Abstract
A cDNA encoding a protein with 456 amino acids whose sequence shows considerable similarity to plant acyltransferases was identified among 750 Clarkia breweri flower expressed sequence tags. The cDNA was expressed in Escherichia coli, and the protein produced was shown to encode the enzyme benzoyl-coenzyme A (CoA):benzyl alcohol benzoyl transferase (BEBT). BEBT catalyzes the formation of benzylbenzoate, a minor constituent of the C. breweri floral aroma, but it also has activity with a number of other alcohols and acyl CoAs. The BEBT gene is expressed in different parts of the flowers with maximal RNA transcript levels in the stigma, and no expression was observed in the leaves under normal conditions. However, BEBT expression was induced in damaged leaves, reaching a maximum 6 h after damage occurred. We also show here that a closely related tobacco (Nicotiana tabacum) gene previously shown to be induced in leaves after being challenged by phytopathogenic bacteria also has BEBT activity, whereas the most similar protein to BEBT in the Arabidopsis proteome does not use benzoyl CoA as a substrate and instead can use acetyl CoA to catalyze the formation of cis-3-hexen-1-yl acetate, a green-leaf volatile.
Floral scents are often rich in volatile esters. In Clarkia breweri ([Gray] Greene; Onagraceae), an annual plant native to California, for example, benzylacetate constitutes 20% to 40% (w/w) of the total scent output (depending on the particular C. breweri line), whereas the two other esters present in its aroma, benzylbenzoate and methylsalicylate, each contribute about 5% to the total volatile output (Raguso and Pichersky, 1995).
Many plants also emit volatile esters from leaves damaged by herbivores. The most commonly reported volatile esters in this class are those derived from the octadecanoid fatty acids, such as cis-3-hexen-1-yl acetate (Ozawa et al., 2000; Arimura et al., 2001; Mattiacci et al., 2001), and methylsalicylate, mostly likely derived from the phenylpropanoid pathway (Lee et al., 1995) or the shikimate pathway (Wildermuth et al., 2001), is also common (Pare and Tumlinson, 1996; Van Poecke et al., 2001). Total volatiles (including, but not limited to, esters) emitted from injured leaves have been shown to function as orientation cues for predatory wasps and mites that feed upon herbivorous insects (Turlings et al., 1990; De Moraes et al., 1998). In addition, some of these volatiles may serve as antimicrobial or antifungal agents to prevent further spread of disease (Hamiltonkemp et al., 1992; Croft et al., 1993; Deng et al., 1993).
We have previously reported the characterization of the enzyme acetyl CoA:benzyl alcohol acetyl transferase (BEAT), which is responsible for the production of the floral volatile benzylacetate in C. breweri flowers (Dudareva et al., 1998a). The concurrent characterization of BEAT and several other structurally similar enzymes has led to the recognition of a novel class of evolutionarily related acyltransferases (EC 2.3.1.x) commonly referred to as the BAHD family of acyltransferases (St-Pierre and De Luca, 2000). There are approximately 60 BAHD gene family members in the model organism Arabidopsis, although to date, the substrates and products of the enzymes encoded by these genes have not been determined. Members of the BAHD family have been identified in other plant species through expressed sequence tag (EST) database construction and analyses, and in a few cases, the biochemical function has been determined as well. For example, benzoyl-CoA:anthranilate N-benzo-yltransferase (HCBT) from carnation (Dianthus caryophyllus) is expressed during infection with Fusarium oxysporum or Phytophthora parasitica and produces several different benzoylated and coumaroylated anthranilide phytoalexin derivatives (Yang et al., 1997). Other BAHD proteins that are also known to be involved in plant defense are TAT and DBAT from Taxus cuspidata, which catalyze the acetylation and benzoylation, respectively, of taxol precursors (Walker and Croteau, 2000a, 2000b; Walker et al., 2000), MAT and DAT from Catharanthus roseus, which catalyze the acetylation of precursors of the alkaloid vindoline (Power et al., 1990; St-Pierre et al., 1998; Laflamme et al., 2001), and SALAT, which is involved in morphine biosynthesis (Grothe et al., 2001). BAHD enzymes involved in caffeoylation and malonylation of anthocyanin pigments have also been reported (Fujiwara et al., 1997, 1998; Yonekura-Sakakibara et al., 2000; Suzuki et al., 2001) as well as those involved in volatile production in strawberry (Fragaria spp.) fruit (Aharoni et al., 2000). It should be noted that the enzymes responsible for the synthesis of methyl esters such as methylsalicylate and methyljasmonate belong to a different family of enzymes (Ross et al., 1999; Seo et al., 2001).
Here, we report the characterization of the gene and enzyme for the biosynthesis of the floral volatile benzylbenzoate in C. breweri. The gene for benzoyl-CoA:benzyl alcohol benzoyl transferase (BEBT) is expressed in flowers, but it is also expressed in leaf tissue after damage. The structure of BEBT indicates that it belongs to the BAHD family of acyltransferases, and its wide substrate specificity may allow it to catalyze the formation of other esters as well.
RESULTS
Benzylbenzoate-Forming Activity in Flowers
We have previously shown that flowers of C. breweri emit benzylbenzoate (Raguso and Pichersky, 1995). The emission of this volatile is highest from the stigma, with petals contributing most of the rest (Dudareva et al., 1998b). Total floral emission peaks 36 h postanthesis, with a second minor peak 72 h postanthesis.
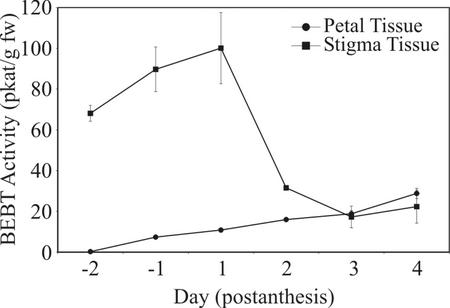
To look at the synthesis of benzylbenzoate, we developed an assay (see “Materials and Methods”) to detect the enzymatic activity of BEBT, the hypothetical enzyme (Croteau, 1977; Dudareva et al., 1998b) that would catalyze the formation of benzylbenzoate from benzyl alcohol and benzoyl CoA (Fig. 1). Crude extracts of stigma and petal tissues from flowers of different ages were assayed. BEBT activity was detected in both petals and stigma. In the stigma, it was high already in unopened flowers, and it peaked on d 1 of anthesis (Fig. 2). On d 2, when the stigma becomes receptive, there was a precipitous drop in BEBT activity of about 3-fold, and activity remained low on d 3 and 4. In petals, BEBT activity was much lower than in stigma but it gradually increased until d 4, at which time BEBT levels were similar to those in the stigma (Fig. 2).
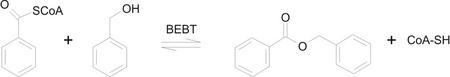
Figure 1.
The reaction catalyzed by BEBT.
Figure 2.
BEBT enzymatic activity in different floral tissues during the lifespan of the flower. Petal and stigma tissues were collected from C. breweri flowers daily starting 2 d before flower opening (d −2) and ending on d 4 postanthesis. For each data point, tissues from three different plants were combined for each assay, at least three independent assays were conducted, and the mean was obtained. ●, Petals; ▪, stigma. pkat, Picomoles of product per second.
Isolation of a cDNA Clone Encoding BEBT
We have recently undertaken the sequencing of approximately 750 EST cDNAs from a C. breweri flower tissue cDNA library. Because the acylation reaction performed by BEBT is similar in principle to the acylation reactions performed by BAHD-type enzymes, we searched the database with BAHD-type protein sequences, including C. breweri BEAT and putative BAHD proteins from Arabidopsis, using the BLAST2 program (Altschul et al., 1990). The search identified one cDNA with homology to BAHD acyltransferase sequences. Because this cDNA was incomplete on both the 5′ and 3′ ends, we performed 5′- and 3′-RACE experiments to obtain a full-length clone. The complete cDNA, which we tentatively designated BEBT, has an open reading frame encoding a protein with 456 amino acid residues and a calculated molecular mass of 50.6 kD. Protein sequence comparisons indicate that BEBT is most similar (72% identity) to a protein encoded by the tobacco (Nicotiana tabacum) hypersensitive response cDNA HSR201 (Czernic et al., 1996) and to two BAHD-like Arabidopsis proteins (protein ID nos. CAC01898.1 and AAF01587.1) that are 57% and 54% identical, respectively (Fig. 3). In addition, proteins encoded by a cDNA from cantaloupe (Cucumis melo) and a gene from rice (Oryza sativa) identified during the sequencing of the rice genome are also very similar to BEBT, being 53% and 63% identical, respectively, to BEBT. The proteins encoded by these genes all share the salient features of the BAHD protein family, most notably the HXXXD motif in the center of the protein believed to be involved in catalysis, and the DFGWG motif of unknown function near the C terminus (St-Pierre and De Luca, 2000).
Figure 3.
Sequence comparisons of BEBT and BEAT from C. breweri, HSR201 from tobacco, and the two most similar proteins to BEBT from Arabidopsis. The BEBT gene accession number is AF500200, and the gene accession number of HSR201 is AF500202. The two Arabidopsis acyltransferase proteins in this figure are identified by their protein accession numbers and correspond to genes from two BAC clones (accession nos. AL391151 and AC009895, respectively). A, Protein sequence alignments of all five proteins using the ClustalX program. Amino acids shaded in black represent identical matches; gray shaded boxes represent conservative changes. The HXXXD motif is indicated with arrowheads; the DFGWG motif is indicated with asterisks. B, Maximum parsimony tree based on protein sequence alignments from A using the program PAUP*.
Enzymatic Characterization of C. breweri BEBT Expressed in Escherichia coli
To determine whether the isolated C. breweri cDNA indeed encoded an enzyme with BEBT activity, we subcloned the complete open reading frame of BEBT into the expression vector pET11a, transformed E. coli cells with the recombinant vector, and induced the expression of BEBT with IPTG. Because E. coli strain BL21(DE3) pLysS, the standard strain for expressing heterologous genes from pET vectors, has a low level of acyltransferase activity derived from the chloramphenicol O-acetyltransferase gene, we used E. coli strain B834 instead. Strain B834 cells have no activity with either acetyl CoA or benzoyl CoA regardless of whether they carry a pET expression vector and regardless of whether they have been treated with IPTG (data not shown).
The spent media of BEBT-expressing E. coli cultures were extracted with pentane and analyzed by gas chromatography-mass spectroscopy (GC-MS), revealing a wide variety of acylated esters carrying acyl moieties ranging from C2-C6 (Table I). In contrast, spent media of induced cultures carrying a pET11a plasmid with no insert, used as a control, contained none of these esters. Because the ester composition in the spent media of BEAT-expressing E. coli cultures is >98% benzylacetate (Dudareva et al., 1998a), it indicates that BEBT, unlike BEAT, may accept several acyl-CoA substrates in addition to acetyl-CoA. Although the products that formed in E. coli when BEBT was expressed clearly depended both on the affinity of the BEBT enzyme to potential substrates and the availability of these substrates in E. coli and although the substrate availability and concentration in the plant cell are likely to be very different from the situation in E. coli, nonetheless these preliminary results demonstrated that BEBT possessed ester-forming activity.
Table I.
Volatile esters extracted from the spent media of E. coli cells expressing BEBTa
| Esters | Ester/Spent Medium |
|---|---|
| ng mL−1 | |
| Acetyl | |
| n-Butyl acetate | 5,576.8 ± 947.8 |
| n-Pentyl acetate | 73.7 ± 17.6 |
| Benzyl acetate | 568.8 ± 139.9 |
| Phenethyl acetate | 180.0 ± 90.6 |
| Butyryl | |
| Ethyl butyrate | 680.6 ± 285.5 |
| n-Butyl butyrate | 1,658.3 ± 326.4 |
| Hexanoyl | |
| Ethyl hexanoate | 734.2 ± 67.6 |
| n-Butyl hexanoate | 530.5 ± 92.4 |
| Others | |
| n-Butyl propanoate | 70.8 ± 17.5 |
| Ethyl octanoate | 479.0 ± 204.4 |
Only the compounds that constituted >1% of the total volatiles present and that were not found in the controls are shown.
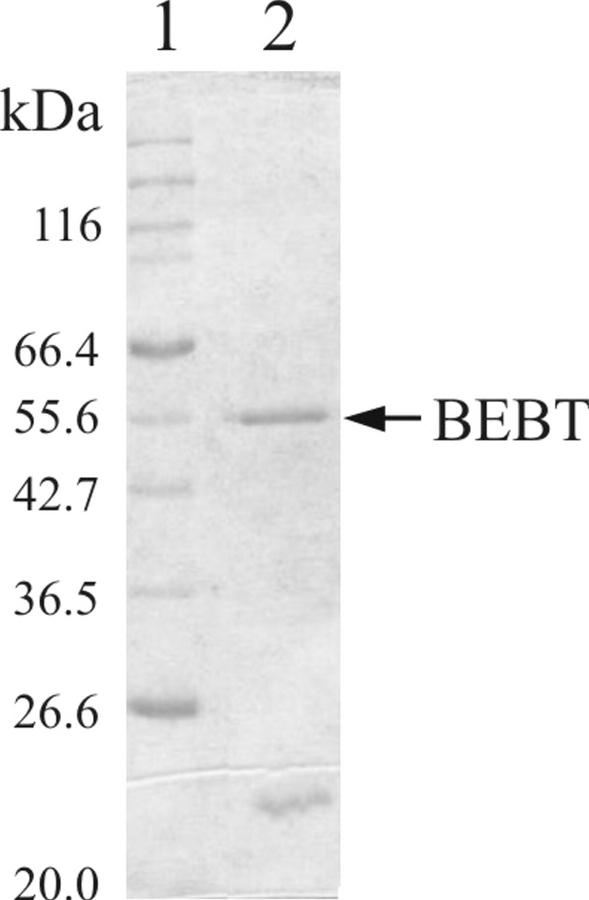
To characterize the biochemical properties of BEBT in vitro, we purified the complete, non-fusion BEBT enzyme from the crude E. coli extract by anion-exchange chromatography on DEAE, followed by another anion-exchange column, Mono Q, and finally by size-exclusion chromatography on a Q-Superose column (Fig. 4), as described in “Materials and Methods” (we had originally tried to introduce a His tag at either the N- or C-terminal ends of the protein, however both constructs yielded a protein with a dramatically lower activity). After the Q-Superose purification step, BEBT was essentially separated from all but one other protein with a molecular mass of 22.6 kD, which constituted about 40% of the total protein concentration. Although the latter protein was present in the Q-Superose fraction with the highest BEBT activity, its concentration in this and adjacent fractions did not correlate well with the levels of BEBT activities in these fractions, whereas BEBT protein concentrations and activity levels did (data not shown). It is noteworthy that BEBT migrated on SDS-PAGE as a protein with a molecular mass of 55 kD, whereas its calculated molecular mass is 50.6 kD. BEAT protein similarly has a calculated molecular mass of 48.2 kD, but migrates on a gel as a protein with a molecular mass of 58 kD (Dudareva et al., 1998a).
Figure 4.
Purification of C. breweri BEBT produced in E. coli. Lane 1, Mr markers. Lane 2, Q-Superose fraction with the highest levels of BEBT-specific activity.
In initial assays of BEBT activity, we used a variety of alcohols with benzoyl CoA and acetyl CoA. With acetyl CoA, benzyl alcohol was the preferred substrate, but other alcohols could also be used (data not shown). However, the Km value for acetyl CoA, 818 μm (Table II), strongly suggests that acetyl CoA is not commonly used by BEBT as the acyl donor in vivo. With benzoyl CoA, for which a Km value of 20.5 μm was determined, BEBT could use benzyl alcohol (with a Km value of 46.8) and a variety of other alcohols, some of them with higher rates (at a substrate concentration of 1 mm), particularly octanol and cinnamyl alcohol (Table III). BEBT had a Km value for cinnamyl alcohol of 97.8 μm (Table II). We also examined the activity of BEBT with other acyl CoA substrates by performing competition experiments (Beuerle and Pichersky, 2002a) of these nonradioactive CoA esters with radioactive acetyl CoA. Benzoyl CoA and cinnamoyl CoA were better competitors than either butanoyl CoA or hexanoyl CoA. The Km value for cinnamoyl CoA was determined to be 464 μm (Table II). Coumaroyl CoA, on the other hand, did not act as a competitor and could not serve as a substrate. On the basis of the these results and the measured Km values, we concluded that benzoyl CoA and benzyl alcohol are likely to be in vivo substrates of BEBT. It is possible that other alcohols similar to benzyl alcohol (such as octanol) and other acyl CoA esters similar to benzoyl CoA (such as cinnamoyl CoA) may also be acted upon by BEBT, but at present, we are not aware of other possible ester products in C. breweri besides benzylbenzoate that could result from catalysis by BEBT.
Table II.
Kinetic parameters of BEBT
| Km | Kcat | Kcat/Km | |
|---|---|---|---|
| μm | s−1 | nm−1 s−1 | |
| Benzyl alcohol (with benzoyl CoA) | 46.8 | 4 | 85.2 |
| Cinnamyl alcohol (with benzoyl CoA) | 97.8 | 4.6 | 47.2 |
| Benzoyl CoA (with benzyl alcohol) | 20.5 | 2.5 | 124 |
| Cinnamoyl CoA (with benzyl alcohol) | 464 | 0.4 | 0.9 |
| Acetyl CoA (with benzyl alcohol) | 818 | 17.5 | 21.5 |
Table III.
Relative activity of BEBT with a variety of substrates
| Alcohol Substrate | Benzoyl CoA as the Acyl Substrate | Acetyl CoA as the Acyl Substrate |
|---|---|---|
| Benzyl alcohol | 100a | 1001 |
| ρ-Hydroxy benzyl alcohol | 0 | 0 |
| m-Hydroxy benzyl alcohol | 113 | 8.7 |
| Cinnamyl alcohol | 172.6 | 8.4 |
| 2-Phenyl ethanol | 56.9 | 12.1 |
| Anthranilate | 0 | 0 |
| cis-3-Hexen-1-ol | 149.9 | 50.5 |
| Geraniol | 190.8 | 45.4 |
| Linalool | 0 | 0.6 |
| Ethanol | 0 | 0 |
| 1-Butanol | 107.6 | 35.5 |
| Octanol | 205.2 | 71.5 |
| Hexanol | 148.2 | 32.9 |
Activity with benzylalcohol was set at 100%, and other activities are shown as a percentage of this activity. All alcohol substrates were tested at a concentration of 1 mm. The specific activity of BEBT with benzoyl CoA and benzyl alcohol as substrates was 17.1 nkat mg−1 protein, and the specific activity of BEBT with acetyl CoA and benzyl alcohol as substrates was 1.4 nkat mg−1 protein.
The pH optima of BEBT was determined to be pH 7.7, whereas only 39% and 69% of maximal activity were observed at pH of 6.5 and of 9.0, respectively. The enzyme was 100% stable for 30 min at 30°C and 80% stable for 30 min at 37°C. After incubation at 50°C for 30 min, the enzyme was completely inactivated. The effects of monovalent and divalent cations were also tested. None were found to be stimulatory, however Mg2+, Ca2+, Co2+, Zn2+, and Cu2+ had a strong inhibitory effect (50%–100% inhibition at a final concentration of 5 mm). The apparent molecular mass of the protein as determined by size-exclusion chromatography was approximately 47 to 49.5 kD. Because the predicted molecular mass of the BEBT protein is 50.6 kD, we concluded that BEBT is a monomeric enzyme.
Organ Specificity and Developmental Changes of BEBT Expression in C. breweri
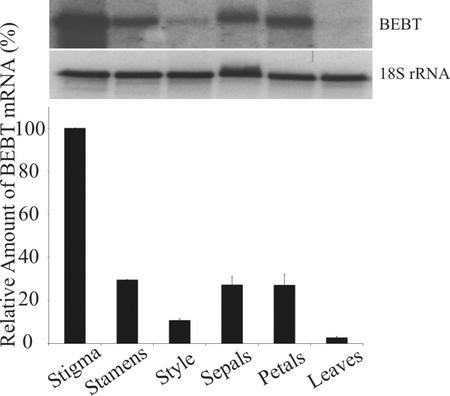
RNA gel-blot analysis with d 1 postanthesis flowers and leaf tissue was carried out to examine the tissue(s) in which the BEBT gene is expressed (Fig. 5). The highest steady-state levels of BEBT transcripts were observed in the stigma. Stamens, sepals, and petals had approximately the same amount of transcripts, although it was approximately 30% of that found in the stigma. Transcript levels in style tissue were 10% of those found in the stigma, whereas healthy leaves had little discernible expression of BEBT (Fig. 5).
Figure 5.
RNA gel-blot analysis of the relative abundance of BEBT mRNA transcripts in the stigma, stamen, style, sepals, petals, and leaves of C. breweri plants. Tissues were harvested from mature plants and floral tissue samples were taken from flowers on d 1 of anthesis. Lanes were loaded with 4 μg of total RNA. After probing with the BEBT probe and quantitation of the results, each blot was rehybridized with an 18S rDNA probe to normalize samples. A sample blot is shown above. The graphical representation below represents an average of three independent experiments.
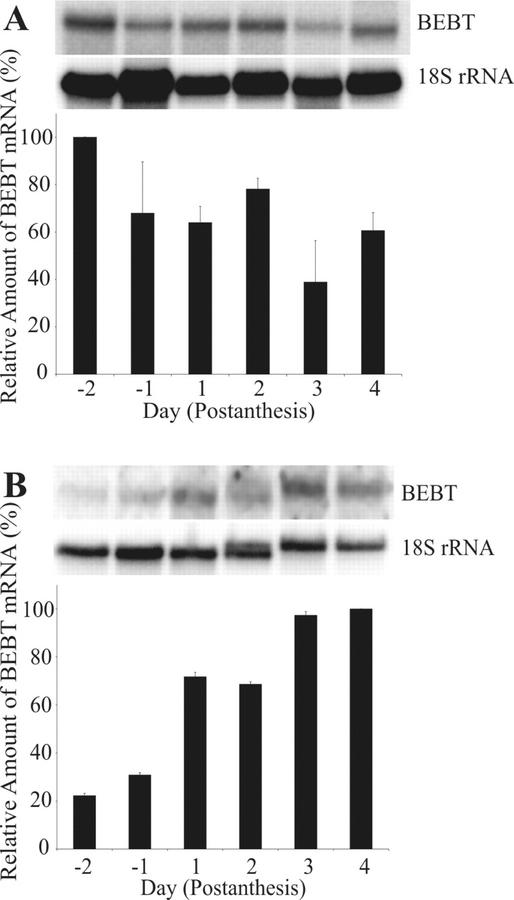
The levels of BEBT transcripts were examined over the life span of the flower by comparing the stigma, the organ with the highest levels of BEBT transcripts, to the petals, the largest organ (by mass) of the flower. The stigmata of flower buds 2 d before anthesis contained the highest levels of BEBT transcripts, and these levels subsequently dropped but remained within 40% to 60% of their initial value by d 3 to 4 postanthesis (Fig. 6A). In contrast, BEBT mRNA transcript levels from petal tissue steadily increased throughout the lifespan of the flower, reaching a 5-fold increase on d 4 postanthesis as compared with d −2 (Fig. 6B). However, BEBT mRNA levels in petals at any given day were always severalfold lower than those in the stigma.
Figure 6.
Expression of BEBT in petal and stigma tissues of C. breweri flowers during floral development. A, RNA gel-blot analysis of the relative abundance of BEBT mRNA in stigma tissue. Stigma tissue was collected daily from flowers starting 2 d before flower opening and continuing until d 4 postanthesis. B, RNA gel-blot analysis of the relative abundance of BEBT mRNA in petal tissue. Petal tissue was collected at the same times as the tissues in A. For all experiments, each lane was loaded with 4 μg of total RNA. After hybridization with the BEBT probe and quantitation of the results, blots were stripped and reprobed with an 18S rDNA probe to normalize samples. In each panel, a sample blot is shown above, and the graphical representation below represents an average of two independent experiments.
Quantitation of BEBT Protein Levels in the Stigma by Immunoblotting
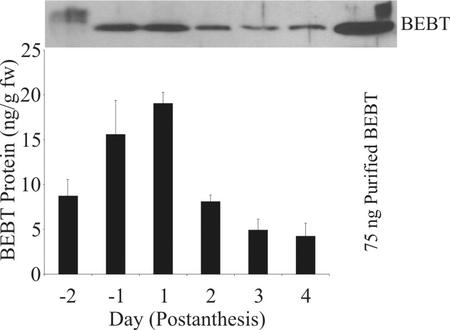
BEBT protein levels were quantified in the stigma by SDS-PAGE followed by immunoblots of stigma samples together with samples of known amounts of BEBT purified from E. coli (Fig. 7, top). In stigma extracts from all stages of development, with the exception of d −2 stigma, the antibodies made against purified C. breweri BEBT produced in E. coli (see “Materials and Methods”) recognized a single protein that migrated on the gel to the same position as did BEBT produced in E. coli. In d −2 stigma, the anti-BEBT antibodies recognized an unfocused protein band migrating slightly more slowly than E. coli-produced BEBT (Fig. 7, top). The reasons for the aberrant behavior of BEBT in d −2 stigma in SDS-PAGE could not be determined. The quantitative results of the immunoblots (Fig. 7, bottom) indicated that BEBT protein levels followed the measured levels of BEBT enzymatic activity (Fig. 2), dropping equally rapidly on d 2 postanthesis, the day when the stigma lobes open up and the stigma becomes receptive, compared with d 1 postanthesis.
Figure 7.
Variation of the levels of BEBT protein over the lifespan of the stigma. Stigma tissue was collected daily from flowers starting 2 d before flower opening, and continuing until d 4 postanthesis. Samples were run on SDS-PAGE, and the gels were blotted onto filters. The filters were first probed with anti-BEBT antibodies, followed by incubation with secondary antibodies conjugated to alkaline phosphatase. Bands were visualized by chemiluminescence. A sample blot is shown above, and the graphical representation below represents an average of two independent experiments.
Characterizaton of the Activities of the Proteins Encoded by Tobacco HSR201 cDNA and by Another BEBT-Related cDNA from Arabidopsis
The sequence of the protein encoded by the HSR201 cDNA from tobacco is the most similar to BEBT of the protein sequences currently found in the databases (Fig. 3). It has previously been shown that HSR201 is expressed in damaged leaves of tobacco after infection with the microorganism Pseudomonas solanacearum (Czernic et al., 1996). To analyze the function of the protein encoded by HSR201, we isolated total RNA from tobacco leaves 6 h after damaging them with a razor blade, and cloned the open reading frame of HSR201 into the pCR T7 TOPO-CT expression vector, using oligonucleotides based on the published sequence. The HSR201 protein obtained in the E. coli expression system had BEBT activity with Km values for benzoyl CoA and benzyl alcohol of 35 and 19 μm, respectively, similar to the corresponding Km values of BEBT. The HSR201 protein showed no activity with acetyl CoA at concentrations <1 mm.
We also examined the activities of the proteins encoded by the two Arabidopsis genes whose proteins show the highest similarity to BEBT. cDNAs encoding CAC01898.1 and AAF01587.1 were expressed in E. coli. No activity could be detected for protein CAC01898.1 with a variety of alcohols and either acetyl CoA or benzoyl CoA. Protein AAF01587.1 had no detectable activity with benzoyl CoA at concentrations <1 mm but exhibited strong activity with acetyl CoA and cis-3-hexen-1-ol and related alcohols (e.g. hexanol and octanol). AAF01587.1, which we have designated CHAT (for acetyl CoA:cis-3-hexen-1-ol acetyl transferase) had a Km value of 10.5 μm for acetyl CoA and 165 μm for cis-3-hexen-1-ol.
BEBT Expression in Wounded Leaves
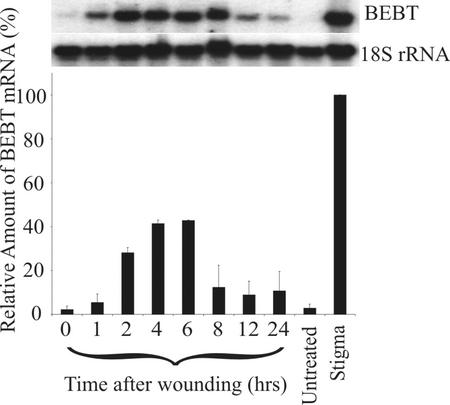
Because HSR201 proved to encode BEBT and its expression was known to be induced in leaves upon wounding, we examined the expression of C. breweri BEBT in wounded leaves. Total RNA was extracted from damaged leaves at intervals of 0, 1, 2, 4, 6, 8, 12, and 24 h after wounding, and the RNA samples were analyzed by gel blotting and probing with a BEBT probe (Fig. 8). BEBT mRNA levels rose dramatically from 2 to 6 h after wounding, with peak expression evident after 6 h, decreasing afterward. It should be noted that BEBT transcript levels in wounded leaves were still roughly one-half of that found in the stigma of d 1 flowers.
Figure 8.
RNA gel-blot analysis of the relative expression of BEBT in wounded and non-wounded leaf tissues of C. breweri. Mature leaves were harvested either before (untreated) or after mechanical wounding. Leaves were wounded by making two parallel incisions approximately 8 mm long to each side of the midvein with a sterile razor blade, and tissue was collected at 0, 1, 2, 4, 6, 8, 12, and 24 h after wounding. RNA from stigma tissue (included for comparison) was collected from d 1 flowers. Total RNA (4 μg) was loaded onto each lane of the gel. After probing with the BEBT probe and quantitation of the results, the blots were then stripped and reprobed with an 18S rDNA probe to normalize samples. A sample blot is shown above, and the graphical representation below represents an average of two independent experiments.
DISCUSSION
BEBT, a BAHD-Type Acyltransferase, Is Responsible for Benzylbenzoate Synthesis in C. breweri Flowers
The BAHD family of acyl transferases is a recently discovered group of enzymes (St-Pierre and De Luca, 2000). The function and substrate specificity of only a few representatives, which include enzymes that catalyze the formation of both volatile and nonvolatile esters, have so far been determined. Even so, it is clear that a hallmark of these enzymes is the relatively wide range of alcohols that they can react with. On the other hand, the range of acyl-CoA substrates has not yet been extensively tested, due mostly to the lack of availability of these compounds. However, at least in one case, that of HCBT from carnation, it was shown that the enzyme has a somewhat lower Km value for cinammoyl CoA than for benzoyl CoA, even though benzoyl CoA is the substrate used in vivo (Yang et al., 1997).
The products detected in the spent media of E. coli cells expressing BEBT (Table I) show that BEBT can use several medium-chain acyl CoA substrates and a variety of mostly primary alcohols in addition to benzoyl CoA and benzyl alcohol. However, what products are formed in E. coli depend on the presence and concentrations of the respective substrates vis-à-vis the affinity of BEBT for those substrates. The possibility that the in planta substrates of BEBT are not found in E. coli is highly likely.
The Km values of BEBT for the substrates benzyl alcohol and benzoyl CoA—46.8 and 20.5 μm, respectively—are in a range that is similar to those of BEAT and several other BAHD acyltransferases for their substrates, and they strongly argue that BEBT is responsible for the synthesis of benzylbenzoate, at least in flowers. For comparison, the enzyme benzoyl-CoA:taxane 2α-O-benzoyltransferase from Taxus spp. was reported to have a Km value of 300 μm for benzoyl CoA (Walker and Croteau, 2000b). The enzyme HCBT, another BAHD benzoyltransferase, has a Km value of 50 μm for benzoyl CoA, similar to that of BEBT (Yang et al., 1997). There is currently no data on the concentrations of benzoyl CoA in C. breweri or any other species known to make benzoate esters, although benzoic acid:CoA ligase has recently been identified and partially purified from C. breweri (Beuerle and Pichersky, 2002a). On the other hand, the Km values of BEBT for acetyl-CoA and cinnamoyl CoA—818 μm and 464 μm, respectively—suggest that these acyl CoAs are not commonly used by BEBT. In addition, the ratio of Kcat/Km (benzoyl CoA) to Kcat/Km (acetyl CoA) shows that BEBT preferentially uses benzoyl CoA to acetyl CoA by a factor of 5.8.
Additional support for the role of the protein encoded by the BEBT gene in benzylbenzoate production in flowers comes from the general correlation between BEBT mRNA levels and BEBT enzyme activity in the different floral parts (Figs. 2 and 4) and from the correlation between BEBT enzymatic activity in the stigma and the amount of BEBT protein as determined by immunoblotting with antibodies made against the protein encoded by the BEBT gene (Figs. 2 and 6).
Although BEBT transcript levels and BEBT enzymatic activity levels in the stigma are always higher than those in the petals, nonetheless the levels of BEBT enzymatic activity in the stigma show an interesting pattern of change relative to the BEBT mRNA levels. BEBT activity levels show a moderate buildup until d 1 of anthesis followed by quick decline, even though BEBT mRNA levels do not show such a corresponding decline. The buildup in BEBT activity levels until d 1 of anthesis suggests that the turn-over of the BEBT protein is slow (as is the case for other scent enzymes (Dudareva and Pichersky, 2000), and therefore BEBT protein continues to accumulate even though BEBT steady-state transcript levels are relatively stable. The precipitous decline in BEBT enzymatic levels (per gram fresh weight) on d 2 of anthesis is clearly attributable to a decrease in total BEBT protein (Fig. 7), and this decline is not caused by a similar decline in the steady-state levels of BEBT mRNA (Fig. 6A). Day 2 of anthesis is the day in which the stigma opens up and becomes receptive, a process that involves rapid cell expansion and consequently a reduction in protein content per cell (Pichersky et al., 1994). However, even when BEBT activity levels and BEBT protein levels are plotted after normalization to protein content (data not shown), a substantial drop of 50% in both is still observed from d 1 to 2. This reduction in BEBT activity, therefore, must involve a mechanism of specific protein degradation and may be related to the yet-undetermined function of benzylbenzoate in the stigma.
Acyltransferase Activities in Wounded Leaves
It is well established that fatty acid-derived aldehyde, alcohol, and ester “green-leaf” volatiles are emitted upon injury (Walling, 2000). Our results showing the induction of BEBT in wounded leaves and the induction of Arabidopsis CHAT by wounding (J.C. D'Auria, unpublished data) point to the possible role of such acetyltransferases in the defense response of leaves to injury. Although CHAT may be involved in the biosynthesis of cis-3-hexenyl acetate, it is likely that BEBT activity in the leaf is responsible mostly for the production of benzylbenzoate. Other species are also known to produce this compound in leaves. For example, leaves of rice produce benzylbenzoate in response to oviposition by the insect whitebacked planthopper (Sogatella furcifera), and it was shown that benzylbenzoate has ovicidal properties (Seino et al., 1996). Our results showing that the tobacco hypersensitive-response cDNA HSR201 encodes BEBT suggest that tobacco leaves may also synthesize benzylbenzoate or a related compound under stress. Other esters that are known to be synthesized in damaged leaves include ρ-coumaroyl- and feruloyltyramine in tomato (Lycopersicon esculentum; Pearce et al., 1998) and 5-caffeoylquinic (chlorogenic) acid and its derivatives in lettuce (Lactuca sativa; TomasBarberan et al., 1997). It is worth noting that RNA-blot analysis indicated that the BEBT-related gene BEAT, which encodes an enzyme that catalyzes the formation of benzyl acetate, is not generally expressed in C. breweri leaves (Dudareva et al., 1998a) nor is it induced upon damage (N. Dudareva and E. Pichersky, unpublished data).
Evolution of BEBT and CHAT
Although BEBT and HSR201 have strong affinity to benzoyl CoA and low affinity to acetyl CoA, they are closely related to CHAT, which has a much stronger affinity to acetyl CoA than to benzoyl CoA. A similar situation was reported for two closely related acyltransferases from Taxus spp., one that uses acetyl CoA and the other benzoyl CoA (Walker and Croteau, 2001). This suggests that the specificity for the acyl CoA moiety can evolve relatively easily in the BAHD family of enzymes.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Details of the construction of true-breeding Clarkia breweri stocks and growth conditions are as described (Raguso and Pichersky, 1995). Arabidopsis ecotype Columbia plants were used of for cloning CHAT. Tobacco (Nicotiana tabacum) seeds were germinated on Murashige and Skoog medium plates and grown for 2 weeks. Seedlings were then transplanted to soil (Sunshine mix no. 2, Sun Gro Horticulture, Bellevue, WA) and grown under 12-h light/12-h dark light conditions in a growth chamber.
Reagents
All solvents and reagents were molecular biology grade or reagent grade and were obtained from Sigma (St. Louis), Aldrich (Milwaukee), or Fisher Scientific (Loughborough, Leicestershire, UK). Radiolabeled [acety-1-14C]CoA was purchased from ICN (Irvine, CA). [7-14C]Benzoyl-CoA was enzymatically synthesized from [7-14C]benzoic acid (16.6 mCi mmol−1; Sigma) as previously described (Beuerle and Pichersky, 2002b).
Enzyme Extraction and Assay
Crude protein extracts were prepared by macerating tissue parts in a microcentrifuge tube with a Teflon-coated homogenizer in the presence of ice-cold buffer containing 50 mm Bis-Tris-HCl, pH 7.0, 5 mm dithioerythritol (DTE), 1% (w/v) polyvinylpyrrolidone-40, and 10% (v/v) glycerol. A ratio of extraction buffer:tissue fresh weight of 10:1 (v/w) was used. The slurry was then centrifuged for 10 min at 4°C, and the supernatant was transferred to a new tube. For each time point, flower tissue and leaves from three different plants were combined, and assays were repeated at least twice.
Assay samples were prepared by adding the following to a 0.6-mL microcentrifuge tube: 5.0 μL of crude extract, 10.0 μL of 5× assay buffer (250 mm Bis-Tris Propane, pH 7.7), 1.0 μL of 50 mm alcohol substrate dissolved in dimethyl sulfoxide, 1.0 μL of [7-14C]benzoyl-CoA, and 33 μL of water to bring the assay volume to 50 μL. Acetyltransferase assays for both BEBT and CHAT were performed as above with the substitution of 0.25 μL of [acetyl-1-14C]CoA (50 mCi mmol−1) for the radiolabeled benzoyl-CoA. The assays were carried out at room temperature for 30 min. The radioactively labeled acylated product was extracted twice with 180 μL of hexane, and the total organic phase was transferred to a scintillation vial with 2 mL of nonaqueous scintillation fluid (Econo-Safe, Research Products International, Mount Prospect, IL) and counted in a liquid-scintillation counter (model LS6500, Beckman Coulter, Fullerton, CA). The raw data (counts per minute) were converted to picokatals based on the specific activity of the radiolabeled substrate and the known efficiency of the scintillation counter used. Assays in which no alcohol substrate was added were performed to test background thioesterase activity. In addition, boiled enzyme extracts were substituted for intact enzyme to test nonenzymatic breakdown of the CoA thioester via reaction with reducing agents in the enzyme extract. The identity of the products were verified by GC-MS as described previously using authentic standards (Dudareva et al., 1998a).
BEBT cDNA Identification
A total of 735 cDNAs were randomly and automatically isolated and sequenced similar to those previously reported for a basil (Ocimum basilicum) EST database (Gang et al., 2001). The C. breweri flower EST database can be found at: https://sativa.biology.lsa.umich.edu/blast/blast.html (login, clarkia; password, breweri). The BEBT cDNA was identified by BLAST searches with other BAHD sequences.
Cloning of BEBT and Its Homologs from Arabidopsis and Tobacco
The BEBT cDNA clone identified in the C. breweri EST database was incomplete. RACE was used to obtain the missing sequence from either end (Czernic et al., 1996; Martz et al., 1998). Two specific primers were designed (forward primer, 5′-CCCATTCGACGACCTGGCTCACCGG-3′; backward primer, 5′-CCCAGCTCGTGTCACGTCTGAAACCAAG-3′) and used in 5′- and 3′-RACE, respectively. First-strand cDNAs for the RACE experiments were made from total RNA of 1-d-old C. breweri flowers. On the basis of the sequences obtained, the full-length cDNA was amplified from first-strand cDNA using the forward primer 5′-AATGGCGCATGATCAATCTCTCT-3′ and the backward primer 5′-TCTCATCAAAGGGAAGACTGTGT-3′. The resulting 1,375-nucleotide (nt) DNA fragment was subcloned into a TA cloning vector (pCRT7/CT TOPO-TA, Invitrogen, Carlsbad, CA). The sequences of independent clones were determined in their entirety on both strands. For further expression analysis, the open reading frame of BEBT was subcloned into the pET-T7(11a) expression vector (Novagen, Madison, WI).
For the gene residing on BAC clone AC009895 (CHAT), a full-length cDNA was obtained by RT-PCR using total RNA extracted from Arabidopsis flower tissue using the forward primer 5′-AATGGACCATCAAGTGTC-TCTGC-3′ and the backward primer 5′-TCATCCTTTAGACACATT-TAGCACTCC-3′. The resulting 1,366-nt DNA fragment was cloned as described above for BEBT. All attempts to clone the gene residing on the BAC clone AL391151 via RT-PCR using total RNA from various plant tissues were unsuccessful. Because this gene has only one intron, the putative open reading frame was amplified from genomic DNA, and the introns were spliced out as described by Nam et al. (1999) by using the following two pairs of primers (front pair, forward primer 5′-AATGTCCGGGTCACTCACGTT and backward primer 5′-GCATTTCAAGCGCGTGACCTGCATGAGCAT-3′; back pair, forward primer 5′-CCTTTGATGCTCATGCAGGTCACGCGCTTGAAATGC-3′ and backward primer 5′-TTACAGAGAAGACATGATCAA-3′). The resulting 1,387-nt DNA fragment was again cloned as described above.
The full-length cDNA clone HSR201 from tobacco was cloned by RT-PCR using first-strand cDNA obtained from total RNA extracted from mechanically wounded tobacco leaves 6 h after wounding. Wounds were made by making two lateral incisions to each side of the midvein with a sterile razor blade. The primers used in the RT-PCR were the forward primer 5′-AATGGATTCAAAGCAATCATCAGA-3′ and the backward primer 5′-TCAAAGGGCAGGTCTGATAATG-3′ and resulted in a DNA fragment that was 1,384 nt in length. This fragment was also cloned into a TA cloning vector (pCRT7/CT TOPO-TA, Invitrogen) for expression analysis.
Sequence Alignment and Phylogenetic Analysis
Amino acid sequence alignments were generated using the ClustalX computer program (Thompson et al., 1997). Aligned sequences were analyzed using the PAUP* program (Phylogenetic Analysis Using Parsimony, Sinaur Associates, Sunderland, MA), with amino acids treated as unordered characters, and resulted in a single shortest tree shown in a PAUP* phylogram format. A neighbor-joining tree was also generated. It showed the same branches as that in the maximum-parsimony tree.
Protein Production in Escherichia coli and BEBT Purification
BEBT expression in E. coli was induced as previously described (Nam et al., 1999) with the following minor modifications: All expression constructs in this study were transformed into the E. coli cell line B834. E. coli cells were grown to an OD600 of 0.4 and then induced with 1 mm IPTG. After harvesting, the cells were resuspended in lysis buffer containing 50 mm Tris-HCl (pH 8.0), 10 mm NaCl, 1 mm EDTA, 5 mm DTE, 1 mm phenylmethylsulfonyl fluoride, and 10% (v/v) glycerol, lysed by sonification (3× for 30 s), and centrifuged at 10,000g for 15 min.
Soluble BEBT protein was first loaded onto DEAE anion-exchange column (10 mL of DE53, Whatman, Clifton, NJ) pre-equilibrated with buffer A (50 mm Bis-Tris, pH 7.5, 10% [v/v] glycerol, and 5 mm DTE) at a flow rate of 1 mL min−1 on an FPLC apparatus (Amersham Pharmacia Biotech AB, Uppsala). After washing off unbound materials from the column with buffer A, BEBT was eluted with a linear gradient of 0 to 0.5 mm KCl in buffer A. BEBT activity eluted in the 0.25 to 0.3 mm range. Fractions with the highest BEBT activity (10 mL) were pooled and diluted with buffer A to a final volume of 40 mL and loaded onto a Mono-Q FPLC column (0.5-cm diameter × 6.0 cm; Amersham Pharmacia Biotech AB). BEBT was eluted with a 100 to 500 mm KCl linear gradient with 3-mL fractions being collected. BEBT activity eluted with 130 to 140 mm KCl. Peak fractions from Mono-Q were examined by SDS-PAGE gel electrophoresis followed by Coomassie Brilliant Blue or silver staining of the gel. BEBT protein concentration was determined as previously described (Bradford, 1976).
Preparation of BEBT Antibodies and Protein Gel Blots
C. breweri BEBT protein produced in E. coli was purified from inclusion bodies as previously described (Dudareva et al., 1996). The final step in the purification consisted of electrophoresis on SDS-PAGE gel, staining with Coomassie Blue, and excision of the gel segment where the separated BEBT was located. Antibodies were prepared by Cocalico Biologicals (Reamstown, PA) by injecting macerated gel fragments containing the purified BEBT and following the company's protocol. Protein gel blots and quantitation of results were performed as described in Dudareva et al. (1996).
Extraction of Volatile Esters from the Spent Media of E. coli and GC-MS Analysis
Extraction of volatile esters from the spent media of E. coli expressing BEBT or containing pET11a vector without any insert were performed and analyzed by GC-MS as reported by Dudareva et al. (1998a) with the substitution of pentane for hexane as the solvent.
Molecular Mass Estimation
Peak BEBT activity from the Mono-Q fractions was pooled, concentrated down to 200 μL, and run on a gel filtration column (Q-Superose, Amersham Pharmacia Biotech AB) precalibrated with molecular mass markers as previously described (Ross et al., 1999).
Determination of Kinetic Parameters for BEBT and CHAT
In all kinetics studies, appropriate enzyme concentration and incubation times were chosen so that the reaction velocity was linear during the incubation time period. To determine the Km value for each substrate, one substrate concentration was fixed at a saturated level and the concentration of the other substrate to be measured was varied. Lineweaver-Burk, Eadie-Hofstee, and Hanes plots were constructed and an average of their Km values were used to report the apparent Km value.
Temperature Effect on BEBT Activity
BEBT protein from the Mono-Q fraction was incubated at temperatures ranging from 4°C to 65°C for 30 min and then chilled on ice. Samples incubated at each temperature were then used for the enzyme assays. At least four independent assays were performed for each point, and an average was taken.
pH Optimum of BEBT Stability
The optimum pH for BEBT activity was determined using Bis-Tris propane as the buffering system ranging from 6.5 to 9.0. Results presented are an average of at least four independent assays.
Effectors
Enzyme assays were performed with one of the following cations present in the assay buffer at a final concentration of 5 mm: K+, Na+, Ca2+, Cu2+, Fe2+, Mg2+, Mn2+, Zn2+, and Co2+. Results presented are an average of at least four independent experiments.
RNA Extraction and Northern-Blot Analysis
Total RNA was isolated from both C. breweri and Arabidopsis tissues using the RNeasy plant mini kit (Qiagen), using buffer RLC for C. breweri and RLT for Arabidopsis. For the leaf-wounding treatments of C. breweri leaves, two parallel incisions of 8 to 10 mm were made on either side of the midvein using a sterile razor blade. Leaf tissue was collected at 0, 1, 2, 4, 6, 8, 12, and 24 h after the initial incision. In all cases, 100 mg of tissue was used, and the protocol described in the kit was followed. To reduce the amounts of polysaccharides extracted from C. breweri leaves using the RNeasy kit, 20 μL of 50% high Mr PEG (15,000–20,000) was added to the microcentrifuge tube after the RLC buffer was added. This mixture of tissue, buffer, and PEG was incubated for 10 min at room temperature. After this, all steps were performed as directed in the kit manual.
Total RNA (4 μg) from C. breweri tissues was resolved on 1% (w/v) agarose-formaldehyde gels, blotted to Hybond-N+ nylon membranes, hybridized, and stripped as described by Sambrook et al. (1989) and as per the instructions in the Hybond-N+ manual (Amersham Pharmacia Biotech AB). Probes were synthesized using the rediprime II kit (Amersham Pharmacia Biotech AB) from a PCR-amplified fragment of BEBT using the full-length primers used previously for cloning. Hybridization signals were counted in a phosphor imager (Bio-Rad), stripped, and reprobed with a 18S rRNA probe to normalize mRNA levels and control for discrepancies in concentration readings given by spectrophotometer.
ACKNOWLEDGMENTS
We thank Dr. Ulrich Matern for providing us with a small amount of [14C]benzoyl CoA used in the initial characterization of BEBT and Drs. Till Beuerle and Yoko Sekiwa for the synthesis of [14C]benzoyl CoA and several other substrates used in this study.
Footnotes
This work was supported by the National Science Foundation (grant no. MCB–9974463), by Novartis Agribusiness Biotechnology Research, Inc., and by a National Institutes of Health training grant fellowship in genetics to J.C.D. (grant no. 5 T32 GM07544).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.006460.
LITERATURE CITED
- Aharoni A, Keizer LCP, Bouwmeester HJ, Sun ZK, Alvarez-Huerta M, Verhoeven HA, Blaas J, van Houwelingen A, De Vos RCH, van der Voet H et al. Identification of the SAAT gene involved in strawberry flavor biogenesis by use of DNA microarrays. Plant Cell. 2000;12:647–661. doi: 10.1105/tpc.12.5.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arimura G, Ozawa R, Horiuchi J, Nishioka T, Takabayashi J. Plant-plant interactions mediated by volatiles emitted from plants infested by spider mites. Biochem Syst Ecol. 2001;29:1049–1061. [Google Scholar]
- Beuerle T, Pichersky E. Purification and characterization of benzoate:coenzyme A ligase from Clarkia breweri. Arch Biochem Biophys. 2002a;400:258–264. doi: 10.1016/S0003-9861(02)00026-7. [DOI] [PubMed] [Google Scholar]
- Beuerle T, Pichersky E. Enzymatic synthesis and purification of aromatic coenzyme A esters. Anal Biochem. 2002b;302:305–312. doi: 10.1006/abio.2001.5574. [DOI] [PubMed] [Google Scholar]
- Bradford MM. Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Croft KPC, Juttner F, Slusarenko AJ. Volatile products of the lipoxygenase pathway evolved from Phaseolus vulgaris (L.) leaves inoculated with Pseudomonas syringae pv phaseolicola. Plant Physiol. 1993;101:13–24. doi: 10.1104/pp.101.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau R. Biosynthesis of benzaldehyde, benzyl alcohol, and benzyl benzoate from benzoic acid in cranberry. J Food Biochem. 1977;1:317–326. [Google Scholar]
- Czernic P, Huang HC, Marco Y. Characterization of hsr201 and hsr515, two tobacco genes preferentially expressed during the hypersensitive reaction provoked by phytopathogenic bacteria. Plant Mol Biol. 1996;31:255–265. doi: 10.1007/BF00021788. [DOI] [PubMed] [Google Scholar]
- De Moraes CM, Lewis WJ, Pare PW, Alborn HT, Tumlinson JH. Herbivore-infested plants selectively attract parasitoids. Nature. 1998;393:570–573. [Google Scholar]
- Deng WL, Hamiltonkemp TR, Nielsen MT, Andersen RA, Collins GB, Hildebrand DF. Effects of 6-carbon aldehydes and alcohols on bacterial proliferation. J Agric Food Chem. 1993;41:506–510. [Google Scholar]
- Dudareva N, Cseke L, Blanc VM, Pichersky E. Evolution of floral scent in Clarkia: novel patterns of S-linalool synthase gene expression in the C. breweri flower. Plant Cell. 1996;8:1137–1148. doi: 10.1105/tpc.8.7.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva N, D'Auria JC, Nam KH, Raguso RA, Pichersky E. Acetyl-CoA:benzylalcohol acetyltransferase: an enzyme involved in floral scent production in Clarkia breweri. Plant J. 1998a;14:297–304. doi: 10.1046/j.1365-313x.1998.00121.x. [DOI] [PubMed] [Google Scholar]
- Dudareva N, Pichersky E. Biochemical and molecular genetic aspects of floral scents. Plant Physiol. 2000;122:627–633. doi: 10.1104/pp.122.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva N, Raguso RA, Wang JH, Ross EJ, Pichersky E. Floral scent production in Clarkia breweri: III. Enzymatic synthesis and emission of benzenoid esters. Plant Physiol. 1998b;116:599–604. doi: 10.1104/pp.116.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara H, Tanaka Y, Fukui Y, Nakao M, Ashikari T, Kusumi T. Anthocyanin 5-aromatic acyltransferase from Gentiana triflora: purification, characterization and its role in anthocyanin biosynthesis. Eur J Biochem. 1997;249:45–51. doi: 10.1111/j.1432-1033.1997.t01-1-00045.x. [DOI] [PubMed] [Google Scholar]
- Fujiwara H, Tanaka Y, Yonekura-Sakakibara K, Fukuchi-Mizutani M, Nakao M, Fukui Y, Yamaguchi M, Ashikari T, Kusumi T. cDNA cloning, gene expression and subcellular localization of anthocyanin 5-aromatic acyltransferase from Gentiana triflora. Plant J. 1998;16:421–431. doi: 10.1046/j.1365-313x.1998.00312.x. [DOI] [PubMed] [Google Scholar]
- Gang DR, Wang JH, Dudareva N, Nam KH, Simon JE, Lewinsohn E, Pichersky E. An investigation of the storage and biosynthesis of phenylpropenes in sweet basil. Plant Physiol. 2001;125:539–555. doi: 10.1104/pp.125.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grothe T, Lenz R, Kutchan TM. Molecular characterization of the salutardinol 7-O-acetyltransferase involved in morphine biosynthesis in opium poppy Papaver somniferum. J Biol Chem. 2001;276:30717–30723. doi: 10.1074/jbc.M102688200. [DOI] [PubMed] [Google Scholar]
- Hamiltonkemp TR, McCracken CT, Loughrin JH, Andersen RA, Hildebrand DF. Effects of some natural volatile compounds on the pathogenic fungi Alternaria alternata and Botrytis cinerea. J Chem Ecol. 1992;18:1083–1091. doi: 10.1007/BF00980064. [DOI] [PubMed] [Google Scholar]
- Laflamme P, St-Pierre B, De Luca V. Molecular and biochemical analysis of a Madagascar periwinkle root-specific minovincinine-19-hydroxy-O-acetyltransferase. Plant Physiol. 2001;125:189–198. doi: 10.1104/pp.125.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HI, Leon J, Raskin I. Biosynthesis and metabolism of salicylic acid. Proc Natl Acad Sci USA. 1995;92:4076–4079. doi: 10.1073/pnas.92.10.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martz F, Maury S, Pincon G, Legrand M. cDNA cloning, substrate specificity and expression study of tobacco caffeoyl-CoA 3-O-methyl-transferase, a lignin biosynthetic enzyme. Plant Mol Biol. 1998;36:427–437. doi: 10.1023/a:1005969825070. [DOI] [PubMed] [Google Scholar]
- Mattiacci L, Rocca BA, Scascighini N, D'Alessandro M, Hern A, Dorn S. Systemically induced plant volatiles emitted at the time of “danger”. J Chem Ecol. 2001;27:2233–2252. doi: 10.1023/a:1012278804105. [DOI] [PubMed] [Google Scholar]
- Nam KH, Dudareva N, Pichersky E. Characterization of benzylalcohol acetyltransferases in scented and non-scented Clarkia species. Plant Cell Physiol. 1999;40:916–923. doi: 10.1093/oxfordjournals.pcp.a029623. [DOI] [PubMed] [Google Scholar]
- Ozawa R, Shimoda T, Kawaguchi M, Arimura G, Horiuchi J, Nishioka T, Takabayashi J. Lotus japonicus infested with herbivorous mites emits volatile compounds that attract predatory mites. J Plant Res. 2000;113:427–433. [Google Scholar]
- Pare PW, Tumlinson JH. Plant volatile signals in response to herbivore feeding. Fla Entomol. 1996;79:93–103. [Google Scholar]
- Pearce G, Marchand PA, Griswold J, Lewis NG, Ryan CA. Accumulation of feruloyltyramine and ρ-coumaroyltyramine in tomato leaves in response to wounding. Phytochemistry. 1998;47:659–664. [Google Scholar]
- Pichersky E, Raguso RA, Lewinsohn E, Croteau R. Flower scent production in Clarkia (Onagraceae): I. Localization and developmental modulation of monoterpenes emission and linalool synthase activity. Plant Physiol. 1994;106:1533–1540. doi: 10.1104/pp.106.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power R, Kurz WGW, Deluca V. Purification and characterization of acetyl coenzyme A:deacetylvindoline 4-O-acetyltransferase from Catharanthus roseus. Arch Biochem Biophys. 1990;279:370–376. doi: 10.1016/0003-9861(90)90504-r. [DOI] [PubMed] [Google Scholar]
- Raguso RA, Pichersky E. Floral volatiles from Clarkia breweri and C. Concinna (Onagraceae): recent evolution of floral scent and moth pollination. Plant System Evol. 1995;194:55–67. [Google Scholar]
- Ross JR, Nam KH, D'Auria JC, Pichersky E. S-Adenosyl-l-methionine:salicylic acid carboxyl methyltransferase, an enzyme involved in floral scent production and plant defense, represents a new class of plant methyltransferases. Arch Biochem Biophys. 1999;367:9–16. doi: 10.1006/abbi.1999.1255. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Seino Y, Suzuki Y, Sogawa K. An ovicidal substance produced by rice plants in response to oviposition by the whitebacked planthopper, Sogatella furcifera (Horvath) (Homoptera: Delphacidae) Appl Entomol Zool. 1996;31:467–473. [Google Scholar]
- Seo HS, Song JT, Cheong JJ, Lee YH, Lee YW, Hwang I, Lee JS, Choi YD. Jasmonic acid carboxyl methyltransferase: a key enzyme for jasmonate-regulated plant responses. Proc Natl Acad Sci USA. 2001;98:4788–4793. doi: 10.1073/pnas.081557298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre B, De Luca V. Evolution of acyltransferase genes: origin and diversification of the BAHD superfamily of acyltransferases involved in secondary metabolism. In: John RI, Romeo T, Varin L, De Luca V, editors. Recent Advances in Phytochemistry Evolution of Metabolic Pathways. Vol. 34. Oxford: Elsevier Science Publishing; 2000. pp. 285–315. [Google Scholar]
- St-Pierre B, Laflamme P, Alarco AM, De Luca V. The terminal O-acetyltransferase involved in vindoline biosynthesis defines a new class of proteins responsible for coenzyme A-dependent acyl transfer. Plant J. 1998;14:703–713. doi: 10.1046/j.1365-313x.1998.00174.x. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Nakayama T, Yonekura-Sakakibara K, Fukui Y, Nakamura N, Nakao M, Tanaka Y, Yamaguchi M, Kusumi T, Nishino T. Malonyl-CoA:anthocyanin 5-O-glucoside‴-O-malonyltransferase from scarlet sage (Salvia splendens) flowers. J Biol Chem. 2001;276:49013–49019. doi: 10.1074/jbc.M108444200. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TomasBarberan FA, LoaizaVelarde J, Bonfanti A, Saltveit ME. Early wound- and ethylene-induced changes in phenylpropanoid metabolism in harvested lettuce. J Am Soc Hortic Sci. 1997;122:399–404. [Google Scholar]
- Turlings TCJ, Tumlinson JH, Lewis WJ. Exploitation of herbivore-induced plant odors by host-seeking parasitic wasps. Science. 1990;250:1251–1253. doi: 10.1126/science.250.4985.1251. [DOI] [PubMed] [Google Scholar]
- Van Poecke RMP, Posthumus MA, Dicke M. Herbivore-induced volatile production by Arabidopsis thaliana leads to attraction of the parasitoid Cotesia rubecula: chemical, behavioral, and gene-expression analysis. J Chem Ecol. 2001;27:1911–1928. doi: 10.1023/a:1012213116515. [DOI] [PubMed] [Google Scholar]
- Walker K, Croteau R. Molecular cloning of a 10-deacetylbaccatin III-10-O-acetyl transferase cDNA from Taxus and functional expression in Escherichia coli. Proc Natl Acad Sci USA. 2000a;97:583–587. doi: 10.1073/pnas.97.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker K, Croteau R. Taxol biosynthesis: molecular cloning of a benzoyl-CoA:taxane 2α-O-benzoyltransferase cDNA from Taxus and functional expression in Escherichia coli. Proc Natl Acad Sci USA. 2000b;97:13591–13596. doi: 10.1073/pnas.250491997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker K, Croteau R. Taxol biosynthetic genes. Phytochemistry. 2001;58:1–7. doi: 10.1016/s0031-9422(01)00160-1. [DOI] [PubMed] [Google Scholar]
- Walker K, Schoendorf A, Croteau R. Molecular cloning of a taxa-4(20),11(12)-dien-5 α-ol-O-acetyl transferase cDNA from Taxus and functional expression in Escherichia coli. Arch Biochem Biophys. 2000;374:371–380. doi: 10.1006/abbi.1999.1609. [DOI] [PubMed] [Google Scholar]
- Walling LL. The myriad plant responses to herbivores. J Plant Growth Regul. 2000;19:195–216. doi: 10.1007/s003440000026. [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature. 2001;414:562–565. doi: 10.1038/35107108. [DOI] [PubMed] [Google Scholar]
- Yang Q, Reinhard K, Schiltz E, Matern U. Characterization and heterologous expression of hydroxycinnamoyl/benzoyl-CoA:anthranilate N-hydroxycinnamoyl/benzoyltransferase from elicited cell cultures of carnation, Dianthus caryophyllus L. Plant Mol Biol. 1997;35:777–789. doi: 10.1023/a:1005878622437. [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Tanaka Y, Fukuchi-Mizutani M, Fujiwara H, Fukui Y, Ashikari T, Murakami Y, Yamaguchi M, Kusumi T. Molecular and biochemical characterization of a novel hydroxycinnamoyl-CoA:anthocyanin 3-O-glucoside-6"-O-acyltransferase from Perilla frutescens. Plant Cell Physiol. 2000;41:495–502. doi: 10.1093/pcp/41.4.495. [DOI] [PubMed] [Google Scholar]