cell plates are formed asymmetrically challenges our current model of plant cytokinesis in which cell plate formation is symmetric and centrifugal. Thus, in addition to the specific function of PLE and HYA, all cytokinesis-defective mutants point to a general novel characteristic of cell plate formation. The idea of asymmetric cell plate formation is supported by a recent paper of Cutler and Ehrhardt (2002) using three-dimensional live-cell imaging.
The Number of Nuclei Influences Microtubule Structures and Polarity
The cytokinesis defects of ple and hya mutants do not inhibit multiple rounds of nuclear divisions. Thus, these multinucleated cells share some aspects of syncytia where free nuclear divisions occur uncoupled from cytokinesis and cellularization (Otegui and Staehelin, 2000b). Studies of MTs during the cellularization process of endosperm syncytia indicate that the formation of phragmoplasts is a primary difference between conventional and nonconventional cytokinesis. It has been shown that in higher plants, the nuclear envelope acts as a microtubule organizing center. Whereas the phragmoplasts of somatic cells derive from anaphase spindle remnants, the syncytial cells build miniphragmoplasts from MTs, which radiate from the microtubule organizing centers of adjacent nuclei (Otegui and Staehelin, 2000b).
Our study indicates that the presence of multiple nuclei has a dramatic impact on the different MT arrays including short diffuse cytoplasmic, irregular perinuclear, misplaced cortical, PPBs, spindle, and phragmoplast MTs. The diverse aberrant MT arrays become more dramatic with increasing the number of interfering nuclei in the cells of ple and hya mutants. Nothing equivalent to the syncytial-type miniphragmoplasts or radiating nuclear MTs develop, indicating that missing PLE and HYA action does not turn somatic-type into a syncytial-type cytokinesis. The MT phenotypes of ple and hya mutants suggest the possibility that a defect in MT dynamic or positioning may be enough to hinder proper cell wall formation. Once nuclei of daughter cells are incompletely separated, their interfering signals lead to a cellular disaster impeding all events that rely on the coordination between the nucleus, the cytoskeletal elements, and the cell periphery.
A morphological process that requires tight control between the nucleus and cell cortex is the positioning and emergence of root hairs. The syncytial epidermis trichoblasts are still able to form root hairs; thus, tip growth is not inhibited. But the position and the number of root hairs on one trichoblast vary. In rare cases, root hairs bifurcate, indicative of defects in tip growth polarity. Similar phenotypes have been observed by disruption the MT cytoskeleton dynamics with taxol (Bibikova et al., 1999) or α-tubulin reduction (Bao et al., 2001). The root hair phenotype might indicate a MT-associated function for PLE and HYA. Thus, in addition to the regulation of cytokinesis, PLE and HYA could help to elucidate the importance of the interplay between the nucleus, cytoskeletal structures, and cell morphogenesis.
Threshold Model of PLE and HYA Action
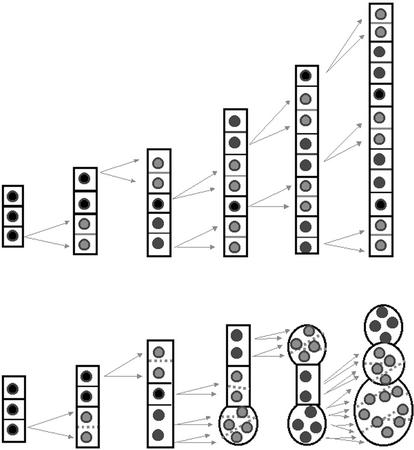
In summary, we propose that a certain threshold activity of the PLE and HYA gene products is needed to stabilize cytokinetic structures. If these stabilizers are not present or functional or if a certain threshold could not be synthesized, cell wall formation cannot be completed (Fig. 10).
Figure 10.
Schematic representation of cell division pattern within a cell file of wild-type and cytokinesis-defective root meristems. The diagram illustrates our interpretation of the cytokinesis defects in roots. The ordered cell files of wild-type roots are generated through transverse divisions in the meristem that has been already established during embryogenesis. After germination, cells start to divide. Our model proposes that the first defect in mutant root meristems may not yet change the normal morphogenesis of the affected daughter cells. But during subsequent rounds of divisions, the syncytial status of daughter cells leads to a cellular disaster. Instead of adding only one new daughter cell into a cell file, the syncytial nuclei divide simultaneously and positional signals between the nuclei and cytoskeletal elements get misinterpreted leading to visible morphological changes. This cellular disaster does not prevent further cell divisions as cells continue their division program up to five times.
This model explains the rare embryonic phenotypes of the mutants, because their threshold level may be lower during embryogenesis or under growth conditions that reduce cell division such as growth in soil or on nutrient medium without Suc. This model would also allow another possible interpretation for the organ-specific phenotype, because our alleles may not be null mutants and the residual activity of the proteins reaches a threshold that is sufficient for embryogenesis. It also explains the radial gradient of phenotypic severity from the outside (epidermis) to the inside of the root (stele). In stele cells, the spatial constraints do not allow a dramatic volume increase even if daughter cells are not completely isolated by a new cell wall. On the other hand, these cells have a smaller diameter, thus, a smaller cell plate is needed for a complete separation. This model is also supported by the observation that cell division appears to be slowed down in multinucleated cells (Assaad et al., 1996; our histochemical results). Thus, as soon as multiple nuclei are debating cellular decisions, the timing of cytokinesis is disturbed.
Isolation of PLE and HYA will complement the increasing number of cytokinesis-specific genes and will provide a more detailed picture of the structural basis of cytokinesis and its regulation.
MATERIALS AND METHODS
Plant Material, Growth Condition, and Mutant Screening
Wild-type Arabidopsis accessions (Landsberg erecta [Ler], Columbia [Col], and Wassilewskija [Ws]) were obtained from the Arabidopsis Stock Center (Columbus, Ohio). Seeds were sterilized, seedlings were grown and screened for abnormal root morphology on vertical nutrient agar plates, and plants were propagated on soil according to Hauser et al. (1995) and Hauser and Bauer (2000). The mutants were isolated because of their wavy growth pattern and irregularly expanded roots among approximately 5,000 EMS and 175,000 T-DNA mutagenized (K. Feldmann, University of Arizona [Tucson] and DuPont [Wilmington, DE]) Arabidopsis M2 and T4 seed pools, respectively.
Phenotypic Characterization and Microscopic Analyses
The differential interference contrast microscopy (Nomarski) was performed with cleared preparations. Roots were fixed with methanol:acetic acid (MA = 3:1, v/v) and treated with Herr's solution (chloralhydrate:phenol:lactic acid:xylol:carnation oil = 2:2:2:1:2, v/v). Transverse root sections were obtained as described by Hauser et al. (1995).
nDNA of MA fixed roots was stained with 0.5 μg mL−1 DAPI in phosphate-buffered saline with Tween 20 (PBT) (130 mm NaCl, 10 mm phosphate buffer, pH 7, and 0.1% [v/v] Tween) for 10 min. Excess stain was removed by several washes with PBT followed by a glycerin series (30% [v/v], 50% [v/v], and 70% [v/v] glycerin in PBT). Seedlings were mounted in Citifluor (Plano, Wetzlar, Germany).
Double staining for callose and DNA was performed with MA-fixed plant material. After washes with sterile water, DNA was stained with 2 μm DAPI in water for 5 min and washed with water for 1 min. Callose staining was conducted with 0.5% (w/v) Aniline blue in water for 10 min followed by 2-min washes with 2 mm Na2PO4, pH 7.5. Seedlings were mounted in 0.2% (w/v) Aniline blue, 100 mm Tris-HCl, pH 9, and 50% (v/v) glycerol on microscope slides. Roots were analyzed with a confocal laser scanning microscope (Axiovert, Zeiss, Jena, Germany) under UV with the DAPI filter set (365-nm excitation and 420-nm emission). Pictures were taken with a 167 mt camera (Contax, Japan) using R-100 film (Konica, Japan). The slides were scanned and arranged using Adobe Photoshop (Adobe Systems, Mountain View, CA). Histochemical GUS staining was done as described by Hauser and Bauer (2000).
For confocal microscopy, seedlings were fixed in MA and stained with 0.5 μm YO-PRO (Molecular Probes, Eugene, OR) in PBT solution for 10 min. Washing and mounting was similar to the DAPI staining protocol. For YO-PRO staining of mature embryos, dry seeds were fixed in MA at 12 to 48 h, washed twice with water for 10 min, and incubated in 0.5 μm YO-PRO for 2 h. The seeds were kept in water until the testa was dissected under the microscope, and the embryos were mounted in 70% (v/v) glycerin. YO-PRO stains predominately DNA with weaker staining of RNA and the cell wall. Pictures were taken using filters for 488-nm excitation on a confocal laser scanning microscope (MSE 600, Bio-Rad, Hercules, CA) with an argon/krypton laser. Pictures were saved as PIC files translated with the Paint Shop Pro3 (batch conversion of raw file formats, v89a-nominated with 768 width and 512 length and 76-byte hadder size) into TIFF files and manipulated in Adobe Photoshop. The size of the root meristem was defined as by Hauser and Bauer (2000).
For resin embedding, root tips were fixed with 2.5% (v/v) glutaraldehyde in 5 mm Na-cacodylate buffer (pH 6.8) for 1.5 h under low vacuum. After six 10-min washes in 5 mm Na-cacodylate buffer (pH 6.8), the roots were post-fixed with 1% (w/v) osmium tetroxide in 5 mm Na-cacodylate buffer (pH 6.8) for 2 h. After several washes, samples were dehydrated in an ethanol series starting with 30% (v/v) for 30 min, 60% (v/v), 75% (v/v), and 90% (v/v) each for 1 h, and 100% overnight. Media exchange started with two additional 100% (v/v) ethanol steps, followed by one 100% (v/v) ethanol:propylene oxide (1:1, v/v) and one pure propylene oxide step, each for 1 h. Media infiltration continued with a 3-h incubation of a propylene oxide:Spurr's resin (3:1, v/v) mixture and an overnight incubation with propylene oxide:Spurr's resin (1:1, v/v). The seedlings were allowed to equilibrate for 24 h in a propylene:Spurr's resin (1:3, v/v) mixture and with pure Spurr's resin two times each for 24 h. Polymerization was performed for 8 h at 70°C. Semithin (1- to 2-μm) sections of embedded roots were stained about 30 s to 1 min. with 1% (w/v) aqueous toluidine blue mixed with 2% (w/v) Na2CO3 in 1:1 ratio at 50°C to 60°C. After washing, staining continued with 0.1% (w/v) aqueous basic fuchsine for about 30 s at the same temperature. Cell walls and polysaccharides stained bright red, nucleoli were dark blue, and the nucleoplasm and the cytoplasm were purple. For transmission electron microscopy (TEM), ultrathin (80- to 90-nm) sections were cut from the Spurr's resin embedded roots and stained under N2 atmosphere with uranyl acetate for 1 h and with lead citrate for 5 min. Sections were visualized using a Zeiss TEM 900. The images were digitized using an image analyzer (Q500MC, Leica Microsystems, Wetzlar, Germany) with Q500MC software and the monochromatic camera system CF8/1 FMC.
Immunolocalization of Microtubules
Before fixation, whole seedlings were incubated in microtubule stabilization buffer (MTSB; 50 mm PIPES, 5 mm EGTA, and 5 mm MgSO4, pH 7.0) with 10% (v/v) dimethyl sulfoxide for 15 min at room temperature (RT). This solution was then exchanged with freshly prepared fixative (4% [w/v] p-formaldehyde in MTSB, 5% [v/v] dimethyl sulfoxide, and 0.1% [v/v] glutaraldehyde). After at least 1 h of fixation, the seedlings were washed three times with MTSB for total of 30 min. The roots were transferred to aminopropyltri-ethoxysilan (Sigma, St. Louis)-treated microscope slides. After desiccation, the roots were rehydrated for 10 min with MTSB and 0.1% (v/v) Nonident P40 (Sigma). Slides were carefully treated with 300 μL of 2% (w/v) driselase (Sigma) in MTSB for 30 min to 1 h, followed by washing three times with MTSB and 0.1% (v/v) Triton X-100 for about 30 min at RT. After three additional washing steps with MTSB for about 30 min, roots were blocked with 300 μL of 3% (w/v) bovine serum albumin (BSA; Sigma A-7030) in MTSB in a humid chamber for 1 h at RT, followed by an incubation with 300 μL of a 1:100 (v/v) dilution of the rat monoclonal YOL1/34 (anti-yeast-α-tubulin subunit antibody; MCA78S, Serotec, Oxford) in 3% (w/v) BSA/MTSB in the humid chamber overnight at RT. Slides were carefully washed three times with MTSB for 1 h then incubated with 300 μL of a 1:100 (v/v) dilution of the fluorescein-isothiocyanate-labeled secondary anti-rat antibody (Sigma F-1763) in 3% (w/v) BSA/MTSB in a dark, wet chamber for about 4 h. Slides were washed three times with MTSB for 60 min and mounted in Citifluor. The slides were keep at 4°C in the dark until microscopic examination. CLSM was performed with a Bio-Rad MSE 600 at 488 nm. Pictures were taken, stored, and manipulated as described above.
Genetic Analyses and Mapping
Homozygous plants were used for pair wise crosses between the different cytokinesis mutants to determine complementation groups. The chromosomal locations of the ple and hya genes were determined by measuring the recombination frequency between the mutant genes and microsatellite markers (Bell and Ecker, 1994; modified as Hauser et al., 1995) and CAPS (Konieczny and Ausubel, 1993). More than 600 F2 plants from crosses of ple-3, hya-3, hya-1, and hya-2 to Col were used for mapping. For double mutant analysis, homozygous hya-1, hya-2, and ple-1 plants were used to pollinate homozygous ple-1, ple-2, and hya-1 plants, respectively. F1 and F2 progeny of those crosses were analyzed for their root phenotype on vertical agar plates. Among the progeny of F2 plants with [ple/ple.HYA/ple] and [PLE/ple.hya/hya] genotypes we were able to obtain a higher frequency of double mutants which were use to analyze their cellular phenotypes.
Genomic DNA for PCR analysis was isolated according to Hauser et al. (1998b). We designed a new forward primer for the microsatellite nga129 5′-CATAATCGAATCGGACACGAC-3′ resulting in 59-bp-smaller PCR fragments for all ecotypes. In addition, the SSLP marker AthS0191 was used (http://www.Arabidopsis.org/servlets/TairObject?id=18&type=marker). The CAPS marker ASBII detects a HinfI Col/Ws polymorphism (Niyogi et al., 1993). For double mutant analysis, a closer molecular marker was developed from the Trp β-synthase 1 gene (TSB1; Last et al., 1991). The primers TSBI-F 5′-GTGCCGAGGTGATGCTTAGG-3′ and the TSBI-R 5′-GTCACCCAATCTCTTATCGCTTCA-3′ amplify a 162-bp fragment in Col; the slightly smaller Ws fragment is distinguishable on native 5% (w/v) PAGE. This marker is located between ple and ASBII and has a recombination frequency with PLE of about 5% (w/v).
ACKNOWLEDGMENTS
We thank Edi Schneeberger and Gernot Resch for technical assistance. We are specially obliged to Christian Schöfer for his permission and kind instruction in the use of the ultramicrotome, to Mirjana Ilijin-Jug for helping with the sections, to Milada Ciamporová and Martina Weber for instructions on the TEM, and to Otmar Hohenwarter for the use of the confocal laser scanning microscope. We thank Dieter Schweizer, Keith Roberts, Chun-Ming Liu, Farhah Assaad, Wolfgang Lukowitz, and Katharina Schneider for stimulating discussions. We also thank Pablo Scolnik and the DuPont de Nemours company for the T-DNA mutagenized lines. The ple-1 allele was kindly provided by John Schiefelbein. We are obliged to John Celenza for the CYCB1;1:CDB:GUS marker line. Finally, we thank Josef Glössl for his attentive support.
Footnotes
The initial stages of this work were supported by the National Science Foundation (to P.N.B.) and by a Schrödinger-Auslandsstipendium from the Austrian Science Foundation (grant no. J0676–MOB to M.-T.H.). S.M. was supported by the Austrian National Bank (Jubiläumsfondprojekt no. 5598) and by a European Grant (no. PL-960217).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.004416.
LITERATURE CITED
- Assaad FF, Huet Y, Mayer U, Jürgens G. The cytokinesis gene KEULE encodes a Sec1 protein that binds the syntaxin KNOLLE. J Cell Biol. 2001;152:531–544. doi: 10.1083/jcb.152.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaad FF, Mayer U, Wanner G, Jürgens G. The KEULE gene is involved in cytokinesis in Arabidopsis. Mol Gen Genet. 1996;253:267–277. doi: 10.1007/pl00008594. [DOI] [PubMed] [Google Scholar]
- Bao Y, Kost B, Chua NH. Reduced expression of alpha-tubulin genes in Arabidopsis thaliana specifically affects root growth and morphology, root hair development and root gravitropism. Plant J. 2001;28:145–157. doi: 10.1046/j.1365-313x.2001.01142.x. [DOI] [PubMed] [Google Scholar]
- Baskin TI. On the constancy of cell division rate in the root meristem. Plant Mol Biol. 2000;43:545–554. doi: 10.1023/a:1006383921517. [DOI] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Bhamidipati A, Lewis SA, Cowan NJ. ADP ribosylation factor-like protein 2 (Arl2) regulates the interaction of tubulin-folding cofactor D with native tubulin. J Cell Biol. 2000;149:1087–1096. doi: 10.1083/jcb.149.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova TN, Blancaflor EB, Gilroy S. Microtubules regulate tip growth and orientation in root hairs of Arabidopsis thaliana. Plant J. 1999;17:657–665. doi: 10.1046/j.1365-313x.1999.00415.x. [DOI] [PubMed] [Google Scholar]
- Chen YC, McCormick S. Sidecar pollen, an Arabidopsis thaliana male gametophytic mutant with aberrant cell divisions during pollen development. Development. 1996;122:3243–3253. doi: 10.1242/dev.122.10.3243. [DOI] [PubMed] [Google Scholar]
- Christensen CA, Subramanian S, Drews GN. Identification of gametophytic mutations affecting female gametophyte development in Arabidopsis. Dev Biol. 1998;202:136–151. doi: 10.1006/dbio.1998.8980. [DOI] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW. Polarized cytokinesis in vacuolate cells of Arabidopsis. Proc Natl Acad Sci USA. 2002;99:2812–2817. doi: 10.1073/pnas.052712299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. Cellular organisation of the Arabidopsis thaliana root. Development. 1993;119:71–84. doi: 10.1242/dev.119.1.71. [DOI] [PubMed] [Google Scholar]
- Field C, Li R, Oegema K. Cytokinesis in eukaryotes: a mechanistic comparison. Curr Opin Cell Biol. 1999;11:68–80. doi: 10.1016/s0955-0674(99)80009-x. [DOI] [PubMed] [Google Scholar]
- Fujie M, Kuroiwa H, Kawano S, Kuroiwa T. Studies on the behavior of organelles and their nucleoids in the root apical meristem of Arabidopsis thaliana (L.) Col. Planta. 1993;189:443–452. doi: 10.1007/BF00194444. [DOI] [PubMed] [Google Scholar]
- Glotzer M. The mechanism and control of cytokinesis. Curr Opin Cell Biol. 1997;9:815–823. doi: 10.1016/s0955-0674(97)80082-8. [DOI] [PubMed] [Google Scholar]
- Hauser BA, He JQ, Park SO, Gasser CS. TSO1 is a novel protein that modulates cytokinesis and cell expansion in Arabidopsis. Development. 2000;127:2219–2226. doi: 10.1242/dev.127.10.2219. [DOI] [PubMed] [Google Scholar]
- Hauser BA, Villanueva JM, Gasser CS. Arabidopsis TSO1 regulates directional processes in cells during floral organogenesis. Genetics. 1998a;150:411–423. doi: 10.1093/genetics/150.1.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser M-T, Adhami F, Dorner M, Fuchs E, Glössl J. Generation of co-dominant PCR based markers by duplex analysis on high resolution gels. Plant J. 1998b;16:117–125. doi: 10.1046/j.1365-313x.1998.00271.x. [DOI] [PubMed] [Google Scholar]
- Hauser M-T, Bauer E. Histochemical analysis of root meristem activity in Arabidopsis thaliana using a cyclin:GUS (β-glucuronidase) marker line. Plant Soil. 2000;226:1–10. [Google Scholar]
- Hauser MT, Morikami A, Benfey PN. Conditional root expansion mutants of Arabidopsis. Development. 1995;121:1237–1252. doi: 10.1242/dev.121.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- His I, Driouich A, Nicol F, Jauneau A, Hoefte H. Altered pectin composition in primary cell walls of korrigan, a dwarf mutant of Arabidopsis deficient in a membrane bound endo-1,4-glucanase. Planta. 2001;212:348–358. doi: 10.1007/s004250000437. [DOI] [PubMed] [Google Scholar]
- Hülskamp M, Parekh NS, Grini P, Schneitz K, Zimmermann I, Lolle SJ, Pruitt RE. The STUD gene is required for male-specific cytokinesis after telophase II of meiosis in Arabidopsis thaliana. Dev Biol. 1997;187:114–124. doi: 10.1006/dbio.1997.8554. [DOI] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Kosambi DD. The estimation of map distances from recombination values. Ann Eugen. 1944;12:172–175. [Google Scholar]
- Lane DR, Wiedemeier A, Peng L, Hofte H, Vernhettes S, Desprez T, Hocart CH, Birch RJ, Baskin TI, Burn JE et al. Temperature-sensitive alleles of RSW2 link the KORRIGAN endo-1,4-β-glucanase to cellulose synthesis and cytokinesis in Arabidopsis. Plant Physiol. 2001;126:278–288. doi: 10.1104/pp.126.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Last RL, Bissinger PH, Mahoney DJ, Radwanski ER, Fink GR. Tryptophan mutants in Arabidopsis the consequences of duplicated tryptophan synthase beta genes. Plant Cell. 1991;3:345–358. doi: 10.1105/tpc.3.4.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister C, Dean C. Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 1993;4:745–750. doi: 10.1046/j.1365-313x.1996.10040733.x. [DOI] [PubMed] [Google Scholar]
- Liu CM, Meinke DW. The titan mutants of Arabidopsis are disrupted in mitosis and cell cycle control during seed development. Plant J. 1998;16:21–31. doi: 10.1046/j.1365-313x.1998.00268.x. [DOI] [PubMed] [Google Scholar]
- Liu Z, Running MP, Meyerowitz EM. TSO1 functions in cell division during Arabidopsis flower development. Development. 1997;124:665–672. doi: 10.1242/dev.124.3.665. [DOI] [PubMed] [Google Scholar]
- Lukowitz W, Mayer U, Jürgens G. Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell. 1996;84:61–71. doi: 10.1016/s0092-8674(00)80993-9. [DOI] [PubMed] [Google Scholar]
- Lukowitz W, Nickle TC, Meinke DW, Last RL, Conklin PL, Somerville CR. Arabidopsis cyt1 mutants are deficient in a mannose-1-phosphate guanylyltransferase and point to a requirement of N-linked glycosylation for cellulose biosynthesis. Proc Natl Acad Sci USA. 2001;98:2262–2267. doi: 10.1073/pnas.051625798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer U, Herzog U, Berger F, Inzé D, Jurgens G. Mutations in the pilz group genes disrupt the microtubule cytoskeleton and uncouple cell cycle progression from cell division in Arabidopsis embryo and endosperm. Eur J Cell Biol. 1999;78:100–108. doi: 10.1016/S0171-9335(99)80011-9. [DOI] [PubMed] [Google Scholar]
- McElver J, Patton D, Rumbaugh M, Liu CM, Yang LY, Meinke D. The TITAN5 gene of Arabidopsis encodes a protein related to the ADP ribosylation factor family of GTP binding proteins. Plant Cell. 2000;12:1379–1392. doi: 10.1105/tpc.12.8.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacry P, Mayer U, Jürgens G. Genetic dissection of cytokinesis. Plant Mol Biol. 2000;43:719–733. doi: 10.1023/a:1006457723760. [DOI] [PubMed] [Google Scholar]
- Niyogi KK, Last RL, Fink GG, Keith B. Suppressor of trp1 fluorescence identify a new Arabidopsis gene, TRP4, encoding the anthranilate synthase beta subunit. Plant Cell. 1993;5:1011–1027. doi: 10.1105/tpc.5.9.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui M, Staehelin LA. Cytokinesis in flowering plants: more than one way to divide a cell. Curr Opin Plant Biol. 2000a;3:493–502. doi: 10.1016/s1369-5266(00)00119-9. [DOI] [PubMed] [Google Scholar]
- Otegui M, Staehelin LA. Syncytial-type cell plates: a novel kind of cell plate involved in endosperm cellularization of Arabidopsis. Plant Cell. 2000b;12:933–947. doi: 10.1105/tpc.12.6.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui MS, Mastronarde DN, Kang BH, Bednarek SY, Staehelin LA. Three-dimensional analysis of syncytial-type cell plates during endosperm cellularization visualized by high resolution electron tomography. Plant Cell. 2001;13:2033–2051. doi: 10.1105/TPC.010150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SK, Howden R, Twell D. The Arabidopsis thaliana gametophytic mutation gemini pollen1 disrupts microspore polarity, division asymmetry and pollen cell fate. Development. 1998;125:3789–3799. doi: 10.1242/dev.125.19.3789. [DOI] [PubMed] [Google Scholar]
- Park SK, Twell D. Novel patterns of ectopic cell plate growth and lipid body distribution in the Arabidopsis gemini pollen1 mutant. Plant Physiol. 2001;126:899–909. doi: 10.1104/pp.126.2.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radcliffe PA, Vardy L, Toda T. A conserved small GTP-binding protein Alp41 is essential for the cofactor-dependent biogenesis of microtubules in fission yeast. FEBS Lett. 2000;468:84–88. doi: 10.1016/s0014-5793(00)01202-3. [DOI] [PubMed] [Google Scholar]
- Samuels AL, Giddings TH, Staehelin LA. Cytokinesis in tobacco BY-2 and root tip cells: a new model of cell plate formation in higher plants. J Cell Biol. 1995;130:1345–1357. doi: 10.1083/jcb.130.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Kato T, Kakegawa K, Ishii T, Liu YG, Awano T, Takabe K, Nishiyama Y, Kuga S, Sato S et al. Role of the putative membrane-bound endo-1,4-beta-glucanase KORRIGAN in cell elongation and cellulose synthesis in Arabidopsis thaliana. Plant Cell Physiol. 2001;42:251–263. doi: 10.1093/pcp/pce045. [DOI] [PubMed] [Google Scholar]
- Scheres B, Wolkenfelt H, Willemsen V, Terlouw M, Lawson E, Dean C, Weisbeek P. Embryonic origin of the Arabidopsis primary root and root meristem initials. Development. 1994;120:2475–2487. [Google Scholar]
- Song JY, Leung T, Ehler LK, Wang C, Liu Z. Regulation of meristem organization and cell division by TSO1, an Arabidopsis gene with cysteine-rich repeats. Development. 2000;127:2207–2217. doi: 10.1242/dev.127.10.2207. [DOI] [PubMed] [Google Scholar]
- Spielman M, Preuss D, Li FL, Browne WE, Scott RJ, Dickinson HG. TETRASPORE is required for male meiotic cytokinesis in Arabidopsis thaliana. Development. 1997;124:2645–2657. doi: 10.1242/dev.124.13.2645. [DOI] [PubMed] [Google Scholar]
- Tzafrir I, McElver JA, Liu CM, Yang LJ, Wu JQ, Martinez A, Patton DA, Meinke DW. Diversity of TITAN functions in Arabidopsis seed development. Plant Physiol. 2002;128:38–51. [PMC free article] [PubMed] [Google Scholar]
- Vantard M, Cowling R, Delichere C. Cell cycle regulation of the microtubular cytoskeleton. Plant Mol Biol. 2000;43:691–703. doi: 10.1023/a:1006346107807. [DOI] [PubMed] [Google Scholar]
- Waizenegger I, Lukowitz W, Assaad F, Schwarz H, Jürgens G, Mayer U. The Arabidopsis KNOLLE and KEULE genes interact to promote vesicle fusion during cytokinesis. Curr Biol. 2000;10:1371–1374. doi: 10.1016/s0960-9822(00)00775-2. [DOI] [PubMed] [Google Scholar]
- Webb MC, Gunning BES. Embryo sac development in Arabidopsis thaliana: I. Megasporogenesis, including the microtubular cytoskeleton. Sex Plant Reprod. 1990;3:244–256. [Google Scholar]
- Webb MC, Gunning BES. Embryo sac development in Arabidopsis thaliana: II. The cytoskeleton during megagametogenesis. Sex Plant Reprod. 1994;7:153–163. [Google Scholar]
- Zuo J, Niu QW, Nishizawa N, Wu Y, Kost B, Chua NH. KORRIGAN, an Arabidopsis endo-1,4-β-glucanase, localizes to the cell plate by polarized targeting and is essential for cytokinesis. Plant Cell. 2000;12:1137–1152. doi: 10.1105/tpc.12.7.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]