Abstract
Glutathione S-transferase (GST) gene expression was examined in several Triticum species, differing in genome constitution and ploidy level, to determine genome contribution to GST expression in cultivated, hexaploid bread wheat (Triticum aestivum). Two tandemly duplicated tau class GST genes (TtGSTU1 and TtGSTU2) were isolated from a single bacterial artificial chromosome clone in a library constructed from the diploid wheat and D genome progenitor to cultivated wheat, Triticum tauschii. The genes are very similar in genomic structure and their encoded proteins are 95% identical. Gene-specific reverse transcriptase-polymerase chain reaction analysis revealed differential transcript accumulation of TtGSTU1 and TtGSTU2 in roots and shoots. Expression of both genes was induced by herbicide safeners, 2,4-dichlorophenoxyacetic acid and abscisic acid, in the shoots of T. tauschii; however, expression of TtGSTU1 was always higher than TtGSTU2. In untreated seedlings, TtGSTU1 was expressed in both shoots and roots, whereas TtGSTU2 expression was only detected in roots. RNA gel-blot analysis of ditelosomic, aneuploid lines that are deficient for 6AS, 6BS, or 6DS chromosome arms of cultivated, hexaploid bread wheat showed differential genome contribution to safener-induced GST expression in shoots compared with roots. The GST genes from the D genome of hexaploid wheat contribute most to safener-induced expression in the shoots, whereas GSTs from the B and D genomes contribute to safener-induced expression in the roots.
Glutathione S-transferases (GSTs) belong to multigene families common to all plants (Edwards et al., 2000; McGonigle et al., 2000). They are well known for their responses to numerous endogenous and xenobiotic stresses, and glutathione conjugation of toxic electrophilic molecules. The roles of GST proteins in endogenous plant metabolism, as well as their role in stress tolerance, have yet to be clearly defined. GST gene expression is induced after exposure to many stresses, including biotic stresses such as pathogen attack and fungal elicitors, and abiotic stresses such as heat shock, cold, high salt, UV light exposure, heavy metals, and herbicides. Phytohormone treatments such as ethylene, auxins, abscisic acid (ABA), methyl jasmonate, and salicylic acid have also been shown to induce expression of GSTs (for review, see Marrs, 1996; Dixon et al., 1998; Edwards et al., 2000). Induction of GST expression by so many diverse stimuli implies that plant GSTs are critical in plant response to stress, either by participating in the signal transduction process and/or detoxifying harmful compounds produced in response to or as a result of a given stress. It is likely that GST gene expression is induced by conditions that lead to oxidative stress (Polidoros and Scandalios, 1999). The encoded GST proteins play an important but poorly understood role in plant response to stress, possibly through the central role of antioxidant function. GST enzymatic activity could involve direct glutathione conjugation to toxic electrophilic molecules, or gluta-thione-dependent peroxidase activity, using glutathione as reductant for the detoxification of toxic oxygen species, oxygen radicals, and lipid peroxides formed during or after plant stress (Dixon et al., 1998; Edwards et al., 2000). One biochemical function of GST proteins that is well defined is their role in herbicide metabolism in crops. GSTs are the predominant detoxification enzymes in maize (Zea mays) and cereal crops that are responsible for metabolism of triazine herbicides, acetamide herbicides, and certain graminicides, such as fenoxaprop-ethyl in wheat (Edwards and Cole, 1996; Riechers et al., 1996b, 1997b; Cummins et al., 1997). Herbicide-detoxifying GSTs have been well characterized in maize and soybean (Glycine max; Fuerst et al., 1993; Irzyk and Fuerst, 1993, 1997; Jepson et al., 1994; Andrews et al., 1997; Droog, 1997; Dixon et al., 1998), but have also been identified and partially characterized in wheat (Cummins et al., 1997; Riechers et al., 1997b).
Herbicide safeners are chemical compounds that increase the tolerance of certain grass crops (e.g. maize, grain sorghum [Sorghum bicolor], wheat, rice [Oryza sativa]) to herbicides (Hatzios and Hoagland, 1989). Herbicide safeners protect the crop plant by increasing herbicide metabolism and detoxification pathways (Fuerst and Gronwald, 1986; Hatzios, 1991; Farago et al., 1994; Riechers et al., 1996a; Davies and Caseley, 1999). The increase in metabolism results from an increase in the activity of herbicide detoxification enzymes, such as GSTs, cytochrome P450-dependent monooxygenases, and glucosyltransferases (Gronwald et al., 1987; Hatzios, 1991; Cole, 1994; Kreuz et al., 1996). Despite the widespread agronomic use of safeners and information about their effects on GST and P-450 enzymatic activity, there is little information on the precise molecular mechanism for safener induction of the genes that encode these herbicide-detoxifying enzymes. A safener-binding protein and its activity have been characterized in maize seedlings, along with its gene expression patterns (Walton and Casida, 1995; Scott-Craig et al., 1998). An area that is especially lacking in information is the identification and characterization of important regulatory sequences present in the promoters of safener-responsive GST genes, and the transcription factors that bind to these DNA sequences in agronomically important grasses such as maize, rice, and wheat. Molecular analyses of plant GST genomic sequences have focused mainly on dicot species, and have examined GST gene expression in response to pathogen attack, plant hormones, or stress treatments. Promoter regulatory elements that confer increased GST gene expression in response to auxins, ethylene, salicylic acid, hydrogen peroxide, heat shock, heavy metals, and plant pathogens have been identified in several dicot species including potato (Solanum tuberosum), tobacco (Nicotiana tabacum), Arabidopsis, soybean, and carnation (Dianthus caryophyllus; Martini et al., 1993; Itzhaki et al., 1994; Ulmasov et al., 1994; Droog et al., 1995; Maxson and Woodson, 1996; Strittmatter et al., 1996; Chen and Singh, 1999; Johnson et al., 2001). Safener-responsive expression of GSTs and their cDNA sequences have been reported for maize, wheat, and rice (Jepson et al., 1994; Irzyk and Fuerst, 1997; Riechers et al., 1997a; Wu et al., 1999). However, among these safener-responsive genes in monocot crops, only the maize GST-27 promoter has been partially characterized (Robertson et al., 2000).
Our studies have utilized the diploid wheat Triticum tauschii (synonymous with Aegilops tauschii and Aegilops squarrosa) as a model plant and genome to understand regulation of GST expression in grass crops with large and/or polyploid genomes (Keller and Feuillet, 2000). Previous research focused on a herbicide safener-induced GST isozyme that was purified from T. tauschii using anion-exchange and affinity chromatography, and was biochemically characterized (Riechers et al., 1997b). This safener-inducible GST isozyme can use the chloroacetamide herbicide dimethenamid as a substrate (Riechers et al., 1997b), where its conjugation with reduced glutathione results in metabolic detoxification of the herbicide (Dixon et al., 1998). In subsequent studies, a corresponding cDNA was isolated from T. tauschii and was used to map the homoeologous GST genes to a chromosome arm in cultivated, hexaploid bread wheat (Triticum aestivum) and to a linkage group in barley (Hordeum vulgare; Riechers et al., 1997a, 1998). Here, we report the analysis of genomic sequences for the safener-inducible, tau class GST genes (TtGSTU1 and TtGSTU2) and characterization of their expression profiles in T. tauschii. We also utilized ditelosomic, aneuploid wheat lines and various wheat species that differ in genome constitution and/or ploidy level to determine genome contributions to expression patterns in cultivated, hexaploid bread wheat. Our results provide novel evidence for differential genome contributions to constitutive and inducible gene expression in organs of hexaploid wheat, and differences in expression between closely related gene family members that are organized as a tandem repeat in a large grass genome.
RESULTS
Isolation and Sequence Analysis of GST Genes from T. tauschii
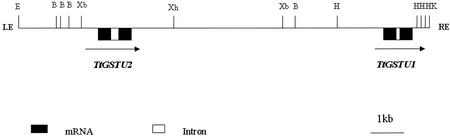
Three different GST genes were isolated by screening a T. tauschii large DNA insert genomic library (Moullet et al., 1999). The genomic library was screened with the safener-inducible GST TSI-1 (renamed TtGSTU1; Edwards and Dixon, 2000) cDNA that had been isolated previously from T. tauschii (Riechers et al., 1997a). A total of four positively hybridizing genomic clones were obtained and further analyzed. DNA gel-blot analysis of the four bacterial artificial chromosome (BAC) clones digested with different restriction enzymes showed that BAC 1 (insert size of 150 kb) and BAC 4 (insert size of 130 kb) contained overlapping genomic fragments, and each BAC clone appeared to contain at least two GST genes. Subsequent experiments focused on analyzing only BAC 1. BAC 1 contained an approximately 14-kb EcoRI/KpnI-hybridizing fragment. An XhoI site within the 14-kb fragment was utilized to separate this fragment into two smaller fragments of 8.5 and 5.3 kb. Each smaller fragment contained one of two tandemly repeated GST genes, TtGSTU2 and TtGSTU1 (accession no. AY013753; Fig. 1). These results are consistent with DNA gel-blot analysis (using XbaI as the restriction enzyme) of the cultivated, hexaploid bread wheat genome (Riechers et al., 1998), where it was hypothesized that the homoeologous GST genes were represented by at least two copies in each of the three wheat genomes. Sequence analysis of the entire 14-kb fragment showed that the coding region and untranslated regions (UTRs) of TtGSTU1 are identical to the TtGSTU1 cDNA (Riechers et al., 1997a). Both of the tandemly duplicated genes contain an intron that interrupts the coding region at the same location, although the length and sequence of the intron varies between the two genes. TtGSTU1 has a single intron of 99 bp, whereas TtGSTU2 has a 319-bp intron (Fig. 1), suggesting that these are tau class, or type III, GST genes (Droog, 1997; Edwards and Dixon, 2000; Edwards et al., 2000).
Figure 1.
Restriction map of the 14-kb region of the BAC 1 clone from T. tauschii, containing the two tandemly duplicated GST genes. Restriction sites are: B, BamHI; E, EcoRI; H, HindIII; K, KpnI; Xb, XbaI; and Xh, XhoI. LE, Left end (position 0); RE, right end (position 13, 710).
DNA gel-blot and sequence analysis of BAC 3 showed that it contained only the TtGSTU1 gene, and was not analyzed further. DNA gel-blot analysis of the BAC 2 clone, digested with several restriction enzymes, showed a single, weakly hybridizing band (data not shown). BAC 2 was found to contain a related, yet divergent, GST-like sequence, which was named TtGSTU3 (accession no. AY013754). The nucleotide sequence of the TtGSTU3 gene's coding region is 76% identical to the corresponding region of the TtGSTU1 gene, but this gene apparently does not contain an intron.
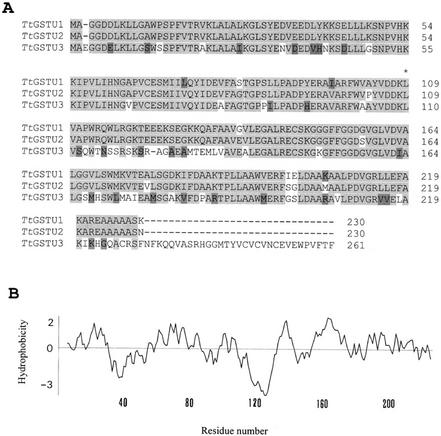
Comparison of the deduced amino acid sequences of the three GST genes showed that the TtGSTU1 and TtGSTU2 proteins are 95% identical and 96% similar, whereas the TtGSTU1 and TtGSTU3 proteins are 68% identical and 80% similar (Fig. 2A). There is a 31-amino acid residue extension at the C terminus of the TtGSTU3-encoded protein, compared with both TtGSTU1 and TtGSTU2 (Fig. 2A). These three GSTs belong to the tau class of plant GSTs, according to the classification system proposed by Droog (1997) and Edwards et al. (2000). The encoded proteins contain the triplet of amino acid residues His-Lys-Lys, which are conserved only in the tau class GST sequences, and are located at position 53 to 55 (numbering of amino acid residues is according to the TtGSTU1 sequence throughout the text, unless stated otherwise). The conserved triplet of His-Asn-Gly in the tau class is also conserved in these three GSTs at position 61 to 63 (Fig. 2A). TtGSTU3 is a unique GST, however, in that it appears to be a tau class GST protein based on its deduced amino acid sequence, but its gene sequence does not contain a single intron, which is characteristic of tau class GST genes.
Figure 2.
Comparison of tau class GST proteins from T. tauschii. A, Alignment of the deduced amino acid sequences of TtGSTU1, TtGSTU2, and TtGSTU3. Identical amino acids and conservatively substituted residues are shaded with light and dark gray, respectively. The amino acid residue where exon 2 of TtGSTU1 and TtGSTU2 starts is indicated by an asterisk above the sequences. GenBank accession numbers are as follows: TtGSTU1 and TtGSTU2 (AY013753), and TtGSTU3 (AY013754). B, Hydropathy profile of the TtGSTU1 protein, as determined by the method of Kyte and Doolittle (1982) using a window of 10 amino acid residues.
The hydropathy profiles of the TtGSTU1 and TtGSTU2 proteins, as determined by the method of Kyte and Doolittle (1982), are almost identical. Both proteins contain a strong hydrophilic region around amino acids 110 to 130 (Fig. 2B, only TtGSTU1 is shown). This hydrophilic region is located where exon 2 of each gene starts (at amino acid 109 in TtGSTU1 and TtGSTU2; Fig. 2A). The function of this hydrophilic region is not known, but may play an important role in substrate binding and/or substrate specificity. Alternatively, this region may play a role in determining the intracellular localization of the GST proteins. This region in TtGSTU1 and TtGSTU2 is very similar in sequence to the same region of the mouse (Mus musculus) mGSTA4-4 protein (amino acid residues 110–120), which also contains several charged residues and was shown to be important for a proposed electrostatic interaction with the plasma membrane of mouse liver hepatocytes (Singh et al., 2002). Hydropathy analysis of the TtGSTU3 protein showed minor differences relative to TtGSTU1, although slightly less hydrophilic character was noted around amino acids 110 to 130 (Fig. 2A).
Repeat Elements Are Dispersed in the Intergenic Regions of the 14-kb Contiguous Sequence of BAC 1
Two open reading frames (encoding the proteins TtGSTU1 and TtGSTU2) were found within the completely sequenced 14-kb interval from BAC 1. A BLAST search with this 14-kb interval identified sequences with similarity to several repeat elements from barley, maize, and the diploid wheat Triticum monococcum. The 14-kb interval contains about 2.6 kb of sequence upstream of the TATA box of TtGSTU2 (Fig. 1) until the EcoRI site (and start of the 14-kb contiguous sequence) is reached. In this region, about 1 kb of sequence (bp 1–1,039 of accession no. AY013753) was highly homologous to the long terminal repeat (LTR) of several retrotransposons, including the Angela-type retrotransposons from T. monococcum (Wicker et al., 2001). This 1-kb portion also shares 82% nucleotide identity with a portion of the LTR of the BARE-1 copia-like retroelement from barley (Manninen and Schulman, 1993).
Approximately 8 kb of sequence (bp 3,889–11, 859 of accession no. AY013753) is located between the TtGSTU2 and TtGSTU1 genes (Fig. 1). Within this 8-kb intergenic sequence, there are three regions that show homology to the LTRs of several retrotransposons from cereal species. Sequences from position 5,261 to 5,890 are similar to the LTRs of Angela-type retrotransposons (Wicker et al., 2001), and also shares about 83% identity with the barley BARE-1 copia-like retroelement over a 170-bp region (Manninen and Schulman, 1993). Position 8,062 to 8,451 was also similar to the LTRs of Angela-type retrotransposons (Wicker et al., 2001) and also shares 86% identity with the BARE-1 copia-like retroelement in barley over a 130-bp region (Manninen and Schulman, 1993). Position 8,775 to 9,124 shares about 90% nucleotide identity with LTRs of the BARE-1-like retrotransposons Angela-2, Angela-3, and Angela-4 (Wicker et al., 2001).
An 868-bp sequence flanks the 3′ end of the TtGSTU1 mRNA until the KpnI restriction site (and end of the 14-kb contiguous sequence) is reached. A BLAST search with this sequence showed that a region of about 660 bp (toward the 3′ end) shares 95% nucleotide identity with intron 10 of the T. tauschii starch synthase I gene (Li et al., 1999), which may indicate the presence of a repeat family that is present throughout different parts of the wheat genome. This region also shares homology with the intron of the maize ACCase gene (accession no. U90128), which was noted to contain a number of retroelements, such as colonist-1 and colonist-2.
5′-Flanking Regions of TtGSTU1 and TtGSTU2
In comparison with the 5′-UTR present in the TtGSTU1 cDNA, the start site for transcription initiation was set approximately 90 bp upstream of the MET start codon for both GST genes (Fig. 3). The major distinguishing feature of the 5′-UTRs in the two genes is an “AC” dinucleotide simple sequence repeat, present just upstream of the translational start site. The TtGSTU1 genomic sequence contains eight copies of the AC repeat, whereas the TtGSTU2 gene has five AC repeats (Fig. 3). An appropriately placed TATA box can be easily recognized 36 bp 5′ to the transcription start site in both genes. Comparison of alignments of the nucleotide sequences of the promoters of TtGSTU1 and TtGSTU2 revealed that the two genes are very similar for about 800 nucleotides upstream of the TATA boxes, except for two large gaps that were noted: a 22-bp gap in the TtGSTU1 promoter at position −195, and a large gap in the TtGSTU2 promoter at position −363 (Fig. 3).
Figure 3.
Comparison of the 5′-flanking sequences of the two tandem GST genes, TtGSTU1 and TtGSTU2, isolated from the BAC 1 clone. Numbering is determined from the putative transcription start site by comparison of the 5′-UTR sequence of the TtGSTU1 gene with the TtGSTU1 cDNA. Potential transcriptional regulatory elements identified by homology searches are underlined in bold and labeled accordingly.
Preliminary analysis of the UTRs and promoter regions using a plant transcription factor homology database (Higo et al., 1999) identified several potential cis-acting regulatory elements. Sequences similar to the TATA box of many eukaryotic promoters were found at position −36 of both TtGSTU1 and TtGSTU2. The sequence RYACGTGGYR (R = A/G and Y = C/T), which was identified as an ABA-responsive element (ABRE) in Arabidopsis (Iwasaki et al., 1995), was found in both TtGSTU1 and TtGSTU2 promoters at −230 and −249, respectively (Fig. 3). An ethylene-responsive enhancer element AWTTCAAA (W = A/T) identified in the carnation GSTI gene (Itzhaki et al., 1994) and a fruit-ripening gene (Montgomery et al., 1993) was located in the minus strand of the TtGSTU1 promoter at −525 (Fig. 3). The sequences of TGTCTC (Ulmasov et al., 1995a) and CATATG (Xu et al., 1997), which were related to auxin-responsive expression, were found in TtGSTU1 at −1,118 and −119 (Fig. 3). The auxin-responsive element TGTCTC was also found in the minus strand of TtGSTU2 at −2,078 (not shown). A G box-like sequence (Menkens et al., 1995) containing the palindromic hexamer TAGCTA was found at position −308 in the TtGSTU1 gene and position −351 in the TtGSTU2 gene (Fig. 3). This same sequence in the TtGSTU2 gene is actually a palindromic octamer, GTAGCTAC.
Expression of GST Genes in T. tauschii following Treatment with Safeners and Hormones
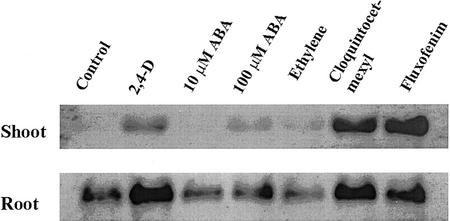
RNA gel-blot analysis showed that constitutive GST expression was detected in roots of T. tauschii, but expression in shoots was low or not detectable (Fig. 4). GST expression was highly induced by the safeners cloquintocet-mexyl and fluxofenim in T. tauschii shoots and roots. Induction of GST expression by these safeners in wheat shoots is consistent with previous results (Riechers et al., 1998). Because potential ABA-, ethylene-, and auxin-responsive regulatory elements were identified (by homology searches) in the promoters of the GST genes (Fig. 3), we also examined expression in response to these plant hormones. The synthetic auxin herbicide 2,4-D highly induced GST expression in roots and also induced expression in shoots, although to lower levels than the safeners in shoot tissue. ABA at a relatively high concentration (100 versus 10 μm) slightly increased GST expression in T. tauschii shoots (Fig. 4). Ethylene had no effect on GST expression in T. tauschii shoots or roots (Fig. 4). The fact that both safeners caused the greatest increase in GST expression, relative to the plant hormones, suggests that safeners may be tapping into a different regulatory pathway for induction of GST expression, or that the signal for induction may be stronger and/or longer lasting for a safener relative to the plant hormones examined.
Figure 4.
RNA gel-blot analysis of GST expression in T. tauschii after treatment with herbicide safeners and plant hormones. Total RNA (10 μg lane−1) was analyzed, and the blot was probed with the digoxigenin (DIG)-labeled TtGSTU1 cDNA coding region from T. tauschii. Etiolated seedlings were treated for 48 h with aqueous solutions containing 100 μm 2,4-dichlorophenoxyacetic acid (2,4-D), 10 μm or 100 μm ABA, 20 μg mL−1 2-chloroethyl-phosphonic acid for ethylene treatment, and 10 μm of the safeners cloquintocet-mexyl and fluxofenim. All treatment solutions were applied as vermiculite drenches to the seedlings 72 h after transferring pots to room temperature; the seedlings were exposed to the treatments for 48 h, then shoots and roots were harvested (5-d total growth period). Uniform loading was verified by comparing RNA intensities after ethidium bromide staining and by hybridizing the blots with a wheat actin probe (data not shown).
Differential Expression of TtGSTU1 and TtGSTU2 in T. tauschii
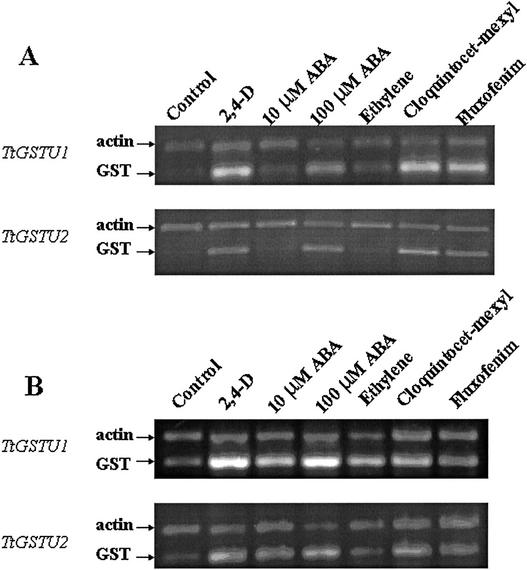
Due to the high degree of similarity between the two GST genes at the nucleic acid level, we used semiquantitative reverse transcriptase (RT)-PCR (Kinoshita et al., 1992) to detect individual expression patterns of the two tightly linked GST genes in roots and shoots of T. tauschii. Primers were designed (Table I) that would selectively amplify only one of the two tandemly duplicated GST genes. Constitutive GST expression in both roots and shoots was contributed mainly by TtGSTU1, and this gene also seems to be the most highly induced and expressed in safener-treated seedlings (Fig. 5). TtGSTU1 was also induced by 100 μm ABA and 2,4-D in shoots (Fig. 5). TtGSTU2 expression was not detectable in control shoots, but was weakly expressed in control roots. However, TtGSTU2 expression could be detected in control shoots when amplifying for 35 cycles (data not shown), compared with the 25 cycles shown in Figure 5. TtGSTU2 expression was induced by safeners, 100 μm ABA, and 2,4-D in both roots and shoots, although the level of expression was always less than that of TtGSTU1 (Fig. 5). Ethylene had little effect on TtGSTU1 and TtGSTU2 expression in either shoots or roots (Fig. 5).
Table I.
Primers for RT-PCR
| Name | Sequence | PCR Product Size |
|---|---|---|
| bp | ||
| 5′Cons | 5′-aagggcctgagctacgag-3′ | – |
| 3′TtGSTU1 | 5′-tgctggcggctcacttg-3′ | 622 |
| 3′TtGSTU2 | 5′-gtgtgctggctcagttag-3′ | 622 |
| 3′TtGSTU3 | 5′-gcatcaagcgagccgaaac-3′ | 527 |
| Actin forward | 5′-ctggactcacaccttctacaacgagctccgtgt-3′ | – |
| Actin reverse | 5′-atccagacactgtacttcctt-3′ | 765 |
Figure 5.
Semiquantitative RT-PCR analysis of individual GST gene expression in T. tauschii. Expression was analyzed in shoots (A) and roots (B) of T. tauschii. Seedling growth conditions and treatments, and total RNA samples were the same as those used for RNA gel-blot analysis shown in Figure 4. Total RNA (5 μg) was used to synthesize first strand cDNA, and a fraction (1/20) of the first strand cDNA was used as template for PCR amplification of individual gene transcripts. Ethidium bromide-stained RT-PCR products were separated in 1.2% (w/v) agarose gels and analyzed with 1D image analysis software (Eastman-Kodak, Rochester, NY). The wheat actin gene was used as a constitutively expressed control gene and loading control.
We could not detect the expression of TtGSTU3 by RT-PCR in either untreated T. tauschii seedlings or treated seedlings (data not shown), which suggests that TtGSTU3 may be transcriptionally inactive (a pseudogene), or has an expression pattern entirely different from TtGSTU1 and TtGSTU2. This might include expression in other tissues or organs, or possibly under different stress conditions.
GST Expression in Cultivated, Hexaploid Bread Wheat and Other Triticum Spp.
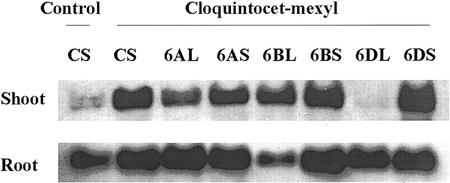
In addition to examining GST expression in T. tauschii, we also investigated GST expression in cultivated, hexaploid bread wheat (ABD genomes). Homoeologous GST genes were previously mapped to the short arms of group 6 chromosomes in cultivated, hexaploid bread wheat via Southern hybridization (Riechers et al., 1998), using the T. tauschii TtGSTU1 cDNA to probe ditelosomic, aneuploid wheat lines of group 6 chromosomes that are missing either the short or long arm of chromosome 6A, 6B, or 6D. RNA gel-blot analyses of these same lines were conducted, again using the TtGSTU1 cDNA as a probe. It is clear that in safener (cloquintocet-mexyl)-treated wheat shoots, removal of the tandem GST genes in the chromosome 6DL line (missing the short arm only) almost completely eliminated GST expression (Fig. 6). However, removal of the homoeologous GST genes in the chromosome 6AL and 6BL lines (missing the short arms of 6A and 6B, respectively) had relatively minor effects on GST expression levels, even though the 6A or 6B GST alleles were no longer present.
Figure 6.
RNA gel-blot analysis of GST expression in chromosome group 6 ditelosomic, aneuploid wheat lines. GST expression was analyzed in aneuploid lines derived from cultivated, hexaploid bread wheat cv Chinese Spring, in both roots and shoots. Treatments included the control (unsafened) or 10 μm of the safener cloquintocet-mexyl for 48 h as described in Figure 4. Total RNA (10 μg lane−1) was analyzed, and the blot was probed with the DIG-labeled TtGSTU1 cDNA coding region from T. tauschii. L, Long arm only (missing the short arm); S, short arm only (missing the long arm); CS, cultivated, hexaploid bread wheat cv Chinese Spring base genotype.
GST expression in cultivated, hexaploid bread wheat roots showed a different pattern when individual group 6 chromosome arms were removed (Fig. 6). When the short arms of chromosomes 6A and 6D were removed (in lines 6AL and 6DL), there was no detectable difference in the level of GST expression in safener-treated roots. However, when the short arm of chromosome 6B was removed (in the 6BL line), there was a significant decrease in GST gene expression. This suggests that in safener-treated cultivated, hexaploid bread wheat roots, the GST allele(s) on chromosome 6BS is a major contributor to GST expression; in contrast, the GST alleles on chromosome 6DS are most important in safener-treated cultivated, hexaploid bread wheat shoots and the GST alleles on chromosomes 6AS and 6BS appear to be minor contributors in shoots.
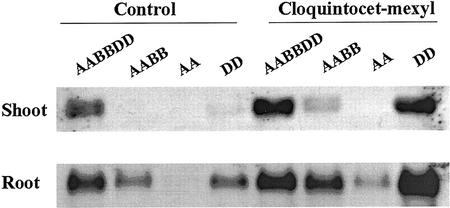
This difference in genome contribution to GST gene expression in roots and shoots is also noted in Figure 7, where several Triticum spp. were analyzed for their response to the safener cloquintocet-mexyl in roots and shoots. Triticum spp. were chosen that differ in genome constitution (diploids A only or D only, and tetraploid AB) and were compared with cultivated, hexaploid bread wheat (ABD genomes). GSTs were highly induced in the shoots of cultivated, hexaploid bread wheat (AABBDD) and T. tauschii (DD), although there was also a relatively high constitutive level of GST mRNA in cultivated, hexaploid bread wheat cv Chinese Spring (AABBDD; Fig. 7). In contrast, there was no induction of GST expression in the shoots of T. monococcum (AA) and relatively minor induction in Triticum turgidum subsp. durum (AABB) shoots (Fig. 7). However, the safener increased GST expression in the roots of all four wheat species, although the lowest induction occurred in T. monococcum roots (Fig. 7). These results confirm our previous observation that GST genes in the D genome of wheat are contributing the vast majority of safener-induced expression in shoots; however, in roots, it appears that GST genes in both the B and D genomes are the major contributors to GST expression (Figs. 6 and 7).
Figure 7.
RNA gel-blot analysis of wheat genome contribution to GST expression. Four wheat species were examined, each differing in genome composition and/or ploidy level. Wheat seedlings were either unsafened (control) or treated with 10 μm of the safener cloquintocet-mexyl for 48 h as described in Figure 4. Total RNA (10 μg per lane) was analyzed, and the blot was probed with the DIG-labeled TtGSTU1 cDNA coding region from T. tauschii. AABBDD, Cultivated, hexaploid bread wheat; AABB, T. turgidum subsp. durum; AA, T. monococcum; DD, T. tauschii.
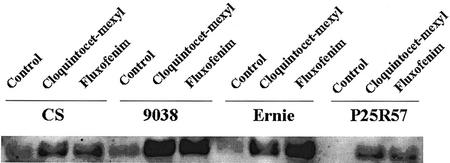
Cultivated, hexaploid bread wheat cv Chinese Spring was used in all of our expression studies because it is the base genotype from which the ditelosomic aneuploids were derived (Sears, 1954). There was a relatively high level of constitutive GST expression in shoots of cv Chinese Spring, compared with T. turgidum subsp. durum, T. monococcum, and T. tauschii, yet it also showed safener induction (Figs. 6 and 7). To further investigate this finding, three other varieties of cultivated, hexaploid bread wheat (CITR 9038, Ernie, and P25R57) were used to compare and contrast with cv Chinese Spring. These four wheat varieties differed in both basal level of GST expression and safener induction in shoots in response to the two safeners cloquintocet-mexyl and fluxofenim (Fig. 8). The varieties Chinese Spring and CITR 9038 showed the highest basal levels of GST expression, whereas there was no detectable basal GST expression in P25R57. GST expression was highly induced by both safeners in CITR 9038, which is consistent with the large increases in GST enzyme activity (in response to both safeners) reported previously for this wheat line (Riechers et al., 1996b). The variety Ernie showed a higher level of induction of GST expression after treatment with both safeners relative to the variety P25R57 (Fig. 8). These varying ranges of constitutive and safener-induced GST mRNA levels among wheat varieties and genetic lines are consistent with the broad range of constitutive and safener-increased GST activity levels reported previously (Riechers et al., 1996b), and provide further evidence that there is a great amount of genetic diversity present for GST expression levels in wheat germplasm.
Figure 8.
Cultivated, hexaploid bread wheat varietal response to safeners and differential GST expression. Wheat seedlings were either unsafened (control) or treated with 10 μm of the safener cloquintocet-mexyl or fluxofenim for 48 h as described in Figure 4. CS, Chinese Spring.
Although it was not determined which genome(s) is contributing the constitutive expression observed in cultivated, hexaploid bread wheat cv Chinese Spring shoots (Figs. 6 and 7), the variable constitutive and safener-induced expression levels observed in cultivated, hexaploid bread wheat varieties (Fig. 8) suggest genome complexities indicative of further evolution and diversification of homoeologous GST gene family members after their duplication by polyploidization of the A, B, and D genomes in cultivated, hexaploid bread wheat. The diversification of TtGSTU1 and/or TtGSTU2 expression patterns in both T. tauschii (by a duplication event) and cultivated, hexaploid bread wheat (by polyploidization) is consistent with a relaxation of purifying selection in duplicate genes of recent origin (Lynch and Conery, 2000; Hofer and Ellis, 2002).
DISCUSSION
Genomic Organization
Screening a T. tauschii BAC genomic library (average insert size of 119 kb; Moullet et al., 1999) allowed us to isolate the BAC 1 clone (insert size of 150 kb) containing the two tandemly repeated GST genes TtGSTU1 and TtGSTU2, both contained within a 14-kb restriction fragment. Within this 14-kb fragment, there were only two open reading frames, both coding for the two highly homologous tau class GSTs. These two GSTs contain the entire transcription units in two exons interrupted by one intron, with complete conservation of intron position. However, the length and nucleotide sequences of the two genes' introns are different. Similar GST gene clusters have been identified in the carnation genome (Itzhaki and Woodson, 1993) as well as in the Arabidopsis genome (Arabidopsis Genome Initiative, 2000; Edwards et al., 2000). In contrast to TtGSTU1 and TtGSTU2, the two tightly linked carnation GST genes were obtained from a single lambda phage clone, and contained 10 exons and nine introns (Itzhaki and Woodson, 1993), characteristic of zeta class plant GSTs. These two GST genes showed conservation in their intron positions and also the length and nucleotide sequences of the introns (Itzhaki and Woodson, 1993).
The recent findings from carnation, Arabidopsis, and wheat all indicate that GSTs from the same subclass are grouped on chromosomes as tandem duplications (Edwards et al., 2000). Comparisons of TtGSTU1 and TtGSTU2 sequences with rice genome sequences from GenBank and the draft recently completed by the Beijing Genomics Institute (http://btn.genomics.org.cn/rice) identified at least two clusters of tandemly duplicated tau class GST genes in rice. One of these clusters contained 20 closely related tau class GST genes on a single BAC clone from rice chromosome 10 (accession no. AC091680). At least 12 of these 20 genes were expressed in leaves, roots, or callus tissue based on their identity with rice GST cDNAs or ESTs. Another tandem duplication of tau class GST genes was found on a rice BAC clone from chromosome 1 (accession no. AP003450). Among these 22 rice tau class GST genes, both TtGSTU1 and TtGSTU2 show the highest similarity to the same OsGSTU4 gene present within the 20-gene cluster on chromosome 10, suggesting that the duplication event leading to TtGSTU1 and TtGSTU2 may have occurred after the divergence of the rice and wheat genomes. The high degree of similarity between TtGSTU1 and TtGSTU2 in both the coding and 5′-flanking regions (except for a few insertions or deletions in their promoters; Fig. 3) supports the notion that these genes arose due to a recent duplication event. TtGSTU1 and TtGSTU2 map to wheat chromosome 6 (Riechers et al., 1998), which is proposed to be syntenic with rice chromosome 2 (Moore et al., 1995; Moore, 2000). Our finding that the rice genes with the highest similarity to TtGSTU1 and TtGSTU2 map to chromosome 10 may suggest a lack of microsynteny for these regions (Bennetzen, 2000; Tarchini et al., 2000), or that the true rice orthologs of the wheat GST genes may be present in the estimated 8% of the rice genome not covered by the draft sequence from the Beijing Genomics Institute.
Among cereal crops, rice appears to have a predominance of tau class GST genes, which was also reported for hexaploid wheat (Cummins et al., 1997; Edwards and Dixon, 2000) but in contrast with maize, which was noted to have more phi class GST genes (Edwards and Dixon, 2000). However, these observations in wheat and maize have been based mainly on sequence analysis and immunological characterization of GST enzymes that possess herbicide detoxification activities. A complete sequence analysis of the entire rice, wheat, and maize genomes would be necessary to confirm this preliminary speculation on the relative abundance of specific GST subclasses in cereal crops.
BLAST searches showed that the intergenic regions of the 14-kb fragment contained sequences similar to the LTRs of retrotransposons, which is in accord with previous reports that showed a large portion of the wheat genome consists of repetitive DNA elements (Wicker et al., 2001). Certain types of retrotransposons are preferentially located near the centromeres of the chromosomes of grass species, such as wheat, barley, sorghum, and maize (Kumar and Bennetzen, 1999). Mapping the chromosomal location of a homologous GST in barley with the TtGSTU1 cDNA showed that it is located very close to the centromere of chromosome 6HS (Riechers et al., 1998). Based on these results, we postulate that the precise chromosomal location of TtGSTU1 and TtGSTU2 in T. tauschii and cultivated, hexaploid bread wheat may be the same as in barley (i.e. near the centromere of chromosome 6DS). This would also be consistent with our finding of the retrotransposon-like sequences in the intergenic regions of the BAC 1 clone from T. tauschii.
Expression Analyses in T. tauschii
Although TtGSTU1 and TtGSTU2 have similar gene structures and share very high identities at the amino acid and DNA sequence levels, they display different expression patterns in the roots and shoots of T. tauschii seedlings, as well as in response to various chemical inducers. Our RT-PCR results show that TtGSTU1 is constitutively expressed in both roots and shoots of 5-d-old T. tauschii seedlings grown under control (untreated) conditions. Using RNA gel-blot and immunoblot analyses, a similar expression pattern was reported in maize for the phi class GST-29 gene (ZmGSTF1), which was found to be constitutively expressed in a number of maize tissues (Jepson et al., 1994; Holt et al., 1995). Unlike maize GST-29, which showed a minimal increase in transcript levels upon herbicide safener treatment (Jepson et al., 1994), TtGSTU1 was strongly induced by herbicide safeners in shoots. Using RT-PCR and gene-specific primers, TtGSTU2 transcripts were detected in control roots, but not shoots, of 5-d-old etiolated seedlings. The phi class maize GST-27 gene (ZmGSTF2), like TtGSTU2, was constitutively expressed in roots, and no expression was detected in other tissues (Jepson et al., 1994; Holt et al., 1995). Furthermore, herbicide safener treatments caused dramatic increases in the expression of both TtGSTU1 and TtGSTU2 (Fig. 5) and the maize GST-27 gene (Jepson et al., 1994; Holt et al., 1995; Irzyk and Fuerst, 1997). These genes also showed induction in response to treatments with high levels of 2,4-D in maize leaves (Jepson et al., 1994) and T. tauschii shoots (Fig. 4), although the level of induction was always lower than with the safeners in these aerial tissues. Induction of expression in response to ethylene was not detected for the GSTs in T. tauschii (Fig. 4) or the maize GST-27 gene, except at very high concentrations that also led to phytotoxicity (Jepson et al., 1994).
These similar expression profiles imply that the promoters of these inducible wheat and maize GST genes may contain similar safener-responsive regulatory elements. An alignment of the promoters of TtGSTU1 and TtGSTU2 with the maize GST-27 promoter (reported in patent no. WO 93/01294, Bridges et al., 1993; GenBank accession no. A32436) did not reveal any conserved regions, and a search with the plant transcription factor homology database (Higo et al., 1999) only identified several CCAAT box sequences within the first 2 kb of the maize GST-27 promoter (D. Riechers and S. Moose, unpublished data). Because TtGSTU1 and TtGSTU2 are tau class GSTs, and the maize GST-27 is a phi class GST, there may be different safener-responsive regulatory elements in their promoters. Detailed functional analyses of the promoter sequences for these safener-inducible GSTs in maize and wheat may identify important regulatory elements that govern safener-induced expression.
In comparison with two wheat GSTs reported in the literature, TtGSTU1 shares 30% amino acid sequence identity with the phi class GstA1 protein (Mauch and Dudler, 1993) and 25% amino acid sequence identity with the zeta class TA-GSTZ1 protein (Subramaniam et al., 1999). GstA1 was further characterized for its expression patterns in wheat, detected at both the mRNA and protein levels (Mauch and Dudler, 1993). GstA1 was induced in response to challenge by pathogens and a cell-free fungal extract, but not by xenobiotics (Mauch and Dudler, 1993), suggesting a role for this gene in plant defense reactions against pathogen attack. In contrast, both TtGSTU1 and TtGSTU2 were highly induced by safener treatment in T. tauschii, as well as the synthetic auxin 2,4-D (and the phytohormone ABA to a limited extent). These results suggest that the tau class TtGSTU1 and TtGSTU2 proteins have important roles in xenobiotic metabolism in wheat, and may also have significant yet undefined roles in response to plant stresses.
Expression Analyses in Cultivated, Hexaploid Bread Wheat and Other Triticum Spp.
In addition to examining GST expression in the diploid wheat T. tauschii, gene expression was investigated in cultivated, hexaploid bread wheat, other diploid and tetraploid wheat species, as well as in ditelosomic aneuploid wheat lines that are missing individual arms of group 6 chromosomes. GST loci were previously mapped to the short arms of chromosomes 6A, 6B, and 6D in cultivated, hexaploid bread wheat, using the TtGSTU1 cDNA as a probe (Riechers et al., 1998). This allowed us to use the same probe to detect homoeologous GST transcripts in ditelosomic, aneuploid lines of cultivated, hexaploid bread wheat. The results demonstrated that GST expression in safener-treated wheat shoots was mainly contributed by GSTs from the D genome, whereas GSTs from both the B and D genomes contribute to safener-induced GST expression in wheat roots. The GST gene(s) on chromosome 6AS are not expressed to a significant extent in either control or safener-treated roots or shoots of T. monococcum, and also do not appear to be significant contributors to expression in safener-treated cultivated, hexaploid bread wheat seedlings. The most straightforward explanation to describe these expression patterns in wheat shoots is that the tandem GST genes on chromosome 6DS contain safener-responsive element(s) in their promoters or UTRs that are lacking in GST genes on chromosomes 6AS and 6BS. However, our data with ditelosomic, aneuploid wheat lines do not rule out the possibility that a regulatory factor could also be located on chromosome 6DS that controls the response to safeners in wheat shoots.
RNA gel-blot analysis of Triticum spp. with different genome constitutions showed that safener treatment dramatically increased GST expression in the shoots of cultivated, hexaploid bread wheat (AABBDD) and T. tauschii (DD), but not in T. monococcum (AA) or T. turgidum subsp. durum (AABB). These results are consistent with the results of expression analyses from the ditelosomic, aneuploid wheat lines of group 6 chromosomes, clearly indicating that GST genes in the D genome of wheat contribute most toward safener-induced GST expression in shoots. When assaying for GST enzyme activity in different Triticum spp. with a herbicide substrate, much higher GST activities were present in safener-treated shoots of both cultivated, hexaploid bread wheat and T. tauschii, relative to T. turgidum subsp. durum or other wheat species that lack the D genome (Edwards and Cole, 1996; Riechers et al., 1996b). The results of these studies confirm that the D genome is an important source of GST isozymes that are involved in the safener response in hexaploid and diploid wheats containing the D genome (Riechers et al., 1996b, 1997b).
Expression analyses in safener-treated roots from various Triticum spp. were also consistent with results found with the ditelosomic, aneuploid wheat lines, although the results were different from those observed in shoots. Safener treatment increased GST transcript levels in the roots of species or lines that contain the B or D genomes. Thus, GST genes in the B and D genomes contribute to safener-induced GST expression in wheat roots, whereas GST genes from the D genome contribute most toward expression in safener-treated shoots. This pattern of genome- and organ-specific expression of GSTs implies that the genes' promoters or untranslated sequences may contain different transcriptional regulatory elements that control gene expression in wheat roots versus shoots. Interestingly, comparisons of diploid wheats (T. monococcum and T. tauschii) with cultivated, hexaploid bread wheat indicate that genome contributions to safener-induced GST expression appear to be conserved following the polyploidization of cultivated wheat (Figs. 6 and 7). However, differences in GST expression among cultivated, hexaploid bread wheat varieties show that constitutive and safener-induced GST expression is variable within cultivated, hexaploid wheat (Fig. 8; Riechers et al., 1996b). The recent GST gene duplication event that led to TtGSTU1 and TtGSTU2 may have permitted the diversification of gene expression patterns in T. tauschii (Lynch and Conery, 2000; Hofer and Ellis, 2002). Further duplication of these genes by the poly-ploidization events that led to cultivated, hexaploid bread wheat may have allowed for their continued evolution within cultivated wheat (as noted by varietal differences in Fig. 8). Future work will be aimed at characterizing the structure and expression patterns of the TtGSTU1 and TtGSTU2 gene homoeologs in T. monococcum and cultivated, hexaploid bread wheat. Additional functional studies of these homoeologous genes, which differ in their expression response to herbicide safeners, will help identify the regulatory elements and factors that are important for herbicide safener-induced GST gene expression in wheat.
MATERIALS AND METHODS
Screening of a Triticum tauschii BAC Library
High-density filters of BAC clones (Moullet et al., 1999) were screened with the coding region of the cDNA encoding the safener-induced TtGSTU1 (Riechers et al., 1997a), previously isolated from T. tauschii. DNA hybridization and washing conditions were as reported by Lagudah et al. (1991). Plasmid DNA from individual BAC clones was isolated using the alkaline lysis method and DNA insert sizes were estimated by pulse field gel electrophoresis according to Moullet et al. (1999).
DNA Sequencing and Analysis
DNA fragments containing sequences of interest were subcloned into pBluescript SK+ (Stratagene, La Jolla, CA). Sequencing was conducted on both strands and reactions were performed at the sequencing center at the University of Illinois (Urbana) using the Big Dye kit (Perkin-Elmer Applied Biosystems, Foster City, CA) and an ABI Prism 377 (ABI, Sunnyvale, CA). DNA sequence analyses and amino acid alignments were performed using the AlignX tool of Vector NTI Suite V.6 software (InforMax, Inc., Bethesda, MD).
Plant Material
For RNA extraction and analysis, seeds were planted in plastic pots containing vermiculite. Pots were watered to saturation with deionized water, covered with aluminum foil, and subjected to prechilling at 4°C for 5 d to increase and synchronize seed germination. Pots were then removed from the cold and incubated at room temperature without light for a total of 5 d. For safener and plant hormone treatments, pots were transferred to room temperature, watered with deionized water, and incubated for 3 d. Then, the pots were watered with 10 μm cloquintocet-mexyl, 10 μm fluxofenim, 100 μm 2,4-D, 10 μm or 100 μm ABA, or 20 μg mL−1 2-chloroethyl-phosphonic acid (Sigma, St. Louis) for ethylene treatments, and incubated for another 48 h at room temperature. Roots and etiolated shoots were harvested separately, frozen in liquid nitrogen, and stored at −80°C until RNA extraction.
RNA Gel-Blot Analysis
Total RNA was isolated from plant tissues using TRIzol total RNA isolation reagent (Invitrogen Life Technologies, Carlsbad, CA) according to the manufacturer's instructions. The following manufacturer-recommended modification was used: Equal volumes of a high salt solution (1.2 m sodium citrate and 0.8 m NaCl) and isopropanol were added during the RNA precipitation step to decrease polysaccharide contamination and obtain >90% pure RNA (A260/A280 > 1.8). RNA was denatured at 55°C in the present of formamide and formaldehyde and separated by electrophoresis in 1.2% (w/v) agarose gels (containing 0.4 m formaldehyde). Equal loading among wells was verified by ethidium bromide staining. RNA was transferred to Nytran N membranes (Schleicher & Schuell, Keene, NH) by capillary blotting in 10× SSC overnight. RNA was fixed to the membrane by cross-linking on an UV Stratalinker (Stratagene). Blots were prehybridized in DIG Easy Hyb buffer (Roche Applied Science, Indianapolis) for 4 h and hybridized to a DIG-labeled TtGSTU1 cDNA coding region probe (Riechers et al., 1997a) overnight at 50°C. The blots were washed at a final stringency in 0.1× SSC and 0.1% (w/v) SDS at 65°C. Blots were developed with CDP-Star chemiluminescent substrate, then exposed to Hyperfilm (Amersham Biosciences, Piscataway, NJ).
Semiquantitative RT-PCR
Total RNA (5 g) was annealed to an oligo(dT)12-18 primer (Invitrogen Life Technologies), then first strand cDNAs were synthesized using Superscript II RT (Invitrogen Life Technologies). The genomic sequences for TtGSTU1, TtGSTU2, and TtGSTU3 were used to design primers for subsequent PCR amplification. Gene-specific primers were designed based on the comparisons of DNA and deduced amino acid sequences of the three GST genes. The forward primer was the same for the three GST genes, and was located at the amino acids 28 to 33 of TtGSTU1 and TtGSTU2, or 29 to 34 of TtGSTU3 (Table I). The reverse primers were designed to be gene specific, and were located near the C-terminal sequences of the open reading frames (Table I). The wheat actin transcript served as an internal, constitutively expressed loading control. The two actin primers used for RT-PCR (Table I) were designed from conserved sequences in the rice (Oryza sativa) actin gene (GenBank accession no. X16280). The reaction mixture contained 1 μL of first strand cDNA, 0.2 mm dNTPs, 1.0 mm MgCl2, 0.4 μm each primer, and 1.25 units of Taq polymerase (Invitrogen Life Technologies) in a total volume of 25 μL. PCR cycling conditions were as follows: an initial denaturation step at 95°C for 10 min, 25 amplification cycles (95°C for 50 s, 65°C for 30 s, and 72°C for 2.5 min), and a final elongation step at 72°C for 10 min. For semiquantitative RT-PCR, linearity for each amplification was confirmed (Kinoshita et al., 1992; Riechers and Timko, 1999). Specificity of amplification for each GST gene was verified by using plasmid controls containing each individual gene fragment under the same PCR conditions as described above (data not shown).
ACKNOWLEDGMENTS
We thank Dr. Lynn Holappa for maintaining the GST cDNA plasmid construct, Dr. Stephen Jones for helpful discussions concerning the research and manuscript, and Larry Boze for assistance in growing wheat plants in the greenhouse.
Footnotes
This work was supported in part by the Cooperative State Research, Education, and Extension Service (project no. ILLU–15–0357).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.004796.
LITERATURE CITED
- Andrews CJ, Skipsey M, Townson JK, Morris C, Jepson I, Edwards R. Glutathione transferase activities toward herbicides used selectively in soybean. Pestic Sci. 1997;51:213–222. [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Bennetzen JL. Comparative sequence analysis of plant nuclear genomes: microcolinearity and its many exceptions. Plant Cell. 2000;12:1021–1029. doi: 10.1105/tpc.12.7.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges IG, Brifht SWJ, Greenland AJ, Holt DC, Jepson I, Schuch WW, inventors. (1993) Plant-derived enzyme and DNA sequences, and uses thereof. International Patent Application No. PCT/GB92/01187.
- Chen W, Singh KB. The auxin, hydrogen peroxide and salicylic acid induced expression of the Arabidopsis GST6promoter is mediated in part by an ocs element. Plant J. 1999;19:667–677. doi: 10.1046/j.1365-313x.1999.00560.x. [DOI] [PubMed] [Google Scholar]
- Cole DJ. Detoxification and activation of agrochemicals in plants. Pestic Sci. 1994;42:209–222. [Google Scholar]
- Cummins I, Cole DJ, Edwards R. Purification of multiple glutathione transferases involved in herbicide detoxification from wheat (Triticum aestivumL.) treated with the safener fenchlorazole-ethyl. Pestic Biochem Physiol. 1997;59:35–49. [Google Scholar]
- Davies J, Caseley JC. Herbicide safeners: a review. Pestic Sci. 1999;55:1043–1058. [Google Scholar]
- Dixon DP, Cummins I, Cole DJ, Edwards R. Glutathione-mediated detoxification systems in plants. Curr Opin Plant Biol. 1998;1:258–266. doi: 10.1016/s1369-5266(98)80114-3. [DOI] [PubMed] [Google Scholar]
- Droog F. Plant glutathione S-transferases, a tale of theta and tau. J Plant Growth Regul. 1997;16:95–107. [Google Scholar]
- Droog F, Spek A, van der Kooy A, de Ruyter A, Hoge H, Libbenga K, Hooykaas P, van der Zaal B. Promoter analysis of the auxin-regulated tobacco glutathione S-transferase genes Nt103-1 and Nt103-35. Plant Mol Biol. 1995;29:413–429. doi: 10.1007/BF00020974. [DOI] [PubMed] [Google Scholar]
- Edwards R, Cole DJ. Glutathione transferases in wheat (Triticum) species with activity toward fenoxaprop-ethyl and other herbicides. Pestic Biochem Physiol. 1996;54:96–104. [Google Scholar]
- Edwards R, Dixon DP. The role of glutathione transferases in herbicide metabolism. In: Cobb AH, Kirkwood RC, editors. Herbicides and Their Mechanisms of Action. Sheffield, UK: Sheffield Academic Press; 2000. pp. 38–71. [Google Scholar]
- Edwards R, Dixon DP, Walbot V. Plant glutathione S-transferases: enzymes with multiple functions in sickness and in health. Trends Plant Sci. 2000;5:193–198. doi: 10.1016/s1360-1385(00)01601-0. [DOI] [PubMed] [Google Scholar]
- Farago S, Brunhold C, Kreuz K. Herbicide safeners and glutathione metabolism. Physiol Plant. 1994;91:537–542. [Google Scholar]
- Fuerst EP, Gronwald JW. Induction of rapid metabolism of metolachlor in sorghum (Sorghum bicolor) shoots by CGA-92194 and other antidotes. Weed Sci. 1986;34:354–361. [Google Scholar]
- Fuerst EP, Irzyk GP, Miller KD. Partial characterization of glutathione S-transferase isozymes induced by the herbicide safener benoxacor in maize. Plant Physiol. 1993;102:795–802. doi: 10.1104/pp.102.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronwald JW, Fuerst EP, Eberlein CV, Egli MA. Effect of herbicide antidotes on glutathione content and glutathione S-transferase activity of sorghum shoots. Pestic Biochem Physiol. 1987;29:66–76. [Google Scholar]
- Hatzios KK. An overview of the mechanisms of action of herbicide safeners. Z Naturforsch. 1991;46c:819–827. [Google Scholar]
- Hatzios KK, Hoagland RE. Crop Safeners for Herbicides. Development, Uses, and Mechanisms of Action. San Diego: Academic Press; 1989. [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer J, Ellis N. Conservation and diversification of gene function in plant development. Curr Opin Plant Biol. 2002;5:56–61. doi: 10.1016/s1369-5266(01)00228-x. [DOI] [PubMed] [Google Scholar]
- Holt DC, Lay VJ, Clarke ED, Dinsmore A, Jepson I, Bright SWJ, Greenland AJ. Characterization of the safener-induced glutathione S-transferase isoform II from maize. Planta. 1995;196:295–302. doi: 10.1007/BF00201388. [DOI] [PubMed] [Google Scholar]
- Irzyk GP, Fuerst EP. Purification and characterization of a glutathione S-transferase from benoxacor-treated maize (Zea mays) Plant Physiol. 1993;102:803–810. doi: 10.1104/pp.102.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irzyk GP, Fuerst EP. Characterization and induction of maize glutathione S-transferases involved in herbicide detoxification. In: Hatzios KK, editor. Regulation of Enzymatic Systems Detoxifying Xenobiotics in Plants. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1997. pp. 155–170. [Google Scholar]
- Itzhaki H, Maxson JM, Woodson WR. An ethylene-responsive enhancer element is involved in the senescence-related expression of the carnation glutathione S-transferase (GST1) gene. Proc Natl Acad Sci USA. 1994;91:8925–8929. doi: 10.1073/pnas.91.19.8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzhaki H, Woodson WR. Characterization of an ethylene-responsive glutathione S-transferase gene cluster in carnation. Plant Mol Biol. 1993;22:43–58. doi: 10.1007/BF00038994. [DOI] [PubMed] [Google Scholar]
- Iwasaki T, Yamaguchi-Shinozaki K, Shinozaki K. Identification of a cis-regulatory region of a gene in Arabidopsis thalianawhose induction by dehydration is mediated by abscisic acid and requires protein synthesis. Mol Gen Genet. 1995;247:391–398. doi: 10.1007/BF00293139. [DOI] [PubMed] [Google Scholar]
- Jepson I, Lay VJ, Holt DC, Bright SWJ, Greenland AJ. Cloning and characterization of maize herbicide safener-induced cDNAs encoding subunits of glutathione S-transferase isoforms I, II and IV. Plant Mol Biol. 1994;26:1855–1866. doi: 10.1007/BF00019498. [DOI] [PubMed] [Google Scholar]
- Johnson C, Boden E, Desai M, Pascuzzi P, Arias J. In vivotarget promoter-binding activities of a xenobiotic stress-activated TGA factor. Plant J. 2001;28:237–243. doi: 10.1046/j.1365-313x.2001.01147.x. [DOI] [PubMed] [Google Scholar]
- Keller B, Feuillet C. Colinearity and gene density in grass genomes. Trends Plant Sci. 2000;5:246–251. doi: 10.1016/s1360-1385(00)01629-0. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Imamura J, Nagai H, Shimotohno K. Quantification of gene expression over a wide range by the polymerase chain reaction. Anal Biochem. 1992;206:231–235. doi: 10.1016/0003-2697(92)90358-e. [DOI] [PubMed] [Google Scholar]
- Kreuz K, Tommasini R, Martinoia E. Old enzymes for a new job. Herbicide detoxification in plants. Plant Physiol. 1996;111:349–353. doi: 10.1104/pp.111.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Bennetzen J. Plant retrotransposons. Annu Rev Genet. 1999;33:479–532. doi: 10.1146/annurev.genet.33.1.479. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle R. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lagudah ES, Appels R, McNeil D, Brown AHD. The molecular-genetic analysis of Triticum tauschii, the D genome donor to hexaploid wheat. Genome. 1991;34:375–386. [Google Scholar]
- Li Z, Rahman S, Kosar-Hashemi B, Mouille G, Appels R, Morell MK. Cloning and characterization of a gene encoding wheat starch synthase I. Theor Appl Genet. 1999;98:1208–1216. [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Manninen I, Schulman AH. BARE-1, a copia-like retroelement in barley (Hordeum vulgareL.) Plant Mol Biol. 1993;22:829–846. doi: 10.1007/BF00027369. [DOI] [PubMed] [Google Scholar]
- Marrs KA. The functions and regulation of glutathione S-transferases in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:127–158. doi: 10.1146/annurev.arplant.47.1.127. [DOI] [PubMed] [Google Scholar]
- Martini N, Egen M, Runtz I, Strittmatter G. Promoter sequences of a potato pathogenesis-related gene mediate transcriptional activation selectively upon fungal infection. Mol Gen Genet. 1993;236:179–186. doi: 10.1007/BF00277110. [DOI] [PubMed] [Google Scholar]
- Mauch F, Dudler R. Differential induction of distinct glutathione S-transferases of wheat by xenobiotics and by pathogen attack. Plant Physiol. 1993;102:1193–1201. doi: 10.1104/pp.102.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxson JM, Woodson WR. Cloning of a DNA-binding protein that interacts with the ethylene-responsive enhancer element of the carnation GST1gene. Plant Mol Biol. 1996;31:751–759. doi: 10.1007/BF00019463. [DOI] [PubMed] [Google Scholar]
- McGonigle B, Keeler SJ, Lau S-MC, Koeppe MK, O'Keefe DP. A genomics approach to the comprehensive analysis of the glutathione S-transferase gene family in soybean and maize. Plant Physiol. 2000;124:1105–1120. doi: 10.1104/pp.124.3.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menkens AE, Schindler U, Cashmore AR. The G-box: a ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem Sci. 1995;20:506–510. doi: 10.1016/s0968-0004(00)89118-5. [DOI] [PubMed] [Google Scholar]
- Montgomery J, Goldman S, Deikman J, Margossian L, Fischer RL. Identification of an ethylene-responsive region in the promoter of a fruit ripening gene. Proc Natl Acad Sci USA. 1993;90:5939–5943. doi: 10.1073/pnas.90.13.5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore G. Cereal chromosome structure, evolution, and pairing. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:195–222. doi: 10.1146/annurev.arplant.51.1.195. [DOI] [PubMed] [Google Scholar]
- Moore G, Devos KM, Wang Z, Gale MD. Cereal Genome Evolution. Grasses, line up and form a circle. Curr Biol. 1995;5:737–739. doi: 10.1016/s0960-9822(95)00148-5. [DOI] [PubMed] [Google Scholar]
- Moullet O, Zhang HB, Lagudah ES. Construction and characterisation of a large DNA insert library from the D genome of wheat. Theor Appl Genet. 1999;99:305–313. [Google Scholar]
- Polidoros AN, Scandalios JG. Role of hydrogen peroxide and different classes of antioxidants in the regulation of catalase and glutathione S-transferase gene expression in maize (Zea maysL.) Physiol Plant. 1999;106:112–120. [Google Scholar]
- Riechers DE, Fuerst EP, Miller KD. Initial metabolism of dimethenamid in safened and unsafened wheat shoots. J Agric Food Chem. 1996a;44:1558–1564. [Google Scholar]
- Riechers DE, Irzyk GP, Fuerst EP, Jones SS. Nucleotide sequence of a cDNA encoding a safener-induced glutathione S-transferase (accession no. AF004358) from Triticum tauschii (PGR 97-110) Plant Physiol. 1997a;114:1568. doi: 10.1104/pp.114.4.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechers DE, Irzyk GP, Jones SS, Fuerst EP. Partial characterization of glutathione S-transferases from wheat (Triticum spp.) and purification of a safener-induced glutathione S-transferase from Triticum tauschii. Plant Physiol. 1997b;114:1461–1470. doi: 10.1104/pp.114.4.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechers DE, Kleinhofs A, Irzyk GP, Jones SS. Chromosomal location and expression of a herbicide safener-regulated glutathione S-transferase gene in Triticum aestivum and linkage relations in Hordeum vulgare. Genome. 1998;41:368–372. [Google Scholar]
- Riechers DE, Timko MP. Structure and expression of the gene family encoding putrescine N-methyltransferase in Nicotiana tabacum: new clues to the evolutionary origin of cultivated tobacco. Plant Mol Biol. 1999;41:387–401. doi: 10.1023/a:1006342018991. [DOI] [PubMed] [Google Scholar]
- Riechers DE, Yang K, Irzyk GP, Jones SS, Fuerst EP. Variability of glutathione S-transferase levels and dimethenamid tolerance in safener-treated wheat and wheat relatives. Pestic Biochem Physiol. 1996b;56:88–101. [Google Scholar]
- Robertson N, Paine JA, Sonnewald U, Jepson I. Expression of the chemically inducible maize GST-27promoter in potato. Potato Res. 2000;43:335–345. [Google Scholar]
- Scott-Craig JS, Casida JE, Poduje L, Walton JD. Herbicide safener-binding protein of maize. Purification, cloning, and expression of an encoding cDNA. Plant Physiol. 1998;116:1083–1089. doi: 10.1104/pp.116.3.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears ER. The aneuploids of common wheat. Mo Agric Exp Stn Res Bull. 1954;572:1–59. [Google Scholar]
- Singh SP, Janecki AJ, Srivastava SK, Awasthi S, Awasthi YC, Xia SJ, Zimniak P. Membrane association of glutathione S-transferase mGSTA4-4, an enzyme that metabolizes lipid peroxidation products. J Biol Chem. 2002;277:4232–4239. doi: 10.1074/jbc.M109678200. [DOI] [PubMed] [Google Scholar]
- Strittmatter G, Gheysen G, Gianinazzi-Pearson V, Hahn K, Niebel A, Rohde W, Tacke E. Infections with various types of organisms stimulate transcription from a short promoter fragment of the potato gst1gene. Mol Plant-Microbe Interact. 1996;9:68–73. doi: 10.1094/mpmi-9-0068. [DOI] [PubMed] [Google Scholar]
- Subramaniam K, Ye Z, Buechley G, Shaner G, Solomos T, Ueng PP. Isolation of a zeta class wheat glutathione S-transferase gene. Biochim Biophys Acta. 1999;1447:348–356. doi: 10.1016/s0167-4781(99)00176-1. [DOI] [PubMed] [Google Scholar]
- Tarchini R, Biddle P, Wineland R, Tingey S, Rafalski A. The complete sequence of 340 kb of DNA around the rice Adh1-Adh2region reveals interrupted colinearity with maize chromosome 4. Plant Cell. 2000;12:381–391. doi: 10.1105/tpc.12.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle T. The ocs element in the soybean GH2/4promoter is activated by both active and inactive auxin and salicylic acid analogues. Plant Mol Biol. 1994;26:1055–1064. doi: 10.1007/BF00040688. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Liu ZB, Hagen G, Guilfoyle TJ. Composite structure of auxin response elements. Plant Cell. 1995a;7:1611–1623. doi: 10.1105/tpc.7.10.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Ohmiya A, Hagen G, Guilfoyle T. The soybean GH2/4 gene that encodes a glutathione S-transferase has a promoter that is activated by a wide range of chemical agents. Plant Physiol. 1995b;108:919–927. doi: 10.1104/pp.108.3.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton JD, Casida JE. Specific binding of a dichloroacetamide herbicide safener in maize at a site that also binds thiocarbamate and chloroacetanilide herbicides. Plant Physiol. 1995;109:213–219. doi: 10.1104/pp.109.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Stein N, Albar L, Feuillet C, Schlagenhauf E, Keller B. Analysis of a contiguous 211 kb sequence in diploid wheat (Triticum monococcumL.) reveals multiple mechanisms of genome evolution. Plant J. 2001;26:307–316. doi: 10.1046/j.1365-313x.2001.01028.x. [DOI] [PubMed] [Google Scholar]
- Wu J, Cramer CL, Hatzios KK. Characterization of two cDNAs encoding glutathione S-transferases in rice and induction of their transcripts by the herbicide safener fenclorim. Physiol Plant. 1999;105:102–108. [Google Scholar]
- Xu N, Hagen G, Guilfoyle T. Multiple auxin response modules in the soybean SAUR 15Apromoter. Plant Sci. 1997;126:193–201. [Google Scholar]