Abstract
Proanthocyanidin (PA), or condensed tannin, is a polymeric flavanol that accumulates in a number of tissues in a wide variety of plants. In Arabidopsis, we found that PA precursors (detected histochemically using OsO4) accumulate in the endothelial cell layer of the seed coat from the two-terminal cell stage of embryo development onwards. To understand how PA is made, we screened mature seed pools of T-DNA-tagged Arabidopsis lines to identify mutants defective in the synthesis of PA and found six tds (tannin-deficient seed) complementation groups defective in PA synthesis. Mutations in these loci disrupt the amount (tds1, tds2, tds3, tds5, and tds6) or location and amount of PA (tds4) in the endothelial cell layer. The PA intermediate epicatechin has been identified in wild type and mutants tds1, tds2, tds3, and tds5 (which do not produce PA) and tds6 (6% of wild-type PA), whereas tds4 (2% of wild-type PA) produces an unidentified dimethylaminocinnamaldehyde-reacting compound, indicating that the mutations may be acting on genes beyond leucoanthocyanidin reductase, the first enzymatic reduction step dedicated to PA synthesis. Two other mutants were identified, an allele of tt7, which has a spotted pattern of PA deposition and produces only 8% of the wild-type level of type PA as propelargonidin, and an allele of tt8 producing no PA. Spotted patterns of PA deposition observed in seed of mutants tds4 and tt7-3 result from altered PA composition and distribution in the cell. Our mutant screen, which was not exhaustive, suggests that the cooperation of many genes is required for successful PA accumulation.
Flavonoids are a diverse group of plant secondary metabolites that accumulate in a wide variety of plant tissues and include anthocyanins, flavonols, and the polymeric flavanols known as proanthocyanidins (PAs; Fig. 1). Like their flavanol constituents, PAs are rich in hydrophobic aromatic rings and hydroxyl groups that can interact with biological molecules, particularly proteins, by hydrogen bonds and hydrophobic interactions. Because PAs are polymeric, their interaction with proteins is much stronger than that of monomeric flavanols, presumably because of a “chelate” effect where polymeric PAs can interact with large proteins at multiple sites, increasing the strength of overall molecular interaction as well as minimizing dissociation of the complex once it is formed (Fersht, 1985). This strong interaction of PAs with proteins is probably the basis of their main role in plants and their uses by man.
Figure 1.
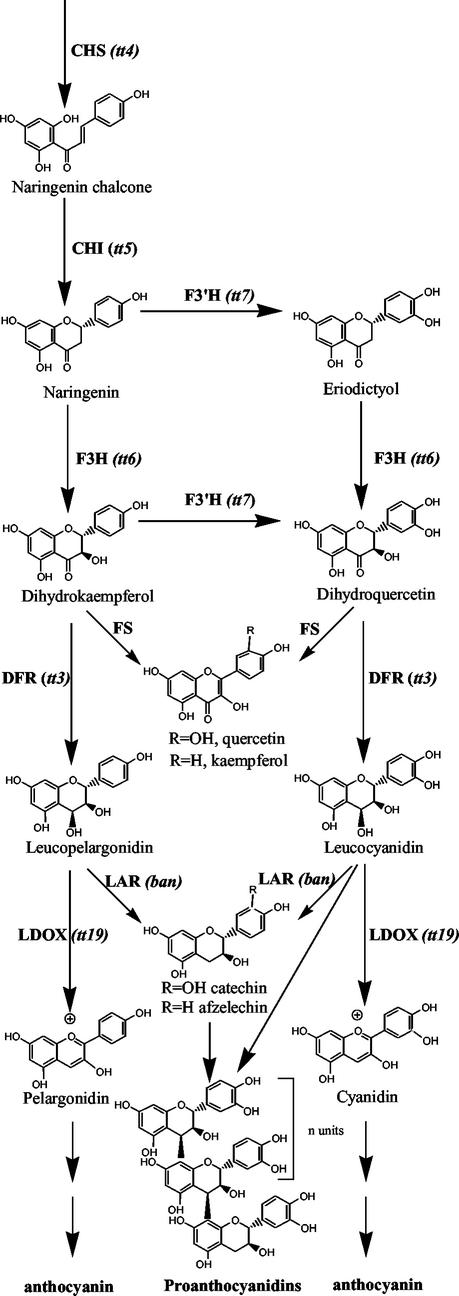
The anthocyanin and PA biosynthetic pathways. Enzymatic steps altered in the transparent testa mutants referred to in the text are shown. There is no evidence that Arabidopsis produces tri-hydroxylated intermediates or end products in PA or anthocyanin biosynthesis, so this branch of the pathway is not shown. All steps analyzed so far, with the possible exception of flavonol synthase (FS), are encoded by a single gene in Arabidopsis. CHI, Chalcone isomerase; F3H, flavanone 3-hyroxylase; F3′H, flavanone 3′-hydroxylase; DFR, dihydroflavonol reductase; LDOX, leucoanthocyanidin dioxygenase.
Because PA content of dietary plants can have both positive and negative effects on animal nutrition (Waghorn and Jones, 1989), an important agricultural objective is to manipulate the level of PA in pasture legumes (Morris and Robbins, 1997). When present in bloat-safe forage legumes, PA can bind to and cause the precipitation of dietary proteins, inhibiting the formation of stable proteinaceous foams, thereby preventing bloat (Tanner et al., 1995). Increasing the PA content of pastures such as alfalfa (Medicago sativa) and white clover (Trifolium repens) may decrease the incidence of bloat. Unfortunately, the presence of large amounts of PA can also reduce the palatability of forage legumes (Kumar and Singh, 1984), so in some cases it may be desirable to reduce PA contents of pastures. An understanding of the biochemical steps involved in PA biosynthesis and its regulation is an important prerequisite for successful manipulation of PA biosynthesis.
PA biosynthesis shares the common flavonoid pathway with the anthocyanins until after the flavan-3,4-diol step (e.g. leucocyanidin in Fig. 1). This common pathway has been dissected genetically and biochemically in a number of plants including Arabidopsis. Many of the genes common to both PA and anthocyanin biosynthesis, such as chalcone synthase, chalcone isomerase, flavonoid 3′-hydroxylase (F3′H), and dihydroflavonol reductase (DFR), have been cloned from the Arabidopsis tt (transparent testa) mutants (Feinbaum and Ausubel, 1988; Shirley et al., 1992; Schoenbohm et al., 2000); however, little is known of the genes or proteins involved in the synthesis of PA. The first committed step to PA biosynthesis, and the only one to be enzymically characterized, is catalyzed by the enzyme leucoanthocyanidin reductase (LAR). Reactions catalyzing the conversion of (+)-3,4-cis-leucocyanidin to (+)-catechin have been described in a number of plants including Douglas fir (Pseudotsuga menziesii), barley (Hordeum vulgare), Onobrychis viciifolia, and other legumes (Stafford and Lester, 1984; Tanner and Kristiansen, 1993; Singh et al., 1997; Skadhauge et al., 1997). PA is thought to be synthesized by sequential addition of an intermediate derived from a flavan-3,4-diol (e.g. leucocyanidin) to a flavan-3-ol-initiating unit (e.g. catechin) or a preexisting chain (Stafford, 1989). This reaction sequence makes the pathway unusual in that a molecule further down the pathway, i.e. catechin, is required to initiate polymer synthesis, but then the immediate precursors (leucocyanidins) of this initiator are sufficient to sustain polymer synthesis. To achieve high degrees of polymerization of PA, it would be important that the flux to leucocyanidin, the extending unit, be much greater than that through to catechin. Alternatively, the synthesis of a small number of initiating units may precede in time a much greater flux of extending units, which means that the enzyme LAR may only need to be active for a short time at the beginning of PA biosynthesis. Leucocyanidin sits at a three-way junction in the anthocyanin/PA pathway. The enzyme leucoanthocyanidin dioxygenase commits it to the anthocyanin pathway, whereas LAR synthesizes catechin, the initiating unit of the PA polymer. The third fate of leucocyanidin is to become an extension unit of the growing PA polymer. Condensation of leucocyanidin and catechin can be achieved in vitro nonenzymatically to yield a polymer that resembles PA synthesized in vivo (Delcour et al., 1983).
Nevertheless, study of barley PA mutants suggest that at least one condensing enzyme may be required in vivo (Jende-Strid, 1993) and there is ample precedent for the existence of enzymes catalyzing reactions at high rates that can occur nonenzymatically (Fersht, 1985). The existence of such condensing enzymes has yet to be demonstrated. The elaboration of PA is made more complex by the possibility of combinatorial incorporation of two discrete isomeric suites of monomers into the polymer, namely the 2,3-cis and 2,3-trans isomers, exemplified by epicatechin and catechin, respectively, and the one-, two-, or three-hydroxylated B-ring isomers, exemplified by afzelechin, catechin, and gallocatechin, respectively. Both the composition and degree of polymerization of PA may change during development. For example, in O. viciifolia leaves, a shift in PA composition from 83% to 48% 2,3-cis isomers is observed, and the proportion of prodelphinidin units changes from 60% to 90% with increasing leaf maturity (Koupai-Abyazani et al., 1993b). The origin of the stereo-isomerization and the mechanism by which this change in PA composition occurs is not known.
PA accumulates, and is probably polymerized, within the vacuole, so both the initiating and extension units of the polymer must be transported into the vacuole by transporters. A single transporter could facilitate transport of all monomers of PA synthesis, but multiple specific transporters that discriminate between initiator and extender units, 2,3-cis and 2′3-trans isomers, and the B-ring hydroxylation isomers would give more control over the composition and Mr of the final PA polymer. Although the vacuole is the ultimate site of PA accumulation in the cell, PA-containing provacuoles have been observed to originate from the rough endoplasmic reticulum (Chafe and Durzan, 1973; Baur and Walkinshaw, 1974; Parham and Kaustinen, 1977). These small provacuoles appear to undergo fusion with a larger vacuole in the cell. The formation of PA-containing provacuoles and vacuoles has been used to study the biogenesis of vacuoles (Hilling and Amelunxen, 1985). Vacuoles destined to play a role in pigment accumulation in the cell are distinct from those destined for protein storage or lytic functions. The tonoplast of vacuoles destined for storage of vegetative storage proteins or for pigment accumulation are marked by the presence of the vacuolar tonoplast intrinsic protein δ-TIP alone, or a combination of δ- plus γ-TIP (Jauh et al., 1999). Presumably, vacuoles similar in function to pigment-containing vacuoles are the site of PA accumulation in cells.
In Arabidopsis, mutants affecting flavonoid synthesis have been identified, and include the tt mutants (Koornneef, 1990; Shirley et al., 1992, 1995). Many of the tt mutants were identified in mutant populations of Arabidopsis because of altered seed coat color or alterations to dormancy and germination characteristics (Debeaujon et al., 2000; Winkel-Shirley, 2001). The BAN (BANYULS) gene has been cloned (Devic et al., 1999), which appears to be specific to the PA biosynthetic pathway. A mutant of this gene was readily identified because it accumulated elevated levels of anthocyanin, making the developing seed appear pink very early on in development, and then turning dark brown upon maturation of the seed. This phenotype is presumably due to diversion of monomers destined for PA to the anthocyanin pathway and subsequent oxidative processes in the seed coat (Albert et al., 1997). This mutant is unable to synthesize flavan-3-ols (Devic et al., 1999). Due to the phenotype of the mutant and the sequence homology of BAN to DFR enzymes, BAN is a candidate LAR-encoding gene. However, LAR or DFR enzymatic activity of BAN has yet to be demonstrated and the BAN protein is only 20% identical to the LAR enzyme from Desmodium uncinatum (G.J. Tanner, K. Francki, S. Abrahams, P.J. Larkin, J.M. Watson, and A.R. Ashton, unpublished data).
More recently, Debeaujon et al. (2001) reported the cloning of TT12, which encodes a transporter-like protein that appears to be required for PA accumulation in vacuoles of the seed coat endothelium. This is distinctly different from the glutathione S-transfe-rase-mediated mechanism proposed for transport of anthocyanin into the vacuole (Mueller et al., 2000). There are also a number of other tt mutants now described that might be specifically involved in PA synthesis because the tt12 phenotype is epistatic to tt9, tt10, tt13, and tt14 (Debeaujon et al., 2001); however, these mutants have not been defined biochemically.
The cloning of TT8 and TT2, which are regulators of BAN and DFR gene expression in Arabidopsis seed, has been described (Nesi et al., 2000, 2001). The TT8 gene encodes a basic helix-loop-helix domain protein that is required for expression of BAN and DFR mRNA in Arabidopsis siliques (Nesi et al., 2000), whereas TT2 is an R2R3 MYB protein that is required for expression of DFR, LDOX, BAN, and TT12 mRNA (Nesi et al., 2001). The expression of PA in the seed coat endothelium appears to be defined by the limited expression pattern of the TT2 gene (Nesi et al., 2001). Whether TT2 and TT8 also act together to regulate later steps in PA biosynthesis is not known. In contrast to PA, the synthesis of anthocyanin is more widespread in Arabidopsis, occurring in leaves, flowers, siliques, and four cell layers in maternal tissue of the developing seed (Devic et al., 1999). This indicates that although anthocyanin and PA may be synthesized from a common pathway, the PA-specific genes are regulated differently than the anthocyanin-specific genes.
An alternative way of identifying Arabidopsis mutants specifically affected in the PA branch of the flavonoid pathway is to use the aromatic aldehyde reagent p-dimethylaminocinnamaldehyde (DMACA),which specifically reacts with PA polymers, small oligomers, and the immediate PA precursors, the flavan-3,4-diols and flavan-3-ols (McMurrough and McDowell, 1978; Delcour and Janssens de Varabeke, 1985; Garcia-Florenciano et al., 1989; Treutter, 1989). In this paper, we describe a simple screening method using DMACA stain on seed pools of Arabidopsis T-DNA-tagged lines to identify mutants of PA biosynthesis. Using thin-layer chromatography (TLC) and HPLC, these mutants have been characterized biochemically, and are shown to be specific to the PA pathway. The only flavan-3-ol detected in wild type and mutants tds1, tds2, tds3-1, tds5, and tds6 is epicatechin. The tds4 mutant accumulates an unidentified flavan-3-ol. The mutants tds4 and tt7-3 show altered levels and patterns of PA deposition in the cell, presumably due to the incorporation of unusual intermediates into the PA. The preliminary biochemical analyses of the tds mutants presented here suggest that Arabidopsis is a good model in which to dissect the PA pathway, and to obtain the genes responsible for the synthesis of PA.
RESULTS
Isolation of Mutants and Allelic Complementation Analyses
To identify the steps involved in PA biosynthesis, we screened T-DNA-tagged mutants available from seed stock centers using DMACA stain to detect seeds with altered PA accumulation. Mutants specific for the PA pathway should have normal anthocyanin but altered PA content. After identifying pools containing mutant seed, individual plants were grown from duplicate unstained seed pools, and their seeds were collected and then stained with DMACA. Ten individual mutants with either reduced PA or an altered pattern of accumulation of PA were identified from the screen. Allelic complementation tests were done to determine the number of loci represented by the mutants. The results of the complementation analysis and a summary of the mutant phenotypes appear in Table I. The 10 mutants fall into eight complementation groups, representing a mutation frequency of PA-free mutants of at least one in 1,900 mutants screened. The frequency of anthocyanin-positive PA-free mutants was one in 2,700. Not all of the mutants initially observed in the pools of seed were actually isolated because some were represented by only one to two seeds in the initial stained pool that might have failed to germinate or grow to maturity in the duplicate pool. tds1, tds2, and tds3-1 are from the Feldmann collection of mutants in the Ws-2 background, tds3-2 and tds4 are from Institut National de la Recherche Agronomique in the Ws-4 background, and tds5, tds6, tt7-3, tt8-4, and tt8-5 are from the Weigel mutant collection in the Col-7 background.
Table I.
Characteristics of the tds mutants
| Background | Mutation | Seed Color | Leaf Ant | Seed PA | Flavanol | Flavonols (Seed)
|
|
|---|---|---|---|---|---|---|---|
| K | Q | ||||||
| Wassilewskija (Ws)-2 | tds1 | Tan | + | –a | + | + | + |
| Ws-2 | tds2 | Tan | +b | –a | + | + | + |
| Ws-2 | tds3-1 | Tan | + | – | + | + | + |
| tds3-2 | |||||||
| Ws-4 | tds4 | Yellow | –/+ | Spotty | +b | + | + |
| Columbia (Col)-7 | tds5 | Tan | + | –a | + | + | + |
| Col-7 | tds6 | Tan | + | +/–b | + | + | + |
| Col-7 | tt8-4 | Yellow | + | – | – | + | + |
| tt8-5 | |||||||
| Col-7 | tt7-3 | Tan | +b | Spotty | +b | + | – |
| Enkheim | ban | Brown | + | – | – | + | + |
| Landsberg erecta (dfr−) | tt3 | Yellow | – | – | – | + | + |
| Landsberg erecta (chs−) | tt4 | Yellow | – | – | – | – | – |
| Wild type | Tan/dark tan | + | + | + | + | + | |
K, Kaempferol; Q, quercetin.
Positive at the basal end of the seed.
Present but not wild type.
One of the mutants shared the same phenotype as tt7-1, which is mutated in the F3′H gene (Schoenbohm et al., 2000). Allelism tests confirmed that this mutant was an allele at the TT7 locus and it was named tt7-3. Similarly, because of the phenotype, two of the mutants were crossed to tt8-1, and found to be alleles at the TT8 locus, and they were named tt8-4 and tt8-5. The mutants ban (Devic et al., 1999), tt1, and tt2 (Shirley et al., 1995) were specifically of interest because of their potential role in PA biosynthesis, and crosses were performed between ban (F36), tt1-1, and tt2-1 and each new mutant to determine allelism. None of these new mutants was allelic to ban, tt1-1, or tt2-1 and so they have been named tds (tannin-deficient seed).
Reciprocal crosses between the mutants and wild type revealed that all of the F1 testa exhibited phenotypes conferred by the maternal parent. All F2 seed displayed wild-type phenotype. Segregation of the mutant phenotype was observed in F3 seed. These results are consistent with gene expression in maternal tissue and the inheritance of the PA-free phenotype as a recessive trait. The segregation of the mutant and wild-type phenotypes in F3 seed after crossing to wild type was determined. The segregation of marker genes such as kanamycin or herbicide resistance in the F3 generation was also assessed, independently of the mutant phenotype. Three of the mutant phenotypes, tds4, tt7-3, and tds5, were shown to segregate independently of the resistance markers, indicating that they result from a spontaneous mutation or partial T-DNA insertion. The DNA flanking the T-DNA in the tt8-4 mutant allele was obtained by plasmid rescue and the interrupted gene encodes the bHLH protein previously described by Nesi et al. (2000).
Description of the tds Mutant Phenotypes with DMACA Staining
Unstained seed of mutants tds1, tds2, tds3-1, tds5, and tds6 are tan in color and slightly paler than wild-type seed grown under the same conditions, whereas tds4 is pale yellow in color (not shown). The color of seed changes with time after harvesting, due to oxidative processes in the seed, in a similar way to that described previously for some tt mutants (Debeaujon et al., 2001). Figure 2 shows the phenotype of mature seed of the mutants stained with DMACA. Mutants tds1, tds2, and tds5 are DMACA negative apart from a small area at the basal end of the seed (Fig. 2, B, C, and F), which appears to accumulate PA. tds3-1 is uniformly DMACA negative, including the basal end of the seed (Fig. 2D). The tds6 mutant has a slight DMACA reaction (Fig. 2G), although clearly distinguishable from wild type (Fig. 2A). Mutants tds4 and tt7-3 (Fig. 2, E and I) both show an altered pattern of PA deposition, tt7-3 showing solid spots, whereas tds4 has a more patchy staining pattern with DMACA. An enlarged image of tds4 and tt7-3 DMACA-stained seed is also shown (Fig. 2J). In addition to the spots of PA, tt7-3 also has spots of anthocyanin (not shown). We also observed this spotty pattern of PA and anthocyanin accumulation for tt7-1 in the Landsberg ecotype (Koornneef et al., 1982). tt8-4 is also yellow when not stained (not shown) and is uniformly DMACA negative (Fig. 2H), including the cells of the basal end of the seed. Mature tt4 and ban seed was also stained and found to be DMACA negative (Fig. 2K), shown in a mixed pool with wild type and tds2 for comparison.
Figure 2.
Wild-type and mutant mature seed stained with DMACA. A through I, Pools of mature seed including Ws-2, tds1, tds2, tds3-1, tds4, tds5, tds6, tt8-4, and tt7-3 showing differences in staining with DMACA. J and K, Enlarged ∼images comparing Ws-2, tds4, and tt7-3 (J) and Ws-2, ban, tt4, and tds2 (K). Bars = 0.05 mm (A through I) and 0.025 mm (J and K).
Because only the seed coat produces PA, we were able to harvest whole siliques to investigate the accumulation of PA in Arabidopsis seeds. Siliques from all stages of development were removed from plants and placed directly into DMACA stain to visualize DMACA reacting PA and precursors of PA. Developing seeds were excised and those at the walking stick stage of development are shown in Figure 3. In contrast to the DMACA reaction of mature seed (Fig. 2), the developing seed from all our mutants except tt8-4 stained positively with DMACA (Fig. 3). Some differences in the intensity of staining were observed, both between mutants and compared with wild type. The mutants tt7-3 and tds4 (Fig. 3, E and I) showed the isolated patches of DMACA staining, observed in mature seeds, throughout their development.
Figure 3.
Wild-type and mutant developing seed stained with DMACA. A through I, Pools of developing seed dissected from siliques, including Ws-2, tds1, tds2, tds3-1, tds4, tds5, tds6, tt8-4, and tt7-3, showing DMACA-reacting PA intermediates present in all except tt8-4. Bar = 0.05 mm.
Qualitative and Quantitative Assessment of Anthocyanin
Because anthocyanin and PA synthesis share the same sequence of reactions to the common intermediate leucocyanidin (Fig. 1), it was important to determine if the mutants were specific to the PA branch of the pathway. Mutants of specific interest would be expected to be anthocyanin positive and PA free. For this reason, analyses of tt7-3, tt8-4, tt3-1, and ban (F36) mutants were included in this study as a reference point for the identification of the tds mutant phenotype (Koornneef, 1990; Shirley et al., 1992; Albert et al., 1997; Devic et al., 1999; Nesi et al., 2000; Schoenbohm et al., 2000).
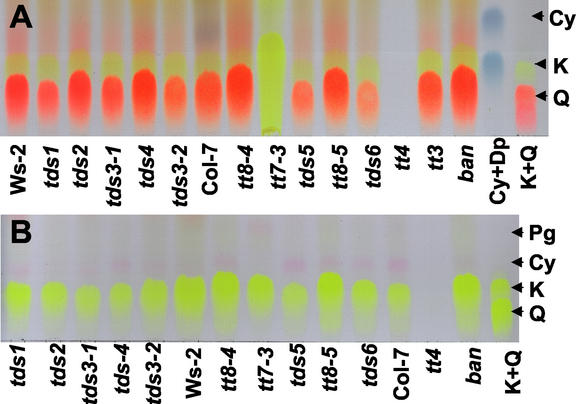
TLC is a useful way of comparing whole seed and leaf extracts to visualize flavonols and anthocyanins in the same sample. Although both flavonols and anthocyanins are easily visualized on TLC, the Natural Products (NP) stain reagent differentially stains flavonols that contain two adjacent hydroxyl groups on the B ring orange (e.g. quercetin) and one hydroxyl group on the B ring yellow (e.g. kaempferol; Wagner, 1984). The NP stain can also enhance the appearance of anthocyanins, which are present at lower concentrations than the flavonols in the tissues analyzed. Because flavonols and anthocyanins are present in tissues as glycosylated, acylated, or other derivatives, it is useful to acid hydrolyze extracts to convert these modified intermediates to their common aglycone form (shown in Fig. 1). Figure 4 shows acid-hydrolyzed seed (Fig. 4A) and leaf anthocyanin (Fig. 4B) extracts. The flavonols kaempferol and quercetin were present in the seeds of most mutants (Fig. 4A), including tt3 (which lacks DFR) and ban. The exceptions were tt4, which lacks chalcone synthase and would not be expected to contain these intermediates, and tt7-3, which accumulates only the monohydroxylated kaempferol (Fig. 4, A and B). The large amount of flavonols present in the seed extract prevented detection of anthocyanins in these samples. Leaf anthocyanin extracts contain only kaempferol (Fig. 4B), and not quercetin, indicating a difference in the expression of F3′H or flavonol synthase genes between leaves and seeds of Arabidopsis. Cyanidin is clearly visible on the TLC of acid hydrolyzed leaf anthocyanin extracts (Fig. 4B). Analysis using TLC showed that most of the mutants accumulated wild-type levels of the flavonols kaempferol and quercetin and, therefore, were mutated in genes acting at or beyond DFR in the pathway.
Figure 4.
TLC of anthocyanin extracts from mature seed and leaves. A, TLC of acid-hydrolyzed seed anthocyanin extracts showing flavonols, kaempferol (K, yellow), and quercetin (Q, orange) when sprayed with NP reagent. All mutants have both K and Q, except tt4, which has neither, and tt7-3, which has K only. Anthocyanidins are not evident on seed TLC. B, TLC of acid-hydrolyzed leaf anthocyanin extracts, not sprayed with NP reagent, showing K present in leaves, but not Q, and pink cyanidin (Cy) in all mutants except tt4 and tt7-3, which has pelargonidin (Pg).
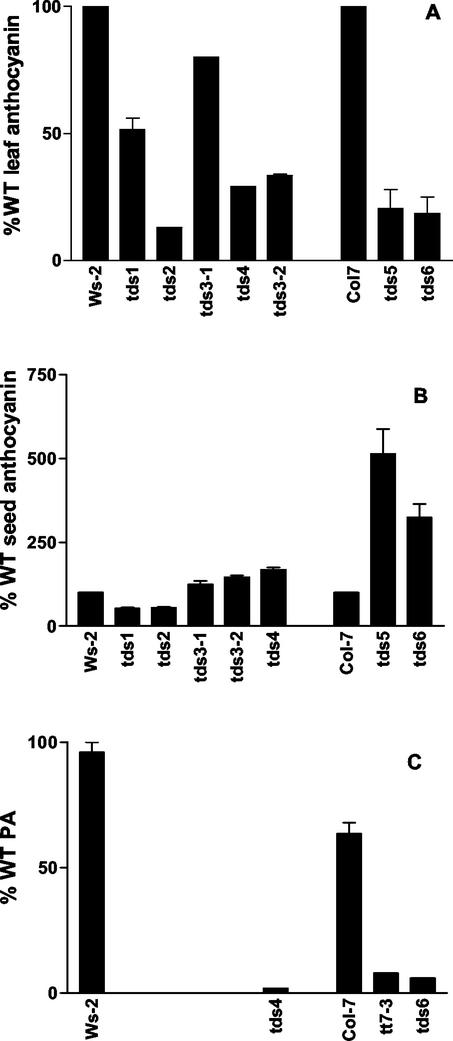
The visible spectra of anthocyanins extracted from leaves and mature seeds were used to determine quantitative and qualitative differences in anthocyanin accumulation. All of the mutants produced 20% to 80% of wild-type anthocyanin in leaf material, as shown in Figure 5A. Two of the mutants, tds1 and tds2, showed a decrease in concentration of anthocyanin in mature seeds (Fig. 5B), but all other mutants accumulated 20% to 500% more anthocyanin than wild type in seeds (Fig. 5B). Leaves produced more anthocyanin than seeds, per gram fresh weight of material (results not shown); conversely, the seeds tended to accumulate larger amounts of flavonols.
Figure 5.
Quantitation of anthocyanin and PA. A, Quantitation of leaf anthocyanin as a percentage of wild-type values, measured in duplicate. B, Mature seed anthocyanin shown as a percentage of wild type, measured in duplicate. C, PA measured in mature seed for Ws-2 tds4, Col-7, tt7-3, and tds6, measured in duplicate. Results shown as a percentage relative to Ws-2 wild type. Error bars = sd.
Analysis of PA and Its Intermediates
Because developing seed of wild type and tds mutants possessed DMACA-reacting compounds (Fig. 3) and the mature seed of tds1, tds2, tds3-1, and tds5 did not (Fig. 2), PA was extracted from developing siliques and mature seed and analyzed using TLC and HPLC to identify the DMACA-reacting compounds. Measurable amounts of PA were extracted from mature seed of Ws-2, Col-7, tds4, tds6, and tt7-3, shown in Figure 5C. Mature seed of other mutants did not accumulate measurable amounts of PA and are not shown. Col-7 accumulated 70% of the PA of Ws-2 wild-type mature seed when grown under the same conditions. The two mutants tds4 and tt7-3 that show a patchy or spotted pattern of PA accumulation also showed a marked decrease in extractable PA, 2% and 8% that of their wild type, respectively. The mutant tds6, which is slightly positive when stained with DMACA (Fig. 2), accumulated 6% of the wild-type level of PA in the mature seed. Acid hydrolysis of Ws-2 PA produced cyanidin only (results not shown).
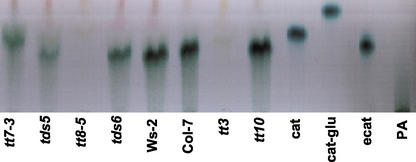
Extracts were also prepared from developing Arabidopsis siliques, which included seeds up to the late heart stage or walking stick stage of embryo development. The same extract was used to analyze both PA precursors and PA polymer. PA polymer was not detected in the developing seeds of any sample, including wild type. The fraction containing PA precursors was separated using TLC and sprayed with DMACA reagent, shown in Figure 6. A compound was detected in tds1, tds2, tds3-1, tds4 (not shown), wild type, tds5, and tds6 mutants (Fig. 6) that was absent from tt8-4 and tt3 control extracts. This compound had the same RF as authentic epicatechin. Because tt3 and tt8-4, both of which lack DFR activity, did not accumulate this intermediate, it might be related to steps downstream of DFR in the PA synthesis pathway (Fig. 1). Extracts from tt7-3 contained a faster migrating compound than that of wild type, which may correspond to afzelechin or epiafzelechin; however, standards of these compounds are not available for identification of these intermediates by comparison of RF values. In this solvent, the monohydroxylated flavonoid isomers migrate faster than the corresponding dihydroxylated isomers consistent with our tentative identification of this compound as afzelechin or epiafzelechin.
Figure 6.
TLC analyses of ethyl acetate fractions of PA extracts from developing siliques. The ethyl acetate fraction contains PA intermediates that react with DMACA. Mutants tt8-4, tt7-3, tds5, and tds6 are compared with wild type, tt10, and tt3 as positive and negative controls. Authentic standards of catechin (cat), epicatechin (e-cat), catechin glucoside (cat-glu), and O. viciifolia PA are shown. tt8-5 and tt3 lack the DMACA-reacting intermediate, and tt7-3 has an alternative intermediate, possibly afzelechin or epiafzelechin, due to the lack of F3′H activity. Polymers of PA are not observed in the soluble fraction of PA extracts from developing mutant or wild-type siliques.
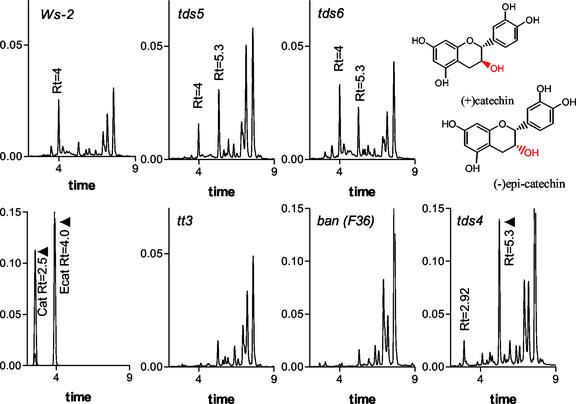
The extracts containing PA precursors were also analyzed using HPLC and chromatograms are shown in Figure 7. A peak with retention time of 4 min was observed in tds1, tds2, tds3-1 (not shown), Ws-2, tds5, and tds6 samples (Fig. 7). Extracts of tt3 and ban did not have this compound, so it might be a product of enzymatic steps beyond DFR and BAN in the pathway. This compound was purified by preparative HPLC methods and found to be DMACA positive and comigrated with the single DMACA-reacting bands in samples except tt7-3 on the TLC (Fig. 6). HPLC mass spectrometry was used to determine the compounds molecular mass as 291.2 D, which is the same as both protonated catechin and epicatechin. Because this compound comigrates with epicatechin standard on both TLC and HPLC (Figs. 6 and 7), has the same molecular mass as epicatechin, and, like epicatechin, reacts strongly with DMACA, yielding a blue product, it is likely to be epicatechin. In addition to this peak, other mutants produce novel intermediates, or enhanced amounts of intermediates relative to wild type. The mutant tds4 produced a unique compound with a retention time of 2.92 min and tds4, tds5, and tds6 produced an unidentified compound with a retention time of 5.3 min, approximately 2- to 4-fold greater than wild type (Fig. 7).
Figure 7.
HPLC analyses of ethyl acetate fractions of PA extracts from developing siliques. The peak appearing in the Ws-2, tds5, and tds6 traces at 4 min has the same retention time as the epicatechin standard and is DMACA positive, corresponding to the intermediate observed on TLC plates. This peak was purified from Ws-2 and analyzed by HPLC mass spectroscopy. This intermediate is not observed in tt3 or ban extracts. The differences in structure and retention times of the stereoisomers catechin and epicatechin are shown. The traces of extracts from tds1, tds2, and tds3-1 siliques are similar to tds5 and tds6 and are not shown.
Microscopic Examination of the Localization of PA in Wild Type, tds4, and tt7-3
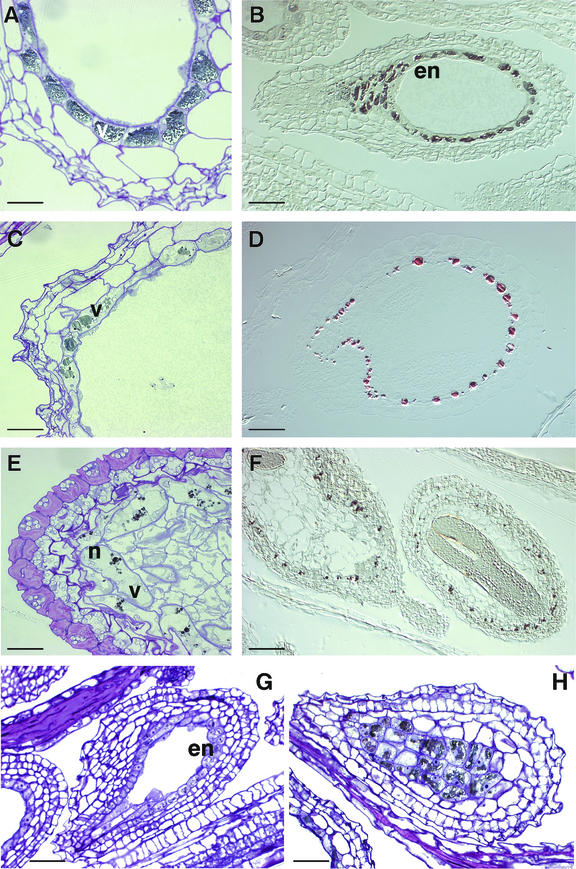
When whole wild-type Arabidopsis seedlings were stained with DMACA, only the developing seed coat gave a positive reaction for PA (not shown). Sections of developing seeds were treated with OsO4 to detect the accumulation of PA or its precursors, and then lightly counterstained with toluidine blue to show cell structure in the tissue. Figure 8 shows that PA is synthesized in wild-type Arabidopsis in the endothelial layer of the testa (Fig. 8, A and B). PA, or its precursors, was visible as a gray or black deposit in endothelial cells from as early as the two-terminal cell stage of embryo development (Fig. 8G), which coincides with 18 h after flowering (Mansfield, 1994). PA appears in all cells of this layer (Fig. 8H). In wild-type seeds, the vacuole containing PA occupies almost the entire cell contents (Fig. 8A), giving the general appearance of containing PA uniformly throughout the endothelial layer of the seed coat. In tt7-3, the PA appears as discrete spots within cells (Fig. 8, C and D), reminiscent of the pattern seen with DMACA stain at lower magnification in whole seeds (Fig. 2I). Higher magnification showed this staining to occur in the vacuole of the cell (Fig. 8C). PA was visible in tds4 later in development than for wild type and was associated with small provacuolar bodies that did not appear to fuse with each other or the main vacuole (Fig. 8, E and F). Generally, more mature seeds were difficult to section due to the treatment with osmium tetroxide and are not shown.
Figure 8.
Microscopic sections of OsO4-treated developing seeds showing altered PA accumulation in tt7-3 and tds4. A, B, G, and H, PA accumulation in the endothelial layer of Ws-2 developing seeds, shown as a gray deposit within the vacuole. PA intermediates were evident at the two-terminal cell stage of development (G) and continued to accumulate in the heart stage (A and B). C and D, tt7-3 at the heart stage of development showing small round inclusions of PA in vacuoles. E and F, tds4 at the torpedo stage of development, showing very small PA inclusions or provacuoles, distinct from the main vacuole of the cell (E). en, Endothelium; v, vacuole; n, nucleus. Bar = 30 (A, C, and E), 40 (B, D, and F), and 140 (G, H) μm.
DISCUSSION
In some species, the PA biosynthesis pathway is summarized as an interconnecting pathway or confusing network of grids of all possible routes to the end products (Stafford, 1989). An added complication is the variety of hydroxylation patterns and stereoisomers that may be incorporated into PA at different stages of development in different species (Koupai-Abyazani et al., 1993b). Using a simple staining method on T-DNA-tagged lines available from seed stock centers, we have identified six TDS genes in Arabidopsis, which when mutated cause defects in the synthesis of PA, as well as mutants in two known genes, TT7 and TT8. In both wild type and five of the tds mutants (tds1, 2, 3, 5, and 6), the only PA precursor to accumulate is epicatechin, suggesting that in Arabidopsis, the pathway might be simpler than in other species because only one stereoisomer was identified. Because our screen uncovered eight genes among the 10 mutants we found, and because we did not find new alleles of known PA-deficient mutants such as ban and tt2, it is likely that we have not identified mutants for all of the genes required for PA biosynthesis in Arabidopsis. To date, some 21 tt mutants have been identified (Winkel-Shirley, 2001). From analyses of double mutants, the tt12 mutation is epistatic to tt9, tt10, tt13, and tt14 (Debeaujon et al., 2001). From the observations of vanillin staining, it appears that tt10 and tt14 possess PA intermediates (Debeaujon et al., 2000) and that tt9, tt11, tt13, and tt15 (Debeaujon et al., 2001) lack PA or its precursors in the seed body. These tt mutants are of particular interest because they may be allelic to the tds mutants described in this paper.
Because PA is colorless and cannot be detected visually in the seed, a chemical test is necessary to screen specifically for PA mutants. Kristensen and Aastrup (1986) used the aromatic aldehyde vanillin to identify 10 ant loci in barley acting on seed PA biosynthesis. From analysis of enzyme activities and intermediates, ant 19 might encode LAR in barley (Jende-Strid, 1991) and ant 26 might encode a putative condensing enzyme, responsible for the condensation of monomers into polymers (Jende-Strid, 1993). The function of other anthocyanin-positive PA-free barley mutants such as ant25, ant27, and ant28 is not known, but their existence indicates that at least five genes are necessary for PA synthesis in the barley seed coat.
Although vanillin is commonly used to detect PA, a number of other stains are available for the detection of PA and its immediate precursors such as leucocyanidin and epicatechin. The sensitivity of the DMACA stain (another aromatic aldehyde) is some 4-fold greater than vanillin, and has been used to detect PA in plant species previously thought to lack PA when tested using vanillin stain (Li et al., 1996). Another advantage of DMACA over vanillin is that DMACA stains PA blue-purple, whereas vanillin-stained PA is pink and can be confused with pink anthocyanin pigments in tissues containing both compounds. Furthermore lignin in plant tissues can also stain reddish in the strong HCl of the DMACA and vanillin reagents.
Our observation and isolation of PA-specific mutants using DMACA stain on mature dry seed might seem anomalous because the DMACA-positive PA precursors (epicatechin and leucocyanidin) that could be present in mature mutant seed are likely to mask mutants defective in steps beyond the synthesis of epicatechin. This might be due to the leaching of flavan-3-ols from the seed before the DMACA staining was observed to be complete. Koupai-Abyazani et al. (1993a) analyzed the leachate of alfalfa seeds and demonstrated that within 6 h, most of the catechin and epicatechin that accumulates in alfalfa seeds had leached into the surrounding solution. Similar leaching of PA intermediates has been observed in white clover and O. viciifolia (Young and Paterson, 1980; Prevost et al., 1990). During the screening process, the seed was stained for 7 to 10 d to ensure complete staining of all seed in the pool because only one or two unstained seed in the original pools might be mistaken for mutants. We reason that mutants in PA synthesis have been identified in Arabidopsis seed pools due to lack of DMACA staining because of the leaching of monomers from the seed during the staining process, revealing the existence of mutants in steps beyond the synthesis of epicatechin. The biochemical analyses presented here confirm that the tds mutants are blocked in PA synthesis after the production of flavan-3-ols.
OsO4 is widely used in microscopy as a fixative and stain because it can be reduced by a variety of biological molecules, covalently linking to or cross-linking them (White et al., 1976). We found that OsO4 intensely stains the PA-containing inner endothelial layer of wild-type Arabidopsis seed coats but not the PA-deficient mutants. The ability of OsO4 to detect PA in tissue sections was used to reveal altered subcellular patterns of PA accumulation in tds4 and tt7-3. The mutant tt7-3 was identified due to its spotted pattern of PA accumulation. The seeds were spotted because the PA did not fill the whole cell but was confined to the middle of the cell. The TT7 gene encodes the enzyme flavonoid 3′-hydroxylase that adds a second hydroxyl to the 4′-hydroxylated B ring to start the cyanidin stream of flavonoid intermediates. Although this spotted PA accumulation in tt7 mutants has not been described previously, we observed the same PA-spotted phenotype in the original tt7-1 allele, so the phenotype is likely to be a result of mutations in the F3′H gene, rather than in other genes. The identity of tt7-3 was confirmed using TLC analyses and also allelic complementation to tt7-1. Acid hydrolysis of wild-type Arabidopsis PA produced cyanidin only, indicating that intermediates with one hydroxyl group on the B ring are not normally utilized for PA synthesis, whereas hydrolyzed PA of tt7-3 produced only pelargonidin, something that is quite rare in any plant (Stafford, 1989). In addition to the restricted accumulation and composition of PA, the tt7-3 seed accumulates only 6% of the PA of wild type. This low level of PA in tt7-3 may reflect a limited capacity of existing PA enzymes to utilize leucopelargonidin as a substrate for PA synthesis in the seed of Arabidopsis. The accumulation of lower levels of anthocyanin in leaves of tt7-1 plants (Koornneef et al., 1982) shows that the enzymes of anthocyanin biosynthesis are also less active with the pelargonidin stream of intermediates. Two enzymes, F3H and DFR, are common to the anthocyanin and PA pathway. The differences observed in microscopic sections between cells accumulating propelargonidin versus procyanidin PA in tt7-3 and wild type, respectively, might also indicate physical properties of the two types of PA molecules. We have observed a spotted PA pattern in the testa of other previously described mutants; for example, in tt6 mutants, which lack flavanone 3-hydroxylase activity (Wisman et al., 1998). Developing seeds of the ban mutant also go through a phase showing spots of anthocyanin (Albert et al., 1997), only later appearing to be uniformly anthocyanin positive.
The tds4 mutant was identified because the seed PA had a patchy, filamentous distribution pattern. The tds4 mutant accumulated a DMACA-reacting intermediate that is not epicatechin, and we were able to extract some PA from mature seed of this mutant. The limited amount of PA observed in tds4 is likely to be made using this alternative intermediate. Microscopy revealed that the PA intermediate (detected with OsO4) was confined to small vesicles or provacuoles and did not accumulate in the main vacuole. The occurrence of PA in small vacuoles in tds4 is unlikely to be due to the low level of PA produced in this mutant because tds6, which accumulates a similar low level of PA, does not form a patchy pattern of PA when stained with DMACA. It is intriguing to note that PA-containing provacuoles that undergo fusion with the main vacuole have been reported (Chafe and Durzan, 1973; Baur and Walkinshaw, 1974; Parham and Kaustinen, 1977; Hilling and Amelunxen, 1985). These results suggest that the pathway of PA accumulation may be much more complex than a minimal pathway that involves membrane-bound transporters transporting monomeric units into a preexisting vacuole. The presence of pro-vacuolar intermediates suggests the need for the complex membrane targeting, and vesicle formation and fusion like those that accompany events in the secretory pathway (Bethke and Jones, 2000; Hadlington and Denecke, 2000).
Staining of whole Arabidopsis plants with DMACA showed that PA is only present in the seed coat. Our observations suggest that PA accumulation might occur in two stages during seed development. TLC analysis shows that the chain-initiating unit epicatechin, but no other PA monomers or oligomers, were present in pooled siliques from flowering until the walking stick stage of development at 4 to 5 d after flowering (Mansfield, 1994). OsO4-reactive material, presumably epicatechin, was also visible in sections of the two-terminal cell stage of embryo development, approximately 18 h after flowering (Mansfield, 1994), so monomer synthesis must begin before this stage. However, extractable quantities of PA polymers were not observed until much later in seed development. Notably, even in the dry seed, substantial amounts of epicatechin remain, indicating that the addition of extension units might occur preferentially to growing PA chains rather than to monomeric epicatechin. A similar observation was made in barley (Kristiansen, 1984; Jende-Strid, 1993), where the amount of catechin in developing barley seeds increases to approximately 100 nmol seed−1 within 16 to 18 d after flowering, then the level falls to 40 nmol seed−1 by 28 d after flowering.
The leucocyanidin extension units are not detectable either in wild-type or mutant seeds at any stage of development, but this may reflect the low steady-state concentration of this reactive intermediate. Because leucocyanidin is the precursor of both epicatechin and anthocyanin, the occurrence of both these products shows that leucocyanidin must also be present, both during the early stages of PA accumulation as well as the later stages when the leucocyanidin extension units are added to the epicatechin-initiating unit. LAR, which converts leucocyanidin to catechin, need only be present during the initial phase, whereas the preceding enzymes in the pathway including DFR need to be present during the polymerization phase as well. The BAN gene that might encode LAR is, in fact, expressed in the early stage of PA synthesis (Devic et al., 1999) as might be expected of the LAR gene. The temporal separation of PA initiation from polymerization might also provide a means of identifying genes specific to either stage. For example, transporters specific for epicatechin might be expressed at an early stage, whereas a leucocyanidin transporter would only be required during the polymerization phase. Notably, the transcript of the MATE transporter-encoding gene TT12 is only present during the early stages of embryo development (Nesi et al., 2001), consistent with a possible role in epicatechin transport. Similarly, enzymes, if any, involved in polymerization should only be present at the later stages. The cue for initiation of expression of genes responsible for polymerization might be the onset of seed desiccation at around 9 d after flowering, a key regulatory point for seed development. Polymerization could be enzyme catalyzed, but also might occur spontaneously as a direct result of the physical changes that take place during desiccation of the seed. It is possible that stereospecific polymerization to form PA occurs as a result of a dirigent protein, as may occur for lignin biosynthesis (Davin and Lewis, 2000). One or more of our four tds mutants that make epicatechin, which presumably have leucocyanidin available for chain extension and yet do not produce PA, are potential candidates for condensing/polymerizing enzymes.
Biochemical analysis indicated that Arabidopsis normally produces only one flavan-3-ol, epicatechin, and not the mixture of intermediates observed in other species. Assuming that epicatechin is the product of LAR, then we have identified five genetically encoded steps in addition to LAR that are necessary for PA accumulation in Arabidopsis. Thorough biochemical analysis of Arabidopsis PA has not been reported, but the presence of a pool of epicatechin suggests that Arabidopsis PA might be made using this flavan-3-ol isomer as an initiating unit for polymer synthesis. Although the enzymic synthesis of (+)-catechin from (+)3,4-cis-leucocyanidin has been demonstrated (Tanner and Kristiansen, 1993), the origin of (−)-epicatechin and other (−) isomers is not known. In the flavonoid pathway, the earliest step at which alternative 3-hydroxyl isomers might be formed is at the flavanone 3-hydroxylase enzyme step, when the 3-hydroxyl group is first incorporated to form dihydrokaempferol or dihydroquercetin. In a study of flavan-3-ol synthesis in extracts from leaves of O. viciifolia, Singh et al. (1997) were able to demonstrate the synthesis of catechin and gallocatechin, but unable to demonstrate the synthesis of epicatechin or epigallocatechin from dihydroquercetin, even though O. viciifolia leaf polymers were composed of roughly 50% epigallocatechin and only 30% gallocatechin. They proposed that the (−) isomers are synthesized by an unspecified alternative route in the flavonoid pathway in O. viciifolia because there was no evidence for their direct synthesis nor an epimerase activity acting on (+)-dihydroquercetin or its known products (Fig. 1). Analysis of the specificities of Arabidopsis enzyme activities may be one way in which to establish the origin of the (−)-isomers because Arabidopsis has only a single copy of F3′H, DFR, and BAN genes, and accumulates epicatechin, not catechin.
Two of the mutants, tds1 and tds2, showed a decrease in concentration of anthocyanin in mature seeds, whereas there appears to be a small increase in seed anthocyanin in mutants tds3-1 and tds4, and a larger increase in mutants tds5 and tds6. The ban mutant also accumulates 1.5 times more anthocyanin than its wild type (Devic et al., 1999), suggesting that substrates such as leucocyanidin normally destined for PA synthesis might be diverted to anthocyanin in mutants unable to make PA. If BAN serves the function of LAR in Arabidopsis, then the browning observed in ban mutants might be a result of oxidation of excess leucocyanidin, rather than browning of anthocyanin as suggested by Devic et al. (1999). The blockage downstream of BAN in the tds mutants does not seem to cause a large excess of epicatechin or any other PA intermediate to accumulate. Because the tds mutants are able to use some of the leucocyanidin to make epicatechin, and some appear to have enhanced levels of anthocyanin, then little or no leucocyanidin remains to oxidize and cause browning. Quantitation of the relative amounts of PA, monomer, and anthocyanin in wild-type and mutant seeds needs to be done to determine whether or not this is the case. It should be noted that some of the quantitative differences observed between the tds mutants might be due to their differing genetic backgrounds.
The ban mutant was used for comparison in the analysis of the tds mutants because it is the only previously characterized Arabidopsis mutant that contains anthocyanin but not PA. It is clear from this study that ban is unable to accumulate any PA precursors, and that tds1, tds2, tds3-1, tds5, and tds6 mutants are able to synthesize epicatechin, and that tds4 is able to synthesize a DMACA-reacting intermediate that is not epicatechin. This analysis places the tds mutants after LAR in the pathway. Importantly, leucocyanidin does not appear to accumulate in wild-type, ban, or tds mutant seeds, at any stage of development. Although the precise role of BAN in PA synthesis is yet to be determined, it seems that BAN is involved in PA synthesis at the branch between anthocyanin and PA synthesis (Devic et al., 1999), and that, if it is not LAR [i.e. capable of synthesizing (+)-catechin from (+)-3,4-cis-leucocya-nidin], then it might perform the stereospecific synthesis of epicatechin. The substrates required to test this suggestion are not currently available and would need to be chemically synthesized. By sequence similarity, BAN is most closely related to DFR, and both are members of the Reductase Epimerase Dehydrogenase family of proteins that include epimerases (Labesse et al., 1994; Jornvall et al., 1995). Potentially, BAN could be an enzyme that epimerizes the 3-OH group, either at the dihydroquercetin or leucocyanidin step. We recently have purified, cloned, and expressed the LAR enzyme from D. uncinatum in Escherichia coli (G.J. Tanner, K.T. Francki, S. Abrahams, P.J. Larkin, J.M. Watson, and A.R. Ashton, unpublished data) and find that it is only very distantly related to BAN (20% amino acid identity). The enzymatic characterization of BAN would determine whether BAN also has LAR activity or constitutes another step in the PA pathway.
Because the tds mutants are able to make flavan-3-ol, they appear virtually the same as wild type until after the walking stick stage of embryo development. Only then does the difference between mutant and wild type become clear. Because it can be difficult to reliably distinguish between the tds mutants on phenotype alone, the identification of double mutants to assist in assigning an order to gene action in the PA pathway is difficult to achieve with certainty. Where it is possible to identify a tds/tds double mutant with certainty, they are being sought to determine the epistatic relationships between the mutants. The tds mutants being in different genetic backgrounds may also cause some difficulties in identifying the tds/tds double mutants.
The tds mutants might be defective in epimerization, transport, or condensation of PA monomers, due to a lack of enzymes that normally act to produce PA. Given that anthocyanin and PA are often located in different cell types within a plant at different stages of development, it is likely that a set of regulators exist that control only the expression of PA biosynthesis. These regulators might act to control genes responsible for flavan-3-ol formation, transport into the vacuole, or polymerization of flavan-3-ols and flavan-3,4-diols into PA. The existing tds mutants will be valuable in dissecting the PA pathway, particularly when the TDS genes have been cloned.
MATERIALS AND METHODS
PA Mutant Screen
Seed pools from Feldmann (6,500), Institut National de la Recherche Agronomique (second and third set, 3,900), and Weigel (first set, 8,600) sets of mutants, available through the Arabidopsis Biological Resource Center (Ohio State University, Columbus), consisting of pooled seed from 100 T-DNA-tagged mutant lines were screened in the first round. Seed was stained with DMACA reagent (2% [w/v] DMACA in 3 m HCl/50% [w/v] methanol) for 1 week, and then washed three times with 70% (v/v) ethanol. The stained pools were then examined for seed showing altered PA expression using a microscope. A second round of screening consisted of staining five pools of 20 that made up the initial pool of 100. When mutant seeds were seen in these two rounds of screening, seed was then grown from selected pools of 20. Seed was sterilized using 0.1% (w/v) mercuric chloride for 15 min, washed three times with water, germinated on Murashige and Skoog media, and then transferred to soil after 2 weeks. Plants were grown in 16-h day at 22°C and 8-h night at 18°C in a growth cabinet. Whole siliques were stained with DMACA reagent and the individual PA-free mutants from the seed pools identified.
Genetic Analysis of Mutants
The mutants were backcrossed with either wild-type Col-7, Ws-2, or Ws-4. After selfing the F1 plants, F2 seed was collected and stained with DMACA to examine PA phenotype. After selfing the F2 plants, F3 seed was collected and stained with DMACA to determine segregation of the mutant phenotype. The same process was used for crosses between mutants to determine allelism. Samples of F2 seed were also germinated on Murashige and Skoog containing either kanamycin (50 μg mL−1) or Basta (5 μg mL−1) to assess the segregation of marker genes (nptII or BAR) of the T-DNA. Samples of seed from ban (accession no. F36), tt1, tt2, tt3, tt7, and tt8 mutants (accession nos. cs82, cs83, cs84, cs88, and cs111, respectively) were obtained from the Arabidopsis Biological Resource Center.
Anthocyanin and PA Extraction
Leaves and developing siliques collected from at least 10 plants of each type analyzed were frozen in liquid N2 and stored at −80°C. Samples were ground in liquid N2 and anthocyanin and PA extracted using 0.37% (w/v) HCl in methanol or 70% (w/v) acetone containing 0.1% (w/v) ascorbate, respectively, for 16 to 18 h at 4°C. This was repeated two times, for 2 h each extraction. The crude anthocyanin preparations were extracted further using Folch partitioning (Folch et al., 1951) with chloroform/water to remove chlorophyll (two times), and then extracted with hexane (two times). To simplify interpretation of chromatograms, glycosides were removed by acid hydrolysis and the free aglycones examined. Samples were hydrolyzed by adding an equal volume of 37% (w/v) HCl and boiling for 15 min. Boiled samples were then extracted into pentan-2-ol, which was evaporated under vacuum centrifugation. Samples were dissolved in 0.37% (w/v) HCl in methanol, spotted onto 0.1-mm cellulose TLC plates (Merck, Rahway, NJ), and developed using A & F no. 9 (HCl:formic acid:water, 19:40:41 [v/v]; Andersen and Francis, 1985). Dried plates were sprayed with 1% (w/v) methanolic diphenylboryloxyethylamine (NP stain), followed by 5% (w/v) ethanolic polyethylene glycol 4,000, and then analyzed for anthocyanins and flavonols. Images of the plates were recorded in visible light with an HPScanJet 4C/T scanner (Hewlett-Packard, Palo Alto, CA) or photographed under UV illumination at 310 and 365 nm.
The acetone fraction of PA extracts was treated with ethyl acetate to partition the monomers and small oligomers into the ethyl acetate phase from PA polymers that remain in the aqueous phase (Nonaka et al., 1983, 1985). Both fractions were then extracted with hexane (three times) and then chloroform. The ethyl acetate fractions were spotted directly onto cellulose TLC plates, and developed using s-butanol:water:acetic acid:chloroform (70:20:10:10 [v/v]; Kristiansen, 1984). Dried plates were sprayed with DMACA reagent diluted 20-fold in methanol and analyzed for flavan-3-ols. PA samples were depolymerized and converted to anthocyanidins by acid hydrolysis and then analyzed as for anthocyanin samples.
Measurement of Anthocyanin and PA Content
PA monomer and polymer was quantitated using DMACA reagent in a 96-well plate reader (Molecular Devices, Spectra MAX 340 PC, Sunnyvale, CA). Standard curves were prepared by serial dilution of catechin monomer, trimer, and condensed tannin (isolated from Onobrychis viciifolia and quantitated by weight) standards (Tanner et al., 1995). The plate was scanned between 600 and 700 nm for a peak at 640 nm within 15 min of the addition of DMACA reagent. Samples containing PA showed a precipitate after 2 to 3 h, whereas small polymer standards did not. This method was also used to detect PA contamination of anthocyanin preparations.
Leaf anthocyanin extracts were scanned from 410 to 600 nm to determine the anthocyanin absorbance peak at about 520 to 530 nm. It was found that mutants such as tt3 gave a broad peak between 510 and 530 nm, even though they lack anthocyanin. For the purposes of calculation, the optical density (OD) at 600 nm was subtracted from the peak anthocyanin absorbance value. Seed anthocyanin concentrations were calculated using OD λmax − OD 600 nm g−1 fresh weight of material. Anthocyanin extracts were analyzed similarly, with anthocyanin being expressed for both leaf and seed extracts as a percentage relative to wild-type levels, because this value was constant for replicate experiments performed at different times. Ws-2 and Col-7 wild types were found to have different λmax values (scans not shown) and so each mutant is compared with its wild type.
An aliquot of the ethyl acetate extract was dried by vacuum centrifugation at room temperature. The residue was dissolved in 100 μL of water and analyzed by HPLC on a Goldpack (Activon, Sydney) 3- × 0.46-cm-i.d. column packed with 3 μm Exsil 100A, ODS C18 packing and eluted at 2 mL min−1 with a gradient from solvent A (2% [v/v] aqueous acetic acid) to 60% (w/v) solvent B (methanol) over 10 min, and returning to starting conditions over 5 min, with the detector set at 280 nm. The void volume of the column and system was 500 μL. Peaks of interest from wild-type Ws-2 seeds were repurified as described above but using water as solvent A, and the mass determined using HPLC mass spectrometry.
Microscopy
Fresh siliques were harvested and placed directly into DMACA reagent for 16 to 18 h, rinsed three times with 70% (w/v) ethanol, and then photographed at 6.3× magnification. Mature dry seed was stained similarly for 7 to 10 d until all seed were stained in wild-type samples. Samples for sectioning were fixed in glutaraldehyde, treated with OsO4 (Nielson and Griffith, 1978), lightly counterstained with toluidine blue, dehydrated, embedded, and then cut in 0.5- and 1-μm sections. Images of sections at 20× and 63× magnification were obtained with or without a Nomarsky filter.
Materials
Authentic standards of kaempferol, quercetin, myrcetin, naringenin, pelargonidin, cyanidin, delphinidin, catechin, and epicatechin were purchased from commercial suppliers. Leucocyanidin was prepared using published methods (Tanner and Kristiansen, 1993).
ACKNOWLEDGMENTS
We wish to thank the Plant Industry Horticulture Unit (Urbrae, South Australia, Australia) for access to the HPLC-mass spectrometer and Mark Downey for assistance with the mass determination of epicatechin, Celia Miller for assistance with microscopy, and Carl Davies for help with figure preparation.
Footnotes
This work was supported by Pioneer Hi-Bred and by Meat and Livestock Australia.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.006189.
LITERATURE CITED
- Albert S, Delseny M, Devic M. BANYULS, a novel negative regulator of flavonoid biosynthesis in the Arabidopsis seed coat. Plant J. 1997;11:289–299. doi: 10.1046/j.1365-313x.1997.11020289.x. [DOI] [PubMed] [Google Scholar]
- Andersen OM, Francis GW. Simultaneous analysis of anthocyanins and anthocyanidins on cellulose thin layers. J Chromatogr. 1985;318:450–454. [Google Scholar]
- Baur PS, Walkinshaw CH. Fine structure of tannin accumulations in callus cultures of Pinus elliotti (slash pine) Can J Bot. 1974;52:615–619. [Google Scholar]
- Bethke PC, Jones RL. Vacuoles and prevacuolar compartments. Curr Opin Plant Biol. 2000;3:469–475. doi: 10.1016/s1369-5266(00)00115-1. [DOI] [PubMed] [Google Scholar]
- Chafe SC, Durzan DJ. Tannin inclusions in cell suspension cultures of white spruce. Planta. 1973;113:251–262. doi: 10.1007/BF00390512. [DOI] [PubMed] [Google Scholar]
- Davin LB, Lewis NG. Dirigent proteins and dirigent sites explain the mystery of specificity of radical precursor coupling in lignan and lignin biosynthesis. Plant Physiol. 2000;123:453–461. doi: 10.1104/pp.123.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Leon-Kloosterziel KM, Koornneef M. Influence of the testa on seed dormancy, germination and longevity in Arabidopsis. Plant Physiol. 2000;122:403–413. doi: 10.1104/pp.122.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Peeters AJM, Leon-Kloosterziel KM, Koornneef M. The TRANSPARENT TESTA 12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell. 2001;13:853–871. doi: 10.1105/tpc.13.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcour JA, Ferreira D, Roux DG. Synthesis of condensed tannins: IX. The condensation sequence of leucocyanidin with (+) catechin and the resultant procyanidin. J Chem Soc Perkin Trans. 1983;1:1711–1717. [Google Scholar]
- Delcour JA, Janssens de Varabeke D. A new colourimetric assay for flavanoids in pilsner beers. J Inst Brew. 1985;91:37–40. [Google Scholar]
- Devic M, Guilleminot J, Debeaujon I, Bechtold N, Bensaude E, Koornneef M, Pelletier G, Delseny M. The BANYULS gene encodes a DFR-like protein and is a marker of early seed coat development. Plant J. 1999;19:387–398. doi: 10.1046/j.1365-313x.1999.00529.x. [DOI] [PubMed] [Google Scholar]
- Feinbaum RL, Ausubel FM. Transcriptional regulation of the Arabidopsis thaliana chalcone synthase gene. Mol Cell Biol. 1988;8:1985–1992. doi: 10.1128/mcb.8.5.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A. Enzyme Structure and Mechanism. New York: WH Freeman and Company; 1985. Chemical catalysis; pp. 47–97. [Google Scholar]
- Folch J, Less M, Sloane-Stanley GHB. A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem. 1951;226:497–509. [PubMed] [Google Scholar]
- Garcia-Florenciano E, Barcelo AR, Sabater F, Munoz R. The spectrophotometric determination of indole-3-methonol in small samples with p-dimethylaminocinnamaldehyde on the basis of the formation of an azafulvenium salt. Anal Biochem. 1989;183:172–176. doi: 10.1016/0003-2697(89)90185-1. [DOI] [PubMed] [Google Scholar]
- Hadlington JL, Denecke J. Sorting of soluble proteins in the secretory pathway of plants. Curr Opin Plant Biol. 2000;3:461–468. doi: 10.1016/s1369-5266(00)00114-x. [DOI] [PubMed] [Google Scholar]
- Hilling B, Amelunxen F. On the development of the vacuole: II Further evidence for endoplasmic reticulum origin. Eur J Cell Biol. 1985;38:195–200. [Google Scholar]
- Jauh GY, Phillips TE, Rogers JC. Tonoplast intrinsic protein isoforms as markers for vacuolar functions. Plant Cell. 1999;11:1867–1882. doi: 10.1105/tpc.11.10.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jende-Strid B. Gene-enzyme relations in the pathway of flavonoid biosynthesis in barley. Theor Appl Genet. 1991;81:668–674. doi: 10.1007/BF00226735. [DOI] [PubMed] [Google Scholar]
- Jende-Strid B. Genetic control of flavonoid biosynthesis in barley. Heriditas. 1993;119:187–204. [Google Scholar]
- Jornvall H, Persson B, Krook M, Atrian S, Gonzalez-Duarte R, Jeffery J, Ghosh D. Short-chain dehydrogenases/reductases (SDR) Biochemistry. 1995;34:6003–6013. doi: 10.1021/bi00018a001. [DOI] [PubMed] [Google Scholar]
- Koornneef M. Mutations affecting the testa colour in Arabidopsis. Arabid Inf Serv. 1990;27:1–4. [Google Scholar]
- Koornneef M, Luiten W, de Vlaming P, Schram AW. A gene controlling flavonoid-3′-hydroxylation in Arabidopsis. Arabid Inf Serv. 1982;19:113–115. [Google Scholar]
- Koupai-Abyazani MR, McCallum J, Muir AD, Lees GL, Bohm BA, Towers GHN, Gruber MY. Purification and characterisation of a proanthocyanidin polymer from seed alfalfa (Medicago sativa Cv. Beaver) J Agric Food Chem. 1993a;41:565–569. [Google Scholar]
- Koupai-Abyazani MR, McCallum J, Muir AD, Lees GL, Bohm BA, Towers GHN, Gruber MY. Developmental changes in the composition of proanthocyanidins from leaves of Sanfoin (Onobrychus viciifolia Scop.) as determined by HPLC analysis. J Agric Food Chem. 1993b;41:1066–1070. [Google Scholar]
- Kristiansen K. Biosynthesis of procyanidins in barley: genetic control of the conversion of dihydroquercetin to catechin and procyanidins. Carlsberg Res Commun. 1984;49:503–524. [Google Scholar]
- Kristensen H, Aastrup S. A non-destructive screening method for proanthocyanidin-free barley mutants. Carlsberg Res Commun. 1986;51:509–513. [Google Scholar]
- Kumar R, Singh M. Tannins: their adverse role in ruminant nutrition. J Agric Food Chem. 1984;32:447–453. [Google Scholar]
- Labesse G, Vidal-Cros A, Chomilier J, Gaudry M, Mornon J-P. Structural comparisons lead to the definition of a new superfamily of NAD (P)(H)-accepting oxidoreductases: the single-domain reductases/epimerases/dehydrogenases (the “RED” family) Biochem J. 1994;304:95–99. doi: 10.1042/bj3040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YG, Tanner G, Larkin P. The DMACA-HCl protocol and the threshold proanthocyanidin content for bloat safety in forage legumes. J Sci Food Agric. 1996;70:89–101. [Google Scholar]
- Mansfield SG. Embryogenesis: introduction. In: Bowman J, editor. Arabidopsis: an Atlas of Morphology and Development. New York: Springer; 1994. pp. 351–361. [Google Scholar]
- McMurrough I, McDowell J. Chromatographic separation and automated analysis of flavanols. Anal Biochem. 1978;91:92–100. doi: 10.1016/0003-2697(78)90819-9. [DOI] [PubMed] [Google Scholar]
- Morris P, Robbins MP. Manipulating condensed tannins in forage legumes. In: McKersie BD, Brown DCW, editors. Biotechnology and the Improvement of Forage Legumes. Wallingford, Oxon, UK: CAB International; 1997. pp. 147–173. [Google Scholar]
- Mueller L, Goodman CD, Silady RA, Walbot V. AN9, a petunia glutathione S-transferase required for anthocyanin sequestration is a flavonoid-binding protein. Plant Physiol. 2000;123:1561–1570. doi: 10.1104/pp.123.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell. 2000;12:1863–1878. doi: 10.1105/tpc.12.10.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L. The Arabidopsis TT2 gene encodes an R2R3 domain protein that acts as a key determinant for proanthocyandidn accumulation in developing seed. Plant Cell. 2001;13:2099–2114. doi: 10.1105/TPC.010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielson AJ, Griffith WP. Tissue fixation and staining with osmium tetroxide: the role of phenolic compounds. J Histochem Cytochem. 1978;26:138. doi: 10.1177/26.2.75221. [DOI] [PubMed] [Google Scholar]
- Nonaka G, Muta M, Nishioka I. Myricatin, a galloylflavanonol sulfate and prodelphinidin gallates from Myrica rubra. Phytochemistry. 1983;22:237–241. [Google Scholar]
- Nonaka G, Nishimura H, Nishioka I. Tannins and related compounds: XXVI. Isolation and structures of stenophyllanins A, B and C, novel tannins from Quercus stenophylla. J Chem Soc Perkin Trans. 1985;1:163–172. [Google Scholar]
- Parham RA, Kaustinen HM. On the site of tannin synthesis in plant cells. Bot Gaz. 1977;138:465–467. [Google Scholar]
- Prevost D, Jain DK, Bordeleau LM. Growth inhibition of rhizobia isolated from arctic legumes (Austragalus and Oxytropis spp.) and sainfoin (Onobrychis viciifolia) by sainfoin seed diffusate. Phytoprotection. 1990;71:113–119. [Google Scholar]
- Schoenbohm C, Martens S, Eder C, Forkmann G, Weisshaar B. Identification of the Arabidopsis thaliana flavanoid-3′-hydroxylase gene and functional expression of the encoded P450 enzyme. Biol Chem. 2000;381:749–753. doi: 10.1515/BC.2000.095. [DOI] [PubMed] [Google Scholar]
- Shirley BW, Hanley S, Goodman HM. Effects of ionising radiation on a plant genome: analysis of two Arabidopsis transparent testa mutants. Plant Cell. 1992;4:333–347. doi: 10.1105/tpc.4.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM. Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 1995;8:659–671. doi: 10.1046/j.1365-313x.1995.08050659.x. [DOI] [PubMed] [Google Scholar]
- Singh S, McCallum J, Gruber M, Towers GHN, Muir AD, Bohn BA, Koupai-Abyazani MR, Glass ADM. Biosynthesis of flavan-3-ols by leaf extracts of Onobrychis viciifolia. Phytochemistry. 1997;44:425–432. [Google Scholar]
- Skadhauge B, Gruber M, Thomsen KK, von Wettstein D. Leucocyanidin reductase activity and accumulation of proanthocyanidins in developing legume tissues. Am J Bot. 1997;84:497–503. [Google Scholar]
- Stafford HA. The enzymology of proanthocyanidin biosynthesis. In: Hemingway RW, Karchesy JJ, editors. Chemistry and Significance of Condensed Tannins. New York: Plenum Press; 1989. pp. 345–368. [Google Scholar]
- Stafford HA, Lester HH. Flavan-3-ol biosynthesis; the conversion of (+)-dihydroquercetin and flavan-3,4-cis-diol (leucocyanidin) to (+) catechin by reductases extracted from cell suspension cultures of Douglas fir. Plant Physiol. 1984;76:184–186. doi: 10.1104/pp.76.1.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner G, Kristiansen KN. Synthesis of 3,4-cis-[3H]-leucocyanidin and enzymatic reduction to catechin. Anal Biochem. 1993;209:274–277. doi: 10.1006/abio.1993.1119. [DOI] [PubMed] [Google Scholar]
- Tanner GJ, Moate PJ, Davis LH, Laby RH, Yuguang L, Larkin PA. Proanthocyanidins (condensed tannin) destabilise plant protein foams in a dose dependent manner. Aust J Agric Res. 1995;46:1101–1109. [Google Scholar]
- Treutter D. Chemical reaction detection of catechins and proanthocyanidins with 4-dimethylaminocinnamaldehyde. J Chromatogr. 1989;467:185–193. [Google Scholar]
- Waghorn GC, Jones WT. Bloat in cattle: 46. Potential of dock (Rumex obtusifolius) as an anti bloat for cattle. N Z J Agric Res. 1989;32:227–235. [Google Scholar]
- Wagner H. Flavanoid drugs. In: Wagner H, Bladt S, Zgainski EM, editors. Plant Drug Analysis: a Thin Layer Chromatography Atlas. Berlin: Springer-Verlag; 1984. pp. 163–193. [Google Scholar]
- White DL, Andrews SB, Faller JW, Barrnett RJ. The chemical nature of osmium tetroxide fixation and staining of membranes by x-ray photoelectron spectroscopy. Biochim Biophys Acta. 1976;436:577–592. doi: 10.1016/0005-2736(76)90442-9. [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. Flavonoid biosynthesis. A colourful model for genetics, biochemistry, cell biology and biotechnology. Plant Physiol. 2001;126:485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisman E, Hartmann U, Sagasser M, Baumann E, Palme K, Hahlbrock K, Saedler H, Weisshaar B. Knockout mutants from an En-1 mutagenised Arabidopsis thaliana population generate phenylpropanoid biosynthesis phenotypes. Proc Natl Acad Sci USA. 1998;95:12432–12437. doi: 10.1073/pnas.95.21.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young H, Paterson VJ. Condensed tannins from white clover seed diffusate. Phytochemistry. 1980;19:159–160. [Google Scholar]