Abstract
Cytochrome P450 enzymes of the closely related CYP90 and CYP85 families catalyze essential oxidative reactions in the biosynthesis of brassinosteroid (BR) hormones. Arabidopsis CYP90B1/DWF4 and CYP90A1/CPD are responsible for respective C-22 and C-23 hydroxylation of the steroid side chain and CYP85A1 catalyzes C-6 oxidation of 6-deoxo intermediates, whereas the functions of CYP90C1/ROT3, CYP90D1, and CYP85A2 are still unknown. Semiquantitative reverse transcriptase-polymerase chain reaction analyses show that transcript levels of CYP85 and CYP90 genes are down-regulated by brassinolide, the end product of the BR biosynthesis pathway. Feedback control of the CYP90C1, CYP90D1, and CYP85A2 genes by brassinolide suggests that the corresponding enzymes might also participate in BR synthesis. CYP85 and CYP90 mRNAs show strong and transient accumulation during the 1st week of seedling development, as well as characteristic organ-specific distribution. Transcripts of CYP90A1 and CYP85A2 are preferentially represented in shoots and CYP90C1, CYP90D1, and CYP85A1 mRNAs are more abundant in roots, whereas CYP90B1 is ubiquitously expressed. Remarkably, the spatial pattern of CYP90A1 expression is maintained in the BR-insensitive cbb2 mutant, indicating the independence of organ-specific and BR-dependent regulation. Quantitative gas chromatography-mass spectrometry analysis of endogenous BRs in shoots and roots of Arabidopsis, pea (Pisum sativum), and tomato (Lycopersicon esculentum) reveal similar partitioning patterns of BR intermediates in these species. Inverse correlation between CYP90A1/CPD transcript levels and the amounts of the CYP90A1 substrate 6-deoxocathasterone in shoots and roots suggests that transcriptional regulation plays an important role in controlling BR biosynthesis.
Brassinosteroids (BRs) are plant steroid hormones that influence a wide range of important developmental processes, including germination, cell elongation, differentiation of vascular elements, photomorphogenesis, and pollen fertility (Clouse and Sasse, 1998; Steber and McCourt, 2001). The maintenance of optimal local BR concentrations by coordinated biosynthetic and inactivation mechanisms, in combination with the differential responsiveness of target cells and tissues, enables the proper regulation of these physiological functions during plant development. The pathways of BR biosynthesis have been elucidated by a series of detailed biochemical studies. Brassinolide (BL), the biologically most active BR, is synthesized from campesterol via either early or late C-6 oxidation routes (Fig. 1). The BR biosynthesis pathways are conserved between Catharanthus roseus, Arabidopsis, pea (Pisum sativum), tomato (Lycopersicon esculentum), and rice (Oryza sativa), although a limitation of early C-6 oxidation has been observed in some of these species (Fujioka and Sakurai, 1997; Fujioka et al., 2000; Noguchi et al., 2000; Nomura et al., 2001).
Figure 1.
The pathway of BR biosynthesis. Black arrows represent conversion steps with confirmed or assumed involvement of cytochrome P450 monooxygenases. Identified Arabidopsis P450 enzymes of the pathway are indicated. Numbering of the carbon positions oxidized in BRs is given at the structural formula of campesterol.
Molecular genetic analysis of BR-deficient mutants has identified several BR biosynthesis genes that, with the exception of DET2 (DEETIOLATED2; Li et al., 1996) were all found to encode cytochrome P450 monooxygenases (for review, see Bishop and Yokota, 2001). Arabidopsis DWF4 (DWARF4), a protein classified as CYP90B1 according to the international cytochrome P450 nomenclature (Nelson et al., 1996; http://drnelson.utmem.edu/CytochromeP450.html), was shown by BR intermediate feeding to catalyze C-22 hydroxylation of the steroid side chain (Choe et al., 1998). Likewise, rescue of the Arabidopsis cpd (constitutive photomorphogenesis and dwarfism) mutant revealed that CPD/CYP90A1, another member of the CYP90 family, functions as C-23 steroid hydroxylase (Szekeres et al., 1996). Rescue of the dwarf phenotypes of cpd and dwf4 mutants by BR intermediates indicates that CYP90A1 and CYP90B1 are responsible for the C-23 and C-22 side chain hydroxylation reactions in both the early and late C-6 oxidation pathways of BR biosynthesis. Mutation of the Arabidopsis ROT3 (ROTUNDIFOLIA3) gene, encoding CYP90C1, results in defective cell elongation and reduced leaf expansion. Because of the apparent lack of phenotypic rescue with externally supplied BRs, the role of CYP90C1 in BR biosynthesis remained unclear (Kim et al., 1998, 1999). Similarly, due to the lack of mutants, no function has been assigned for CYP90D1, the fourth Arabidopsis gene of the CYP90 family.
C-6 oxidation of BR intermediates is catalyzed by an enzyme of the CYP85 family (Bishop et al., 1996), as was demonstrated in vitro with yeast (Saccharomyces cerevisiae)-expressed CYP85A1 of both tomato (DWARF) and Arabidopsis (Bishop et al., 1999; Shimada et al., 2001). In these assays, CYP85A1 oxidized only the late biosynthetic intermediates 6-deoxotea-sterone, 3-dehydro-6-deoxoteasterone, 6-deoxotypha-sterol, and 6-deoxocastasterone, but did not catalyze the C-6 oxidation of campestanol, a substrate of CYP90B1 (Fig. 1). The function of CYP85A2, the second member of the CYP85 family in Arabidopsis, is so far unclear. Two further oxidative reactions, namely C-2 hydroxylation and the formation of BL by Bayer-Villiger lactonization of the steroid B ring, are also thought to be catalyzed by yet unidentified cytochrome P450 enzymes (Asami and Yoshida, 1999). Recently, Kang et al. (2001) have detected steroid C-2 hydroxylase activity of DDWF1 (dark-induced DWF-like protein 1), a pea P450 designated CYP92A6. Because the Arabidopsis genome does not encode any member of the CYP92 family, in this plant, the C-2 hydroxylation reaction is probably performed by a different cytochrome P450 enzyme.
The regulatory mechanisms of BR homeostasis are little understood. Noguchi et al. (1999, 2000) observed the accumulation of BL and its precursors, as well as up-regulation of the DWF4 and CPD transcripts, in the BR-insensitive bri1 mutant of Arabidopsis, suggesting a role for BRI1 in the regulation of BR biosynthesis. Furthermore, BL treatment of Arabidopsis seedlings markedly decreased the steady-state level of CPD mRNA, and this transcriptional response was shown to require de novo protein synthesis (Mathur et al., 1998). These results suggest that BR synthesis is controlled by an elaborate feedback regulation, one that shows analogy to the negative control of GA biosynthesis genes by GAs (Yamaguchi and Kamiya, 2000).
The cellular concentration of active BRs is also influenced by the catabolism of BL and/or its precursors. In feeding experiments, the activation-tagged Arabidopsis bas1-D mutant overexpressing BAS1/CYP72B1 was found to accumulate biologically inactive 26-hydroxybrassinolide (Neff et al., 1999). The dwarf phenotypes of bas1-D and chibi2, another activation-tagged Arabidopsis mutant with high CYP72C1 level (Nagatani et al., 1998; Bishop and Yokota, 2001), are very similar to those of the BR-deficient mutants. As compared with the wild type, the BR-deficient and -insensitive Arabidopsis mutants contain diminished BAS1 transcript levels, indicating that BL may induce expression of the corresponding catabolic enzyme (Choe et al., 2001).
So far, only limited information is available about the temporal and spatial control of the genes responsible for BR biosynthesis. Strong CPD expression was detected during the 1st week of seedling development, and in transgenic plants, a CPD promoter-driven GUS reporter fusion showed activity in cotyledons, leaves, and floral organs, but not in roots (Mathur et al., 1998). Similar GUS histochemical (G.J. Bishop, unpublished data) and in situ hybridization assays (Pien et al., 2001) revealed that tomato DWARF promoter activity is localized mainly in the apical and root meristem regions, whereas ROT3 is expressed in all organs and cell types of Arabidopsis seedlings (Kim et al., 1999). Thus, further studies are required to elucidate how and to what extent differential expression of particular BR biosynthesis genes affects active hormone levels and intermediate partitioning during plant development.
In this paper, we report that genes of the closely related CYP85 and CYP90 cytochrome P450 families implicated in BR biosynthesis are coordinately regulated by BL. Feedback control of the genes encoding ROT3/CYP90C1 and CYP90D1 suggests that these enzymes may also be involved in BR synthesis. Although all CYP85 and CYP90 genes are strongly expressed during the 1st week of seedling development, their transcripts have characteristically different accumulation patterns in the shoots and roots of seedlings and fully developed plants. The expression level of the CPD/CYP90A1 gene shows correlation with the spatial partitioning of the CYP90A1 substrate 6-deoxocatha-sterone, suggesting that transcriptional control of the CYP85 and CYP90 genes can contribute to the regulation of BR biosynthesis.
RESULTS
Cytochrome P450 Monooxygenases in BR Biosynthesis Are Evolutionarily Related
All cytochrome P450 enzymes of Arabidopsis with known function in BR biosynthesis belong to either the CYP85 or CYP90 families. Protein sequence comparison based on BLAST homology analysis (Altschul et al., 1990) revealed that these two P450 families are closely related, sharing approximately 35% amino acid sequence identity. We found that the CYP85 and CYP90 proteins also show high levels (about 30%) of sequence identity with ent-kaurenoic acid oxidases, members of the CYP88 P450 family involved in GA biosynthesis (Helliwell et al., 2001). In contrast, the two Arabidopsis CYP72 hydroxylases responsible for BR inactivation are only distantly related to the P450s of BR biosynthesis, featuring less than 20% sequence identity with any member of the CYP90 and 85 families. A phylogenetic tree generated by the ClustalW multiple alignment program (Thompson et al., 1994) shows the close relationship between BR-biosynthetic and CYP88 P450s, as compared with CYP72 proteins, in Arabidopsis (Fig. 2A).
Figure 2.
Structural relationship between selected Arabidopsis cytochrome P450 proteins and their genes. A, Unrooted cladogram based on the primary structure of P450 families involved in BR biosynthesis (CYP85 and CYP90), BR catabolism (CYP72), and GA biosynthesis (CYP88). Amino acid identity values, as compared with CPD/CYP90A1, are given in brackets. B, Exon/intron structure of the genes encoding CYP85A1 (AB009048), CYP85A2 (AP002060), CPD/CYP90A1 (X87367), DWF4/CYP90B1 (AL132979), ROT3/CYP90C1 (Z99708), CYP90D1 (AP001307), CYP88A3 (AC000098), CYP88A4 (AC005700), BAS1/CYP72B1 (AC003105), and CHIBI2/CYP72C1 (AC007651). Exon sizes are given in bp.
The analysis of exon-intron organization of the same P450 genes uncovered similar relationships (Fig. 2B). Each intron of the CYP85, CYP90 and CYP88 genes was found at one of eight conserved positions, whereas CYP72B1 and CYP72C1 showed a different exon-intron pattern. The close relationship indicated by both protein and gene structure analyses, together with the similarity of enzymatic functions, suggest that during their evolution, the CYP85 and CYP90 families diverged after their specialization to steroid substrates.
Coordinated Feedback Regulation of CYP85 and CYP90 Genes
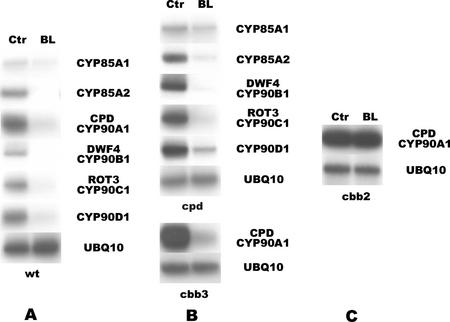
It was demonstrated previously that transcription of the CPD gene is down-regulated by BL, the end product of BR biosynthesis (Mathur et al., 1998). Therefore, we were interested in determining whether the transcript levels of other CYP90 or CYP85 transcripts are similarly regulated by this phytohormone. Because of the low abundance of these P450 mRNAs, in these experiments, the steady-state transcript levels were monitored by more sensitive semiquantitative RT-PCR, rather than northern hybridization. BL treatment reduced the amount of CPD/CYP90A1, DWF4/CYP90B1, ROT3/CYP90C1, CYP90D1, CYP85A1, and CYP85A2 transcripts to approximately 10% or less of the level detected in untreated control seedlings (Fig. 3A). These data show that in Arabidopsis all CYP85 and CYP90 gene activities are controlled by BR-dependent feedback regulation.
Figure 3.
Effect of BL on the steady-state mRNA levels of BR-biosynthetic P450s. Reverse transcriptase (RT)-PCR products obtained from total RNA of 7-d-old seedlings incubated for 4 h in the presence (BL) or absence (Ctr) of 100 nm BL. A, Wild type; B, BR-deficient cpd and cbb3 mutants; C, BR-insensitive cbb2 mutant. UBQ10 was used as internal control.
To confirm our results, we also analyzed the relative amounts of CYP85 and CYP90 transcripts in BL-treated and untreated Arabidopsis mutants impaired in BR biosynthesis or perception. Compared with wild-type plants, the mRNA levels of CYP85 and all four CYP90 genes were significantly higher, indicating derepressed expression, in the BR-deficient cpd and cbb3 mutants (cbb3 being allelic to cpd; Kauschmann et al., 1996). The amounts of CYP85 and CYP90 transcripts were reduced in these mutants upon external application of BL, but remained somewhat higher than in BL-treated wild-type plants (Fig. 3B). In the BR-insensitive cbb2 mutant (Kauschmann et al., 1996), however, BL had no effect on the expression of CPD (Fig. 3C) or any other BR-responsive CYP85 or CYP90 genes (data not shown). This result indicates that BR-mediated feedback regulation of the CYP85 and CYP90 genes is dependent on the function of the BRI1 Leu-rich repeat receptor kinase (Li and Chory, 1997), which has been inactivated in the cbb2 mutant.
Regulation of CYP85 and CYP90 mRNA Levels during Germination and Seedling Development
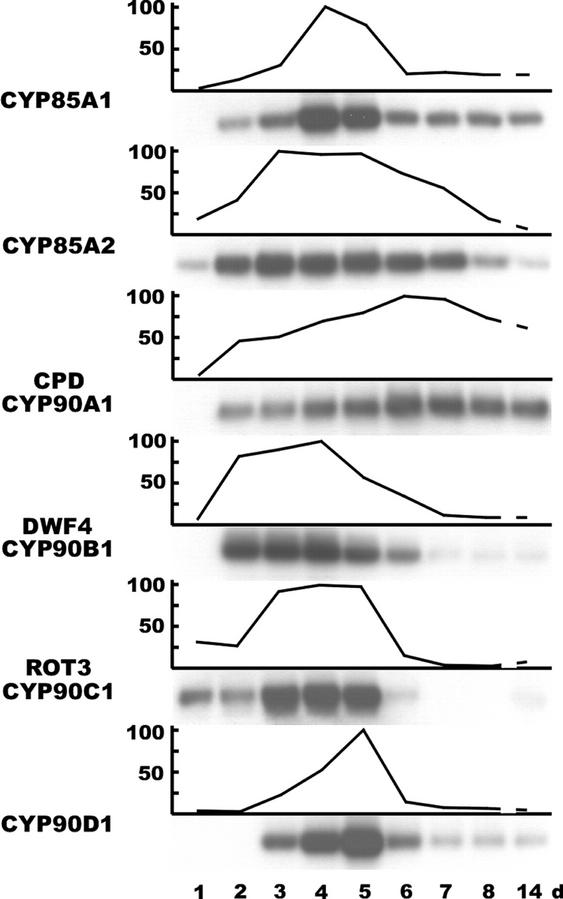
To gain better insight into the regulation of BR-biosynthetic P450 genes during the early stages of plant development, we determined the relative amounts of CYP85 and CYP90 transcripts by RT-PCR in seedlings and young plants throughout the first 8 d after imbibition and after 2 weeks of development (Fig. 4). At the earliest, transcripts of the CYP85A2 and ROT3 genes were already detectable from the 1st d of germination. Each CYP85 and CYP90 mRNA reached a peak level of abundance during the 1st week of seedling development but, with the exception of CPD, their levels declined to about 10% or less of the maximum values by the end of this period. Subsequently, between d 8 and 14 of the time course, only little or no change was detectable in the transcript levels. Although individual CYP85 and CYP90 genes featured different temporal expression profiles, their transient induction during the 1st week after germination suggests the requirement of BR biosynthesis enzymes during the early stages of seedling development.
Figure 4.
Changes in transcript levels of CYP85 and CYP90 genes during germination and seedling development. RT-PCR products prepared from total RNA of developing wild-type seedlings and young plants (1 through 8 and 14 d after imbibition). Quantitative data are plotted as percentage of the highest value measured during the experimental period.
Differential Regulation of CYP85 and CYP90 Transcript Levels in Shoots and Roots
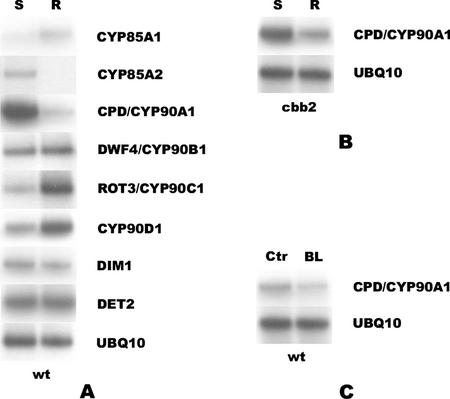
Previously, we reported that CPD expression is much stronger in the aerial parts than the roots of Arabidopsis (Mathur et al., 1998). To obtain information on the organ-specific regulation of CYP85 and CYP90 genes, their transcript levels were compared by RT-PCR in roots and shoots (representing combined cotyledon and hypocotyl tissues) of 7-d-old seedlings. CPD and CYP85A2 were found preferentially expressed in cotyledons and hypocotyls, whereas the expression of CYP85A1, ROT3, and CYP90D1 was stronger in roots (Fig. 5A). Intriguingly, the highly homologous CYP85A1 and CYP85A2 genes exhibited different spatial expression patterns, whereas the closely related ROT3 and CYP90D1 displayed similar ones. We also used RT-PCR assays to determine the expression of the DET2 and DIM1 genes that encode non-P450-type enzymes (Li et al., 1996; Klahre et al., 1998) acting upstream of CYP85 and CYP90 monooxygenases in BR synthesis. In contrast to most CYP85 and CYP90 messages, the DIM1 and DET2 transcripts were equally abundant in the shoots and roots of the seedlings.
Figure 5.
Differential accumulation of BR-biosynthetic P450 mRNAs in shoots and roots. A, Transcript levels in shoots (S) and roots (R) of wild-type seedlings. B, CPD/CYP90A1 transcript levels in shoots (S) and roots (R) of BR-insensitive cbb2 seedlings. C, CPD/CYP90A1 transcript levels in roots (R) of wild-type seedlings incubated for 4 h in the presence (BL) or absence (Ctr) of 100 nm BL. RT-PCR products were obtained from total RNA of 7-d-old seedlings. UBQ10 was used as internal control.
The shoot to root ratios of mRNA levels in 7- and 20-d-old Arabidopsis plants are shown in Table I. These values indicate a preferential accumulation of CYP85A2 and CPD transcripts in the shoots of 7-d-old seedlings. The data obtained from 20-d-old plants reflect the same distribution pattern as those of the young seedlings but, probably due to the lower activity of CYP85 and CYP90 genes in older plants, less pronounced organ-specific differences in CYP85A1, CYP85A2, and CPD expression were detected.
Table I.
Shoot to root ratio of transcript levels of genes involved in BR biosynthesis
| Transcript | 7-d-Old Arabidopsis | 20-d-Old Arabidopsis |
|---|---|---|
| CYP85A1 | 0.26 | 0.60 |
| CYP85A2 | 12.12 | 8.66 |
| CYP90A1 | 8.33 | 3.23 |
| CYP90B1 | 0.95 | 1.09 |
| CYP90C1 | 0.31 | 0.32 |
| CYP90D1 | 0.32 | 0.28 |
| DIM1 | 1.02 | 0.98 |
| DET2 | 0.90 | 0.95 |
To clarify whether BR regulation is required for organ-specific accumulation, we also assayed levels of the CPD transcript in shoots and roots of BR-insensitive cbb2 seedlings. As it is shown in Figure 5B, the difference between the amounts of the CPD transcript in shoots and roots was found similar to that observed in the wild type (Fig. 5A). We also found that the low CPD transcript level in the roots of wild-type seedlings could be further decreased by BL treatment (Fig. 5C). These data indicate that the organ-specific control of CPD expression acts independently from the hormonal feedback regulation.
Endogenous BR Levels in Shoots and Roots of Arabidopsis, Pea, and Tomato
Differential expression of CYP85 and CYP90 genes in the roots and aerial parts of the plant might influence BR biosynthesis and BR levels. To see if this was the case, we determined the amounts of endogenous BRs in the shoots and roots of Arabidopsis using quantitative gas chromatography (GC)-mass spectrometry analysis. In addition, similar analyses were performed on roots and shoots of pea and tomato to ascertain the conservation of organ-specific BR distribution. The data revealed a differential accumulation of BR biosynthesis intermediates in the aerial and underground parts of these plants (Table II). Despite the varying levels of particular BR forms in the three species, the pattern of their organ-specific partitioning was found to be very similar. The early intermediates 6-deoxocathasterone, 6-deoxoteasterone, 3-dehydro-6-deoxoteasterone, and 6-deoxotyphasterol were preferentially represented in the roots, whereas 6-deoxocastasterone and castasterone, synthesized later in the pathway, were more abundant in the shoots. The intermediates of the early C-6 oxidation pathway were at or below the detection level in all samples, whereas BL could only be observed in the Arabidopsis samples and the roots of pea. In Arabidopsis, the amount of 6-deoxocathasterone was more than 2-fold higher in the roots, which were shown to contain low CPD transcript level (Table II; Fig. 5A). Because CPD/CYP90A1 catalyzes the conversion of 6-deoxocathasterone to 6-deoxoteasterone, the accumulation of its substrate in the root indicates a low conversion rate in this organ and, hence, a good correlation between transcript abundance and the actual enzyme activity.
Table II.
Endogenous BR levels in shoots and roots of Arabidopsis, pea, and tomato
| BR | BR Level
|
|||||
|---|---|---|---|---|---|---|
| 20-d-Old Arabidopsis
|
15-d-Old pea
|
36-d-Old tomato
|
||||
| Shoots | Roots | Shoots | Roots | Shoots | Roots | |
| ng kg−1 fresh wt | ||||||
| 6-Deoxocathasterone | 790 | 1,800 | 670 | 5,100 | 640 | 2,800 |
| 6-Deoxoteasterone | 100 | 170 | 85 | 330 | 150 | 120 |
| 3-Dehydro-6-deoxoteasterone | 180 | 320 | 73 | 670 | 16 | 62 |
| 6-Deoxotyphasterol | 540 | 970 | 1,700 | 4,300 | 250 | 480 |
| 6-Deoxocastasterone | 220 | 90 | 11,700 | 630 | 330 | 180 |
| Cathasterone | NDa | ND | ND | ND | ND | ND |
| Teasterone | ND | ND | ND | 2 | ND | ND |
| 3-Dehydroteasterone | ND | ND | ND | ND | ND | ND |
| Typhasterol | ND | ND | ND | ND | ND | ND |
| Castasterone | 150 | 35 | 690 | 38 | 140 | 11 |
| Brassinolide | Trace | Trace | ND | 24 | ND | ND |
ND, Not detected.
DISCUSSION
Ensuring optimal physiological levels of active BRs requires a sensitive regulation of their biosynthesis. A recent comparative analysis of endogenous BR levels in Arabidopsis, pea, and tomato suggested similar control mechanisms in these species and indicated that C-22 and C-23 hydroxylation and C-6 oxidation are likely rate-limiting reactions of the pathway (Nomura et al., 2001). All of these oxidative steps are catalyzed by cytochrome P450-type enzymes of the CYP90 and CYP85 families; therefore, the levels of these enzymes are expected to influence the efficiency of BR synthesis.
Previously, we reported that the activity of Arabidopsis CPD, encoding the C-23-hydroxylase CYP90A1, is negatively controlled by BRs at the transcriptional level (Mathur et al., 1998). Our present data show that DWF4, coding for the C-22 steroid side chain hydroxylase CYP90B1 acting immediately upstream of CPD, is also down-regulated by BL. In the pathway of GA synthesis, similar transcriptional feedback mechanisms have been identified and shown to modulate the expression of GA 20-oxidase and GA 3β-hydroxylase genes (Yamaguchi and Kamiya, 2000). We found that in Arabidopsis, in addition to CPD and DWF4, the remaining CYP90 and CYP85 genes are also subject to feedback regulation by BL. Considering the highly similar primary structure of CYP85 and CYP90 proteins and that all of them with identified enzymatic functions participate in BR biosynthesis (Bishop and Yokota, 2001; Shimada et al., 2001), the BR-repressible expression of the CYP85A2, ROT3, and CYP90D1 genes strongly suggests a role for their P450 products in BR biosynthesis.
CYP85A2, sharing 82% amino acid sequence identity with CYP85A1, may represent a second Arabidopsis enzyme with steroid C-6 oxidase activity. At least a partial redundancy of this function in Arabidopsis is suggested by the lack of BR-deficient dwarf mutants defective in CYP85A1. Because P450 monooxygenases of the BR pathway are known to accept multiple substrates (Choe et al., 2001; Shimada et al., 2001), these two enzymes may also differ in their substrate preferences. In vivo feeding experiments using radiolabeled precursors revealed an early and a late C-6 oxidation of campestanol and 6-deoxocasta-sterone, respectively (Choi et al., 1997), whereas less efficient conversion of 6-deoxotyphasterol to typhasterol was also demonstrated (Noguchi et al., 2000). In yeast expression systems, CYP85A1 of both tomato (DWARF) and Arabidopsis was shown to oxidize 6-deoxocastasterone. The Arabidopsis enzyme also utilized the upstream intermediates 6-deoxoteas-terone, 3-dehydro-6-deoxoteasterone, and 6-deoxoty-phasterol, but not campestanol and 6-deoxocathaste-rone (Bishop et al., 1999; Shimada et al., 2001). If the lack of campestanol conversion was not due to its limited uptake by the yeast cells, this early C-6 oxidation would require the action of another enzyme for which CYP85A2 is a likely candidate.
The possible role(s) of the closely related (53% amino acid identity) ROT3/CYP90C1 and CYP90D1 proteins in BR synthesis is unclear. The rot3 mutant phenotype (Kim et al., 1998) is much weaker than those of the other BR biosynthesis mutants, which may indicate that CYP90C1 has overlapping function with another enzyme, possibly CYP90D1. In Arabidopsis, there are two potentially P450-mediated reactions in BR synthesis for which the genes have not yet been identified, namely the C-2 hydroxylation reaction and the Bayer-Villiger lactonization step converting castasterone to BL (Asami and Yoshida, 1999). Therefore, it seems conceivable that CYP90C1 and/or CYP90D1 might participate in one of these enzymatic reactions.
Transcript levels of the CYP85 and CYP90 genes were found to change in a wide range, from about 10% of the wild-type amount in BL-treated plants to 5 times the wild-type value in BR-deficient mutants. Thus, under normal developmental conditions, BR biosynthesis can be efficiently controlled through feedback regulation of these genes because their expression is partially repressed at physiological BR concentrations. The similarity of BR response suggests that the activity of CYP85 and CYP90 genes might be controlled by the same transcriptional regulators that modulate CPD expression (Mathur et al., 1998). The BR response of CPD, and probably all other feedback-controlled CYP85 and CYP90 genes, requires an intact BR perception mechanism. Down-regulation of CPD was abolished in mutants deficient in the BRI1 BR receptor function, just as in bin2, another BR-insensitive mutant (Li et al., 2001).
Differential organ-specific expression of the CYP85 and CYP90 genes may provide another means of controlling BR biosynthesis. This regulation appears to be independent of BR action because: (a) BR insensitivity does not interfere with shoot-specific accumulation of the CPD transcript, (b) low root levels of this mRNA further decrease upon BL treatment, and (c) CYP85 and CYP90 genes, displaying similar steroid responses, show different preferences for shoot- and root-specific expression. Transcripts of CYP85A1 were detected primarily in the roots, whereas those of CYP85A2 accumulated preferentially in the shoots. The differential organ specificity can be seen as further indication for the different functions of the two Arabidopsis CYP85 enzymes. In a recent microarray-based transcript analysis of 142 Arabidopsis cytochrome P450 genes, Xu et al. (2002) have shown that both CYP90A1 and CYP85A1 are preferentially expressed in the aerial portion of 30-d-old plants. In the case of the CYP85A1 transcript, the difference between the shoot versus root ratio detected by these authors and our organ specificity data is likely caused by hybridization of the array probe with both the CYP85A1 and CYP85A2 mRNAs. These transcripts share 82% sequence homology, which is above the claimed 70% distinction limit of these microarray assays (Xu et al., 2002). In contrast to genes of the CYP85 and CYP90 families, DIM1 and DET2, encoding enzymes acting farther upstream in the BR pathway were found to be ubiquitously expressed.
In addition to the differences in transcript levels, we have also detected differential distribution of BR biosynthesis intermediates between the aerial and underground organs. We found that in Arabidopsis 6-deoxotyphasterol and earlier precursors were more abundant in the roots, whereas the level of 6-deoxocastasterone and castasterone was higher in the shoots. A similar pattern of organ-specific intermediate accumulation could be observed in pea and tomato, indicating that analogous mechanisms may regulate BR distribution in these plants. In accordance with these findings, C-27 BRs (i.e. 28-norca-stasterone and its precursors) have also been shown recently to be differentially partitioned between the shoots and roots of tomato (Yokota et al., 2001). The potential significance of higher early intermediate levels in roots and the accumulation of 6-deoxo-castasterone and castasterone in shoots is unclear, but worthy of further investigation. The distribution of CYP85 and CYP90 transcripts suggests that roots actively participate in BR synthesis. Because in several plant species root development is inhibited at sub-nanomolar BR concentrations (Clouse and Sasse, 1998), suppression of metabolic flow to biologically active BR forms might help to maintain the low hormone level in this organ. With the sensitivity of our GC-selected ion monitoring analysis, BL could only be detected in Arabidopsis and pea, but not in tomato, where castasterone is thought to be the only active BR (Yokota et al., 1997; Nomura et al., 2001). In Arabidopsis roots, the low level of CPD expression was found to coincide with the accumulation of 6-deoxocathasterone, the substrate of CPD/CYP90A1. This seems to indicate a role for transcriptional regulation in determining the abundance and activity of CYP90A1, and perhaps also other P450 enzymes of the BR pathway.
Considering the importance of BRs in regulating early developmental functions, high-level expression of the BR-biosynthetic CYP85 and CYP90 genes in germinating seeds and young seedlings implies that, in addition to BL accumulation in the seeds (Fujioka et al., 1998), efficient de novo synthesis might be required for ensuring the optimal hormone concentration. Although in the whole plant, the activity of these genes declines after the seedling stage, strong expression may be maintained in differentiating regions, as it was shown in the case of CPD (Mathur et al., 1998). Transcriptional activity of the genes involved in BR metabolism is controlled by multiple physiological factors. High level of active hormone results in the repression of biosynthetic P450 genes and the induction of BAS1 responsible for BR catabolism (Choe et al., 2001). In addition to their feedback regulation, the activities of CYP85 and CYP90 genes are also subject to organ-specific and developmental control. Furthermore, a recent DNA microarray analysis revealed that the expression of several genes required for the synthesis of early sterol intermediates in the BR pathway are down-regulated by light (Ma et al., 2001). Therefore, it is reasonable to believe that these transcriptional mechanisms are crucial for adjusting the optimal levels and maintaining the homeostasis of active BRs.
MATERIALS AND METHODS
Plant Material and Growth Conditions
In vitro cultures of wild-type Arabidopsis (ecotype Columbia-0), the BR-deficient cpd (Szekeres et al., 1996), cbb3, and BR-insensitive cbb2 (Kauschmann et al., 1996) mutants were grown from surface-sterilized seeds on Murashige and Skoog medium supplemented with 0.5% (w/v) Suc and 0.2% (w/v) Phytagel (Sigma, St. Louis) at 22°C, under 14-h-light/10-h-dark cycles. BR treatments were carried out in the same Murashige and Skoog liquid medium supplemented with 100 nm BL (CIDtech Research Inc., Missisauga, ON, Canada), whereas hormone-free control samples received only the ethanol carried in with the BL stock solution (0.01% [v/v]). The Arabidopsis plants used for organ-specific mRNA and BR analyses were grown under similar conditions in Gamborg's B5 liquid medium (Gamborg et al., 1968) with continuous shaking at 50 rpm. Seeds of pea (Pisum sativum L. cv Torsdag) and tomato (Lycopersicon esculentum Mill. cv Sekaiichi) were sown in vermiculite and the germinated plants grown in the greenhouse under natural light (13-h day, 11-h night). Five-day-old pea seedlings and 22-d-old tomato plants were then grown hydroponically in the same greenhouse using Tadano and Tanaka (1980) liquid medium for further 10 and 14 d, respectively.
Analysis of Steady-State Transcript Levels
Steady-state mRNA levels were analyzed by semiquantitative and quantitative RT-PCR assays according to Chelly and Kahn (1994) with minor modifications. Total RNA was isolated from 1 g of fresh plant material using TRI Reagent (Sigma). After treatment with RNase-free DNaseI, cDNA was prepared from 5 μg of RNA with Ready-To-Go T-Primed First-Strand Kit (Pharmacia Biotech, Piscataway, NJ). One-tenth of the cDNA obtained was PCR amplified within the linear range of accuracy by specific primers spanning 250- to 300-bp regions near the 3′ ends of the translated sequences. One percent of the RT-PCR products was labeled with [α-32P]dCTP using a single detection primer that was three nucleotides longer in the 3′ direction than one of the amplification primers. Signal intensities were detected by autoradiography after size separation on a 2% (w/v) agarose gel and quantitatively evaluated using a PhosphorImager 445 SI (Molecular Dynamics Inc., Sunnyvale, CA). The constitutively expressed UBQ10 mRNA (Sun and Callis, 1997) was used as internal control. The cDNA-specific PCR primers used are given in Table III. The number of amplification cycles was 15 for UBQ10, 20 for CPD, DIM1 and DET2, and 25 for CYP85A1, CYP85A2, ROT3, and CYP90D1.
Table III.
Primers used in the RT-PCR analyses
| Transcript | Accession No. | Nucleotide Sequence (from 5′ to 3′) |
|---|---|---|
| CYP85A1 | AB009048 | GGACGTGAAGTCAATGAAGTTCACT |
| TTCCTTACCAGGACAAAGCCTTGTC | ||
| CYP85A2 | AP002060 | CGAACCGCTCACTCTCGACGAT |
| AATAGCTCTTTGATTCTAAGCTCT | ||
| CPD/CYP90A1 | X87367 | GAATGGAGTGATTACAAGTC |
| GTGAACACATTAGAAGGGCCTG | ||
| DWF4/CYP90B1 | AL132979 | GAAGGAACTAGGAGAGTCAG |
| CCACGTCGAAAAACTACCACTTC | ||
| ROT3/CYP90C1 | Z99708 | GGAGATGAAGAGGCGTAAATTGGA |
| GCAAATACTGCTGTTTGCCGATCC | ||
| CYP90D1 | AP001307 | GTCAAATTCCTCTCTGATTCTCCTG |
| TCGAGACCAGGGCACAATCTCTGAC | ||
| DIM1 | AB025631 | CTCGAATGGGTCCACCGCGAAATG |
| CATACAATTCACCATTAAACATTC | ||
| DET2 | AC007661 | AATCTCCTCAATGGTTATATC |
| CGTGTACAGAAAAAATCCAATACC | ||
| UBQ10 | AL161503 | GGACCAGCAGCGTCTCATCTTCGCT |
| CTTATTCATCAGGGATTATACAAG |
Quantitative Determination of Endogenous BR Levels
Twenty-day-old Arabidopsis, 15-d-old pea (n = 192), and 36-d-old tomato (n = 147) plants were separated into shoots (130, 289, and 241 g fresh weight, respectively) and roots (115, 293, and 78 g fresh weight, respectively). BR extraction and analysis were carried out as has been described by Nomura et al. (2001). In brief, methanol extracts of these tissues were subjected to solvent partitioning and purified by LH-20 chromatography and then reversed phase HPLC. Before LH-20 chromatography, charcoal chromatography was applied to the Arabidopsis shoot extracts; silica gel chromatography was applied to the extracts of pea shoots, pea roots, and tomato roots; and both silica gel and charcoal chromatography were applied to the tomato shoot extract. Quantitative analyses of BRs were conducted by GC-mass spectrometry/selected ion monitoring, using a JMS AX 505W instrument (JEOL, Tokyo).
ACKNOWLEDGMENTS
We are thankful to Thomas Altmann (Max-Planck-Institut für Molekulare Pflanzenphysiologie, Golm, Germany) for seeds of the cbb2 and cbb3 mutants, and Suguru Takatsuto (Joetsu University, Joetsu-shi, Japan) for providing the deuterium-labeled BR standards.
Footnotes
This work was supported by the Hungarian National Research Foundation (Országos Tudományos Kutatás: Alap [OTKA], grant no. T 32432), by the Human Frontiers Science Program (grant no. RG00162–2000 to G.J.B, C.K., and T.Y.), by the Japan Society for the Promotion of Science (Grand-in-Aid for Scientific Research no. 11460057 to T.Y and postdoctoral fellowship to T.N.), by scientific exchange programs between the Deutsche Forschungsgemeinschaft and the Hungarian Academy of Sciences (project nos. 436–UNG-113/143 and D–132), by scientific exchange programs between the Deutsches Zentrum für Luft und Raumfahrt e.V. and the Hungarian Science and Technology Foundation (project nos. UNG–027–99 and D–7/99), and by the British Council (support to G.J.B.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.005439.
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Asami T, Yoshida S. Brassinosteroid biosynthesis inhibitors. Trends Plant Sci. 1999;4:348–353. doi: 10.1016/s1360-1385(99)01456-9. [DOI] [PubMed] [Google Scholar]
- Bishop GJ, Harrison K, Jones JDG. The tomato dwarf gene isolated by heterologous transposon tagging encodes the first member of a new cytochrome P450 family. Plant Cell. 1996;8:959–969. doi: 10.1105/tpc.8.6.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop GJ, Nomura T, Yokota T, Harrison K, Noguchi T, Fujioka S, Takatsuto S, Jones JDG, Kamiya Y. The tomato DWARF enzyme catalyses C-6 oxidation in brassinosteroid biosynthesis. Proc Natl Acad Sci USA. 1999;96:1761–1766. doi: 10.1073/pnas.96.4.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop GJ, Yokota T. Plants steroid hormones, brassinosteroids: current highlights of molecular aspects on their synthesis/metabolism, transport, perception and response. Plant Cell Physiol. 2001;42:114–120. doi: 10.1093/pcp/pce018. [DOI] [PubMed] [Google Scholar]
- Chelly J, Kahn A. RT-PCR and mRNA quantification. In: Mullis KB, Ferré F, Gibbs RA, editors. The Polymerase Chain Reaction. Boston: Birkhäuser; 1994. pp. 97–109. [Google Scholar]
- Choe S, Fujioka S, Noguchi T, Takatsuto S, Yoshida S, Feldmann KA. Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J. 2001;26:573–582. doi: 10.1046/j.1365-313x.2001.01055.x. [DOI] [PubMed] [Google Scholar]
- Choe SW, Dilkes BP, Fujioka S, Takatsuto S, Sakurai A, Feldmann KA. The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22α-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell. 1998;10:231–243. doi: 10.1105/tpc.10.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YH, Fujioka S, Nomura T, Harada A, Yokota T, Takatsuto S, Sakurai A. An alternative brassinolide biosynthetic pathway via late C-6 oxidation. Phytochemistry. 1997;44:609–613. [Google Scholar]
- Clouse SD, Sasse JM. Brassinosteroids: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:427–451. doi: 10.1146/annurev.arplant.49.1.427. [DOI] [PubMed] [Google Scholar]
- Fujioka S, Noguchi T, Watanabe T, Takatsuto S, Yoshida S. Biosynthesis of brassinosteroids in cultured cells of Catharanthus roseus. Phytochemistry. 2000;53:549–553. doi: 10.1016/s0031-9422(99)00582-8. [DOI] [PubMed] [Google Scholar]
- Fujioka S, Noguchi T, Yokota T, Takatsuto S, Yoshida S. Brassinosteroids in Arabidopsis thaliana. Phytochemistry. 1998;48:595–599. doi: 10.1016/s0031-9422(98)00065-x. [DOI] [PubMed] [Google Scholar]
- Fujioka S, Sakurai A. Biosynthesis and metabolism of brassinosteroids. Physiol Plant. 1997;100:710–715. [Google Scholar]
- Gamborg OL, Miller RA, Ojima K. Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res. 1968;50:151–158. doi: 10.1016/0014-4827(68)90403-5. [DOI] [PubMed] [Google Scholar]
- Helliwell CA, Chandler PM, Poole A, Dennis ES, Peacock WJ. The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc Natl Acad Sci USA. 2001;98:2065–2070. doi: 10.1073/pnas.041588998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J-G, Yun J, Kim D-H, Chung K-S, Fujioka S, Kim J-I, Dae H-W, Yoshida S, Takatsuto S, Song P-S et al. Light and brassinosteroid signals are integrated via a dark-induced small G protein in etiolated seedling growth. Cell. 2001;105:625–636. doi: 10.1016/s0092-8674(01)00370-1. [DOI] [PubMed] [Google Scholar]
- Kauschmann A, Jessop A, Koncz C, Szekeres M, Willmitzer L, Altmann T. Genetic evidence for an essential role of brassinosteroids in plant development. Plant J. 1996;9:701–713. [Google Scholar]
- Kim G-T, Tsukaya H, Saito Y, Uchimiya H. Changes in the shapes of leaves and flowers upon overexpression of cytochrome P450 in Arabidopsis. Proc Natl Acad Sci USA. 1999;96:9433–9437. doi: 10.1073/pnas.96.16.9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G-T, Tsukaya H, Uchimiya H. The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes Dev. 1998;12:2381–2391. doi: 10.1101/gad.12.15.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahre U, Noguchi T, Fujioka S, Takatsuto S, Yokota T, Nomura T, Yoshida S, Chua N-H. The Arabidopsis DIMINUTO/DWARF1 gene encodes a protein involved in steroid synthesis. Plant Cell. 1998;10:1677–1690. doi: 10.1105/tpc.10.10.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J. A role for brassinosteroids in light-dependent development of Arabidopsis. Science. 1996;272:398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH, Vafeados D, Chory J. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 2001;127:14–22. doi: 10.1104/pp.127.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H, Deng XW. Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell. 2001;13:2589–2607. doi: 10.1105/tpc.010229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Molnár G, Fujioka S, Takatsuto S, Sakurai A, Yokota T, Adam G, Voigt B, Nagy F, Maas C et al. Transcription of the Arabidopsis CPD gene, encoding a steroidogenic cytochrome P450, is negatively controlled by brassinosteroids. Plant J. 1998;14:593–602. doi: 10.1046/j.1365-313x.1998.00158.x. [DOI] [PubMed] [Google Scholar]
- Nagatani A, Nakamura M, Yokota T, Tanaka S, Mochizuki N. Light-responses of plants and phytohormones. Plant Cell Physiol. 1998;39:S-9. [Google Scholar]
- Neff MM, Nguyen SM, Malancharuvil EJ, Fujioka S, Noguchi T, Seto H, Tsubuki M, Honda T, Takatsuto S, Yoshida S et al. BAS1: a gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc Natl Acad Sci USA. 1999;96:15316–15323. doi: 10.1073/pnas.96.26.15316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DR, Koymans L, Kamataki T, Stegeman JJ, Feyereisen R, Waxman DJ, Waterman MR, Gotoh O, Coon MJ, Estabrook RW et al. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics. 1996;6:1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Tax FE, Yoshida S, Feldmann K. Biosynthetic pathways of brassinolide in Arabidopsis. Plant Physiol. 2000;124:201–209. doi: 10.1104/pp.124.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann KA, Tax FE. Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 1999;121:743–752. doi: 10.1104/pp.121.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Sato T, Bishop GJ, Kamiya Y, Takatsuto S, Yokota T. Accumulation of 6-deoxocathasterone and 6-deoxocathasterone in Arabidopsis, pea and tomato is suggestive of common rate-limiting steps in brassinosteroid biosynthesis. Phytochemistry. 2001;57:171–178. doi: 10.1016/s0031-9422(00)00440-4. [DOI] [PubMed] [Google Scholar]
- Pien S, Wyrzykowska J, Fleming AJ. Novel marker genes for early leaf development indicate spatial regulation of carbohydrate metabolism within the apical meristem. Plant J. 2001;25:663–674. doi: 10.1046/j.1365-313x.2001.01002.x. [DOI] [PubMed] [Google Scholar]
- Shimada Y, Fujioka S, Miyauchi N, Kushiro M, Takatsuto S, Nomura T, Yokota T, Kamiya Y, Bishop GJ, Yoshida S. Brassinosteroid-6-oxidases from Arabidopsis and tomato catalyze multiple C-6 oxidations in brassinosteroid biosynthesis. Plant Physiol. 2001;126:770–779. doi: 10.1104/pp.126.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber CM, McCourt P. A role for brassinosteroids in germination in Arabidopsis. Plant Physiol. 2001;125:763–769. doi: 10.1104/pp.125.2.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C-W, Callis J. Independent modulation of Arabidopsis thaliana polyubiquitin mRNAs in different organs and in response to environmental changes. Plant J. 1997;11:1017–1027. doi: 10.1046/j.1365-313x.1997.11051017.x. [DOI] [PubMed] [Google Scholar]
- Szekeres M, Németh K, Koncz-Kálmán Z, Mathur J, Kauschmann A, Altmann T, Rédei GP, Nagy F, Schell J, Koncz C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- Tadano T, Tanaka A. The effect of low phosphate concentration in culture medium on early growth of several crop plants. Jpn J Soil Sci Plant Nutr. 1980;51:399–404. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Bak S, Decker A, Paquette SM, Feyereisen R, Galbraith DW. Microarray-based analysis of gene expression in very large gene families: the cytochrome P450 gene superfamily of Arabidopsis thaliana. Gene. 2002;272:61–74. doi: 10.1016/s0378-1119(01)00516-9. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Kamiya Y. Gibberellin biosynthesis: its regulation by endogenous and environmental signals. Plant Cell Physiol. 2000;41:251–257. doi: 10.1093/pcp/41.3.251. [DOI] [PubMed] [Google Scholar]
- Yokota T, Nomura T, Nakayama M. Identification of brassinosteroids that appear to be derived from campesterol and cholesterol in tomato shoots. Plant Cell Physiol. 1997;38:1291–1294. [Google Scholar]
- Yokota T, Sato T, Takeuchi Y, Nomura T, Uno K, Watanabe T, Takatsuto S. Roots and shoots of tomato produce 6-deoxo-28-norcastasterone, 6-deoxo-28-nortyphasterol and 6-deoxo-28-norcastasterone, possible precursors of 28-norcastasterone. Phytochemistry. 2001;58:233–238. doi: 10.1016/s0031-9422(01)00237-0. [DOI] [PubMed] [Google Scholar]