Abstract
Advances in high-throughput technologies, such as ChIP–chip, and the completion of human and mouse genomic sequences now allow analysis of the mechanisms of gene regulation on a systems level. In this study, we have developed a computational genomics approach (termed ChIPModules), which begins with experimentally determined binding sites and integrates positional weight matrices constructed from transcription factor binding sites, a comparative genomics approach, and statistical learning methods to identify transcriptional regulatory modules. We began with E2F1 binding site information obtained from ChIP–chip analyses of ENCODE regions, from both HeLa and MCF7 cells. Our approach not only distinguished targets from nontargets with a high specificity, but it also identified five regulatory modules for E2F1. One of the identified modules predicted a colocalization of E2F1 and AP-2α on a set of target promoters with an intersite distance of <270 bp. We tested this prediction using ChIP–chip assays with arrays containing ∼14,000 human promoters. We found that both E2F1 and AP-2α bind within the predicted distance to a large number of human promoters, demonstrating the strength of our sequence-based, unbiased, and universal protocol. Finally, we have used our ChIPModules approach to develop a database that includes thousands of computationally identified and/or experimentally verified E2F1 target promoters.
The completion of human and mouse genome sequences and the increasing availability of gene annotations have made it possible for bioinformaticians to develop new approaches to analyze important biological problems. One such problem is the attempt to catalog the complete set of target genes for each of the ∼2000 transcription factors in the human genome. Numerous computational tools have been developed to facilitate the identification of transcription factor binding sites (TFBSs). One such current strategy is the application of ab initio motif discovery algorithms that search for recurring patterns in a given set of related sequences. Examples of this type of strategy include MEME (Bailey and Gribskov 1997), AlignACE (Roth et al. 1998), and Gibbs Motifs Sampler (Thompson et al. 2003). Another approach is to search for known binding sites based on a precompiled library of all previously characterized motifs or positional weight matrices (PWMs). MATCH (Kel et al. 2003) using the TRANSFAC database (Wingender et al. 2000) and MSCAN (Alkema et al. 2004) using JASPAR (Sandelin et al. 2004a) are two broadly used approaches. Unfortunately, using a strictly bioinformatics-based approach to identify target genes of transcription factors is still extremely challenging because most TFBSs are degenerate sequences that occur quite frequently in the mammalian genome.
Recently, computational strategies have been used in combination with data generated from high-throughput techniques such as gene expression and ChIP–chip. Although computational tools such as MDScan (Liu et al. 2002) and MarsMotifs (Smith et al. 2005) have aided experimental biologists in the discovery of regulatory information, a large false-positive prediction rate is still a major problem. One reason for the high false-positive rate is that some strategies fail to take into consideration other factors that might contribute to functional regulatory networks. Recently, several improvements have been designed to reduce spurious predictions (see Elnitski et al. 2006 for a review of several different computational approaches for identifying TFBS). One improvement applies a comparative genomics approach (phylogenetic footprinting) and is based on the assumption that orthologous genes will be subject to the same regulatory mechanisms in different species. The other improvement expands the analysis beyond the search for a single motif to the identification of cis-regulatory modules (CRMs) and is based on the concept that the biochemical specificity of transcription is generated by combinatorial interactions between transcription factors. To aid in identifying cis-regulatory modules for coexpressed genes, several bioinformatics tools, such as ModuleSearcher and ModuleScanner (Aerts et al. 2003), CREME (Sharan et al. 2003), oPOSSUM (Ho-Sui et al. 2005), CONFAC (Karanam and Moreno 2004), and ROVER (Haverty et al. 2004), have been developed. Also, researchers such as Jin et al. (2004) and Cheng et al. (2006) have combined phylogenetic footprinting and prior knowledge of interacting transcription factor partners to identify Estrogen Receptor alpha target genes from ChIP–chip experimental data and to generate predictions of substantially better specificity than analysis of isolated binding sites in promoter sequences.
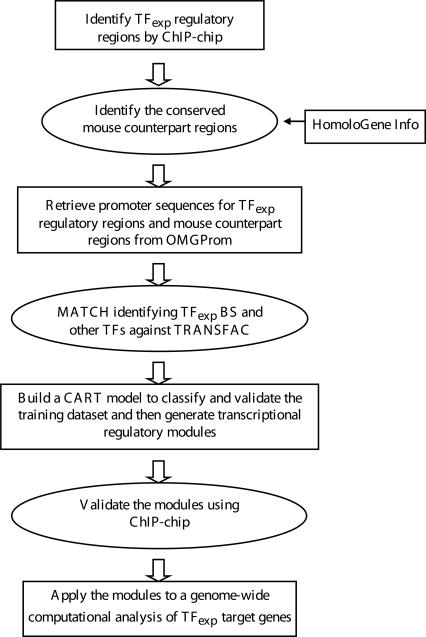
In this study, we have developed a computational genomics approach called ChIPModules (summarized in Figure 1 and Supplemental Figure S5) to identify cis-regulatory modules for human transcription factors. Of critical importance, we began with a set of experimentally identified binding sites for the factor of interest, which in this study was composed of E2F1 binding sites identified by ChIP–chip in 30 Mb of the human genome. We employed PWMs and evolutionary conservation to refine the set of E2F1 sites and then searched for sites for other factors that occur within a short distance of these E2F1 sites. The predicted ChIPModules were then confirmed experimentally using ChIP–chip assays and arrays that contained tens of thousands of human promoters. Finally, we compiled a database that includes both experimentally and computationally identified E2F1 target promoters. The strength of our approach is that it is sequenced-based and unbiased, and can be applied to any set of ChIP–chip experimental data.
Figure 1.
Flow chart showing the ChIPModules approach. Shown is a schematic indicating the steps needed to develop a database of target promoters for a particular site-specific human transcription factor. The approach begins with a set of experimentally defined binding sites (TFexp), refines the set to include only those sites conserved in the orthologous mouse promoters, searches for nearby binding sites for other factors, builds a CART model to generate a high confidence set of co-occurring binding sites, validates the colocalization of the factors using additional ChIP–chip assays, and then searches for the validated ChIPModules in a large promoter database. A detailed description of each step can be found in Supplemental Figure S5.
Results
Collection of the data sets
An outline of our combined experimental and computational ChIPModules approach is shown in Figure 1, and a detailed description of each step is provided in Supplemental Figure S5. As described below, we have applied this approach to analyze E2F1 data sets derived from ChIP–chip experiments performed with two cancer cell lines, HeLa and MCF7 (Bieda et al. 2006). These previous ChIP–chip experiments employed high-density oligonucleotide arrays (termed ENCODE arrays) on which 44 regions of the human genome (The ENCODE Project Consortium 2004) were tiled at a density of one 50-mer every 38 bp. Each region spanned from 500 kb to 1.9 Mb, but repeat regions were not tiled on the array, leading to a total of ∼380,000 probes on the array, which represent the nonrepetitive portion of ∼30 Mb (1%) of the human genome.
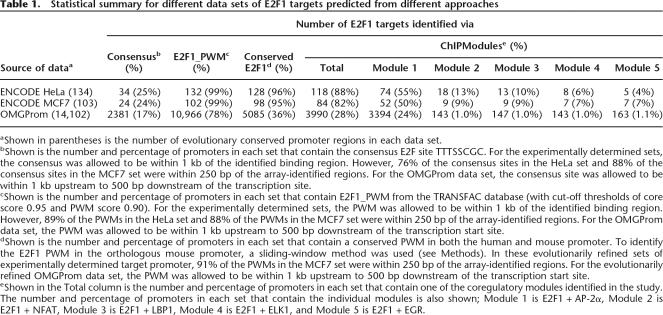
To identify transcriptional regulatory modules, we first defined a training data set (termed ENCODE HeLa) that includes a set of E2F1 binding sites and a set of non-E2F1 target promoters. The E2F1 binding sites were identified by ChIP–chip assays on ENCODE arrays using HeLa cells and were the L1 set, defined as being present in the top 2% of the array data and having a P-value < 0.0001 (Bieda et al. 2006). Using these 205 sites, we identified 134 regulatory regions that were conserved between the human and mouse genomes. As a set of non-E2F1-bound promoters, we selected 98 regions from the ENCODE arrays that did not show enrichment in the E2F1 ChIP–chip assays but were conserved between the human and mouse genomes. Each negative control region was between 500 bp and 1 kb (the approximate length of the E2F1 positive regulatory regions identified by ChIP–chip) and corresponded to a sequence that fell within 5 kb upstream to 2 kb downstream of the start site of a gene. Therefore, for the following analyses, we began with a set of 134 E2F1 target promoter sequences and a set of 98 non-E2F1 target promoter sequences for the initial training data set (Table 1, Source of Data column). As a second training data set (ENCODE MCF7), we used 148 positive regulatory regions identified in E2F1 ChIP–chip experiments using the MCF7 cell line (which were refined to 103 conserved human/mouse regions). We also used a set of 14,102 promoters from the OMGProm database (Palaniswamy et al. 2005) to examine the specificity of the model built from the approaches described in this study.
Table 1.
Statistical summary for different data sets of E2F1 targets predicted from different approaches
aShown in parentheses is the number of evolutionary conserved promoter regions in each data set.
bShown is the number and percentage of promoters in each set that contain the consensus E2F site TTTSSCGC. For the experimentally determined sets, the consensus was allowed to be within 1 kb of the identified binding region. However, 76% of the consensus sites in the HeLa set and 88% of the consensus sites in the MCF7 set were within 250 bp of the array-identified regions. For the OMGProm data set, the consensus site was allowed to be within 1 kb upstream to 500 bp downstream of the transcription site.
cShown is the number and percentage of promoters in each set that contain E2F1_PWM from the TRANSFAC database (with cut-off thresholds of core score 0.95 and PWM score 0.90). For the experimentally determined sets, the PWM was allowed to be within 1 kb of the identified binding region. However, 89% of the PWMs in the HeLa set and 88% of the PWMs in the MCF7 set were within 250 bp of the array-identified regions. For the OMGProm data set, the PWM was allowed to be within 1 kb upstream to 500 bp downstream of the transcription start site.
dShown is the number and percentage of promoters in each set that contain a conserved PWM in both the human and mouse promoter. To identify the E2F1 PWM in the orthologous mouse promoter, a sliding-window method was used (see Methods). In these evolutionarily refined sets of experimentally determined target promoter, 91% of the PWMs in the MCF7 set were within 250 bp of the array-identified regions. For the evolutionarily refined OMGProm data set, the PWM was allowed to be within 1 kb upstream to 500 bp downstream of the transcription start site.
eShown in the Total column is the number and percentage of promoters in each set that contain one of the coregulatory modules identified in the study. The number and percentage of promoters in each set that contain the individual modules is also shown; Module 1 is E2F1 + AP-2α, Module 2 is E2F1 + NFAT, Module 3 is E2F1 + LBP1, Module 4 is E2F1 + ELK1, and Module 5 is E2F1 + EGR.
Refinement of the data sets
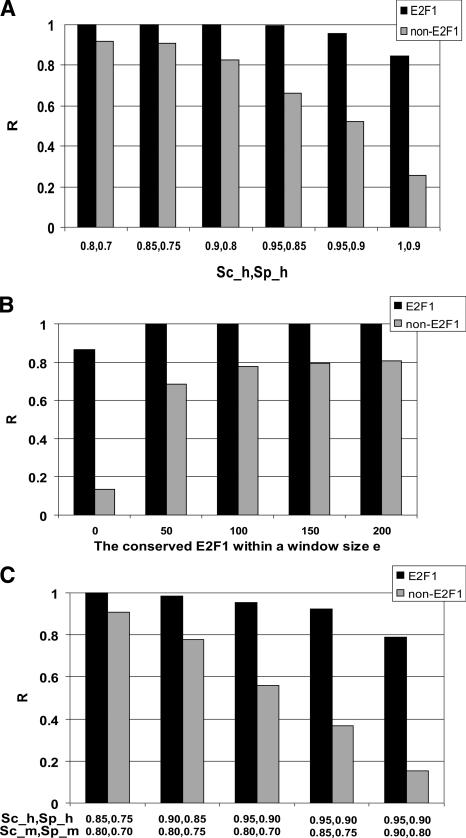
A site bound by a transcription factor is usually modeled by either a consensus sequence or a PWM. To determine the percentage of experimentally determined E2F1 target promoters that contain an E2F consensus site, we searched for the sequence TTTSSCGC within or near the peaks identified using ENCODE arrays. We found that only 34 of the 134 (∼25%) experimentally identified E2F1 target promoters from HeLa cells and only 24 of the 103 (24%) experimentally identified E2F1 target promoters from MCF7 cells contained E2F consensus sites (Table 1, Consensus column). Because it is clear from many lines of experimental evidence that factors such as E2F1 can bind to sequences divergent from the consensus (Tao et al. 1997; Wells et al. 2002; Lavrrar and Farnham 2004), we also used a PWM to identify binding motifs. For the analysis shown here, we used the PWM E2F1_Q3 from the TRANSFAC database (Wingender et al. 2000), which was compiled from 13 experimentally verified human E2F1 binding sites; however, similar results were obtained using alternative E2F1 PWMs from TRANSFAC (data not shown, see Supplemental Figure S3). To define optimal cut-off values for matches in the human promoters to the core of the consensus E2F1 site (Sc_h) and to the E2F1 PWM (Sp_h), we used several combinations of Sc_h (from 0.8 to 1.0) and Sp_h (0.7–0.9) (Fig. 2A). As expected, as the cut-off values increase (i.e., become more stringent), the number of predicted E2F1 target promoters decreases in both the HeLa E2F1 target and the nontarget data sets, with the least predicted number of identified promoters at Sc_h = 1.0 and Sp_h = 0.9. Cut-off values were chosen to be 0.95 for Sc_h and 0.90 for Sp_h, because 99% of the experimentally defined E2F1 target promoters, but only 50% of the negative control set, was positive using these values. Applying these cut-off values to the HeLa and MCF7 data sets, we found that ∼99% of the experimentally identified E2F1 binding sites (in both the HeLa and MCF7 data sets) contained a good match to the E2F1 PWM (Table 1; E2F1 PWM column). However, 78% of the promoters in the OMGProm also contained an E2F1 site using these same parameters. Clearly, additional refinement of the analyses used to predict E2F1 target promoters is needed.
Figure 2.
(A) A histogram graphical representation of the prediction rate (R) that a promoter contains an E2F1 binding site vs. several combinations of the match to the core Sc_h) or E2F1 PWM Sp_h) in the promoter. The values of 0.95 for Sc_h and 0.9 for Sp_h were chosen for the further analyses. (B) A histogram graphical representation of the prediction rate (R) that a human promoter contains an E2F1 binding site vs. a window size (e) for identifying an E2F1 site in the orthologous mouse promoter, using an Sc_m of 0.8 and an Sp_m of 0.7. (C) A histogram graphical representation of the prediction rate (R) that a human promoter contains an E2F1 binding site vs. several combinations of the match to the core sequences (Sc_h and Sc_m) and to the PWMs (Sp_h,, Sp_m) for both human and mouse promoters, using a window size of 100 bp. The values of 0.95 for Sc_h, 0.9 for Sp_H, 0.8 for Sc_m, and 0.7 for Sp_m were chosen for the further analyses.
The next step in our ChIPModules approach was to determine which of the orthologous mouse promoters also contained E2F1 PWMs. For this, we employed a sliding window of various sizes to measure the conservation of the predicted E2F1 sites for each pair of orthologous human and mouse promoter sequences in both the E2F1 sets (HeLa and MCF7) and in the control non-E2F1 set. There are two variables in this experiment: (1) a window size that defines the distance of the E2F1 site identified in the mouse promoter from the exact position it would be predicted to be based on the alignment of the mouse and human promoter regions and (2) the mouse score cut-off values for the match to the consensus E2F site and the E2F1 PWM (Sc_m and Sp_m). We first determined an optimal window size using fixed mouse cut-off values; then we determined optimal cut-off values for mouse scores using the optimized window size. We tested a window size varying from 0 to 200 bp (Fig. 2B) and found that the target set showed a significantly higher prediction of E2F1 binding in the mouse promoters than did the nontarget set, using cut-off values of Sc_m at 0.8 and Sp_m at 0.7. We chose a window size of 100 bp for our next set of analyses; smaller window sizes would have begun reducing the number of promoters in the nontarget training set to a size that would be too small for further analyses. To determine the optimized mouse score cut-off values, we next examined various combinations of human (Sc_h from 0.8 to 1.0; Sp_h from 0.7 to 0.9) and mouse (Sc_m from 0.8 to 0.9; Sp_m from 0.7 to 0.9) cut-off values using a window size of 100 bp (Fig. 2C). We found that at the cut-off values of Sc_h of 0.95, Sp_h of 0.9, Sc_m of 0.8, and Sp_m of 0.7, 128 of 134 E2F1 target promoters in the HeLa set and 98 of the 103 E2F1 target promoters in the MCF7 set are conserved. The use of these parameters has retained >95% of the experimentally identified target promoters but has reduced the number of predicted E2F1 target promoters in the OMGProm database from 78% to only 36% (Table 1, Conserved E2F1 column).
Identifying transcriptional regulatory modules
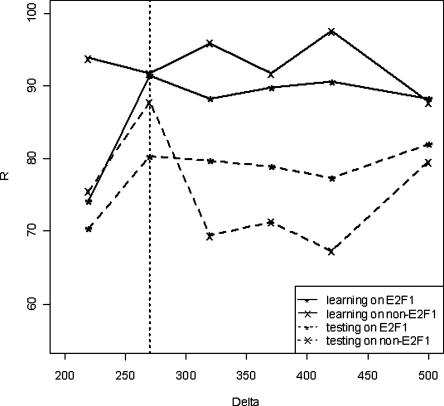
Having refined our set of target promoters, we next searched for binding sites of other transcription factors that were near the identified E2F1 sites in both the human and mouse orthologous promoter pairs. We began with the refined HeLa data set of 128 target promoters and 49 nontarget promoters (having predicted pseudo-E2F1 binding sites). When searching for other transcription factor binding sites, a conservation score of 0.6 was used because several studies (Suzuki et al. 2004; Jin et al. 2006) have shown at least 60% identity for human–mouse orthologous pairs within 2 kb of transcription start sites; a biologically relevant binding site should be conserved at least as well as the surrounding sequence. A set of ∼300 TFBS (i.e., ∼300 different PWMs) from the TRANSFAC database was queried to find those colocalizing near the E2F1 sites using a range of Delta (Δ) values from 220 to 500 bps with an interval of 50 bps. For each Δ value, we first identified a set of PWMs that were found at a higher frequency near the E2F1 PWMs in the E2F1 target promoter set than near the E2F1 PWMs in the nontarget set, using a P-value < 0.05. These PWMs were then used as predictor variables to construct a classification and regression tree (CART) model. For each Δ value, we constructed an optimized model based on the prediction rates on learning samples and testing samples (90% of the training data was used to build the model and 10% was left out of the training set for testing the model; a 10-fold cross-validation was used, see Methods). By evaluating the performance from CART models, the best Δ value (i.e., the optimal value when performance in all four categories is considered) was determined to be 270 bps (Fig. 3). At a Δ value of 270 bps, 24 PWMs were found to be present at a significantly higher frequency (P < 0.05) near the E2F1 binding sites in the E2F1 target promoters than near the predicted pseudo-E2F1 sites in the nontarget promoters and were thus considered to be overrepresented motifs. Of these 24 motifs used for constructing the CART model, the CART model was able to infer 5 transcriptional regulatory modules (E2F1 + AP-2α, E2F1 + NFAT, E2F1 + LBP1, E2F1 + ELK1, and E2F1 + EGR), using a 10-fold cross validation and the “Gini” splitting tree method (see Supplemental Fig. S4 and Supplemental Tables S2A and S2B).
Figure 3.
A graphical representation of the prediction rate (R) vs. Δ values (obtained from the CART results) for different groups of data. (Solid “*” line) Data for E2F1 on training data; (solid “x” line) data for non-E2F1 on training data; (broken “*” line) data for E2F1 on testing data after 10-fold cross validation; (broken “x” line) data for non-E2F1 on testing data after 10-fold cross validation. At a Δ of 270 bp, the prediction rates for E2F1 and non-E2F1 targets on both training and testing samples perform the best.
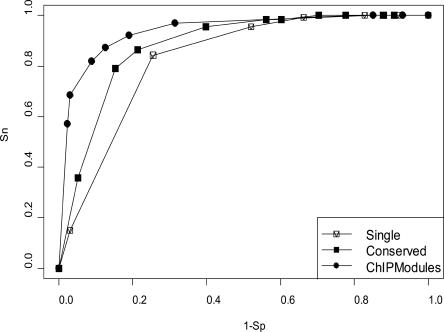
To evaluate the performance of our ChIPModules approach, we used a receiver operating characteristic (ROC) curve, which is a representation of the trade-offs between sensitivity (Sn, the true positive rate; vertical coordinate) and specificity (1 − the false negative rate of Sp; the horizontal coordinate). A ROC curve of a good classifier model will be as close as possible to the upper-left corner of the chart, indicating a high number of true positives and at the same time a small number of false positives. We plotted a ROC curve for the E2F1ENCODE HeLa data set using threedifferent approaches: the presence of an E2F1_PWM in a human promoter (Single), the conservation of E2F1 PWMs in orthologous mouse/human promoters (Conserved), and our ChIPModules approach (Fig. 4). The results clearly show that ChIPModules performed the best among all three approaches with a true positive rate (Sn) of 0.9 and a false positive rate (1 − Sp) of only 0.1.
Figure 4.
ROC curves show that the ChIPModules approach, which has a true positive Sn value of 0.9 and a false positive 1 − Sp value of 0.1, performs better than using only the presence of an E2F1 PWM in a human promoter (Single) or the presence of a conserved E2F1 PWM in both human and mouse promoters (Conserved).
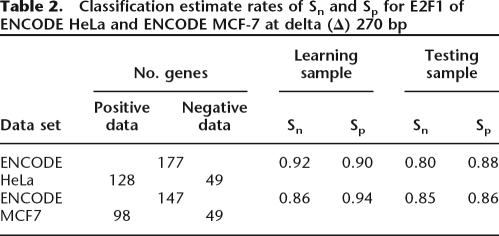
After establishing these parameters for the HeLa ENCODE data, we repeated the process for the MCF7 ENCODE data. The classification results of the CART model using both the HeLa and MCF7 ENCODE ChIP–chip data are listed in Table 2. Both Sn and Sp from 10-fold cross-validation testing samples are >80%. Furthermore, the model predicted ∼90% of E2F1 targets to have at least one of these five modules (Sn of 0.92 in ENCODE HeLa and Sn of 0.86 in ENCODE MCF7 data sets) and ∼90% of non-E2F1 targets to lack any of these modules (Sp of 0.90 in HeLa and Sp of 0.94 in MCF7 data sets).
Table 2.
Classification estimate rates of Sn and Sp for E2F1 of ENCODE HeLa and ENCODE MCF-7 at delta (Δ) 270 bp
The performance of our model was further assessed by using it to identify E2F1 target promoters in the OMGProm database, in comparison to classification of a promoter as an E2F1 target based on the presence of a consensus sequence (TTTSSCGC), or the presence of a conserved E2F1_PWM. Results of these comparisons can be found in Table 1. In all three data sets, the E2F1_PWM predicted the highest rate of putative E2F1 binding sites. Although the 99% PWM predictions for the experimentally determined HeLa and MCF7 sites are perhaps reasonable, this model predicted that as high as 78% of all promoters are E2F1 targets when the PWM is applied to the OMGProm data set. Thus, the E2F1_PWM appears to suffer from an unacceptably high false-positive rate. Although inclusion of the conservation information did enhance the difference in the percentage of targets identified in the experimentally determined sets versus the complete OMGProm set, the ChIPModules approach not only captured 88% of the E2F1 targets from ENCODE HeLa and 82% for ENCODE MCF7 data sets, it also substantially reduced the (assumed) false positive rate for the OMGProm data set.
Experimental validation of the ChIPModules approach
Using our ChIPModules approach, we estimate that 28% of all promoters in the OMGProm database will be possibly bound by E2F1 plus one of the other 5 identified factors (Table 1). By far the highest percentage combination was E2F1 + AP-2α; our ChIPModules approach suggests that ∼24% of the set of OMGProm would be bound by both E2F1 and AP-2α. This prediction can be tested using antibodies to E2F1 and AP-2α in ChIP–chip experiments. Unfortunately, a microarray that exactly corresponds to the set of conserved human and mouse promoters from the OMGProm is not available. However, we did have available a microarray that contains ∼14,000 human promoter regions, each 5 kb in length and represented by 50 oligomers spaced on average 110 bp apart. Although many of these ∼14,000 regions represent the promoter sequences of known genes, some are simply the region surrounding the 5′-most end of a cloned transcript and might not correspond to the actual promoter of that gene. Also, many promoters will be in silenced chromatin in any particular cell type and therefore will not be available for binding by E2F1 and AP-2α. Nevertheless, we would expect that, if our predictions concerning the colocalization of E2F1 and AP-2α are correct, then a large percentage of the experimentally determined E2F1 target promoters should also be bound by AP-2α.
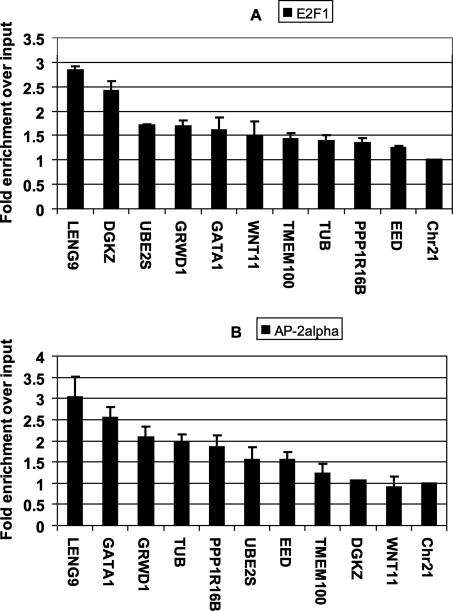
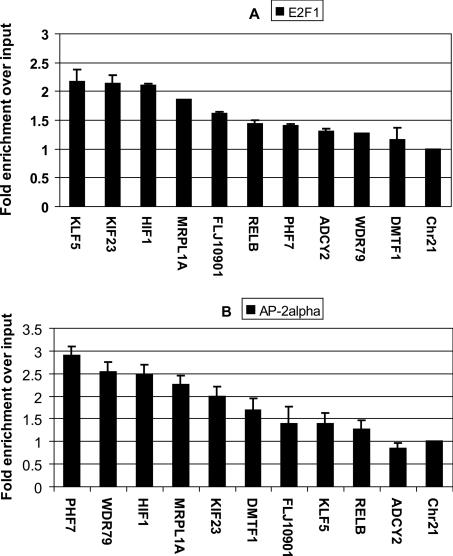
We performed ChIP assays with an antibody that recognizes E2F1 and an antibody that recognizes AP-2α, prepared amplicons from the ChIP samples and a portion of the input chromatin, and applied labeled amplicons to the promoter microarray. After hybridization and scanning, the experimental antibody hybridization signals were divided by the signal from the total input to provide a fold enrichment value for each oligomer on the array. One method to identify target promoters would be to use the median or mean values of the set of 50 oligomers for each promoter to rank all 14,000 promoters. Because a binding region identified by ChIP–chip is only ∼500–1500 bp in length and the promoter regions on the array are 5 kb, it is possible that a promoter could show very high enrichment for both E2F1 and AP-2α, but the sites could be several kb apart. Our ChIPModules approach predicts that the E2F1 and AP-2α binding sites should be within 270 bp of each other. To test our predictions concerning the colocalization of E2F1 and AP-2α on target promoters accurately, it is critical to know the position of the E2F1 and AP-2α sites within the 5 kb regions. Therefore, we developed a computational peak finding program (peaksPicking, see Supplemental Methods and Supplemental Fig. S2) to identify E2F1 and AP-2α binding regions. Using this peak finding program (which requires at least 5 consecutive probes for a region to be called a peak), we first identified the peaks on three E2F1 ChIP–chip arrays and three AP-2α ChIP–chip arrays (Table 3). We then identified the peaks that were called in at least two of three E2F1 arrays (resulting in 2267 E2F1 binding sites) and peaks that were called in at least two of the three AP-2α arrays (resulting in 2624 AP-2α binding sites); a list of these promoters can be found in Supplemental Table S1. Finally, we compared the list of E2F1 peaks and AP-2α peaks and found that of the 2267 promoters bound by E2F1, 925 of them (41%) were also bound by AP-2α, with <270 bp between the two binding sites. Interestingly, the percentage of overlap between the sets of E2F1 and AP-2α promoters does not greatly increase when the distance between the two binding regions is allowed to increase from 270 to 1000 bp (Table 3). This percentage of the overlap promoters is quite significant (P < 10−6), when compared with the <5% overlap called by chance alone when the binding regions are allowed to be up to 1000 bp apart. Thus, experimental ChIP–chip analyses support the identification of ChIPModule 1. To confirm the array data, we randomly chose a set of 10 promoters identified by the arrays as being E2F1 and AP-2α target genes and performed PCR reactions using amplicons prepared from E2F1 and AP-2α ChIP samples; a region of chromosome 21 was used as a negative control. All ten of the promoters showed a higher enrichment of E2F1 than did the negative control; nine of the 10 promoters showed a higher enrichment of AP-2α than did the negative control (Fig. 5).
Table 3.
Promoters bound by both E2F1 and AP-2α
aAllowed gap distance between two binding regions of E2F1 and AP-2α.
bRandom sampling of the data points from the E2F1 and AP-2α arrays was used to generate 1000 random data sets, then 590 peaks were identified at the top 10% level. The overlap targets were then determined using the peaks from the random data sets (a P-value was estimated to be <10−6).
Figure 5.
PCR confirmations of E2F1 (A) and AP-2α (B) binding to a set of 10 randomly selected promoters identified in the ChIP–chip assays. A region of chromosome 21 is used as a negative control. All fold enrichments were compared with the enrichment of the total input and normalized to the negative control. The signals were within the linear range of the assay, providing a semiquantitative analysis. The error bars were calculated based on the standard deviation from two samples of confirmation.
We also used the DAVID program (Dennis et al. 2003) to categorize functionally the E2F1 target genes. We compared the set of genes bound by both E2F1 and AP-2α to the set of genes bound by E2F1 but not AP-2α. Of 925 genes bound by both E2F1 and AP-2α, 509 (55%) had gene ontology information in the DAVID database and were used in our analysis, and of 1342 genes bound by E2F1 but not AP-2α, 759 (57%) had gene ontology information in the DAVID database and were used in our analysis. In the E2F1 plus AP-2α as well as the E2F1 without AP-2α classes, there are 6 categories that compose the majority of the genes; the % of each category and the P-value is shown in Supplemental Table S4. We also randomized the ∼14,000 promoters on the array and chose three sets of 1000 for a similar DAVID analysis. We found that only two categories of genes were enriched in the E2F1 targets, as compared with the randomized sets; these are nucleic acid binding proteins and nuclear proteins. Although the specific genes regulated by E2F1 alone versus E2F1 plus AP-2α are different, the two classes of target genes are both enriched in transcription factors. These results suggest that the major role of E2F1 in the cell is to regulate other transcription factors. Interestingly, although E2F1 was first identified as a factor critical for transcription of cell cycle-regulated genes (Dimova and Dyson 2005 and references therein), the category of cell cycle-regulated genes was only 4% (E2F1 + AP-2α) or 5% (E2F1 − AP-2α) of all E2F1 target genes, suggesting that regulation of the cell cycle is only one of the many functions of E2F1.
Experimental validation of computationally predicted ChIPModule 1 promoters
We began our studies by experimentally identifying E2F1 target promoters using ChIP–chip analysis of the ENCODE regions, which contain ∼400 genes. Using this small data set, we identified 5 ChIPModules and experimentally tested one of these predicted modules (ChIPModule 1; E2F + AP-2α) using an array that contained 14,000 promoters. From these validation ChIP–chip results, we selected a high confidence (the top 10% peaks level) set of experimentally identified E2F1 and AP-2α promoters and found that a large number of these promoters were in fact bound by both factors, with all sites being within 270 bp of each other. To determine whether our ChIPModules approach would have predicted the experimentally identified promoters, we examined the set of 925 commonly bound promoters identified in Table 3 as both E2F1 and AP-2α target promoters in the ChIP–chip data sets. Of these 925 promoters, 587 have human and mouse orthologous pairs but only 502 have conserved E2F1 binding sites and thus could be used for our ChIPModules approach. Of these 502 promoters, 359 (72%) would have been predicted to be in ChIPModule 1 (i.e., bound by both E2F1 and AP-2α) by the ChIPModules approach (see Supplemental Table S2C).
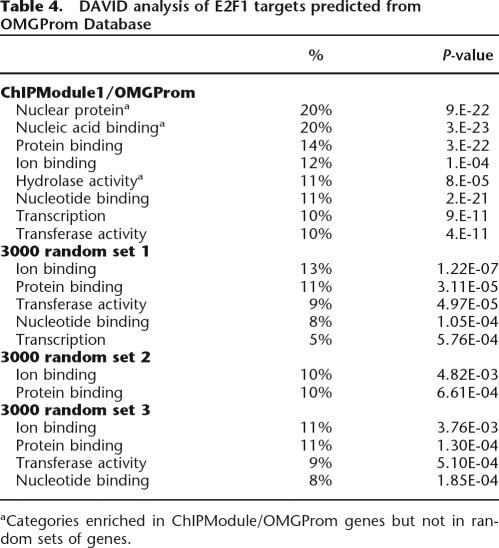
On the basis of the success of our experimental confirmations of the predicted association of E2F1 and AP-2α and the fact that over 70% of the new set of experimentally identified E2F1 and AP-2α target promoters would have been predicted by our ChIPModules approach, we felt confident that we could apply the ChIPModules approach to a large promoter data set (see Supplemental Table S3). Using this approach, we classified 3990 promoters from the OMGProm database for inclusion in the E2F1 + AP-2α ChIPModule 1 (Table 1). A detailed annotation of these 3990 regulatory regions, indicating the predicted binding sites of E2F1 and AP-2α within each of the promoters, is provided in Supplemental Table S2D. We then used the DAVID analysis program to characterize the computationally predicted set of E2F1 + AP-2α promoters. As shown in Table 4, we found that this set of computationally identified E2F1 + AP-2α promoters was very similar to that obtained using DAVID to analyze the experimentally identified E2F1 + AP-2α promoters (see Supplemental Table S4); six of the eight highest categories are found to constitute the same percentages of promoters in both sets. It is possible that the additional category (hydrolase activity) in the computationally identified ChIPModule 1 set from the OMGProm database was not identified in the ChIP–chip assays because of differences in the sets of promoters on the arrays versus in the OMGProm database. To confirm that the computationally identified ChIPModule 1 promoters are in fact bound by E2F1 and AP-2α, we randomly selected 10 predicted ChIPModule 1 promoters (from the OMGProm database) and performed PCR analysis of amplicons prepared from E2F1 and AP-2α ChIP samples. Most of the promoters show higher E2F1 and AP-2α binding than the negative control primer set (Fig. 6).
Table 4.
DAVID analysis of E2F1 targets predicted from OMGProm Database
aCategories enriched in ChIPModule/OMGProm genes but not in random sets of genes.
Figure 6.
PCR confirmations of E2F1 (A) and AP-2α (B) binding to a set of 10 randomly selected promoters predicted by our ChIPModules approach from the OMGProm data set. A region of chromosome 21 is used as a negative control. All fold enrichments were compared to the enrichment of the total input and normalized to the negative control. The signals were within the linear range of the assay, providing a semiquantitative analysis. The error bars were calculated based on the standard deviation from two samples of confirmation.
Discussion
In this study, we have developed a computational genomics approach, termed ChIPModules, to identify transcriptional regulatory modules from ChIP–chip data. ChIPModules integrates PWMs constructed for specific transcription factors, comparative genomics of conserved transcription factor binding sites between the human and mouse orthologous gene pairs, and a robust statistical method termed CART. Importantly, this computational approach begins with, and is then validated by, ChIP–chip experimental assays. Although intensive computational studies have recently focused on ChIP–chip data (Liu et al. 2002; Martin et al. 2004; Zhou and Wong 2004; Gupta and Liu 2005; Hong et al. 2005; Smith et al. 2005; Wang et al. 2005), most of the approaches are limited to motif discovery or improving on existing models and lack a focus on combinatorial regulation among transcription factors. Although Wang et al. (2005) and Zhou and Wong (2004) did focus on predicting combinatorial regulation modules, they did not perform experimental validation of their predictions. Our approach not only systematically infers the combinatorial interaction between a specific transcription factor and its partners from the ChIP–chip data but also incorporates a follow-up ChIP–chip validation step to assess the accuracy of our predictions. Importantly, using the ChIPModules approach we have identified thousands of promoters that are predicted to be cobound by E2F1 and one of five other factors.
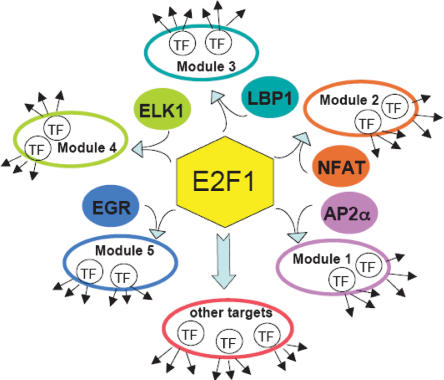
E2F1 is a key regulator in cell cycle progression (Bell and Ryan 2004), has been characterized as both an oncogene and a tumor suppressor gene, and has been extensively studied in our laboratory (Weinmann et al. 2002; Wells et al. 2003) and other laboratories (Ren et al. 2002; Mundle and Saberwal 2003 and references therein). In this study, we used a small set of E2F1 target promoters identified from ChIP–chip studies of the ENCODE regions, discovered five regulatory modules that suggested coregulation by E2F1 and another factor, and then experimentally demonstrated that ChIPModules could successfully classify E2F1 targets. We then used the OMGProm database to determine a large set of predicted E2F1 target promoters. Among these 3990 computationally predicted E2F1 target genes, 3394 were classified into Module 1 (E2F1 + AP-2α), 143 were classified into Module 2 (E2F1 + NFAT), 147 were classified into Module 3 (E2F1 + LBP1), 143 were classified into Module 4 (E2F1 + ELK1), and 163 were classified into Module 5 (E2F1 + EGR). Interestingly, a previous study (Tabach et al. 2005) demonstrated that the transcription factor ELK1, which we identified in Module 4, co-occurs significantly and has synergistic effects with E2F in transformation assays. The most prevalent regulatory module that we identified links E2F1 with AP-2α, which has been characterized as a tumor suppressor gene in breast cancer (Pellikainen et al. 2002; Douglas et al. 2004). We experimentally validated this predicted module using a ChIP–chip approach and identified at least ∼900 promoters that demonstrate binding of E2F1 and AP-2α within 270 bp of each other. The fact that a significant portion of the E2F1-bound promoters are also bound by AP-2α verifies that our approach indeed has the potential to reveal a logic-based regulatory network by modeling combinatorial interactions. Although the specific genes bound by E2F1 alone versus E2F1 plus one of the identified factors in the 5 modules are different, transcription factors are E2F1 targets in all classes. These results suggest that the major role of E2F1 in the cell is to cooperate with and regulate other transcription factors (Fig. 7).
Figure 7.
E2F1 cooperates with and regulates other transcription factors. Shown is a schematic indicating the five different modules identified in this study using E2F1 ChIP–chip data. DAVID analysis of the OMPGProm database-identified promoters (Supplemental Table S3) revealed that transcription factors are a predominant category of target genes in all five E2F1 ChIPModules: Nucleic acid binding proteins constituted ∼20% of ChIPModule 1, 26% of ChIPModule 2, 10% of ChIPModule 3, 26% of ChIPModule 4, and 22% ChIPModule 5.
Although prior to our study there had not been a genome-wide bioinformatics-based analysis of E2F1 target promoters, several previous studies have identified other factors that may coregulate promoters along with E2F family members (Elkon et al. 2003; Cam et al. 2004; Das et al. 2006). For example, Cam et al. (2004) used MDScan (Liu et al. 2002) to identify a motif for NRF1 in a set of experimentally determined E2F4 binding sites. Also, using ChIP–chip data Elkon et al. (2003) predicted functional links between E2F1 and NF-Y, CREB, and NRF-1. There are several possible reasons why the previous studies identified a different set of interacting factors than we identified: (1) the previous ChIP–chip experiments were performed using different cell lines and different promoter arrays; (2) the previous training data sets included E2F4 targets, whereas we only used E2F1 binding site data; (3) we applied a comparative genomics approach; and (4) the different statistical methods applied in the previous approaches may result in different interacting partners being identified. The latter point may be the most critical difference. For instance, we also analyzed our E2F1 target promoters using the program oPOSSUM (Ho-Sui et al. 2005) and found that the ELK1 motif, but not the other ChIPModule motifs, was identified (from a list of the top 15 motifs with P-values < 0.2). It is not clear why AP-2α, NFAT, LBP1, and EGR were not identified using oPOSSUM; however, a major difference is that we have used an advanced classification model CART and only consider the transcription factor motifs that fall within a short distance of the E2F1 PWMs. It is possible that the modules predicted by oPOSSUM and by previous studies (Elkon et al. 2003; Das et al. 2006) are also correct; unfortunately experimental validation of these predictions have not yet been performed. In addition to experimentally verifying that E2F1 and AP-2α bind to the same promoters using ChIP–chip assays, we have also performed two additional specificity controls. First, we performed ChIP–chip experiments with ENCODE arrays using an antibody to OCT4, identified OCT4 binding sites, and then used the experimentally identified OCT4 binding sites in a CART analysis to find colocalizing motifs. Although we did find several motifs that colocalize with the OCT4 binding sites (V.X. Jin, H. O’Geen, and P.J. Farnham, in prep.), AP-2α was not one of the identified motifs. This demonstrates that AP-2α will not be identified as a partner for all transcription factors, providing specificity to our computational identification of E2F1 and AP-2α modules. Second, we performed an additional ChIP–chip analysis using human promoter arrays and an antibody to the transcription factor ZNF217. We found that only 277 (12%) of the 2264 E2F1 were bound by ZNF217 (S.R. Krig, V.X. Jin, and P.J. Farnham, unpubl.). Thus, not all transcription factors colocalize with E2F1, providing specificity for our experimentally determined colocalization of E2F1 and AP-2α. However, we realize that our experiments do not conclusively demonstrate that E2F1 and AP-2α are both bound to a given promoter at the same time. Studies examining co-occupancy of promoters by these two factors will require future experimental analyses such as SeqChIP (Geisberg and Struhl 2004).
The technique of ChIP–chip provides strong in vivo evidence of the recruitment of a specific factor to DNA. However, there are several different mechanisms by which a factor can be recruited to the DNA and be identified in a ChIP–chip assay (Kato et al. 2004). Three types of interactions between a transcription factor and its binding site on DNA include: (1) direct binding of a transcription factor to a high affinity consensus site; (2) piggy-back binding, in which a transcription factor is recruited via protein–protein interactions with another factor that directly interacts with the DNA at a specific motif, and (3) partner-binding, in which a transcription factor directly binds to a low affinity binding site on DNA but specificity is achieved via interaction with another nearby factor bound to a specific motif. A model built based on the identification of a single transcription factor binding to its consensus site systematically eliminates identification of the second and third class of binding sites. However, our ChIPModules approach specifically identifies the third class of target promoters. Previous experimental studies have shown that E2F family members can regulate transcription via cooperation with other DNA binding factors. For example, the studies of Giangrande et al. (2004) suggest that any promoter containing an E box paired with an E2F element is a potential target of E2F3. Also, Schlisio et al. (2002) showed that E2F2 and E2F3, but not E2F1, could interact with YY1 to activate the Cdc6 promoter. Finally, on the basis of our studies of promoters bound specifically by E2F1 but not other E2Fs (Wells et al. 2002), we had previously suggested that the ability of E2F1 to activate promoters that lack an E2F consensus site requires both E2F1/DNA interactions and protein–protein interactions between E2F1 and a factor that binds adjacent to the nonconsensus E2F site (Lavrrar and Farnham 2004). However, all of these previous studies focused on a small set of promoters and did not address the global importance of the identified cooperative interactions. Our current studies suggest that perhaps 80% of all E2F1 target promoters in the HeLa and MCF7 tumor cell lines are regulated by partner-binding, with half of these belonging to ChIPModule 1, E2F1 + AP-2α. Further studies of E2F1 target genes using our ChIPModules approach and data sets derived from ChIP–chip assays of normal and tumor tissues are in progress.
Methods
Promoter sequence retrieval
Orthologous promoter sequences, corresponding attributes, and annotation data were retrieved from an integrated information resource [http://bioinformatics.med.ohio-state.edu/OMGProm) (Palaniswamy et al. 2005)]. Briefly, the OMGProm data were obtained via an efficient data-mining pipeline, which collects experimentally substantiated full-length mRNA/5′UTRs, first exons, and promoters from GenBank (Benson et al. 2003), DBTSS (Suzuki et al. 2002), and EPD (Schmid et al. 2004). A 5′ flanking region of 1 kb upstream to 1 kb downstream of each target gene was designated as a promoter sequence because it is the most extended promoter sequence for each target. Each promoter sequence was then aligned to a mouse orthologous promoter sequence of 10 kb upstream to 10 kb downstream of the transcriptional start site for the orthologous mouse gene by the program ClustalW (Thompson et al. 1994), where the aligned portion of sequences was used to identify the conservation for the orthologous pairs.
Identification of transcription factor binding sites
TFBSs for other factors were identified by the MATCH (Kel et al. 2003) program, using the PWMs from the TRANSFAC database (Wingender et al. 2000). For each pair of human and mouse orthologous promoters, we searched for ∼300 family transcription factors (TFs) with ∼500 PWMs corresponding to known human transcription factors using the “minFN_good83.prf” profile (profile of cut-off values with minimum number of false-negative predictions) of MATCH. Each predicted TFBS was determined by 4 parameters: (1) human core score (Sc_h); (2) human PWM score (Sp_h); (3) mouse core score (Sc_m); and (4) mouse PWM core score (Sp_m). The core and PWM scores, ranging from 0 (worst) to 1 (best), reflect the similarity of predicted sites to the core of the consensus and to the full consensus sequence. We used a sliding-window method similar to the method used by Sandelin et al. (2004b) to measure the degree of conservation of a predicted specific TFBS in a pair of orthologous sequences. A site (denoted M) is considered to be conserved if there is at least one site for a given factor in the orthologous sequences within a given window size (denoted e) and the scores are greater than a threshold (T), where T is a user defined parameter. The conservation of other TFBSs is determined by the percentage of identical base-pairs from the ClustalW aligned sequences.
Identifying combinatorial interactions of transcription factors
The set of experimentally defined TFexp identified by ChIP–chip (in this case, E2F1-bound regions) is denoted as C1, which specifies a set of promoter sequences of n regions, C1 = pro. After we identified the mostly like E2F1 binding site (as described above) for each member of set C1, we focused on searching for other neighborhood TFs within a defined distance Δ on either side of the E2F1 binding site, with Δ ranging from 220 bp to 500 bp. We also did the same procedure for the negative control set C2 (C2 = {C21, C22, . . . , C2m}) of m nonspecific TFregulated promoter sequences (described above). A Fisher’s exact test (two-tailed) was used to calculate the P-value to evaluate the significance of each motif overrepresented in C1 as compared with C2. A set S (S = {TF1, TF2, . . . , TFk}) of the k candidate motifs with a P-value less than a threshold pt (a user defined parameter) was selected to use in the CART model (described below).
CART
CART (Breiman et al. 1984) analysis was employed to develop a classification model for separating the specific TFexp set C1 from the nonspecific TF set C2. The approach is an advanced data-mining tool and it partitions data into discrete classes using user-defined feature variables as predictor variables. To build our CART model, we used the set S of M candidate TFs selected by the above method to produce a binary matrix D for the data sets C1 and C2, where each binding site was considered as a binary variable, such that it was either 1 or 0, depending on its presence within a −Δ bp to +Δ bp region of a specific TFexp (formula 1):
where D is a binary matrix of TFs, yi is the class label for C1 (=0) and C2 (=1), xik is the binary value of TFk that represents presence (=1) or absence (=0) of its binding site within the neighborhood of TF α, N is the number of promoters, M is the number of TFs. The “Gini” method was selected as the splitting method for growing the tree (formula 2):
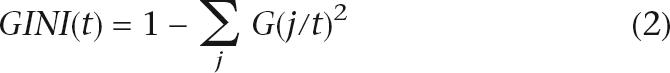 |
where G(j/t) is the relative part of class j at node t.
Our analysis was performed on the commercially available CART software (Salford Systems, San Diego, CA). We used 10-fold cross-validation to estimate the balance of the tree structure produced by CART. The total number of samples are divided into 10 subsamples Z1, Z2, . . . , Z10 of almost equal sizes of N1, N2, . . . , N10. A tree is computed 10 times, each time leaving out one of the subsamples from the computations and using that subsample as a test sample for cross-validation, so that each subsample is used (10 – 1) times in the learning sample and just once as the test sample. This estimate is computed as in formula 3:
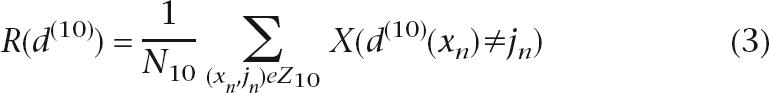 |
where R is the prediction rate, X is the indicator function: X = 1 if the statement X(d(10)(xn)≠jn) is true, X = 0 if the statement X(d(10)(xn)≠jn) is false, and d(10)(x) is the classifier computed from the sub sample Z − Z10.
ROC curve
A ROC curve, which graphically depicts the performance of a classification method for different costs, was employed in evaluating the classifications in our approach. In the curve, the vertical coordinate is a true positive rate termed as sensitivity (Sn), and the horizontal coordinate is a false positive rate termed as 1 − specificity (1 - Sp).
 |
 |
where both TP (a true positive) and TN (a true negative) are correct classifications; both FP (a false positive) and FN (a false negative) are incorrectly classifications.
ChIP–chip assays
MCF7 cells were grown at 37°C in a humidified 5% CO2 incubator in Dulbecco’s Modified Eagle Medium supplemented with 2 mM glutamine, 1% Penicillin/Streptomycin, and 10% fetal bovine serum. ChIP assays were performed as previously described with minor modifications (Weinmann and Farnham 2002). A complete protocol can be found on our Web site at http://genomics.ucdavis.edu/farnham/ and in Oberley and Farnham (2003). Antibodies used in this study include E2F1 (KH20/KH95) (Upstate Biotechnology, cat# 05–379), AP-2α (c-18)x (Santa Cruz Biotechnology cat# sc-184X), and rabbit IgG (Alpha Diagnostic, cat# 210–561–9515). The secondary rabbit anti-mouse IgG (cat# 55436) was purchased from MP Biomedicals. For analysis of the ChIP samples prior to amplicon generation and to confirm target promoters identified by ChIP–chip or ChIPModules, immunoprecipitates were dissolved in 50 μl of water. Standard PCR reactions using 2 μl of the immunoprecipitated DNA were performed. PCR products were separated by electrophoresis through 1.5% agarose gels and visualized by ethidium bromide intercalation. For details concerning the generation of amplicons from ChIP samples, see http://genomics.ucdavis.edu/farnham/ and Bieda et al. (2006). Amplicons were then sent to NimbleGen Systems, Inc. (Madison, WI), where they were hybridized to the 5-kb human promoter array created there. The 5-kb human promoter array design is a two-array set, containing 5.0 kb of each promoter region. Where individual 5.0 kb regions overlap, they are merged into a single larger region, preventing redundancy of coverage. The promoter regions thus range in size from 5.0 kb to 50 kb. These regions are tiled at a 110-bp interval, using variable length probes with a target Tm of 76°C. Only promoter array 2, representing promoter regions on chromosome 11 through chromosome 23, was used for hybridization and the data were extracted according to standard operating procedures by NimbleGen Systems Inc.
DAVID analysis
Functional annotations were performed using the program Database for Annotation, Visualization, and Integrated Discovery (DAVID) 2.1 (Dennis et al. 2003; see also http://apps1.niaid.nih.gov/david/). DAVID is a Web-based, client/server application that allows users to access a relational database of functional annotation. Functional annotations are derived primarily from LocusLink at the National Center for Biotechnology Information (NCBI). DAVID uses LocusLink accession numbers to link gene accessioning systems like GenBank, UniGene, and Affymetrix identifiers to biological annotations including gene names and aliases, functional summaries, Gene Ontologies, protein domains, and biochemical and signal transduction pathways. The same parameters were used for all analyses presented in this study. These parameters were Gene Ontology Molecular Function term, level 2; Interpro name in the Protein Domains section; and SP_PIR_Keywords in the Functional Categories section. After performing the analysis, all categories that represented <5% of the total number of genes were eliminated. In addition, redundant terms (e.g., transcriptional regulation and transcription factor activity) and noninformative terms (e.g., multigene family) were also eliminated.
Acknowledgments
This work was supported in part by Public Health Service grant CA45250, HG003129, and DK067889. As part of our analyses, we used ChIP–chip data collected as part of the ENCODE Project Consortium (Bieda et al. 2006). We thank the members of the Farnham laboratory for helpful discussion and Celina Mojica for excellent technical assistance. Finally, we thank the ENCODE Project Consortium for discussion and support.
Footnotes
[Supplemental material is available online at www.genome.org. The E2F1 and AP-2α ChIP–chip data are deposited in GEO (GEO series # GSE5175, which includes GPL3930 and GSM116738–GSM116742)].
Article published online before print. Article and publication data are at http://www.genome.org/cgi/doi/10.1101/gr.5520206
References
- Aerts S., Van Loo P., Thijs G., Moreau Y., De Moor B., Van Loo P., Thijs G., Moreau Y., De Moor B., Thijs G., Moreau Y., De Moor B., Moreau Y., De Moor B., De Moor B. Computational detection of cis-regulatory modules. Bioinformatics. 2003;19:ii5–ii14. doi: 10.1093/bioinformatics/btg1052. [DOI] [PubMed] [Google Scholar]
- Alkema W.B., Johansson O., Lagergren J., Wasserman W.W., Johansson O., Lagergren J., Wasserman W.W., Lagergren J., Wasserman W.W., Wasserman W.W. MSCAN: Identification of functional clusters of transcription factor binding sites. Nucleic Acids Res. 2004;32:W195–W198. doi: 10.1093/nar/gkh387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L., Gribskov M., Gribskov M. Score distributions for simultaneous matching to multiple motifs. J. Comput. Biol. 1997;4:45–59. doi: 10.1089/cmb.1997.4.45. [DOI] [PubMed] [Google Scholar]
- Bell L.A., Ryan K.M., Ryan K.M. Life and death decisions by E2F-1. Cell Death Differ. 2004;11:137–142. doi: 10.1038/sj.cdd.4401324. [DOI] [PubMed] [Google Scholar]
- Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L., Lipman D.J., Ostell J., Wheeler D.L., Ostell J., Wheeler D.L., Wheeler D.L. GenBank. Nucleic Acids Res. 2003;31:23–27. doi: 10.1093/nar/gkg057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieda M., Xu X., Singer M., Green R., Farnham P.J., Xu X., Singer M., Green R., Farnham P.J., Singer M., Green R., Farnham P.J., Green R., Farnham P.J., Farnham P.J. Unbiased location analysis of E2F1 binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 2006;16:595–605. doi: 10.1101/gr.4887606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L., Friedman J.H., Olshen R.A., Stone C.J., Friedman J.H., Olshen R.A., Stone C.J., Olshen R.A., Stone C.J., Stone C.J. Classification and regression trees. Chapman & Hall; New York: 1984. [Google Scholar]
- Cam H., Balciunaite E., Blais A., Spektor A., Scarpulla R.C., Young R., Kluger Y., Dynlacht B.D., Balciunaite E., Blais A., Spektor A., Scarpulla R.C., Young R., Kluger Y., Dynlacht B.D., Blais A., Spektor A., Scarpulla R.C., Young R., Kluger Y., Dynlacht B.D., Spektor A., Scarpulla R.C., Young R., Kluger Y., Dynlacht B.D., Scarpulla R.C., Young R., Kluger Y., Dynlacht B.D., Young R., Kluger Y., Dynlacht B.D., Kluger Y., Dynlacht B.D., Dynlacht B.D. A common set of gene regulatory networks links metabolism and growth inhibition. Mol. Cell. 2004;16:399–411. doi: 10.1016/j.molcel.2004.09.037. [DOI] [PubMed] [Google Scholar]
- Cheng A.S., Jin V.X., Fan M., Smith L.T., Liyanarachchi S., Yan P.S., Leu Y.W., Chan M.W., Plass C., Nephew K.P., Jin V.X., Fan M., Smith L.T., Liyanarachchi S., Yan P.S., Leu Y.W., Chan M.W., Plass C., Nephew K.P., Fan M., Smith L.T., Liyanarachchi S., Yan P.S., Leu Y.W., Chan M.W., Plass C., Nephew K.P., Smith L.T., Liyanarachchi S., Yan P.S., Leu Y.W., Chan M.W., Plass C., Nephew K.P., Liyanarachchi S., Yan P.S., Leu Y.W., Chan M.W., Plass C., Nephew K.P., Yan P.S., Leu Y.W., Chan M.W., Plass C., Nephew K.P., Leu Y.W., Chan M.W., Plass C., Nephew K.P., Chan M.W., Plass C., Nephew K.P., Plass C., Nephew K.P., Nephew K.P., et al. Combinatorial analysis of transcription factor partners reveals recruitment of c-Myc to estrogen receptor-α responsive promoters. Mol. Cell. 2006;21:393–404. doi: 10.1016/j.molcel.2005.12.016. [DOI] [PubMed] [Google Scholar]
- Das D., Nahle Z., Zhang M.Q., Nahle Z., Zhang M.Q., Zhang M.Q. Adaptively inferring human transcriptional subnetworks. Mol. Syst. Biol. 2006;2:E1–E14. doi: 10.1038/msb4100067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G.J., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A., Yang J., Gao W., Lane H.C., Lempicki R.A., Gao W., Lane H.C., Lempicki R.A., Lane H.C., Lempicki R.A., Lempicki R.A. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- Dimova D., Dyson N., Dyson N. The E2F transcriptional network: Old acquaintances with new faces. Oncogene. 2005;24:2810–2826. doi: 10.1038/sj.onc.1208612. [DOI] [PubMed] [Google Scholar]
- Douglas D.B., Akiyama Y., Carraway H., Belinsky S.A., Esteller M., Gabrielson E., Weitzman S., Williams T., Herman J.G., Baylin S.B., Akiyama Y., Carraway H., Belinsky S.A., Esteller M., Gabrielson E., Weitzman S., Williams T., Herman J.G., Baylin S.B., Carraway H., Belinsky S.A., Esteller M., Gabrielson E., Weitzman S., Williams T., Herman J.G., Baylin S.B., Belinsky S.A., Esteller M., Gabrielson E., Weitzman S., Williams T., Herman J.G., Baylin S.B., Esteller M., Gabrielson E., Weitzman S., Williams T., Herman J.G., Baylin S.B., Gabrielson E., Weitzman S., Williams T., Herman J.G., Baylin S.B., Weitzman S., Williams T., Herman J.G., Baylin S.B., Williams T., Herman J.G., Baylin S.B., Herman J.G., Baylin S.B., Baylin S.B. Hypermethylation of a small CpGuanine-rich region correlates with loss of activator protein-2α expression during progression of breast cancer. Cancer Res. 2004;64:1611–1624. doi: 10.1158/0008-5472.can-0318-2. [DOI] [PubMed] [Google Scholar]
- Elkon R., Linhart C., Sharan R., Shamir R., Shiloh Y., Linhart C., Sharan R., Shamir R., Shiloh Y., Sharan R., Shamir R., Shiloh Y., Shamir R., Shiloh Y., Shiloh Y. Genome-wide in silico identification of transcriptional regulators controlling the cell cycle in human cells. Genome Res. 2003;13:773–780. doi: 10.1101/gr.947203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elnitski L., Jin V.X., Farnham P.J., Jones S.J.M., Jin V.X., Farnham P.J., Jones S.J.M., Farnham P.J., Jones S.J.M., Jones S.J.M. Locating mammalian transcription factor binding sites: A survey of computational and experimental techniques. Genome Res. 2006 doi: 10.1101/gr.4140006. (this issue) [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium The ENCODE (ENCyclopedia of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- Geisberg J.V., Struhl K., Struhl K. Quantitative sequential chromatin immunoprecipitation, a method for analyzing co-occupancy of proteins at genomic regions in vivo. Nucleic Acids Res. 2004;32:e151. doi: 10.1093/nar/gnh148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giangrande P.H., Zhu W., Rempel R.E., Laakso N., Nevins J.R., Zhu W., Rempel R.E., Laakso N., Nevins J.R., Rempel R.E., Laakso N., Nevins J.R., Laakso N., Nevins J.R., Nevins J.R. Combinatorial gene control involving E2F and E Box family members. EMBO J. 2004;23:1336–1347. doi: 10.1038/sj.emboj.7600134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M., Liu J.S., Liu J.S. De novo cis-regulatory module elicitation for eukaryotic genomes. Proc. Natl. Acad. Sci. 2005;102:7079–7084. doi: 10.1073/pnas.0408743102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haverty P.M., Hansen U., Weng Z., Hansen U., Weng Z., Weng Z. Computational inference of transcriptional regulatory networks from expression profiling and transcription factor binding site identification. Nucleic Acids Res. 2004;32:179–188. doi: 10.1093/nar/gkh183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong P., Liu X.S., Zhou Q., Lu X., Liu J.S., Wong W.H., Liu X.S., Zhou Q., Lu X., Liu J.S., Wong W.H., Zhou Q., Lu X., Liu J.S., Wong W.H., Lu X., Liu J.S., Wong W.H., Liu J.S., Wong W.H., Wong W.H. A boosting approach for motif modeling using ChIP–chip data. Bioinformatics. 2005;21:2636–2643. doi: 10.1093/bioinformatics/bti402. [DOI] [PubMed] [Google Scholar]
- Ho-Sui S.J., Mortimer J., Arenillas D.J., Brumm J., Walsh C.J., Kennedy B.P., Wasserman W.W., Mortimer J., Arenillas D.J., Brumm J., Walsh C.J., Kennedy B.P., Wasserman W.W., Arenillas D.J., Brumm J., Walsh C.J., Kennedy B.P., Wasserman W.W., Brumm J., Walsh C.J., Kennedy B.P., Wasserman W.W., Walsh C.J., Kennedy B.P., Wasserman W.W., Kennedy B.P., Wasserman W.W., Wasserman W.W. oPOSSUM: Identification of over-represented transcription factor binding sites in co-expressed genes. Nucleic Acids Res. 2005;33:3154–3164. doi: 10.1093/nar/gki624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin V.X., Leu Y.W., Liyanarachchi S., Sun H., Fan M., Nephew K.P., Huang T.H., Davuluri R.V., Leu Y.W., Liyanarachchi S., Sun H., Fan M., Nephew K.P., Huang T.H., Davuluri R.V., Liyanarachchi S., Sun H., Fan M., Nephew K.P., Huang T.H., Davuluri R.V., Sun H., Fan M., Nephew K.P., Huang T.H., Davuluri R.V., Fan M., Nephew K.P., Huang T.H., Davuluri R.V., Nephew K.P., Huang T.H., Davuluri R.V., Huang T.H., Davuluri R.V., Davuluri R.V. Identifying estrogen receptor α target genes using integrated computational genomics and chromatin immunoprecipitation microarray. Nucleic Acids Res. 2004;32:6627–6635. doi: 10.1093/nar/gkh1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin V.X., Singer G.A., Agosto-Perez F.J., Liyanarachchi S., Davuluri R.V., Singer G.A., Agosto-Perez F.J., Liyanarachchi S., Davuluri R.V., Agosto-Perez F.J., Liyanarachchi S., Davuluri R.V., Liyanarachchi S., Davuluri R.V., Davuluri R.V. Genome-wide analysis of core promoter elements from conserved human and mouse orthologous pairs. BMC Bioinformatics. 2006;7:114. doi: 10.1186/1471-2105-7-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanam S., Moreno C.S., Moreno C.S. CONFAC: Automated application of comparative genomic promoter analysis to DNA microarray data sets. Nucleic Acids Res. 2004;32:W475–W484. doi: 10.1093/nar/gkh353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Hata N., Banerjee N., Futcher B., Zhang M.Q., Hata N., Banerjee N., Futcher B., Zhang M.Q., Banerjee N., Futcher B., Zhang M.Q., Futcher B., Zhang M.Q., Zhang M.Q. Identifying combinatorial regulation of transcription factors and binding motifs. Genome Biol. 2004;5:R56. doi: 10.1186/gb-2004-5-8-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kel A.E., Gossling E., Reuter I., Cheremushkin E., Kel-Margoulis O.V., Wingender E., Gossling E., Reuter I., Cheremushkin E., Kel-Margoulis O.V., Wingender E., Reuter I., Cheremushkin E., Kel-Margoulis O.V., Wingender E., Cheremushkin E., Kel-Margoulis O.V., Wingender E., Kel-Margoulis O.V., Wingender E., Wingender E. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavrrar J.L., Farnham P.J., Farnham P.J. The use of transient chromatin immunoprecipitation assays to test models for E2F1-specific transcriptional activation. J. Biol. Chem. 2004;279:46343–46349. doi: 10.1074/jbc.M402692200. [DOI] [PubMed] [Google Scholar]
- Liu X.S., Brutlag D.L., Liu J.S., Brutlag D.L., Liu J.S., Liu J.S. An algorithm for finding protein-DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat. Biotechnol. 2002;20:835–839. doi: 10.1038/nbt717. [DOI] [PubMed] [Google Scholar]
- Martin C.F., Fu Y., Yu L., Chen J., Hansen U., Weng Z., Fu Y., Yu L., Chen J., Hansen U., Weng Z., Yu L., Chen J., Hansen U., Weng Z., Chen J., Hansen U., Weng Z., Hansen U., Weng Z., Weng Z. Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res. 2004;32:1372–1381. doi: 10.1093/nar/gkh299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundle S.D., Saberwal S., Saberwal S. Evolving intricacies and implications of E2F1 regulation. FASEB J. 2003;17:569–574. doi: 10.1096/fj.02-0431rev. [DOI] [PubMed] [Google Scholar]
- Oberley M.J., Farnham P.J., Farnham P.J. Probing chromatin immunoprecipitates with CpG island microarrays to identify genomic sites occupied by DNA-binding proteins. Methods Enzymol. 2003;371:577–596. doi: 10.1016/S0076-6879(03)71043-X. [DOI] [PubMed] [Google Scholar]
- Palaniswamy S.K., Jin V.X., Sun H., Davuluri R.V., Jin V.X., Sun H., Davuluri R.V., Sun H., Davuluri R.V., Davuluri R.V. OMGProm: An integrated resource of orthologous mammalian gene promoters. Bioinformatics. 2005;21:835–836. doi: 10.1093/bioinformatics/bti119. [DOI] [PubMed] [Google Scholar]
- Pellikainen J., Kataja V., Ropponen K., Kellokoski J., Pietilainen T., Bohm J., Eskelinen M., Kosma V.M., Kataja V., Ropponen K., Kellokoski J., Pietilainen T., Bohm J., Eskelinen M., Kosma V.M., Ropponen K., Kellokoski J., Pietilainen T., Bohm J., Eskelinen M., Kosma V.M., Kellokoski J., Pietilainen T., Bohm J., Eskelinen M., Kosma V.M., Pietilainen T., Bohm J., Eskelinen M., Kosma V.M., Bohm J., Eskelinen M., Kosma V.M., Eskelinen M., Kosma V.M., Kosma V.M. Reduced nuclear expression of transcription factor AP-2 associates with aggressive breast cancer. Clin. Cancer Res. 2002;8:3487–3495. [PubMed] [Google Scholar]
- Ren B., Cam H., Takahashi Y., Volkert T., Terragni J., Young R.A., Dynlacht B.D., Cam H., Takahashi Y., Volkert T., Terragni J., Young R.A., Dynlacht B.D., Takahashi Y., Volkert T., Terragni J., Young R.A., Dynlacht B.D., Volkert T., Terragni J., Young R.A., Dynlacht B.D., Terragni J., Young R.A., Dynlacht B.D., Young R.A., Dynlacht B.D., Dynlacht B.D. E2F integrates cell cycle progression with DNA repair, replication, and G2/M checkpoints. Genes & Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth F.P., Hughes J.D., Estep P.W., Church G.M., Hughes J.D., Estep P.W., Church G.M., Estep P.W., Church G.M., Church G.M. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat. Biotechnol. 1998;16:939–945. doi: 10.1038/nbt1098-939. [DOI] [PubMed] [Google Scholar]
- Sandelin A., Alkema W., Engstrom P., Wasserman W.W., Lenhard B., Alkema W., Engstrom P., Wasserman W.W., Lenhard B., Engstrom P., Wasserman W.W., Lenhard B., Wasserman W.W., Lenhard B., Lenhard B. JASPAR: An open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004a;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A., Wasserman W.W., Lenhard B., Wasserman W.W., Lenhard B., Lenhard B. ConSite: Web-based prediction of regulatory elements using cross-species comparison. Nucleic Acids Res. 2004b;32:W249–W252. doi: 10.1093/nar/gkh372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlisio S., Halperin T., Vidal M., Nevins J.R., Halperin T., Vidal M., Nevins J.R., Vidal M., Nevins J.R., Nevins J.R. Interaction of YY1 with E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. EMBO J. 2002;21:5775–5786. doi: 10.1093/emboj/cdf577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid C.D., Praz V., Delorenzi M., Perier R., Bucher P., Praz V., Delorenzi M., Perier R., Bucher P., Delorenzi M., Perier R., Bucher P., Perier R., Bucher P., Bucher P. The Eukaryotic Promoter Database EPD: The impact of in silico primer extension. Nucleic Acids Res. 2004;32:D82–D85. doi: 10.1093/nar/gkh122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan R., Ovcharenko I., Ben-Hur A., Karp R.M., Ovcharenko I., Ben-Hur A., Karp R.M., Ben-Hur A., Karp R.M., Karp R.M. CREME: A framework for identifying cis-regulatory modules in human–mouse conserved segments. Bioinformatics. 2003;19:i283–i291. doi: 10.1093/bioinformatics/btg1039. [DOI] [PubMed] [Google Scholar]
- Smith A.D., Sumazin P., Das D., Zhang M.Q., Sumazin P., Das D., Zhang M.Q., Das D., Zhang M.Q., Zhang M.Q. Mining ChIP–chip data for transcription factor and cofactor binding sites. Bioinformatics. 2005;21:i403–i412. doi: 10.1093/bioinformatics/bti1043. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Yamashita R., Nakai N., Sugano S., Yamashita R., Nakai N., Sugano S., Nakai N., Sugano S., Sugano S. DBTSS: DataBase of Human Transcriptional Start Sites and full-length cDNAs. Nucleic Acids Res. 2002;30:328–331. doi: 10.1093/nar/30.1.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Yamashita R., Shirota M., Sakakibara Y., Chiba J., Sugano J.M., Nakai K., Sugano S., Yamashita R., Shirota M., Sakakibara Y., Chiba J., Sugano J.M., Nakai K., Sugano S., Shirota M., Sakakibara Y., Chiba J., Sugano J.M., Nakai K., Sugano S., Sakakibara Y., Chiba J., Sugano J.M., Nakai K., Sugano S., Chiba J., Sugano J.M., Nakai K., Sugano S., Sugano J.M., Nakai K., Sugano S., Nakai K., Sugano S., Sugano S. Sequence comparison of human and mouse genes reveals a homologous block structure in the promoter regions. Genome Res. 2004;14:1711–1718. doi: 10.1101/gr.2435604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabach Y., Milyavsky M., Shats I., Brosh R., Zuk O., Yitzhaky A., Mantovani R., Domany E., Rotter V., Pilpel Y., Milyavsky M., Shats I., Brosh R., Zuk O., Yitzhaky A., Mantovani R., Domany E., Rotter V., Pilpel Y., Shats I., Brosh R., Zuk O., Yitzhaky A., Mantovani R., Domany E., Rotter V., Pilpel Y., Brosh R., Zuk O., Yitzhaky A., Mantovani R., Domany E., Rotter V., Pilpel Y., Zuk O., Yitzhaky A., Mantovani R., Domany E., Rotter V., Pilpel Y., Yitzhaky A., Mantovani R., Domany E., Rotter V., Pilpel Y., Mantovani R., Domany E., Rotter V., Pilpel Y., Domany E., Rotter V., Pilpel Y., Rotter V., Pilpel Y., Pilpel Y. The promoters of human cell cycle genes integrate signals from two tumor suppressive pathways during cellular transformation. Mol. Syst. Biol. 2005;1:E1–E15. doi: 10.1038/msb4100030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Kassatly R., Cress W.D., Horowitz J.M., Kassatly R., Cress W.D., Horowitz J.M., Cress W.D., Horowitz J.M., Horowitz J.M. Subunit composition determines E2F DNA-binding site specificity. Mol. Cell. Biol. 1997;17:6994–7007. doi: 10.1128/mcb.17.12.6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J., Higgins D.G., Gibson T.J., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson W., Rouchka E.C., Lawrence C.E., Rouchka E.C., Lawrence C.E., Lawrence C.E. Gibbs Recursive Sampler: Finding transcription factor binding sites. Nucleic Acids Res. 2003;31:3580–3585. doi: 10.1093/nar/gkg608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Cherry J.M., Nochomovitz Y., Jolly E., Botstein D., Li H., Cherry J.M., Nochomovitz Y., Jolly E., Botstein D., Li H., Nochomovitz Y., Jolly E., Botstein D., Li H., Jolly E., Botstein D., Li H., Botstein D., Li H., Li H. Inference of combinatorial regulation in yeast transcriptional networks: A case study of sporulation. Proc. Natl. Acad. Sci. 2005;102:1998–2003. doi: 10.1073/pnas.0405537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann A.S., Farnham P.J., Farnham P.J. Identification of unknown target genes of human transcription factors through the use of chromatin immunoprecipitation. Methods. 2002;26:37–47. doi: 10.1016/S1046-2023(02)00006-3. [DOI] [PubMed] [Google Scholar]
- Weinmann A.S., Yan P.S., Oberley M.J., Huang T.H.-M., Farnham P.J., Yan P.S., Oberley M.J., Huang T.H.-M., Farnham P.J., Oberley M.J., Huang T.H.-M., Farnham P.J., Huang T.H.-M., Farnham P.J., Farnham P.J. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes & Dev. 2002;16:235–244. doi: 10.1101/gad.943102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J., Graveel C.R., Bartley S.M., Madore S.J., Farnham P.J., Graveel C.R., Bartley S.M., Madore S.J., Farnham P.J., Bartley S.M., Madore S.J., Farnham P.J., Madore S.J., Farnham P.J., Farnham P.J. The identification of E2F1-specific genes. Proc. Natl. Acad. Sci. 2002;99:3890–3895. doi: 10.1073/pnas.062047499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J., Yan P.S., Cechvala M., Huang T., Farnham P.J., Yan P.S., Cechvala M., Huang T., Farnham P.J., Cechvala M., Huang T., Farnham P.J., Huang T., Farnham P.J., Farnham P.J. Identification of novel pRb binding sites using CpG microarrays suggests that E2F recruits pRb to specific genomic sties during S phase. Oncogene. 2003;22:1445–1460. doi: 10.1038/sj.onc.1206264. [DOI] [PubMed] [Google Scholar]
- Wingender E., Chen X., Hehl R., Karas H., Matys V., Meinhardt T., Pruss M., Reuter I., Schacherer F., Chen X., Hehl R., Karas H., Matys V., Meinhardt T., Pruss M., Reuter I., Schacherer F., Hehl R., Karas H., Matys V., Meinhardt T., Pruss M., Reuter I., Schacherer F., Karas H., Matys V., Meinhardt T., Pruss M., Reuter I., Schacherer F., Matys V., Meinhardt T., Pruss M., Reuter I., Schacherer F., Meinhardt T., Pruss M., Reuter I., Schacherer F., Pruss M., Reuter I., Schacherer F., Reuter I., Schacherer F., Schacherer F. TRANSFAC: An integrated system for gene expression regulation. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Wong W.H., Wong W.H. CisModule: De novo discovery of cis-regulatory modules by hierarchical mixture modeling. Proc. Natl. Acad. Sci. 2004;101:12114–12119. doi: 10.1073/pnas.0402858101. [DOI] [PMC free article] [PubMed] [Google Scholar]