Abstract
Saturated and unsaturated N-acylethanolamines (NAEs) occur in desiccated seeds primarily as 16C and 18C species with N-palmitoylethanolamine and N-linoleoylethanolamine (NAE 18:2) being most abundant. Here, we examined the metabolic fate of NAEs in vitro and in vivo in imbibed cotton (Gossypium hirsutum) seeds. When synthetic [1-14C]N-palmitoylethanolamine was used as a substrate, free fatty acids (FFA) were produced by extracts of imbibed cottonseeds. When synthetic [1-14C]NAE 18:2 was used as a substrate, FFA and an additional lipid product(s) were formed. On the basis of polarity, we presumed that the unidentified lipid was a product of the lipoxygenase (LOX) pathway and that inclusion of the characteristic LOX inhibitors nordihydroguaiaretic acid and eicosatetraynoic acid reduced its formation in vitro and in vivo. The conversion of NAE 18:2 in imbibed cottonseed extracts to 12-oxo-13-hydroxy-N-(9Z)-octadecanoylethanolamine was confirmed by gas chromatography-mass spectrometry, indicating the presence of 13-LOX and 13-allene oxide synthase, which metabolized NAE 18:2. Cell fractionation studies showed that the NAE amidohydrolase, responsible for FFA production, was associated mostly with microsomes, whereas LOX, responsible for NAE 18:2-oxylipin production, was distributed in cytosol-enriched fractions and microsomes. The highest activity toward NAE by amidohydrolase was observed 4 to 8 h after imbibition and by LOX 8 h after imbibition. Our results collectively indicate that two pathways exist for NAE metabolism during seed imbibition: one to hydrolyze NAEs in a manner similar to the inactivation of endocannabinoid mediators in animal systems and the other to form novel NAE-derived oxylipins. The rapid depletion of NAEs by these pathways continues to point to a role for NAE metabolites in seed germination.
In mammalian cells, N-acylethanolamines (NAEs) have varied physiological roles. N-Arachidonylethanolamine (anandamide), a type of NAE in mammalian brain tissue, is an endogenous ligand for the cannabinoid receptor and modulates neurotransmission. Anandamide also can activate vanilloid (capsaicin) receptors and function as an endogenous analgesic (Pertwee, 2001), and appears to be involved in neuroprotection (Hansen et al., 2000; Van der Stelt et al., 2001). In other animal tissues, NAEs have been implicated in immunomodulation (Buckley et al., 2000), synchronization of embryo development (Paria and Dey, 2000), and induction of apoptosis (Sarker et al., 2000). These endogenous bioactive molecules termed “endocannabinoids” are hydrolyzed by fatty acid amidohydrolase (AHase) to terminate their signaling functions.
In plants NAEs are present in substantial amounts in desiccated cotton (Gossypium hirsutum) seeds (1.6 μg−1 g fresh weight), and their levels decline after a few hours of imbibition (Chapman et al., 1999). Individual NAEs were identified predominantly as 16C and 18C species with N-palmitoylethanolamine (NAE 16:0) and N-linoleoylethanolamine (NAE 18:2) being the most abundant. NAEs in both plant and animal cells are derived from N-acylphosphatidylethanolamines (NAPEs), a minor membrane lipid constituent of cellular membranes (Schmid et al., 1990; Chapman, 2000). NAEs are produced by the action of a phospholipase D (PLD). In plants NAEs were produced in cell suspensions (Chapman et al., 1998) and leaves (Tripathy et al., 1999) in minutes after pathogen elicitor perception, raising the possibility that these molecules function in plant defense signaling. Exogenous NAE 14:0 at submicromolar concentrations was sufficient to activate Phe ammonia-lyase (PAL) expression in cell suspensions and leaves of tobacco (Nicotiana tabacum; Tripathy et al., 1999). The occurrence of NAEs in seeds with substantially different structural properties than those found in elicitor treated leaves and their rapid depletion during seed imbibition (Chapman, 2000) suggest that these lipids may have a role in the regulation of seed germination.
Recent studies have shown that NAE 18:2 and NAE 18:3 could be converted into hydroperoxy NAE by purified soybean (Glycine max) lipoxygenase-1 (LOX; Van der Stelt et al., 1997, 2000). The hydroperoxides of NAE 18:2 and NAE 18:3 could subsequently be converted by alfalfa (Medicago sativa) hydroperoxide (HPO) lyase and flax (Linum usitatissimum) seed allene oxide synthase (AOS) into novel oxylipins (Van der Stelt et al., 2000). N-Arachidonoylethanolamine (anandamide), the mammalian neurotransmitter, could be converted into hydroperoxy NAE by purified 5-LOX from barley (Hordeum vulgare) and tomato (Lycopersicon esculentum; Van Zadelhoff et al., 1998). As an alternative, NAE AHase, also designated fatty acid amide hydrolase (FAAH; Cravatt et al., 2001), acts upon NAE to produce free fatty acid (FFA) and ethanolamine, and in mammalian cells this pathway is responsible for inactivation of endocannabinoid lipid mediators (Schmid, 2000). Thus, there are two possible enzymatic pathways (LOX and NAE AHase) that might be responsible for the observed decline in NAEs during seed imbibition.
To begin to understand the role of NAEs in seeds, we investigated the metabolic fate of NAEs in cottonseeds upon imbibition, germination, and during postgerminative growth, a period previously noted to be active in NAPE/NAE metabolism (Chapman, 2000). Our results indicate that there are two pathways capable of metabolizing NAEs in seeds: a LOX-mediated pathway selective for unsaturated NAEs (e.g. NAE 18:2) and a NAE AHase activity, which uses both unsaturated and saturated NAEs. Both enzymatic pathways were most active in imbibed seeds consistent with depletion of NAEs in vivo and at a time period just preceding or coincident with radicle emergence, suggesting NAE metabolism may play a role in the regulation of seed germination. These results will provide the basis for future studies aimed at understanding the functional role of NAE metabolism in seed germination and seedling growth.
RESULTS
Identification of NAE Metabolites and Subcellular Distribution of the Enzymes
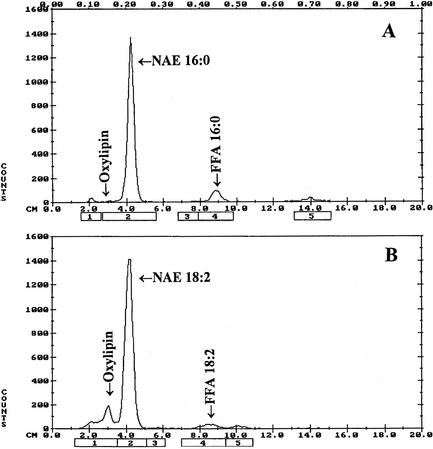
Several types of NAEs are prevalent in the desiccated seeds of plants (Chapman et al., 1999). For example, the total NAE content in desiccated cottonseeds was approximately 1,600 ng g−1 fresh weight, of which NAE 18:2 was approximately 940 ng g−1 fresh weight and NAE 16:0 was about 380 ng g−1 fresh weight. The levels of these NAEs declined sharply within few hours of seed imbibition to 200 ng for NAE 18:2 and to about 160 ng for NAE 16:0. Here, these two most abundant NAEs in desiccated seeds, NAE 16:0 and NAE 18:2, were used to evaluate the metabolic fate of NAE in imbibing seeds. When [1-14C]16:0 NAE was used as substrate, there was only one product (comigrating with palmitic acid, Rf ≅ 0.48) formed by extracts of imbibed cottonseeds (Fig. 1A). When [1-14C]18:2 NAE was used as a substrate there were two apparent products formed (Fig. 1B): one comigrating with linoleic acid (Rf ≅ 0.46) and the other more polar, near the origin (Rf ≅ 0.12), was tentatively identified as NAE oxylipin. These data suggested that endogenous NAEs in desiccated seeds were metabolized by two pathways: one producing FFAs and other likely producing NAE-derived oxylipins.
Figure 1.
Representative chromatograms of radioactive lipids separated on thin-layer chromatography (TLC) plates. For the assay, 100 μm [14C]NAE with 20,000 dpm in 50 mm MES buffer (pH 6.5) was used. Reactions were initiated by adding 400 μL of microsomes and were incubated at 30°C for 1 h with shaking. Lipids are extracted as described in “Materials and Methods,” and distribution of radioactivity on TLC plates was evaluated by radiometric scanning (Bioscan system 200 image scanner). When synthetic NAE 16:0 was used as a substrate, FFA was produced (A) and when synthetic NAE 18:2 was used as a substrate, FFA and an additional lipid product(s) were formed (B).
The subcellular distribution of NAE-LOX (NAE oxylipin formation) and NAE AHase (FFA formation) differed in cell fractions prepared from 4-h imbibed cottonseeds (Tables I and II). The presumed NAE-oxylipin formation was distributed both in membrane and cytosol-enriched fractions, whereas the enzyme responsible for FFA formation was localized almost exclusively to microsomes (Tables I and II). AHase activity toward NAE 18:2 was higher than AHase activity toward NAE 16:0, whereas formation of NAE oxylipin from NAE 18:2 was considerably higher than the corresponding AHase activity. These relative activities toward the different NAEs are consistent with the more rapid consumption of NAE 18:2 in vivo during seed imbibition.
Table I.
Subcellular distribution of NAE-LOX (NAE oxylipin formation) and NAE amidohydrolase (FFA formation) activities in 4-h imbibed cottonseed cell fractions utilizing NAE 16:0 as substrate
| Cell Fraction | FFA
|
Oxylipin
|
||
|---|---|---|---|---|
| Total activity | Specific activity | Total activity | Specific activity | |
| nmol h−1 | nmol h−1 mg−1 protein | nmol h−1 | nmol h−1 mg−1 protein | |
| 10,000g Supernatant | 51.04 ± 1.42 | 0.53 ± 0.01 | 0 | 0 |
| 10,000g Pellet | 0 | 0 | 0 | 0 |
| 150,000g Supernatant | 0 | 0 | 0 | 0 |
| 150,000g Pellet | 10.68 ± 0.51 | 0.63 ± 0.03 | 0 | 0 |
Cell fractions were prepared in (and pellets resuspended in) 100 mm potassium-phosphate (pH 7.2), 10 mm KCl, 1 mm EDTA, 1 mm EGTA, and 400 mm Suc. For assays, 100 μm [14C]NAE 16:0 (20,000 dpm) in 50 mm MES buffer (pH 6.5) was used. Reactions were initiated by adding 400 μL of respective cell fraction in a total volume of 800 μL. The data are means and sd of three replicates and are representative of three experiments.
Table II.
Subcellular distribution of NAE-LOX (NAE oxylipin formation) and NAE amidohydrolase (FFA formation) activities in 4-h imbibed cottonseed cell fractions utilizing NAE 18:2 as substrate
| Cell Fraction | FFA
|
Oxylipin
|
||
|---|---|---|---|---|
| Total activity | Specific activity | Total activity | Specific activity | |
| nmol h−1 | nmol h−1 mg−1 protein | nmol h−1 | nmol h−1 mg−1 protein | |
| 10,000g Supernatant | 33.06 ± 2.37 | 0.35 ± 0.02 | 780.97 ± 3.08 | 8.16 ± 0.03 |
| 10,000g Pellet | 0 | 0 | 18.09 ± 1.67 | 0.46 ± 0.04 |
| 150,000g Supernatant | 0 | 0 | 435.49 ± 2.87 | 7.01 ± 0.05 |
| 150,000g Pellet | 19.46 ± 2.27 | 1.15 ± 0.13 | 126.16 ± 1.56 | 7.44 ± 0.08 |
Cell fractions were prepared in (and pellets resuspended in) 100 mm potassium-phosphate (pH 7.2), 10 mm KCl, 1 mm EDTA, 1 mm EGTA, and 400 mm Suc. For assays, 100 μm [14C]NAE 18:2 (20,000 dpm) in 50 mm MES buffer (pH 6.5) was used. Reactions were initiated by adding 400 μL of respective cell fraction in a total volume of 800 μL. The data are means and sd of three replicates and are representative of three experiments.
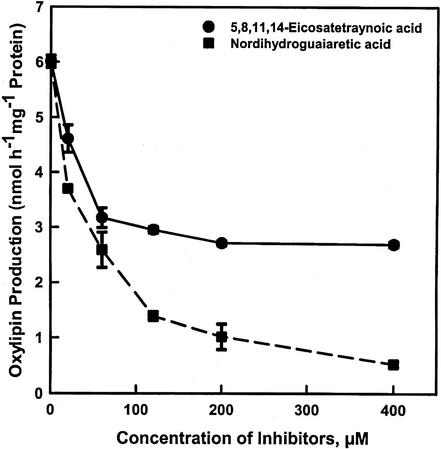
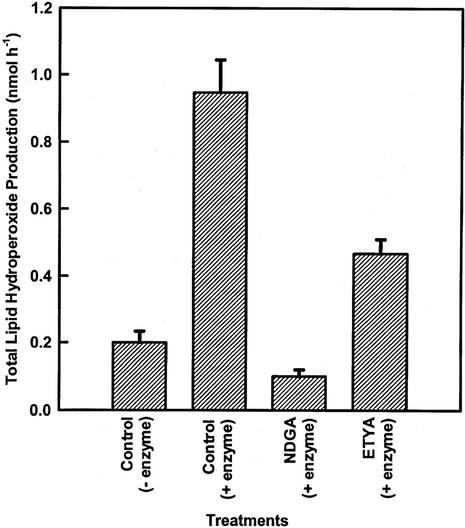
To test whether the oxylipins were formed by the LOX pathway, the influence of two widely used LOX inhibitors on their formation was determined (Fig. 2). Both 5,8,11,14-eicosatetraynoic acid (ETYA) and nordihydroguaiaretic acid (NDGA) reduced NAE-oxylipin formation in a concentration-dependent manner. NDGA appeared to be a more potent inhibitor of NAE 18:2-LOX than ETYA, particularly at higher concentrations. As an alternative, NAE 18:2-dependent lipid peroxide formation was estimated spectrophotometrically (Fig. 3). Consistent with the above results, inclusion of both inhibitors reduced the formation of NAE 18:2 lipid hydroperoxide. The small amount of lipid peroxide detected in the absence of enzyme (control-enzyme) was likely attributable to the spontaneous oxidation of NAE 18:2 during assay reactions, because no lipid peroxide was detected when NAE 18:2 was omitted from reactions (not shown). These data indicate that the polar product in the incubation is formed by the LOX pathway.
Figure 2.
The effects of LOX inhibitors on the metabolism of NAE 18:2 in vitro. The amount of NAE-oxylipin was determined by incubating (1 h) synthetic NAE 18:2 with a 150,000gmax (60 min) supernatant of imbibed cottonseeds. Total lipids were extracted from the reaction mixture and were separated by TLC (hexane:ethyl acetate:methanol, 60:40:5; v/v). Identification and quantification of radiolabeled lipids were performed by radiometric scanning. ETYA is a dual-specific inhibitor, affecting both LOX and cyclooxygenases, and is irreversible (Grullich et al., 2001). NDGA is a classical inhibitor of different LOXs (Kulkarni and Sajan, 1999). There was almost complete inhibition of oxylipin production at 400 μm NDGA and 50% inhibition at 400 μm ETYA. The data points are means and sd of three replicates of one experiment.
Figure 3.
Determination of LOX activity was performed with a lipid hydroperoxide assay kit. Control, 80 nmol of NAE without enzyme showed the natural hydroperoxidation, which is 0.200 nmol h−1; control with enzyme (3.33 mg protein per assay), the total activity was 0.947 nmol h−1; NDGA (100 μm) with enzyme showed the effect of a classical LOX inhibitor. This inhibitor inhibited the effect of natural peroxidation as well which was also observed in other radiolabeled experiments; and ETYA (100 μm) with enzyme showed the expected effect of LOX inhibitor. There was 50% inhibition, which was also observed in radiolabeled NAE 18:2 substrate metabolism experiments. Experiments without synthetic substrate was also carried out to confirm the absence of lipid hydroperoxide in the cell extract itself. Also, an experiment without EDTA was carried out to investigate any possible role of EDTA. All of those experiments were negative. Determination of LOX activity was performed with a commercially available lipid hydroperoxide (LPO) assay kit (catalog no. 705002, Cayman Chemical). For each assay, 80 nmol of NAE 18:2 was used as substrate and incubated with crude extract for 1 h at 30°C with shaking (110 rpm). The lipid peroxides that were formed were extracted from the samples into chloroform and quantified by measuring A500 compared with the standard lipid hydroperoxide (13-hydroperoxy octadecadienoic acid). The data points are means and sd of three replicates of one experiment.
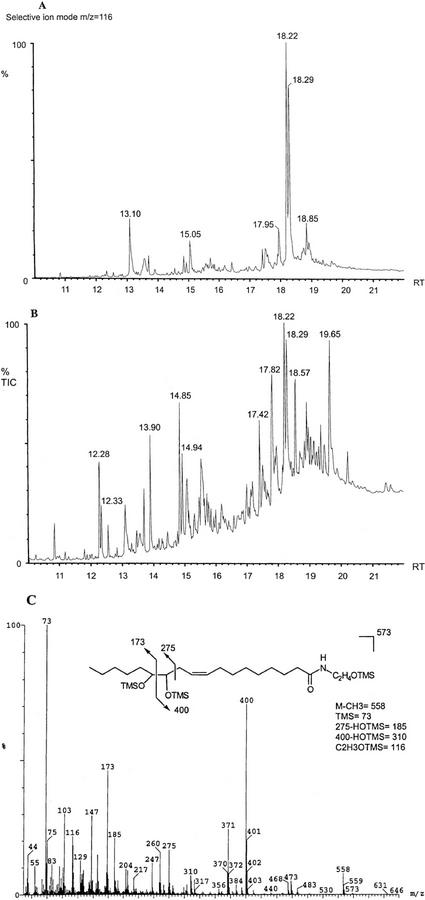
To elucidate the structure of the polar compound, gas chromatography-mass spectrometry (GC/MS) analysis was performed (Fig. 4). Selective ion monitoring at m/z 116 (diagnostic of ethanolamine containing lipids) revealed the presence of two oxygenated NAE 18:2 metabolites in incubations of cottonseed extracts incubated with NAE 18:2, with retention times of 18.22 and 18.29 min, respectively (Fig. 4A). These compounds were identified as trimethylsilylated, reduced α-ketols (diastereomers) 12-oxo-13-hydroxy-N-(9Z)-octadecenoylethanolamine by their electron impact mass spectrum (Fig. 4C). Predictable fragmentation ions including the molecular ion [M+] (m/z 573) were clearly identifiable, and spectra were comparable with those recorded in previous studies (Van der Stelt et al., 2000). Only α-ketols originating from 13-hydroperoxy NAE (18:2) were detected, indicating that imbibed cottonseeds contained both a 13-LOX and 13-AOS that metabolized NAE 18:2. Of interest also was the identification of the α-ketols 12-oxo-13-hydroxy-(9Z)-octadecenoic acid in the total ion chromatogram at retention times 14.85 and 14.94 (Fig. 4B, diastereomers), which can be explained by the subsequent actions of NAE AHase, LOX, and AOS. These results most importantly demonstrate unequivocally that NAE 18:2 was metabolized by 13-LOX (and 13-AOS) in extracts of imbibed cottonseeds, raising the possibility that a new class of oxylipins may be involved in seed germination.
Figure 4.
A, Single-ion chromatogram at m/z 116 (characteristic fragment for the ethanolamine group) of NAE metabolites after incubating with 150,000g supernatant of cottonseeds. The peaks with RT = 18.22 and 18.29 are α-ketols of 13-hydroperoxy NAE (two diastereomers) = 12-oxo-13-hydroxy-N-(9Z)-octadecenoylethanolamine. B, Total ion chromatogram of derivatized lipid products after incubating NAE 18:2 with the 150,000g supernatant of imbibed cottonseeds. Several peaks were identified by MS: 12.28 min, C18:2 linoleic acid; 12.33 min, C18:1 oleic acid; 12.55 min, C18:0 stearic acid; 13.90 min, 13-HPOD (13-hydroperoxy octadecadienoic acid); 14.85 and 14.94 min, α-ketols of 13-HPOD; 18.22 and 18.29 min, α-ketols of 13-hydroperoxy NAE = 12-oxo-13-hydroxy-N-(9Z)-octadecenoylethanolamine. C, Electron impact mass spectrum of fully reduced TMS-ethers of 12-oxo-13-hydroxy-N-(9Z)-octadecenoylethanolamine, the compound eluting with retention time of 18.22 min (A and B).
Inclusion of FAAH inhibitors to study sensitivity of the cottonseed FFA producing enzyme showed a concentration-dependent effect (Table III). There was approximately a 40% inhibition of FFA production with 10 mm phenylmethylsulfonyl fluoride, a potent inhibitor of FAAH activity consistent with the catalytic mechanism of a Ser hydrolase (Wiley et al., 2000). In contrast, there was only 10% inhibition by 10 μm arachidonyl trifluoromethyl ketone (ATMK), an analog of anandamide. ATMK is a potent inhibitor of mammalian anandamide hydrolysis showing complete inhibition at 7.5 μm (Koutek et al., 1994). The difference in sensitivity to ATMK may indicate a different property of cottonseed NAE AHase or may simply be a reflection of the lack of arachidonyl fatty acid derivatives in higher plant tissues.
Table III.
The effects of FAAH inhibitors on metabolism of NAE 18:2 in vitro
| Concentrations | Specific Activity | Relative Inhibition |
|---|---|---|
| nmol h−1 mg−1 Protein | % | |
| PMSF | ||
| 0 mm | 1.88 ± 0.20 | 0.0 |
| 0.01 mm | 1.90 ± 0.42 | −1.1 |
| 0.1 mm | 1.68 ± 0.32 | 10.6 |
| 1 mm | 1.62 ± 0.12 | 13.8 |
| 10 mm | 1.14 ± 0.11 | 39.4 |
| ATMK | ||
| 0 μm | 1.79 ± 0.30 | 0.0 |
| 0.01 μm | 1.71 ± 0.35 | 4.5 |
| 0.1 μm | 1.60 ± 0.26 | 10.6 |
| 1 μm | 1.64 ± 0.28 | 8.4 |
| 10 μm | 1.63 ± 0.10 | 8.9 |
The amounts of FFA formation was determined by incubating (1 h) synthetic NAE 18:2 with 150,000gmax (60 min) microsomes from 10,000gmax (30 min) supernatant. The radiometric analysis showed arachidonyl trifluoromethyl ketone (ATMK) was more potent than phenylmethylsulfonyl fluoride (PMSF). Both inhibitors had similar effect when NAE 16:0 was utilized as substrate. The data are means and sd of three replicates and are representative of two experiments.
Developmental Changes in NAE Metabolism
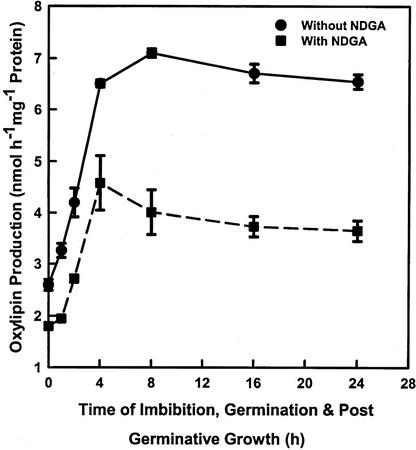
The capacity for NAE oxylipin formation by cytosol-enriched fractions increased during seed imbibition to the highest levels (7 nmol h−1 mg−1 protein) by 8 h after commencing imbibition (Fig. 5). Activity remained at this level throughout the first 24 h of postgerminative growth. Under these conditions, cottonseeds germinate at 12 to 18 h after commencing imbibition, and lipid mobilization (marked by glyoxylate cycle enzymes) is most active in 24- to 48-h-old seedlings (for summary, see Chapman and Sprinkle, 1996). As before, inclusion of NDGA helped to confirm that the activity was attributable to LOX-like enzyme.
Figure 5.
Time course of NAE 18:2 metabolism in cytosol-enriched fractions isolated at various times of seed imbibition (up to 4 h), germination (at about 12 h), and postgerminative growth. The amount of oxylipin production was determined by incubating 100 μm synthetic NAE 18:2 (20,000 dpm) with 400 μL of the supernatant (150,000gmax supernatant of 10,000gmax supernatant) in a final volume of 800 μL with shaking for 1 h at 30°C. Lipids were extracted as described in “Materials and Methods.” Identification and quantification of radiolabeled NAE-lipids were performed by radiometric scanning. The maximum specific activity of “LOX” was at 8 h (just before seed germination). When 100 μm NDGA was used, there was almost 50% inhibition of oxylipin production. It indicates that the product of NAE 18:2 was most likely attributable to enzymatic action of LOX. The data points are means and sd of three replicates of one representative experiment repeated for three times.
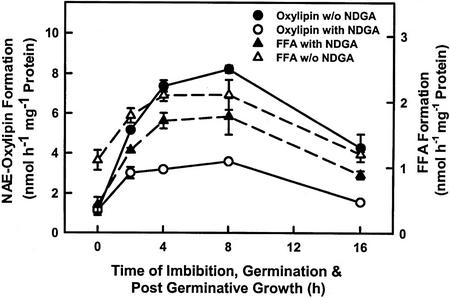
The developmental change in cytosolic NAE oxylipin formation was somewhat different from that associated with microsomes (Fig. 6). Microsomal NAE oxylipin formation, like that associated with the cytosol-enriched fractions, increased during imbibition to its highest levels by 8 h after commencing imbibition; however, the membrane-associated activity dropped substantially by 16 h (Fig. 6) and was undetectable by 24 h (not shown). The developmental change in membrane-associated NAE oxylipin formation paralleled that of NAE AHase (Fig. 6). In the case of microsomes, NDGA had a profound effect on NAE oxylipin formation (indicative of a LOX-mediated pathway) but had only a modest effect on AHase activity. Overall, these results indicate that metabolism of NAEs is most active during seed imbibition, just before seed germination, and well before the period of lipid mobilization for postgerminative seedling growth. This conclusion is consistent with the time period of NAE depletion in vivo (Chapman et al., 1999).
Figure 6.
Time course of NAE 18:2 metabolism in microsomes isolated at various stages of cottonseed imbibition, germination, and postgerminative seedling growth. Both AHase and LOX activities were detected in microsomes (see also Tables I and II). Both activities increased before and decreased after seed germination. NDGA reduced substantial oxylipin formation, whereas there was minimal effect on FFA production in vitro. The highest specific activities of LOX and NAE AHase were at 8 h and 4 to 8 h, respectively, after commencing imbibition (just before seed germination, 12–18 h). The data points are means and sd of three replicates within a given experiment and are representative of three experiments.
Enzymatic Properties of AHase and NAE Oxylipin Formation
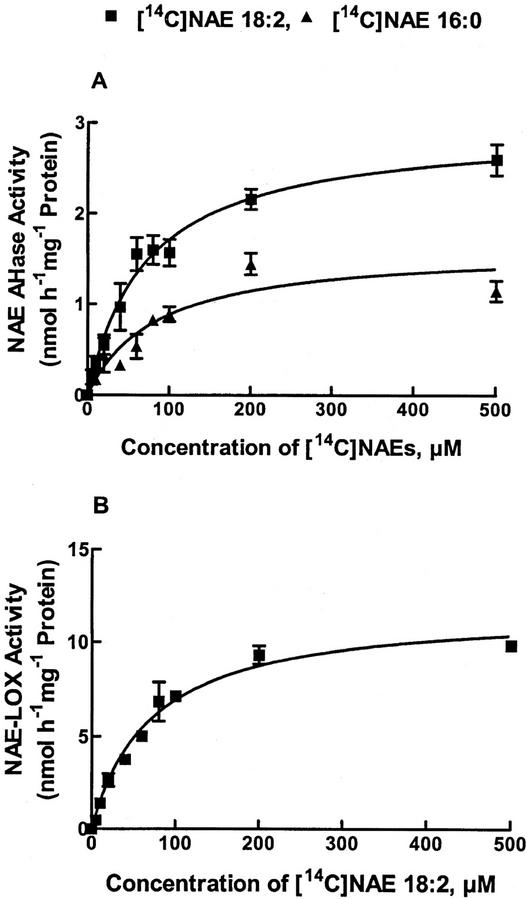
We compared the enzymatic properties of the NAE AHase pathway and NAE-LOX-mediated pathway in cottonseed microsomes to estimate the relative capacity of each pathway to contribute to the metabolism of the predominant seed NAEs (Fig. 7; Table IV). Both pathways exhibited typical Michaelis-Menten kinetics when initial velocity measurements were made at increasing NAE concentrations (Fig. 7). For the NAE AHase, the apparent Km values were similar for NAE 16:0 and NAE 18:2 (Km values for NAE 16:0 and NAE 18:2 are 83 μm and 74 μm, respectively), although the apparent Vmax values estimated for NAE 18:2 were nearly twice that as for NAE 16:0 (Vmax values for NAE 16:0 and NAE 18:2 are 1.6 and 3.0 nmol h−1 mg−1 protein, respectively). NAE oxylipin formation from NAE 18:2 also exhibited typical saturation kinetics at increasing NAE concentrations (Fig. 7B). The apparent Km value (70 μm) was similar to that estimated for the NAE AHase, indicating similar substrate affinities for both pathways. On the other hand, the apparent Vmax (12 nmol h−1 mg−1 protein) was four times that of the NAE AHase-mediated pathway, indicating a greater capacity for NAE oxylipin formation than for NAE hydrolysis. Although comparisons of kinetic parameters cannot be interpreted to indicate relative metabolic flux, these data do indicate that the capacity for NAE consumption by these two pathways estimated in vitro exceeds that required for NAE depletion in vivo. The degradation rates for NAE 18:2 and NAE 16:0 were calculated to be 15 and 4.5 ng h−1 seed−1, respectively, in vivo (Chapman et al., 1999); whereas the rates were 160 and 80 ng h−1 seed−1 for NAE 18:2 and NAE 16:0, respectively, in vitro through NAE AHase pathway, based on data presented in Table IV. The conversion rate for NAE 18:2 through NAE-LOX pathway was 20 μg h−1 seed−1 in vitro (Table IV), indicating this pathway was relatively more capable of contributing to the metabolism of polyunsaturated NAE (18:2) than was AHase.
Figure 7.
Concentration-dependent formation of FFA from NAE substrate in cottonseed microsomes under initial velocity conditions. The apparent Km and Vmax for NAE 16:0 were estimated to be 83 μm and 1.6 nmol h−1 mg−1 protein, respectively. The apparent Km and Vmax for NAE 18:2 were estimated to be 74 μm and 3.0 nmol h−1 mg−1 protein, respectively. Concentration-dependent formation of oxylipin from NAE 18:2 incubating the same cell fraction showed the apparent Km and Vmax to be 70 μm and 12 nmol h−1 mg−1 protein, respectively. In A, NAE 16:0 and NAE 18:2 were the substrates, and in B, NAE 18:2 was the substrate. Lines represent nonlinear regression fits of the data using Michaelis-Menten equation (Prism software, v3.0, GraphPad Software, San Diego). Kinetic parameters were estimated from regression analyses. Although the microsomal NAE AHase appears to have similar affinities for saturated and polyunsaturated NAE species, the maximum rate of product formation from NAE 18:2 is about twice that for NAE 16:0. Similarly in B, the enzyme has similar affinity of 70 μm with four times greater production rate of conversion of NAE 18:2 into NAE-LOX product. Data points are averages of triplicate samples within a representative experiment.
Table IV.
Summary of kinetic parameters of NAE utilizing enzymes in cottonseed microsomes
| Substrate | Pathway | Km | Vmax |
|---|---|---|---|
| μm | nmol h−1 mg−1 protein | ||
| NAE 16:0 | AHase | 83.43 | 1.62 |
| NAE 18:2 | AHase | 73.65 | 2.97 |
| NAE 18:2 | NAE-LOX | 69.91 | 11.82 |
Parameters were estimated by fitting the data in Fig. 8 to the Michaelis-Menten equation (GraphPad Prism software, version 3.0).
NAE Metabolism in Vivo
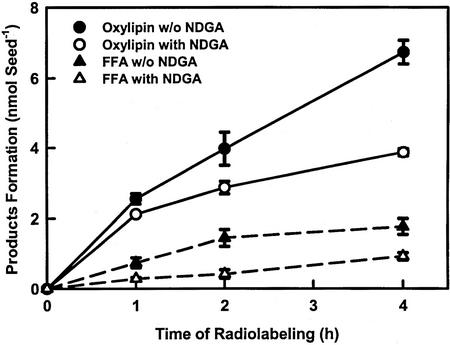
NAE was metabolized in vivo by both the NAE-LOX pathway and NAE AHase pathway during seed imbibition (Fig. 8). Radiotracer experiments with imbibing seeds showed that [1-14C]18:2 NAE was converted to NAE oxylipin and FFA in a time-dependent manner. In the presence of the LOX inhibitor, NDGA there was reduction of NAE-oxylipin formation, which was particularly evident after 4 h. Production of FFA from NAE 18:2 also was reduced in the presence of the LOX inhibitor, which may suggest these two pathways are somewhat interdependent. Application of NAE 18:2 or NDGA under these conditions did not influence seed germination. The metabolic results in vivo are consistent with those obtained in vitro and confirm that NAE 18:2 is metabolized by LOX and AHase pathways, which together likely account for the in vivo depletion of NAE 18:2 during seed imbibition (Chapman et al., 1999).
Figure 8.
Metabolism of [14C]NAE 18:2 in vivo by imbibing cottonseeds. Seed coats were removed from imbibing (4 h) seeds, which were then incubated 30 min with (or without, dimethyl sulfoxide-only control) LOX inhibitor (5 μL of 16 mm NDGA per seed) before application of radiolabeled NAE 18:2 (0.1 μCi seed−1, 2.04 mCi mmol−1). Imbibed seeds were incubated on moist filter paper in covered petri dishes for additional 1, 2, and 4 h in the dark. Lipids were extracted and separated by TLC as described in “Materials and Methods,” and distribution of radioactivity was evaluated by radiometric scanning (Bioscan system 200 image scanner). The data points are means and sd of four replicates within a single experiment. Additional experiments showed identical trends, although the efficiency of incorporation of radiolabel varied somewhat from experiment to experiment. Both oxylipin and FFA production increased with time. The conversion of NAE 18:2 to NAE-oxylipin was reduced by application of a classical LOX inhibitor, NDGA (see Fig. 2). NDGA had some effect on production of FFA, as well. For clarity, the amount of radioactivity in NAE 18:2 is not included but represented nearly all of the remaining proportion of radioactive lipid.
DISCUSSION
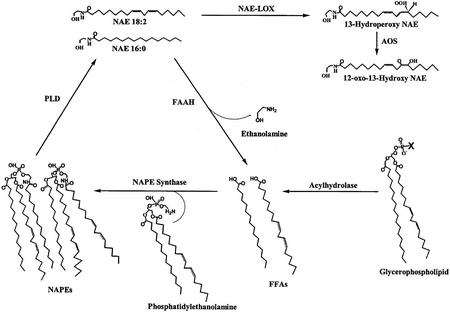
The recent identification and quantification of NAEs in desiccated seeds and their disappearance after 4 h of imbibition (Chapman et al., 1999) raised the question as to the metabolic fate of these compounds. Preliminary evidence suggested that these NAEs could be hydrolyzed in imbibed seeds by an NAE AHase activity (Chapman et al., 1999), similar to the FAAH found in some animal systems. However, here, a detailed evaluation now indicates a more complicated scheme for NAE metabolism in seeds than originally anticipated (Fig. 9). NAEs with saturated fatty acid constituents, like the endogenous NAE 16:0, are indeed hydrolyzed by an AHase activity, whereas polyunsaturated NAEs (NAE 18:2) appear to be metabolized by two pathways, the NAE AHase pathway and a LOX-mediated pathway. The AHase pathway leads to the formation of FFAs, which could be reincorporated into NAPE, the precursor for NAEs (see Fig. 9). In fact, NAPE biosynthesis was shown to be increased during seed imbibition, germination, and early postgerminative growth (Sandoval et al., 1995; Chapman and Sprinkle, 1996) as judged by increases in lipid levels and enzyme activity.
Figure 9.
Proposed scheme for the metabolism of NAEs in seeds and seedlings. NAPEs are hydrolyzed by PLD to yield saturated and unsaturated species of NAE. FAAH hydrolyzes NAEs to FFAs and ethanolamine. As an alternative, some polyunsaturated NAEs are metabolized by 13-LOX and 13-AOS to yield NAE oxylipins (13-hydroperoxy NAE, 13-hydroperoxy octadecadienoylethanolamine and 12-oxo-13-hydroxy NAE, 12-oxo-13-hydroxy octadecenoylethanolamine). FFAs formed from hydrolysis of NAE or glycerophospholipids can be incorporated directly into the N-position of NAPE (Chapman, 2000; Rawyler and Braendle, 2001) by NAPE synthase. This overall cycle could be used for signal transduction (formation of NAE lipid mediators) or to scavenge FFAs (for membrane protection) depending upon cellular demands. The “X” in the glycerophospholipid molecule represents the head group of any phospholipid class (e.g. Ser, choline, ethanolamine, etc.).
Metabolism of NAEs during seed imbibition by a membrane-associated AHase activity is reminiscent of the mechanism for NAE activation in animal systems. The FAAH has been cloned from a number of mammalian tissues and shown to encode an enzyme with amidase and esterase activities of broad substrate specificity including several fatty acid amides and acylglycerols (Cravatt et al., 1996). More extensive studies of substrate specificity for the purified recombinant rat enzyme recently showed that it was capable of hydrolyzing a wide array of unsaturated, and to a less extent saturated, fatty acid primary amides (Boger et al., 2000). As the chain length of fatty acid (saturated) constituents decreased, the rate of hydrolysis increase (Ueda et al., 2000). Mammalian FAAH inhibitors had a relatively modest effect on cottonseed NAE AHase enzyme(s) (Table III). This may indicate that plant NAE AHases are diverged from the mammalian counterparts to reflect a specificity for NAEs abundant in plant tissues. However, Ueda et al. (2001) recently reported a new enzyme from lung tissues, designated NAE 16:0 hydrolase, that was much less sensitive to inhibition of FAAH, suggesting that animal systems may contain more than one NAE AHase. Additional work will be required at the biochemical and molecular levels to more fully understand the nature of NAE AHase activity(ies) in plant systems, and to define what specific role this pathway plays in seed germination.
Recent evidence demonstrated that purified plant LOX, AOS, and HPO lyase could metabolize synthetic NAEs to generate novel oxylipins (Van der Stelt et al., 2000), raising the possibility that plants might catalyze these reactions in vivo. Here, we provide several lines of evidence that support this concept and highlight seed imbibition and germination as a period intensely involved in formation of these metabolites. The time course of NAE 18:2 metabolism in cytosol-enriched fractions isolated at various times of seed imbibition, germination, and postgerminative growth showed the maximum specific activity of LOX (7 nmol h−1 mg−1 protein) at 8 h after imbibition, which is just before seed germination (Fig. 5). Microsomes isolated at the same developmental stages similarly showed the same time period for the highest specific activity of LOX (8 nmol h−1 mg−1 protein) and NAE AHase (2 nmol h−1 mg−1 protein; Fig. 6) consistent with results in vivo (Chapman et al., 1999).
The fate of NAE-derived oxylipins is unclear at this point, but it is tempting to speculate these NAE oxylipins may have a role of their own during seed germination. For example, Feussner and coworkers (2001) recently proposed that 13-LOX-mediated pathway is associated with a “priming” function in oilseeds (Brassica napus) for postgerminative triacylglycerol mobilization. It should be emphasized, however, that here the timing of the most intensive NAE metabolism of cottonseed NAEs preceded radicle emergence and lipid mobilization (Figs. 5 and 6; also see Chapman and Sprinkle, 1996), so these NAE-derived oxylipins may play other roles perhaps as lipid mediators involved in the regulation of seed germination. In any case, the role of NAE metabolism in imbibing seeds is not specific to oilseeds because a similar depletion of seed NAEs was observed in non-oilseeds (e.g. pea [Pisum sativum]; Chapman et al., 1999).
MATERIALS AND METHODS
Chemicals
[1-14C]Palmitic acid (53 mCi−1 mmol in ethanol) and [1-14C]linoleic acid (53 mCi mmol−1 in ethanol) were from PerkinElmer Life Sciences (Boston).
Chemical Synthesis of NAEs
Specific NAE types were synthesized from respective radiolabeled FFA by first producing the fatty acyl chloride (Hillard et al., 1995). The FFA was dissolved in dichloromethane and then mixed with dimethylformamide (1 mol equivalent) and oxalyl chloride (1.2 mol equivalent). The fatty acyl chloride was mixed with a 10-fold excess of ethanolamine to convert the acyl chloride to the corresponding NAE. Products were extracted in dichloromethane and purified by TLC. Yield and purity of NAEs were estimated by radiometric scanning (System 200 image scanner, Bioscan, Washington, DC). The yield was routinely 65% to 70% (starting from FFA), and purity after TLC was >99%. Radiospecific activity was calculated from the original 14C-labeled FFA and adjusted accordingly with non-radiolabeled synthetic NAE produced by the same method.
Plant Material
Cotton (Gossypium hirsutum L. Stoneville 7A glandless) seeds were provided by Dr. R.B. Turley (Cotton Physiology and Genetics Laboratory, U.S. Department of Agriculture-Agricultural Research Service, Stoneville, MS). For all of the experiments, seeds were surface-sterilized with 20% (v/v) commercial bleach (sodium hypochlorite) solution for 5 min. Seeds were rinsed several times and imbibed in distilled water (in the dark) for 4 h at 30°C with aeration. For time-course experiments, imbibed seeds were placed in filter paper scrolls as previously described (Chapman and Trelease, 1991a) and germinated and grown in the dark (30°C).
Preparation of Cellular Fractions
Cell fractions were prepared by differential centrifugation as described (Chapman and Trelease, 1991b) with some modifications. In brief, seeds imbibed for 4 h were chopped with a steel blade on ice in homogenization medium containing 100 mm potassium-phosphate (pH 7.2), 10 mm KCl, 1 mm EDTA, 1 mm EGTA, and 400 mm Suc. The homogenates were filtered through four layers of cheesecloth and centrifuged at 650gmax (4°C) for 10 min in a centrifuge (RC 5C, SS 34 rotor, Sorvall, Newton, CT). The 650gmax supernatant was centrifuged at 10,000gmax (4°C) for 30 min in the same centrifuge. The resulting supernatant was centrifuged at 150,000gmax (4°C) for 60 min in Sorvall Discovery 90 model ultracentrifuge centrifuge (Ti45 rotor, Beckman Coulter, Fullerton, CA). Microsomes (150,000gmax pellet) were resuspended in homogenization medium (0.3 mL per original gram fresh weight). Protein concentration was estimated according to Bradford (1976) using bovine serum albumin as the standard.
Lipid Extractions and Analysis
For enzymatic assays in vitro, 100 μm (20,000 dpm) of [14C]NAE substrate (combined with nonradioactive NAE and radiolabeled on the carbonyl carbon) was suspended by sonication in 50 mm MES buffer (pH 6.5). Reactions were initiated by adding 400-μL cell fractions. Reactions were terminated, and lipids were extracted into chloroform according Bligh and Dyer (1959) modified to eliminate endogenous PLD activity (Chapman and Moore, 1993). In brief, reactions were stopped by adding 2 mL of hot 2-propanol (70°C) to 800 μL of the aqueous assay reaction mixture and heated at 70°C for 30 min. One milliliter of chloroform was added to the mixtures, and lipids were extracted at 4°C overnight. One milliliter of chloroform and 2 mL of KCl (1 m) were added to induce phase separation. The aqueous layer was aspirated off, and the organic layer was washed twice with 2 mL of 1 m KCl and once with deionized water (MilliQ UF plus). The organic phase was collected and dried under nitrogen. Lipid classes were separated by TLC (hexane:ethyl acetate:methanol, 60:40:5; v/v). Identification and quantification of radiolabeled lipids were performed by radiometric scanning (Bioscan system 200 image scanner) and comigration with known standards.
NAE Metabolism in Vivo
For radiolabeling experiments in vivo, seed coats were removed from imbibed (4 h) seeds, which were then preincubated for 30 min with (or without, dimethyl sulfoxide-only control) LOX inhibitor (5 μL of 16 mm NDGA per seed) before application of radiolabeled NAE 18:2. Synthetic NAE 18:2 (0.1 μCi, 2.04 mCi−1 mmol) was applied in a small volume to each seed. Imbibed seeds were incubated for various time periods in the dark on moistened filter paper in 100- × 15-mm covered petri dishes. Radiolabeled lipids were extracted and analyzed as described above.
Spectrophotometric Assay of Lipid Hydroperoxide Formation
Determination of LOX activity also was performed with a commercially available lipid hydroperoxide (LPO) assay kit (catalog no. 705002, Cayman Chemical, Ann Arbor, MI). For each assay, 80 nmol of NAE 18:2 was used as substrate and incubated with crude extract for 1 h at 30°C with shaking (110 rpm). The lipid peroxides that were formed were extracted from the samples into chloroform and quantified by measuring A500 compared with the standard lipid hydroperoxide (13-hydroperoxy octadecadienoic acid). Lipid peroxide formation in imbibed cottonseed extracts was protein, temperature, and NAE 18:2 dependent.
Identification of NAE Oxylipins by GC/MS
NAE-derived oxylipins were evaluated by GC/MS as previously described (Van der Stelt et al., 2000), except lipids were reduced with NaBH4 instead of NaBD4. In brief, imbibed cottonseed cell fractions, prepared as described above, were diluted 1:1 in 50 mm potassium phosphate buffer, pH 6, and incubated with 100 μm NAE 18:2 for 2 h at room temperature. Lipid products were extracted, reduced with NaBH4, methylated, and trimethylsilylated before identification by GC/MS using the conditions described previously (Van der Stelt et al., 2000).
ACKNOWLEDGMENTS
We thank Dr. Swati Tripathy (University of North Texas) and Dr. Ivo Feussner (Institute of Crop Genetics, Gatersleben, Germany) for helpful advice regarding NAEs and LOX, respectively.
Footnotes
This work was supported by the United States Department of Agriculture-National Research Initiative Competitive Grants Program (grant no. 99–35304–8002).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.004689.
LITERATURE CITED
- Bligh EG, Dyer WG. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Boger DL, Fecik RA, Patterson JE, Miyauchi H, Patricelli MP, Cravatt BF. Fatty acid amide hydrolase substrate specificity. Bioorg Med Chem Lett. 2000;10:2613–2616. doi: 10.1016/s0960-894x(00)00528-x. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for quantification of microgram quantities of protein utilizing the principle of protein-dye bind. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Buckley NE, McCoy KL, Mezey E, Bonner T, Zimmer A, Felder CC, Glass M, Zimmer A. Immunomodulation by cannabinoids is absent in mice deficient for cannabinoid CB2 receptor. Eur J Pharmacol. 2000;396:141–149. doi: 10.1016/s0014-2999(00)00211-9. [DOI] [PubMed] [Google Scholar]
- Chapman KD. Emerging physiological roles for N-acylphosphatidylethanolamine metabolism in plants: signal transduction and membrane protection. Chem Phys Lipids. 2000;108:221–230. doi: 10.1016/s0009-3084(00)00198-5. [DOI] [PubMed] [Google Scholar]
- Chapman KD, Moore TS., Jr N-Acylphosphatidylethanolamine synthesis in plants: occurrence, molecular composition, and phospholipid origin. Arch Biochem Biophys. 1993;301:21–33. doi: 10.1006/abbi.1993.1110. [DOI] [PubMed] [Google Scholar]
- Chapman KD, Sprinkle WB. Developmental, tissue-specific and environmental factors regulate the biosynthesis of N-acylphosphatidylethanolamine in cotton (Gossypium hirsutum L.) Plant Physiol. 1996;149:277–284. [Google Scholar]
- Chapman KD, Trelease RN. Acquisition of membrane lipids by differentiating glyoxysomes: role of lipid bodies. J Cell Biol. 1991a;115:995–1007. doi: 10.1083/jcb.115.4.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman KD, Trelease RN. Intracellular localization of phosphotidylcholine and phosphotidylethanolamine synthesis in cotyledons of cotton seedlings. Plant Physiol. 1991b;95:69–76. doi: 10.1104/pp.95.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman KD, Tripathy S, Venables BJ, Desouza AD. N-Acylethanolamines: formation and molecular composition of a new class of plant lipids. Plant Physiol. 1998;116:1163–1168. doi: 10.1104/pp.116.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman KD, Venables BJ, Markovic R, Blair RW, Bettinger C. N-Acylethanolamines in seeds: quantification of molecular species and their degradation upon imbibition. Plant Physiol. 1999;120:1157–1164. doi: 10.1104/pp.120.4.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cravatt BF, Demarest K, Patricelli MP, Bracey MH, Giang DK, Martin BR, Lichtman AH. Supersensitivity to anandamide and enhanced endogenous cannabinoid signaling in mice lacking fatty acid amide hydrolase. Proc Natl Acad Sci USA. 2001;98:9371–9376. doi: 10.1073/pnas.161191698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cravatt BF, Giang DK, Mayfield SP, Boger DL, Lerner RA, Gilula NB. Molecular characterization of an enzyme that degrades neuromodulatory fatty-acid amides. Nature. 1996;84:83–87. doi: 10.1038/384083a0. [DOI] [PubMed] [Google Scholar]
- Feussner I, Kuhn H, Wasternack C. Lipoxygenase-dependent degradation of storage lipids. Trends Plant Sci. 2001;6:268–273. doi: 10.1016/s1360-1385(01)01950-1. [DOI] [PubMed] [Google Scholar]
- Grullich C, Duvoisin RM, Wiedmann M, Leyen K. Inhibition of 15-lipoxygenase leads to delayed organelle degradation in the reticulocyte. FEBS Lett. 2001;489:51–54. doi: 10.1016/s0014-5793(01)02080-4. [DOI] [PubMed] [Google Scholar]
- Hansen HS, Moesgaard B, Hansen HH, Petersen G. N-Acylethanolamines and precursor phospholipids: relation to cell injury. Chem Phys Lipids. 2000;108:135–150. doi: 10.1016/s0009-3084(00)00192-4. [DOI] [PubMed] [Google Scholar]
- Hillard CJ, Edgemond WS, Campbell WB. Characterization of ligand binding to the cannabinoid receptor of rat brain membranes using a novel method: application to anandamide. J Neurochem. 1995;64:677–683. doi: 10.1046/j.1471-4159.1995.64020677.x. [DOI] [PubMed] [Google Scholar]
- Koutek B, Prestwick GD, Howlett AC, Chin SA, Salehani D, Akhavan N, Deutsch DG. Inhibitors of arachidonoyl ethanolamide hydrolysis. J Biol Chem. 1994;269:22937–22940. [PubMed] [Google Scholar]
- Kulkarni AP, Sajan M. Lipoxygenase: another pathway for glutathione conjugation of xenobiotics. A study with human term placental lipoxygenase and ethacrynic acid. Arch Biochem Biophys. 1999;371:220–227. doi: 10.1006/abbi.1999.1439. [DOI] [PubMed] [Google Scholar]
- Paria BC, Dey SK. Ligand-receptor signaling with endocannabinoids in preimplantation embryo development and implantation. Chem Phys Lipids. 2000;108:211–220. doi: 10.1016/s0009-3084(00)00197-3. [DOI] [PubMed] [Google Scholar]
- Pertwee RG. Cannabinoid receptors and pain. Prog Neurobiol. 2001;63:569–611. doi: 10.1016/s0301-0082(00)00031-9. [DOI] [PubMed] [Google Scholar]
- Rawyler AJ, Braendle RA. N-Acylphosphatidylethanolamine accumulation in potato cells upon energy shortage caused by anoxia or respiratory inhibitors. Plant Physiol. 2001;127:240–251. doi: 10.1104/pp.127.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval JA, Huang Z-H, Garrett DC, Gage DA, Chapman KD. N-Acylphosphatidylethanolamine in dry and imbibing cotton (Gossypium hirsutum L.) seeds: amounts, molecular species and enzymatic synthesis. Plant Physiol. 1995;109:269–275. doi: 10.1104/pp.109.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarker KP, Obara S, Nakata M, Kitajima I, Maruyama I. Anandamide induces apoptosis of PC-12 cells: involvement of superoxide and caspase-3. FEBS Lett. 2000;472:39–44. doi: 10.1016/s0014-5793(00)01425-3. [DOI] [PubMed] [Google Scholar]
- Schmid HHO. Pathways and mechanisms of N-acylethanolamine biosynthesis: can anandamide be generated selectively? Chem Phys Lipids. 2000;108:71–87. doi: 10.1016/s0009-3084(00)00188-2. [DOI] [PubMed] [Google Scholar]
- Schmid HHO, Schmid PC, Nataranjan V. N-Acylated glycerophospholipids and their derivatives. Prog Lipid Res. 1990;29:1–43. doi: 10.1016/0163-7827(90)90004-5. [DOI] [PubMed] [Google Scholar]
- Tripathy S, Venables BJ, Chapman KD. N-Acylethanolamines in signal transduction of elicitor perception: attenuation of alkalinization response and activation of defense gene expression. Plant Physiol. 1999;121:1299–1308. doi: 10.1104/pp.121.4.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda N, Puffenbarger RA, Yamamoto S, Deutsch DG. The fatty acid amide hydrolase (FAAH) Chem Phys Lipids. 2000;108:107–121. doi: 10.1016/s0009-3084(00)00190-0. [DOI] [PubMed] [Google Scholar]
- Ueda N, Yamanaka K, Yamamoto S. Purification and characterization of an acid amidase selective for N-palmitoylethanolamine, a putative endogenous anti-inflammatory substance. J Biol Chem. 2001;276:35552–35557. doi: 10.1074/jbc.M106261200. [DOI] [PubMed] [Google Scholar]
- Van der Stelt M, Nieuwenhuizen WF, Veldink GA, Vliegenthart JFG. Dioxygenation of N-linoleoyl amides by soybean lipoxygenase-1. FEBS Lett. 1997;411:287–290. doi: 10.1016/s0014-5793(97)00718-7. [DOI] [PubMed] [Google Scholar]
- Van der Stelt M, Noordermeer MA, Kiss T, Van Zadelhoff G, Merghart B, Veldink GA, Vliegenthart JFG. Formation of a new class of oxylipins from N-acyl(ethanol) amines by the lipoxygenase pathway. Eur J Biochem. 2000;267:2000–2007. doi: 10.1046/j.1432-1327.2000.01203.x. [DOI] [PubMed] [Google Scholar]
- Van der Stelt M, Veldhuis WB, Van Haaften GW, Fezza F, Bisogno T, Bar PR, Veldink GA, Vliegenthart JFG, Di Marzo V, Nicolay K. Exogenous anandamide protects rat brain against acute neuronal injury in vivo. J Neurosci. 2001;21:8765–8771. doi: 10.1523/JNEUROSCI.21-22-08765.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Zadelhoff G, Veldink GA, Vliegenthart JFG. With anandamide as substrate plant 5-lipoxygenases behave like 11-lipoxygenase. Biochem Biophys Res Commun. 1998;248:33–38. doi: 10.1006/bbrc.1998.8910. [DOI] [PubMed] [Google Scholar]
- Wiley JL, Dewey MA, Jefferson RG, Winckler RL, Bridgen DT, Willoughby KA, Martin BR. Influence of phenylmethylsulfonyl fluoride on anandamide brain levels and pharmacological effects. Life Sci. 2000;67:1573–1583. doi: 10.1016/s0024-3205(00)00749-9. [DOI] [PubMed] [Google Scholar]