Abstract
Tyrosine (Tyr) phosphorylation represents an important biochemical mechanism to regulate many cellular processes. No Tyr kinase has been cloned so far in plants. Dual-specificity kinases are reported in plants and the function of these kinases remains unknown. A 1.7-kb cDNA that encodes serine/threonine/Tyr (STY) kinase was isolated by screening peanut (Arachis hypogaea) expression library using the anti-phospho-Tyr antibody. The histidine-tagged recombinant kinase histidine-6-STY predominantly autophosphorylated on Tyr and phosphorylated the histone primarily on threonine. Genomic DNA gel-blot analysis revealed that STY kinase is a member of a small multigene family. The transcript of STY kinase is accumulated in the mid-maturation stage of seed development, suggesting a role in the signaling of storage of seed reserves. The STY kinase mRNA expression, as well as kinase activity, markedly increased in response to cold and salt treatments; however, no change in the protein level was observed, suggesting a posttranslational activation mechanism. The activation of the STY kinase is detected after 12 to 48 h of cold and salt treatments, which indicates that the kinase may not participate in the initial response to abiotic stresses, but may play a possible role in the adaptive process to adverse conditions. The transcript levels and kinase activity were unaltered with abscisic acid treatment, suggesting an abscisic acid-independent cold and salt signaling pathway. Here, we report the first identification of a non-MAP kinase cascade dual-specificity kinase involved in abiotic stress and seed development.
Phosphorylation of Ser, Thr, and Tyr residues on target proteins by protein kinases represents an important biochemical mechanism to regulate enzyme activities and many other cellular processes (Hunter, 1987). Although an increasing number of Ser/Thr kinases have been cloned, the existence of protein Tyr kinases is yet to be demonstrated in plants. Nevertheless, protein Tyr phosphorylation has been reported in wheat (Triticum aestivum; Suzuki and Shinshi, 1995), coconut (Cocos nucifera; Islas-Flores et al., 1998), and pea (Pisum sativum; Barizza et al., 1999). Phosphorylation of cyclin-dependent protein kinases at Tyr-15 is mediated by Wee1 and related kinases (Sun et al., 1999). Actin in Mimosa pudica is heavily phosphorylated at the Tyr and the extent of phosphorylation correlates with the bending of petiole (Kazuhisa et al., 2000). Despite the apparent absence of classical Tyr kinases, dual-specificity kinases that carry out Tyr phosphorylation are reported in plants. Dual-specificity kinases ATN1, APK1, and ADK1 from Arabidopsis (Hirayama and Oka, 1992; Ali et al., 1994; Tregear et al., 1996) and GmPK6 from soybean (Glycine max; Feng et al., 1993) have been isolated, but the function of these kinases remains unknown.
Plant growth is greatly affected by abiotic factors like low temperature and salinity. There is increasing evidence that genes involved in signal transduction are shown to be up-regulated in response to cold acclimation (Thomashow, 1999). Furthermore, several of these genes are also regulated by salt stress (Kurkela and Borg-Franck, 1992; Yamaguchi-Shinozaki and Shinozaki, 1994). Mitogen-activated protein (MAP) kinases and their upstream kinases have been shown to be activated under various stresses (Zhang and Klessig, 2001). In addition, SNF1 kinases (Halford and Hardie, 1998) and calcium-dependent protein kinases (Stone and Walker, 1995) are shown to be induced by abiotic stresses.
Here, we report the identification of a new class of dual-specificity kinase, Ser/Thr/Tyr (STY) kinase, which is distinct from the above-mentioned kinases, involved in abiotic stress response and seed development. STY kinase is a structural mosaic of Ser/Thr and Tyr kinases that autophosphorylates predominantly on Tyr, and phosphorylated the histone primarily on Thr. The isolated kinase is regulated by cold and salt stresses by posttranslational activation mechanism.
RESULTS
In Silico Analysis of the STY Kinase cDNA Clone
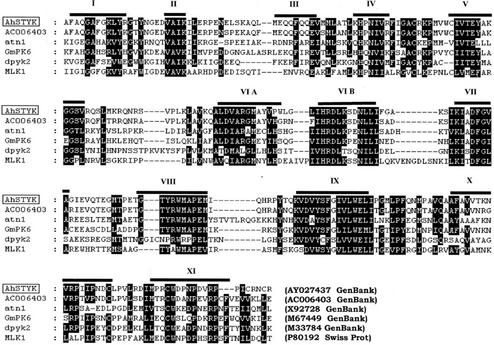
Sequence analysis of the largest clone obtained after screening of peanut (Arachis hypogaea) expression library with anti-phospho-Tyr antibodies revealed a 1,740-bp gene that encodes a protein with an open reading frame (ORF) of 411 amino acids with a theoretical pI of 8.54 and molecular mass of 46.1 kD (GenBank accession no. AY027437). The ORF is flanked by a 200-bp 5′-untranslated region (UTR) and a 309-bp 3′-UTR with stop codons in all three reading frames upstream of the initiating ATG, and a poly(A+) tail at the end of a 3′-UTR, indicating that the isolated clone represents full-length cDNA. The STY kinase polypeptide contains all 11 subdomains for protein kinases (Hanks and Hunter, 1995) and the kinase catalytic domain spans from 133 to 378 amino acids. In subdomain VIB, STY kinase contains a KPM sequence motif that shows homology to Ser/Thr kinases. In subdomain VIII, RWM sequence motif is more closely related to Tyr kinases. In subdomain XI, the dipeptide CW characteristic of all the Tyr kinases is also found in the STY kinase protein sequence.
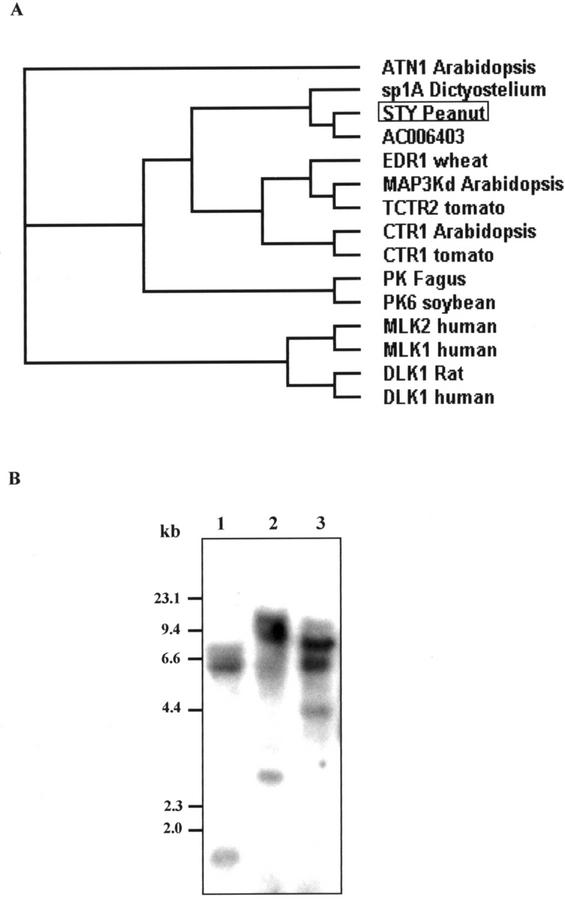
A BLAST search revealed that the STY kinase showed 75% homology with a putative protein kinase from Arabidopsis. Four other protein kinases, namely Arabidopsis ATN1 (43%), soybean GmPK6 (42%), Dictyostelium discoideum sp1A (37%), and human MLK1 (35%) shared identities greater than 30% with STY kinase over the catalytic domain. Multiple sequence alignment of STY kinase protein sequence with the related sequences is depicted in Figure 1. The most striking common motifs are located in the core of the catalytic domain and are as follows: the conserved sequences of subdomains V (TEY), VIB (HRDL), VII (DFGVAGI), and VIII (TYRWMAPE). In analogy with the MAP kinases, STY kinase also has the TEY motif in subdomain V. MAP kinases have the same motif between VII and VIII domains. The non-kinase domains do not show homology to any of the sequences in the database. The sequence similarity of the STY kinase revealed homology to Src (Schmidt-Ruppin A-2) Tyr kinases from chicken (Gallus gallus) and human (Weijland et al., 1997; Xu et al., 1999). Five potential glycosylation sites are found throughout the protein sequence (Asn-X-S/T). The hydropathy plot of STY kinase revealed a single transmembrane domain (316–332). The transmembrane domain has a Leu-/Ile-rich repeat (LRR). There is another LRR between 253 and 267 amino acids. Figure 2A is the molecular phylogenic estimation of the most parsimonious tree, which revealed that the STY kinase is closely related to a putative protein kinase from Arabidopsis and Dictyostelium discoideum non-receptor Tyr kinase (sp1A).
Figure 1.
Sequence alignment of peanut STY kinase catalytic domain sequence and those of the five most closely related sequences in the databases, namely AC006403 (putative protein kinase) and ATN1 from Arabidopsis, GmPK6 from soybean, DPYK1 from Dictyostelium discoideum, and MLK1 from human (Homo sapiens). Black boxes indicate positions at which the residues are identical, and gray boxes highlight residues that are similar. The positions of the catalytic subdomains are indicated with roman numerals. Databanks and database accession numbers are shown within brackets. Sequences were aligned using ClustalW and GeneDoc.
Figure 2.
A, Phylogenetic tree based on an alignment of peanut STY kinase (STY, accession no. AY027437) with putative protein kinase from Arabidopsis (accession no. AC006403) and 13 related proteins. GenBank accession numbers are: ATN 1 (Arabidopsis), S61766; sp1A (D. discoideum), P18160; EDR 1 (barley [Hordeum vulgare]), AAG31142; MAP3K (Arabidopsis), AA7459; TCTR2 (Lycopersicum esculentum), T06576; CTR1 (Arabidopsis), Q05609; CTR1 (L. esculentum), AAD10057; K (Fagus sylvatica), CAA66149; PK6 (soybean), S29851; MLK1 (human), P80192; MLK2 (human), Q02779; DLK (Rattus norvegicus), JC53399; and DLK (human), NP 00472. Protein sequences were aligned using the ClustalW. Distance trees were calculated using the neighbor-joining method. The lengths of the branches are proportional to the degree of divergence and thus correspond to the statistical significance of the phylogeny between the protein sequences. B, Southern-blot analysis STY kinase. Genomic DNA (20 μg) from immature peanut seed was digested with EcoRI (lane 1), SacI (lane 2), and BamHI (lane 3), separated on 0.8% (w/v) agarose gel, and probed with full-length STY kinase cDNA at high-stringency conditions (hybridization at 65°C, and washes at 65°C and 0.1× SSC).
Molecular organization of the STY kinase gene in peanut was determined by Southern analysis (Fig. 2B). Peanut genomic DNA was digested with various restriction enzymes and probed with a 1.7-kb full-length STY kinase gene. Digestion of the STY kinase gene with BamHI that cleaves within the probe region resulted in three hybridization signals. The digestions with EcoRI and SacI that do not cut within the probe also produced three hybridization signals. However, additional weak signals were observed, suggesting the presence of a homologous gene(s) in the genome of peanut. A simple hybridization pattern appears to be consistent with the STY kinase being a small multigene family.
Dual-Specificity Kinase Activity of STY Kinase
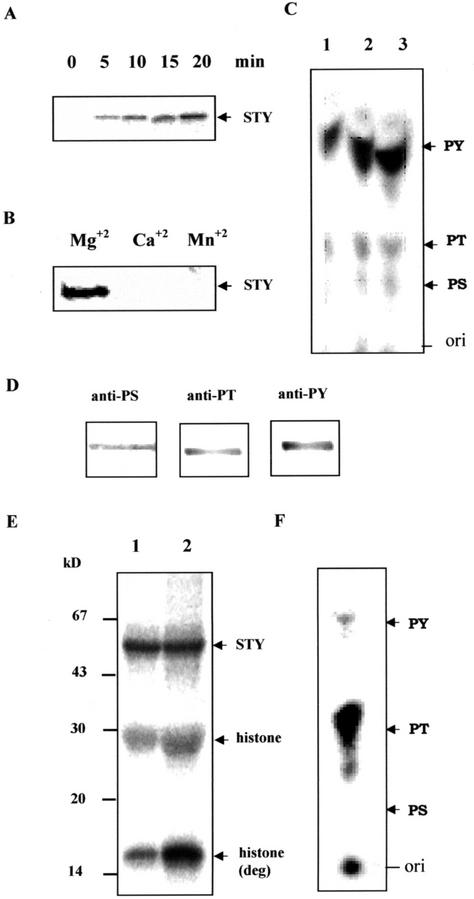
The cDNA corresponding to the ORF was subcloned into pRSET C and expressed in Escherichia coli as an N-terminal fusion protein of oligo-His. The His-6-STY was purified by a nickel affinity column and shown to have a molecular mass of 52 kD, which was 6 kD more than that of theoretical molecular mass. The higher molecular mass of the protein could be due to posttranslational modifications or aberrant mobility of His tag proteins. To determine autophosphorylation kinetics, the STY kinase was incubated with [γ-32P]ATP in an in vitro kinase assay for various time intervals, and maximum phosphorylation was observed in 20 min (Fig. 3A). The autophosphorylation activity remained the same even at 60 min (data not shown). The stoichiometry of autophosphorylation was calculated to be 3 ± 0.38 mol of phosphate incorporated per mol of STY kinase, which was obtained from 0.1 mm ATP concentration under standard assay conditions. The reaction was linear with the amount of protein (data not shown). The reaction was dependent on Mg2+; however, no phosphorylation was observed either with Ca2+ or Mn2+ (Fig. 3B). Phosphoamino acid analysis of autophosphorylated protein indicated that the STY kinase predominantly phosphorylated Tyr (>80%) but less on phospho-Ser and phospho-Thr (Fig. 3C). This was further confirmed by performing immunoblotting with monoclonal antibodies for all three phosphoamino acids (Fig. 3D). When histone H1 (type III-S) was used as an exogenous substrate, we detected phosphorylation predominantly in one of its degradation product (15 kD) in addition to the protein (Fig. 3E). However, recombinant protein did not phosphorylate substrates such as enolase, casein, and aprotinin, suggesting that the STY kinase is not a promiscuous kinase (data not shown). Phosphoamino acid analysis of histone phosphorylation by the STY kinase indicated that the protein phosphorylated the substrate maximally at Thr and less at Tyr. However, phospho-Ser was not detected in the autoradiogram (Fig. 3F).
Figure 3.
A, Time course of autophosphorylation of STY kinase. B, Effect of divalent cations (10 mm) on the autophosphorylation of STY kinase. C, Phosphoamino acid analysis of autophosphorylated STY kinase. Recombinant STY kinase was autophosphorylated, resolved on 12% (w/v) SDS-PAGE, and transferred onto a polyvinylidene difluoride membrane. The reaction product was hydrolyzed and separated by silica thin-layer chromatography (TLC) as described in “Materials and Methods.” The positions of the origin (ori), phospho-Ser (PS), phospho-Thr (PT), and phospho-Tyr (PY) are indicated along the right side of the TLC. Increasing amounts of hydrolyzed phosphoamino acids were spotted in lanes 1 through 3. D, Autophosphorylated protein was electrophoretically transferred onto a nitrocellulose membrane, and was reacted with the anti-phospho-Ser, anti-phospho-Thr, and anti-phospho-Tyr monoclonal antibodies. E, Five (lane 1) and 10 (lane 2) μg of histone III S was subjected to phosphorylation by STY kinase (750 ng) and the amount of phosphorylated histone was visualized by autoradiography. Molecular mass standards are indicated in the left in kilodaltons. F, Phosphoamino acid analysis of histone-III S phosphorylation by STY kinase.
Expression of STY Kinase in Peanut
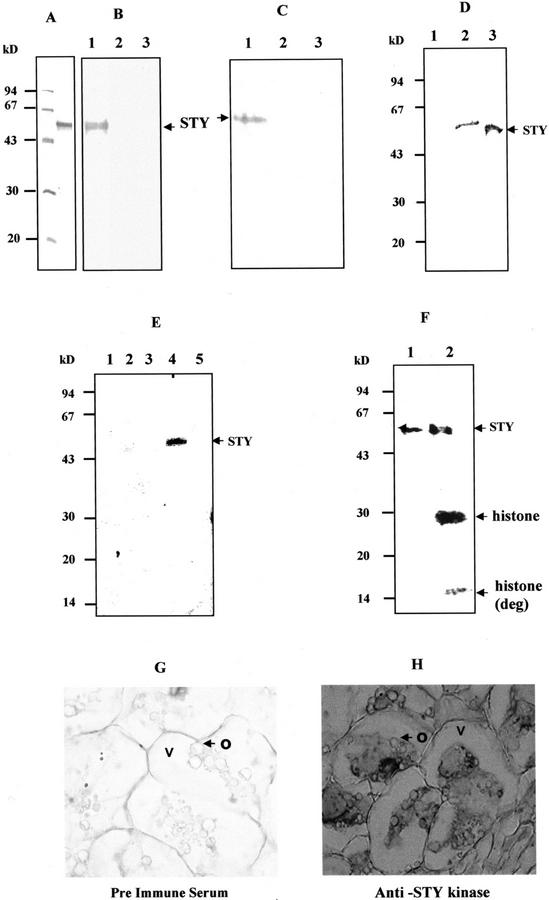
To study the specificity of the antibodies raised against recombinant protein, the antibodies were affinity purified and used for western-blot analysis. His-6-STY was found to cross-react with the affinity-purified immune serum but not with pre-immune serum and immune serum that had been pre-incubated with His-6-STY (Fig. 4, A and B). As shown in the Figure 4C, the monospecific immune serum detected a major protein band of 52 kD from the total protein extracts of immature peanut. The protein was not detected with pre-immune serum and blocked immune serum (Fig. 4C). As a consequence, immunoblotting of microsomal and soluble proteins of immature peanut with STY kinase specific antibodies detected a 52-kD protein in the cytosol (Fig. 4D). Immunoprecipitation and immune complex kinase assay of the soluble and membrane proteins of peanut cotyledons resulted in a 52-kD protein present in the cytosol but not in the membranes (Fig. 4E). The higher molecular mass of the protein could be due to posttranslational modifications. Immune complex kinase assay of the soluble and membrane proteins of peanut with pre-immune serum did not result in any phosphorylated proteins. In addition, immune complex kinase assay of the boiled peanut protein with affinity-purified anti-STY kinase antibodies did not result in labeled proteins (data not shown). Immunoprecipitation of the peanut cytosol followed by immune complex kinase assay with histone revealed that the STY kinase is present as an active kinase in vivo (Fig. 4F). Immunohistochemical analysis indicated that STY kinase is localized in the cytoplasm of seed, corroborating immunoblotting and immunoprecipitation data. The STY kinase antibodies did not immunodecorate the oil bodies of the seed (Fig. 4, G and H).
Figure 4.
Specificity of the antibodies and Intracellular localization of STY kinase. A, Recombinant His-6-STY was purified by nickel affinity column, run on 12% (w/v) SDS-PAGE, and stained with Coomassie Brilliant Blue R-250. Molecular mass standards are indicated on the left. B, Each lane of the 12% (w/v) SDS-polyacrylamide gel contains 200 ng of the recombinant His-6-STY protein. The primary antibodies used were affinity-purified immune serum raised against His-6-STY protein (lane 1), pre-immune serum (lane 2), and blocked immune serum (lane 3). C, Each lane of the 12% (w/v) SDS-PAGE contains 50 μg of total protein extracted from peanut immature seed. The primary antibodies used were the same as described in B. The primary antibodies used were affinity-purified immune serum raised against His-6-STY protein (lane 1), pre-immune serum (lane 2), and blocked immune serum (lane 3). D, Fifty micrograms of peanut membranes (lane 1), cytosol (lane 2), and 500 ng of STY kinase (lane 3) were subjected to immunoblotting using anti-STY kinase antibodies. E, Two-hundred fifty micrograms of CHAPS-solubilized membrane (lanes 1 and 2) and soluble (lanes 3 and 4) fractions were immunoprecipitated using pre-immune serum (lanes 1 and 3) and anti-STY kinase antiserum (lanes 2 and 4) as described in “Materials and Methods” and subjected to phosphorylation. Boiled plant protein is immunoprecipitated using anti-STY kinase antibodies (lane 5). The antibodies were used at 1:100 (v/v) dilution. F, Immunoprecipitation and immune complex assay of STY kinase (lane 1) and histone phosphorylation of immune complex (lane 2) of peanut cytosol. Cytosol (250 μg) was subjected to immunoprecipitation with 1:100 (v/v) dilution of the affinity-purified anti-STY kinase antibodies. G and H, Immunohistochemical localization of STY kinase in peanut seed. Transverse section of seed was incubated with 1:500 (v/v) diluted pre-immune serum (G) or with anti-STY kinase antibodies (H). Tissue sections were incubated with peroxidase-coupled secondary antibodies and developed using 3,3′-diaminobenzidine as a chromogenic substrate. Magnification: ×63. O, Oil bodies; V, vacuole.
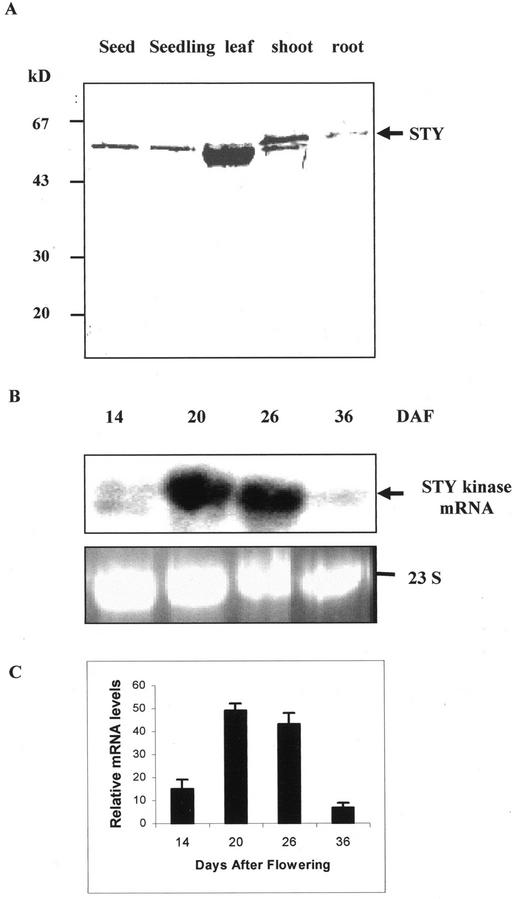
Immunoblot analysis was performed to determine the expression of STY kinase protein levels in different organs of peanut plant. STY kinase protein was detected in all organs of the plant with higher levels in the shoot as compared with other organs studied (Fig. 5A). The developmental expression of the STY kinase gene was examined by RNA gel-blot analysis (Fig. 5B). A high level of mRNA expression was detected at 20 to 26 d after flowering (DAF) that was approximately a 5-fold increase as compared with that of 14 DAF. The levels of mRNA expression drastically reduced at 36 DAF. The size of transcript was consistent with the sequence data and confirms that the isolated cDNA was full length.
Figure 5.
A, Tissue-specific immunoblot analysis of different organs of peanut probed with antibodies raised against the recombinant STY kinase. The band at approximately 50 kD in the leaf and shoot lane corresponds to the large subunit of Rubisco, to which either the primary or secondary antibodies bound nonspecifically. Molecular mass standards are indicated on the left in kilodaltons. B, Developmental stage RNA gel-blot analysis. Four different seed developmental stages based on DAF were analyzed. For the rRNA panel, size-separated total RNA (20 μg) was stained with ethidium bromide to show that similar amounts of RNA were loaded per lane. The prominent band is 23 S rRNA. C, Relative mRNA levels were densitometrically estimated, and histograms were constructed with the data from Figure 5B. Error bars represent the sd of three experiments.
Transcriptional and Posttranslational Activation of STY Kinase by Low Temperature and High Salt
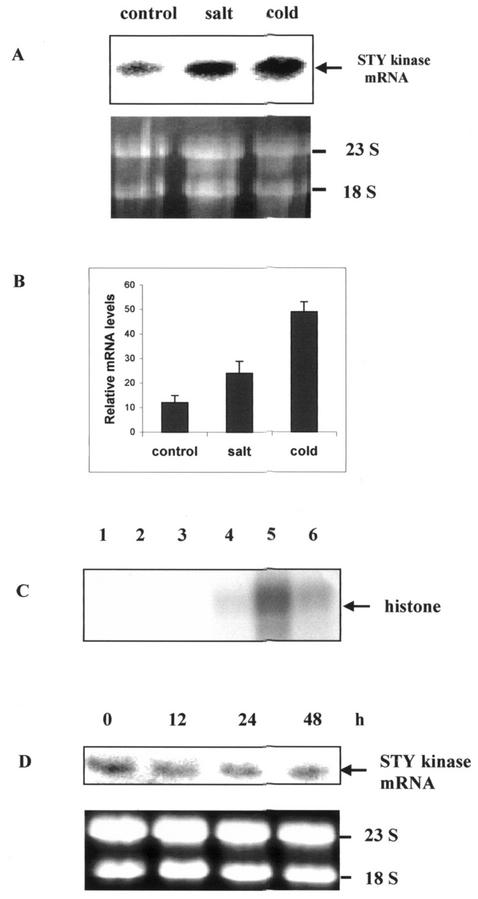
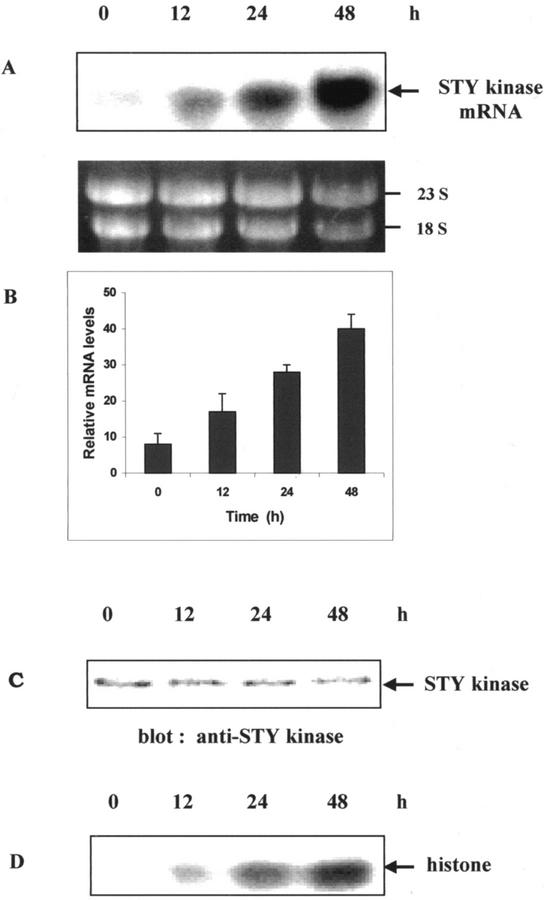
To analyze the expression of STY kinase under various stress conditions, northern-blot analyses and histone kinase activities were carried out and found that the mRNA level and histone kinase activity of STY kinase increased when peanut seedlings were under cold or high-salinity stress conditions. The magnitude of transcript accumulation and kinase activity was higher with cold treatment (4°C) than with NaCl (250 mm) for 24 h (Fig. 6, A–D). Immunoprecipitation and immune complex kinase assays with cold- and salt-treated seedlings with pre-immune serum did not result in any labeled protein (Fig. 6D). We did not observe significant change in the expression of STY kinase when seedlings were subjected to various hormonal treatments (indole acetic acid, benzyl amino purine, gibberillic acid, and ABA; data not shown). There was no change in mRNA levels when peanut seedlings were treated with 100 μm ABA (Fig. 6E). Upon cold treatment, there was a time-dependent up-regulation of mRNA levels (Fig. 7, A and B). In 48 h, the level of mRNA expression was very high. Cold-treated seedlings were immunoblotted with anti-STY kinase antibodies and a 52-kD protein was specifically decorated (Fig. 7C). However, there was no change in the steady-state protein levels with cold-treated seedlings. When aliquots of the same seedlings were immunoprecipitated and subjected to histone kinase assay, time-dependent increase in the histone kinase activity was observed (Fig. 7D). There was no significant change in the mRNA levels and kinase activity with cold treatment at the early time points such as 1 and 6 h (data not shown).
Figure 6.
A, Two-day-old seedlings of peanuts were grown in the dark at 25°C, and treated either with 250 mm NaCl or with 4°C, and harvested after 24 h. Untreated samples were used as controls. Total RNAs were isolated from the peanut seedlings and subjected to northern analysis using STY kinase full-length cDNA as the probe. B, Relative mRNA levels were densitometrically estimated, and histograms were constructed with the data from Figure 6A. Error bars represent the sd of four experiments. C, Two-hundred fifty micrograms of the protein from unstressed, cold-, and salt-stressed seedlings were immunoprecipitated with pre-immune serum (lanes 1–3) and with affinity-purified anti-STY kinase antiserum (lanes 4–6). Histone kinase assay was performed with unstressed (lanes 1 and 4), cold- (lanes 2 and 5), and salt- (lanes 3 and 6) stressed seedlings. D, Two-day-old seedlings of peanuts were grown in the dark at 25°C, and were treated with 100 μm abscisic acid (ABA) and harvested after the indicated time. Total RNAs were isolated from the peanut seedlings and subjected to northern analysis using STY kinase full-length cDNA as a probe. For the rRNA panel, size-separated total RNA (20 μg) was stained with ethidium bromide to show that similar amounts of RNA were loaded per lane. The prominent bands are rRNAs.
Figure 7.
Two-day-old seedlings of peanuts were grown in the dark at 25°C, and were transferred to 4°C and harvested after the indicated time. A, Total RNAs were isolated from the peanut seedlings and subjected to northern analysis using STY kinase full-length cDNA as a probe. For the rRNA panel, size-separated total RNA (20 μg) was stained with ethidium bromide to show that similar amounts of RNA were loaded per lane. The prominent bands are rRNAs. B, Relative mRNA levels were densitometrically estimated, and histograms were constructed with the data from Figure 7A. Error bars represent the sd of three independent experiments. C, Protein extracts (25 μg) from untreated control (0 min) or seedlings treated with cold (4°C) were subjected to immunoblot analysis with STY kinase antibodies at the indicated time periods. D, Protein extracts (250 μg) from untreated control (0 min) or seedlings treated with cold (4°C) were immunoprecipitated with 1:100 (v/v) dilution of anti-STY kinase antibodies. Kinase activity of the immune complex was subsequently determined using histone III-S as an exogenous substrate.
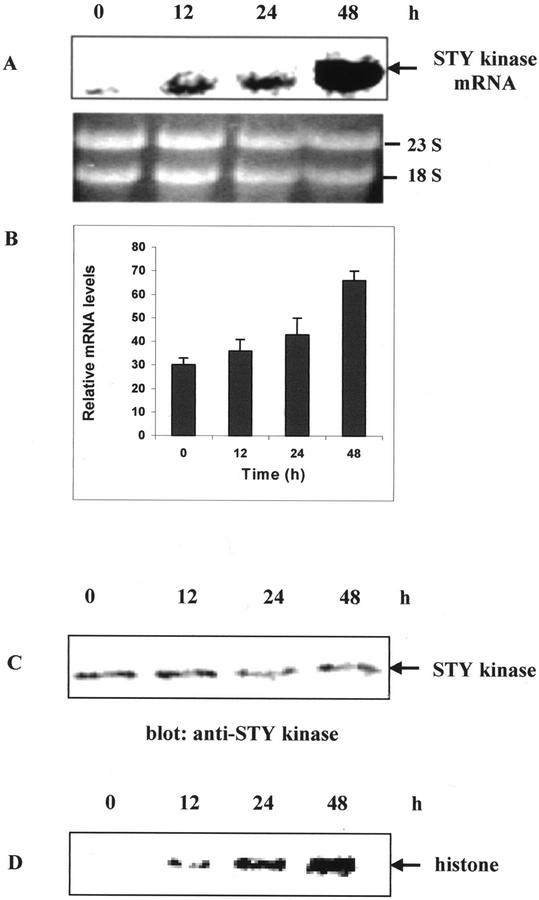
When peanut seedlings were subjected to 250 mm NaCl, the mRNA levels increased proportionately with time and reached maximum at 48 h (Fig. 8, A and B). Untreated seedlings and seedlings treated at the early time point (6 h) showed a basal mRNA expression and kinase activity. Salt-treated seedlings were subjected to immunoblot analysis and a 52-kD protein is specifically decorated; however, there was no change in the steady-state protein levels (Fig. 8C). There was a time-dependent increase in the histone kinase activity with salt-treated seedlings (Fig. 8D). The other abiotic stresses such as high temperature and dehydration (20% [w/v] polyethylene glycol) treatments did not have much effect on STY kinase mRNA expression and kinase activity (data not shown). These results indicate that the STY kinase is regulated by cold and salt treatments in an ABA-independent manner by posttranslational activation mechanism.
Figure 8.
Two-day-old seedlings of peanuts were grown in the dark at 25°C, and were treated with 250 mm NaCl at 25°C and harvested after the indicated time. A, Time course of mRNA expression of salt-treated seedlings. B, Densitometry scan of relative mRNA levels obtained from Figure 8A. Error bars represent the sd of two independent experiments. C, Immunoblot of salt-treated seedlings with STY kinase antibodies at the indicated time periods. D, Protein extracts (250 μg) from untreated control (0 min) or seedlings treated with cold (4°C) were immunoprecipitated with 1:100 (v/v) dilution of anti-STY kinase antibodies. Kinase activity of the immune complex was subsequently determined using histone III-S as an exogenous substrate.
DISCUSSION
This study describes molecular characterization of the developmentally regulated and stress-related dual-specificity kinase from peanut. Protein Tyr kinases play a vital role in the regulation of cell growth, differentiation, and development in animal systems (Sun and Tonks, 1994). On the other hand, no Tyr kinase has been cloned from plants. The translation product of the isolated cDNA shows homology to Tyr kinases in the database. The autophosphorylated kinase is recognized by anti-p-Tyr antibody, indicating that STY kinase autophosphorylates on the Tyr. Additional evidence for Tyr phosphorylation was from phosphoamino acid analysis. The methods of screening expression libraries with anti-p-Tyr antibodies were successfully employed in mammalian systems (Letwin et al., 1992), yeast (Saccharomyces cerevisiae; Stern et al., 1991), and D. discoideum (Tan and Spudich, 1990). The significant feature of the STY kinase cDNA is that the gene product has all the sequence attributes of protein Ser/Thr kinases, even though it was isolated by anti-p-Tyr antibody. Earlier attempts to clone plant cDNAs for classical Tyr kinases using anti-p-Tyr antibodies resulted in dual-specificity protein kinases (Ali et al., 1994). This raises an intriguing possibility that the dual-specificity kinases alone are responsible for Tyr phosphorylation in plants. Arabidopsis ATN1 and soybean GmPK6 are the dual-specificity kinases that are not studied in detail. Arabidopsis ADK1 has been overexpressed and it is shown to autophosphorylate maximally on Thr than Tyr. Peanut STY kinase is the first plant protein kinase shown to autophosphorylate predominantly on Tyr. We have also demonstrated the dual specificity of the STY kinase, even in substrate phosphorylation with histone. Except for the MAP kinases (MEK1), the physiological significance of other dual-specificity kinases is yet to be determined.
The STY kinase sequence has striking features of potential functional significance. There are two LRRs in the STY kinase protein sequence. LRRs are shown to be involved in signal transduction during developmental and environmental cues (Ellis et al., 2000). The presence of LRRs in the transmembrane domain suggests a possible role of STY kinase in plant defense.
The MAP kinase cascade is involved in stress signaling in plants. Besides MAP kinases, other stress-activated plant kinases have been reported wherein the phosphorylation of Ser and Thr, but not Tyr, is responsible for activation of these kinases. These belong to calcium-dependent protein kinases (Sheen, 1996) and the SNF1 family of kinases (Halford and Hardie, 1998). Peanut STY is a novel kinase that does not belong to any of the category of kinases reported so far and thus forms a new division in the plant kinase family. STY kinase is the first report of a non-MAP kinase cascade dual-specificity kinase involved in abiotic stresses. The activation of the STY kinase is detected after 12 to 48 h of cold and salt treatments, which indicates that the kinase may not participate in the initial response to abiotic stresses, but suggesting a possible role in the adaptive process to adverse conditions.
STY kinase is not only induced by abiotic stresses but also responds to developmental cues. The transcript of STY kinase is accumulated in the mid-cotyledonary stage of seed development, where the metabolism of proteins and lipids is highly active. It is possible that peanut STY kinase could also be involved in the signal transduction mechanisms related to storage of metabolites. The regulation of cold-inducible genes (RC12A, RC12B, COR6.6, COR15, COR78, DREB1, RD29A, and KIN1) during Arabidopsis development has been studied in detail, and the results indicated that the expression of these genes is regulated in different tissues both in stressed and unstressed conditions (Yamaguchi-Shinozaki and Shinozaki, 1993; Baker et al., 1994; Wang and Cutler, 1995; Xu et al., 1995; Medina et al., 2001). Low-temperature-responsive genes (BLT101, BLT4.9, and BLT14) have been reported in barley, and these genes are also regulated during development (Pearce et al., 1998). It is possible that many genes regulated during plant development are also regulated by environmental stresses. It has been reported earlier that some of the salt-inducible genes are also induced by cold stress (Kurkela and Borg-Franck, 1992; Yamaguchi-Shinozaki and Shinozaki, 1994).
Cold and salt stresses increased STY kinase mRNA levels. The activation of STY kinase preceded the increase in mRNA levels, suggesting that the STY kinase gene might be a target of the kinase pathway. This pattern is reminiscent of positive feedback loop that occurs in the activation of several protein kinase cascades. However, an increase of STY kinase transcripts was not correlated with an increase in the protein levels. It could be possible that the transcripts accumulate but are not translated. Alternatively, the translation of the STY kinase transcripts and steady-state protein levels of STY kinase may stay constant. A similar observation was made for alfalfa (Medicago sativa) MAP kinase (MMK4; Jonak et al., 1996). In addition, the transcripts of glycogen synthase kinase-3 were accumulated after wounding in alfalfa and the protein levels remained constant; however, there was an increase in kinase activity (Jonak et al., 2000). These results indicated that protein amounts were tightly regulated by complex intracellular mechanisms in some protein kinases involved in stress response and adaptation.
Stress adaptation in plants is mediated either by ABA-dependent or -independent pathways (Ishitani et al., 1987). ABA has an important role in mediating responses to environmental stresses, including cold and salt stresses. Exogenously applied ABA is also able to induce expression of several cold and salt stress genes (Shinozaki and Yamaguchi-Shinozaki, 1997). When peanut seedlings were treated with ABA, no changes in the mRNA levels or kinase activity were observed. These experiments indicate that the cold- and salt-induced activation of STY kinase is independent of ABA. The expression of several cold- or salt-induced genes is independent of ABA, indicating the presence of ABA-dependent and -independent pathways for low-temperature and salt responses.
Because peanut is a chilling-tolerant species, identification of the STY kinase as a cold stress-responsive gene may enable to genetically engineer the chilling-sensitive plants for chilling tolerance. Also, involvement of STY kinase in seed development offers opportunities to manipulate seed development and maturation. The identification of STY kinase as a stress-responsive and developmentally regulated dual-specificity kinase may provide new insights in unraveling plant signal transduction pathways.
MATERIALS AND METHODS
Plant Material and Treatments
Field-grown developing peanut (Arachis hypogaea) cotyledons were harvested at various stages after flowering and used either fresh or frozen at −80°C. Toward stress treatments, peanut seeds were surface sterilized and grown in water-saturated filter paper (Whatman, Clifton, NJ) in petri dishes for 2 d. Cold (4°C) and salt (250 mm NaCl) treatments were given at various time intervals. Towards hormonal treatment of seedlings, 100 μm indole acetic acid, benzyl amino purine, gibberillic acid, and ABA were used.
Construction and Screening of cDNA Library
A seed-specific cDNA library of peanut was constructed in λ-ZAP II (Stratagene, La Jolla, CA). To produce anti-phospho-Tyr antibodies, rabbit polyclonal antiserum was raised against a polymer of phospho-Tyr, Ala, Gly, and keyhole limpet hemocyanin and affinity purified as described (Kornbluth et al., 1988). The expression library was screened using anti-phospho-Tyr antibodies. A total of 2 × 106 plaques were screened. Positive plaques were purified by three additional screenings with the polyclonal antibodies and parallel with a monoclonal anti-p-Tyr antibody (Sigma, St. Louis).
Sequencing and Sequence Analysis
Plasmid DNA was isolated from the positive clones by alkaline lysis (Sambrook et al., 1989), and was sequenced on both strands using the Biotech Taq cycle sequencing kit (Amersham-Pharmacia Biotech, Uppsala) on an automated sequencer (Applied Biosystems 377, PE-Applied Biosystems, Foster City, CA). Sequence analysis was performed using BLAST search (http://www.ncbi.nlm.nih.gov/BLAST). Multiple Sequence alignment obtained using the ClustalW (http://www.ebi.ac.uk/) were displayed using the program GENEDOC (http://bioinformer.ebi.ac.uk/newsletter/archives/1/genedoc.shtml). Molecular phylogenies were computed by PHYLIP (Phylogeny Inference Package, version 3.52, Department of Genetics, University of Washington, Seattle) and tree files generated by the above methods were displayed and printed using TREEVIEW.
Genomic Southern and Northern Hybridization Analyses
Towards Southern analysis, high-Mr genomic DNA was isolated from peanut cotyledons (Murray and Thompson, 1980). Twenty micrograms of DNA was digested with EcoRI, SacI, and BamHI, fractionated on 0.8% (w/v) agarose gel, and transferred onto a Hybond N+ nylon membrane (Southern, 1975). For northern analysis, total RNA was extracted from cotyledons at four different stages of development and stress-induced seedlings (Verwoerd et al., 1989). Aliquots of total RNA (20 μg) were run on 1% (w/v) agarose gels containing 19% (v/v) formaldehyde, 40 mm MOPS, 10 mm sodium acetate, 1 mm EDTA, and transferred to Hybond N+ membrane (Amersham-Pharmacia Biotech). Southern and northern hybridizations were carried out in aqueous hybridization buffer (phosphate buffer [pH 7.2], 7% [w/v] SDS, and 20 mm EDTA) with a random-primed 32P-labeled STY kinase cDNA as a probe at 65°C. Blots were washed in 0.2× SSC and 0.1% (w/v) SDS at 65°C. Equal loading of RNA was confirmed by visual comparison of ethidium bromide-stained 18 S and 23 S rRNAs of the agarose gel.
Expression and Purification of His-6-STY Kinase Fusion Protein
The cDNA spanning for coding region of STY kinase (lacking 11 amino acids from the N terminus) was subcloned into the His-tagged fusion protein expression vector, pRSET C, at bglII and kpn1 cloning sites. The resultant construct was expressed in Escherichia coli BL-21 (pLysS). The fusion protein was induced with 0.4 mm isopropyl-1-thio-β-d-galactopyranoside for 4 h. The recombinant protein was induced in large scale and purified by nickel-nitrilotriacetic acid agarose chromatography (Qiagen USA, Valencia, CA). Purified fractions containing the eluted protein were analyzed by 12% (w/v) SDS-PAGE followed by Coomassie Blue staining (Laemmli, 1970).
In Vitro Kinase Assays
Autophosphorylation assay was performed by incubating 0.5 μg of purified recombinant STY kinase in reaction buffer (50 mm Tris-HCl [pH 7.5] and 10 mm MgCl2) in the presence of 25 μm [γ-32P]ATP (3,000 dpm pmol−1) at 30°C for 20 min. The reaction was stopped by the addition of SDS-PAGE loading buffer, and kinase activity was detected by autoradiography after 12% (w/v) SDS-PAGE. Substrate phosphorylation assay mixture consisted of 750 ng of STY kinase, 5 μg of histone III-S (Sigma), 50 mm Tris-HCl (pH 7.5), and 10 mm MgCl2, and the incubation was carried out at 30°C for 20 min. Phosphorylated products were separated by 12% (w/v) SDS-PAGE and the labeled proteins were detected by autoradiography.
Phosphoamino Acid Analysis
The purified STY kinase was labeled in vitro with [γ-32P]ATP as described above and electroblotted onto a polyvinylidine difluoride membrane. After autoradiography, radioactive band of interest was excised and hydrolyzed in 200 μL of 6 m HCl for 2 h at 110°C. The hydrolyzate was dried in a Speed-Vac concentrator and resuspended in 20 μL of water containing 1 mg mL−1 of each of the phosphoamino acid markers such as phospho-Ser, phospho-Thr, and phospho-Tyr (Sigma). Two microliters of the hydrolyzate was analyzed by ascending silica TLC (Merck, Rahway, NJ) using a solvent system containing a mixture of ethanol and ammonia (3.5:1.6 [v/v]; Munoz and Marshall, 1990). The position of phosphoamino acid markers was detected by ninhydrin staining of the silica-TLC plate (0.25% [w/v] ninhydrin in acetone). The plate was then exposed for autoradiography to locate the position of the 32P-labeled amino acids.
Immunoblotting
Two New Zealand white male rabbits were subcutaneously immunized at 2-week intervals with 0.5 to 1.0 mg of fusion protein. The polyclonal antiserum was purified by protein A-Sepharose column chromatography. Subcellular fractionation of peanut cotyledon was performed (Tumaney et al., 2001) and protein concentrations were determined by the Bradford method (Bradford, 1976) using bovine serum albumin as the standard. For immunodetection of STY kinase proteins, the soluble and membrane fractions were isolated from immature peanuts and separated on 12% (w/v) SDS-polyacrylamide gels. The proteins were transferred onto a nitrocellulose membrane (Towbin et al., 1979) and the blots were then probed with anti-STY kinase antibodies (1:2,000 dilution [v/v]) for 60 min. Blots were washed in phosphate-buffered saline with 0.05% Tween-20, and incubated for 60 min with peroxidase-coupled secondary antibodies. Proteins were detected by using 3,3′-diaminobenzidine as a chromogenic substrate.
Immunoprecipitation and Immune Complex Kinase Assay
The soluble and membrane proteins (200 μg) from immature peanuts were precleared with 25 μL of protein A-agarose (Bangalore Genei, Bangalore, India) beads for 1 h. The agarose beads were removed by centrifugation and 2 μg of affinity-purified antibody was added to the extracts and incubated with agitation for 6 h at 4°C. After brief centrifugation, the precipitate was washed three times with buffer (250 mm Tris-HCl [pH 7.5] and 10 mm MgCl2). The immunoprecipitated proteins were phosphorylated as described before.
Immunohistochemistry and Microscopy
Peanut tissues were fixed in formaldehyde:acetic acid:70% (v/v) ethanol (5:5:90 [v/v]), dehydrated in ethanol followed by infiltration with paraffin in n-butanol, and microtome sectioned (0.5 μm). The sections were mounted on glass slides, deparaffinized with xylene, rehydrated, and incubated with 0.6% (v/v) hydrogen peroxide in methanol. The sections were then incubated in a blocking solution containing 5% (v/v) goat serum and 1% (w/v) bovine serum albumin in phosphate-buffered saline for 60 min at room temperature. The slides were placed in a humidified chamber and sections were covered with 2 μg mL−1 of affinity-purified anti-STY kinase antibodies diluted in phosphate-buffered saline with 1% (w/v) bovine serum albumin for 1 h. Control for specificity included 2 μg mL−1 rabbit pre-immune serum in place of primary antibodies. The sections were then incubated with 0.05% (w/v) 3,3′-diaminobenzidine in 0.1% (v/v) hydrogen peroxide to produce a brown reaction product. Images were captured using a Zeiss-Axioscope microscope (Zeiss, Jena, Germany).
GenBank Accession Number
The GenBank accession number for the nucleotide sequence reported in the article is AY027437.
Footnotes
This research was supported by a grant from the Council of Scientific and Industrial Research (New Delhi, India).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.005173.
LITERATURE CITED
- Ali N, Halfter U, Chua NH. Cloning and biochemical characterization of a plant protein kinase that phosphorylates serine, threonine and tyrosine. J Biol Chem. 1994;269:31626–31629. [PubMed] [Google Scholar]
- Baker SS, Wilhelm KS, Thomashow MF. The 5′-region of Arabidopsis thaliana cor15ahas cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol Biol. 1994;24:701–713. doi: 10.1007/BF00029852. [DOI] [PubMed] [Google Scholar]
- Barizza E, Schiavo FL, Terzi M, Filippini F. Evidence suggesting protein tyrosine phosphorylation in plants depends on the developmental conditions. FEBS Lett. 1999;447:191–194. doi: 10.1016/s0014-5793(99)00272-0. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Ellis J, Dodds P, Pryor T. The generation of plant disease resistance gene specificities. Trends Plant Sci. 2000;5:373–379. doi: 10.1016/s1360-1385(00)01694-0. [DOI] [PubMed] [Google Scholar]
- Feng XH, Zhao Y, Bottino PJ, Kung SD. Cloning and characterization of a novel member of protein kinase family from soybean. Biochim Biophys Acta. 1993;1172:200–204. doi: 10.1016/0167-4781(93)90295-o. [DOI] [PubMed] [Google Scholar]
- Halford NG, Hardie DG. SNF1-related protein kinases: global regulators of carbon metabolism in plants? Plant Mol Biol. 1998;37:735–748. doi: 10.1023/a:1006024231305. [DOI] [PubMed] [Google Scholar]
- Hanks SK, Hunter T. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- Hirayama T, Oka A. Novel protein kinase of Arabidopsis thaliana(APK1) that phosphorylates tyrosine, serine and threonine. Plant Mol Biol. 1992;20:653–662. doi: 10.1007/BF00046450. [DOI] [PubMed] [Google Scholar]
- Hunter T. A thousand and one protein kinases. Cell. 1987;50:823–829. doi: 10.1016/0092-8674(87)90509-5. [DOI] [PubMed] [Google Scholar]
- Ishitani M, Xiong L, Stevenson B, Zhu JK. Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell. 1987;9:935–949. doi: 10.1105/tpc.9.11.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islas-Flores I, Oropeza C, Hernandez-Sotomayor SMT. Protein phosphorylation during coconut zygotic embryo development. Plant Physiol. 1998;118:257–263. doi: 10.1104/pp.118.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonak C, Beisteiner D, Beyerly J, Hirt H. Wound-induced expression and activation of WIG, a novel glycogen synthase kinase 3. Plant Cell. 2000;12:1467–1476. doi: 10.1105/tpc.12.8.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonak C, Kiegrel S, Ligternik W, Barker PJ, Huskisson NS, Hirt H. Stress signaling in plants: A mitogen activated protein kinase pathway is activated by cold and drought. Proc Natl Acad Sci USA. 1996;93:11274–11279. doi: 10.1073/pnas.93.20.11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazuhisa K, Yoshiro K, Masanori Y, Nobuyuki K, Masazumi S, Takahide T. Tyrosine phosphorylation in plant bending. Nature. 2000;407:37–38. doi: 10.1038/35024149. [DOI] [PubMed] [Google Scholar]
- Kornbluth S, Paulson KE, Hanafusa H. Novel tyrosine kinase identified by phosphotyrosine antibody screening of cDNA libraries. Mol Cell Biol. 1988;8:5541–5544. doi: 10.1128/mcb.8.12.5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurkela S, Borg-Franck M. Structure and expression of Kin2, one of two cold- and ABA-induced genes of Arabidopsis thaliana. Plant Mol Biol. 1992;19:689–692. doi: 10.1007/BF00026794. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Letwin K, Mizzen L, Motro B, Ben-David Y, Bernstein A, Pawson T. A mammalian dual specificity protein kinase, Nek1, is related to the NIMA cell cycle regulator and highly expressed in meiotic germ cells. EMBO J. 1992;11:3521–3531. doi: 10.1002/j.1460-2075.1992.tb05435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina J, Catala R, Salinas J. Developmental and stress regulation of RC12A and RC12B, two cold-inducible genes of Arabidopsis encoding highly conserved hydrophobic proteins. Plant Physiol. 2001;125:1655–1666. doi: 10.1104/pp.125.4.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz G, Marshall SH. An alternative method for a fast separation of phosphotyrosine. Anal Biochem. 1990;190:233–237. doi: 10.1016/0003-2697(90)90185-c. [DOI] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce RS, Houlston CE, Atherton KM, Rixon JE, Harrison P, Hughes MA, Dunn M. Localization of expression of three cold-induced genes, blt101, blt4.9, and blt14, in different tissues of the crown and developing leaves of cold-acclimated cultivated barley. Plant Physiol. 1998;117:787–795. doi: 10.1104/pp.117.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Vol. 1. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. Extraction and purification of plasmid DNA; pp. 25–28. [Google Scholar]
- Sheen J. Ca+2dependent protein kinases a stress signal transduction in plants. Science. 1996;274:900–1902. doi: 10.1126/science.274.5294.1900. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Gene expression and signal transduction in water-stress response. Plant Physiol. 1997;115:327–334. doi: 10.1104/pp.115.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;218:23–31. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stern DF, Zheng P, Beidler DR, Zerillo C. Spk1, a new kinase from Saccharomyces cerevisiae, phosphorylates proteins on serine, threonine and tyrosine. Mol Cell Biol. 1991;11:987–1001. doi: 10.1128/mcb.11.2.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JM, Walker JC. Plant protein kinase families and signal transduction. Plant Physiol. 1995;108:451–457. doi: 10.1104/pp.108.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Tonks NK. The coordinated action of protein tyrosine phosphatases and kinases in cell signaling. Trends Biochem Sci. 1994;19:480–485. doi: 10.1016/0968-0004(94)90134-1. [DOI] [PubMed] [Google Scholar]
- Sun Y, Dilkes BP, Zhang C, Dante RA, Carneiro NP, Lowe KS, Jung R, Gordon-Kamm WJ, Larkins BA. Characterization of maize (Zea maysL.) Wee 1 and activity in developing endosperm. Proc Natl Acad Sci USA. 1999;96:4180–4185. doi: 10.1073/pnas.96.7.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Shinshi H. Transient activation and tyrosine phosphorylation of a protein kinase in tobacco cells treated with a fungal elicitor. Plant Cell. 1995;7:639–647. doi: 10.1105/tpc.7.5.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JL, Spudich JA. Developmentally regulated protein-tyrosine kinase genes in Dictyostelium discoideum. Mol Cell Biol. 1990;10:3578–3583. doi: 10.1128/mcb.10.7.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tregear JW, Jouannic S, Schwebel-Dugue N, Kreis M. An unusual protein kinase displaying characteristics of both the serine/threonine kinase and tyrosine families is encoded by the Arabidopsis thalianagene ATN1. Plant Sci. 1996;117:107–119. [Google Scholar]
- Tumaney AW, Shekar S, Rajasekharan R. Identification, purification, and characterization of monoacylglycerol acyltransferase from developing peanut cotyledons. J Biol Chem. 2001;276:10847–10852. doi: 10.1074/jbc.m100005200. [DOI] [PubMed] [Google Scholar]
- Verwoerd TC, Dekker BMM, Hoekema A. A small scale procedure for rapid isolation of plant RNAs. Nucleic Acids Res. 1989;17:2362–2369. doi: 10.1093/nar/17.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Cutler AJ. Promoters from kin1 and cor 6.6, two Arabidopsis thalianalow-temperature- and ABA-inducible genes, direct strong beta-glucuronidase expression in guard cells, pollen and young developing seeds. Plant Mol Biol. 1995;28:619–634. doi: 10.1007/BF00021188. [DOI] [PubMed] [Google Scholar]
- Weijland A, Williams JC, Neubauer G, Courtneidge SA, Wierenga RK, Superti-Furga G. Src regulated by C-terminal phosphorylation is monomeric. Proc Natl Acad Sci USA. 1997;94:3590–3595. doi: 10.1073/pnas.94.8.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Doshi A, Lei M, Eck MJ, Harrison SC. Crystal structures of c-Src reveal features of its autoinhibitory mechanism. Mol Cell. 1999;3:629–638. doi: 10.1016/s1097-2765(00)80356-1. [DOI] [PubMed] [Google Scholar]
- Xu W, Purugganan MM, Polisensky DH, Antosiewicz DM, Fry SC, Braam J. Arabidopsis TCH4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. Plant Cell. 1995;7:1555–1567. doi: 10.1105/tpc.7.10.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. The plant hormone abscisic acid mediates the drought-induced expression but not the seed specific expression of rd22, a gene responsive to dehydration-stress in Arabidopsis thaliana. Mol Gen Genet. 1993;238:17–25. doi: 10.1007/BF00279525. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsisgene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell. 1994;9:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Klessig DF. MAPK cascades in plant defense signaling. Trends Plant Sci. 2001;6:520–527. doi: 10.1016/s1360-1385(01)02103-3. [DOI] [PubMed] [Google Scholar]