Abstract
Identification of regulatory molecules that determine the extent and direction of expansion is necessary to understand how cell morphogenesis is controlled in plants. We recently identified COB (COBRA) as a key regulator of the orientation of cell expansion in the root. Analysis of the Arabidopsis genome sequence indicated that COB belongs to a multigene family consisting of 12 members, all predicted to encode glycosylphosphatidylinositol-anchored proteins. All but two of the COBL (COB-like) genes are expressed in most organs examined, suggesting possible redundancy. Sequence comparisons, phylogenetic analyses, and exon-intron positions revealed that the COB family is composed of two main subgroups sharing a common architecture, one subgroup being characterized by an additional N-terminal domain. Identification of expressed sequence tags corresponding to potential orthologs in other plant species suggested that COB-related functions are required in all vascular plants. Together, these results indicate that COB family members are likely to be important new players at the plasma membrane-cell wall interface.
Cell morphogenesis is heavily influenced and restricted by the presence of an extracellular network of carbohydrates and proteins, the cell wall. Far from being an inert and stable exoskeleton, the primary wall is a highly dynamic extracellular matrix characterized by plastic, elastic, and viscous physical properties. These properties are conferred by the nature of the different constituent polymers, a load-bearing cellulose/xyloglucan (hemicellulose) array, and a compression-resistant pectin gel (Roberts, 1994). The developmental regulation of cell wall dynamics required for cell expansion and cell shape modeling must be directed by proteins capable of organizing, loosening, and rearranging the different polysaccharide networks and incorporating newly synthesized material. More than 17% of Arabidopsis genes contain a signal peptide sequence and over 400 proteins are annotated as being localized in the cell wall (Arabidopsis Genome Initiative [AGI], 2000), suggesting the possibility that more than 1,000 genes are implicated in wall biogenesis and modification (Carpita et al., 2001).
Proteins localized to the cell wall, such as the Hyp-, Pro-, or Gly-rich proteins, arabinogalactan proteins, and expansins, were originally identified through assays for biochemical and/or biophysical activities. More recently, forward and reverse genetic approaches have led to the identification of new classes of cell wall modifiers such as cellulose synthases (Arioli et al., 1998; Fagard et al., 2000), endo-β-1,4-glucanases (Nicol et al., 1998), and wall-associated kinases (Kohorn, 2001). By analyzing Arabidopsis mutants exhibiting abnormal cell expansion during root development (Benfey et al., 1993; Hauser et al., 1995), COBRA was identified as an essential player in the regulation of the orientation of cell expansion (Schindelman et al., 2001). Reduced levels of crystalline cellulose microfibrils in the mutant suggested a role for COBRA in cellulose deposition or crystallization (Schindelman et al., 2001).
The COBRA protein is predicted to be anchored to the external plasma membrane leaflet by a glycosylphosphatidylinositol (GPI) moiety. Addition of the GPI anchor is performed in the endoplasmic reticulum and implies the cleavage of a hydrophobic C-terminal peptide and the subsequent linkage of a preassembled GPI anchor via an amide bond onto the last amino acid residue remaining after the cleavage, called the attachment or ω-site (Udenfriend and Kodukula, 1995). This posttranslational modification has been commonly associated with cell surface proteins in animal cells and yeast (Saccharomyces cerevisiae), but has been discovered only recently in plants (Youl et al., 1998; Oxley and Bacic, 1999; Sherrier et al., 1999; Svetek et al., 1999). In animals, the GPI anchor is frequently associated with polar protein sorting, and proteins containing this modification are found in microdomains at the cell surface (Simons and Ikonen, 1997; Friedrichson and Kurzchalia, 1998; Varma and Mayor, 1998). Hence, in addition to providing a means of attachment to the plasma membrane, GPI linkage allows for the cotargeting of unrelated proteins to the same membrane subdomain. Moreover, the GPI anchor can be cleaved by specific phospholipases, which results in the release of the protein from the membrane. This free protein could potentially serve as a signal or as a diffusible enzyme or structural component. Immunolocalization of the COBRA protein indicated that it is present in discrete regions along the longitudinal sides of elongating root cells (Schindelman et al., 2001). One model for its action is that COBRA regulates the polarity of cell expansion by influencing the cellulose microfibril network and defining expansion-resistant areas in regions of the elongating cell.
Completion of the Arabidopsis genome sequence has enabled large-scale analysis of some cell wall protein families such as expansins (Lee et al., 2001; Li et al., 2002), arabinogalactan proteins (Gaspar et al., 2001), and cellulose synthases (Richmond and Somerville, 2000). These families tend to be large. For example, the α-expansins consist of 26 family members (Lee et al., 2001) and the cellulose synthase family is represented by at least 10 CesA-type genes and 41 Csl (cellulose synthase-like) genes (Richmond and Somerville, 2000; Carpita et al., 2001; Desprez et al., 2002). Here, we report that COBRA is part of a new multigene family with the identification of 11 new family members. A phylogenetic analysis of the COB family in Arabidopsis and in other plant species indicates that this family is biphyletic and that COB-related functions are likely to be required in all vascular plants. Moreover, reverse transcriptase (RT)-PCR analysis of all the COB-like genes revealed, with two exceptions, overlapping expression patterns, indicating the possibility of functional redundancy between the different genes. This redundancy is further suggested by the conditional nature of the cob phenotype. Altogether, these results suggest that the COB family members are new players at the plasma membrane-cell wall interface, possibly involved in the development of various organs and tissues.
RESULTS
COBRA Belongs to a Multigene Family in Arabidopsis
A search of Arabidopsis databases with the COBRA sequence (Schindelman et al., 2001) identified 11 homologs. We have named these new members of the COB family COBL (COB-like; Table I). One gene, dl4100c, was previously identified as a cDNA encoding a cell wall protein and named AtSEB1 (Gy et al., 1998). Genomic and deduced amino acid sequences of the COB-like genes were retrieved from the National Center for Biotechnological Information (NCBI) and the Munich Information Center for Protein Sequences (MIPS). The accuracy of the predicted splice sites was verified using the GeneMark.hmm program and by aligning the available EST or cDNA sequence with the genomic sequence. In three cases, this analysis suggested that the current annotation is either incorrect or incomplete.
Table I.
COB gene family members in Arabidopsis
| Gene Name (The Arabidopsis Information Resource [TAIR]) | Gene Code (AGI) | Locus (GenBank) | Length | Protein (GenBank) | Length | Expressed Sequence Tag (EST)/cDNA (TAIR/The Institute for Genomics Research) | Family Name |
|---|---|---|---|---|---|---|---|
| bp | amino acids | ||||||
| F21M12.17 | At1g09790 | AC000132.1 | 1,332 | AAB60732 | 443 | + | COBL6 |
| F21N10.4 | At1g—– | AC027033.3 | 1,335 | AAG12670 | 444 | − | COBL3 |
| F14P3.14 | At3g02210 | AC009755.7 | 1,359 | AAF02128 | 452 | + | COBL1 |
| MUH15.2 | At3g16860 | AP001308.1 | 1,962 | BAB00585 | 653 | ++ | COBL8 |
| K10D20.12 | At3g20580 | AP000410.1 | 2,019 | BAB01166 | 672 | − | COBL10 |
| K17E7.12 | At3g29810 | AP000736.3 | 1,335 | BAB02996 | 444 | − | COBL2 |
| dI4100c | At4g16120 | AL161543.2 | 1,986 | CAA74765 | 661 | ++ | COBL7 |
| T24A18.6 | At4g27110 | AL035680.1 | 2,154 | CAB38841 | 717 | − | COBL11 |
| F14F8.10 | At5g15630 | AL391144.1 | 1,296 | CAC01762 | 431 | + | COBL4 |
| K21P3.15 | At5g49270 | AB016872.1 | 1,992 | BAB10345 | 663 | − | COBL9 |
| MSL3.4 | At5g60920 | AF319663.1 | 1,371 | AAK56072 | 456 | +++ | COB |
| MSL3.7 | At5g60950 | AB008269.1 | 615 | BAB10644 | 204 | + | COBL5 |
Gene names, genome codes, accession nos., gene and protein lengths are indicated. +, No. of corresponding EST/cDNAs identified by public databases searches; –, no EST was found. No AGI code has been attributed to F21N10.4. COB-like (COBL) family names are proposed.
In COBL4 (F14F8.10), a 55-bp first exon and a 74-bp first intron, as well as 53 bp at the beginning of exon 2, were identified. This resulted in the addition of 36 amino acids encoding a putative signal peptide. A similar misannotation was found and previously corrected for the COBRA sequence (Schindelman et al., 2001). A surprising situation was that of COBL2 (K17E7.12) and COBL3 (F21N10.4): Their genomic sequences are 100% identical, but their annotation is different. By aligning the exons of the other COB-like genes, we proposed a new prediction for these two loci, which is a composite of the original ones: The 5′ one-half corresponds to the original prediction for COBL3 and the 3′ region corresponds to that of COBL2. Identification of ESTs or cDNA will be required to confirm this organization.
The annotation for COBL6 (F21M12.17) was tentatively improved using GeneMark.hmm and by examining the alignment with other COB-like sequences. The original second exon was shortened by 33 bp, which became part of the first original intron. The original second, third, and fourth introns, as well as the third and fourth exons, were converted into a single 529-bp intron. The original fifth intron was shortened by 81 bp, which became part of the new third exon. Finally, an EST corresponding to the 3′ end allowed us to convert the last predicted intron into an exon, introducing a new stop codon. COBL5 (MSL3.7) appears to encode a truncated protein because of an in-frame mutation that introduces a stop codon. The availability of an EST that covers this region confirmed this in-frame mutation and indicated that this truncated gene is actively transcribed. The predicted/corrected coding regions and deduced protein lengths are given in Table I.
Chromosomal Distribution of the COB Family Genes Suggests Both Ancient and Recent Duplications
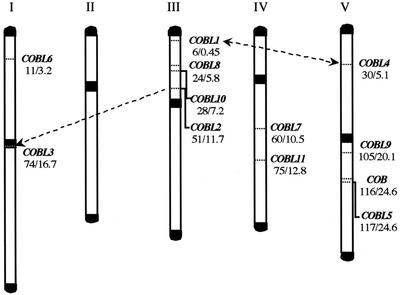
Recent studies have revealed that the Arabidopsis genome contains numerous duplicated areas representing 58% of its total size (AGI, 2000). Subsequent gene loss, smaller duplications, and local rearrangements have resulted in the present complicated organization (Blanc et al., 2000; Vision et al., 2000). To see if the expansion of the COB family was likely to have been a result of segmental duplications, we determined the chromosomal distribution of the 12 COB homologs using the Arabidopsis Sequence Map Overview at TAIR (Fig. 1). A group of four genes (COBL1, COBL8, COBL10, and COBL2) is located on the upper arm of chromosome III and another group of three genes (COBL9, COB, and COBL5) is found on the lower arm of chromosome V. Neither appears to be tightly clustered. COBL3 is located in the centromeric region of chromosome I. COBL6, COBL7, COBL11, and COBL4 are distributed on the upper arm of chromosome I, lower arm of chromosome IV, and upper arm of chromosome V, respectively (Fig. 1).
Figure 1.
Location of the COB homologs on the Arabidopsis chromosomes. The five chromosomes are labeled by roman numerals. The telomeric and centromeric regions are represented in black. For each gene, its genetic and physical distance from the top of the chromosome is given in centiMorgans (left no.) and megabases (right no.), respectively. Identified duplication events are represented by dashed arrows.
COBL1 and COBL4 reside within a large, duplicated segment present in the upper arm of chromosomes III and V, respectively (double-headed arrow, Fig. 1). This duplication event was dated around 170 million years ago (Vision et al., 2000). The complete identity observed between COBL2 and COBL3 suggests a very recent duplication. Analysis of the surrounding sequences indicated that this identity extends over at least eight kilobases: Downstream of both COB-like genes is the same pseudogene (K17E7.13/F21N10.5), followed by the same predicted gene (K17E7.14/F21N10.6), although F21N10.6 is interrupted by a series of transposons. Upstream of COBL2 is another pseudogene (K17E7.11) and upstream of COBL3 are a large number of transposons of different types (Ty3, AtMU2, AtCOPIAs, and ARNOLDs) as well as minisatellite repeats, both found in centromeric regions. Together, these observations suggest that a DNA translocation, from a small area (containing COBL2) located north of the centromeric region of chromosome III to the centromeric region of chromosome I, took place relatively recently (arrow, Fig. 1).
The COBRA Family Consists of Two Subgroups of Putative GPI-Anchored Proteins
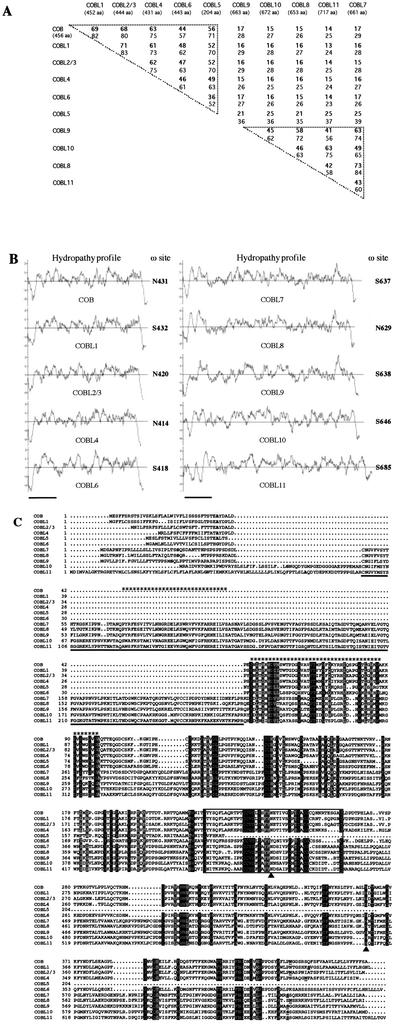
Based on their length, the COB-like proteins can be subdivided into two subgroups, one about 45% longer than the other. One subgroup (the five deduced proteins corresponding to COBL1, COBL2/3, COBL4, COBL5, and COBL6) has a structure very similar to that of COBRA, whereas the other subgroup (COBL8, COBL9, COBL10, and COBL11) shows higher similarity to COBL7. Pair-wise comparisons of the protein sequences confirmed the existence of two distinct subgroups (Fig. 2A). Between subgroups, the identity ranges only from 13% to 25%. Comparisons among the COB subgroup genes showed identity in the range of 36% to 71%. Within the COBL7 subgroup, the proteins are 41% to 73% identical.
Figure 2.
Features of the COB family proteins. A, Degree of similarity between the COB family proteins. Values indicate the percentage of identity (bold nos.) and similarity obtained by pair-wise comparisons between the full-length deduced protein sequences. COBL3 has not been included because it is 100% identical to COBL2. The upper triangle represents COBRA intrasubgroup homologies, the lower represents those for the COBL7 subfamily. B, Hydropathy plot and GPI prediction for the COB family proteins. The hydropathic profile of each member of the family has been analyzed using the Kyte and Doolittle method. The vertical axis represents the degree of hydrophilicity (positive values) or hydrophobicity (negative values). The horizontal axis represents the length of the protein in amino acids (bars represent 100 amino acids). The potential GPI modification was predicted using big-PI predictor and the most likely cleavage site (ω) positions are indicated to the right of each hydropathy profile. C, Sequence alignment of the COB family proteins. The alignment generated by MULTALIN has been edited manually. Gray and black shading indicate conservative and identical residues, respectively, found in at least 85% of the sequences analyzed. Periods represent gaps introduced in the sequences for optimal alignment. Bold letters in this N-terminal domain indicate predicted signal peptide cleavage sites. The 170-amino acid underlined sequence represents the N-terminal domain specific to the COBL7 subclass. A Cys-rich domain highly conserved across the whole family is boxed (CCVS domain). In the C terminus, underlined and bold residues correspond to the predicted cleavage (ω) sites. Two conserved consensus N-glycosylation sites are indicated by a black triangle. The two HMM-predicted putative cellulose-binding sites are indicated by asterisk stretches on the top of the alignment.
To determine if the COBL proteins are likely to have a GPI anchor similar to COBRA (Schindelman et al., 2001), we analyzed their hydrophobicity (Kyte and Doolittle, 1982). The truncated COBL5 protein was not included in this analysis. As shown in Figure 2B, all amino acid sequences examined display a similar profile, with a central hydrophilic portion located between two hydrophobic regions. The N-terminal region corresponds to the signal peptide required for targeting to the endoplasmic reticulum and the hydrophobic C terminus is consistent with the presence of a cleaved propeptide required for GPI linkage. The abnormally long N-terminal hydrophobic region encoded in COBL11 (which would result in a double signal peptide but would not interfere with the GPI prediction; see below) may suggest another erroneous annotation that we could not verify by comparison to an EST.
In addition to the hydrophobic C terminus, GPI addition requires more specific sequence requirements corresponding to the following essential motifs: an unstructured linker region of variable length upstream of the cleavage ω-site, a region of small side chain residues including the ω-site, and a spacer region before the hydrophobic tail (Udenfriend and Kodukula, 1995; Eisenhaber et al., 1999). These features (and others related to protein tertiary structure) have been integrated into a prediction algorithm, big-PI predictor (Eisenhaber et al., 1999), which we used to evaluate the different COB-like proteins. Among the 11 proteins, the program found eight to have a significant potential for GPI modification using the default parameters and proposed a possible GPI addition for the remaining three proteins (COBL7, COBL9, and COBL10). Moreover, when PSORT (Nakai and Horton, 1999) was used to predict GPI modification, all the COB family members were found to be good candidates for GPI addition. The potential ω-cleavage sites predicted by the big-PI software are indicated in Figure 2B. It appears likely, therefore, that all the COB-like sequences except COBL5 encode GPI-anchored proteins.
Alignment of the different protein sequences showed that the N- and C-terminal regions, despite their functional importance, displayed only low similarity, suggesting that there is no strong selective pressure on these areas as long as their hydrophobic nature is conserved (Fig. 2C). Analysis using the ProDom program (Corpet et al., 2000) suggested that the proteins of the COB family, based on their homology and predicted protein structure, are made up of different domains of unknown function, characterized by the conservation of several residues including Cys, Gly, and aromatic amino acids, especially Trp. The COBL7 subgroup is characterized by an N-terminal region of 170 amino acids (underlined in Fig. 2C). All COB family members seem to share three domains: The first domain in common follows the specific 170-amino acid region in the COBL7 subgroup and corresponds to the N-terminal domain in the COB subgroup. The second common region, the CCVS domain (thin box, Fig. 2C), is particularly rich in Cys and contains a consensus N-glycosylation site, a posttranslational modification frequently associated with GPI-anchored proteins and more generally with extracellular proteins. A second conserved N-glycosylation site is present in the third and C-terminal domain, which starts with a short region of poor overall similarity (but with higher subclass similarity) and could represent a spacer region between the CCVS domain and the conserved region preceding the GPI ω-cleavage site.
Phylogenetic Analysis Identifies Subdivisions in the Two COB Family Subgroups
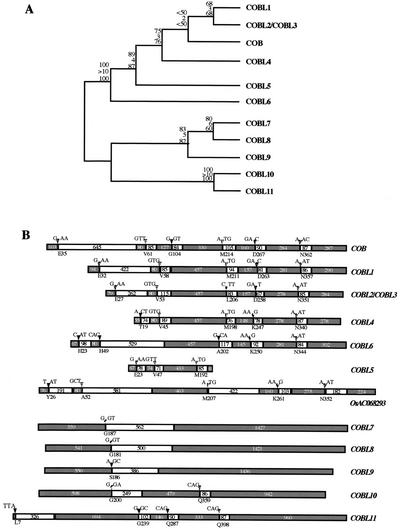
To determine the evolutionary relationships among the COB gene family, a phylogenetic analysis was performed using the full protein sequences. The tree generated by a heuristic search forms two well-defined clades identical to the previously described subgroups (Fig. 3A). The phylogenetic tree could not be rooted because no outgroup could be identified (see below). The topology of the tree, which was largely supported by high bootstrap, Bremer, and jackknife values, was in agreement with the division of the family into two subgroups based on overall protein length, identity, and domain structure. The robust resolution in the tree also enabled us to make further subdivisions within each clade. The COB clade was resolved into four branches: COB/COBL1-3, COBL4, COBL5, and COBL6. The clade of longer proteins has two branches: COBL7-9 and COBL10-11. Further subdivisions could not be proposed due to the high similarity among closely related sequences.
Figure 3.
Phylogenetic relationships in the COB gene family. A, Phylogenetic tree generated from the alignment of COB and COBL proteins. The tree is a single parsimony tree of 1,442 steps and has a consistency index of 0.89 and retention index of 0.86. Values at nodes indicate jackknife (top), Bremer support (middle), and bootstrap values (bottom). B, Exon-intron structure of the COB family genes. If not corrected, exon-intron positions are as predicted by NCBI and MIPS. Exons are indicated as black boxes and introns are indicated as white boxes. Exon and intron sizes are indicated with the number of bases within each box. The beginning of each intron is indicated by a small triangle: Identical filling shades are used for conserved introns, whereas an empty triangle denotes a non-conserved intron. Nucleotide triplets indicate the frame interrupted by each intron; the corresponding amino acid and its position in the deduced protein sequence are indicated below the schematic gene. COBL11's first exon is 21 bp long. The predicted structure of the rice (Oryza sativa) gene AC68293 is also shown.
To further test the phylogenetic analysis, the 12 COB-related sequences were analyzed for the distribution of introns and conservation of splice sites (Fig. 3B). Analysis of the exon-intron organization was in agreement with the division of the COB family into two subgroups. With the exception of the first intron (the second in COBL6), intron length varied only to a small degree, which might indicate a selective pressure to conserve this structure. The exon-intron organization is largely conserved within the two subgroups, but the number of introns differs, ranging from five to six (excluding the truncated COBL5) in the COB subgroup and from one to three in the COBL7 subgroup. Analysis of intron position and reading frame is entirely congruent with the cladistic analysis of amino acid sequences because COB-related genes of the two clades with similar exon-intron structure were grouped together on the phylogenetic tree.
Organ-Specific Expression of the COBRA Family Members
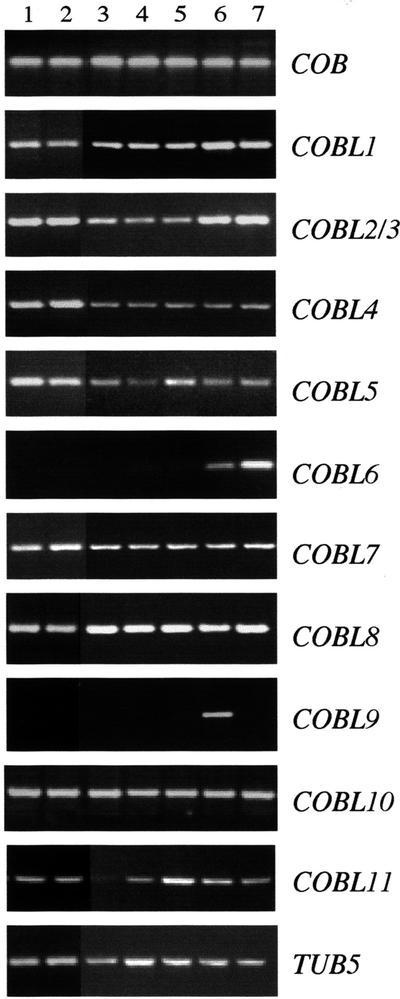
Expression data associated with the origin of an EST or cDNA can be a good preliminary source of information. Some of the ESTs corresponding to COBL1, COBL5, COBL7, and COBL8 came from libraries made from developing seeds and from siliques (COBL6) or from roots (COBL7 and COBL8). If the representation of a given gene in EST databases can be used to estimate the relative abundance of the corresponding mRNA, then the COBRA gene probably has the strongest expression level with more than 60% of the identified ESTs for any of the COB family members.
To determine the organ-specific expression pattern of each member of the COB family, RT-PCR analysis was performed on RNA isolated from 2-week-old roots and aerial organs and 7-week-old roots, rosette leaves, cauline leaves, flowers, and siliques. For most of the genes analyzed, transcripts were detected in all organs tested (Fig. 4). As reported previously (Schindelman et al., 2001), the COBRA gene is expressed in the root as well as in different aerial organs. The transcript of COBL6 was only detected in flowers and siliques and expression of COBL9 was restricted to flowers. With these two exceptions, no clear functional specificity could be inferred from this expression analysis. Overlapping expression domains may indicate that the activity of some COBL protein is functionally redundant. In addition, genes of both subclasses, which may encode complementary functions at the plasma membrane-cell wall interface, may be expressed in the same tissue. In situ hybridizations, as well as functional analyses, will help to clarify this eventuality.
Figure 4.
Expression of the COB-related genes analyzed by RT-PCR. RT-PCR reactions were performed with specific primers to the 3′ end of the last exon and the 3′-untranslated region, on cDNA prepared from 2-week-old roots (1) and aerial parts (2), 7-week-old roots (3), rosette leaves (4), cauline leaves (5), flowers (6), and siliques (7). After 30 to 40 amplification cycles, products were run on a 2% (w/v) agarose gel, and their identity was confirmed by sequencing. Amplification of the tubulin 5 transcript was used as an internal control.
The COB Family Arose and Diversified Early in Vascular Plants
Among the publicly available plant EST databases, more than 200 homologs of COBRA and COB-like genes from 17 different genera associated with 20 species could be identified. Orthology with the different COB family members was determined by cladistic analysis based on the previously established tree (see “Materials and Methods”). Redundant ESTs from the same species were eliminated and analysis was performed on ESTs encoding proteins of at least 140 amino acids in length. Alignment with COB and COB-like proteins revealed that a large majority of these selected ESTs covered the first common domain to all Arabidopsis COB family proteins. This area, which is located between the N-terminal COBL7 subgroup-specific and the CCVS domains (compare with Fig. 2C), was used for the phylogenetic analysis. COB family ortholog status of each EST was determined by the unambiguous presence of the EST sequence in a clade with the Arabidopsis gene. For the purposes of establishing hypotheses of membership in an ortholog class, when an EST did not occur in a clade, it was related to the nearest more basal Arabidopsis COB gene family member. In accordance, potential orthologs in dicots and monocots were identified for COB, COBL1-4, and the COBL7 subgroup and are presented with their putative orthologous Arabidopsis proteins in Figure 5A. Homologous ESTs with high similarity encoding smaller protein fragments were also found in Brassica napus, Lycopersicon hirsutum, Lycopersicon pennelii, Gossypium arboreum, Mesenbryanthemum crystallinum, Secale cereale, Triticum monococcum, and Pinus taeda. ESTs showing low similarity were also found in the moss Physcomitrella patens (data not shown). Unfortunately, these sequences were too short to be included in our phylogenetic analysis. Shotgun sequencing of the rice genome recently revealed the presence of five potential COB homologs in rice, two showing similarity with the COB subgroup (including OsAC068923, Fig. 2B) and three with the COBL7 subgroup. Hence, orthologs of the two Arabidopsis subgroups have been found, indicating that both types of COB-like proteins coexist in other plant species. This phylogenetic analysis also revealed the presence of a clade consisting exclusively of ESTs from grass plants. This clade does not appear to have an ortholog in Arabidopsis and would constitute a sister clade of COB/COBL1-3 and COBL4 (Fig. 5A). One member of this potential COBL grass clade, OsAC068923, corresponds to the predicted coding region of a gene recently sequenced on chromosome 10 of the rice genome. Analysis of the exon-intron organization (Fig. 2B) revealed that this rice gene, with only five introns, is likely to be more related to COBL1 than to COB or COBL2-4.
Figure 5.
Identification of COB and COBL orthologs in other plant species. A, Phylogenetic relationship between Arabidopsis COB family members and ESTs from other species showing similarity with the N-terminal or central domain of the COB or COBL7 subgroups, respectively. A putative grass-specific clade is indicated by a bracket. Notes indicate that very similar ESTs corresponding to independent genes and unresolved by the phylogenetic matrix were found: aLjAW720520, StBE471885, MtBE205436, MtBE205049, LeAW033217; bMtBE203991; cGmAW278341; and dSbBG649393. The phylogenetic tree is a strict consensus of 33 trees with tree length = 1,444, consistency index = 0.74, and retention index = 0.79. The tree was rooted at the break between COB/COBL1-6 versus COBL7-11. B, Alignment of Arabidopsis COB family members and potential orthologous sequences in the CCVS domain area. Shading is as indicated in Figure 2C. Boxed is an example of a conserved Thr that could be used as a diagnostic residue for COBL7 orthologs. Eu, Euphorbia esula; Gm, Glycine max; Gh, Gossypium hirsutum; Hv, Hordeum vulgare; Le, Lycopersicon esculentum; Lj, Lotus japonicum; Ms, Medicago sativa; Mt, Medicago truncatula; Os, rice; Pt, Populus tremula; Sb, Sorghum bicolor; St, Solanum tuberosum; Zm, Zea mays.
Alignment of the Arabidopsis COB and COBL sequences with some of their potential orthologs overlapping in the CCVS domain also showed a strong conservation of this area in both dicots and monocots (Fig. 5B). Moreover, the relative divergence between the two subgroups defined in Arabidopsis is also quite apparent in the other plant species. Such conservation was also observed in other protein domains (data not shown). In most cases, the similarity was higher between members of the same orthologous group than, for example, homologs in the same species. Given the similarity observed for members of the same orthologous group, independent of their originating species, one way to refine orthology assignments of ESTs released in the future is to find clade-specific residues in well-conserved domains. In the CCVS domain, a Thr (boxed in Fig. 5B) conserved exclusively in members of the COBL7 clade would represent such a diagnostic residue for COBL7 orthologs.
Database queries, including iterative profile searches, in prokaryotes, fungi, and other eukaryotes, and using either full-length proteins or motifs such as the CCVS domain presented above, did not lead to the identification of COB homologs outside the plant kingdom. The absence of an outgroup and more distantly related COBL sequences did not permit a more thorough evolutionary analysis.
DISCUSSION
COB and Its Arabidopsis Homologs Define a New Gene Family in Plants
Analysis of the Arabidopsis genome indicated that 11 COB-related genes homologous to COBRA (Schindelman et al., 2001) are present in this model plant. The revisions in their annotations reinforced their overall similarity as well as their potential to represent GPI-anchored proteins like COBRA (see below). Phylogenetic analyses based on sequence comparisons as well as on exon-intron structure showed that the 12 COB family members can be divided into two clades, one showing strong similarity to COB (COBL1–6) and the other being very similar to COBL7 (AtSEB1; COBL7–11). The existence of an extra N-terminal domain in the COBL7 through 11 subgroup lends further support to the division into two clades. The phylogenetic analysis also suggested possible divisions within the two main COB family subgroups. However, it remains to be demonstrated whether these subgroups indicate divergent functions and can be used to propose a durable classification of this new family in plants.
Several authors have suggested that phylogenetic trees can be used to determine orthology of genes in gene families (Burglin, 1994; Ruvkun and Hobert, 1998). We have used this approach to determine provisional ortholog status for ESTs that encode proteins longer than 140 amino acids. EST searches retrieved orthologs for both subgroups of the COB family, suggesting that it arose before the split of the monocot and dicot phyla. In addition, the existence of a COB-orthologous EST from P. taeda indicates that COB-related function was already present in gymnosperms. To our surprise, we found a well-supported clade of COB orthologs that seems specific to the grasses. This divergent grass lineage could indicate the existence of a distinct COB-related function in monocots or, alternatively, it could reflect a loss of this ortholog in Arabidopsis and possibly in all dicots. The existence of potential grass orthologs for COBL2/3 and COBL4 suggests that divergence in the COB subclass happened before the separation between monocots and dicots. Therefore, the possibility that the whole COB/COBL1-4 subgroup evolved more rapidly in Arabidopsis, generating several closely related isoforms divergent from the grass COBL lineage, can be ruled out. Analysis of the rice AC068923 gene structure suggests that this grass clade shares a most recent common ancestor with COBL1-3.
The bootstrap, Bremer, and jackknife values supporting the branch between COB and COBL4 are rather low, which would be consistent with a topology in which COBRA is a sister clade of COBL4. If this were the case, it could be alternatively hypothesized that COB appeared more recently, after the separation between monocots and dicots. The appearance of a sixth intron in COB, which is absent in its closer relatives, supports this possibility. New releases of grass ESTs, as well as the various grass genome-sequencing projects, will permit a test of this hypothesis by determining, for example, whether orthologs of COBRA exist in monocots. Knowing the function of the Arabidopsis gene, this sort of EST analysis could be used to predict the function of the corresponding orthologous gene in its respective species.
We did not find ESTs or genomic sequences showing significant similarity to members of the COB family outside the plant kingdom, suggesting that this family is restricted to plants. The existence of homologous ESTs in the moss P. patens indicates that this gene family already existed in lower plants. It would also be interesting to determine whether non-plant cellulose-synthesizing organisms such as the bacteria Acetobacter xylinum or the slime mold Dictyostelium discoideum (Delmer, 1999) contain COB-related genes.
COB Family Members Are New Potential Players at the Plasma Membrane-Cell Wall Interface
With the completion of the Arabidopsis genome sequence, determining the function of all members of a gene family is a major challenge. In the COB family, only COBRA has a known function in regulating the orientation of cell expansion during root development (Schindelman et al., 2001). The reduction of crystalline cellulose in cob and the association of the COB protein with the plasma membrane, most likely via a GPI anchor, suggest that COBRA plays a regulatory role probably by influencing either the crystallization or deposition of cellulose microfibrils in the cell wall of expanding root cells. All but one of the COB-like proteins are also good candidates to be GPI anchored. Although three of them were not predicted to be GPI modified using the default parameters of the big-PI predictor program, it should be noted that the software was primarily designed and tested on yeast and worm datasets. The availability of sequences for plant proteins with demonstrated GPI modifications (Youl et al., 1998; Oxley and Bacic, 1999; Schultz et al., 2000; Takos et al., 2000) should aid in optimizing this already powerful GPI prediction software for use with plant proteins. Altogether, the expression data and the GPI modification prediction suggest that some members of the COB family may be expressed in the same cells and targeted to the same area of the plasma membrane.
Domain Structure of the COBL Proteins. A First Step toward Function?
Except for COBL6 and COBL9, which appear to be specifically expressed during flower development, expression analysis of the other COB-like genes revealed overlapping expression patterns. The expression patterns suggest there may be a certain redundancy among the COB family members that could explain the conditional nature of the cobra phenotype (Hauser et al., 1995; Schindelman et al., 2001). An appealing hypothesis, largely based on the phenotypic analysis of cob, is that the other COB family members play a role in regulating cell wall dynamics. Homology searches of protein domain databases such as Interpro (Apweiler et al., 2001) did not reveal any common motifs between COB-like proteins and any other proteins, including characterized cell wall-associated proteins. However, alignment of the COB-like proteins revealed conservation of Gly and Pro residues, which are characteristic of some extracellular proteins. A set of conserved Cys could be involved in disulfide bond formation or binding of metal ions. This could explain the original and probably erroneous annotation of a truncated clone of COBRA that was able to complement a yeast mutant deficient in a phytochelatin synthase (Leuchter et al., 1998).
A Hidden Markov model-based program, Superfamily (Gough et al., 2001), identified a part of the COBL7-11-specific N-terminal domain as having a weak similarity to the family II of cellulose-binding domains (Fig. 2C). In addition, a similar cellulose-binding domain was predicted in the first common domain of the COB family (Fig. 2C). These two regions, and especially the latter one, are characterized by the conservation of aromatic residues, which have been shown to be critical for cellulose binding. In particular, Trp has been shown to confer a higher affinity for crystalline cellulose as compared with Phe or Tyr (Linder and Teeri, 1997). Moreover, conservation of similarly non-clustered Trp residues has been found in cellulose-binding domains of microbial cellulases (Gilkes et al., 1991). Interestingly, the residue mutated in the cob-3 mutant, leading to the cell expansion defect, is a Trp (W55) conserved in four of the five COB-subgroup proteins, which is present in the second putative cellulose-binding site. Thus, the two subgroups in the Arabidopsis COB family could be characterized by one (COB/COBL1-6) and two (COBL7-11) potential cellulose-binding sites, respectively. Members of each subgroup could fulfill complementary functions, acting separately or possibly in concert in cellulose microfibril elaboration.
The data presented here will prove valuable in designing and interpreting biochemical and functional analyses that we are presently implementing to unravel the specificity of all COB family members. Our analysis indicates that members of this newly identified family are likely to play important roles at the plasma membrane-cell wall interface and that the functions associated with the different COB-like genes will help elucidate the relationships between cell wall dynamics, cell expansion, and plant morphogenesis.
MATERIALS AND METHODS
Plant Growth Conditions
Arabidopsis plants of the Columbia ecotype were grown as described previously (Benfey et al., 1993). cob-1 and cob-3 are in the Columbia and Wassilewskija ecotypes, respectively.
Sequence Retrieval, Alignment, and Comparison
Gene, protein, EST, and cDNA sequences were identified by searching public databases available at NCBI (http://www.ncbi.nlm.nih.gov), TAIR (http://www.Arabidopsis.org), The Institute for Genomic Research (http://www.tigr.org/tdb/agi/), and MIPS (http://mips.gsf.de/proj/thal/; Schoof et al., 2002) with the BLAST algorithms (Altschul et al., 1990, 1997). Exon-intron splice sites were verified using the GeneMark.hmm program (Lukashin and Borodovsky, 1998) and the available EST/cDNA sequences, as well as by eye. Sequences were aligned using the MULTALIN program (http://prodes.toulouse.inra.fr/multalin/; Corpet, 1988), and pair-wise comparisons were performed with ClustalW (Thompson et al., 1994). The ProDom database was used to designate possible domain arrangements in the COB family proteins (http://protein.toulouse.inra.fr/prodom/; Corpet et al., 2000). Signal peptide and GPI prediction were done using SignalP (http://www.cbs.dtu.dk/services/SignalP; Nielsen et al., 1997) and big-PI predictor (http://mendel.imp.univie.ac.at/gpi/index_ content.html; Eisenhaber et al., 2000), respectively. Hydropathy plots were generated at http://bioinformatics.weizmann.ac.il/hydroph using a Kyte and Doolittle method.
Phylogenetic Analysis
Sequences obtained from BLAST searches were compiled after corrections into a NEXUS matrix using PAUP 4.0b4a (Swofford, 1999). Parsimony searches were performed using a heuristic search with tree bisection and reconnection swapping and 100 random taxon addition iterations to ensure adequate tree space search. Jackknife and bootstrap values were generated using PAUP4.0b4a (Swofford, 1999) with 1,000 resampling replicates used for each analysis. Bremer support values were generated using AutoDecay (Eriksson, 1997). The matrix used to generate this tree is available online at http://research.amnh.org/molecular/index.html under “Cobra.matrix” and has 792 total amino acid characters.
Over 200 ESTs were retrieved by BLAST searches from public databases. Translated ESTs of greater than 140 amino acids were selected for further analysis. A large majority of these translated ESTs covered the beginning of the N-terminal domain of COB and COBL1-6, which also corresponds to the central domain of COBL7-11. Translated EST sequences were placed into a data file and aligned using Clustal. EST sequences that were identical in the matrix were removed as indicated, and phylogenetic analysis was performed using parsimony as described previously. The tree generated by PAUP 4.0 can be found at http://www.amnh.org.research.molecularlabs and had the following statistics: steps = 1,444, number of trees = 33, consistency index = 0.74, retention index = 0.79, and rescaled consistency index = 0.58. Strict consensus trees of the multiple parsimony trees were constructed and the COB family ortholog status of each EST was determined by its unambiguous presence in a clade with the Arabidopsis gene.
RT-PCR Analysis
Organ-specific expression of the different COB homologs was analyzed by nonquantitative RT-PCR in 2-week-old roots and rosette leaves as well as roots, rosette and cauline leaves, flowers, and siliques of 7-week-old plants. Flowers and siliques were collected at different developmental stages and pooled. Samples were ground in liquid nitrogen and total RNA was isolated using the RNeasy kit (Qiagen USA, Valencia, CA) according to the manufacturer's instructions. For RT-PCR experiments, 1 μg of total RNA (as determined by UV spectrophotometry) was treated with 1 unit of Rnase-free Dnase for 30 min at room temperature to remove any residual genomic DNA contamination. cDNAs were generated using the Thermoscript RT-PCR system (Life Technologies/Gibco-BRL, Cleveland). PCR reactions were carried out with Taq polymerase (Boehringer Mannheim/Roche, Basel) and primers were designed against the extremity of the last exon and the beginning of the 3′-untranslated region. The gene-specific primers used are as follows (forward/reverse): COBRA (COB), caacggtggttcccgtcac/cgtttataccactccgccgtaacc; F14P3.14 (COBL1), cgcacaaatcagtcggttccc/gagaacaaagaagtggtagcc; K17E7.12/F21N10.4 (COBL2/3), gtccaaacattgcaacctcgc/gtcacaatacatacatagcatgc; F14F8.10 (COBL4), ctaccaaactctgcacaaggg/gtacagagtcattgatcaatggc; MSL3.7 (COBL5), gaataactgcagccctaatgacc/ctcaagtctttgattttgtagc; F21M12.17 (COBL6), ggtgatgaatgtgttatgcc/gaagcatggaacaatgtaggttc; AtSEB1 (COBL7), atgagaagtagccaacaccg/ggtaacatatcttcatagcacc; MUH15.2 (COBL8), ccacgagcaacagtcacagg/cgaaattcaagaatcacacgg; K21P3.15 (COBL9), gtggtggcagacgaaatggg/ggttttctgcttttcgtctgcc; K10D20.12 (COBL10), agagctcagggcatagacgc/caatgatataacaatctgctcc; and T24A18.6 (COBL11), ttccgggatgagattatccg/caatctgctttagttcctttccg. The product of each reaction was run on a 2% (w/v) agarose gel and sequenced to confirm its identity. The primer couple designed to amplify the transcript of COBL10 also amplified a larger band corresponding to the unrelated gene At3g19370.
ACKNOWLEDGMENTS
We acknowledge Casey Roehrig for her excellent technical assistance and help in figure preparation. We thank Joanna Chiu for use of some reagents. We also thank Alice Paquette, Anita Fernandez, and Kenneth Birnbaum for critical reading of the manuscript.
Footnotes
This work was supported by the National Science Foundation (grants to P.N.B.), by the Graduate School of Arts and Sciences at New York University (Dean's Dissertation Fellowship to G.S.), and in part by the Lewis B. and Dorothy Cullman Program for Molecular Systematic Studies and the Ambrose Monell Collection for Molecular and Microbial Research (to R.D.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.007468.
LITERATURE CITED
- AGI. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti T, Corpet F, Croning MDR et al. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 2001;29:37–40. doi: 10.1093/nar/29.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Hofte H, Plazinski J, Birch R et al. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science. 1998;279:717–720. doi: 10.1126/science.279.5351.717. [DOI] [PubMed] [Google Scholar]
- Benfey PN, Linstead PJ, Roberts K, Schiefelbein JW, Hauser MT, Aeschbacher RA. Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development. 1993;119:57–70. doi: 10.1242/dev.119.Supplement.57. [DOI] [PubMed] [Google Scholar]
- Blanc G, Barakat A, Guyot R, Cooke R, Delseny M. Extensive duplication and reshuffling in the Arabidopsisgenome. Plant Cell. 2000;12:1093–1101. doi: 10.1105/tpc.12.7.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burglin TR. A Caenorhabditis elegans prosperohomologue defines a novel domain. Trends Biochem Sci. 1994;19:70–71. doi: 10.1016/0968-0004(94)90035-3. [DOI] [PubMed] [Google Scholar]
- Carpita N, Tierney M, Campbell M. Molecular biology of the plant cell wall: searching for the genes that define structure, architecture and dynamics. Plant Mol Biol. 2001;47:1–5. [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F, Servant F, Gouzy J, Kahn D. ProDom and ProDom-CG: tools for protein domain analysis and whole genome comparisons. Nucleic Acids Res. 2000;28:267–269. doi: 10.1093/nar/28.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmer DP. Cellulose biosynthesis: exciting times for a difficult field of study. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:245–276. doi: 10.1146/annurev.arplant.50.1.245. [DOI] [PubMed] [Google Scholar]
- Desprez T, Vernhettes S, Fagard M, Refregier G, Desnos T, Aletti E, Py N, Pelletier S, Hofte H. Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6. Plant Physiol. 2002;128:482–490. doi: 10.1104/pp.010822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhaber B, Bork P, Eisenhaber F. Prediction of potential GPI-modification sites in proprotein sequences. J Mol Biol. 1999;292:741–758. doi: 10.1006/jmbi.1999.3069. [DOI] [PubMed] [Google Scholar]
- Eisenhaber B, Bork P, Yuan Y, Loeffler G, Eisenhaber F. Automated annotation of GPI anchor sites: case study C. elegans. Trends Biol Sci. 2000;25:340–341. doi: 10.1016/s0968-0004(00)01601-7. [DOI] [PubMed] [Google Scholar]
- Eriksson T. AutoDecay Version 2.9.8 (hypercard stock disk distributed by the author). Botoniska Institutionen, Stockholm University; 1997. [Google Scholar]
- Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, McCann M, Rayon C, Vernhettes S, Hofte H. PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell. 2000;12:2409–2424. doi: 10.1105/tpc.12.12.2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichson T, Kurzchalia TV. Microdomains of GPI-anchored proteins in living cells revealed by crosslinking. Nature. 1998;394:802–805. doi: 10.1038/29570. [DOI] [PubMed] [Google Scholar]
- Gaspar Y, Johnson KL, McKenna JA, Bacic A, Schultz CJ. The complex structures of arabinogalactan-proteins and the journey towards understanding function. Plant Mol Biol. 2001;47:161–176. [PubMed] [Google Scholar]
- Gilkes NR, Henrissat B, Kilburn DG, Miller RC, Warren RAJ. Domains in microbial β-1,4-glucanases: sequence conservation, function and enzyme families. Microbiol Rev. 1991;55:303–315. doi: 10.1128/mr.55.2.303-315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough J, Karplus K, Hughey R, Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent proteins of known structure. J Mol Biol. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- Gy I, Aubourg S, Sherson S, Cobbett CS, Cheron A, Kreis M, Lecharny A. Analysis of a 14-kb fragment containing a putative cell wall gene and a candidate for the ARA1, arabinose kinase, gene from chromosome IV of Arabidopsis thaliana. Gene. 1998;16:201–210. doi: 10.1016/s0378-1119(98)00049-3. [DOI] [PubMed] [Google Scholar]
- Hauser MT, Morikami A, Benfey PN. Conditional root expansion mutants of Arabidopsis. Development. 1995;121:1237–1252. doi: 10.1242/dev.121.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohorn BD. Plasma membrane-cell wall contacts. Plant Physiol. 2001;124:31–38. doi: 10.1104/pp.124.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lee Y, Choi D, Kende H. Expansins: ever-expanding numbers and functions. Curr Opin Plant Biol. 2001;4:527–532. doi: 10.1016/s1369-5266(00)00211-9. [DOI] [PubMed] [Google Scholar]
- Leuchter R, Wolf K, Zimmermann M. Isolation of an Arabidopsis cDNA complementing a Schizosaccharomyces pombemutant deficient in phytochelatin synthesis. Plant Physiol. 1998;117:1526. [Google Scholar]
- Li Y, Darley CP, Ongaro V, Fleming A, Schipper O, Baldauf SL, McQueen-Mason SJ. Plant expansins are a complex multigene family with an ancient evolutionary origin. Plant Physiol. 2002;128:854–864. doi: 10.1104/pp.010658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder M, Teeri T. The roles and function of cellulose-binding domains. J Biotechnol. 1997;57:15–28. doi: 10.1016/s0168-1656(97)00088-6. [DOI] [PubMed] [Google Scholar]
- Lukashin AV, Borodovsky M. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- Nicol F, His I, Jauneau A, Vernhettes S, Canut H, Hofte H. A plasma membrane-bound putative endo-1,4-beta-d-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 1998;17:5563–5576. doi: 10.1093/emboj/17.19.5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Oxley D, Bacic A. Structure of the glycosylphosphatidylinositol anchor of an arabinogalactan protein from Pyrus communissuspension-cultured cells. Proc Natl Acad Sci USA. 1999;96:14246–14251. doi: 10.1073/pnas.96.25.14246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond TA, Somerville CR. The cellulose synthase superfamily. Plant Physiol. 2000;124:495–498. doi: 10.1104/pp.124.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts K. The plant extracellular matrix in a new expansive mood. Curr Opin Cell Biol. 1994;6:688–694. doi: 10.1016/0955-0674(94)90095-7. [DOI] [PubMed] [Google Scholar]
- Ruvkun G, Hobert O. The taxonomy of developmental control in Caenorhabditis elegans. Science. 1998;282:2033–2041. doi: 10.1126/science.282.5396.2033. [DOI] [PubMed] [Google Scholar]
- Schindelman G, Morikami A, Jung J, Baskin TI, Carpita NC, Derbyshire P, McCann MC, Benfey PN. COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev. 2001;15:1115–1127. doi: 10.1101/gad.879101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H, Zaccaria P, Gundlach H, Lemcke K, Rudd S, Kolesov G, Arnold R, Mewes HW, Mayer KFX. MIPS Arabidopsis thalianaDatabase (MAtDB): an integrated biological knowledge resource based on the first complete plant genome. Nucleic Acids Res. 2002;30:91–93. doi: 10.1093/nar/30.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz CJ, Johnson KL, Currie G, Bacic A. The classical arabinogalactan protein gene family of Arabidopsis. Plant Cell. 2000;12:1751–1768. doi: 10.1105/tpc.12.9.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrier DJ, Prime TA, Dupree P. Glycosylphosphatidylinositol-anchored cell-surface proteins from Arabidopsis. Electrophoresis. 1999;20:2027–2035. doi: 10.1002/(SICI)1522-2683(19990701)20:10<2027::AID-ELPS2027>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- Svetek J, Yadav MP, Nothnagel EA. Presence of a glycosylphosphatidylinositol lipid anchor on rose arabinogalactan proteins. J Biol Chem. 1999;274:14724–14733. doi: 10.1074/jbc.274.21.14724. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP 4.01: Phylogenetic Analysis Using Parsimony (and Other Methods), Version 4.01. Sunderland, MA: Sinauer; 1999. [Google Scholar]
- Takos AM, Dry IB, Soole KL. Glycosyl-phosphatidylinositol-anchor addition signals are processed in Nicotiana tabacum. Plant J. 2000;21:43–52. doi: 10.1046/j.1365-313x.2000.00651.x. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udenfriend S, Kodukula K. How glycosyl-phosphatidylinositol-anchored membrane-proteins are made. Annu Rev Biochem. 1995;64:563–591. doi: 10.1146/annurev.bi.64.070195.003023. [DOI] [PubMed] [Google Scholar]
- Varma R, Mayor S. GPI-anchored proteins are organized in submicron domains at the cell surface. Nature. 1998;394:798–801. doi: 10.1038/29563. [DOI] [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis. Nature. 2000;290:2114–2117. doi: 10.1126/science.290.5499.2114. [DOI] [PubMed] [Google Scholar]
- Youl JJ, Bacic A, Oxley D. Arabinogalactan-proteins from Nicotiana alata and Pyrus communiscontain glycosylphosphatidylinositol membrane anchors. Proc Natl Acad Sci USA. 1998;95:7921–7926. doi: 10.1073/pnas.95.14.7921. [DOI] [PMC free article] [PubMed] [Google Scholar]