Abstract
Here, we report the isolation and characterization of a strong dominant-negative phytochrome A (phyA) mutation (phyA-300D) in Arabidopsis. This mutation carries a single amino acid substitution at residue 631, from valine to methionine (V631M), in the core region within the C-terminal half of PHYA. This PHYA core region contains two protein-interactive motifs, PAS1 and PAS2. Val-631 is located within the PAS1 motif. The phyA-V631M mutant protein is photochemically active and accumulates to a level similar to wild type in dark-grown seedlings. Overexpression of PHYA-V631M in a wild-type background results in a dominant-negative interference with endogenous wild-type phyA, whereas PHYA-V631M in a phyA null mutant background is inactive. To investigate the specificity of this mutation within the phytochrome family, the corresponding amino acid substitution (V664M) was created in the PHYTOCHROME B (PHYB) polypeptide. We found that the phyB-V664M mutant protein is physiologically active in phyB mutant and causes no interfering effect in a wild-type background. Together, our results reveal a unique feature in phyA signal propagation through the C-terminal core region.
One of the most important environmental factors affecting plant growth and survival is light. Plants employ arrays of photoreceptors to detect and respond to a broad spectrum of light (Kendrick and Kronenberg, 1994; McNellis and Deng, 1995). One of the best-studied groups of photoreceptors is the red (R)/far red (FR) light-absorbing phytochromes (Quail, 1997). In Arabidopsis, there is a family of five phytochromes whose apoproteins are encoded by a multigene family (PHYA–PHYE; Sharrock and Quail, 1989; Clack et al., 1994). Among the five phytochromes, the best characterized is the FR-absorbing phytochrome A (phyA). Like all phytochromes, phyA is capable of a photoreversible conformational change between the Pr and the Pfr forms in response to light. Whereas the Pfr form is rapidly degraded, the Pr form of phyA is stable in the cytosol. Upon exposure to R light, the Pr form is converted to the Pfr form, whereas exposure to FR light converts the Pfr form back to the Pr form. This photoconversion correlates with the absorption spectra of these forms, where Pr absorbs maximally at 666 nm and Pfr absorbs maximally at 730 nm (Quail, 1997). It is currently hypothesized that both the Pfr and the photocycled Pr forms of phyA are the active species (Shinomura et al., 2000) and possess kinase activity (Yeh and Lagarias, 1998). Upon light perception, phyA and phytochrome B (phyB) migrate into the nucleus, where they presumably trigger downstream events (Kircher et al., 1999).
Since the discovery of the phytochrome system, the search has been on to identify the signal transduction mechanism through which light perception is coupled to changes in gene expression and cell physiology that control growth and development. Use of both biochemical and genetic means have identified candidate downstream-signaling components of the phytochrome pathway. Microinjection into hypocotyl cells of the phytochrome-deficient aurea mutant of tomato (Lycopersicon esculentum) has been used to biochemically assay the activities of various pharmacological agents and putative signaling intermediates (Bowler et al., 1994). Such studies suggest that heterotrimeric G proteins Ca2+-calmodulin and cyclic-guanosine-5′ monophosphate may mediate phytochrome-induced responses. A reverse genetic approach has recently implicated the involvement of the only G-α protein in Arabidopsis in light inhibition of hypocotyl elongation (Okamoto et al., 2001).
Both yeast (Saccharomyces cerevisiae) two-hybrid protein-protein interaction screens and mutational approaches have resulted in identification of over a dozen putative downstream components and interacting partners of phytochromes (for review, see Quail, 2002; Wang and Deng, 2002). Those putative signaling components include both cytoplasmic and nuclear factors, some of which are transcription factors directly acting on light-responsive promoters. For phyA, the majority of the signaling components identified so far are nuclear localized. This is consistent with the view that light-activated phyA rapidly migrates into nucleus (Kircher et al., 1999) and triggers most of the signaling events there.
A plethora of studies have revealed that phytochrome molecules contain multiple functional domains (for review, see Quail, 1997; Park et al., 2000). The N-terminal half contains the chromophore attachment site and is responsible for the observed spectrum specificity of phyA and phyB. The C-terminal half seems to be responsible for dimerization and downstream signal transmission (Jordan et al., 1996). A region in the C-terminal half, designated as the “core region” or “Quail box,” was revealed to be critically important for phytochrome signal propagation (Quail et al., 1995; Park et al., 2000). This core region in phyA contains two PAS motifs, presumed to be involved in protein-protein interaction. This region was shown to be important for interaction with the reported phytochrome-interactive partners (Ni et al., 1998; Choi et al., 1999). Although it was reported that the C-terminal half of phyA and phyB could be interchangeable for some R and FR responses (Quail et al., 1995), some aspect of distinction in the phyA and phyB signal propagation through their C-terminal core regions to downstream targets might be expected because there are a number of unique downstream components specific to either phyA or phyB.
Here, we report that in a mutant screen for an etiolated phenotype under continuous FR light (FRc), we have recovered a strong dominant-negative mutation of PHYA. This dominant interfering phenotype observed in the heterozygous state is even stronger than those previously reported for transgenic seedlings overexpressing phyA dominant-negative mutant forms (Boylan et al., 1994; Emmler et al., 1995). Characterization of this new phyA mutant revealed an insight specific to phyA-signaling propagation through its C-terminal domain.
RESULTS
Isolation of phyA-300D, a Strong Dominant-Negative Allele of PHYA
In an effort to isolate mutants impaired specifically in phyA signaling, long hypocotyl mutant seedlings specific for FRc were screened from several available T-DNA insertion mutagenized collections. Seven elongated hypocotyl mutants were identified (Table I). Genetic complementation tests revealed that most of the mutations defined new alleles of known genes; however, one new locus, designated FHY4, was defined by a single-mutant allele (fhy4-1).
Table I.
Summary of the mutant screen
| Isolate Name | Allele | Nature of Mutation |
|---|---|---|
| 128 | fhy3-128 | Recessive |
| 6476 | fhy3-6476 | Recessive |
| A2GO5 | phyA-300D | Dominant |
| 135 | phyA-301 | Recessive |
| 158 | hy5-158 | Recessive |
| CS19960 | fhy1-19960 | Recessive |
| 149 | fhy1-149 | Recessive |
| A2GO6 | fhy4-1 | Recessive |
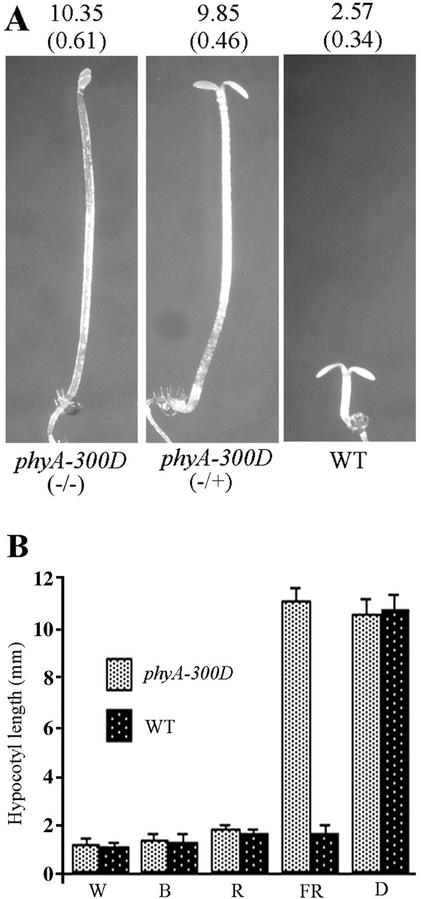
One of the mutants, now designated phyA-300D, exhibited a 3:1 mutant:wild-type phenotype segregation in a backcross to wild type (Table II). This result suggests that the phyA-300D mutation behaves as a dominant trait. This was further supported by the fact that during our allelism test between phyA-300D and various known phyA-signaling mutants, the F1 progenies always displayed long hypocotyl phenotypes, contrasting to those of a normal recessive phyA allele (Table II; data not shown). In the F2 populations, it was revealed that phyA-300D complements all other mutations except phyA, indicating that this may be a new allele of PHYA. As shown in Figure 1A, seedlings heterozygous for the phyA-300D allele possess long hypocotyls under FRc, similar to that of the homozygous seedlings. However, phyA-300D seedlings grown under other light wavelengths displayed normal de-etiolated phenotypes, suggesting that the phyA-300D phenotype is FRc specific (Fig. 1B).
Table II.
Complementation analysis of phyA mutants
| Cross | Generation | Seedlings with Short Hypocotyl | Seedlings with Long Hypocotyl |
|---|---|---|---|
| phyA-300D × WT | F1 | 0 | 69 |
| F2 | 188 | 677 | |
| phyA-300D × phyA-1 | F1 | 0 | 2 |
| F2 | 0 | 400 | |
| phyA-205 × WT | F1 | 0 | 14 |
| F2 | 105 | 370 | |
| phyA-205 × phyA-300D | F1 | 0 | 9 |
| F2 | 0 | 200 | |
| phyA-301 × WT | F1 | 30 | 0 |
F1 and F2 progeny of the complementation crosses were grown in continuous far red light for 6 d before scoring seedling phenotypes and their segregation.
Figure 1.
Phenotype characterization of the dominant phyA-300D mutant. A, Three-day-old FRc-grown phyA-300D homozygote (−/−), heterozygote (−/+), and wild-type (WT) seedlings. Average hypocotyl lengths (in millimeters) are shown above each seedling (n = 25) with sd shown in parentheses. B, Hypocotyl length of wild-type (WT) and phyA-300D seedlings grown in continuous white (W) and blue (B) light and Rc (R) and FRc (FR), or in darkness (D) for 6 d. Each column represents a mean value of the hypocotyl length (in millimeters) taken from 25 seedlings with the sd indicated by an error bar.
To further verify this dominant mutant as a phyA allele, mapping analysis of the phyA-300D mutation was carried out (see “Materials and Methods”). As expected, phyA-300D was mapped to chromosome 1 near the PHYA locus (data not shown).
phyA-300D Is Caused by a Single Amino Acid Change at Residue 631
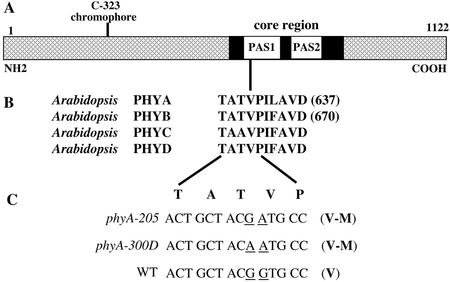
To determine the exact nature of the phyA-300D mutation, overlapping regions covering the PHYA gene from the mutant background were PCR amplified and sequenced. This sequence analysis revealed two nucleotide changes, both from guanine (G) to adenine (A), affecting two adjacent codons of the PHYA open reading frame (Fig. 2). The first G to A mutation is silent, altering the codon 630 from ACG to ACA, both encoding the same amino acid, Thr. The second G to A nucleotide change is a missense mutation, which alters the codon at position 631 from GTG (encoding for Val) to ATG (encoding for Met). Thus, the overall consequence in phyA-300D is a Val to Met change at amino acid 631 (V631M) in the PHYA polypeptide (Fig. 2C). This Val-631 is located within the first of the two PAS motifs, within the core region or Quail box of the phytochrome C-terminal half (Fig. 2). The core region of phyA has been demonstrated to be critical in phytochrome signal propagation to downstream targets (Quail et al., 1995). However, all those reported mutations had been described as recessive in nature (Xu et al., 1995).
Figure 2.
The structural features of phyA and the molecular lesion of the phyA-300D mutation. A, Diagram of the Arabidopsis PHYA protein. The chromophore-binding site (C-323), core region, and its two PAS motifs are indicated. B, The conserved Val and its surrounding amino acid residues among four Arabidopsis phytochromes. Note that the Val is invariant among all known phyA species from higher plants, although it can be substituted by Ala in all other phytochromes among different higher plant species. The single-letter codes of amino acids are shown in bold. C, Nucleotide sequence comparison of the PHYA gene in phyA-205, phyA-300D, and wild type (WT) around the Val-631 region. Amino acids (in bold single-letter code) are shown above their codons. In both mutant alleles, there is a single identical amino acid residue mutation (V631M) in the translated proteins.
phyA-205 Is an Independent Allele of PHYA with the Same Amino Acid Alteration as phyA-300D
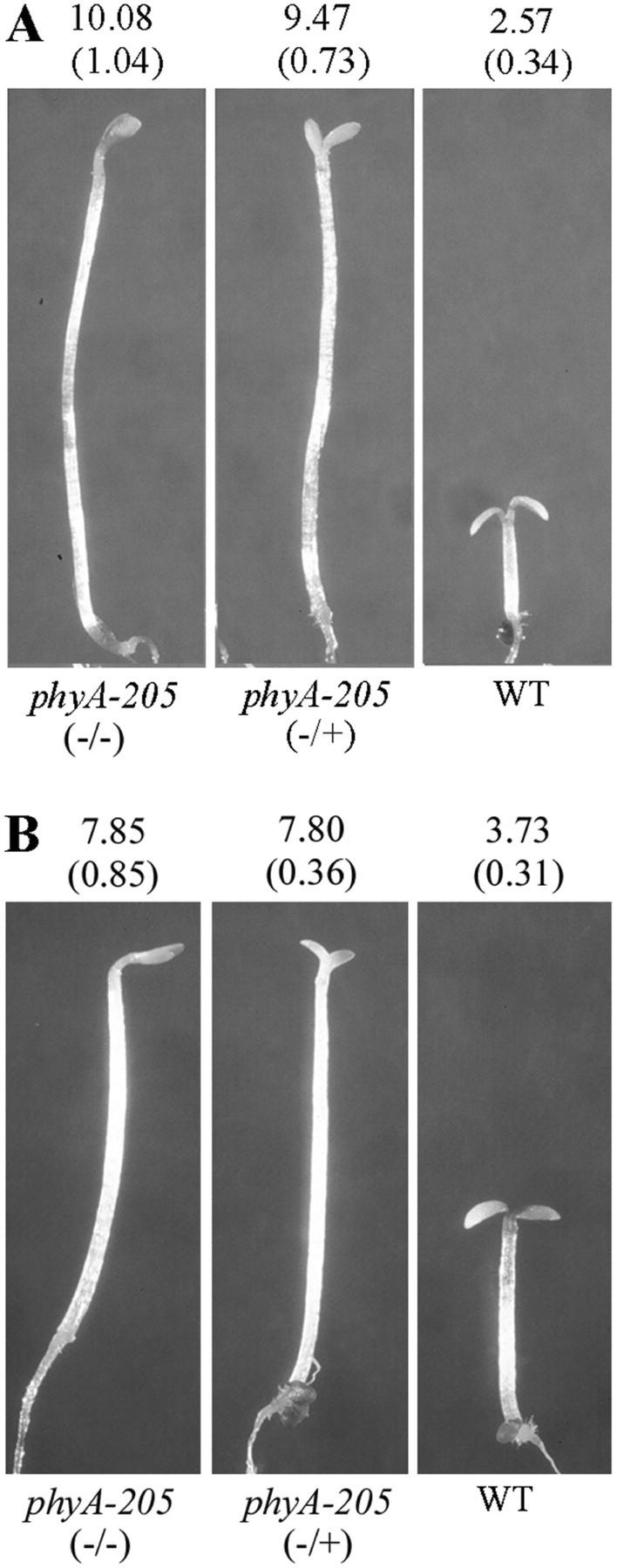
It is intriguing that a V631M mutation of PHYA was previously reported for another allele, phyA-205 (Reed et al., 1994). However, at that time phyA-205 was described as a recessive mutation with an intermediate phenotype. We further examined the segregation property of the phyA-205 backcross with wild type and our results clearly indicated that it is a strong dominant mutation (Table II). Furthermore, phyA-205 failed to complement phyA-300D in both the F1 and F2 populations (Table II), as expected from their allelism. As shown in Figure 3A, seedlings heterozygous for the phyA-205 mutation showed long hypocotyls under FRc similar to that of homozygous mutant seedlings (Fig. 3A). Further sequence analysis of the phyA-205 mutant confirmed that it contains the reported single G-to-A mutation in codon 631 (Reed et al., 1994; Fig. 2C). Thus, there are two independent mutations, phyA-300D and phyA-205, both resulting in the same V631M mutation in the PHYA protein (Fig. 2C). This result is consistent with the conclusion that the V631M mutation caused a dominant-negative phenotype for phyA.
Figure 3.
The phyA-205 has a dominant-negative phenotype and is not dependent on Suc concentration. Average hypocotyl lengths (in millimeters) are shown above each seedling (n = 25) with the sd shown in parentheses. A, Phenotypic comparison of 3-d-old FR light-grown phyA-205 homozygous (−/−), heterozygous (−/+), and wild-type (WT) seedlings on 0.3% (w/v) Suc medium. B, Phenotypic comparison of 3-d-old FR light-grown phyA-205 homozygous (−/−), heterozygous (−/+), and wild-type (WT) seedlings on 2% (w/v) Suc medium.
Because the original characterization of phyA-205 used growth media (GM) supplemented with 2% (w/v) Suc (Reed et al., 1994) and our studies used 0.3% (w/v) Suc, we examined the effect of Suc concentration on hypocotyl length in the phyA-205 mutant. As shown in Figure 3, the high concentration of Suc indeed inhibited hypocotyl elongation under FRc. However, this inhibition was observed in both phyA-205 homozygous and heterozygous seedlings. At either concentration of Suc, the heterozygous and homozygous mutant seedlings have similar hypocotyl lengths under the same FRc growth condition. phyA-300D homozygous and heterozygous mutant seedlings similarly also displayed this Suc inhibition of hypocotyl elongation (data not shown). This effect of Suc was apparently not unique to the dominant mutations, because both phyA-1 and phyA-301 (recessive mutations) displayed a similar decreased hypocotyl length under the higher Suc concentration.
The PHYA-V631M Is Sufficient to Confer a Dominant-Negative Interference to phyA-Mediated FRc Inhibition of Hypocotyl Elongation
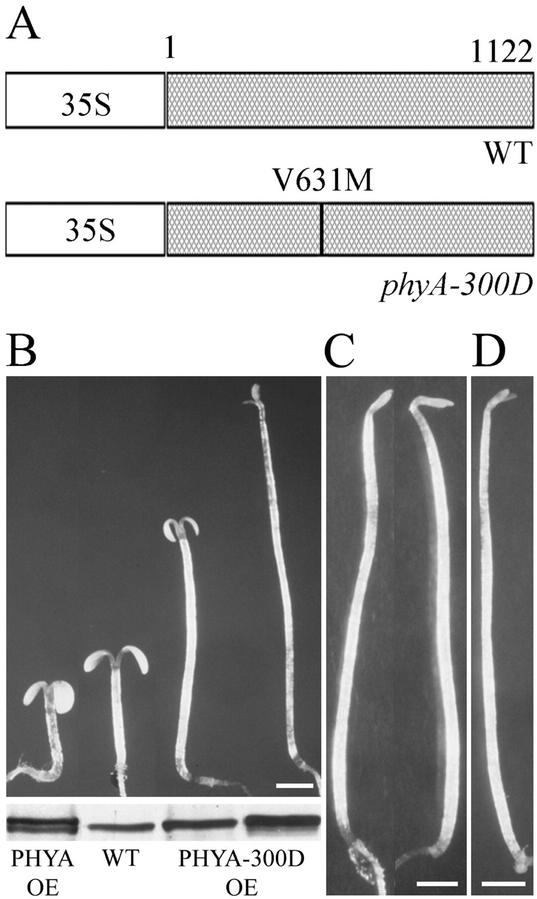
To test whether a V631M mutation in PHYA is sufficient to confer a FR-specific dominant-negative phenotype, we introduced a 35S promoter-driven PHYA-V631M and wild-type PHYA into wild-type Arabidopsis and a null mutant (phyA-101; Fig. 4A). When grown in FRc, transgenic seedlings expressing PHYA-V631M (from the phyA-300D allele) exhibited an etiolated phenotype (Fig. 4B). When grown complete darkness, there is no observable difference between transgenic and wild-type seedlings (Fig. 4C). However, overexpression of PHYA-V631M in a phyA null mutant did not rescue its mutant phenotype (Fig. 4D), further confirming the loss-of-function nature of the PHYA-V631M protein. Thus, our result implies that not only did phyA-V631M lose its ability to respond to FR light, but that the presence of PHYA-V631M is sufficient to interfere with endogenous wild-type phyA function in a dominant-negative fashion.
Figure 4.
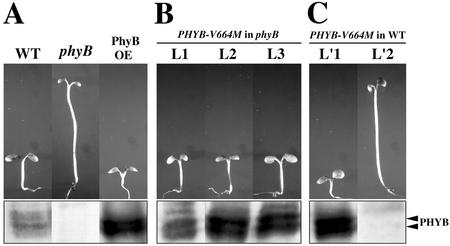
Expression of PHYA-V631M mutant protein is sufficient to cause a dominant interfering effect on endogenous phyA signaling. A, Diagrams of the transformation cassettes. The wild-type PHYA (WT) and PHYA-V631M full-length coding regions were driven by the cauliflower mosaic virus 35S promoter. B, Morphological comparison of 3-d-old FR light-grown seedlings of wild-type PHYA overexpressor (PHYA OE), wild-type (WT), and two mutant PHYA-V631M overexpressor (PHYA-300D OE) lines. PHYA protein immunoblots are shown in the bottom panel. C, Morphological comparison of 3-d-old dark-grown seedlings with PHYA-V631M overexpressor transgene in wild-type background (left) and the wild-type control (right). D, Morphology of 3-d-old FR light-grown seedling with PHYA-V631M overexpressor transgene in a phyA null mutant background. Scale bars in B through D = 1 mm.
Interestingly, the degree of dominant-negative interference of the transgenic plants depends on mutant PHYA-V631M protein dose. As shown in Figure 4B, a high-expression level of mutant PHYA-V631M resulted in seedlings with an extremely elongated hypocotyl and closed cotyledons, resembling that of the phyA null mutants, whereas a lower expression level resulted in an intermediate phenotype, a long hypocotyl, and open but small cotyledons (Fig. 4B). As expected, expression of wild-type PHYA resulted in seedlings with short hypocotyls and open cotyledons (Fig. 4B), similar to the reported phyA overexpressor phenotype (Boylan et al., 1994).
The phyA-V631M Abundance Is Not Reduced in Mutant Seedlings Grown in Darkness But Is Slightly Higher under FR Light
As a first step to determine a possible molecular basis of the phyA-V631M mutation effect, the phyA protein levels of the phyA-300D and wild-type seedlings grown in darkness and in FRc were examined using immunoblot analysis. As shown in Figure 5, the level of phyA protein in dark-grown phyA-300D seedlings is as abundant as in wild-type seedlings. Interestingly, phyA-300D grown under FRc accumulated to about 2-fold higher levels of phyA protein than that of wild type. This result rules out the possibility that the phyA-300D phenotype is a result of lower PHYA levels under FR light. It should be noted that the dominant mutant phenotype of phyA-300D could be a result of increased levels of the mutant PHYA-V631M protein. However, this hypothesis would be argued against by the observation that the phyA-105 mutant, which has a single amino acid change, accumulated to similar high levels of the PHYA-V631M mutant protein in FRc, and still exhibited completely recessive properties (Xu et al., 1995).
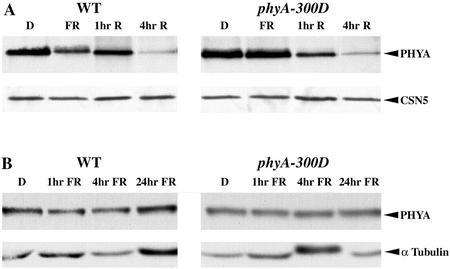
Figure 5.
Characterization of wild-type and PHYA-V631M stability under R and FR light. A, phyA levels in 3-d-old dark (D) and FR light-grown wild-type Columbia (WT) and phyA-300D, and in 3-d-old dark-grown seedlings exposed to Rc for 1 or 4 h (hr; top). The immunoblots were probed with a phyA-specific polyclonal antibody. An equal amount of total protein was loaded in each lane, as verified by CSN5 immunoblot (bottom). B, phyA levels in 3-d-old dark-grown (D) wild-type Columbia (WT), and phyA-300D seedlings and in 3-d-old dark-grown seedlings exposed to FRc light for 1, 4, or 24 h (hr). An equal amount of total protein was loaded in each lane, as verified by immunoblot using anti-tubulin antibodies (bottom).
phyA-V631M Possesses Normal Light Triggered Degradation Kinetics and Photochemical Properties
It is known that phyA accumulates in darkness as the Pr form and is rapidly degraded upon exposure to light, because of the susceptible nature of the phyA Pfr form to proteasome-mediated protein degradation. Therefore, we examined the degradation kinetics of the phyA-V631M mutant protein compared with wild-type phyA protein upon exposure to continuous R light (Rc) or FRc. Dark-grown mutant and wild-type seedlings were exposed to Rc for 0, 1, and 4 h and harvested for immunoblot analysis. The degradation kinetics shown in Figure 5A indicates that there is no significant difference between the phyA-V631M mutant and wild-type phyA proteins. As expected, FRc treatment of dark-grown mutant or wild-type seedlings does not cause observable degradation of the phyA proteins (Fig. 5B). This result suggests that the mutant phyA-V631M protein was able to photoconvert from Pr to Pfr upon exposure to Rc, and only the Pfr form of phyA-V631M was labile and subjected to rapid degradation.
To test whether phyA-V631M protein retains typical phyA R-FR reversibility, the differential absorbance spectra for phyA-300D and wild-type seedlings were examined. To this end, phyA-300D, phyA-101 (a phyA null allele), and wild-type seedlings were grown in darkness for 3 d and extracts were prepared under green safelight as described in “Materials and Methods.” This assay revealed that phyA-V631M has an essentially identical spectral property as wild type (data not shown).
The PHYA-V631M Protein Retains Dimerization Capability
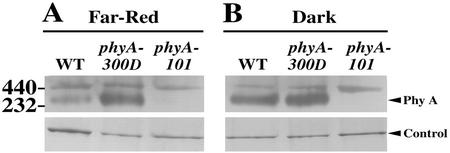
The Val residue at position 631 is located in the first PAS motif of PHYA that overlaps with the phyA dimerization region. Thus, we examined the ability of PHYA-V631M to form dimers in vivo. To this end, crude extracts from homozygous mutant and wild-type seedlings grown in darkness or FRc were subjected to native gel electrophoresis followed by immunoblot analysis (Fig. 6). The migration and amount of PHYA-V631M dimer from phyA-300D seedlings under native gel conditions are comparable with that of wild-type plants, indicating that the PHYA-V631M can effectively dimerize in vivo.
Figure 6.
Native gel analysis of phyA dimerization in FR and dark-grown wild-type and mutant seedlings. Extracts were separated on a 4% to 20% (w/v) gradient non-denaturing gel and probed with a polyclonal PHYA antibody. The phyA dimer is marked by a triangle on the right. Equal loading was ensured by visualizing a protein band staining intensity (marked by control). The positions of two mass markers are indicated on the left side. The phyA-101 is a null mutant control. A, FR light-grown seedlings (5 d); B, dark-grown seedlings (5 d).
Overexpression of PHYB-V664M in Wild-Type Plants Failed to Interfere with phyB Signaling
The PHYA and PHYB polypeptides are highly homologous (Sharrock and Quail, 1989). The region surrounding the Val-631 of PHYA and the corresponding region in PHYB is highly conserved (Fig. 2B). For PHYB, its Val residue at position 664 is equivalent to the Val at position 631 of PHYA. Thus, it is of interest to determine whether a corresponding PHYB-V664M mutant protein would confer a similar dominant-negative effect on phyB function. For this reason, we specifically created a corresponding PHYB-V664M mutant transgene under 35S promoter control. The transgene was transformed into wild-type Arabidopsis and a phyB null mutant (phyB-101; Fig. 7A). Homozygous transgenic lines were obtained from each construct, and their hypocotyl elongation under Rc was examined. Contrasting to PHYA-V631M, the PHYB-V664M mutant protein is clearly functional and active in both the phyB null mutant and wild-type seedlings in the lines tested. As shown in Figure 7B, PHYB-V664M can rescue the phyB null phenotype when overexpressed in the phyB-101 mutant background. Furthermore, transgenic seedlings overexpressing the PHYB-V664M in a wild-type background exhibited a hyper-photomorphogenic phenotype with shorter hypocotyls than those of wild type (Fig. 7C), similar to seedlings overexpressing wild-type PHYB (Fig. 7A). Several PHYB-V664M transgenic lines that showed longer hypocotyl under Rc turned out to be cosuppression lines and had an undetectable level of phyB protein. This result indicates that PHYB-V664M behaves differently from PHYA-V631M. Our results imply that the mutant PHYB-V664M protein neither reduces its ability to respond to Rc, nor is capable of interfering with wild-type phyB signaling. Thus, the role of Val-631 in phyA defined in this work is specific to phyA and not applicable to phyB.
Figure 7.
The PHYB-V664M protein is physiologically active and does not confer an interfering effect on phyB signaling. The 3-d-old seedlings are shown on top, and their corresponding PHYB protein immunoblots are shown on the bottom. A, Three-day-old Rc light-grown wild type (WT), PHYB overexpressor (PHYB OE) in wild-type background, and the phyB-101 null mutant. B, Three-day-old Rc light-grown seedlings of three independent PHYB-V664M overexpressor lines (L1, L2, and L3 of PHYB-V644M OE) in the phyB-101 null mutant background. C, Three-day-old Rc light-grown seedlings of two different PHYB V644M overexpressor (PHYB-V644M OE) lines in a wild-type background. L′1 is an overexpressor line for PHYB-V664M, and L′2 is a cosuppression line.
DISCUSSION
PHYA-V631M Has a Strong Dominant-Negative Interfering Activity
Several lines of evidence presented in this report definitively show that PHYA-V631M is capable of conferring a strong dominant-negative interfering effect on normal phyA signaling. First, two independent alleles with the same V631M amino acid change exhibited a strong long hypocotyl phenotype under FR in the heterozygous state. The hypocotyl length of heterozygous mutant seedlings is nearly identical to that of their homozygous mutant seedlings. The only distinction between the heterozygous and homozygous seedlings is the slightly more advanced cotyledon development in the heterozygous mutants. The heterozygous seedlings exhibit opening and slight enlargement of the cotyledons after extended growth under FRc. Second, introduction of the PHYA-V631M transgene into wild-type Arabidopsis confirmed that PHYA-V631M is sufficient to confer the dominant-negative interfering effect. Our results imply that not only does the mutant PHYA-V631M protein reduce its ability to respond to FRc, it is also able to interfere with the ability of endogenous phyA to mediate FRc responses in a dominant-negative fashion. The PHYA-V631M is clearly photochemically active but not functional in mediating FRc responses, because its homozygous seedlings exhibited a phenotype essentially indistinguishable from that of the phyA null mutations. Furthermore, overexpression of PHYA-V631M in a phyA null mutant background also failed to show any phenotypic rescue (Fig. 4D).
Although a large number of phyA mutations within the two PAS motifs have been described, all of them were reported as recessive (Reed et al., 1994; Xu et al., 1995). In fact, only one report described a slightly weak semidominant effect of phyA mutations under low-FR photon fluence conditions and with no sugar in the GM (Whitelam et al., 1993). In this case, heterozygous PHYA/phyA seedlings for two alleles, at least one is a null allele, exhibited slightly longer hypocotyls (3 versus 1 mm in wild type) under FRc but were still significantly shorter than those of phyA homozygous mutants (12–13 mm) under the same growth condition. However, this weak semidominant effect of phyA mutations is largely growth-condition dependent and was not observed by other researchers using different growth conditions (Reed et al., 1994; Xu et al., 1995). We also failed to observe a semidominant effect of the same null phyA-1 allele (Whitelam et al., 1993) under our growth conditions, whereas the dominant effect observed in PHYA-V631M is very strong and consistent under our experimental conditions.
On the other hand, a dominant interfering effect has been reported for transgenic seedlings overexpressing several heterologous phyA species. For example, overexpression of three distinct oat (Avena sativa) phyA mutant forms (major deletions in very N- or C-terminal half) in Arabidopsis resulted in a strong dominant-negative interfering effect over endogenous Arabidopsis phyA function (Boylan et al., 1994). Overexpression of an amino-terminal deletion of rice (Oryza sativa) phyA in tobacco seedlings also resulted in a dominant interfering effect (Emmler et al., 1995). Last, overexpression of an intact fern phyA in Arabidopsis also caused a weak dominant interfering effect (Okamoto et al., 1997). A general explanation for those dominant interfering effects is that those mutant phyA molecules (oat or rice) or the too-diverged fern phyA may not be able to mediate a productive interaction with phyA-signaling partners, and thus, titrate out the available phyA-signaling partners. Nevertheless, comparing the degrees of the dominant-negative interfering phenotypes in those overexpressing lines, the phyA-V631M heterozygous mutant seedlings exhibited possibly the strongest interfering phenotype. Furthermore, this strong dominant-negative interfering phenotype in phyA-300D and phyA-205 is caused by a single amino acid substitution.
The Role of the PAS-Like Domain in phyA- Signaling Activity
Molecular and biochemical characterization of phyA indicates that the apoprotein folds into two major structural domains: a globular NH2-terminal domain cradling the covalently attached chromophore in a hydrophobic pocket, and a more extended C-terminal domain, with a short flexible hinge region connecting the two (Quail, 1997; Park et al., 2000). Evidence also indicates that the determinants of the photosensory specificity of phyA and phyB to FRc and Rc reside in the NH2-terminal domains. In addition, the proximal end of the C-terminal domain, designated the core region or Quail box of the polypeptide, contains determinants necessary for regulatory activity or for the effective communication of perceived light signals to the cellular transduction circuitry. It is generally assumed that the phytochrome molecules, rather than directly binding to target gene promoters, relay information via signaling intermediates or directly to DNA-binding transcription factors (Quail, 2002; Wang and Deng, 2002).
The V631M mutation in phyA-300D and phyA-205 falls within the “core region” of the PHYA protein (Fig. 2A). This structural domain has shown a propensity for light-signaling mutations (Quail, 1997). The severity of the homozygous phyA-300D mutant phenotype reveals a critical role of the Val residue within this core region for phyA function. This region of the PHYA protein has been shown to be a “hot spot” for missense mutations resulting in regulatory mutants that produce PHYA protein, but cannot transduce the light signal to elicit the photomorphogenic response. However, phyA-V631M is the first case where a strong dominant-negative interfering phenotype has been described, whereas other mutations in this region have been reported as recessive (Reed et al., 1994; Xu et al., 1995).
This core region of PHYA contains two PAS-like motifs and may be involved in phytochrome signal transduction by contacting downstream partners (Fig. 2A; Ni et al., 1998). Previous studies have shown that this core region of phytochrome interacts directly with phytochrome partners including NDPK2 (Choi et al., 1999) and PIF3 (Ni et al., 1998), supporting the conclusion that the PAS motifs or the core region defines protein-protein interactive surfaces. One possible explanation of the negative interference of the PHYA-V631M on phyA signaling is that the phyA-V631M mutant protein interacts too strongly with downstream positively acting components of the pathway. These stronger than wild-type interactions with downstream components for PHYA-V631M would compromise the capability of the activated target protein to initiate downstream events. In heterozygous plants, the PHYA-V631M protein would titrate out the downstream phyA target because of the higher affinity between PHYA-V631M and the target, thus, effectively preventing wild-type phyA from interacting with the target and blocking the proper phyA-mediated event. However, this is certainly not the only explanation, and there are other alternative possibilities. One alternative is that PHYA-V631M could have a stronger affinity to form a functionally inactive heterodimer with the wild-type PHYA. In heterozygous plants, this higher affinity to form nonfunctional PHYA-V631M::PHYA heterodimers could prevent the formation of active wild-type phyA homodimers. As a result, the heterozygous plants would fail to respond to FRc. A third possibility is that the PHYA-V631M homodimer and its heterodimer with wild-type PHYA might have an elevated Pfr to Pf form dark reversion rate, which could potentially account for the interfering phenotype. An important task of future work will be to determine which mode of action is used in plants.
The phyA-V631M Dominant-Negative Effect Is Not Applicable to phyB
Although there is a high-sequence homology between the Arabidopsis PHYA and PHYB apoproteins surrounding PHYA amino acid Val-631 (Fig. 2A), we demonstrated that the corresponding Val to Met mutation (V664M) in Arabidopsis phyB does not trigger a similar dominant-negative effect on phyB signaling. The PHYB-V664M mutant protein was active in both phyB null mutant and wild-type plants when introduced into the proper backgrounds. These results indicate that Val-631 is critical for PHYA-mediated FR signaling but not applicable to PHYB-mediated signaling. This specificity to phyA is also consistent with the observation that all known phyA proteins, including those from both monocots and dicots, have the invariant Val at the corresponding position (Xu et al., 1995). However, the Val residue is not absolutely conserved in other higher plant phytochromes, and in some cases, the corresponding position has changed to Ala (Xu et al., 1995). This observation is consistent with the conclusion that the role of Val-631 in phyA as defined by the V631M mutation is unique to phyA, and not applicable to phyB. Thus, it defines a key distinction in the manner through which phyA and phyB transduce light signal via their C-terminal core regions.
MATERIALS AND METHODS
Plant Materials, Mutant Screen, and Mapping
Arabidopsis mutants phyA-1, phyA-101, phyA-205, and phyB-101 (Landsberg erecta ecotype) were described previously (Whitelam et al., 1993; Reed et al., 1994). Several available T-DNA insertion mutagenized collections were used, including those from the Arabidopsis Stock Center and Dr. Steven Dellaporta (Yale University; Galbiati et al., 2000). Either T2 or T3 seeds were screened for mutant seedlings exhibiting a complete (long hypocotyl and closed cotyledons) or partial etiolated (long hypocotyl and open cotyledons) phenotype after 3 d of growth under FRc. Seed sterilization and plating were essentially the same as previously described (Hsieh et al., 2000) except the GM was supplemented with 0.3% (w/v) Suc. To improve germination rate, the seeds on the plates were exposed to 24 h of white light after vernalization for 2 d at 4°C in the dark. The light conditions used were: Rc (100 μmol m−2 s−1), continuous blue light (50 μmol m−2 s−1), continuous white light (200 μmol m−2 s−1), and FRc (175 μmol m−2 s−1). Growth chambers (E-30 LED 2/3, Percival Scientific, Perry, IA) were maintained at 22°C constant temperature.
For mapping, mutant phyA-300D (Columbia ecotype) was crossed to wild-type Landsberg erecta ecotype. F2 progenies were grown on 0.3% (w/v) GM plates and screened under FR for a short hypocotyl, whereas F3 progeny were grown on 0.3% (w/v) GM plates and screened under FR light for a long hypocotyl. DNA from individual homozygous mutant seedlings was prepared and used for PCR-based simple sequence length polymorphism or cleaved-amplified polymorphic sequence mapping (Bell and Ecker, 1994; Konieczny and Ausubel, 1993).
Native Protein Gel and Immunoblot Analyses
Five-day-old FR or dark-grown seedlings were collected under green safelight for protein analysis. Seedlings were frozen in liquid nitrogen and ground with a mortar and pestle in 100 μL of grinding buffer (10% [w/v] glycerol, 400 mm Suc, 50 mm Tris, pH 8.0, and 1 mm phenylmethylsulfonyl fluoride). For native gel analysis, 20 μg of each sample was run on a 4% to 20% (w/v) non-denaturing gradient gel (Tris-Gly, pH 8.0). After electrophoresis, proteins were transferred to Immobilon-P membrane (Millipore, Bedford, MA) for immunoblot analysis. For the PHYA protein level, protein was extracted from 3-d-old dark-grown seedlings and from dark-grown seedlings subjected to Rc or FRc for the specified period of time. Protein concentration was determined by Bradford assay (Bio-Rad, Hercules, CA), loaded equally to SDS-PAGE, and transferred to nylon membrane. The protein-blot analysis was performed according to procedures described (Okamoto et al., 2001). Type-selected antibodies for phyA (Xu et al., 1995) and phyB (Ni et al., 1998) were used.
Phytochrome Difference Spectrum
Arabidopsis protein extracts were prepared from 6-d-old dark-grown seedlings as described (Weller et al., 1996). The extract was dispensed into a spectrophotometer ultra-microcuvette and placed in a FR-LED chamber (200 μmol m−2 s−1) for 360 s to induce photoconversion of phytochrome to the Pr form. After this exposure, the cuvette was placed into the chamber of a spectrophotometer (DU-70, Beckman Coulter, Fullerton, CA), and the baseline was taken. The cuvette was then exposed to R light for 180 s. R light was generated by using a light projector (Ektagraphic III AMT, Eastman Kodak, Rochester, NY) with a Kodak extra bright lamp module. The light passed though a Plexiglas filter (CBS Red 650, Westlake Plastics Co., Lenni, PA) and was reflected into the chamber of the spectrophotometer using a mirror. After R light exposure, an absorbance versus wavelength plot was generated, with readings taken every nanometer from 500 to 800 nm.
DNA Sequencing of the PHYA Gene
Genomic DNA was isolated from phyA-300D and phyA-205 mutants. A series of primers was synthesized based on the published PHYA sequence (Dehesh et al., 1994), and PCR was used to amplify the segments of the PHYA gene from the phyA-300D and phyA-205 genomic DNA. PCR products were purified using a PCR purification kit (Qiagen USA, Valencia, CA) and sequenced.
Construction of Full-Length PHYA and PHYB Overexpressing Plasmids and Arabidopsis Transformation
Total RNA was extracted from 6-d-old dark-grown seedlings of either wild type or the phyA-300D mutant using Trizol reagent (Invitrogen, Carlsbad, CA). cDNA was synthesized using a reverse transcriptase kit (Advantage RT for PCR kit, CLONTECH Laboratories, Palo Alto, CA). The entire coding region of PHYA was divided into two fragments for PCR amplification. The first fragment used a primer, 5′-GCG TCG ACA TGT CAG GCT CTA GGC CGA C-3′, which covers the translation start codon of PHYA with an incorporated SalI cleavage site, and the second primer was 5′-AAT TTT GAG ATC ATT TAG CTT CG-3′. The second fragment used a primer covering the stop codon of PHYA with an incorporated SmaI cleavage site 5′-CGC CCG GGC TAC TTG TTT GCT GCA GCG-3′, and the second primer was 5′-GAA TAC CAC ATG GAT TCA ACG-3′. The two PCR fragments were cloned into the pCR 2.1 TOPO cloning vector (Invitrogen). A unique internal enzyme site (Bst 1107) and a XhoI site within the TOPO vector were used to ligate the two PCR fragments to construct the full-length coding region for either the mutant or wild-type PHYA gene. Using the incorporated SalI site and a SacI site within the TOPO cloning vector, the full-length PHYA gene was ligated into the binary vector pPZPY122 (Yamamoto et al., 1998), and the sequence was confirmed.
The wild-type full-coding region of PHYB was generated using a similar RT-PCR method as for PHYA above except that it used one primer set. The forward primer (5′-GCC CCG GGA TGG TTT CCG GAG TCG GG-3′) was designed over the translation start codon of PHYB with an incorporated SmaI cleavage site. The reverse primer (5′-CGT CTA GAA CTA ATA TGG CAT CAT CAG CAT C-3′) was designed over the stop codon of PHYB with an incorporated XbaI site. The generated PCR product was cloned into the pCR 2.1 TOPO cloning vector (Invitrogen) and sequenced. This wild-type PHYB cDNA clone was used as the template for generating the mutant PHYB construct via PCR that specifically incorporates an A-to-G nucleotide on codon 664 to convert Val to Met. As a result, a full-length PHYB-V664M mutant coding region was produced. Using the incorporated SmaI site and a KpnI site within the TOPO cloning vector, the full-length phyB genes were ligated into the binary vector pPZPY122 (using the filled-in XbaI and KpnI sites within the vector; Yamamoto et al., 1998).
All PHYA and PHYB binary constructs were electroporated into the Agrobacterium tumefaciens strain GV3101 (MP90) and transformed into the proper Arabidopsis strains using the floral dip method (Clough and Bent, 1998).
ACKNOWLEDGMENTS
We thank Haiyang Wang for reading and commenting on the manuscript and Dr. Peter Quail for PHYA and PHYB antibodies. We thank Steve Dellaporta for making his T-DNA tagged Arabidopsis collection available for the mutant screen. We are extremely grateful to Haiyang Wang for his help in PHYA cloning and to Matthew J. Terry for his guidance involving the difference spectra experiments.
Footnotes
This work was supported by the National Institutes of Health (grant no. GM47850 to X.W.D.). X.W.D. was a National Science Foundation Presidential Faculty Fellow, and J.H. is a National Institutes of Health predoctoral trainee.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.005264.
LITERATURE CITED
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Bowler C, Neuhaus G, Yamagata H, Chua NH. Cyclic CMP and calcium mediate phytochrome phototransduction. Cell. 1994;77:73–81. doi: 10.1016/0092-8674(94)90236-4. [DOI] [PubMed] [Google Scholar]
- Boylan MT, Douglas N, Quail PH. Dominant negative suppression of Arabidopsis photoresponses by mutant phytochrome A sequences identify spatially discrete regulatory domains in the photoreceptor. Plant Cell. 1994;6:449–460. doi: 10.1105/tpc.6.3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi G, Yi H, Lee J, Kwon Y, Soh M, Shin B, Luka Z, Hahn T, Song PS. Phytochrome signaling is mediated through nucleoside diphosphate kinase 2. Nature. 1999;401:610–613. doi: 10.1038/44176. [DOI] [PubMed] [Google Scholar]
- Clack T, Mathews S, Sharrock RA. The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of phyD and phyE. Plant Mol Biol. 1994;25:413–427. doi: 10.1007/BF00043870. [DOI] [PubMed] [Google Scholar]
- Clough S, Bent A. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Dehesh K, Franci C, Sharrock RA, Somers DE, Welsch JA, Quail PH. The Arabidopsis phytochrome A gene has multiple transcription start sites and a promoter sequence motif homologous to the repressor element of monocot phytochrome A genes. Photochem Photobiol. 1994;59:379–384. doi: 10.1111/j.1751-1097.1994.tb05051.x. [DOI] [PubMed] [Google Scholar]
- Emmler K, Stockhaus J, Chua NH, Schafer E. An amino-terminal deletion of rice phytochrome A results in a dominant negative suppression of tobacco phytochrome A activity in transgenic tobacco seedlings. Planta. 1995;197:103–110. doi: 10.1007/BF00239945. [DOI] [PubMed] [Google Scholar]
- Galbiati M, Moreno MA, Nadzan G, Zourelidou M, Dellaporta SL. Large-scale T-DNA mutagenesis in Arabidopsis for functional genomic analysis. Funct Integr Genomics. 2000;1:25–34. doi: 10.1007/s101420000007. [DOI] [PubMed] [Google Scholar]
- Hsieh HL, Okamoto H, Wang M, Ang LH, Matsui M, Goodman H, Deng XW. FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsis development. Genes Dev. 2000;14:1958–1970. [PMC free article] [PubMed] [Google Scholar]
- Jordan ET, Cherry JR, Walker JM, Vierstra RD. The amino-terminus of phytochrome A contains two distinct functional domains. Plant J. 1996;9:243–257. doi: 10.1046/j.1365-313x.1996.09020243.x. [DOI] [PubMed] [Google Scholar]
- Kendrick RE, Kronenberg GHM, editors. Photomorphogenesis in Plants. Ed 2. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. [Google Scholar]
- Kircher S, Kozma-Bognar L, Kim L, Adam E, Harter K, Schafer E, Nagy F. Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell. 1999;11:1445–1456. doi: 10.1105/tpc.11.8.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- McNellis TW, Deng XW. Light control of seedling morphogenetic pattern. Plant Cell. 1995;7:1749–1761. doi: 10.1105/tpc.7.11.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- Okamoto H, Matsui M, Deng XW. Overexpression of the heterotrimeric G-protein alpha-subunit enhances phytochrome-mediated inhibition of hypocotyl elongation in Arabidopsis. Plant Cell. 2001;13:1639–1652. doi: 10.1105/TPC.010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto H, Sakamoto K, Tomizawa KI, Nagatani A, Wada M. Photoresponses of transgenic Arabidopsis overexpressing the fern Adiantum capillus-veneris PHY1. Plant Physiol. 1997;115:79–85. doi: 10.1104/pp.115.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CM, Bhoo SH, Song PS. Inter-domain crosstalk in the phytochrome molecules. Semin Cell Dev Biol. 2000;11:449–456. doi: 10.1006/scdb.2000.0200. [DOI] [PubMed] [Google Scholar]
- Quail PH. An emerging molecular map of the phytochromes. Plant Cell Environ. 1997;20:657–665. [Google Scholar]
- Quail PH. Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol. 2002;3:85–93. doi: 10.1038/nrm728. [DOI] [PubMed] [Google Scholar]
- Quail PH, Boylan MT, Parks BM, Short TW, Xu Y, Wagner D. Phytochromes: photosensory perception and signal transduction. Science. 1995;268:675–680. doi: 10.1126/science.7732376. [DOI] [PubMed] [Google Scholar]
- Reed JW, Nagatani A, Elich TD, Fagan M, Chory J. Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 1994;104:1139–1149. doi: 10.1104/pp.104.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock RA, Quail PH. Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev. 1989;3:1745–1757. doi: 10.1101/gad.3.11.1745. [DOI] [PubMed] [Google Scholar]
- Shinomura T, Uchida K, Furuya M. Elementary processes of photoperception by phytochrome A for high-irradiance response of hypocotyl elongation in Arabidopsis. Plant Physiol. 2000;122:147–156. doi: 10.1104/pp.122.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Deng XW. Phytochrome signaling. In: Somerville CR, Meyerowitz EM, editors. The Arabidopsis Book. Rockville, MD: American Society of Plant Biologists; 2002. http://www.aspb.org/ . doi/10.1199/tab.0074. http://www.aspb.org/ publications/arabidopsis. publications/arabidopsis. [Google Scholar]
- Weller JL, Terry MJ, Rameau C, Reid JB, Kendrick RE. The phytochrome-deficient pcd1 mutant of pea is unable to convert heme to biliverdin IXa. Plant Cell. 1996;8:55–67. doi: 10.1105/tpc.8.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harberd NP. Phytochrome A null mutants of Arabidopsis display a wild type phenotype in white light. Plant Cell. 1993;5:757–768. doi: 10.1105/tpc.5.7.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Parks BM, Short TW, Quail PH. Missense mutations define a restricted segment in the C-terminal domain of phytochrome A critical to its regulatory activity. Plant Cell. 1995;7:1433–1443. doi: 10.1105/tpc.7.9.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Matsui M, Ang L, Deng XW. Role of a COP1 interactive protein in mediating light-regulated gene expression in Arabidopsis. Plant Cell. 1998;10:1083–1094. doi: 10.1105/tpc.10.7.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh KC, Lagarias JC. Eukaryotic phytochromes: light regulated serine/threonine protein kinases with histidine kinase ancestry. Proc Natl Acad Sci. 1998;95:13976–13981. doi: 10.1073/pnas.95.23.13976. [DOI] [PMC free article] [PubMed] [Google Scholar]