Abstract
Using monoclonal antibodies specific for each apoprotein and full-length purified apoprotein standards, the levels of the five Arabidopsis phytochromes and their patterns of expression in seedlings and mature plants and under different light conditions have been characterized. Phytochrome levels are normalized to the DNA content of the various tissue extracts to approximate normalization to the number of cells in the tissue. One phytochrome, phytochrome A, is highly light labile. The other four phytochromes are much more light stable, although among these, phytochromes B and C are reduced 4- to 5-fold in red- or white-light-grown seedlings compared with dark-grown seedlings. The total amount of extractable phytochrome is 23-fold lower in light-grown than dark-grown tissues, and the percent ratios of the five phytochromes, A:B:C:D:E, are measured as 85:10:2:1.5:1.5 in etiolated seedlings and 5:40:15:15:25 in seedlings grown in continuous white light. The four light-stable phytochromes are present at nearly unchanging levels throughout the course of development of mature rosette and reproductive-stage plants and are present in leaves, stems, roots, and flowers. Phytochrome protein expression patterns over the course of seed germination and under diurnal and circadian light cycles are also characterized. Little cycling in response to photoperiod is observed, and this very low amplitude cycling of some phytochrome proteins is out of phase with previously reported cycling of PHY mRNA levels. These studies indicate that, with the exception of phytochrome A, the family of phytochrome photoreceptors in Arabidopsis constitutes a quite stable and very broadly distributed array of sensory molecules.
The phytochromes are a family of soluble chromoproteins that function in the absorption of red (R) and far-red (FR) light and the transduction of intracellular signals during light-regulated plant development. In early studies, physiological and biochemical experiments indicated that higher plants contain at least two different forms of phytochrome (Hillman, 1967; Abe et al., 1985; Shimazaki and Pratt, 1985; Tokuhisa et al., 1985). These were frequently referred to as type I or “light-labile” phytochrome, which predominated in etiolated tissue, and type II or “light-stable” phytochrome, which predominated in light-grown tissue (for review, see Furuya, 1993; Pratt, 1995). Identification of PHY gene families, first in Arabidopsis and subsequently in many other plant species, and analysis of their phytochrome protein products have generally confirmed this view (Mathews et al., 1995; Mathews and Sharrock, 1997; Alba et al., 2000), although most plants contain more than two forms and the levels and light stabilities of all of the phytochromes from a single species have not been analyzed up to this time.
Complete or partial sequences of PHY genes from a large number of flowering plants have been isolated. Among these, the most intensively studied are those of Arabidopsis, the complete PHY gene complement of which consists of five members, PHYTOCHROME A through PHYTOCHROME E (PHYA–PHYE; Sharrock and Quail, 1989; Clack et al., 1994). Tomato (Lycopersicon esculentum), the only other eudicot from which all of the phytochrome coding sequences have been isolated, also contains five PHY genes (Alba et al., 2000). These are similar to the Arabidopsis genes and, with the exception of the pairs of recently derived phyB-like genes Arabidopsis B/D and tomato B1/B2, are thought to be orthologs of the Arabidopsis genes (Alba et al., 2000). PHY genes sequenced from a variety of higher plants, including both monocots and dicots, all show extensive similarity in sequence and overall structure to the Arabidopsis sequences. Phylogenetic analysis indicates that a duplication event in early seed plant evolution gave rise to two initial PHY gene lineages, PHYA/C and PHYB/D/E, which underwent later duplications in angiosperms to generate the divergent PHYA, PHYC, PHYE, and PHYB/D lineages (Mathews et al., 1995; Mathews and Sharrock, 1997; Alba et al., 2000). Members of these lineages are found in most plants examined, with the exception of PHYE genes, which have not been found in sampled monocots and may have evolved only in dicots after the monocot/dicot divergence or may have been lost in an early monocot progenitor (Mathews and Sharrock, 1996). Hence, available evidence strongly supports the view that the five Arabidopsis PHY genes and their protein products are representative of phytochrome diversity found across a broad range of angiosperm plants.
The phenotypic consequences of mutations in the Arabidopsis PHY genes and in PHY genes from other species have been used to assess the respective roles of these molecules in regulation of R/FR photomorphogenic responses. A single Arabidopsis gene, PHYA, encodes a light-labile type I phytochrome, which has been shown through mutant analysis to function in very low fluence and FR high irradiance responses (Shinomura et al., 1996; Hamazato et al., 1997; Whitelam and Devlin, 1997). Mutations in PHYA genes from tomato, pea (Pisum sativum), and rice (Oryza sativa) result in similar defects in these general classes of phytochrome responses (Weller et al., 1997; Lazarova et al., 1998a; Takano et al., 2001). Somers et al. (1991) showed that the Arabidopsis PHYB and PHYC genes encode proteins that are less abundant than phyA in etiolated tissue and appear to be light stable. These, therefore, resemble type II phytochromes. The abundance and light stability of the phyD and phyE phytochromes have not been analyzed, but these proteins are most closely related in sequence to phyB. Arabidopsis phyB, phyD, and phyE mutants have been isolated and used to demonstrate that all three function in low fluence, R/FR-reversible responses and in shade avoidance responses to altered R to FR ratio (Reed et al., 1993; Aukerman et al., 1997; Devlin et al., 1998, 1999). Again, mutations in several PHYB-related genes from other plant species affect low fluence, R/FR-reversible responses and shade avoidance responses in a similar way (Lopez-Juez et al., 1992; Weller et al., 1995; Childs et al., 1997; Devlin et al., 1997; Lazarova et al., 1998b). Mutations in the Arabidopsis PHYC gene or in PHYC homologs from other species have not yet been described.
Before the complexity of the phytochrome family was known, the expression pattern and distribution of phytochrome were characterized in several different plant species using assays based upon spectroscopy, biological activity, and immunolocalization (for review, see Pratt, 1994). These experiments were limited to addressing only the type I or phyA form of phytochrome or were nonselective among the various forms. In general, phytochrome was found in most tissues tested spectroscopically or immunologically, although significant variation in its localization was seen in different species. Variation was also observed in the phytochrome content of different cell types in tissues from a given plant, and phytochrome was often observed to be most abundant in young, rapidly expanding cells (Pratt, 1994).
Following the description of the five Arabidopsis PHY genes, preliminary characterization of the distributions of the PHY mRNAs in isolated plant organs indicated that these transcripts are relatively constitutive throughout the mature plant and throughout development (Clack et al., 1994). A quantitative analysis of the steady-state levels and distributions of all of the five tomato PHY mRNAs was performed and was again found fairly constitutive expression in seedlings (Hauser et al., 1997, 1998). In these studies, induction of all five transcripts was observed over the course of seed germination, 2- to 3-fold diurnal cycling of the PHYA, PHYB1, and PHYB2 mRNAs was seen, and some variation in expression was seen in mature tomato organs, although all five transcripts were detectable in all tissues examined. Promoter-reporter gene fusion constructs for the Arabidopsis PHYA, PHYB, PHYD, and PHYE and tobacco PHYA and PHYB transcription regulatory regions have also been constructed and their activities monitored in transgenic plants (Adam et al., 1994, 1996; Somers and Quail, 1995a, 1995b; Goosey et al., 1997). In these experiments, examples of well-defined organ and tissue specificities for promoter activity were observed, however all of the PHY promoters tested were expressed in a large number of different cell types and in most plant organs. Variation between Arabidopsis and tobacco for their respective PHYA and PHYB promoter expression patterns was also described. All of these approaches to characterization of PHY gene transcription patterns indicate that, although some regulation of PHY gene promoter activity occurs with respect to different cell types and stages of development, phytochromes are likely to be broadly distributed in a given plant and most, perhaps all, plant cells are R/FR responsive.
Earlier efforts at determining the in vivo levels of the phytochrome apoproteins have been carried out principally using oat or pea seedlings and ELISA, radioimmunoassay, or immunoblot assays for phytochrome (Konomi et al., 1987; Tokuhisa and Quail, 1987; Wang et al., 1992, 1993a, 1993b; Pratt, 1995). However, in none of these cases was it possible to distinguish completely among the different forms of phytochrome present in the extracts because a comprehensive set of phy-specific antibodies was not available. The most comprehensive analyses were performed by Wang et al. (1992, 1993a, 1993b) and used monoclonal antibodies (MAb) specific to each of three phytochrome forms from oat. Levels of these phytochromes were determined in unimbibed oat seeds and in dissected 3-d-old seedlings, and it was concluded that one form, 124-kD phytochrome, was predominant in dark-grown seedlings, whereas the other two, the 123- and 125-kD forms, were more abundant in the light and that there was little evidence for marked differences in the spatial distribution of the three forms.
Several lines of evidence indicate that the level of photoreversible phytochrome in plants is a critical determinant of the sensitivity of the plant to light and the strength of the physiological or developmental response to a given light environment. Overexpression of phyA or phyB phytochromes in transgenic plants has striking effects on growth and development in response to light (Boylan and Quail, 1991; Wagner et al., 1991; Cherry et al., 1992), even at the level of only a doubling of PHYB gene dosage (Wester et al., 1994). The PHYA and PHYB wild-type alleles conversely are incompletely dominant to the phyA and phyB null mutations (Koornneef et al., 1980; Whitelam et al., 1993), indicating again that a 2-fold change in photoreceptor level is physiologically significant. Arabidopsis has been used extensively in the analysis of phytochrome function and phytochrome mutants and MAb that specifically detect each of the five Arabidopsis phytochrome apoproteins have been described (Hirschfeld et al., 1998). We have used these reagents here to investigate the light stabilities, distributions, and levels of this family of photoreceptors in wild-type plants throughout the growth cycle, under different light conditions, and over diurnal and circadian light cycles.
RESULTS
Normalization of Protein Gel Loads to DNA Content of Extracts
Tissues of seedlings grown under different light conditions, of mature plants at various ages, and from different plant organs vary in water content, cell size, and protein content. This raises the question of what basis should be used to compare the levels of the phytochromes present in such different tissues. The amount of phytochrome per gram fresh weight, per seedling or organ, or per milligram of total protein fails to account for the variability in the parameters described above. One useful comparison that would take into account several of these variables is an estimate of the average amount of each phytochrome per cell. As an approximation of this, in our analysis, the amount of extracted plant protein loaded on gels has been normalized to the DNA content of each extract. Frozen tissues were ground in liquid nitrogen, and separate aliquots were extracted and assayed for total protein content, using the standard phytochrome extraction buffer, or for total DNA content. The results of these protein and DNA assays are shown in Table I. The protein to DNA ratios range from 65:1 to 138:1 and, for a given tissue, are highly reproducible. Although there is a 7-fold difference in the amounts of protein and DNA per gram fresh weight of tissue when comparing, for example, dark-grown seedlings to flowers, the protein to DNA ratios among all the samples vary only 2-fold. Lanes of gels throughout this paper were loaded as indicated in the last column of Table I where a dark-grown seedling is set as the standard of a 1× load. Therefore, with the exception of the seed germination experiments, where extracts were loaded on a per seed basis (see below), the immunoblot signals on a given immunoblot in the figures in this paper can be interpreted as indicating the level of that phytochrome protein present in a similar number of DNA or genome equivalents from the various tissue samples tested.
Table I.
Protein to DNA ratios in Arabidopsis seedlings, mature plants, and plant organs
| Tissue | Protein | DNA | Protein-to-DNA Ratio | Relative Immunoblot Gel Loada |
|---|---|---|---|---|
| mg g−1 fresh wt | ||||
| 7-d dark | 2.6 (0.2)b | 0.029 (0.002) | 90 | 1× |
| 7-d R 3 h | 2.6 | 0.028 | 93 | 1× |
| 7-d R 12 h | 3.0 | 0.031 | 97 | 1× |
| 7-d R 24 h | 3.8 | 0.040 | 95 | 1× |
| 7-d WL | 8.2 (0.4) | 0.068 (0.005) | 121 | 1.3× |
| 14-d WL | 10.1 (0.4) | 0.089 (0.004) | 113 | 1.3× |
| 21-d WL | 10.4 (0.4) | 0.084 (0.004) | 119 | 1.3× |
| Leaves | 11.1 (0.7) | 0.092 (0.012) | 121 | 1.3× |
| Roots | 4.7 (0.35) | 0.064 (0.012) | 73 | 0.8× |
| Stems | 8.6 (0.6) | 0.062 (0.009) | 138 | 1.5× |
| Flowers | 18 (0.8) | 0.280 (0.035) | 65 | 0.7× |
The protein to DNA ratio of dark-grown seedlings has arbitrarily been set as 1 and the gel loads normalized to this value.
se for three independent experiments. Only a single determination of these values was performed for the R 3-, 12-, and 24-h conditions.
Light Stabilities of Phytochrome Proteins in Seedlings
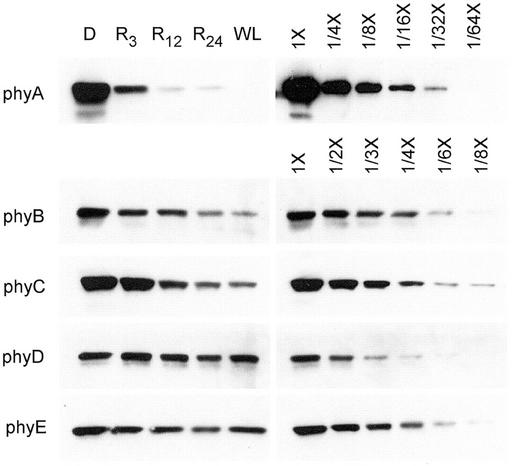
Hirschfeld et al. (1998) described the isolation and characterization of MAb against all five of the Arabidopsis phytochromes. With the exception of the recognition by anti-phyD MAb 2C1 of a low abundance non-phytochrome cross-reacting protein (NPXR) in dark-grown tissue that is described in that work, all available evidence indicates that the anti-phytochrome MAb lines are phytochrome specific. Moreover, it was shown that MAb 2C1 can be used to identify and quantify phyD in dark-grown plant extracts if a 20% (w/v) (NH4)2SO4 fraction rather than a 25% fraction is analyzed (Hirschfeld et al., 1998). To characterize the light stabilities of the Arabidopsis phytochromes, immunoblots of (NH4)2SO4-precipitated fractions of 7-d-old seedlings that were grown in the dark, grown in the dark but irradiated with 3, 12, or 24 h of continuous R light before harvest, or grown in continuous white light (WL) were probed with each of the five anti-phytochrome MAbs. These blots are shown in Figure 1. For the phyD blot, a 20% (w/v) (NH4)2SO4 fraction was prepared to eliminate NPXR from the extract. The seedling extracts were loaded on gels as indicated in Table I, and for each antibody, dilution curves of the dark-grown extracts indicate the response of the assay to relative antigen levels.
Figure 1.
Immunoblot analysis of the light stabilities of the five Arabidopsis phytochromes. Seedlings were grown for 7 d in continuous darkness (D) or WL, or in darkness and then placed under R for 3 h (R3), 12 h (R12), or 24 h (R24) before harvest at 7 d. Extracts were prepared and precipitated with 25% (w/v) (NH4)2SO4 for phyA, B, C, and E or with 20% (w/v) (NH4)2SO4 for phyD. An amount of protein equivalent to 300 μg of total extractable protein was loaded in the D, R3, R12, and R24 lanes and an amount equivalent to 390 μg in the WL lane. For each phytochrome, dilution curves of the darkness extract were included to estimate the -fold difference in the level of that apoprotein under the different light conditions. MAbs used were: phyA, 073d; phyB, B6B3; phyC, C11 and C13; phyD, 2C1; and phyE, 7B3.
In Figure 1, the phyA protein exhibits the strong R-dependent down-regulation that has previously been described for light-labile or type I phytochrome, resulting in a 50- to 100-fold reduction in level by 12 h in R. The phyB and phyC proteins also show significant R-light-induced down-regulation, although the reduction in protein level is more gradual and much less pronounced compared with the phyA down-regulation (Fig. 1). Compared with dark-grown seedlings, levels of phyB and phyC are 4- to 5-fold reduced in seedlings grown either for 6 d in the dark followed by 24 h of R light or for 7 d in continuous WL. The levels of phyD and phyE are not strongly changed upon extended exposure to R or WL, showing at most a 2-fold reduction compared with the dark-grown seedlings (Fig. 1). Thus, phyD and phyE most closely resemble truly light-stable phytochromes. Taken together, the varied responses of the five phytochrome protein levels to R and WL indicate that there is a gradation of light stabilities in this receptor family.
Normalized Phytochrome Levels in Arabidopsis Dark-Grown and Light-Grown Seedlings
It is important to note that, in Figure 1, exposure times for detection of the chemiluminescence signal varied from blot to blot, and although the chemiluminescence signals in the figure accurately reflect the relative levels of each phytochrome protein under the set of conditions being tested, they cannot be used to compare the levels of the different phytochrome proteins with each other. Therefore, the absolute levels of the five phytochromes were determined by comparison with standard curves of purified proteins.
Several protocols for preparation of protein extracts for use in immunoblot analysis of phytochrome have been described. We compared four procedures using Arabidopsis seedling tissues: extraction of fresh tissue into hot SDS buffer (Wang et al., 1992), (NH4)2SO4-precipitated extraction (Somers et al., 1991), the EZ protocol (Martinez-Garcia et al., 1999), and grinding and direct extraction of frozen tissue (Hirschfeld et al., 1998). Extraction of fresh tissue into hot SDS buffer yielded low-quality immunoblots with high backgrounds, and we observed low-phytochrome recovery with the EZ protocol such that the calculated protein levels in the various tissues were approximately 2-fold lower using this method compared with the others. The (NH4)2SO4-precipitated extraction protocol and direct extraction of frozen tissue yielded the most consistent results, and because the (NH4)2SO4-precipitation introduces an additional correction for the fraction of total protein precipitated, the very simple direct extraction method was adopted for most of the quantitative experiments.
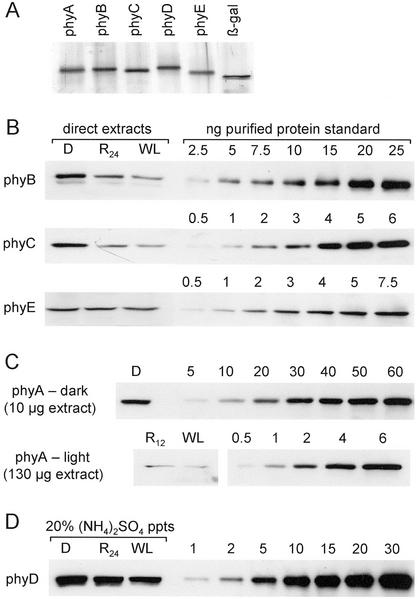
To construct standard curves of purified phytochrome apoproteins, the five Arabidopsis PHY cDNA sequences were expressed from the pET3c vector in Escherichia coli, and the approximately 120-kD phytochrome products were purified from gels by electroelution. Figure 2A shows a silver-stained gel of the five purified proteins. For phytochromes B, C, and E, direct extracts of seedlings grown for 7 d in darkness, for 6 d in darkness followed by 24 h in R light (R24), or for 7 d in continuous WL, were analyzed by immunoblotting with standard curves of each of the purified apoproteins. Figure 2B shows representative blots, which were loaded with 50 μg of the dark-grown or R24 extracts or 65 μg of the WL extract to normalize to DNA content (Table I). For the highly light-labile phyA, a R irradiation of only 12 h was used (R12), and separate blots were prepared for the dark and light extracts because of the large difference in the level of phyA in these samples (Fig. 2C). Measurement of the level of phyD in dark-grown tissue is complicated by cross-reactivity of the anti-phyD 2C1 MAb to a protein (NPXR) found in dark-grown extracts (Hirschfeld et al., 1998). For this reason, the three extracts for phyD quantitation were precipitated with 20% (w/v) (NH4)2SO4, which does not precipitate NPXR protein (Hirschfeld et al., 1998), and these samples were blotted and probed as shown in Figure 2D.
Figure 2.
Quantitation of phytochrome levels in seedling extracts with purified apoprotein standards. A, Silver-stained 6% (w/v) SDS-PAGE gel of 50 ng of each of the E. coli-expressed purified phytochrome apoprotein standards and a commercial preparation of 50 ng of purified β-galactosidase. B, phyB, phyC, and phyE immunoblots of direct extracts of seedlings grown for 7 d in continuous darkness (D) or WL, or for 6 d in the dark followed by 24 h in R (R24) with dilution curves of the purified apoproteins. Fifty micrograms of D and R24 extract protein and 65 μg of WL extract were loaded, and blots were probed with the MAbs described in Figure 1. C, Two separate phyA immunoblots, one with 10 μg of darkness direct extract and one with 130 μg of the R12 or WL direct extracts and appropriate dilution curves of the purified phyA apoprotein, were probed with MAb 073d. D, To eliminate cross-reactivity of the anti-phyD MAb 2C1 with a non-phytochrome protein, 20% (w/v) (NH4)2SO4 precipitated fractions were used instead of direct extracts on the phyD blot. An amount of precipitated protein equivalent to 300 μg of darkness or R24 extract or 390 μg of WL extract was loaded.
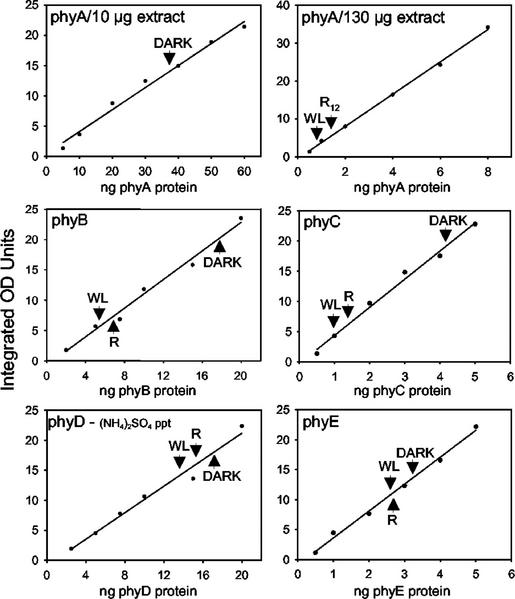
Three independent immunoblots for each of the phytochromes, similar to those shown in Figure 2, B through D, were scanned and quantitated by densitometry. Figure 3 shows curves generated from representative blots and Table II gives the average values determined for nanograms of the five phytochromes per 100 μg of total protein extracted from dark-grown and R-irradiated seedlings and per 130 μg of total protein from WL-grown seedlings (see Table I for normalization to DNA content). The choice of expressing the phytochrome levels using nanograms per 100 μg of total extracted protein from dark-grown seedlings as a base, and all other values relative to this, is arbitrary. It allows phytochrome levels to be compared with each other and, using corrections for the weight, protein content, or estimated number of cells per seedling, can be converted to other units (see “Discussion”). For phyD blots, the amounts of (NH4)2SO4-precipitated protein loaded on gels were the equivalent of 300 μg of total protein from dark-grown seedlings and 390 μg from R- or WL-grown seedlings (Fig. 3). To confirm the comparable nature of the two different types of samples, in a separate experiment, the phyD level in direct extracts of light-grown tissues, where the cross-reacting NPXR protein is absent (Hirschfeld et al., 1998), was measured as 3 ng 100 μg−1 total protein (data not shown).
Figure 3.
Quantitation of phytochrome immunoblots by densitometry. Chemiluminescence signals on films of blots similar to those shown in Figure 2 were scanned and analyzed by densitometry. Densitometry units in integrated OD units are plotted versus the amount of purified apoprotein loaded per lane of the gel. The values obtained for darkness (D), R24 (phyB, C, D, and E), or R12 (phyA), and WL extracts are indicated by arrow heads.
Table II.
Nanograms of phytochrome apoprotein per DNA equivalenta
| Db | R24 | WL | |
|---|---|---|---|
| phyA | 355 (10)c | 2.8d | 0.7 (0.12) |
| phyB | 39.7 (1.7) | 12.3 (3.9) | 7.3 (2.0) |
| phyC | 8.1 (0.6) | 3.1 (0.9) | 2.4 (0.8) |
| phyDe | 5.4 (0.6) | 5.5 (1.8) | 2.9 (0.3) |
| phyE | 6.1 (1.9) | 5.0 (2.4) | 4.8 (1.5) |
DNA equivalents are defined as 100 μg of total protein extracted from dark-grown and R24-grown seedlings and 130 μg of total protein from light-grown seedlings; see Table I for normalization to DNA content.
Seedlings were grown for 7 d in the dark, for 6 d in the dark followed by 24 h in R (R24), or under continuous WL.
se for three independent experiments.
For phyA, the R sample was harvested after 6.5 d in the dark and 12-h irradiation with R. This experiment was performed only once.
The phyD levels were determined from 20% (NH4)2SO4-precipitated extracts.
In dark-grown seedlings, where all five phytochromes are at their highest levels (Figs. 1 and 2), the normalized amounts of the phytochromes per 100 μg total extracted protein are measured as: 355 ng of phyA, 39.7 ng of phyB, 8.1 ng of phyC, 5.4 ng of phyD, and 6.1 ng of phyE. Hence, 85% of phytochrome in the etiolated seedling is phyA, 10% is phyB, and 5% is phyC, D, and E combined. In continuous WL-grown seedlings, the measured levels per 130 μg of total extracted protein are: 0.7 ng of phyA, 7.3 of ng phyB, 2.2 ng of phyC, 2.9 ng of phyD, and 4.8 ng of phyE. This represents a large change in photoreceptor content compared with the results from etiolated seedlings, with approximately 5% phyA, 40% phyB, 25% phyE, and 15% each of phyC and phyD. With reference to overall light stability, these values are consistent with the results in Figure 1 obtained with (NH4)2SO4-precipitated extracts. Phytochrome A, as expected, is a highly light-labile or type I phytochrome, whereas among the type II forms, phyB and phyC have intermediate light stability compared with the most light-stable phyD and phyE forms.
Phytochrome Levels Are Similar in Commonly Used Arabidopsis Ecotypes
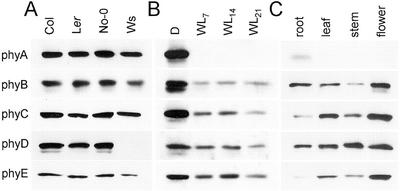
Because various Arabidopsis ecotypes exhibit differences in light-regulated morphological and developmental characteristics and one ecotype, Wassilewskija (Ws), lacks phyD (Aukerman et al., 1997), the levels of the phytochromes in four commonly used ecotypes were examined. Immunoblots of (NH4)2SO4-precipitated extracts of dark-grown 7-d-old seedlings of the Columbia (Col), Landsberg erecta (Ler), Nossen (No-0), and Ws ecotypes probed with the phytochrome-specific MAbs are shown in Figure 4A. Little variation in the expression levels of these proteins is observed among the lines aside from the lack of phyD in the Ws ecotype. All other experiments reported here were performed on the Col ecotype.
Figure 4.
Analysis of phytochrome levels in diverse Arabidopsis ecotypes, in mature rosette plants, and in plant organs. A, Seedlings of Arabidopsis ecotypes Col, Ler, No-0, and Ws were grown in darkness for 7 d, proteins were extracted and precipitated with 25% (w/v) (NH4)2SO4 (for phyA, B, C, and E) or 20% (w/v) (NH4)2SO4 (for phyD), and an amount of precipitated protein equivalent to 300 μg of total extractable protein was fractionated on 6% (w/v) SDS-PAGE gels and blotted. Blots were probed with the MAbs described in Figure 1. B, Seven-day-old seedlings grown in the dark (D) or under continuous WL (WL7) and rosette-stage plants grown under continuous WL for 14 (WL14) or 21 d (WL21) were extracted. Extracts were precipitated with 25% (w/v) (NH4)2SO4 (for phyA, B, C, and E) or 20% (w/v) (NH4)2SO4 (for phyD), and amounts of precipitated protein equivalent to 300 μg (D) or 390 μg (light-grown) of extractable protein were fractionated and immunoblotted as in A above. C, Leaves, inflorescence stems, and flowers were harvested from soil-grown plants, and roots were harvested from liquid grown plantlets. Proteins were extracted and precipitated with 25% (w/v) (NH4)2SO4. Amounts of precipitated protein equivalent to 240 μg (roots), 390 μg (leaves), 450 μg (stems), or 210 μg (flowers) of total extractable protein were fractionated and immunoblotted as in A above.
Phytochrome Levels in Mature Vegetative Tissues and in Plant Organs
To determine whether phytochrome levels change over the transition from seedling to mature plant, the levels of the five phytochrome proteins in 14- and 21-d-old continuous WL-grown Arabidopsis plants were compared with those in 7-d-old dark- and light-grown seedlings. Figure 4B shows that the phyA protein remains very low, below the detection limit on this blot, throughout this time. The levels of phyB, phyC, phyD, and phyE are not strikingly altered in 14- and 21-d-old plants compared with 7-d-old light-grown seedlings. At 21 d, when the plants were beginning to bolt, all four of these receptor proteins are present at equivalent or, at most, 2-fold reduced levels compared with 7-d-old light-grown seedlings, on a per DNA content basis (see Table I).
In Figure 4C, the relative levels of the five phytochrome proteins in four mature plant organs are compared. Extracts of roots, harvested from 14-d-old liquid-grown plants, and rosette leaves, flowering bolt stems, and whole flowers from 28-d-old soil-grown plants were assayed on immunoblots. In each case, the amount of plant extract loaded on the gel was normalized to the DNA content of the extract, as indicated in Table I. After a long exposure to film, the phyA protein is detected weakly in roots and, at very low levels, in flowers. The phyB, phyC, phyD, and phyE proteins are observed in all four plant organs, in each case most abundantly in flowers.
Phytochrome Levels during Germination and Early Seedling Development
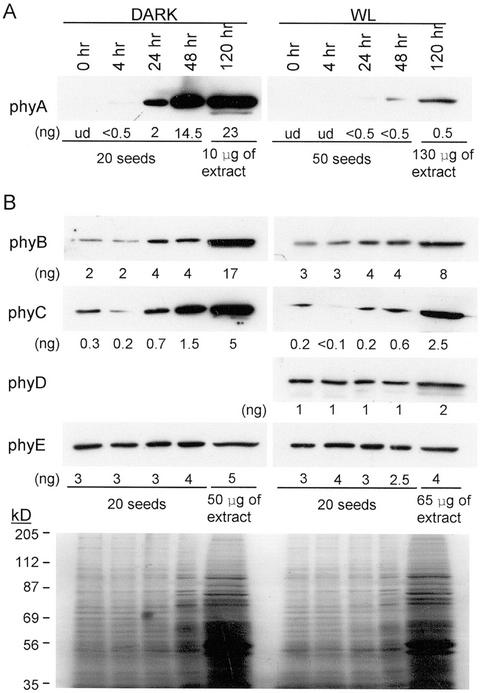
To determine which phytochromes are present during seed germination and early seedling development, the levels of the five Arabidopsis phytochrome proteins over a time course from imbibed seeds through 5 d growth in the dark and in the light were measured as shown in Figure 5. Dry seeds were sterilized (approximately 20 min), then imbibed for 1 h shaking in water in the dark. The 0-h time points in Figure 5 are extracts prepared at the end of this imbibition period. The time course was performed in liquid growth medium, in the dark or in continuous light, to facilitate collection of tissue samples. A parallel experiment done on solid growth medium gave similar results (T. Clack and R.A. Sharrock, unpublished data). The first four lanes of all of the blots shown in Figure 5 contain extract from 20 seeds per lane except the light-grown blot probed for phyA, which contains extract from 50 seedlings per lane to increase the detection sensitivity. The fifth lanes in each blot, containing the 120-h samples, were not loaded on a per seed basis but on a DNA equivalent basis, as in Figure 2, because, by 120 h of development, the quantity of protein in 20 seedlings had increased to the point where it caused the lanes to distort. The stained gel at the bottom of Figure 5B illustrates the protein loads in each lane of the blots shown in Figure 5B. Although not shown here, these blots were compared with standard curves of purified phytochrome apoproteins, as in Figures 2 and 3. The levels of the antigens determined in this way are given below the lanes in the blots in Figure 5. The level of phyD was not assayed in the dark in these experiments because of the detection of the cross-reacting NPXR protein by MAb 2C1 in dark-seedling direct extracts.
Figure 5.
Phytochrome levels in germinating seeds and young seedlings. A, Seeds were germinated in the dark for 0, 4, 24, or 48 h, and the amounts of protein extracted from 20 seeds were loaded in wells of a 6% (w/v) SDS-PAGE gel. One batch of seeds was allowed to grow in the dark for 5 d, the seedlings were extracted, and 10 μg of total protein was loaded in the fifth lane. Seeds were also germinated in continuous WL for 0, 4, 24, or 48 h, and the amount of protein extracted from 50 seeds was loaded on a gel. In the fifth lane of this gel, 130 μg of protein from 5-d-old light-grown seedlings was loaded. After electrophoresis, the gels were blotted and probed with the anti-phyA MAb 073d. B, Seeds were germinated in the dark or in continuous WL for 0, 4, 24, or 48 h, and the amounts of protein extracted from 20 seeds were loaded on 6% (w/v) SDS-PAGE gels. Batches of seeds allowed to grow for 5 d in darkness or continuous light were extracted and 50 or 65 μg of total protein from these seedlings was loaded in the fifth lane of the respective gels. Duplicate identical gels were run, blotted, and probed with MAbs for phyB (B6B3), phyC (C11 and C13), phyD (2C1), and phyE (7B3). Dilution curves of purified apoprotein standards were run next to the lanes shown here and the levels of the phytochromes were measured by scanning densitometry. These levels are given below each lane (ud, undetectable). A representative blot containing the dark and light protein samples and stained with copper stain is shown at the bottom of B to illustrate the relative protein loads. A blot of the dark extracts was not probed for phyD level because of the presence of a cross-reactive protein to that antibody in the dark.
Figure 5A shows that phyA is undetectable in this assay in seeds at the end of the sterilization and 1-h imbibition period (0-h lanes). The seeds used in this experiment were harvested from plants grown in continuous light, and whether phyA accumulates in seeds that are allowed to develop in darkness or under a short photoperiod was not investigated. In the dark, the level of phyA becomes detectable by 4 h after imbibition and rises dramatically over the 5-d time course to reach a level of 230 ng 100 μg−1 extracted protein at d 5. In the light, phyA is detectable by 24 h and reaches a level of 0.5 ng 130 μg−1 extracted protein by d 5 (Fig. 5A). These results are consistent with those in Table II for 7-d-old seedlings, assuming the level of phyA in the dark continues to rise as the seedlings get older. Phytochromes B, D, and E are all present at significant levels (1–3 ng seed−1) in freshly imbibed seeds, whereas phyC is 5- to 10-fold lower (Fig. 5B). On a per seed basis, phyB and phyC increase slightly over the first 48 h of germination and seedling development in the dark but are relatively constant in continuous light. The levels of phyD and phyE do not change over early seedling development. By d 5, the measured levels of all of the phytochromes in Figure 5 are consistent with those measured in 7-d-old seedling in Table II.
Diurnal and Circadian Cycling of Phytochromes
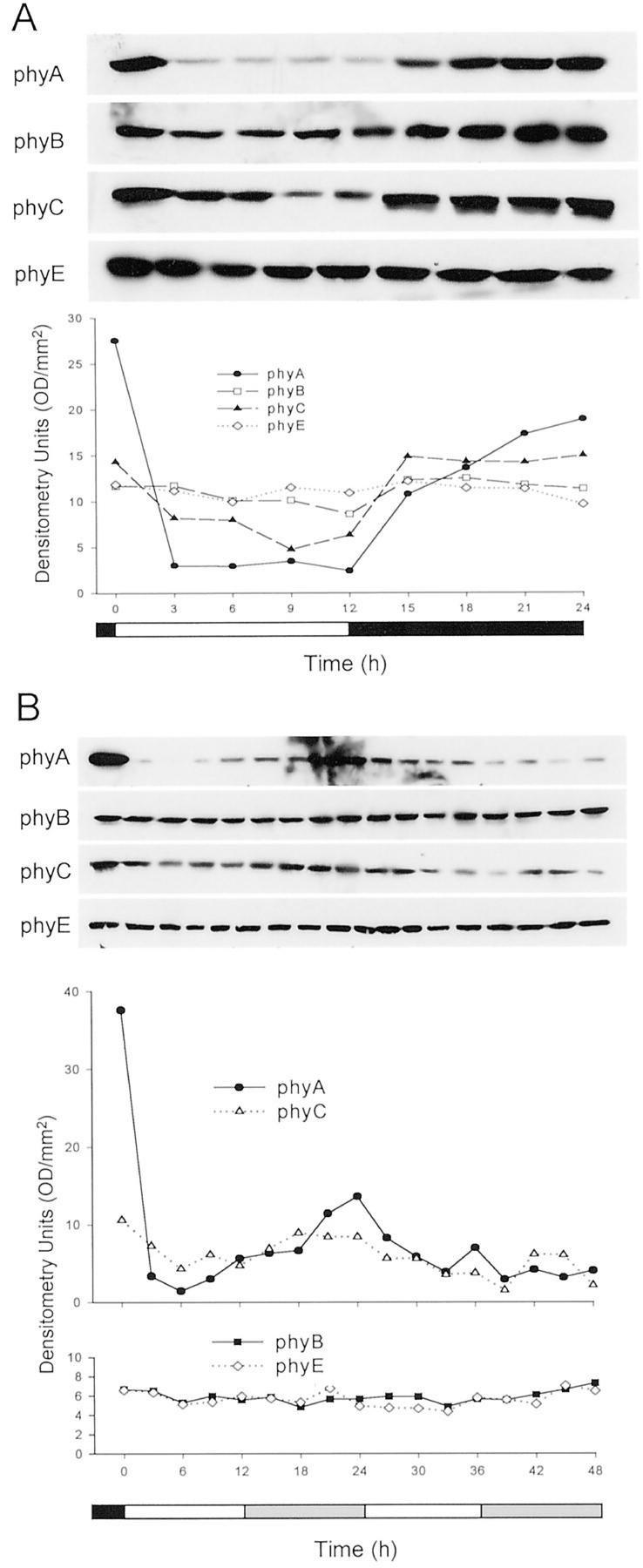
Studies using phy null mutants have demonstrated that phyA, phyB, phyD, and phyE have roles in sensing the input light signals to the circadian clock (Somers et al., 1998; Devlin and Kay, 2000). In addition, pronounced circadian cycling of the activities of the PHYA, PHYB, PHYD, and PHYE promoters has been observed (Kozma Bognar et al., 1999; Toth et al., 2001). To determine whether the levels of the Arabidopsis phyA, B, C, and E apoproteins cycle in a diurnal fashion, seedlings were grown for 5 d under a 12-h WL/12-h dark photoperiod (LD) and harvested at 3-h intervals throughout the 5th d. Protein extracts were prepared, precipitated with 25% (w/v) (NH4)2SO4, and analyzed by immunoblotting. The levels of phyD were not followed in these experiments because of the complication of detection of the NPXR protein by the 2C1 MAb. Figure 6A shows that over a single LD cycle, clear diurnal cycling of the levels of phyA and phyC is evident. Low amplitude cycling of phyB is also indicated, whereas the level of phyE remains constant. The data in Figure 6 are representative of two independent repetitions of this experiment, both of which showed qualitatively similar responses of the four phytochrome proteins. The potential circadian regulation of cycling of the phytochromes was also analyzed. Figure 6B shows steady-state levels of phyA, B, C, and E over a 48-h time course in seedlings that were grown for 5 d under a 12 WL/12 dark photoperiod and transferred to continuous WL. In no case was strong circadian cycling of a phytochrome observed. Again, two independent experiments were performed and data from one experiment are presented in Figure 6B. In both experiments, cycling of phyA at very low protein levels, close to the detection limit of the assay, and cycling of phyC with a peak in the subjective dark and progressive damping over the 48-h time course were seen (Fig. 6B). In these experiments, phyB and phyE did not show significant circadian cycling.
Figure 6.
Diurnal and circadian cycling of phytochrome protein levels. A, Seedlings were grown for 5 d under a 12-h WL/12-h dark photoperiod and harvested at 3-h intervals over the course of the 5th d. Proteins were extracted and precipitated with 25% (w/v) (NH4)2SO4, and amounts of precipitated protein equivalent to 400 μg of extractable protein were fractionated and immunoblotted. Blots were probed with MAbs for phyA (073d), phyB (B6B3), phyC (C11 and C13), and phyE (7B3). X-ray films of chemiluminescence signals were scanned and quantitated by densitometry, and these data are presented graphically in the lower panel. B, Seedlings were grown for 5 d under a 12-h WL/12-h dark photoperiod and transferred to continuous WL. Seedlings were harvested at 3-h intervals, proteins were extracted and precipitated with 25% (w/v) (NH4)2SO4, and these proteins were fractionated, blotted, probed with antibodies, and quantitated as in A.
DISCUSSION
Light is a critical environmental effector of plant growth and development, and it readily penetrates aboveground tissues, so there are few barriers to its detection in cells (Vogelmann, 1994). Therefore, the localization and the physical abundance of a given photoreceptor are likely to be important determinants of its biological activity. In support of this, the expression level of the phyA and phyB phytochromes has been shown to have a quantitative correlation with light responses, both in overexpression assays (Boylan and Quail, 1991; Wagner et al., 1991; Cherry et al., 1992; Wester et al., 1994) and in heterozygotes of null mutations (Koornneef et al., 1980; Whitelam et al., 1993). This suggests that the level of phyA or phyB Pfr formed in the plant is a limiting factor in signaling in these photosensing pathways and that the levels of the phytochrome photoreceptors themselves are critical determinants of photoresponsiveness. We have used type-specific MAb to the five Arabidopsis phytochromes to assess the levels of these molecules in tissue extracts. This analysis does not address the spectral or biological activity of the phytochrome proteins detected, only the abundance of the apoproteins. However, given that overexpressed phytochrome apoprotein in Arabidopsis is converted to correspondingly high levels of spectrally active photoreceptor (Wagner et al., 1991), it is likely that the phytochromobilin chromophore is not limiting in most plant cells and, therefore, that the estimates derived here represent biologically active phytochrome levels.
The Assay
Quantitative analysis of protein levels in plant tissues by immunoblotting is influenced by several considerations including the specificity of the antibodies used, the purity of the proteins used to generate standard curves, and the efficiency of the extraction method. The specificity of the MAb used here has been extensively demonstrated (Hirschfeld et al., 1998). The apoprotein preparations used as standards were highly overexpressed in E. coli cultures and cut out and eluted from denaturing gels. Compared with other methods of preparation we tried, such as affinity chromatography approaches, these samples consistently showed less contamination with extraneous proteins in silver-stained gels. Nevertheless, the concentrations of these phytochrome standards, as determined by protein assay, doubtless reflect small amounts of contaminating protein in each sample. These considerations are likely to affect the quantitation of the phytochromes in relatively small and consistent ways such that the absolute values of nanograms of phytochrome per microgram of protein derived here are very good estimates of their actual levels and the levels of the five forms relative to each other are quite accurate.
We compared several different protocols for preparation of tissue extracts, settling on the method that gave the most consistent and highest recoveries. However, the extraction buffer used in this method does not contain detergent, making it possible that some phytochrome pellets with cellular debris in the centrifugation step of the protocol. Moreover, it has been shown that phyA and phyB are translocated from the cytoplasm to the nucleus in the presence of light (Kircher et al., 1999; Yamaguchi et al., 1999), raising the additional possibility that the extractable amounts of some of the phytochromes could be reduced in light-grown compared with dark-grown tissue as a result of association with pelletable nuclear material (see phyB and phyC in Fig. 1). To address this, experiments were performed in which the pelleted debris from extractions of dark- and light-grown seedlings were taken up in SDS buffer, boiled, and analyzed on immunoblots. Negligible amounts of any of the phytochromes were recovered from the pellets from either tissue (T. Clack and R.A. Sharrock, unpublished data). In addition, experiments in which dark-grown and light-grown seedlings were directly extracted into hot SDS buffer showed similar light-associated reductions of phyB and phyC to those presented here (T. Clack and R.A. Sharrock, unpublished data). Hence, although it remains possible that some phytochrome is nonextractable using current protocols and that this could be differential between dark and light growth conditions, we have no evidence that this has significantly influenced our results.
The amount of each protein extract loaded on the gels used for the immunoblots performed here was normalized to the DNA content of the extract. In doing this, we attempted to normalize the very different tissues assayed to genome equivalents and, in a crude way, to cell numbers. This approach could be compromised by extensive endoreduplication of the nuclear genome or by large changes in the numbers of plastid genomes per cell under different light conditions or at different stages of plant growth. Currently available evidence indicates that, although these parameters vary somewhat in different cell types and at various stages of plant growth, the differences are not large. For instance, using quantitative PCR to follow replication of nuclear, mitochondrial, and chloroplast DNA during leaf growth, Draper and Hays (2000) showed that these DNA molecules exhibit highly parallel rates of replication during growth of cotyledons and fifth true leaves in planta, so that the ratio of the three genomes remains constant throughout leaf development. Endoreduplication of the nuclear genome is observed in many cell types, especially those undergoing differentiation and expansion (Joubes and Chevalier, 2000), and an extra round of endoreduplication often occurs in hypocotyl cells of dark-grown seedlings compared with light-grown seedlings (Gendreau et al., 1997, 1998). This differential cellular polyploidization is phytochrome regulated but affects only some of the cells and leads to only a small change in the overall DNA to cell ratio of this tissue (Gendreau et al., 1998). In our experiments, protein to DNA ratios of seedlings and isolated organs vary only 2-fold between the lowest, flowers, and the highest, stems (see Table I), whereas the protein content per gram fresh weight varies 7-fold.
Phytochrome Contents of Dark-Grown and Light-Grown Seedlings
Given the relative constancy of cellular DNA contents, we believe that normalizing to DNA content of extracts is a meaningful way of comparing values from different tissues or growth conditions. Moreover, by correcting the data for other measured or estimated parameters, these results can be converted to other units. The data in the first column of Table III are taken from Table II and are expressed as nanograms of phytochrome per 100 μg of extracted protein from dark-grown seedlings and nanograms of phytochrome per 130 μg from light-grown (WL) seedlings. We recover 3.7 μg protein/dark-grown seedling and 9.6 μg protein/WL seedling, so the values in the first column also represent nanograms of phytochrome per 27 dark-grown seedlings and nanograms of phytochrome per 13.5 WL seedlings, and the amount of phytochrome/seedling can be directly calculated (Table III). We determined the wet weights of both dark-grown and WL seedlings as 0.8 mg seedling−1 and, therefore, the amount of phytochrome per gram fresh weight can also be directly calculated (Table III).
Table III.
Conversion of nanograms of phytochrome perDNA equivalent in 7-d-old seedlings to other units
| ng DNA equivalent−1a | ng seedling−1b | μg g−1 fresh wtb | pg cell−1b | molecules cell−1 | |
|---|---|---|---|---|---|
| phyA | |||||
| D | 355 | 13 | 16.3 | 0.27 | 1 × 107 |
| WL | 0.7 | 0.05 | 0.06 | 0.005 | 2 × 104 |
| phyB | |||||
| D | 39.7 | 1.5 | 1.8 | 0.03 | 1.2 × 106 |
| WL | 7.3 | 0.54 | 0.68 | 0.005 | 2 × 105 |
| phyC | |||||
| D | 8.1 | 0.30 | 0.38 | 0.006 | 2.5 × 105 |
| WL | 2.4 | 0.18 | 0.22 | 0.0017 | 7 × 104 |
| phyD | |||||
| D | 5.4 | 0.20 | 0.25 | 0.004 | 1.7 × 105 |
| WL | 2.9 | 0.21 | 0.27 | 0.002 | 8.3 × 104 |
| phyE | |||||
| D | 6.1 | 0.23 | 0.28 | 0.005 | 1.9 × 105 |
| WL | 4.8 | 0.36 | 0.44 | 0.003 | 1.4 × 105 |
To estimate the content per cell of each of the phytochromes, four assumptions have been made: (a) that the five phytochromes in the plant are each evenly distributed among all the cells, (b) that the density of dark-grown and WL tissues can be approximated as that of water (1 g cm−3), (c) that an average cell of a light-grown Arabidopsis plant can be roughly estimated as having the dimensions 20 × 20 × 15 μm, giving a volume of 6,000 μm3 (Pyke et al., 1991), and (d) that the DNA per gram fresh weight values in Table I are acceptable indicators of relative cell numbers. All of these assumptions clearly represent simplifications and first approximations but, with that in mind, they yield a useful preliminary picture of phytochrome distribution. Using a density of 1 g cm−3, the 0.8 mg weights of dark-grown and WL seedlings can be converted to volumes of 0.8 mm3 seedling−1. The DNA per gram fresh weight values in Table I indicate that there are 2.3 times as much DNA, and, therefore, 2.3 times as many cells per gram fresh weight in WL cells as dark-grown cells. Because dark-grown and WL seedlings weigh the same and are therefore assumed to have similar volumes, this indicates that there are 2.3 times as many cells per seedling in WL versus dark-grown tissue and, conversely, that dark-grown cells are 2.3 times the size of WL cells. Hence, WL cells are estimated to be 6,000 μm3 and dark-grown cells 14,000 μm3, and seedlings with volumes of 0.8 mm3 are estimated to contain, respectively, 1.3 × 105 cells in WL and 6 × 104 cells in dark-grown seedlings. These values allow the calculation of picograms of phytochrome per cell and, with an average dimeric molecular mass of 250,000 D, number of molecules per cell for each of the phytochromes (Table III). These calculated values indicate that phytochromes are present at low to moderate levels in cells, levels similar to those of hormone and growth factor receptor proteins in animals.
Levels of the Five Phytochromes in Seeds, Seedlings, and Mature Plant Organs
Our results indicate that the original distinction between light-labile type I phytochromes and light-stable type II phytochromes (Furuya, 1993) is valid and useful in its broad outlines. We have shown that: (a) phyA is a uniquely light-labile type I form, which is clearly the most strongly regulated in its expression level; (b) among the four light-stable type II forms, phyB and phyC are significantly reduced upon extended exposure to R light, whereas phyD and phyE are nearly unaffected; (c) the four type II phytochromes are present quite constitutively, being detectable in all samples from imbibed seeds to inflorescence stems and flowers, although significant alteration in some phytochrome levels, such as those of phyB and phyC during early seedling growth, does occur; and (d) in most of the light-grown tissues examined here, phyB is the most abundant of the four type II phytochromes, consistent with its predominant role in the most obvious light-regulated traits. These observations do not markedly change our concept of the phytochrome family. However, they provide a set of quantitative reference values from which physiological, genetic, and molecular experiments on phytochrome function can be interpreted.
The varied responses of the five phytochrome protein levels to light indicate that there is a gradation of light stability in this receptor family, which along with the spectral attributes of the different forms, such as the extent and rate of dark reversion (Eichenberg et al., 2000), may have an important influence on their respective activities. It has long been recognized that the rapid and strong down-regulation of phyA by light may reflect a role for this phytochrome as an antenna for very low fluence light signals. That phyB and phyC are down-regulated to a much lower extent than phyA but in similar fashion to each other indicates that alteration of receptor stability by light may be a general mechanism for controlling the activities of individual phytochromes. It is notable that the level of phyC is reduced severalfold in phyB null mutants, both in dark- and light-grown seedlings (Hirschfeld et al., 1998). These observations and those described here suggest that phyB and phyC may be subject to coordinated turnover.
Using null mutants, phytochromes A, B, and E have been shown to mediate the induction of seed germination by R and FR irradiation and to strongly influence several early seedling growth responses (Parks and Quail, 1993; Reed et al., 1993; Shinomura et al., 1996; Whitelam and Devlin, 1997; Hennig et al., 2002). Roles for the other two phytochromes in germination have not been described and, although phyD mutants are defective in early seedling hypocotyl elongation and cotyledon opening responses (Aukerman et al., 1997), little has been described relating to the roles of phyC or phyE in early seedling growth. In earlier studies, phyA was not detectable on immunoblots of freshly sterilized Arabidopsis seeds or after a 24-h imbibition but was detectable in 2-d-old dark-grown seedlings, whereas phyB was present in seeds at a level similar to that in etiolated seedlings (Shinomura et al., 1994, 1996). This induction of phyA synthesis correlated with the development of photoresponsiveness of seed germination in a phyB mutant line (Shinomura et al., 1996). We also observe that phyA is not detectable in seeds, however it is faintly detectable after 4 h of dark growth, is clearly present after 24 h, and continues to accumulate as the seedling grows. In addition, we find that phyB, D, and E are present at significant levels in freshly imbibed seeds whereas phyC is 5- to 10-fold lower. These phytochromes are either maintained at those levels (phyD and phyE) or accumulate slowly (phyB and phyC) over the course of seedling development, irrespective of whether the seedlings develop in the dark or in the light. The level of phyE is higher in seeds relative to the other phytochromes than at any other time we assayed, perhaps consistent with its novel role in R/FR light control of germination (Hennig et al., 2002).
Circadian Regulation of Phytochrome Levels
Diurnal and circadian cycling of the activities of the PHYA, PHYB, PHYC, PHYD, and PHYE promoters, at levels of from 2- to 8-fold, has been observed using both PHY-LUC reporter genes and analysis of the endogenous mRNA levels (Kozma Bognar et al., 1999; Toth et al., 2001). In those experiments, the PHYC promoter showed a low amplitude rhythm under diurnal LD conditions compared with the others and weak cycling under continuous light or continuous darkness circadian conditions. For all of these promoters, the peak of LUC activity and PHY mRNA abundance was seen during the light phase under light-dark cycles. This contrasts with our findings in that we observe significant diurnal cycling of the phyA and phyC apoproteins and weak cycling of phyB but no significant oscillation, even in continuous light, of phyE. Because of cross-reactivity of our anti-phyD MAb, we did not investigate the cycling of phyD. Moreover, the peaks of the oscillations in the phyA, B, and C apoproteins occur in the dark phase, when the mRNA levels are seen to be low (Toth et al., 2001). This suggests that, although there are fairly robust responses of PHY promoter activities to light cycles, oscillations in the PHY mRNAs are not rapidly converted to corresponding oscillations in protein levels. The diurnal cycling of phyA, B, and C may more closely correlate with light effects on their stabilities than on regulation of their biosynthetic capacity. The identification of the F-box containing family of ZTL/ADO/LKP/FKF proteins as components of the light input pathway to the clock (Nelson et al., 2000; Somers et al., 2000; Jarillo et al., 2001) and the physical interaction of ZTL/ADO1 with phyB (Jarillo et al., 2001) reinforce this possibility. Under LL conditions, which test for circadian regulation, we observe very weak cycling of phyA and phyC, indicating that the mechanisms controlling the levels of these receptors are responsive to the circadian clock.
Comparison with Previous Studies
A pool of anti-phyB MAbs that are now known to cross-react with phyD (Hirschfeld et al., 1998) and the same anti-phyC MAbs used here were used previously to evaluate the light stability of these two proteins (Somers et al., 1991). At that time, it was concluded that phyB and phyC were light stable in comparing dark-grown tissue levels to tissue that had been exposed to 24 h of R light. However, in contrast to this, a later study showed that phyB was reduced 2-fold by 2 h after a 2-min R-light pulse (Anderson et al., 1997). Consistent with this later work, the results shown in Figures 1 through 3 here indicate that phyB and phyC are significantly light-labile compared with highly stable phyD and phyE. With reference to this, Clack et al. (1994) presented evidence that the PHYB and PHYC mRNA levels are unchanged as a fraction of total RNA in dark-grown compared with WL-grown seedlings, suggesting that the down-regulation of the phyB and phyC proteins likely occurs posttranslationally. The levels of the phyB, D, and E proteins in seeds, seedlings, isolated Arabidopsis organs, and over the time course of plant development determined here correlate well with the respective promoter activities of these genes measured in seedlings using RNA blots or promoter-β-glucuronidase fusion transgenes (Clack et al., 1994; Goosey et al., 1997). In contrast, PHYA-β-glucuronidase promoter activity (Somers and Quail, 1995a) and the level of PHYA mRNA (Clack et al., 1994) are higher in the light, relative to the other PHY genes, than would be expected for the low abundance of the phyA protein. This indicates that the phyA level may be controlled posttranslationally in light-grown plants by continuous light-induced proteolytic turnover of this phytochrome form.
Previous efforts at quantifying the different phytochromes in plant tissues have most frequently been performed in peas or oats and are, in general, consistent with our current findings. Konomi et al. (1987) used immunoblotting and ELISA techniques with antibodies selective for phytochrome I (phyA) and phytochrome II (phytochrome purified from light-grown tissue) and standard curves of immunopurified phytochromes I and II to determine the levels of these antigens in dissected pea axes. They found approximately 7 ng of each phytochrome per axis in unimbibed seed and observed increases to 200 and 50 ng axis−1, respectively, for phytochrome I and II after 12 h of imbibition of axes in the dark. The levels of three immunologically distinguishable phytochrome apoproteins in the embryo-containing portions of unimbibed oat seeds and in the shoot, scutellum, and root of 3-d-old oat seedlings were determined more recently by comparing immunoblot signals from tissue extracts to standard curves of each of the partially purified phytochromes (Wang et al., 1992, 1993a, 1993b). The three oat phytochromes were called the 123-, 124-, and 125-kD forms, with the 124-kD form corresponding to phyA. In the dissected embryo portions of oat seeds, the 123-, 124-, and 125-kD phytochromes were measured at 6.1, 1.6, and 1.4 ng embryo−1, respectively (Wang et al., 1992). In etiolated seedlings, the 123-, 124-, and 125-kD proteins were measured at 3.2, 83, and 1.5 ng per 100 μg total extracted protein and in light-grown seedlings at 2.2, <0.5, and 0.5 ng 100 μg−1 total protein (Wang et al., 1993a). The values determined in these studies were clearly derived from different plant species and from different tissues than those presented here. In most cases, the measured quantities of phytochrome were also expressed in units that are not directly comparable with the units used here. Nevertheless, the values are in the same ranges as those we observe, the light lability of oat phyA is of a similar order to that of Arabidopsis, and one of the “light stable” oat phytochromes, the 125-kD form, exhibits a significant reduction in the light relative to the dark, as we describe for Arabidopsis phyB and phyC.
CONCLUSION
The opportunity in Arabidopsis to monitor the levels of all of the members of the phytochrome family is unprecedented, and a comprehensive description of the distributions of these photoreceptors in this model plant will aid in analysis of their functions and mechanisms. Our current findings support an overall picture of a broadly distributed photoreceptor array for R- to FR-light signals in seeds and mature plant organs of Arabidopsis and the functioning of two general types of phytochrome, a single light-labile type I phyA form and multiple light-stable type II forms. The quantitative results presented here are consistent with previous work in both the distantly related dicot pea and the monocot oat. Hence, it is likely that these observations are, in their general implications, applicable across plant genera.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The Arabidopsis Col, Ler, No-0, and Ws ecotypes were obtained from previously described sources (Aukerman et al., 1997). For seedling tissues, seeds were surface-sterilized for 30 min in 15% (v/v) bleach/0.2% (w/v) SDS, rinsed at least five times with sterile water, and plated on sterile filter papers overlaying Murashige and Skoog basal agar medium (Sigma, St. Louis) containing 2% (w/v) Suc and 0.5 g L−1 MES in 100- × 25-mm petri dishes. Plates were kept in the dark at 4°C for 2 to 3 d, treated for 2 h with WL to induce germination, and then placed either in complete darkness or under the described light condition. Light sources for seedling growth were WL from a bank of 40-W cool-white fluorescent bulbs (10 W m−2) or R light from fluorescent bulbs (20WT12/2364, Sylvania, Danvers, MA) filtered through plastic (Roscolene no. 823, Musson Theatrical, Santa Clara, CA; 2.0 W m−2). Radiant output was determined using an IL1400A radiometer equipped with an SEL033/F/W detector (International Light, Newburyport, MA). Rosette and reproductive stage plants were grown on soil under continuous fluorescent light at 20°C in a growth chamber (Conviron, Pembina, ND). Roots were obtained from sterile plantlets grown shaking slowly in liquid Murashige and Skoog medium with 2% (w/v) Suc and 0.5 g L−1 MES under fluorescent lights for 2 weeks. For circadian studies, seeds were sterilized and plated as above and grown in a light-sealed growth chamber at 20°C under a 12-h fluorescent light (35 W m−2)/12-h dark photoperiod. Mature plants were grown on soil under continuous WL at 20°C.
Extraction Conditions and Measurement of Protein to DNA Ratios
The following protocol was adapted from that of Schmidt and Schibler (1995). Seedlings, mature plants, or organs were harvested and immediately frozen in liquid nitrogen. For each sample, 300 mg of tissue was ground to a fine powder under liquid nitrogen in a mortar and pestle, and aliquots of approximately 100 mg were transferred to two tubes and carefully weighed. To one tube, 1.8 mL of protein extraction buffer (50 mm Tris-HCl, pH 8.5, 5 mm EDTA, 75 mm (NH4)2SO4, and 25% [v/v] ethylene glycol) was added, and the tube was vortexed for 2 min and then centrifuged at 12,000g for 10 min. The protein concentrations of 20-, 40-, 60-, 80-, and 100-μl aliquots of the supernatant were determined (Bradford, 1976) using bovine serum albumin as a standard. To the other tube of ground tissue, 0.6 mL of DNA extraction buffer (200 mm Tris-HCl, pH 7.6, 5 mm EDTA, 250 mm NaCl, and 0.5% [w/v] SDS) was added, the tube was vortexed for 2 min, 0.6 mL of phenol/chloroform/isoamyl alcohol 50:49:1(PIC) was added, and the tube was vortexed for an additional 10 min. The samples were centrifuged (12,000g for 5 min) at room temperature, and the aqueous phase was removed. The interface and PIC fraction from this first extraction was re-extracted with an additional 0.6 mL of DNA extraction buffer and the two aqueous phases were mixed. Two volumes of ethanol were added, and nucleic acids were allowed to precipitate overnight at −20°C. The precipitate was collected by centrifugation at 20,000g for 30 min, excess ethanol was aspirated, and the pellet was air-dried briefly. The pellet was dissolved in 0.5 mL of TCS (10 mm Tris, pH 7.5, 05 mm CaCl2, and 1% [w/v] SDS) and transferred to a clean microtube. Proteinase K was added to 40 μg mL−1, and the samples were incubated at 50°C for 30 min. Samples were extracted with PIC, the aqueous phase was collected, and nucleic acids were precipitated with ethanol. The precipitate was collected as described above and dissolved in 100 μl of water. The DNA concentrations of 10-, 20-, and 30-μl aliquots of the sample were measured by staining with Hoechst dye 33258 and determining fluorescence with a Hoefer TKO fluorometer, using calf thymus DNA as a standard.
Protein Extraction and Electrophoresis, Immunoblotting, and Quantification of Immunoblots
For ammonium sulfate-precipitated protein fractions (Figs. 1, 4, and 6), extracts were prepared and precipitated with 25% or 20% (w/v) (NH4)2SO4, as described (Aukerman et al., 1997). For direct extracts (Figs. 2, 3, and 5), 0.25 g of frozen tissue was briefly ground in liquid nitrogen in a mortar and pestle, 0.5 mL of 2× protein extraction buffer plus protease inhibitors (100 mm Tris-HCl pH 8.5, 10 mm EDTA, 150 mm (NH4)2SO4, 50% [v/v] ethylene glycol, 2 μg mL−1 aprotinin, 1 μg mL−1 leupeptin, 1 μg mL−1 pepstatin, 2 mm phenylmethylsulfonyl fluoride, 10 mm iodoacetamide, and 5 μg mL−1 NaHSO3) was added, and the samples were ground for 2 min in the mortar and centrifuged for 5 min at 12,000g in a microfuge at 4°C. A sample of the supernatant was removed, and the protein concentration was determined (Bradford, 1976). The remaining supernatant was mixed with an equal volume of 2× SDS-PAGE sample buffer (Laemmli, 1970) and frozen in liquid nitrogen. Protein extracts were fractionated on SDS-polyacrylamide gels, blotted to nitrocellulose, and probed with MAb as described (Aukerman et al., 1997; Hirschfeld et al., 1998). Phytochromes were detected using the Supersignal West chemiluminescence kit (Pierce, Rockford, IL), exposed x-ray films were scanned on a Fluor-S Multiimager (Bio-Rad, Hercules, CA), and stored image files were analyzed using the Quantity One software package (Bio-Rad). MAbs were anti-phyA 073d, anti-phyB B6B3, anti-phyC C11 and C13, anti-phyD 2C1, and anti-phyE 7B3 (Hirschfeld et al., 1998).
Preparation of Purified Phytochrome Apoprotein Standards
The five Arabidopsis phytochrome apoproteins were expressed in Escherichia coli BL21 (DE3) from full-length cDNA sequences cloned in the pET3c vector as described previously (Somers et al., 1991; Hirschfeld et al., 1998). Cultures of E. coli carrying the pET3c-phyA, B, C, D, or E constructs were induced with isopropylthio-β-galactoside for 4 h, and the cells were collected by centrifugation and suspended in 50 mm Tris-HCl, pH 8.2, 2 mm EDTA. For each cell suspension, lysozyme was added to 0.1 mg mL−1 and Triton X-100 to 0.1% (v/v), and the suspension was incubated for 15 min at 30°C. The suspension was chilled on ice, sonicated, and centrifuged for 15 min at 12,000 rpm. The pellet containing inclusion bodies was washed three times with 0.5% (v/v) Triton X-100, 1 mm EDTA, dissolved in 50 mm Tris-HCl pH 8 m urea, and centrifuged at 20,000 rpm for 15 min to pellet any debris. This preparation was fractionated on a preparative 6% (w/v) SDS gel, proteins were visualized with GelCode E-zinc stain (Pierce, Rockford, IL), and the full-length phytochrome apoprotein band was excised with a razor blade. The excised phytochromes were electroeluted into 25 mm Tris-HCl, pH 8, 192 mm Gly, and 0.01% (w/v) SDS. Protein concentration was determined using a detergent-compatible protein assay (Bio-Rad). For immunoblots, the indicated amounts of the purified phytochrome apoproteins were fractionated on SDS gels in the presence of approximately 10 μg of carrier HELA cell protein. This carrier protein was prepared from cells grown as a monolayer in Dulbecco's modified eagle medium containing 10% (v/v) fetal calf serum (Invitrogen, Carlsbad, CA). Confluent plates of HELA cells were washed with PBS and trypsinized, and the cells collected. The cell pellet was suspended in SDS sample buffer, heated at 95°C for 5 min, and centrifuged at 12,000g for 10 min. These carrier protein preparations were shown to contain no proteins that cross-react with the anti-phy MAbs and were included to aid in the loading of the small amounts of the phytochrome apoprotein standards onto the gels and their electrotransfer to the nitrocellulose membrane.
Footnotes
This work was supported by the National Science Foundation (grant no. IBN–9808801 to R.A.S.). This is journal article no. 2002–25 from the Montana Agricultural Experiment Station, Montana State University (Bozeman).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.005389.
LITERATURE CITED
- Abe H, Yamamoto KT, Nagatani A, Furuya M. Characterization of green tissue-specific phytochrome isolated immunochemically from pea seedlings. Plant Cell Physiol. 1985;26:1387–1399. [Google Scholar]
- Adam E, Kozma-Bognar L, Kolar C, Schafer E, Nagy F. The tissue-specific expression of a tobacco phytochrome B gene. Plant Physiol. 1996;110:1081–1088. doi: 10.1104/pp.110.4.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam E, Szell M, Szekeres M, Schaefer E, Nagy F. The developmental and tissue-specific expression of tobacco phytochrome A genes. Plant J. 1994;6:283–293. [Google Scholar]
- Alba R, Kelmenson PM, Cordonnier-Pratt MM, Pratt LH. The phytochrome gene family in tomato and the rapid differential evolution of this family in angiosperms. Mol Biol Evol. 2000;17:362–373. doi: 10.1093/oxfordjournals.molbev.a026316. [DOI] [PubMed] [Google Scholar]
- Anderson SL, Somers DE, Millar AJ, Hanson K, Chory J, Kay SA. Attenuation of phytochrome A and B signaling pathways by the Arabidopsis circadian clock. Plant Cell. 1997;9:1727–1743. doi: 10.1105/tpc.9.10.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman MJ, Hirschfeld M, Wester L, Weaver M, Clack T, Amasino RM, Sharrock RA. A deletion in the PHYD gene of the Arabidopsis Wassilewskija ecotype defines a role for phytochrome D in red/far-red light sensing. Plant Cell. 1997;9:1317–1326. doi: 10.1105/tpc.9.8.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan MT, Quail PH. Phytochrome A overexpression inhibits hypocotyl elongation in transgenic Arabidopsis. Proc Natl Acad Sci USA. 1991;88:10806–10810. doi: 10.1073/pnas.88.23.10806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cherry JR, Hondred D, Walker JM, Vierstra RD. Phytochrome requires the 6-kDa N-terminal domain for full biological activity. Proc Natl Acad Sci USA. 1992;89:5039–5043. doi: 10.1073/pnas.89.11.5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs KL, Miller FR, Cordonnier-Pratt MM, Pratt LH, Morgan PW, Mullet JE. The sorghum photoperiod sensitivity gene, Ma3, encodes a phytochrome B. Plant Physiol. 1997;113:611–619. doi: 10.1104/pp.113.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clack T, Mathews S, Sharrock RA. The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Mol Biol. 1994;25:413–427. doi: 10.1007/BF00043870. [DOI] [PubMed] [Google Scholar]
- Devlin PF, Kay SA. Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell. 2000;12:2499–2510. doi: 10.1105/tpc.12.12.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Patel SR, Whitelam GC. Phytochrome E influences internode elongation and flowering time in Arabidopsis. Plant Cell. 1998;10:1479–1487. doi: 10.1105/tpc.10.9.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Robson PR, Patel SR, Goosey L, Sharrock RA, Whitelam GC. Phytochrome D acts in the shade-avoidance syndrome in Arabidopsis by controlling elongation growth and flowering time. Plant Physiol. 1999;119:909–915. doi: 10.1104/pp.119.3.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Somers DE, Quail PH, Whitelam GC. The Brassica rapa elongated internode (EIN) gene encodes phytochrome B. Plant Mol Biol. 1997;34:537–547. doi: 10.1023/a:1005880414931. [DOI] [PubMed] [Google Scholar]
- Draper CK, Hays JB. Replication of chloroplast, mitochondrial and nuclear DNA during growth of unirradiated and UVB-irradiated Arabidopsis leaves. Plant J. 2000;23:255–265. doi: 10.1046/j.1365-313x.2000.00776.x. [DOI] [PubMed] [Google Scholar]
- Eichenberg K, Baurle I, Paulo N, Sharrock RA, Rudiger W, Schafer E. Arabidopsis phytochromes C and E have different spectral characteristics from those of phytochromes A and B. FEBS Lett. 2000;470:107–112. doi: 10.1016/s0014-5793(00)01301-6. [DOI] [PubMed] [Google Scholar]
- Furuya M. Phytochromes: their molecular species, gene families, and functions. Annu Rev Plant Physiol. 1993;44:617–645. [Google Scholar]
- Gendreau E, Hofte H, Grandjean O, Brown S, Traas J. Phytochrome controls the number of endoreduplication cycles in the Arabidopsis thaliana hypocotyl. Plant J. 1998;13:221–230. doi: 10.1046/j.1365-313x.1998.00030.x. [DOI] [PubMed] [Google Scholar]
- Gendreau E, Traas J, Desnos T, Grandjean O, Caboche M, Hofte H. Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 1997;114:295–305. doi: 10.1104/pp.114.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goosey L, Palecanda L, Sharrock RA. Differential patterns of expression of the Arabidopsis PHYB, PHYD, and PHYE phytochrome genes. Plant Physiol. 1997;115:959–969. doi: 10.1104/pp.115.3.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamazato F, Shinomura T, Hanzawa H, Chory J, Furuya M. Fluence and wavelength requirements for Arabidopsis CAB gene induction by different phytochromes. Plant Physiol. 1997;115:1533–1540. doi: 10.1104/pp.115.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser BA, Cordonnier-Pratt MM, Pratt LH. Temporal and photoregulated expression of five tomato phytochrome genes. Plant J. 1998;14:431–439. doi: 10.1046/j.1365-313x.1998.00144.x. [DOI] [PubMed] [Google Scholar]
- Hauser BA, Pratt LH, Cordonnier-Pratt MM. Absolute quantification of five phytochrome transcripts in seedlings and mature plants of tomato (Solanum lycopersicum L.) Planta. 1997;201:379–387. doi: 10.1007/s004250050080. [DOI] [PubMed] [Google Scholar]
- Hennig L, Stoddart WM, Dieterle M, Whitelam GC, Schafer E. Phytochrome E controls light-induced germination of Arabidopsis. Plant Physiol. 2002;128:194–200. [PMC free article] [PubMed] [Google Scholar]
- Hillman WS. The physiology of phytochrome. Annu Rev Plant Physiol. 1967;18:301–324. [Google Scholar]
- Hirschfeld M, Tepperman JM, Clack T, Quail PH, Sharrock RA. Coordination of phytochrome levels in phyB mutants of Arabidopsis as revealed by apoprotein-specific monoclonal antibodies. Genetics. 1998;149:523–535. doi: 10.1093/genetics/149.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarillo JA, Capel J, Tang RH, Yang HQ, Alonso JM, Ecker JR, Cashmore AR. An Arabidopsis circadian clock component interacts with both CRY1 and phyB. Nature. 2001;410:487–490. doi: 10.1038/35068589. [DOI] [PubMed] [Google Scholar]
- Joubes J, Chevalier C. Endoreduplication in higher plants. Plant Mol Biol. 2000;43:735–745. doi: 10.1023/a:1006446417196. [DOI] [PubMed] [Google Scholar]
- Kircher S, Kozma-Bognar L, Kim L, Adam E, Harter K, Schafer E, Nagy F. Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell. 1999;11:1445–1456. doi: 10.1105/tpc.11.8.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konomi K, Abe H, Furuya M. Changes in the content of phytochrome I and II apoproteins in embryonic axes of pea seeds during imbibition. Plant Cell Physiol. 1987;28:1443–1451. [Google Scholar]
- Koornneef M, Rolff E, Spruit CJP. Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Z Pflanzenphysiol. 1980;100:147–160. [Google Scholar]
- Kozma Bognar LK, Hall A, Adam E, Thain SC, Nagy F, Millar AJ. The circadian clock controls the expression pattern of the circadian input photoreceptor, phytochrome B. Proc Natl Acad Sci USA. 1999;96:14652–14657. doi: 10.1073/pnas.96.25.14652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structure proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lazarova GI, Kerckhoffs LH, Brandstadter J, Matsui M, Kendrick RE, Cordonnier-Pratt MM, Pratt LH. Molecular analysis of PHYA in wild-type and phytochrome A-deficient mutants of tomato. Plant J. 1998a;14:653–662. doi: 10.1046/j.1365-313x.1998.00164.x. [DOI] [PubMed] [Google Scholar]
- Lazarova GI, Kubota T, Frances S, Peters JL, Hughes MJ, Brandstadter J, Szell M, Matsui M, Kendrick RE, Cordonnier-Pratt MM et al. Characterization of tomato PHYB1 and identification of molecular defects in four mutant alleles. Plant Mol Biol. 1998b;38:1137–1146. doi: 10.1023/a:1006068305454. [DOI] [PubMed] [Google Scholar]
- Lopez-Juez E, Nagatani A, Tomizawa K, Deak M, Kern R, Kendrick RE, Furuya M. The cucumber long hypocotyl mutant lacks a light-stable PHYB-like phytochrome. Plant Cell. 1992;4:241–251. [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia JF, Monte E, Quail PH. A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J. 1999;20:251–257. doi: 10.1046/j.1365-313x.1999.00579.x. [DOI] [PubMed] [Google Scholar]
- Mathews S, Lavin M, Sharrock RA. Evolution of the phytochrome gene family and its utility for phylogenetic analyses of angiosperms. Ann Mo Bot Gard. 1995;82:296–321. [Google Scholar]
- Mathews S, Sharrock RA. The phytochrome gene family in grasses (Poaceae): a phylogeny and evidence that grasses have a subset of the loci found in dicot angiosperms. Mol Biol Evol. 1996;13:1141–1150. doi: 10.1093/oxfordjournals.molbev.a025677. [DOI] [PubMed] [Google Scholar]
- Mathews S, Sharrock RA. Phytochrome gene diversity. Plant Cell Environ. 1997;20:666–671. [Google Scholar]
- Nelson DC, Lasswell J, Rogg LE, Cohen MA, Bartel B. FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell. 2000;101:331–340. doi: 10.1016/s0092-8674(00)80842-9. [DOI] [PubMed] [Google Scholar]
- Parks BM, Quail PH. hy8, a new class of Arabidopsis long hypocotyl mutants deficient in functional phytochrome A. Plant Cell. 1993;5:39–48. doi: 10.1105/tpc.5.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt LH. Distribution and localization of phytochrome within the plant. In: Kendrick RE, Kronenberg GHM, editors. Photomorphogenesis in Plants. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 163–185. [Google Scholar]
- Pratt LH. Phytochromes: differential properties, expression patterns, and molecular evolution. Photochem Photobiol. 1995;61:10–21. [Google Scholar]
- Pyke KA, Marrison JL, Leech RM. Temporal and spatial development of the cells of the expanding first leaf of Arabidopsis thaliana (L.) Heynh. J Exp Bot. 1991;42:1407–1416. [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J. Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell. 1993;5:147–157. doi: 10.1105/tpc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt EE, Schibler U. Cell size regulation, a mechanism that controls cellular RNA accumulation: consequences on regulation of the ubiquitous transcription factors Oct1 and NF-Y and the liver-enriched transcription factor DBP. J Cell Biol. 1995;128:467–483. doi: 10.1083/jcb.128.4.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock RA, Quail PH. Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev. 1989;3:1745–1757. doi: 10.1101/gad.3.11.1745. [DOI] [PubMed] [Google Scholar]
- Shimazaki Y, Pratt LH. Immunochemical detection with rabbit polyclonal and mouse monoclonal antibodies of different pools of phytochrome from etiolated and green Avena shoots. Planta. 1985;164:333–344. doi: 10.1007/BF00402944. [DOI] [PubMed] [Google Scholar]
- Shinomura T, Nagatani A, Chory J, Furuya M. The induction of seed germination in Arabidopsis thaliana is regulated principally by phytochrome B and secondarily by phytochrome A. Plant Physiol. 1994;104:363–371. doi: 10.1104/pp.104.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura T, Nagatani A, Hanzawa H, Kubota M, Watanabe M, Furuya M. Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1996;93:8129–8133. doi: 10.1073/pnas.93.15.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Devlin PF, Kay SA. Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science. 1998;282:1488–1490. doi: 10.1126/science.282.5393.1488. [DOI] [PubMed] [Google Scholar]
- Somers DE, Quail PH. Phytochrome-mediated light regulation of PHYA- and PHYB-GUS transgenes in Arabidopsis thaliana seedlings. Plant Physiol. 1995a;107:523–534. doi: 10.1104/pp.107.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Quail PH. Temporal and spatial expression patterns of PHYA and PHYB genes in Arabidopsis. Plant J. 1995b;7:413–427. doi: 10.1046/j.1365-313x.1995.7030413.x. [DOI] [PubMed] [Google Scholar]
- Somers DE, Schultz TF, Milnamow M, Kay SA. ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell. 2000;101:319–329. doi: 10.1016/s0092-8674(00)80841-7. [DOI] [PubMed] [Google Scholar]
- Somers DE, Sharrock RA, Tepperman JM, Quail PH. The hy3 long hypocotyl mutant of Arabidopsis is deficient in phytochrome B. Plant Cell. 1991;3:1263–1274. doi: 10.1105/tpc.3.12.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano M, Kanegae H, Shinomura T, Miyao A, Hirochika H, Furuya M. Isolation and characterization of rice phytochrome A mutants. Plant Cell. 2001;13:521–534. doi: 10.1105/tpc.13.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuhisa JG, Daniels SM, Quail PH. Phytochrome in green tissue: spectral and immunochemical evidence for two distinct molecular species of phytochrome in light-grown Avena sativa L. Planta. 1985;164:321–332. doi: 10.1007/BF00402943. [DOI] [PubMed] [Google Scholar]
- Tokuhisa JG, Quail PH. The levels of two distinct species of phytochrome are regulated differently during germination in Avena sativa L. Planta. 1987;172:371–377. doi: 10.1007/BF00398666. [DOI] [PubMed] [Google Scholar]
- Toth R, Kevei EE, Hall A, Millar AJ, Nagy F, Kozma-Bognar L. Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol. 2001;127:1607–1616. doi: 10.1104/pp.010467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelmann TC. Light within the plant. In: Kendrick RE, Kronenberg GHM, editors. Photomorphogenesis in Plants. Ed 2. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 491–535. [Google Scholar]
- Wagner D, Tepperman JM, Quail PH. Overexpression of phytochrome B induces a short hypocotyl phenotype in transgenic Arabidopsis. Plant Cell. 1991;3:1275–1288. doi: 10.1105/tpc.3.12.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-C, Cordonnier-Pratt MM, Pratt LH. Detection and quantitation of three phytochromes in umimbibed seeds of Avena sativa L. Photochem Photobiol. 1992;56:709–716. [Google Scholar]
- Wang Y-C, Cordonnier-Pratt MM, Pratt LH. Spatial distribution of three phytochromes in dark- and light-grown Avena sativa L. Planta. 1993a;189:391–396. doi: 10.1007/BF00194436. [DOI] [PubMed] [Google Scholar]
- Wang Y-C, Cordonnier-Pratt MM, Pratt LH. Temporal and light regulation of the expression of three phytochromes in germinating seeds and young seedlings of Avena sativa L. Planta. 1993b;189:384–390. doi: 10.1007/BF00194435. [DOI] [PubMed] [Google Scholar]
- Weller JL, Murfet IC, Reid JB. Pea mutants with reduced sensitivity to far-red light define an important role for phytochrome A in day-length detection. Plant Physiol. 1997;114:1225–1236. doi: 10.1104/pp.114.4.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Nagatani A, Kendrick RE, Murfet IC, Reid JB. New lv mutants of pea are deficient in phytochrome B. Plant Physiol. 1995;108:525–532. doi: 10.1104/pp.108.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wester L, Somers DE, Clack T, Sharrock RA. Transgenic complementation of the hy3 phytochrome B mutation and response to PHYB gene copy number in Arabidopsis. Plant J. 1994;5:261–272. doi: 10.1046/j.1365-313x.1994.05020261.x. [DOI] [PubMed] [Google Scholar]
- Whitelam GC, Devlin PF. Roles of different phytochromes in Arabidopsis photomorphogenesis. Plant Cell Environ. 1997;20:752–758. [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harberd NP. Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell. 1993;5:757–768. doi: 10.1105/tpc.5.7.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi R, Nakamura M, Mochizuki N, Kay SA, Nagatani A. Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J Cell Biol. 1999;145:437–445. doi: 10.1083/jcb.145.3.437. [DOI] [PMC free article] [PubMed] [Google Scholar]