Abstract
The Medicago truncatula expressed sequence tag (EST) database (Gene Index) contains over 140,000 sequences from 30 cDNA libraries. This resource offers the possibility of identifying previously uncharacterized genes and assessing the frequency and tissue specificity of their expression in silico. Because M. truncatula forms symbiotic root nodules, unlike Arabidopsis, this is a particularly important approach in investigating genes specific to nodule development and function in legumes. Our analyses have revealed 340 putative gene products, or tentative consensus sequences (TCs), expressed solely in root nodules. These TCs were represented by two to 379 ESTs. Of these TCs, 3% appear to encode novel proteins, 57% encode proteins with a weak similarity to the GenBank accessions, and 40% encode proteins with strong similarity to the known proteins. Nodule-specific TCs were grouped into nine categories based on the predicted function of their protein products. Besides previously characterized nodulins, other examples of highly abundant nodule-specific transcripts include plantacyanin, agglutinin, embryo-specific protein, and purine permease. Six nodule-specific TCs encode calmodulin-like proteins that possess a unique cleavable transit sequence potentially targeting the protein into the peribacteroid space. Surprisingly, 114 nodule-specific TCs encode small Cys cluster proteins with a cleavable transit peptide. To determine the validity of the in silico analysis, expression of 91 putative nodule-specific TCs was analyzed by macroarray and RNA-blot hybridizations. Nodule-enhanced expression was confirmed experimentally for the TCs composed of five or more ESTs, whereas the results for those TCs containing fewer ESTs were variable.
The rapidly expanding field of genomics provides vast opportunities for evaluating the coordinated functioning and expression of thousands of genes (Lockhart and Winzeler, 2000). The complete sequencing of the Arabidopsis genome (Arabidopsis Genome Initiative [AGI], 2000) and the expansion of functional genomics in this model plant attest to the power of genomic approaches in addressing important questions in plant biology. Large-scale analysis of gene expression in Arabidopsis using cDNA and oligonucleotide arrays has given new insights into photosynthesis (Desprez et al., 1998), biotic and abiotic stresses (Maleck et al., 2000; Schenk et al., 2000; Bohnert et al., 2001; Seki et al., 2001), nitrogen assimilation (Wang et al., 2000), and organ development (Ruan et al., 1998; Girke et al., 2000; Zhu and Wang, 2000). Although Arabidopsis serves as the model system for most plant processes, it suffers from two major weaknesses in consideration of plant-microbe interactions: the absence of symbiotic associations with mycorrhizal fungi and with rhizobia.
In recent years, Medicago truncatula and Lotus japonicus have emerged as model systems for genomic approaches to plant-microbe symbiotic associations (Barker et al., 1990; Handberg and Stougaard, 1992; Cook et al., 1997; Cook, 1999; Oldroyd and Geurts, 2001; Thoquet et al., 2002). Both species possess small genomes, are diploid, have fast generation times, and can be transformed with Agrobacterium tumefaciens and regenerated (Handberg and Stougaard, 1992; Blondon et al., 1994; Handberg et al., 1994; Chabaud et al., 1996; Jiang and Gresshoff, 1997; Stiller et al., 1997; Trinh et al., 1998; Trieu et al., 2000). Currently, both functional and structural genomics approaches are being pursued within each of these species. Covitz et al. (1998) reported the sequencing of about 900 cDNA tags from the M. truncatula root hairs. In addition, hundreds more expressed sequence tags (ESTs) have been isolated and characterized from effective root nodules of L. japonicus and M. truncatula, and a number of genes showing enhanced expression in plant-rhizobium symbiosis were identified (Szczyglowski et al., 1997; Györgyey et al., 2000).
The creation of a large-scale EST database, the M. truncatula Gene Index (MtGI; http://www.tigr.org/tdb/mtgi), from the results of an international effort in high-throughput sequencing, offers the prospect of in silico identification of genes whose expression are specific for or greatly enhanced by symbiosis. Release 4.0 of MtGI was made public in September 2001, and contains over 140,000 sequence entries from 30 non-normalized cDNA libraries representing various vegetative and reproductive organs. Based upon sequence overlap, all ESTs are processed into a nonredundant set of clustered tentative consensus sequences (TCs) and singletons (unique nonoverlapping sequences; Quackenbush et al., 2000). The MtGI database also provides functional annotation and expression summaries (virtual northern) for TCs reflecting the frequency of the corresponding ESTs in each cDNA library. Five of the cDNA libraries selected for MtGI construction are derived from nodules at different developmental stages. The MtGI database becomes a powerful resource for in silico analysis of the nodule transcriptome and discovery of novel nodule-specific genes.
The potential of in silico analysis of EST collections has been demonstrated for a number of plant species (Sasaki et al., 1994; Cooke et al., 1996; Rounsley et al., 1996; Ewing et al., 1999; Ablett et al., 2000; Fernandes et al., 2002; Kruger et al., 2002). In silico-based gene discovery and tissue profiling were performed to study plant fatty acid and lipid metabolism, cell wall biosynthesis, and seed oil production (Van de Loo et al., 1995; Allona et al., 1998; Sterky et al., 1998; Cahoon et al., 1999; Mekhedov et al., 2000; White et al., 2000). Such analyses were made possible through accumulation of large numbers of ESTs, where the gene expression level can be deduced in silico by calculating EST frequencies in different cDNA libraries. Statistical significance of such digital expression profiling applied to representative EST datasets has been validated in several publications (Audic and Claverie, 1997; Ewing et al., 1999; Stekel et al., 2000).
The objectives of our studies were to assess whether Boolean analysis of in silico expression data would be a useful genome-wide approach in identifying novel genes specific to developing and functioning of root nodules. The language of the Boolean formalism (Genoud and Métraux, 1999; Genoud et al., 2001) was applied to reveal a subset of nodule-specific TCs composed of ESTs that were derived exclusively from the nodule cDNA libraries. RNA-blot analysis and macroarrays were used to test the nodule-specific nature of TCs identified through virtual methods.
RESULTS
In Silico Identification of Nodule-Specific TCs
Among 30 cDNA libraries represented in Release 4.0 of MtGI, five were prepared from mRNA extracted from nodules at different developmental stages (Table I). Three major stages of development can be distinguished. The early nodule MtBB library was prepared from emerging nodules attached to the root segments, before detection of N2 fixation (E.-P. Journet, personal communication). R108Mt, GVN, and Nodulated Root libraries represent mature nodules actively fixing N2. It should be noted that the MtBB and the Nodulated Root libraries were prepared from the mixture of roots and nodules and, therefore, potentially contain sequences expressed in root tissues, as well as nodules. Finally, the GVSN library represents senescent nodules. In total, 20,347 EST sequences in MtGI are from nodule libraries, which comprises 14.4% of the 141,501-EST dataset. Given that other cDNA libraries represent all major plant organs (roots, leaves, stems, flowers, pods, and seeds), this number appears to be sufficient for sketching the nodule-specific transcriptome.
Table I.
Sequencing progress of M. truncatula nodule cDNA librariesa
| cDNA Library | Developmental Stage | Total No. of ESTs | No. of ESTs in TCs | No. of Singletons | Library Source |
|---|---|---|---|---|---|
| MtBB | Emerging nodules and adjacent root segments of 21-d-old plants harvested 4 dpib | 7,785 | 7,177 | 608 | Genoscope and Centre National de la Recherche Scientifique-Institut National de la Recherche Agronomique (Castanet-Tolosan cedex, France) |
| R108Mt | Developing young nodules | 386 | 302 | 84 | Institut des Sciences Vegetales (Centre National de la Recherche Scientifique, Gif sur Yvette, France) |
| GVN | Effective root nodules harvested 30 dpi | 6,446 | 5,898 | 548 | University of Minnesota (St. Paul) |
| Nodulated Root | Mixture of roots and nodules | 3,070 | 2,710 | 360 | The Samuel Roberts Noble Foundation (Ardmore, OK) |
| GVSN | Senescent nodules (mixture of 40-dpi nodules harvested 36 h post-shoot removal, and 60-dpi nodules) | 2,660 | 2,393 | 267 | University of Minnesota |
| Total | 20,347 |
As of M. truncatula Gene Index Release 4.0, September 2001. For update, visit www.tigr.org/tdb/mtgi.
dpi, Days post inoculation with Sinorhizobium meliloti.
The language of Boolean formalism was applied to screen MtGI Release 4.0, and to identify those TCs composed of ESTs derived exclusively from MtBB, R108Mt, GVN, Nodulated Root, or GVSN libraries (operator “OR”), but not from any other library (operator “NOT”). This search revealed 340 entries as nodule-specific TCs. All of these TCs are posted on the M. truncatula Consortium Web site (http://www.medicago.org).
Each nodule-specific TC sequence is clustered from individual overlapping ESTs, and, therefore, putatively represents a unique transcript presumably from a single gene. Variability in the number of ESTs comprising each TC likely reflects the differences in abundance of the transcripts from the corresponding genes. Nodule-specific TCs were grouped into four categories based on the number of ESTs contributing to an individual TC contig. Notably, 70% of nodule-specific TCs are represented by two to four ESTs, 17% of the TCs contain five to nine ESTs each, and 7% of the TCs contain 10 to 19 ESTs. Approximately 6% of the TCs contain over 20 ESTs each. Assuming that the number of ESTs comprising a single TC reflects gene expression level, the current categorization of TCs composed of few ESTs as nodule specific may be not final. The likelihood of finding transcripts in non-nodule libraries after deeper sequencing should be considered. This scenario has already proven true for a number of such TCs upon comparison of MtGI Release 3.0 with Release 4.0, which was supplemented with 13,877 additional EST sequences.
In addition to 340 nodule-specific contigs (TCs), the MtGI contains 1,867 singletons also sequenced from nodule libraries. They were not considered for further analysis because their nodule-specific status is questionable due to a limited number of identified transcripts.
All 340 nodule-specific TCs were again analyzed using BLASTX and grouped into three categories based on the statistical significance of their matches to proteins in the GenBank protein database: novel (zero matches in the database), strong similarity (E values less than 10−8), and weak (statistically insignificant) similarity (E values higher than 10−8). Ten (3%) nodule-specific TCs were novel: TC36162, TC29160, TC32908, TC29828, TC40949, TC40984, TC31810, TC35357, TC38228, and TC38832. Because TC36162, TC29160, and TC32908 are represented by a large number of ESTs (16, 8, and 8, respectively), they are apparently expressed at a moderate to rather high level. When the entire GenBank EST database was scanned for the presence of sequences similar to these 10 TCs (TBLASTX analysis), with a single exception, only ESTs from nodule libraries of M. truncatula and M. sativa showed statistically significant degrees of similarity. The exception was TC31810 showing strong similarity not only to nodule ESTs, but also to one EST from M. truncatula leaf and cotyledon cDNA library (E value of 10−43). However, this EST is classified at MtGI as a singleton.
Some 40% (137) of the nodule-specific TCs showed strong similarity to known GenBank sequences, whereas the remaining 57% (193) of the TCs exhibited weak similarity with GenBank sequences (E values higher than 10−8). Within this weak similarity category, a large subgroup of 114 TCs encoded various Cys cluster proteins (CCPs).
Characterization of Nodule-Specific TCs
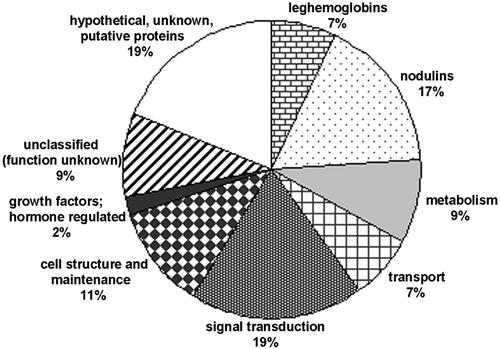
The 137 TCs showing strong similarity to the GenBank protein accessions were subdivided into nine categories based upon the putative function of their strongest BLASTX score (Fig. 1; Tables II and III). Of these TCs, function could be predicted for 76 (55%) TCs. Twenty-three (17%) TCs encoded proteins of unknown function, previously described in legumes as nodule specific, or nodulins (Legocki and Verma, 1980). Twenty-six (19%) TCs displayed strong similarity to hypothetical, unknown, or putative proteins predicted by sequencing of the Arabidopsis and rice genomes. Finally, 12 (9%) TCs corresponded to previously characterized proteins for which the cellular function is not yet understood.
Figure 1.
Distribution of nodule-specific TCs by functional categories. Classification was performed for 137 nodule-specific TCs with strong statistical similarity to GenBank protein sequences (E values lower than 10−8).
Table II.
Nodule-specific TCs of M. truncatula encoding known nodulins
| TC No. | No. of ESTs in TC | Strongest BLASTX Hit | E Value | ESTs from Young/ Mature/ Senescent Nodulesa | Protein Features/Proposed Function |
|---|---|---|---|---|---|
| % | |||||
| TC28588 | 13 | MtN1, M. truncatula(CAA71482) | 10−29 | 77/15/8 | Homolog of plant defense proteins (Gamas et al. 1998) |
| TC40070 | 11 | EnodGRP5, M. sativa(CAB65282) | 10−50 | 0/82/18 | Probable peroxisomal proteinb |
| TC29418 | 4 | MtN9, M. truncatula (CAA77093) | 10−97 | 75/25/0 | Homology to soybean (Glycine max) metalloendo-proteinase (Gamas et al., 1996) |
| TC28970 | 6 | ENOD12 precursor, M. sativa(P30365) | 10−23 | 83/17/0 | Possible destination: vacuole or outside the cellb |
| TC36450 | 5 | MtN16, M. truncatula(CAA75586) | 10−34 | 80/0/20 | Possible destination: endoplasmic reticulumb |
| TC40417 | 6 | ENOD16, M. truncatula (P93328) | 10−98 | 17/66/17 | Phytocyanin-related proteins (Greene et al., 1998) |
| TC36054 | 20 | ENOD18, broad bean (Vicia faba; CAC18556) | 10−75 | 5/90/5 | Possible ATP-binding protein or ATPase (Hohnjec et al., 2000) |
| TC29982 | 2 | MtN19, M. truncatula (CAA75589) | 0.0 | 50/50/0 | Possible plasma membrane proteinb |
| TC28429 | 29 | ENOD20, M. truncatula (P93329) | 10−73 | 52/34/14 | Phytocyanin-related protein (Greene et al., 1998) |
| TC37466 | 2 | MtN20, M. truncatula (CAA75590) | 10−89 | 50/50/0 | Possible peroxisomal or mitochondrial matrix space proteinb |
| TC36819 | 6 | MtN21-like protein, Arabidopsis (CAB53493) | 10−54 | 0/83/17 | Possible integral membrane protein (DUF6 domain signature)b |
| TC36998 | 4 | MtN21, M. truncatula (CAA75575) | 0.0 | 0/100/0 | Same as TC36819 |
| TC40954 | 4 | Nodule-specific protein nms22, M. sativa (T09333) | 10−55 | 0/0/100 | Possible plasma membrane proteinb |
| TC31873 | 8 | MtN22, M. truncatula (CAA75576) | 10−84 | 25/62/13 | Shares homology with nodulin-25; possible destination - vacuole, outside the cell or peribacteroid space (PBSb; Gamas et al., 1996) |
| TC31874 | 84 | MtN22, M. truncatula (CAA75576) | 10−108 | 5/69/26 | Same as TC31873 |
| TC33130 | 3 | Putative, M. truncatula(CAA75573); MtN25 (TBLASTX) (Y15291) | 10−12 | 67/33/0 | No N-terminal signal sequenceb |
| TC35677 | 23 | Nodulin 25, M. truncatula(CAB91091) | 10−140 | 0/73/27 | N-terminal signal sequence may target the protein into the PBSb (Kiss et al., 1990) |
| TC35678 | 2 | Nodulin 25, M. truncatula (CAB91093) | 10−128 | 0/100/0 | Same as TC35677 |
| TC31452 | 2 | Nodulin 25, M. truncatula (CAC33843) | 10−13 | 0/50/50 | Possible peroxisomal or cytosolic proteinb |
| TC30771 | 2 | MtN26, M. truncatula (CAA75574) | 10−11 | 0/100/0 | Possible cytosolic proteinb |
| TC38783 | 2 | MtN27, M. truncatula (CAA75564) | 10−16 | 0/100/0 | No N-terminal signal sequenceb |
| TC35962 | 19 | MtN29, M. truncatula (CAA77088) | 10−37 | 47/42/11 | Possible plasma membrane proteinb |
| TC37619 | 2 | Nodulin-like protein, Arabidopsis (NP_180982) | 10−61 | 0/100/0 | Contains ATP-/GTP-binding site motif Ab |
Percent of ESTs from the young nodule (MtBB), mature nodule (GVN, Nodulated Root, R108Mt), and senescent nodule (GVSN) cDNA libraries, of the total EST no. in the TC contig.
This work; based on analysis of the deduced amino acid sequence.
Table III.
Functional classification of nodule-specific TCs
| TC No. | No. of ESTs in TC | Strongest BLASTX Hit | E Value |
|---|---|---|---|
| Metabolism | |||
| TC36519 | 9 | Peroxidase precursor (AAB48986, M. truncatula) | 10−92 |
| TC40419 | 7 | Carbonic anhydrase (CAH1) (NP_190840, Arabidopsis) | 10−58 |
| TC37526 | 4 | Stearoyl acyl carrier protein desaturase Lldd3A20 (AAD28287, Lupinus luteus) | 10−73 |
| TC30710 | 3 | Lys decarboxylase-like protein (NP_196248, Arabidopsis) | 10−86 |
| TC41286 | 3 | Rubisco small subunit (P16031, Larix laricina) | 10−46 |
| TC31237 | 2 | Chitinase-1 (JC7335, Conus tulipa) | 10−82 |
| TC31585 | 2 | Putative glycosylasparaginase (NP_191968, Arabidopsis) | 10−13 |
| TC34770 | 2 | Phytochelatin synthetase (NP_200900, Arabidopsis) | 10−92 |
| TC34904 | 2 | Nitrilase (Q42965, Nicotiana tabacum) | 10−32 |
| TC34939 | 2 | Putative APG protein (NP_194409, Arabidopsis) | 10−112 |
| TC38721 | 2 | Invertase inhibitor precursor (T03393, N. tabacum) | 10−11 |
| TC41116 | 2 | Putative cytochrome P450 (NP_182075, Arabidopsis) | 10−83 |
| Transport | |||
| TC32516 | 14 | Purine permease (NP_198932, Arabidopsis) | 10−19 |
| TC37150 | 5 | Copper transporter protein (NP_200711, Arabidopsis) | 10−17 |
| TC37507 | 4 | Probable hexose transport protein Hex9 (T10068, Ricinus communis) | 10−19 |
| TC40870 | 4 | 2-on-2 hemoglobin (AAK55409, Arabidopsis) | 10−64 |
| TC30600 | 3 | Sulfate transporter (BAB55634, Arabidopsis) | 10−93 |
| TC30920 | 2 | MATE efflux family protein, putative (NP_187461, Arabidopsis) | 10−24 |
| TC31155 | 2 | ABC transporter-like protein (NP_190919, Arabidopsis) | 10−104 |
| TC34543 | 2 | Amino acid transport protein AAT1 (NP_193844, Arabidopsis) | 10−122 |
| TC42940 | 2 | ABC transporter-like protein (NP_190916, Arabidopsis) | 10−81 |
| Signal transduction | |||
| TC35910 | 14 | Calmodulin (CAA69660, Toxoplasma gondii) | 10−32 |
| TC35911 | 10 | Calmodulin (P02598, Tetrahymena pyriformis) | 10−27 |
| TC35912 | 6 | Calmodulin (P02598, T. pyriformis) | 10−29 |
| TC29198 | 6 | Protein kinase-like protein (NP_195559, Arabidopsis) | 10−23 |
| TC29264 | 6 | Remorin (T07780, Lycopersicum esculentum) | 10−40 |
| TC29680 | 5 | C2H2-type zinc finger protein (NP_187540, Arabidopsis) | 10−13 |
| TC37063 | 5 | Calmodulin (P04464, Triticum aestivum) | 10−13 |
| TC41252 | 4 | Calmodulin (P17928, M. sativa) | 10−26 |
| TC29944 | 4 | Receptor protein kinase-like protein (NP_192429, Arabidopsis) | 10−20 |
| TC33166 | 4 | IRE, protein kinase-like protein (NP_201037, Arabidopsis) | 10−136 |
| TC40528 | 4 | Transcription factor-like protein (NP_195559, Arabidopsis). | 10−129 |
| TC30207 | 3 | His kinase, cytokinin receptor CRE1a (BAB40775, Arabidopsis) | 10−16 |
| TC30711 | 3 | Putative cyclin-dependent kinase inhibitor (NP_199693, Arabidopsis) | 10−32 |
| TC34223 | 3 | Calmodulin (AAD10245, bean [Phaseolus vulgaris]) | 10−17 |
| TC34260 | 3 | Ser/Thr kinase-like protein (NP_194049, Arabidopsis) | 10−49 |
| TC41489 | 3 | Putative Glu-/Asp-binding peptide (NP_171806, Arabidopsis) | 10−48 |
| TC41702 | 3 | His-containing phosphotransfer protein (AAK38843, Catharanthus roseus) | 10−45 |
| TC31111 | 2 | Contains similarity to Pfam domain (CAC70088, Caenorhabditis elegans) | 10−10 |
| TC31168 | 2 | Armadillo repeat-containing protein (AAK60564, N. tabacum) | 10−18 |
| TC31220 | 2 | Phosphatidylinositol transfer-like protein IV (AAK63248, L. japonicus) | 10−24 |
| TC31584 | 2 | Putative bZIP transcription factor (NP_181594, Arabidopsis) | 10−26 |
| TC34514 | 2 | Putative ADP-ribosylation factor (NP_179430, Arabidopsis) | 10−87 |
| TC35401 | 2 | Kinase-like protein (NP_193214, Arabidopsis) | 10−24 |
| TC38167 | 2 | bZIP transcription factor ATB2 (CAA68078, Arabidopsis) | 10−31 |
| TC38181 | 2 | Putative NPK1-related protein kinase 2 (NP_172374, Arabidopsis) | 10−24 |
| TC38237 | 2 | Contains similarity to protein kinase domains (AAD40144, Arabidopsis) | 10−48 |
| TC38318 | 2 | Putative receptor-like protein kinase (BAA96921, Arabidopsis) | 10−72 |
| TC41344 | 2 | Mitogen-activated protein kinase homologue (AAD28617, M. sativa) | 10−139 |
| Cell structure and maintenance | |||
| TC32103 | 40 | Putative bark agglutinin precursor (Q41160, Robinia pseudoacacia) | 10−61 |
| TC36302 | 12 | Putative bark agglutinin precursor (Q41160, R. pseudoacacia) | 10−20 |
| TC28421 | 6 | Cys proteinase (BAB13759, Astragalus sinicus) | 10−161 |
| TC29708 | 5 | UFD1-like protein (NP_193277, Arabidopsis) | 10−22 |
| TC33690 | 4 | Chloroplast nucleoid DNA-binding protein (NP_174430, Arabidopsis) | 10−54 |
| TC33968 | 3 | Cys proteinase (BAB13759, A. sinicus) | 10−87 |
| TC37606 | 3 | BS14b (AAK51151, Arabidopsis) | 10−28 |
| TC41682 | 3 | Lectin-related polypeptide (BAA36416, R. pseudoacacia) | 10−26 |
| TC30949 | 2 | Cys proteinase (BAB13759, A. sinicus) | 10−34 |
| TC31101 | 2 | Anaphase-promoting complex subunit (AY052402.1, Arabidopsis) | 10−39 |
| TC34051 | 2 | Histone H4 homolog (HSWT41, T. aestivum) | 10−38 |
| TC34545 | 2 | Putative cytoskeleton-associated protein (AAF02820, Arabidopsis) | 10−63 |
| TC35317 | 2 | Papain-like Cys proteinase isoform II (AF138266.1, Ipomoea batatas) | 10−104 |
| TC35399 | 2 | Histone deacetylase (CAB37553, Arabidopsis) | 10−155 |
| TC41454 | 2 | Bamacan homolog, chromosome-associated protein (AAD26882, Arabidopsis) | 10−45 |
| Growth factors and hormone regulated | |||
| TC35256 | 2 | Growth factor-like protein (NP_193008, Arabidopsis) | 10−19 |
| TC38102 | 2 | Leginsulin (CAA11040, soybean) | 10−16 |
| TC38244 | 2 | Nt-gh3-deduced protein (AAD32141, N. tabacum) | 10−61 |
| Unclassified; function unknown | |||
| TC32101 | 45 | Basic blue protein, plantacyanin (CAB65280, M. sativa subsp. × varia) | 10−62 |
| TC32092 | 14 | B12D protein (AAD22104, I. batatas) | 10−23 |
| TC36259 | 10 | Contains similarity to embryo-specific protein (BAA97184, Arabidopsis) | 10−29 |
| TC30398 | 3 | Probable wound-induced protein (NP_192765, Arabidopsis) | 10−17 |
| TC34413 | 3 | Similar to salt-inducible membrane protein (AAC17629, Arabidopsis) | 10−21 |
| TC37978 | 3 | Putative RING zinc finger protein (NP_181294, Arabidopsis) | 10−12 |
| TC41606 | 3 | Putative resistance protein (AAG48132, soybean) | 10−39 |
| TC31693 | 2 | Allergen-like protein (NP_193436, Arabidopsis) | 10−19 |
| TC34377 | 2 | Contains similarity to SF16 protein (BAB03067, Arabidopsis) | 10−15 |
| TC35428 | 2 | Contains similarity to embryo-specific protein (BAA97184, Arabidopsis) | 10−25 |
| TC42191 | 2 | Putative PREG1-like negative regulator (BAB09009, Arabidopsis) | 10−59 |
| TC42587 | 2 | Thaumatin-like protein (AAD02499, Arabidopsis) | 10−45 |
| Hypothetical, unknown, putative proteins | |||
| TC40314 | 8 | Hypothetical protein (NP_189170, Arabidopsis) | 10−14 |
| TC41039 | 4 | Putative protein, (NP_195884, Arabidopsis) | 10−26 |
| TC33290 | 3 | Unknown protein (AAD32844, Arabidopsis) | 10−55 |
| TC33893 | 3 | Unknown protein (AAF14673, Arabidopsis) | 10−65 |
| TC34204 | 3 | Unknown protein (NP_187787, Arabidopsis) | 10−11 |
| TC37654 | 3 | Putative protein (NP_566966, Arabidopsis) | 10−09 |
| TC38071 | 3 | Hypothetical protein (BAB03434, rice [Oryza sativa]) | 10−16 |
| TC30927 | 2 | Unknown protein (NP_181485, Arabidopsis) | 10−08 |
| TC31107 | 2 | Putative protein (CAB72187, Arabidopsis) | 10−88 |
| TC31161 | 2 | Unknown protein (NP_569007, Arabidopsis) | 10−52 |
| TC31760 | 2 | Unknown protein (BAB10007, Arabidopsis) | 10−51 |
| TC34143 | 2 | Putative protein (CAA16779, Arabidopsis) | 10−80 |
| TC34629 | 2 | Unknown protein (NP_197347 Arabidopsis) | 10−113 |
| TC34686 | 2 | Hypothetical protein (NP_181906, Arabidopsis) | 10−18 |
| TC38114 | 2 | Unknown protein (AAK93620, Arabidopsis) | 10−28 |
| TC38188 | 2 | Unknown protein (AAK43923, Arabidopsis) | 10−35 |
| TC38847 | 2 | Unknown protein (AAK96788, Arabidopsis) | 10−71 |
| TC39037 | 2 | Putative protein (CAB75447, Arabidopsis) | 10−61 |
| TC40199 | 2 | Putative protein (CAB75911, Arabidopsis) | 10−41 |
| TC41877 | 2 | Hypothetical protein (BAB56065, rice) | 10−14 |
| TC42081 | 2 | Hypothetical protein (AAD31368, Arabidopsis) | 10−74 |
| TC42391 | 2 | Unknown protein (BAB11326, Arabidopsis) | 10−52 |
| TC42548 | 2 | Putative protein (NP_201248, Arabidopsis) | 10−31 |
| TC42604 | 2 | F12A21.16 protein (AAG28905, Arabidopsis) | 10−59 |
| TC42647 | 2 | Hypothetical protein (NP_179295, Arabidopsis) | 10−12 |
| TC31171 | 2 | Putative protein (NP_199431, Arabidopsis) | 10−46 |
Nine of the functionally defined TCs corresponded to leghemoglobins (Lbs). Lb genes are among the most abundantly expressed nodule-specific genes. Each Lb-encoding TC was composed of 13 (TC31876) to 379 (TC35566) ESTs. Nodulin TCs containing the greatest number of ESTs corresponded to MtN22, ENOD20, nodulin-25, ENOD18, MtN29, MtN1, and EnodGRP5, with each containing 84, 29, 23, 20, 19, 13, and 11 ESTs, respectively. The putative functions or cellular locations of the identified nodulins are listed in Table II.
It is worth noting that a number of nodulin TCs contain a high proportion of ESTs from the MtBB library. This library corresponds to early nodule development before N2 fixation. At least 47% of the ESTs in each of TC28588, TC29418, TC28970, TC36450, TC29982, TC28429, TC37466, TC33130, and TC35962 came from the MtBB library, indicating that they are early nodulins induced before the onset of N2 fixation (Nap and Bisseling, 1990). In previous studies (Pichon et al., 1992; Gamas et al., 1996; Greene et al., 1998), these nodulins were also described as being induced early in nodule development.
In comparison, among the 660 Lb ESTs sequenced, only three ESTs originate from the MtBB early nodule library. Lbs would be expected to represent a low number of ESTs in MtBB because they are usually most highly expressed in mature N2-fixing nodules (GVN, R108Mt, and Nodulated Root libraries).
In contrast to early nodulins, all four ESTs comprising TC40954, which is similar to M. sativa nodule-specific protein nms22, are derived from the GVSN library representing senescent nodules. Another nodule-specific TC (TC40868), also sharing some similarity with nms22 (E value of 10−4), contained four GVSN ESTs and one GVN sequence. This information suggests that nms22-like proteins are preferentially expressed during nodule senescence.
Because the function of most nodulins is unresolved, we analyzed the amino acid sequences deduced from their TCs by the PSORT (prediction of protein sorting signals and localization sites) and the Inter-Pro (identification of protein functional domains) programs (Table II). These analyses suggest that homologs of three nodulins, ENOD12 (TC28970), MtN22 (TC31873 and TC31874), and nodulin-25 (TC35677 and TC35678), possess a cleavable N-terminal sequence targeting the protein into the endomembrane system or outside the cell. N-Terminal signal peptides deduced for MtN22-like TCs are identical. Likewise, the deduced N-terminal signal peptides for nodulin-25-like TCs are also identical. MtN22- and nodulin-25-type signal peptides are more similar to each other (48% identity) than to those of the ENOD12-type signal peptide (31% and 25% identity, respectively). The N-terminal sequence of M. sativa nodulin-25 was earlier proposed to target the protein into the PBS of the nodule (Kiss et al., 1990). Several nodulins (encoded by TC29982, TC40954, TC35962, and TC36819) are putative plasma membrane proteins. A homolog of an MtN21-like protein (represented by TC36819), predicted to have an uncleavable N-terminal signal sequence, contains a DUF6 domain signature characteristic of integral membrane proteins. This domain is found in a number of proteins, such as carboxylate/amino acid/amine transporters and phosphate/phosphoenolpyruvate translocator.
Despite the original definition of nodulins as genes expressed exclusively in legume root nodules, eight of the 23 TCs corresponding to the known nodulins also have strong similarities (E values of 10−18 and lower) to genes of nonlegume species. For example, genes similar to nodule-specific TC36819, encoding a nodulin 21-like protein (E value of 10−30), are also found in rice and Arabidopsis (GenBank accession nos. CAB53493 and NP_176984, respectively). Likewise, nodule-specific TC37619 displays strong similarity to an Arabidopsis gene encoding a nodulin-like protein (GenBank accession no. NP_180982.1).
Besides those encoding Lbs, a group of nodule-specific TCs with strong similarity to genes of known function includes 12 (9%) related to metabolism, 9 (7%) related to transport, 28 (19%) related to signal transduction, 15 (11%) related to cell structure/maintenance, and three (2%) related to growth factor/hormone processes (Table III). Among these groups of TCs, those having the greatest number of ESTs encoded peroxidase precursor (nine), carbonic anhydrase (seven), purine permease (14), calmodulins (14 and 10), bark agglutinin precursor (40 and 12), plantacyanin (45), B12D protein (14), and embryo-specific protein (10). The majority of the TCs, however, are composed of four or fewer ESTs.
Two nodule-specific TCs (TC32103 and TC36302) encode proteins that are similar to a bark lectin-related polypeptide/agglutinin of R. pseudoacacia and Cicer arietinum—a protein apparently implicated in nitrogen storage (VandenBosch et al., 1994; van Damme et al., 1995). They are represented by a large number of ESTs (40 and 12, respectively), and, therefore, appear to be relatively abundant messages in nodules. Deduced amino acid sequences of TC32103 and TC36302 are identical at only 41% of the aligned position. Three other TCs also matching R. pesudoacacia/C. arietinum agglutinins have been found in MtGI (TC32259, TC28959, and TC39450). However, the tissue expression patterns deduced from the ESTs composing these TCs indicate that they are not nodule specific.
Basic blue copper protein, or plantacyanin (TC32101), is encoded by another highly expressed nodule-specific TC. The nucleotide sequence of TC32101 is 97% identical to that of nodulin MsNod202 encoding a plantacyanin from M. sativa (Jiménez-Zurdo et al., 2000). Together with two other nodulins, ENOD16 and ENOD20 (Greene et al., 1998), corresponding to nodule-specific TC40417 and TC28429, these proteins contain a copper-binding domain characteristic of a group of plant phytocyanins proposed to be involved in primary defense response (Nersissian et al., 1998). Another copper-related nodule-specific TC appears to encode a putative copper transporter (TC37150).
Two proteins encoded by nodule-specific TC36259 and TC35428, assembled from 10 and two ESTs, respectively, are 77% identical, and are similar to Arabidopsis embryo-specific protein (GenBank accession no. AB019235). Some 40% of the clones comprising TC36259 were sequenced from the early nodule library (MtBB).
One of the unexpected outcomes of in silico survey for nodule-specific TCs was identification of a TC41286 that encodes a protein similar to a Rubisco small subunit, a photosynthesis-related protein normally observed in green tissues. This TC consists of three ESTs derived from the MtBB, Nodulated Root, and GVSN libraries. Surprisingly, the statistical significance of the similarity between the deduced amino acid sequence of TC41286 to Rubisco small subunits of nonleguminous woody plants Betula verrucosa and L. laricina (E values of 10−41 and 10−46, GenBank accession nos. Q96542 and P16031, respectively) is greater than to those found in green tissues of M. truncatula (for example, TC28300; E value of 10−38) or M. sativa (E value of 10−38, GenBank accession no. O65194). TC41286 possesses 57% and 61% amino acid identity with the B. verrucosa and L. laricina accessions, whereas identity with TC28300, apparently encoding a photosynthetically active Rubisco, is only 51%. Comparison of nucleotide sequences of nodule Rubisco-like TC41286 and leaf Rubisco TC28300 revealed that the 64.4% identity between these two forms of Rubisco small subunit starts only from nucleotide 131 of TC41286 and nucleotide 203 of TC28300, whereas 5′ ends of the sequences are much more diverse and share similarity over only a short stretch of 30 nucleotides (data not shown).
Unique Nodule-Specific Calmodulin-Like Proteins
Six nodule-specific TCs with similarity to calmodulins were identified in silico (TC35910, TC35911, TC35912, TC34223, TC41252, and TC37063). The number of ESTs comprising each of these TCs varied from three (TC34223) to 14 (TC35910; Table III). Based upon BLASTX comparisons, the deduced amino acid sequence identity to known calmodulins was lower for the nodule-specific TCs (38%–70%) than for the two TCs encoding typical calmodulins and expressed in various other tissues of M. truncatula (TC31994 and TC35885, 100% identity). Therefore, nodule-specific TCs were named calmodulin-like proteins.
Complete coding sequences (CDS) were obtained for all six of these TCs, and also for two TCs encoding typical calmodulins expressed in various other tissues of M. truncatula (TC31994 and TC35885). To verify the assembly of the contig, at least one representative cDNA clone was completely resequenced for each TCs. Complete cDNA sequences corresponding to all nodule-specific calmodulin-like TCs and to two typical calmodulin TCs are deposited to the GenBank under the accession numbers AF494212 through AF494220.
With two exceptions, the complete CDS length of calmodulin-like TCs and typical calmodulin TCs was comparable (767–983 bp). The CDS for TC34223 was considerably smaller (567 bp), apparently due to an internal deletion. TC37063 appeared to be assembled of two types of clones, identical throughout the entire sequence, but different in length due to an extension of a 3′ region in one of them. Therefore, two versions of TC37063 were proposed, TC37063-s (short, 501 bp) and TC37063-l (long, 781 bp). The 280-bp-long extension at the 3′ end of TC37063-l occurred almost entirely in the 3′-untranslated region; however, the deduced amino acid sequence of TC37063-l is also slightly longer (12 additional amino acids preceding the stop codon). Notably, all TCs, including those encoding typical calmodulins possess relatively long 3′-untranslated regions.
Four nodule-specific TCs encode longer calmodulin-like polypeptides (140–179 amino acids for TC35910, TC35911, TC35912, and TC34223) than the others (TC41252, 116 amino acids, TC37063-s, 103 amino acids; and TC37063-l, 115 amino acids). TC31994 and TC35885 encode calmodulin polypeptides of 149 amino acids, similar to most known calmodulins (Reddy, 2001). Amino acid identity among nodule-specific calmodulin-like proteins varies from 40% to 91%, whereas their identity to typical M. truncatula calmodulins range from 52% to 68% (Table IV).
Table IV.
Percent identity between the deduced amino acid sequences of nodule-specific calmodulin-like proteins and typical calmodulins of M. truncatulaa
| TC No. | TC35910 | TC35911 | TC35912 | TC41252 | TC34223 | TC37063-s | TC37063-l | TC31994 | TC35885 |
|---|---|---|---|---|---|---|---|---|---|
| TC35910 | 100 | 75 | 75 | 58 | 74 | 45 | 45 | 60 | 60 |
| TC35911b | – | 100 | 91 | 56 | 64 | 43 | 40 | 52 | 52 |
| TC35912 | – | – | 100 | 57 | 68 | 43 | 43 | 52 | 52 |
| TC41252 | – | – | – | 100 | 44 | 41 | 40 | 68 | 68 |
| TC34223 | – | – | – | – | 100 | 50 | 50 | 58 | 58 |
| TC37063-s | – | – | – | – | – | 100 | 100 | 68 | 68 |
| TC37063-l | – | – | – | – | – | – | 100 | 65 | 65 |
| TC31994 | – | – | – | – | – | – | – | 100 | 93 |
| TC35885 | – | – | – | – | – | – | – | – | 100 |
TC35910, TC35911, TC35912, TC41252, TC34223, TC37063-s, and TC37063-l encode nodule-specific calmodulin-like proteins. TC31994 and TC35885 encode typical calmodulins expressed in various tissues of M. truncatula.
The amino acid sequence deduced from the original (MtGI) version of TC35911 does not contain a complete signal peptide. However, a sequencing error was revealed upon comparative analysis of individual original ESTs used for assembling TC35911, and from resequencing of some of the clones. An additional nucleotide (C) should be present in position 50. This addition causes a frame shift, a loss of a stop codon at the 5′ end of the original TC35911 version, and results in the translation of a complete N-terminal signal peptide.
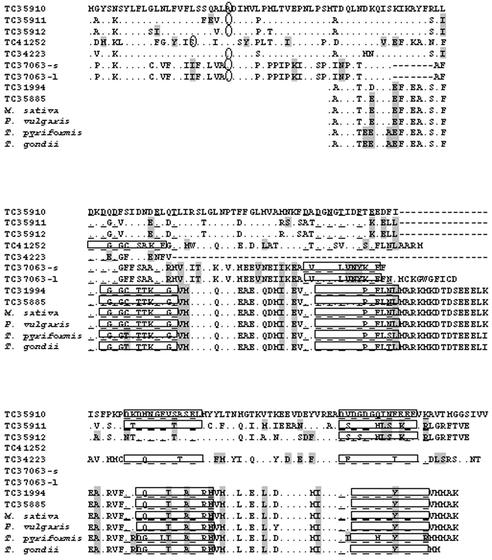
The alignment of the deduced amino acid sequences of nodule-specific calmodulin-like TCs, typical calmodulin TCs, and several calmodulins from other organisms is shown in Figure 2. Typical calmodulins possess four Ca2+-binding domains (EF hand motifs; boxed in Fig. 2), each including several highly conserved residues that form Ca2+-binding sites (underlined amino acids). For example, calmodulins of Medicago sativa, bean, T. pyriformis, T. gondii, and both typical calmodulins of M. truncatula contain all four domains. The Inter-Pro program used to determine Ca2+-binding motifs in nodule-specific calmodulin-like TCs showed that these TCs do not contain all four complete Ca2+-binding domains. The optimal amino acid alignment of calmodulin-like proteins with typical calmodulins produces a gap in the amino acid stretch of four calmodulin-like TCs. This gap occurs in the region corresponding to domain II. Three calmodulin-like proteins (TC35910, TC35911, and TC34223) contain complete domains III and IV only; TC35912 contains domain IV; TC41252 contains domain I, and both versions of TC37063 contain domain II. However, many functionally important amino acid residues in the regions corresponding to the missing complete EF motifs are still conserved in all calmodulin-like TCs.
Figure 2.
Comparison of the deduced amino acid sequences of M. truncatula nodule-specific calmodulin-like proteins (encoded by TC35910, TC35911, TC35912, TC41252, TC34223, TC37063-s, and TC37063-l), typical M. truncatula calmodulins (encoded by TC31994 and TC35885), and calmodulins from Medicago sativa (GenBank accession no. X52398), bean (AAD10245), T. pyriformis (P02598), and T. gondii (Y08373). Comparisons are referenced to the calmodulin-like protein encoded by TC35910. Dots represent identical amino acids. Amino acids shaded in gray possess similar physico-chemical properties. EF hand domains, as predicted by PSORT, are shown in boxes. Underlined amino acids are essential for Ca2+ binding. The circled amino acids in the N-terminal portion of the polypeptides are the last ones in the predicted cleavable signal peptide. Gaps in the sequences (indicated by dashes) are introduced to maintain maximum sequence similarity.
The remarkable unique feature of all nodule-specific calmodulin-like proteins is a conserved 40-amino acid-long N-terminal extension, which is absent from all typical calmodulins. As predicted by PSORT analysis, these N-terminal peptides contain a putative cleavable signal sequences (24 or 18 amino acids long) that potentially target the proteins into the endomembrane system or outside the cell. Typical calmodulins (including M. truncatula TC31994 and TC35885) lack an N-terminal extension encoding a signal sequence. As predicted by PSORT analysis, TC31994 and TC35885 polypeptides are localized in the cytoplasm, typical of the common calmodulins (Zielinski, 1998). Thus, it appears that the subcellular location of these calmodulin-like proteins may be different from that of typical calmodulins.
Interestingly, the signal peptide of calmodulin-like proteins is very similar to those found in nodulin-25 (TC35677 and TC35678). For example, there is 75% similarity between the signal peptides of TC35678 (nodulin-25) and TC35910 (nodule-specific calmodulin-like protein). In both cases, the cleavage site is predicted to occur after the first 24 amino acids of the polypeptide. However, the mature nodulin-25 does not show any similarity to calmodulins and, as determined by Inter-Pro scanning, lacks any EF hand domains. To determine whether any other M. truncatula sequences, besides nodulin-25, contain such an N-terminal signal motif, we searched the MtGI database with the amino acid signal sequences of calmodulin-like proteins (TBLASTN analysis). No other TCs appear to have such a signal peptide. Among the singletons, only two accessions with the similar signal motif were found (AW127197 and BE999027). Both ESTs were sequenced from nodule libraries (GVN and GVSN, respectively), and their closest nucleotide matches are nodule-specific calmodulin-like proteins (BLASTN against MtGI). Moreover, searching the entire National Center for Biotechnology Information (NCBI) protein database did not reveal additional accessions with similar signal motifs (BLASTP analysis).
Nodule-Specific TCs Encoding CCPs
Five types of CCPs showing some similarity to previously described pea (Pisum sativum) nodulin 3 (ENOD3), nodulin 6, and nodulin 14 (Scheres et al., 1990; Kardailsky et al., 1993) have been identified in broad bean (Frühling et al., 2000) and in pea (Kato et al., 2002). These nodule-specific proteins possess two noteworthy features: similar N-terminal secretory signal sequences and conserved Cys-X4-Asp-Cys and Cys-X4-Cys elements in their C-terminal halves. Surprisingly, we found 114 nodule-specific TCs showing some similarity to these previously reported CCPs and also to a hypothetical protein of another legume, Galega orientalis (GenBank accession no. CAB51773). Of these 114 TCs, also named CCPs, 40 TCs are composed of more than five ESTs each, whereas the rest of them are composed of only two to five ESTs. Similar to previously characterized broad bean and pea CCPs, these TCs encode small proteins (60–90 amino acids). Analysis of the deduced amino acid sequences of 114 TCs (several examples are shown in Fig. 3) confirmed the presence of both characteristic features of CCPs. As demonstrated by PSORT analysis, the N terminus of all 114 CCPs contains a putative signal sequence predicted to target the polypeptide out of the cytoplasm. The majority of M. truncatula CCPs contain Cys clusters identical to those of broad bean or pea proteins. However, a few deviations in structure of both Cys clusters were found.
Figure 3.
Comparison of the deduced amino acid sequences of several TCs encoding CCPs with CCPs from other legumes: pea ENOD3 (GenBank accession no. P25225), broad bean CCP2 and CCP5 (GenBank accession nos. AJ243463 and AJ243466), and hypothetical protein of G. orientalis (GenBank accession no. CAB51773). Comparisons are made against pea ENOD3. Dots represent identical amino acids. Amino acids shaded in gray share similar physico-chemical properties. Gaps (indicated by dashes) are introduced to maintain maximum sequence similarity. Conserved Cys clusters are shown in boxes. The circled amino acids are the last ones in the predicted cleavable N-terminal signal sequence. Underlined amino acid in TC39082 is the last one in the predicted signal peptide that is not cleaved.
Three types of changes were found in the first Cys cluster: (a) For 22 of the 114 predicted proteins, the Asp was not conserved; (b) For two predicted proteins (encoded by TC37420 and TC40754), the Cys were separated by three or 12 amino acids instead of 5; and (c) For three predicted proteins, the second Cys was replaced by a Trp or Tyr, possibly due to sequencing errors (Cys is encoded by TGT or TGC, whereas tryptophan is encoded by TGG, and Tyr is encoded by TAT). Therefore, a more correct predominant structure of the first Cys cluster for M. truncatula CCPs would be “Cys-X5-Cys.”
Deviations from the proposed model were also observed in the structure of the second Cys cluster: (a) In six and three of the predicted proteins, two Cys were separated by five or six amino acids instead of four amino acids; and (b) in seven predicted proteins, one of the Cyst was substituted by Phe, Tyr, or Leu. However, similar to the situation with the first cluster, all of these substitutions may be a result of a single nucleotide sequencing error. Our data indicate that the structure of the second Cys cluster can be best described as “Cys-X4–6-Cys.”
Because some ESTs for CCP-encoding TCs were sequenced from the MtBB and GVSN libraries, respectively, expression of CCP genes appears to be induced before the onset of N2 fixation and extends throughout nodule senescence.
Validation of Nodule-Specific TCs Identified in Silico through Macroarrays and RNA Blots
To assess whether genes identified as nodule specific via in silico analysis showed enhanced expression in nodules in vivo, transcript abundance for selected TCs was evaluated by macroarray hybridization and RNA-blot analysis. The 91 TCs chosen for macroarray analysis were composed of a variable number of ESTs: 13 contained 20 or more ESTs, 13 TCs contained 10 to 19 ESTs, 28 TCs contained five to nine ESTs, and 37 TCs contained two to four ESTs. Each TC on the macroarray was represented by two different cDNA clones, and each clone was spotted in duplicate. The experiment evaluated the hybridization intensities for each spot on three different filters probed with radioactively labeled cDNAs derived from nodule, leaf, or root mRNA. Four macroarray hybridizations were performed, each using independently harvested tissue for mRNA extraction. We determined the average nodule-to-root (N:R) and nodule-to-leaf (N:L) ratios of the intensities of hybridization signal for each TC sequence. Table V represents the final N:R and N:L averages from all four experiments. We defined TCs as being nodule enhanced when gene expression in nodules exceeded that in other tissues by at least 2-fold.
Table V.
Experimental evaluation of a nodule-enhanced expression pattern by macroarray hybridization
| TC No. | No. of ESTs in TC | Strongest BLASTX Hit | Average N:R Ratio ± sda | Average N:L Ratio ± sdb |
|---|---|---|---|---|
| TC31874 | 84 | MtN22, M. truncatula (CAA75576) | 33 ± 11 | 57 ± 31 |
| TC31875 | 54 | Lb 2, M. truncatula (P27993) | 127 ± 7 | 107 ± 49 |
| TC32101 | 45 | Basic blue protein, plantacyanin (CAB65280, M. sativa subsp. × varia) | 10 ± 4 | 20 ± 11 |
| TC32103 | 40 | Putative bark agglutinin precursor (Q41160, R. pseudoacacia) | 7 ± 5 | 8 ± 6 |
| TC39528 | 39 | CCP | 9 ± 2 | 6 ± 3 |
| TC35875 | 35 | CCP | 23 ± 19 | 34 ± 4 |
| TC32071 | 32 | N/Ac | 90 ± 50 | 195 ± 55 |
| TC35568 | 28 | CCP | 55 ± 52 | 65 ± 40 |
| TC31903 | 25 | CCP | 27 ± 18 | 53 ± 1 |
| TC35985 | 24 | N/A | 30 ± 15 | 36 ± 25 |
| TC35677 | 23 | Nodulin 25, M. truncatula(CAB91091) | 53 ± 40 | 57 ± 24 |
| TC32321 | 21 | CCP | 18 ± 16 | 8 ± 1 |
| TC36054 | 20 | Early nodulin ENOD18 | 11 ± 6 | 23 ± 2 |
| TC35962 | 19 | MtN29, M. truncatula (CAA77088) | 4 ± 1 | 4 ± 0.7 |
| TC28580 | 19 | CCP | 44 ± 5 | 75 ± 15 |
| TC35570 | 18 | N/A | 98 ± 57 | 84 ± 31 |
| TC36149 | 16 | CCP | 9 ± 3 | 10 ± 1 |
| TC36162 | 16 | No hits foundd | 15 ± 12 | 6 ± 2 |
| TC35910 | 14 | Calmodulin (CAA69660, T. gondii) | 21 ± 17 | 22 ± 18 |
| TC32516 | 14 | Purine permease (NP_198932, Arabidopsis) | 7 ± 3 | 4 ± 0.2 |
| TC32092 | 14 | B12D protein (AAD22104, I. batatas) | 15 ± 6 | 27 ± 34 |
| TC36302 | 12 | Putative bark agglutinin precursor (Q41160, R. pseudoacacia) | 69 ± 2 | 12 ± 7 |
| TC32593 | 11 | N/A | 17 ± 15 | 6 ± 4 |
| TC32650 | 11 | CCP | 14 ± 11 | 8 ± 1 |
| TC35911 | 10 | Calmodulin (P02598, T. pyriformis) | 15 ± 0.8 | 12 ± 5 |
| TC36259 | 10 | Contains similarity to embryo-specific protein (BAA97184, Arabidopsis) | 8 ± 4 | 18 ± 7 |
| TC36530 | 9 | CCP | 67 ± 36 | 92 ± 15 |
| TC32628 | 9 | CCP | 24 ± 4 | 70 ± 57 |
| TC36519 | 9 | Peroxidase precursor (AAB48986, M. truncatula) | 10 ± 0.4 | 6 ± 3 |
| TC40314 | 8 | Hypothetical protein (NP_189170, Arabidopsis) | 14 ± 8 | 5 ± 0.8 |
| TC32908 | 8 | No hits found | 35 ± 29 | 19 ± 3 |
| TC29160 | 8 | No hits found | 8 ± 6 | 4 ± 2 |
| TC40310 | 8 | N/A | 18 ± 12 | 10 ± 4 |
| TC29253 | 8 | N/A | 8 ± 6 | 3 ± 1 |
| TC40328 | 8 | N/A | 120 ± 106 | 91 ± 3 |
| TC40359 | 8 | N/A | 11 ± 4 | 3 ± 0.4 |
| TC29155 | 8 | CCP | 3 ± 1 | 3 ± 0.2 |
| TC36312 | 7 | CCP | 10 ± 3 | 9 ± 3 |
| TC29226 | 7 | CCP | 7 ± 5 | 4 ± 0.2 |
| TC29276 | 7 | N/A | 19 ± 4 | 1.6 ± 1.1 |
| TC31906 | 7 | N/A | 25 ± 19 | 34 ± 19 |
| TC29417 | 6 | CCP | 7 ± 5 | 6 ± 2 |
| TC33096 | 6 | CCP | 24 ± 10 | 25 ± 22 |
| TC29198 | 6 | Protein kinase-like protein (NP_195559, Arabidopsis) | 6 ± 2 | 1.7 ± 2 |
| TC29264 | 6 | Remorin (T07780, L. esculentum) | 7 ± 2 | 3 ± 2 |
| TC33271 | 6 | N/A | 16 ± 6 | 8 ± 0.9 |
| TC28421 | 6 | Cys proteinase (BAB13759, A. sinicus) | 3 ± 1 | 1.7 ± 1.4 |
| TC37087 | 5 | N/A | 22 ± 10 | 1.5 ± 0.7 |
| TC37055 | 5 | N/A | 15 ± 12 | 5 ± 3 |
| TC29708 | 5 | N/A | 3 ± 1 | 1.4 ± 1.2 |
| TC29680 | 5 | UFD1-like protein (NP_193277, Arabidopsis) | 7 ± 3 | 1.3 ± 1.6 |
| TC40868 | 5 | C2H2-type zinc finger protein (NP_187540, Arabidopsis) | 18 ± 8 | 10 ± 7 |
| TC37150 | 5 | N/A | 88 ± 80 | 20 ± 12 |
| TC32266 | 5 | Copper transporter protein (NP_200711, Arabidopsis) | 33 ± 8 | 12 ± 7 |
| TC29975 | 4 | CCP | 10 ± 3 | 3 ± 0.8 |
| TC40842 | 4 | N/A | 62 ± 60 | 14 ± 0.7 |
| TC33762 | 4 | N/A | 16 ± 8 | 11 ± 1.4 |
| TC37311 | 4 | N/A | 48 ± 34 | 33 ± 8 |
| TC29828 | 4 | No hits found | 48 ± 32 | 26 ± 10 |
| TC33869 | 4 | N/A | 10 ± 5 | 5 ± 2 |
| TC30123 | 4 | N/A | 15 ± 9 | 11 ± 6 |
| TC41039 | 4 | Putative protein, (NP_195884, Arabidopsis) | 12 ± 17 | 1.3 ± 1.4 |
| TC33690 | 4 | Chloroplast nucleoid DNA-binding protein (NP_174430, Arabidopsis) | 13 ± 8 | 2 ± 0.8 |
| TC40949 | 4 | No hits found | 11 ± 6 | 1.5 ± 1.1 |
| TC37507 | 4 | Probable hexose transport protein Hex9 (T10068, R. communis) | 7 ± 12 | 1.1 ± 0.8 |
| TC40870 | 4 | 2-on-2 hemoglobin (AAK55409, Arabidopsis) | 15 ± 16 | 6 ± 3 |
| TC41252 | 4 | Calmodulin (P17928, M. sativa) | 23 ± 8 | 4 ± 1.9 |
| TC40984 | 4 | No hits found | 9 ± 7 | 3 ± 1.4 |
| TC30207 | 3 | His kinase, cytokinin receptor CRE1a (BAB40775, Arabidopsis) | 11 ± 18 | 0.9 ± 0.8 |
| TC30443 | 3 | N/A | 47 ± 33 | 6 ± 4 |
| TC33130 | 3 | Putative protein (M. truncatula, CAA75573) | 5 ± 7 | 0.7 ± 0.6 |
| TC33893 | 3 | Unknown protein (AAF14673, Arabidopsis) | 15 ± 10 | 5 ± 3 |
| TC37860 | 3 | N/A | 12 ± 0.3 | 1.7 ± 1.8 |
| TC38013 | 3 | N/A | 23 ± 18 | 4 ± 2 |
| TC34204 | 3 | Unknown protein (NP_187787, Arabidopsis) | 50 ± 28 | 2 ± 0.7 |
| TC30424 | 3 | N/A | 231 ± 187 | 49 ± 22 |
| TC37606 | 3 | BS14b (AAK51151, Arabidopsis) | 13 ± 3 | 1.6 ± 1.5 |
| TC34333 | 3 | N/A | 32 ± 24 | 5 ± 3 |
| TC33807 | 3 | N/A | 10 ± 7 | 2 ± 0.6 |
| TC30711 | 3 | Putative cyclin-dependent kinase inhibitor (NP_199693, Arabidopsis) | 2 ± 1 | 1.1 ± 1.4 |
| TC41544 | 3 | CCP | 20 ± 8 | 4 ± 1.3 |
| TC41489 | 3 | Putative Glu-/Asp-binding peptide (NP_171806, Arabidopsis) | 39 ± 26 | 20 ± 9 |
| TC30710 | 3 | Lys decarboxylase-like protein (NP_196248, Arabidopsis) | 6 ± 2 | 1.8 ± 1.1 |
| TC30398 | 3 | Probable wound-induced protein (NP_192765, Arabidopsis) | 5 ± 1 | 1.1 ± 0.9 |
| TC38071 | 3 | Hypothetical protein (BAB03434, rice) | 4 ± 1.2 | 1.9 ± 2 |
| TC37978 | 3 | Putative RING zinc finger protein (NP_181294, Arabidopsis) | 18 ± 15 | 1.6 ± 1.5 |
| TC39037 | 2 | Putative protein (CAB75447, Arabidopsis) | 10 ± 5 | 1.6 ± 1.5 |
| TC31651 | 2 | CCP | 27 ± 29 | 6 ± 3 |
| TC41877 | 2 | Hypothetical protein (BAB56065, rice) | 7 ± 8 | 4 ± 1.6 |
| TC31584 | 2 | Putative bZIP transcription factor (NP_181594, Arabidopsis) | 5 ± 1.3 | 3 ± 0.3 |
| TC42202 | 2 | N/A | 9 ± 4 | 10 ± 8 |
Average ratio of hybridization intensities (fold increase) between nodule and root probes.
Average ratio of hybridization intensities (fold increase) between nodule and leaf probes.
N/A, Not Assigned; similarity to GenBank entries is above 10−8.
No hits found; no matches in the NCBI protein database.
For all 91 nodule-specific TCs, the average N:R ratio exceeded 2-fold, confirming that expression of all these genes was enhanced in nodules as compared with roots. However, the average N:L ratio was equal to or exceeded 2-fold for only 72 of the TCs, whereas for 19 TCs it was below this value. Of these 19 TCs, three TCs were represented by six or seven ESTs, and the remaining 16 TCs were composed of three to five ESTs each. Overall, the results of the macroarray hybridizations indicate that in silico-based nodule-specific assignment to the TCs may be not correct for TCs composed of five or fewer ESTs. Final verification of the nodule-specific/-enhanced status for such TCs will require more sensitive experimental methods, such as real-time PCR.
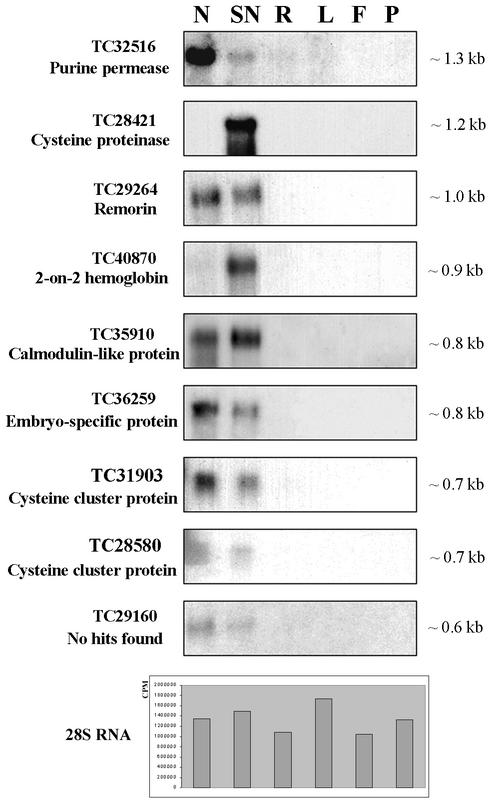
From the 91 TCs selected for macroarrays a subset of nine TCs, each composed of four to 25 ESTs, was examined by RNA-blot analysis. Transcript abundance was evaluated in nodule, senescent nodule, root, leaf, flower, and pod tissues. Equivalent loading of RNA was verified by probing blots with a 28S RNA probe. RNA-blot analysis confirmed the nodule-specific/-enhanced nature of TC32516, TC29264, TC35910, TC36259, TC31903, TC28580, and TC29160 (Fig. 4). Expression of two other nodule-specific TCs (TC40870 and TC28421) dramatically increased during nodule senescence (Fig. 4). Expression of TC28421 (encoding Cys proteinase) was almost undetectable in active N2-fixing nodules by RNA-blot analysis. Not surprisingly, this TC assembly is composed of five ESTs from GVSN (senescent nodule library) and of only one EST from the N2-fixing nodule library (R108Mt). Results of both in silico and in vivo northern analyses for TC28421 indicate why macroarray results did not reveal the enhanced transcript abundance in N2-fixing nodules as compared with leaves. The expression of several other TCs with less than five ESTs each was also examined by RNA-blot analysis (data not shown) and was found to have extremely low levels of hybridization in all tissues. RNA blots could not clearly confirm their in silico classification as nodule specific.
Figure 4.
Northern-blot analysis of selected nodule-specific TCs. Twenty micrograms of total RNA from nodules (N), senescent nodules (SN), roots (R), leaves (L), flowers (F), and pods (P) was separated by gel electrophoresis, transferred onto a nitrocellulose membrane, and hybridized with radioactively labeled cDNA inserts. Inserts represent the clones that belong to nine TCs identified in silico as nodule-specific. Transcript size (kb) is estimated from its electrophoretic mobility. Radioactivity with 28S RNA probe quantified by the AMBIS Radioanalytic Image System (Scanalytics, Billerica, MA) demonstrates the comparative RNA loading.
DISCUSSION
In this report, we have extended the understanding of plant genes involved in symbiotic nitrogen fixation by identifying in silico 340 genes (TCs) that appear to be expressed solely in root nodules. Nodule-specific TCs represent 2.6% of the total TCs annotated in the MtGI. They were identified by applying Boolean search operators to screen 12,925 TCs assembled from over 140,000 ESTs. Nodule-specific TCs are composed of between two and 84 ESTs. Although EST sequencing previously has been successfully used on a limited scale to identify genes that have nodule-enhanced expression (Szczyglowski et al., 1997; Györgyey et al., 2000), this is the first report to employ in silico analysis on a genome-wide scale to identify genes that appear to be specifically related to legume-rhizobium symbiosis. Our analyses revealed several genes with greatly enhanced expression in nodules that were previously overlooked. Moreover, this report is novel in that in silico gene expression data were compared with actual transcript abundance in effective nodules.
Several advantages of an in silico genome-wide approach are immediately evident. Foremost, the number of gene sequences that can be evaluated is virtually unlimited and the analysis is quite rapid. Second, Boolean search operators can simultaneously be applied to ESTs identified from a large number of cDNA libraries reflecting various organs and tissues. Third, the search can be organized to answer a range of questions, such as: (a) which genes are expressed only in selected libraries and not in all others (i.e. nodule specific), (b) which genes are expressed in common in related libraries (i.e. root and shoot meristems), and (c) which genes are represented in all libraries (i.e. constitutively expressed). Last, microarray analysis of gene expression may be limited due to its availability and cost, whereas in silico expression profiling is available to anyone with access to Internet capabilities.
Determining the validity of using in silico expression data as a true reflection of in vivo transcript abundance is extremely important. Audic and Claverie (1997) developed a rigorous statistical test to delineate more precisely and extend the limits within which in silico expression data can be used confidently. To demonstrate the differential expression of a gene, its EST assembly must be composed of more than four ESTs to be considered as having greater than a basal level of expression. Using Audic and Claverie's statistical approach, Ewing et al. (1999) and Mekhedov et al. (2000) analyzed public databases of Arabidopsis and rice to identify genes with differential expression in either selected tissues or between the species. In this work, we attempted to assess the validity of our digital northern data by experimental approach. Preliminary calculations of the probabilities based on formula 1 of Audic and Claverie's analysis (not shown) predicted that nodule-specific TCs of five or fewer ESTs would not have statistically significant differences in expression between nodules and all other tissues. Our experimental data are similar to the prediction. Macroarray results indicate that when a TC sequence identified in silico as being nodule specific is composed of six or more ESTs, the predicted expression profile could usually be verified by physical measurements of transcript abundance on macroarrays or northern blots. In contrast, an in silico-predicted nodule-specific TCs having five or fewer ESTs could not always be confirmed experimentally as actually being expressed in a nodule-specific or -enhanced manner. Our data (Table V; Fig. 4) show that transcript abundance for 19 of 91 in silico nodule-specific TCs chosen for experimental verification was not higher in nodules as compared with leaves (N:L ratio less than 2). Importantly, 16 of these 19 TCs are composed of three to five ESTs only, which implies that the abundance of the corresponding transcripts in these tissues is low. More sensitive experimental approaches will be required to characterize the abundance of their transcripts.
It should be acknowledged that experimental validation of in silico data on macroarrays and RNA blots is complicated by the potential cross hybridization of the closely related sequences. This problem has been already partially addressed in several publications in relation to microarray (Girke et al., 2000; Fernandes et al., 2002) and macroarray (Miller et al., 2002) hybridization systems. For high-density macroarrays, it has been demonstrated that sequences with up to approximately 90% identity show relatively little cross hybridization (Miller et al., 2002). RNA-blot analysis of nodule-specific calmodulin-like TC35910, where a full-length cDNA was used as a probe, was unable detect a hybridization signal in any organ other than nodule or senescent nodule (Fig. 4). This result indicates that no cross hybridization occurred with typical calmodulins, which are expressed elsewhere in the plant. The complete nucleotide sequences of TC35910 and typical calmodulins (TC31994 and TC35885) are identical at 65% and 74%, respectively. This fact indicates that cross hybridization on macroarrays, processed under the same stringency conditions as RNA blots, should not have occurred if the TCs share less than at least 74% identical nucleotides with another closely related TC.
One of the merits of in silico analysis is the opportunity to obtain an overview of the variety of nodule-specific TCs. Although function can be predicted for the protein products of 76 (22%) nodule-specific TCs (TCs for Lbs, TCs related to metabolism, transport, signal transduction, cell structure and maintenance, growth factors, and hormone regulation), 264 (78%) nodule-specific TCs remain functionally uncharacterized. These include TCs from a weak similarity category, TCs for novel proteins and nodulins, unclassified TCs, and TCs similar to hypothetical, unknown, and putative proteins of Arabidopsis and rice. At least 31% of nodule-specific TCs have strong homology to sequences from nonlegume species. These are TCs from a strong similarity category excepting those encoding Lbs and the majority of nodulins. Thus, it appears that a significant proportion of nodule-specific functions are performed by recruiting genes common to all plants. In contrast, approximately one-half of nodule-specific TCs appear to represent the genes unique for legumes. This can be deduced from the fact that corresponding transcripts could not be found in nonlegume species, neither by BLASTX (GenBank protein database) nor by TBLASTX (GenBank EST database) analyses. Legume-specific TCs are those encoding novel proteins, Lbs, and the majority of nodulins (from a strong similarity category), and CCPs (from a weak similarity category). The remaining nodule-specific TCs belonging to the weak similarity category are a potential resource for revealing more legume-specific genes. The fact that complete genomic sequences of Arabidopsis and rice are already available (AGI, 2000; Goff et al., 2002; Yu et al., 2002), and yet these weak similarity TCs do not have statistically significant matches, suggests that these TCs may also encode proteins unique for the legumes. Overall, our data demonstrate that the nodules may be a rich source of genes specific to the legume family.
Boolean search analysis revealed several functionally diverse nodule-specific TCs whose role in nodules was previously overlooked. These include TCs that encode proteins similar to: (a) purine permease, a high-affinity transporter for adenine, cytosine, and purine derivatives (Gillissen et al., 2000); (b) plantacyanin, a plant-specific blue copper protein, apparently involved in the defense response (Nersissian et al., 1998); (c) a homolog of an Arabidopsis embryo-specific protein, whose cellular function is yet to be understood; (d) B12D protein, a protein known to be accumulated in plants during embryo development, seed maturation, and leaf senescence (Aalen et al., 1994; Huang et al., 2001); (e) remorin, a membrane phosphoprotein, suggested to be involved in intercellular communications (Reymond et al., 1996); (f) 2-on-2 hemoglobin, the higher plant homolog of the “truncated” hemoglobins found in bacteria, protozoa, and algae, which possess unique biochemical properties that are likely distinct from those of other plant hemoglobins (Watts et al., 2001); and (g) calmodulin-like proteins.
Calcium is well recognized as a second messenger, playing a vital role in plant responses to biotic and abiotic stimuli (Zielinski, 1998; Snedden and Fromm, 1998; Reddy, 2001). Ca2+ also activates a diverse array of cellular responses affecting plant growth and development. For example, flux in cytoplasmic Ca2+ in root hairs is one of the earliest physiological events occurring in legume-rhizobium interactions (Cárdenas et al., 2000). In the later stages of symbiosis, Ca2+ has been implicated in the functioning of an NH4+ channel of the symbiosome membrane (Tyerman et al., 1995; Streeter, 1998). Ca2+-dependent effects on plant cellular responses are mediated by Ca2+-binding proteins, of which calmodulin is best characterized. Not possessing a catalytic activity of their own, calmodulins interact with various calmodulin-binding proteins, which in turn activate downstream events. Interestingly, among the 340 nodule-specific TCs, we found six calmodulin-like TCs. Although amino acids involved in Ca2+ binding were partially conserved throughout the polypeptide, only two Ca2+-binding domains were complete in the proteins encoded by TC35910, TC35911, and TC34223, and only one was complete in the proteins encoded by TC35912, TC41252, and TC37063. Moreover, unlike typical calmodulins, all nodule-specific calmodulin-like proteins contain a cleavable N-terminal extension. This putative transit sequence is very similar to that found in nodulin-25, which was proposed to be targeted to the PBS (Kiss et al., 1990). PSORT analysis of the calmodulin-like proteins predicted probable targeting of the polypeptides outside the cell. Localization of calmodulin-like proteins to the PBS would also be consistent with Ca2+ modulation of the symbiosome membrane ammonium transporter (Tyerman et al., 1995; Streeter, 1998), and with electron microscopic observations of Ca2+ accumulation inside the PBS (Izmailov et al., 1999).
Another provocative role for nodule calmodulin-like proteins would involve regulation of nodule Glu decarboxylase. This enzyme requires activation by Ca2+-bound calmodulin to convert Glu to γ-aminobutyric acid, which is rapidly accumulated in nodules in response to various stresses (Ling et al., 1994; Serraj et al., 1998). Apyrase, another enzyme known for its calmodulin-binding properties (Hsieh et al., 1996), has also been reported to play an essential role in plant-rhizobium symbiosis (Cohn et al., 2001).
Although this is the first report on plant-encoded calmodulin-like proteins related to legume nodule functioning, a rhizobium-encoded calmodulin-like protein, termed calsymin, has been recently identified in the bean microsymbiont Rhizobium etli (Xi et al., 2000). Similar to nodule-specific expression of plant calmodulin-like proteins, calsymin is expressed in R. etli exclusively during host plant colonization and infection. Moreover, calsymin also appears to be an excreted protein, though its amino acid sequence does not contain a cleavable N-terminal transit peptide. Though calsymin localization and its particular function are unknown, symbiosome structure and nitrogen fixation in nodules formed by the bacterial mutant for calsymin were clearly altered. Overall, the discovery of symbiosis-specific calmodulin-like proteins of both plant and bacterial origin demonstrates the importance of Ca2+-dependent signal transduction processes for functioning of legume root nodules.
A group of 114 nodule-specific TCs was defined as encoding CCPs. The first CCP gene (ENOD3) was reported for pea (Scheres et al., 1990). Later, five more types of CCPs were found and extensively studied in pea (Kato et al., 2002) and in broad bean (Frühling et al., 2000). In silico analysis of the M. truncatula nodule transcriptome shows that this group of proteins is far more extensive than originally thought. The EST content of the various CCP-encoding TCs ranges from two to 39. The fact that 16 of these TCs have more than 10 ESTs suggests that they are highly expressed in effective nodules. Despite significant differences at the nucleotide and even amino acid levels, CCPs are grouped together based on the significant similarity of their N-terminal sequences and the presence of the conserved Cys clusters. N-terminal sequences contain a predicted cleavable signal peptide that could potentially target the polypeptide outside the cell or into the vacuoles or symbiosomes. Similar targeting sequences were reported for CCPs of broad bean and pea, and were proposed to direct the proteins to the PBS or to the vacuoles of the infected cells. It should be mentioned that the signal peptides of other potentially PBS-targeted proteins, such as nodulin-25 and calmodulin-like proteins of M. truncatula, are different from that of CCPs. Cys clusters are known for their metal-binding capacities, which may indicate the involvement of CCPs in binding of molybdenum or iron for nitrogenase, or of cobalt ions required for vitamin B12 synthesis in bacteroids (Scheres et al., 1990; Frühling et al., 2000). The hypothetical relationship of CCPs to bacteroid function may be supported by the fact that expression of CCP genes occurs exclusively in the infected cells of bacteria-/bacteroid-containing nodule zones (Frühling et al., 2000; Kato et al., 2002; M. Fedorova and C.P. Vance, unpublished data).
It is noteworthy that a group of plant defensins, apparently encoding proteinase inhibitors and known for the antifungal activity, also possess several conserved Cys clusters and an N-terminal signal sequence (Maitra and Cushman, 1998; Gao et al., 2000; van der Biezen, 2001). However, amino acid sequence comparisons between known plant defensins and CCPs revealed no similarities (data not shown).
Although we have identified 340 putative nodule-specific genes (nodulins) through an in silico approach, our results need to be viewed conservatively. As originally defined, nodulin genes are those expressed exclusively in nodules (Legocki and Verma, 1980). However, over the last several years, that definition has been modified because a number of nodulin genes show limited expression in other plant organs (de la Peña et al., 1997; Kapranov et al., 1997; Mathesius et al., 2001). The list of such exceptions can be further extended by our survey of the MtGI collection. Several TCs corresponding to the previously known nodulins were not revealed by the Boolean analysis. For example, TC28561 and TC36242, both similar to soybean nodulin 26 (Fortin et al., 1987), and TC29414, similar to soybean early nodulin N93 (Kouchi and Hata, 1993), contain a few ESTs from non-nodule libraries. Of 10 TCs encoding Lb, nine TCs were identified through the Boolean search as being nodule specific. A 10th (TC35564) contains an EST sequenced from the Phytophthora medicaginis-infected root library. Although the possibility of contamination of the infected root RNA with nodule transcripts should not be disregarded, low-level expression of Lb in the infected root tissue should be considered also. Although the genes identified as nodule-specific by our in silico analyses fit the classical definition of nodulins, deeper EST sequencing of individual libraries or more sensitive assessment of transcript abundance may reveal that the so-called nodulins show expression elsewhere in the plant.
It should be also acknowledged that the parameters of Boolean analysis of M. truncatula EST collection used to identify strictly nodule-specific TCs disregards a large group of genes that are also critically involved in nodule functioning and are expressed in a nodule-enhanced, rather than a nodule-specific, manner. For example, TCs encoding Gln synthetase (TC35731), Suc synthase (TC31899), sulfate transporter (TC29347), and hexose transporter (TC29639) would be in this group. Last, 1,867 singletons have been sequenced from nodule libraries. However, because of their apparently low level of expression, it is not possible to confidently assign them a nodule-specific pattern.
A relatively small number of sequences in the database appear to be derived from M. truncatula plastid and mitochondrial genomes. Some of these sequences can be assembled into TCs. However, inspection of the original unprocessed sequence data shows that none of the plastid or mitochondrial-like DNAs, for which the complete sequence is available, have a poly(A+) tail at their 3′ end. We suspect that they may have originated from organellar DNA and not from organellar transcripts. Because nodule libraries were constructed from material collected from tissue containing large numbers of S. meliloti, the possibility exists that some nodule ESTs may have been derived from rhizobium. Therefore, we examined MtGI for the presence of S. meliloti sequences (Galibert et al., 2001) using BLASTN analysis. No significant matches were found.
A number of factors have made the in silico identification of nodule-specific transcripts possible. First, the international community has created a large EST dataset (over 140,000 entries). Second, the ESTs are derived from a collection of libraries constructed from a wide variety of organs, and the data is archived in a relational database. Third, each of the libraries has been sequenced to considerable depth. These factors are extremely important for the validity of an in silico approach, and should be carefully considered for any genome scale analysis.
MATERIALS AND METHODS
Database Analyses, Sequencing, and Sequence Analyses
Structured query language was applied to analyze the MtGI Release 4.0 (http://www.tigr.org/tdb/mtgi) and identify a subset of TCs containing only ESTs from the nodule libraries (MtBB, R108Mt, GVN, Nodulated Root, and GVSN). These nodule-specific TCs were reanalyzed using BLASTX against the NCBI protein database (http://www.ncbi.nlm.nih.gov/BLAST). Additional analysis using TBLASTX was performed for TCs with zero matches in the protein database. The GCG Wisconsin software package (Genetics Computer Group, Madison, WI) was used for sequence analysis and comparisons. Inter-Pro (http://www.ebi.ac.uk/interpro; Apweiler et al., 2001) and PSORT (http://psort.nibb.ac.jp; Nakai and Kanehisa, 1992) software were applied for identification of protein functional domains and prediction of protein sorting signals. TC and clone identification numbers are further given according to the MtGI nomenclature. Additional sequencing for the selected TCs was performed on a 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA) at the Advanced Genetic Analysis Center (University of Minnesota).
Plant Material and Growth Conditions
Seeds of Medicago truncatula [Gaertn.], line A17 of cv Jemalong (T. Huguet, unpublished data), were surface sterilized for 10 min in sulfuric acid, germinated on the petri plates for 2 d, then planted in a sand:vermiculate mix. After planting, seeds were inoculated with Sinorhizobium meliloti 10F51 as described by Egli et al. (1989). Plants were grown in the greenhouse at 24°C, and fertilized weekly with nitrogen-free 0.5× Hoagland solution. One-month-old plants were harvested for nodule, root, and leaf tissue. Flowers and pods were collected from 2-month-old plants. Senescent nodules were obtained 48 h after removal of the shoot material. All tissues were harvested into liquid nitrogen and subsequently used for RNA extraction.
RNA Extraction and RNA-Blot Hybridization
Total RNA was extracted from nodule, senescent nodule, root, leaf, flower, and pod tissues as described by Gregerson et al. (1993). Twenty micrograms of total RNA was separated by electrophoresis through 1.5% (w/v) agarose-formaldehyde gels, transferred to ZetaProbe membrane (Bio-Rad Laboratories, Hercules, CA), and hybridized to 32P-labeled probes as described previously (Pathirana et al., 1992). Probes were made from cDNA inserts of the following clones: pGVSN-12 M18 (0.8 kb) for TC28421, pGVN-51P9 (0.9 kb) for TC32516, pGVSN-13P11 (0.3 and 0.4 kb) for TC40870, pGVN-55I10 (0.9 kb) for TC35910, pGVN-61E23 (0.8 kb) for TC29264, pGVN-64J8 (0.8 kb) for TC36259, N55 (0.6 kb) for TC31903, N71 (0.6 kb) for TC28580, and pGVN-55B21 (0.4 kb) for TC29160.
Macroarray Hybridization
At least two individual clones were evaluated for each TC by macroarray hybridization. cDNA inserts cloned into pBluescript were amplified by PCR of 2 μL of 150-μL overnight bacterial cultures using standard T3 and T7 primers. The quality of each PCR product was examined by gel electrophoresis. Approximately 100 ng of each PCR product was spotted in duplicate onto GeneScreen Plus membranes (NEN Life Science Products, Boston). Each experiment evaluated three membranes hybridized with either 32P-labeled nodule, root, or leaf first strand cDNA probes. Single-stranded probes were synthesized from total RNA using SuperScript II reverse transcriptase (Invitrogen Life Technologies, Carlsbad, CA). The reaction mixture included 7 μL of RNA primer solution [30 μg of total RNA and 0.5 μg of oligo(dT)12–18 primer, annealed by heating to 70°C for 10 min], 4 μL of 5× first strand buffer, 2 μL of 0.1 m dithiothreitol, 1 μL of dNTP mix (2.5 mm dCTP, 2.5 mm dGTP, 2.5 mm dTTP, and 0.0625 mm dATP), 5 μL of [α-32P]dATP (10 mCi mL−1), and 1 μL (200 units) of SuperScript II reverse transcriptase. After 1 h of labeling at 42°C, 1 μL of 5 mm dATP was added, and the incubation was allowed to proceed for additional 30 min. Unincorporated [32P]dATP was removed by passing the mixture through Sephadex G50-G150 columns. 32P incorporation was quantified via liquid scintillation. The final concentration of each probe was adjusted to 106 cpm mL−1 hybridization solution. Membranes were hybridized overnight at 42°C in sodium phosphate buffer containing 50% (v/v) formamide (Gregerson et al., 1993) and subsequently washed at the same temperature with washes of 2× SSC, 0.1% (w/v) SDS; 0.5× SSC, 0.1% (w/v) SDS; and 0.1× SSC, 0.1% (w/v) SDS, for 20 min each wash. The radioactive intensity of the spots on the macroarray was captured by a Phosphor Screen imaging system (Molecular Dynamics/Amersham Biosciences, Piscataway, NJ), and quantified using ImageQuant software. Average N:R and N:L ratios ± sd for each TC were derived from four independent experiments. To ensure the ratios were independent of the amount of the spotted DNA or to the probe binding to vector sequences present at the ends of amplified clones, two types of control experiments were performed. First, different dilutions of a typical amount of PCR products were checked. Four-fold difference in the typical DNA concentration does not alter the ratio results. Second, a PCR-amplified polylinker of the empty pBluescriptSK vector was spotted on the membrane along with other amplified DNAs. No binding of the probe to the polylinker sequence influencing the signal intensities could be detected.
Footnotes
This work was supported by the National Science Foundation (Plant Genome Project no. 9872664) and by the U.S. Department of Agriculture-Agricultural Research Service (grant no. CRIS 3640–21000–014–00D). This is a joint contribution of the U.S. Department of Agriculture-Agricultural Research Service and the Minnesota Agricultural Experimental Station Scientific Journal Series.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.006833.
LITERATURE CITED
- Aalen RB, Opsahlferstad HG, Linnestad C, Olsen OA. Transcripts encoding an oleosin and a dormancy-related protein are present in both the aleurone layer and the embryo of developing barley (Hordeum vulgareL.) seeds. Plant J. 1994;5:385–396. doi: 10.1111/j.1365-313x.1994.00385.x. [DOI] [PubMed] [Google Scholar]
- Ablett E, Seaton G, Scott K, Shelton D, Graham MW, Baverstock P, SladeLee L, Henry R. Analysis of grape ESTs: global gene expression patterns in leaf and berry. Plant Sci. 2000;159:87–95. doi: 10.1016/s0168-9452(00)00335-6. [DOI] [PubMed] [Google Scholar]
- AGI. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Allona I, Quinn M, Shoop E, Swope K, Cyr SS, Carlis J, Riedl J, Retzel E, Campbell MM, Sederoff R et al. Analysis of xylem formation in pine by cDNA sequencing. Proc Natl Acad Sci USA. 1998;95:9693–9698. doi: 10.1073/pnas.95.16.9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MDR et al. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 2001;29:37–40. doi: 10.1093/nar/29.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- Barker DG, Bianchi S, London F, Dattee Y, Duc G, Essad S, Flament P, Gallusci P, Genier G, Guy P et al. Medicago truncatula, a model plant for studying the molecular genetics of the Rhizobium-legume symbiosis. Plant Mol Biol. 1990;8:40–49. [Google Scholar]
- Blondon F, Marie D, Brown S, Kondorosi A. Genome size and base composition in Medicago sativa and M. truncatulaspecies. Genome. 1994;37:264–270. doi: 10.1139/g94-037. [DOI] [PubMed] [Google Scholar]
- Bohnert HJ, Ayoubi P, Borchert C, Bressan RA, Burnap RL, Cushman JC, Cushman MA, Deyholos M, Fischer R, Galbraith DW et al. A genomics approach towards salt stress tolerance. Plant Physiol Biochem. 2001;39:295–311. [Google Scholar]
- Cahoon EB, Carlson TJ, Ripp KG, Schweiger BJ, Cook GA, Hall SE, Kinney AJ. Biosynthetic origin of conjugated double bonds: production of fatty acid components of high-value drying oils in transgenic soybean embryos. Proc Natl Acad Sci USA. 1999;96:12935–12940. doi: 10.1073/pnas.96.22.12935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas L, Holdaway-Clarke TL, Sánchez F, Quinto C, Feijó JA, Kunkel JG, Hepler PK. Ion changes in legume root hairs responding to Nod factors. Plant Physiol. 2000;123:443–451. doi: 10.1104/pp.123.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabaud M, Larsonneau C, Marmouget C, Huguet T. Transformation of barrel medic (Medicago truncatula Gaertn.) by Agrobacterium tumefaciens and regeneration via somatic embryogenesis of transgenic plants with the MtENOD12 nodulin promoter fused to the gusreporter gene. Plant Cell Rep. 1996;15:305–310. doi: 10.1007/BF00232361. [DOI] [PubMed] [Google Scholar]
- Cohn J, Uhm T, Ramu S, Nam Y, Kim D, Penmetsa V, Wood T, Denny R, Young N, Cook DR et al. Differential regulation of a family of apyrase genes from Medicago truncatula. Plant Physiol. 2001;125:2104–2119. doi: 10.1104/pp.125.4.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DR. Medicago truncatula: a model in the making! Curr Opin Plant Biol. 1999;2:301–304. doi: 10.1016/s1369-5266(99)80053-3. [DOI] [PubMed] [Google Scholar]
- Cook DR, VandenBosch K, de Bruijn FJ, Huguet T. Model legumes get the Nod. Plant Cell. 1997;9:275–281. [Google Scholar]
- Cooke R, Raynal M, Laudié M, Grellet F, Delseny M, Morris P-C, Guerrier D, Giraudat J, Quigley F, Clabault G et al. Further progress towards a catalogue of all Arabidopsisgenes: analysis of a set of 5000 non-redundant ESTs. Plant J. 1996;9:101–124. doi: 10.1046/j.1365-313x.1996.09010101.x. [DOI] [PubMed] [Google Scholar]
- Covitz PA, Smith LS, Long SR. Expressed sequence tags from a root-hair enriched Medicago truncatulacDNA library. Plant Physiol. 1998;117:1325–1332. doi: 10.1104/pp.117.4.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Peña TC, Frugier F, McKhann HI, Bauer P, Brown S, Kondorosi A, Crespi M. A carbonic anhydrase gene is induced in the nodule primordium and its cell-specific expression is controlled by the presence of Rhizobiumduring development. Plant J. 1997;11:407–420. doi: 10.1046/j.1365-313x.1997.11030407.x. [DOI] [PubMed] [Google Scholar]
- Desprez T, Amselem J, Caboche M, Höfte H. Differential gene expression in Arabidopsismonitored using cDNA arrays. Plant J. 1998;14:643–652. doi: 10.1046/j.1365-313x.1998.00160.x. [DOI] [PubMed] [Google Scholar]
- Egli MA, Griffith SM, Miller SS, Anderson MP, Vance CP. Nitrogen assimilating enzyme activities and enzyme protein in effective and plant controlled ineffective alfalfa genotypes. Plant Physiol. 1989;91:898–904. doi: 10.1104/pp.91.3.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing RM, Kahla AB, Poirot O, Lopez F, Audic S, Claverie J-M. Large scale statistical analyses of rice ESTs reveal correlated patterns of gene expression. Genome Res. 1999;9:950–959. doi: 10.1101/gr.9.10.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes J, Brendel V, Gai X, Lal S, Chandler VL, Elumalai RP, Galbraith DW, Pierson EA, Walbot V. Comparison of RNA expression profiles based on maize expression sequence tag frequency analysis and micro-array hybridization. Plant Physiol. 2002;128:896–910. doi: 10.1104/pp.010681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin MG, Morrison NA, Verma DP. Nodulin-26, a peribacteroid membrane nodulin is expressed independently of the development of the peribacteroid compartment. Nucleic Acids Res. 1987;15:813–824. doi: 10.1093/nar/15.2.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frühling M, Albus U, Hohnjec N, Geise G, Pühler A, Perlick AM. A small gene family of broad bean codes for late nodulins containing conserved cysteine clusters. Plant Sci. 2000;152:67–77. [Google Scholar]
- Galibert F, Finan TM, Long SR, Puhler A, Abola P, Ampe F, Barloy-Hubler F, Barnett MJ, Becker A, Boistard P et al. The composite genome of the legume symbiont Sinorhizobium meliloti. Science. 2001;293:668–672. doi: 10.1126/science.1060966. [DOI] [PubMed] [Google Scholar]
- Gamas P, Billy de F, Truchet G. Symbiosis-specific expression of two Medicago truncatula nodulin genes, MtN1 and MtN13, encoding products homologous to plant defense proteins. Mol Plant-Microbe Interact. 1998;11:393–403. doi: 10.1094/MPMI.1998.11.5.393. [DOI] [PubMed] [Google Scholar]
- Gamas P, de Calvalho Niebel F, Lescure N, Cullimore JV. Use of a subtractive hybridization approach to identify new Medicago truncatulagenes induced during root nodule development. Mol Plant-Microbe Interact. 1996;9:233–242. doi: 10.1094/mpmi-9-0233. [DOI] [PubMed] [Google Scholar]
- Gao A, Salim SM, Mittanck CA, Wu Y, Woerner BM, Stark DM, Shah DM, Liang J, Rommens CMT. Fungal pathogen protection in potato by expression of a plant defensin peptide. Nat Biotechnol. 2000;18:1307–1310. doi: 10.1038/82436. [DOI] [PubMed] [Google Scholar]
- Genoud T, Métraux JP. Crosstalk in plant cell signaling: structure and function of the genetic network. Trends Plant Sci. 1999;4:503–507. doi: 10.1016/s1360-1385(99)01498-3. [DOI] [PubMed] [Google Scholar]
- Genoud T, Trevino Santa Cruz MB, Métraux JP. Numeric simulation of plant signaling networks. Plant Physiol. 2001;126:1430–1437. doi: 10.1104/pp.126.4.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillissen B, Burkle L, Andre B, Kuhn C, Rentsch D, Brandl B, Frommer WB. A new family of high-affinity transporters for adenine, cytosine, and purine derivatives in Arabidopsis. Plant Cell. 2000;12:291–300. doi: 10.1105/tpc.12.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girke T, Todd J, Ruuska S, White J, Benning C, Ohlrogge J. Microarray analysis of developing Arabidopsisseeds. Plant Physiol. 2000;124:1570–1581. doi: 10.1104/pp.124.4.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan T-H, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- Greene EA, Erard M, Dedieu A, Barker DG. MtENOD16 and 20 are members of a family of phytocyanin-related early nodulins. Plant Mol Biol. 1998;36:775–783. doi: 10.1023/a:1005916821224. [DOI] [PubMed] [Google Scholar]
- Gregerson RG, Miller SS, Twary SN, Gantt JS, Vance CP. Molecular characterization of NADH-dependent glutamate synthase from alfalfa nodules. Plant Cell. 1993;5:215–226. doi: 10.1105/tpc.5.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Györgyey J, Vaubert D, Jiménez-Zurdo JI, Charon C, Troussard L, Kondorosi Á, Kondorosi É. Analysis of Medicago truncatulanodule expressed sequence tags. Mol Plant-Microbe Interact. 2000;13:62–71. doi: 10.1094/MPMI.2000.13.1.62. [DOI] [PubMed] [Google Scholar]
- Handberg K, Stiller J, Thykjaer T, Stougaard J. Transgenic plants: Agrobacterium-mediated transformation of the diploid legume Lotus japonicus. In: Celis J, editor. Cell Biology: A Laboratory Handbook. Orlando, FL: Academic Press; 1994. pp. 119–125. [Google Scholar]
- Handberg K, Stougaard J. Lotus japonicus, and autogamous, diploid legume species for classical and molecular genetics. Plant J. 1992;2:487–496. [Google Scholar]
- Hohnjec N, Küster H, Albus U, Frosch SC, Becker JD, Pühler A, Perlick AM, Frühling M. The broad bean nodulin VfENOD18 is a member of a novel family of plant proteins with homologies to the bacterial MJ0577 superfamily. Mol Gen Genet. 2000;264:241–250. doi: 10.1007/s004380000292. [DOI] [PubMed] [Google Scholar]
- Hsieh HL, Song CJ, Roux SJ. Light-modulated abundance of a mRNA encoding a calmodulin-regulated, chromatin-associated NTPase in pea. Plant Mol Biol. 1996;30:135–147. doi: 10.1007/BF00017808. [DOI] [PubMed] [Google Scholar]
- Huang YJ, To KY, Yap MN, Chiang WJ, Suen DF, Chen SCG. Cloning and characterization of leaf senescence up-regulated genes in sweet potato. Physiol Plant. 2001;113:384–391. doi: 10.1034/j.1399-3054.2001.1130312.x. [DOI] [PubMed] [Google Scholar]
- Izmailov SF, Andreeva IN, Kozharinova GM. Subcellular calcium localization in the root nodules of legumes. Russ J Plant Physiol. 1999;46:93–101. [Google Scholar]
- Jiang Q, Gresshoff PM. Classical and molecular genetics of the model legume Lotus japonicus. Mol Plant-Microbe Interact. 1997;10:59–68. doi: 10.1094/MPMI.1997.10.1.59. [DOI] [PubMed] [Google Scholar]
- Jiménez-Zurdo JI, Frugier F, Crespi MD, Kondorosi A. Expression profiles of 22 novel molecular markers for organogenetic pathways acting in alfalfa nodule development. Mol Plant-Microbe Interact. 2000;13:96–106. doi: 10.1094/MPMI.2000.13.1.96. [DOI] [PubMed] [Google Scholar]
- Kapranov P, Bruijn de FJ, Szczyglowski K. Novel, highly expressed late nodulin gene (LjNOD16) from Lotus japonicus. Plant Physiol. 1997;113:1081–1090. doi: 10.1104/pp.113.4.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I, Yang WC, Zalensky A, van Kammen A, Bisseling T. The pea late nodulin gene PsNOD6 is homologous to the early nodulin gene PsENOD3/14 and is expressed after the leghaemoglobin genes. Plant Mol Biol. 1993;23:1029–1037. doi: 10.1007/BF00021817. [DOI] [PubMed] [Google Scholar]
- Kato T, Kawashima K, Miwa M, Mimura Y, Tamaoki M, Kouchi H, Suganuma N. Expression of genes encoding late nodulins characterized by a putative signal peptide and conserved cysteine residues is reduced in ineffective pea nodules. Mol Plant-Microbe Interact. 2002;15:129–137. doi: 10.1094/MPMI.2002.15.2.129. [DOI] [PubMed] [Google Scholar]
- Kiss GB, Vincze E, Vegh Z, Toth G, Soos J. Identification and cDNA cloning of a new nodule-specific gene, Nms-25 (nodulin-25) of Medicago sativa. Plant Mol Biol. 1990;14:467–475. doi: 10.1007/BF00027493. [DOI] [PubMed] [Google Scholar]
- Kouchi H, Hata S. Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol Gen Genet. 1993;238:106–119. doi: 10.1007/BF00279537. [DOI] [PubMed] [Google Scholar]
- Kruger WM, Pritsch C, Chao S, Muehlbauer GJ. Functional and comparative bioinformatic analysis of expressed genes from wheat spikes infected with Fusarium graminearum. Mol Plant-Microbe Interact. 2002;15:445–455. doi: 10.1094/MPMI.2002.15.5.445. [DOI] [PubMed] [Google Scholar]
- Legocki RP, Verma DP. Identification of “nodule-specific” host proteins (nodulins) involved in the development of rhizobium-legume symbiosis. Cell. 1980;20:153–163. doi: 10.1016/0092-8674(80)90243-3. [DOI] [PubMed] [Google Scholar]
- Ling V, Snedden WA, Shelp BJ, Assmann SM. Analysis of a soluble calmodulin binding protein from fava bean roots: identification of glutamate decarboxylase as a calmodulin-activated enzyme. Plant Cell. 1994;6:1135–1143. doi: 10.1105/tpc.6.8.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart DJ, Winzeler EA. Genomics, gene expression and DNA arrays. Nature. 2000;405:827–836. doi: 10.1038/35015701. [DOI] [PubMed] [Google Scholar]
- Maitra N, Cushman JC. Characterization of a drought-induced soybean cDNA encoding a plant defensin. Plant Physiol. 1998;118:1536. doi: 10.1104/pp.106.2.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleck K, Levine A, Eulgem T, Morgan A, Schmid J, Lawton KA, Dangl JL, Dietrich RA. The transcriptome of Arabidopsis thalianaduring systemic acquired resistance. Nat Genet. 2000;26:403–410. doi: 10.1038/82521. [DOI] [PubMed] [Google Scholar]
- Mathesius U, Keijzers G, Natera SHA, Weinman JJ, Djordjevic MA, Rolfe BG. Establishment of a root proteome reference map for the model legume Medicago truncatulausing the expressed sequence tag database for peptide mass fingerprinting. Proteomics. 2001;1:1424–1440. doi: 10.1002/1615-9861(200111)1:11<1424::AID-PROT1424>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Mekhedov S, Martínez de ilárduya O, Ohlrogge J. Toward a functional catalog of the plant genome. A survey of genes for lipid biosynthesis. Plant Physiol. 2000;122:389–401. doi: 10.1104/pp.122.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller NA, Gong Q, Bryan R, Ruvolo M, Turner LA, LaBrie ST. Cross-hybridization of closely related genes on high-density macroarrays. BioTechniques. 2002;32:620–625. doi: 10.2144/02323pf01. [DOI] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nap J-P, Bisseling T. Nodulin function and nodulin gene regulation in root nodule development. In: Gresshoff PM, editor. The Molecular Biology of Symbiotic Nitrogen Fixation. Boca Raton, FL: CRC Press Inc.; 1990. pp. 181–229. [Google Scholar]
- Nersissian AM, Immoos C, Hill MG, Hart PJ, Williams G, Herrmann RG, Valentine JC. Uclacyanins, stellacyanins, and plantacyanins are distinct subfamilies of phytocyanins: plant specific mononuclear blue copper proteins. Protein Sci. 1998;7:1915–1929. doi: 10.1002/pro.5560070907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GED, Geurts R. Medicago truncatula, going where no plant has gone before. Trends Plant Sci. 2001;6:552–554. doi: 10.1016/s1360-1385(01)02153-7. [DOI] [PubMed] [Google Scholar]
- Pathirana SM, Vance CP, Miller SS, Gannt JS. Alfalfa root nodule phosphoenolpyruvate carboxylase: characterization of the cDNA and expression in effective and plant-controlled ineffective nodules. Plant Mol Biol. 1992;20:437–450. doi: 10.1007/BF00040603. [DOI] [PubMed] [Google Scholar]
- Pichon M, Journet E-P, Dedieu A, de Billy F, Truchet G, Barker DG. Rhizobium melilotielicits transient expression of the early nodulin gene ENOD12 in the differentiating root epidermis of transgenic alfalfa. Plant Cell. 1992;4:1199–1211. doi: 10.1105/tpc.4.10.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush J, Liang F, Holt I, Pertea G, Upton J. The TIGR Gene Indices: reconstruction and representation of expressed gene sequences. Nucleic Acids Res. 2000;28:141–145. doi: 10.1093/nar/28.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy ASN. Calcium: silver bullet in signaling. Plant Sci. 2001;160:381–404. doi: 10.1016/s0168-9452(00)00386-1. [DOI] [PubMed] [Google Scholar]
- Reymond P, Kunz B, Paul-Pletzer K, Grimm R, Eckerskorn C, Farmer EE. Cloning of a cDNA encoding a plasma membrane-associated, uronide binding phosphoprotein with physical properties similar to viral movement proteins. Plant Cell. 1996;8:2265–2276. doi: 10.1105/tpc.8.12.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounsley SD, Glodek A, Sutton G, Adams MD, Sommerville CR, Venter JC, Kerlavage AR. The construction of Arabidopsisexpressed sequence tag assemblies. Plant Physiol. 1996;112:1177–1183. doi: 10.1104/pp.112.3.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y, Gilmore J, Conner T. Towards Arabidopsisgenome analysis-monitoring expression profiles of 1400 genes using cDNA microarrays. Plant J. 1998;15:821–833. doi: 10.1046/j.1365-313x.1998.00254.x. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Song J, Koga-Ban Y, Matsui E, Fang F, Higo H, Nagasaki H, Hori M, Miya M, Murayama-Kayano E et al. Toward cataloguing all rice genes: large-scale sequencing of randomly chosen rice cDNAs from a callus cDNA library. Plant J. 1994;6:615–624. doi: 10.1046/j.1365-313x.1994.6040615.x. [DOI] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Sommerville SC, Manners JM. Coordinated plant defense responses in Arabidopsisrevealed by microarray analysis. Proc Natl Acad Sci USA. 2000;97:11655–11660. doi: 10.1073/pnas.97.21.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B, van Engelen F, van de Knaap E, van de Wiel C, van Kammen A, Bisseling T. Sequential induction of nodulin gene expression in the developing pea nodule. Plant Cell. 1990;2:687–700. doi: 10.1105/tpc.2.8.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K. Monitoring the expression pattern of 1300 Arabidopsisgenes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell. 2001;13:61–72. doi: 10.1105/tpc.13.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serraj R, Shelp BJ, Sinclair TR. Accumulation of gamma-aminobutyric acid in nodulated soybean in response to drought stress. Physiol Plant. 1998;102:79–86. doi: 10.1034/j.1399-3054.1998.1020111.x. [DOI] [PubMed] [Google Scholar]
- Snedden WA, Fromm H. Calmodulin, calmodulin-related proteins and plant responses to the environment. Trends Plant Sci. 1998;3:299–304. [Google Scholar]
- Stekel DJ, Git Y, Falciani F. The comparison of gene expression from multiple cDNA libraries. Genome Res. 2000;10:2055–2061. doi: 10.1101/gr.gr-1325rr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterky F, Regan S, Karlsson J, Hertzberg M, Rohde A, Holmberg A, Amini B, Bhalerao R, Larsson M, Villarroel R et al. Gene discovery in the wood-forming tissues of poplar: analysis of 5,692 expressed sequence tags. Proc Natl Acad Sci USA. 1998;95:13330–13335. doi: 10.1073/pnas.95.22.13330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streeter JG. Effect of elevated calcium concentration in infected cells of soybean (Glycine max (L.) Merr) nodules on nitrogenase activity and N input to the plant. J Exp Bot. 1998;49:997–1003. [Google Scholar]
- Stiller J, Martirani L, Tuppale S, Chian R, Chiurazzi M, Gresshoff PM. High frequency transformation and regeneration of transgenic plants in the model legume Lotus japonicus. J Exp Bot. 1997;48:1357–1365. [Google Scholar]
- Szczyglowski K, Hamburger D, Kapranov P, de Bruijn FJ. Construction of a Lotus japonicuslate nodulin expressed sequence tag library and identification of novel nodule-specific genes. Plant Physiol. 1997;114:1335–1346. doi: 10.1104/pp.114.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoquet P, Gherardi M, Journet EP, Kereszt A, Ane JM, Prosperi JM, Huguet T. The molecular genetic linkage map of the model legume Medicago truncatula: an essential tool for comparative legume genomics and the isolation of agronomically important genes. BioMed Central Plant Biol. 2002;2:1–13. doi: 10.1186/1471-2229-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu AT, Burleigh SH, Kardailsky IV, Maldonado-Mendoza IE, Versaw WK, Blaylock LA, Shin H, Chiou TJ, Katagi H, Dewbre GR et al. Transformation of Medicago truncatula via infiltration of seedlings or flowering plants with Agrobacterium. Plant J. 2000;22:531–541. doi: 10.1046/j.1365-313x.2000.00757.x. [DOI] [PubMed] [Google Scholar]
- Trinh TH, Ratet P, Kondorosi E, Durand P, Kamate K, Bauer P, Kondorosi A. Rapid and efficient transformation of diploid Medicago truncatula and Medicago sativassp falcate lines improved in somatic embryogenesis. Plant Cell Rep. 1998;17:345–355. doi: 10.1007/s002990050405. [DOI] [PubMed] [Google Scholar]
- Tyerman SD, Whitehead LF, Day DA. A channel-like transporter for NH4+ on the symbiotic interface of N2-fixing plants. Nature. 1995;378:629–632. [Google Scholar]
- van Damme EJM, Barre A, Smeets K, Torrekens S, van Leuven F, Rouge P, Peumans WJ. The bark of Robinia pesudoacaciacontains a complex mixture of lectins. Characterization of the proteins and the cDNA clones. Plant Physiol. 1995;107:833–843. doi: 10.1104/pp.107.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Loo FJ, Turner S, Sommerville C. Expressed sequence tags from developing castor seeds. Plant Physiol. 1995;108:1141–1150. doi: 10.1104/pp.108.3.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandenBosch KA, Rodgers LR, Sherrier DJ, Kishinevsky BD. A peanut nodule lectin in infected cells and in vacuoles and the extracellular matrix of nodule parenchyma. Plant Physiol. 1994;104:327–337. doi: 10.1104/pp.104.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Biezen EA. Quest for antimicrobial genes to engineer disease-resistant crops. Trends Plant Sci. 2001;6:89–91. doi: 10.1016/s1360-1385(01)01870-2. [DOI] [PubMed] [Google Scholar]
- Wang RC, Guegler K, LaBrie ST, Crawford NM. Genomic analysis of a nutrient response in Arabidopsisreveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell. 2000;12:1491–1509. doi: 10.1105/tpc.12.8.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts RA, Hunt PW, Hvitved AN, Hargrove MS, Peacock WJ, Dennis ES. A hemoglobin from plant homologous to truncated hemoglobins of microorganisms. Proc Natl Acad Sci USA. 2001;98:10119–10124. doi: 10.1073/pnas.191349198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JA, Todd J, Newman T, Focks N, Girke T, Martínez de la Ilárduya O, Jaworski JG, Ohlrogge JB, Benning C. A new set of Arabidopsisexpressed sequence tags from developing seeds. The metabolic pathway from carbohydrates to seed oil. Plant Physiol. 2000;124:1582–1594. doi: 10.1104/pp.124.4.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi C, Schoeters E, Vanderleyden J, Michiels J. Symbiosis-specific expression of Rhizobium etli casAencoding a secreted calmodulin-related protein. Proc Natl Acad Sci USA. 2000;97:11114–11119. doi: 10.1073/pnas.210181097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GKS, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- Zhu T, Wang X. Large-scale profiling of the Arabidopsistranscriptome. Plant Physiol. 2000;124:1472–1476. doi: 10.1104/pp.124.4.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski RE. Calmodulin and calmodulin-binding proteins in plants. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:697–725. doi: 10.1146/annurev.arplant.49.1.697. [DOI] [PubMed] [Google Scholar]