Abstract
The recent publication of the complete sequence of the Arabidopsis genome allowed us to identify and characterize the last two members of the SHAGGY-like kinase (AtSK) gene family. As a result, the study of the overall spatio-temporal organization of the whole AtSK family in Arabidopsis has become an achievable and necessary aim to understand the role of each SHAGGY-like kinase during plant development. An analysis of the transcript level of the 10 members of the family has been performed using the technique of real-time quantitative reverse transcriptase-polymerase chain reaction. Transcript levels in several organs, under different growth conditions, were analyzed. To calibrate the results obtained, a number of other genes, such as those coding for the two MAP3Kεs and the two MAP4Kαs, as well as the stress response marker RD29A; the small subunit of the Rubisco photosynthetic enzyme Ats1A; the MEDEA chromatin remodeling factor; and the SCARECROW, ASYMMETRIC LEAVES 1, and SUPERMAN transcription factors all involved in key steps of plant development were used. The analysis of our data revealed that eight of the 10 genes of the AtSK family displayed a pseudo-constitutive expression pattern at the organ level. Conversely, AtSK13 responded to osmotic changes and saline treatment, whereas AtSK31 was flower specific and responded to osmotic changes and darkness.
The SHAGGY/GSK3-like kinases are non-receptor Ser-Thr (S/T) kinases playing numerous roles (for review, see Kim and Kimmel, 2000). In animals, they are involved in the determination of cell destiny, resulting in the spatial organization of the body plan. In Drosophila melanogaster, a pool of several isoenzymes called SHAGGY and encoded by a single gene, is involved both in the definition of boundaries between the embryonic segments of the larvae (Siegfried et al., 1992), and in the development of the central and peripheral nervous system (Heitzler and Simpson, 1991). In the sea urchin embryo, the SHAGGY-like enzyme is involved in the definition of the animal/vegetal axis (Emily-Fenouil et al., 1998). In Xenopus laevis embryo, a deficiency for the activity of this kinase results in a defect of the dorso-ventral plan formation, leading to the formation of two heads (He et al., 1995). Finally, in mammals, two enzymes named GSK3α and GSK3β (for glycogen-synthase kinase), encoded by two genes, are involved in the regulation of glycogen metabolism (Oreña et al., 2000), in the stability of the cytoskeleton (Zumbrunn et al., 2001), and in numerous other processes related to oncogenesis (Webster et al., 2001).
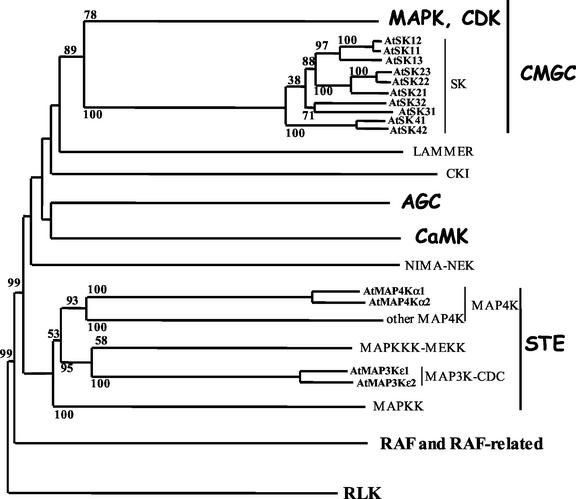
In higher plants, the SHAGGY-like genes are present as small gene families. They have been characterized from a number of plant species (Pay et al., 1993; Tichtinsky et al., 1998; Jonak et al., 2000). Before the completion of the sequencing of the whole Arabidopsis genome, eight genes were known to belong to the SHAGGY-like gene family (AtSK; Jonak et al., 1995; Dornelas et al., 1998, 1999; Tichtinsky et al., 1998). Recently, two additional genes, AtSK13 and AtSK42 (according to the nomenclature of Dornelas et al., 2000), were characterized from the database. The relationship between the 10 Arabidopsis genes, based on the comparison of the sequence of their catalytic domain, is presented in Figure 1. The whole AtSK gene family is divided into four subgroups. AtSK13 forms a subgroup with AtSK11 and AtSK12, whereas AtSK42 forms a subgroup with AtSK41. Altogether, the 10 AtSK genes share between 75.1% and 98.2% identity, and between 90.2% and 99.3% similarity in their catalytic domain. The identity remains high with 50% in the N-terminal region and 65% in the C-terminal region.
Figure 1.
Relationship tree based on the catalytic domain of the Arabidopsis Ser/Thr protein kinases. The sequence-based relationship between AtSK, AtMAP3K, and AtMAP4K, compared with the other groups of S/T kinases, is presented in the tree. In bold are indicated the major groups of S/T kinases, i.e. the receptor-like kinases (RLK) and the RAF kinases (standard characters), as well as the non-receptor S/T kinases, such as CMGC, AGC, CaMK, and STE (comic characters), as defined by Hunter and Plowman (1997). The 10 AtSK genes, the two AtMAP3Kεs, and the two AtMAP4Kαs are typed in bold small characters. The distance tree was constructed using the neighbor-joining method (Saitou and Nei, 1987) via the ClustalX program. We rooted the tree using the sequences of Arabidopsis RLK. The tree, including 52 RAF and 316 non-receptor S/T kinases, has been simplified. The bootstrap values of 100 replicates are only indicated for the AtSK, the AtMAP3Kε, and the AtMAP4Kα proteins.
In plants as in the animal kingdom, the roles of the SHAGGY-like enzymes seem to be numerous. On one hand, Li and Nam (2002) recently reported on the Arabidopsis bin2 mutant containing a semidominant mutation in the AtSK21 catalytic domain, resulting in 30% increase of the kinase activity. bin2 had previously been studied for its insensitivity to the brassinosteroid hormone (Li et al., 2001), and displays a dwarf phenotype, accompanied by curved leaves and an impaired cell elongation. Simultaneously, Perez-Perez et al. (2002) characterized the Arabidopsis ucu1 (ultracurvata1) mutant modified in the same exon and displaying the same phenotype as bin2. On the other hand, Dornelas et al. (2000) reported that Arabidopsis plants expressing antisense for the two SHAGGY-like genes, AtSK11 and AtSK12, displayed defects in flower morphology. The flowers of these plants showed supernumerary petals compared with the wild type. The SHAGGY-like kinases are also involved in the plant response to stress. In Arabidopsis, Piao et al. (2001) reported that AtSK22 conferred resistance to NaCl, whereas in alfalfa (Medicago sativa), Jonak et al. (2000) showed that WIG, a SHAGGY-like homolog, responded to wounding.
In summary, five studies have reported so far on the involvement of four members of the AtSK gene family, three of them in plant development (two specifically in flowers and one in a hormone-dependent developmental process), and one in resistance to abiotic stress. This seems to indicate that the 10 AtSK isoenzymes might each be involved in fundamentally distinct processes, revealing a complex organization of the gene family at the functional level. The aim of our work now is to allocate a biological function to each of these gene members, both to better understand the roles of the SHAGGY-like kinase gene family in plants, and to elucidate the complexity of the functional organization within this gene family.
Because the last two members have been identified recently, the first achievable approach was to perform a comprehensive analysis of the transcription profile of the 10 AtSK genes. SHAGGY/GSK3 kinases, like most of the S/T kinases, are posttranslationally regulated by phosphorylation. Phosphorylation on Ser-9 inhibits the activity of GSK3 (Dajani et al., 2001), whereas phosphorylation on the Tyr-216 increases it (Wang et al., 1994). However, numerous kinase-encoding genes have been shown to respond also at the transcriptional level to different types of treatments, including hormonal (Marcote and Carbonell, 2000;Lindroth et al., 2001), light (Hajouj et al., 2000), sugar (Chikano et al., 2001), wounding (Shin et al., 2001),and pathogen attack (Murillo et al., 2001) stresses, thereby revealing their involvement in these latter biological processes. Furthermore, Jonak et al. (2000) showed that the alfalfa SHAGGY-like kinase WIG responded to wounding at the transcriptional level, a response that has been further confirmed at the posttranslational level. Therefore, specific features of the expression profile of the AtSK genes will reveal putative biological functions for these proteins.
The expression profile of only some of the AtSK genes has already been partially described using techniques such as northern blot under standard growth conditions (Dornelas et al., 1999) or under specific treatments (Piao et al., 1999). However, northern-blotting experiments require both a relatively high expression level for the gene tested, and a specific probe. Aiming at analyzing 10 relatively low-expressed members of a homogeneous gene family all sharing a high percentage of identity, we decided to use the real-time quantitative reverse transcriptase (RT)-PCR technique, which offers both a high sensitivity and a high specificity (Bustin, 2000). In addition, it allowed us to define the absolute level of the targets present in a sample. The accuracy and the reliability of the real-time PCR have previously been reported in many studies on human genotyping and pathogen detection (Greiner et al., 2001; Kariyazono et al., 2001). In plants, it has been used to determine the number of T-DNA insertions in transgenic plants (Ingham et al., 2001), to detect the presence of genetically modified organisms in food (Hernandez et al., 2001), or in turn to quantify the level of transcripts present in plant organs (Lammers et al., 2001; Reintanz et al., 2002). Finally, because it does guaranty a specific detection of each gene of the family, the real-time RT-PCR technique is the best tool to carry out an analysis of the expression level of genes belonging to a very conserved gene family, as illustrated by Yokoyama and Nishitani (2001), on the members of a cell wall enzyme gene family.
This article reports on the comprehensive expression profile of the 10 members of the SHAGGY/GSK3-like kinase gene family in all the main organs of Arabidopsis and under a series of abiotic treatments, including changes of temperature, increase in salt concentration and osmotic pressure, dehydration, leaf wounding, and growth in the dark.
RESULTS
Absolute Levels of the AtSK Transcripts, and Calibration with Relevant Reference Genes
The absolute steady-state transcript level of the 10 AtSK genes was measured, using the technique of real-time RT-PCR (see “Materials and Methods”), in total RNA extracted from seedlings and seven different Arabidopsis organs, namely roots, rosette leaves, cauline leaves, inflorescence stems, flower buds, open flowers, and siliques. In addition, seven treatments were applied to young plants. The first series of treatments corresponded to modifications of temperature (4°C or 40°C). The second series of treatments consisted in dehydrating the plants, or modifying either the salt concentration by adding 150 mm NaCl, or the osmotic pressure by adding 25% (w/v) PEG. Finally, the transcriptional control of leaf wounding was tested, as well as that of the absence of light during several days (see “Materials and Methods”).
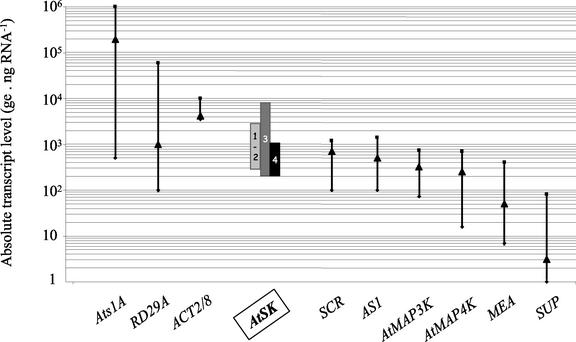
Figure 2 illustrates, for each subgroup of the AtSK gene family (see Fig. 1 for the subgroup definition), the range of the transcript levels observed in the conditions described above. All the members of this gene family displayed a similar steady-state transcript level, with an average around 2,000 copies of transcripts (expressed as genome equivalent [ge]; see “Materials and Methods” for the definition of this unit) per nanogram of total RNA (ge ng RNA−1). However, subgroup 4 exhibits the lowest level, whereas subgroup 3 the highest.
Figure 2.
Range of the absolute steady-state transcript levels for the 21 genes tested. Genes were tested for their steady-state transcript level in eight different organs and seven different growth conditions. The y axis is a log10 representation of the number of ge per nanogram of total RNA (ge ng RNA−1). The four AtSK gene subgroups are represented as vertical boxes. Subgroups 1 and 2 are both represented by the same box. For the other genes, the results are presented as a bar spreading from the highest to the lowest levels observed, with an arrowhead representing the average level. They are organized in descending order from the gene presenting the highest level (Ats1A), to the gene presenting the lowest level (SUP).
About 1,100 S/T kinases have been detected in the genome of Arabidopsis, and two-thirds of them correspond to receptor-like protein kinases. To place the expression profile of the SHAGGY-like kinase genes within this superfamily of S/T kinases, the transcriptional behavior of other non-receptor S/T kinases needed to be surveyed. Therefore, we selected two additional groups of S/T kinases, distant enough from the AtSK at the sequence level to provide information about three dispersed groups of S/T kinases. The first group contains the two MAP3Kε from Arabidopsis, namely AtMAP3Kε1 (Jouannic et al., 2001) and AtMAP3Kε2 (identified by homology to AtMAP3Kε1). These two kinases share 87.6% identity and differ from the other MAP3K at the sequence level (see Fig. 1). The second group corresponds to the two MAP4Kαs namely AtMAP4Kα1 and AtMAP4Kα2, which both are the orthologs of two MAP4Kαs from Brassica napus (Leprince et al., 1999). They share 78.7% identity, and constitute, with the two B. napus MAP4Kα, a small subgroup of the SOK (STE20 oxydative stress kinase) group, closely related to animal and yeast (Saccharomyces cerevisiae) MAP4Ks (Dan et al., 2001). The absolute transcript level of these four genes has been measured using the real-time RT-PCR technique on the same samples as those tested for the AtSK genes. Figure 2 shows that both the AtMAP3Kε and the AtMAP4Kα transcripts were present at levels 6 times lower, on average, than the AtSK transcripts.
In the frame of this study, reference genes used as controls to validate the experimental conditions were needed. The SHAGGY-like proteins are involved in both reproductive and vegetative development. Therefore, to accurately interpret the transcriptional behavior of the SHAGGY-like genes, it was necessary and relevant to test in parallel genes specifically involved in the development of flowers, leaves, and roots. The identity of these genes extended from proteins ubiquitously required to ensure the overall cell growth to proteins specifically involved in discrete processes of plant development. On one hand, the cytoskeleton proteins ACTIN have been chosen as housekeeping references. Although members of the Arabidopsis ACTIN gene family are differentially expressed (Huang et al., 1997), An et al. (1996) demonstrated that the two members, ACT2 and ACT8, display complementary patterns of expression, making their combined expression profile quasiconstitutive. The gene Ats1A coding for one of the small subunits of the Rubisco (Krebbers et al., 1988) has been shown to respond at the transcriptional level to light (Gallagher and Ellis, 1982; Morelli et al., 1985), thereby providing a marker of photosynthesizing tissues. On the other hand, genes specific for flower and seed identities have been tested: the transcription factor SUP (SUPERMAN) is expressed in the floral meristem of Arabidopsis, where it has been shown to control the boundary between the stamen and the carpels (Sakai et al., 1995). Likewise, MEA (MEDEA), a protein homologous to the D. melanogaster chromatin-structure modifier protein Enhancer of Zeste, controls the development of the embryo and the endosperm (Grossniklaus et al., 1998). Therefore, these two genes were used as transcriptional markers of flower identity. The two genes encoding the transcription factors SCR (SCARECROW) and AS1 (ASYMMETRIC LEAVES1) were also tested. Based on the phenotype of scr deficient mutants, SCR plays a very discrete and specific role in the definition of the cortex and the endodermis root cell lines (Di Laurenzio et al., 1996). As for AS1, it codes for an Myb domain protein controlling the size and the morphology of the leaves in Arabidopsis (Byrne et al., 2000). Finally, the gene RD29A, shown to respond to a variety of environmental stresses including dehydration, low temperature, saline treatments, and osmotic changes (Yamaguchi-Shinozaki and Shinozaki, 1993), was used to validate the treatments applied.
These 11 genes have been tested in the same samples as those used for the AtSK genes. For each of the genes tested, Figure 2 illustrates the averaged absolute levels, as well as their range of distribution. First, it shows that the reference genes displayed different averaged absolute transcript levels. The Ats1A and RD29A genes showed a very high level of transcripts (104–105 ge ng RNA−1), whereas MEA and SUP displayed the lowest amount of transcripts (50 ge ng RNA−1 for MEA, and just above the background level for SUP). ACT2/8 transcripts were twice as abundant as the AtSK transcripts. First, these reference genes altogether framed the absolute level of the AtSK genes. Second, all but one gene displayed a broad range in their transcript level, with at least one order of magnitude between the lowest and the highest level. This broad range reflects the variation of levels observed in response to the growth conditions (see further in the text). However, ACT2/ACT8 genes displayed only a maximum of 5-fold variation (between 2.103 and 104 ge ng RNA−1), with most of the samples displaying levels comprised between 4.103 and 6.103 ge ng RNA−1 (data not shown).
Facing so different growth conditions, a standard was needed to calibrate the subsequent data. Therefore, due to both their housekeeping function and their demonstrated stable expression profile, the ACT2/ACT8 genes have been used as a standard. Their absolute transcript levels obtained for each sample will be considered further in the text as a reflection of the amount of cDNA, and the data will be expressed as relative levels of transcripts standardized with the ACTIN2/8 absolute transcript level detected in each sample.
Relative Expression Profile of the 10 AtSK Genes in Arabidopsis Organs
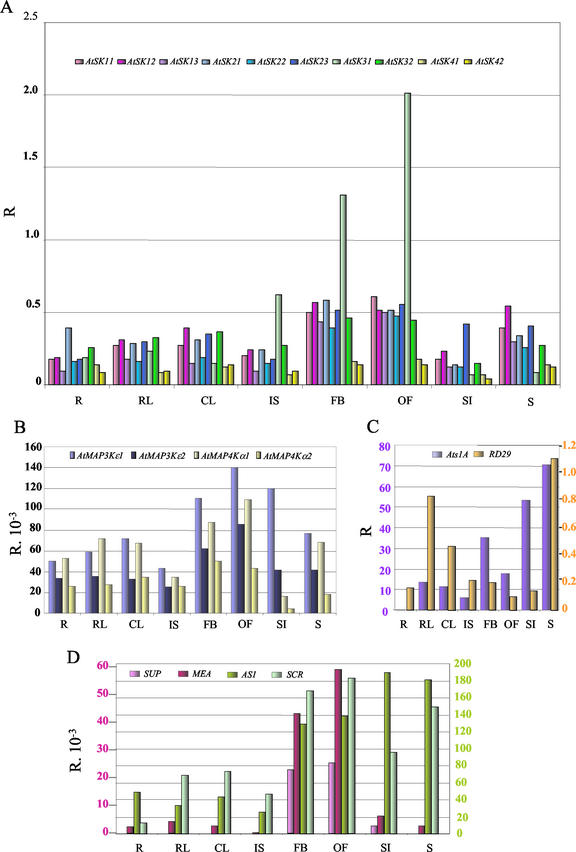
Figure 3 illustrates the relative transcript level of 20 genes standardized with the ACTIN2/8 absolute transcript level in Arabidopsis organs. The results obtained for the 10 AtSK genes are shown in Figure 3A. Each of the 10 genes was shown to be expressed, demonstrating that all the members of this gene family were transcriptionally active. In the growth conditions used, the 10 genes displayed a similar and fairly constant expression pattern, i.e. they were expressed: (a) in all of the eight organs tested, (b) at similar levels in roots, rosette leaves, cauline leaves, inflorescence stems, siliques, and seedlings, and (c) at a higher level (maximum 3-fold) in flower buds and open flowers compared with the other organs. However, AtSK41 and AtSK42 transcript levels were systematically lower than the eight other AtSK genes in most of the organs tested. In addition, AtSK31 displayed a stronger expression in inflorescence organs: Its transcript level is 8 times higher in flower buds and open flowers than in the vegetative organs. It is also 2 to 3 times higher in inflorescence stems.
Figure 3.
Relative expression profile of the 10 AtSK genes compared with 10 reference genes in Arabidopsis organs. The transcript level is represented as a ratio (R) of the absolute value of the studied gene to the absolute value of the ACT2/ACT8 genes. Due to the ratio calculation, the sds are not represented on this figure. The transcript levels have been tested on RNA extracted from roots (R), rosette leaves (RL), cauline leaves (CL), inflorescence stems (IS), flower buds (FB), open flowers (OF), siliques (SI), and seedlings (S). A, Distribution of AtSK transcripts. B, Distribution of the AtMAP3Kε and AtMAP4Kα transcripts. C, Distribution of Ats1A and RD29A transcripts. Two scales are provided on the y axis, each with the color of the corresponding gene. D, Distribution of SUP, MEA, AS1, and SCR transcripts. Like in C, two scales are provided with colors matching the genes.
Figure 3B shows that the two AtMAP3Kε and the two AtMAP4Kα transcripts were detected in all the conditions tested. Altogether, these genes displayed similar expression patterns with a fairly constant transcript level in all the organs tested, except in flower buds and open flowers, where it is slightly increased. Therefore, beside differences in the absolute expression levels mentioned previously (Fig. 2), the relative expression pattern of the above four MAP kinase genes is very similar to that of the AtSK genes, except for AtSK31. Among the 14 S/T kinase genes studied, AtSK31 is the only gene that displayed a significant organ specificity of expression.
Figure 3, C and D, report on the relative transcript level of the six reference genes under the same growth conditions. Figure 3C shows that Ats1A transcripts accumulated at various levels in all the organs tested, except in roots, where they were only detectable at a background level. Hence, Ats1A allowed us to confirm the photosynthetic nature of the aboveground organs tested in this study. In addition, the transcript level of the stress-responding marker RD29A varied to a maximum of 9-fold, which is at least 25 times lower than those observed in stressed plants (see further in the text). This demonstrated that the organs analyzed in this experiment were not under stressing growth conditions. Figure 3D shows that MEA and SUP transcripts were barely detectable in the vegetative organs, but their levels was dramatically increased in flower buds and open flowers compared with vegetative organs. Therefore, in agreement with their functions in embryo and flower development, respectively, MEA and SUP displayed expression patterns extremely specific for reproductive organs. In contrast, AS1 and SCR, each specifically involved in the development of respectively the leaf and the root, displayed similar levels of transcripts in all the organs tested. Therefore, the expression pattern of AS1 and SCR, at the organ level, is reminiscent of that of most AtSK genes, apart from AtSK31, which tends to display, as MEA and SUP, an expression specific for reproductive organs.
Relative Expression Profile of the 10 AtSK Genes under Abiotic Treatments
The relative expression profile of the 10 members of the AtSK gene family, as well as the 10 reference genes, was investigated in plants undergoing abiotic treatments. For each treatment, three-point kinetics was performed. The data are summarized in Table I, and illustrated only for the response-inducing treatments in Figure 4. To emphasize the results, only 2 times variations or more were considered. Among the reference genes, different responses were observed. The transcript level of the stress marker gene RD29A increased up to a 100 times in response to most of the treatments applied, which validated our experimental conditions. However, in the dark, RD29A transcripts were 25 times less abundant than in the light. This reduction is similar to that observed for Ats1A, which displays a 100 times reduction in the same conditions. Most of the genes tested displayed a reduction in their level of transcripts, which is likely to be due to a severe decrease of the metabolic activity of the whole plant, and which, therefore, will not be considered as a relevant response. The Ats1A steady-state level of transcripts seemed also to be positively regulated by NaCl treatment, and negatively regulated by the treatment to PEG. This result is consistent with previous studies showing that the activity of the Rubisco enzyme is inhibited by an increase in the concentration of Pro residue (Sivakumar et al., 2000) and by a hyperosmotic treatment with mannitol (Moreno and Spreitzer, 1999). Conversely, NaCl has a modulating effect on the activity of the Rubisco (Sivakumar et al., 2000). Altogether, the genes RD29A and Ats1A validate the experimental conditions, i.e. temperature, salt, osmotic pressure, dehydration, and darkness. None of the treatments applied to the whole plants are able to modulate the expression pattern of AtMAP3Kε genes, in contrast to AtMAP4Kα genes. Although AtMAP4Kα2 did only weakly respond to PEG treatment, AtMAP4Kα1 transcript levels were highly increased in response to three individual treatments, corresponding to NaCl (7 times), PEG (6.5 times), and wounding (3 times). It is worth noting that genes displaying an extremely low absolute level of transcripts, such as SUP, do display aberrant ratios of induction, which, therefore, are not considered here. Apart from MEA (see further in the text), no significant responses were observed for the other reference genes.
Table I.
Modification rates of the relative transcript levels for the 20 genes tested in the plantlets grown under stress conditions
| Gene | Treatments
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4°C | 40°C | Darkness | Wounding | −Water | +NaCl | +Polyethylene glycol (PEG) | |||||||
| AtSK11 | 0 | 0 | − (2) | 0 | 0 | 0 | 0 | ||||||
| AtSK12 | 0 | 0 | − (5) | 0 | 0 | 0 | 0 | ||||||
| AtSK13 | 0 | 0 | 0 | 0 | + (2) | + (3) | + (4) | ||||||
| AtSK21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| AtSK22 | 0 | 0 | − (2) | 0 | 0 | 0 | 0 | ||||||
| AtSK23 | 0 | 0 | − (3) | 0 | 0 | 0 | 0 | ||||||
| AtSK31 | 0 | 0 | + (4) | 0 | 0 | + (2) | + (2.5) | ||||||
| AtSK32 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| AtSK41 | 0 | 0 | − (4) | 0 | 0 | 0 | 0 | ||||||
| AtSK42 | 0 | 0 | − (4) | 0 | 0 | + (2.5) | + (2) | ||||||
| AtMAP3Kε1 | 0 | 0 | − (4) | 0 | 0 | 0 | 0 | ||||||
| AtMAP3Kε2 | 0 | 0 | − (4) | 0 | 0 | 0 | 0 | ||||||
| AtMAP4Kα1 | 0 | 0 | − (2) | + (3) | 0 | + (7) | + (6.5) | ||||||
| AtMAP4Kα2 | 0 | 0 | − (2) | 0 | 0 | 0 | + (2) | ||||||
| MEA | + (2.5) | 0 | + (8) | 0 | 0 | 0 | 0 | ||||||
| SUP | 0 | 0 | 0 | 0 | 0 | + (2) | + (3) | ||||||
| AS1 | 0 | 0 | − (3) | 0 | 0 | + (2) | + (2) | ||||||
| SCR | 0 | − (2) | − (8) | 0 | 0 | 0 | 0 | ||||||
| Ats1A | − (2) | − (2) | − (100) | 0 | − (2) | + (2.5) | − (4) | ||||||
| RD29A | + (36) | + (100) | − (25) | 0 | + (56) | + (35) | + (30) | ||||||
This table presents the increase or decrease rates of the transcript levels during the treatment kinetics, compared with the T0 value. 0, Variation less than 2-fold. +, Increase; –, decrease; (n), n-fold increase or decrease. More than 2-fold variations are shown in bold. Variations concerning very low expressed genes, such as SUP, may not be significant.
Figure 4.
Relative expression profile of the 10 AtSK genes compared with 10 reference genes in plants undergoing abiotic treatments. Only the results obtained for four treatments (NaCl, PEG, dark, and wounding) of the seven performed in this study are presented. The transcript level is represented as log2 of R′. R′ is defined as the ratio of the Rn value of the studied gene (R = absolute value of the gene/absolute value of ACT2/ACT8−1) at the time Tn, to the R0 value of the same gene at the time T0. The scale is presented in the left margin, whereas the kinetics timepoints are indicated in the right margin. Due to the ratio calculation, the sds are not represented in this figure. The transcriptional response of the 10 AtSK genes is separated from the other 10 reference genes by a vertical bar.
Among the AtSK family, one can classify the 10 genes within two main groups. The first group contains genes that did not significantly respond to any of the treatments applied, i.e. the subgroup I genes AtSK11 and AtSK12, the subgroup II genes, subgroup III gene AtSK32, and subgroup IV gene AtSK41. The second group comprises genes that significantly responded to one or several of the treatments applied. Subgroup I gene AtSK13 displayed a significant increase of its transcript level in response to hyperosmolarity both by addition of NaCl (3 times) and of PEG (4 times). Subgroup IV gene AtSK42 also responded both to NaCl and PEG treatments, but at a lower level, and later (only at 8 h, see Fig. 4) than AtSK13. Although saline treatment moderately increased the level of subgroup III AtSK31 transcripts (2 times only at 8 h, Fig. 4), hyperosmolarity by addition of PEG increased it 2.5 times at both 4 and 8 h. Altogether, AtSK13, AtSK31, and, more moderately, AtSK42, responded positively at the transcriptional level to changes of solutes supply. Interestingly, AtSK31 was the only of the three latter AtSK genes to display an additional response to another treatment. Its transcript level was 4 times higher when the plants were grown in the dark. Likewise, MEA transcripts are 8 times more abundant in plants subjected to 10 d of darkness instead of short-day conditions. Therefore, AtSK31 and MEA are the only two genes of the 20 tested, for which the steady-state level of transcripts is significantly higher under the latter conditions.
DISCUSSION
The work reported in this article illustrates the features and the advantages of the real-time RT-PCR technique. First, it allowed us to estimate the absolute level of transcripts of any type of genes studied, with either a very high or an extremely low level of expression. It is worth noting that no report about quantitative analysis of the SUP or MEA steady-state transcript levels had previously been published, and that genes such as AtMAP3Kε and AtMAP4Kα could not be analyzed quantitatively by another method so far. Second, this technique guaranties the specific detection of the gene studied, even when it belongs to a very conserved gene family. The microarray technology could be a powerful tool, especially when expression profiling is concerned. However, hybridization of chips loaded with full-sized cDNAs or even partial EST would not be suitable for the study of a gene family because of the lack of specificity of the targets. Conversely, due to its high cost, real-time RT-PCR has to remain a technique used for low- to middle-scale studies.
Thanks to the sensitivity and the reliability of this technique, we further described the expression profile of the AtSK genes and the putative functions allocated to individual members of this gene family. First, we have shown that the 10 AtSK genes are expressed at a quasiconstant level, comparable with that of the ACTIN2/8 genes, in all the organs tested. This ubiquitous profile may indicate that the whole AtSK gene family is recruited to play fundamental and basic roles in the cellular functioning of the whole plant. In accordance, Li and Nam (2002) and Perez-Perez et al. (2002), respectively, have recently described that the Arabidopsis bin2 and ucu1 mutants, which are both altered in the catalytic domain of AtSK21, displayed an impairment in the cell elongation along longitudinal axis and in lateral organs.
Second, we showed that three SHAGGY-like genes displayed significant modifications of their steady-state transcript levels in specific organs or in response to specific stresses. In flowers, AtSK31 is overexpressed. Previously, Dornelas et al. (2000) reported that plants expressing antisense of the AtSK11 and AtSK12 genes displayed discrete phenotypic alterations only in flowers. The flowers of these plants presented supernumerary perianth organs and an altered gynecium. Therefore, it seems that at least three SHAGGY-like genes, only two of them belonging to the same subgroup, are involved in flower development or metabolism.
Growth in the dark resulted in an increase of AtSK31 transcript levels. Perez-Perez et al. (2002) have reported that the Arabidopsis mutant ucu1 did not display any etiolated phenotype when grown in the dark, but rather showed a constitutive photomorphogenic response. Therefore, it seems that members of the AtSK gene family are involved in metabolic and developmental responses to darkness. This observation is reminiscent of the function of the SHAGGY enzyme in the control of the circadian rhythm in D. melanogaster (Martinek et al., 2001). Furthermore, Amador et al. (2001) have recently identified the potato (Solanum tuberosum) gene PHOR1, which responds at the transcriptional level to the photoperiod, and whose inhibition results in an early tuberization under short-day conditions. PHOR1 codes for a protein homologous to the D. melanogaster Armadillo, a Segment Polarity protein whose degradation and subcellular localization are controlled by the SHAGGY kinase (Ruel et al., 1999). Altogether, these data tend to indicate that at least two SHAGGY kinases, as well as the signaling pathway they are involved in, play a role in the plant response to light.
AtSK13, and, more moderately AtSK31 and AtSK42, positively responded at the transcriptional level to both an increase of the osmotic pressure and to a saline treatment. The variation of the transcript level in response to addition of NaCl is relevant to the study by Piao et al. (1999, 2001), who, based on yeast mutant complementation experiments and overexpression of the gene in planta, have shown that AtSK22 was involved in NaCl stress resistance in Arabidopsis. Therefore, at least two genes of the SHAGGY-like family may be involved in response, or even resistance as for AtSK22, to saline treatment. In contrast, there is no report so far about a putative involvement of the SHAGGY-like kinases in signaling pathways triggered by osmotic changes. However, numerous kinases are known to be involved in such pathways, such as the MAP kinase module. Here, we showed that an Arabidopsis MAP4K, AtMAP4Kα1, positively responded at the transcriptional level to both saline and PEG treatments, a result that has never been reported so far in plant. The first response is relevant to the description of an MAP cascade induced by high salinity. This cascade comprises an MAP3K (AtMEKK1; Mizoguchi et al., 1996) and an MAP2K (AtMKK2/MEK1, respectively; Morris et al., 1997; Ichimura et al., 1998), the transcript level of which is increased in response to the above treatment. As for the response to the osmotic changes, the induction of AtMAP4Kα1 is reminiscent of the results of Raitt et al. (2000), which showed that in yeast, the “high-osmolarity glycerol” response is mediated by a pathway dependent on the Ste20 MAP4K. Hence, both the SHAGGY-like kinase AtSK13 and the AtMAP4Kα1 might be part of a signaling network involved in the perception of salt and osmotic changes.
In contrast to the AtMAP4Kα genes, none of the AtSK genes did respond to wounding at the transcriptional level. However, Jonak et al. (2000) reported that WIG, an alfalfa gene coding for a SHAGGY-like protein homologous to AtSK32, responded both at the transcriptional and at the posttranslational level to wounding. Under the conditions used, we were unable to detect any induction of AtSK32 transcription along this kinetics. A differential response to abiotic treatments between genes from alfalfa and putative orthologues from other species has already been reported. Davletova et al. (2001) have noticed that a gene coding for a calmodulin-like domain protein kinase from alfalfa differentially responded to heat shock and auxin when compared with its homologs in other plant species. Alfalfa is a member of the Fabaceae family, which has developed, compared with other plant families, specific strategies to respond to wounding and pathogen attack. This might explain differences in the recruitment of SHAGGY-like genes to a wounding response. Alternatively, AtSKs might as well have been recruited for wounding responses by posttranslational modifications of the protein pool, produced from a constant steady-state pool of transcripts.
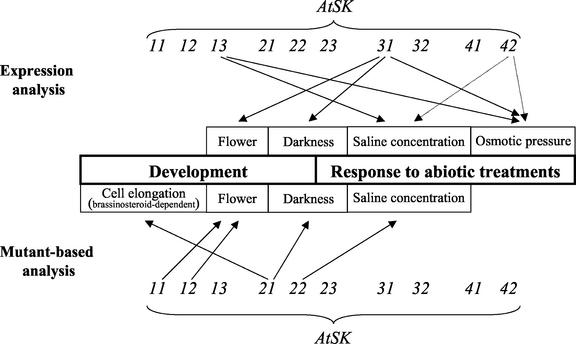
Altogether, these results increased our knowledge about the involvement of the Arabidopsis SHAGGY-like kinases in plant development and in response to stresses. On one hand, they reinforced previous reports on the analysis of shaggy mutants or antisense plants, which showed that one given member of this gene family was involved in plant development (AtSK21; Li and Nam, 2002; Perez-Perez et al., 2002) or in resistance to stress (AtSK22; Piao et al., 2001). We showed here that other genes of the same family, namely AtSK13 and AtSK31, also seem to be involved in the latter processes. On the other hand, the transcriptional induction of AtSK13 in response to osmotic changes allows us to open a new area of investigation on the roles played by these enzymes in the transduction of the signal resulting from a variation of the osmotic pressure. Therefore, plant SHAGGY-like enzymes, like their animal homologs, are likely to be involved in numerous biological processes. Figure 5 illustrates the progressively revealed complexity of the AtSK gene family. Several members seem to participate in the same process, such as AtSK13 and AtSK22 in response to saline treatment, or in turn, AtSK31 and AtSK21 in response to the lack of light. Alternatively, one given member can participate in several processes, such as the flower-specific AtSK31 gene, which responds to both osmotic changes and growth in the dark.
Figure 5.
Summary of the involvement of the AtSK genes in biological processes. The biological processes, which the AtSK genes are involved in, are indicated in boxes in the center of the figure. Each of the AtSK genes is represented above and under the boxes. The arrows indicate that the genes are involved in the indicating biological processes. The bottom one-half of the figure refers to previous studies based on the analysis of mutants or transgenic plants. The top one-half of the figure refers to this study, based on the analysis of the expression pattern of the AtSK genes. The dashed arrows concern slight modifications of the expression level.
Facing such a complexity in the allocation of the AtSK functions, other methods of analysis should be approached. First, the organ level expression profile should be completed by data on cell and tissue specificity. However, in situ hybridization requires the use of probes several hundred base pairs long to provide a signal detectable at the cellular level, which may be difficult to achieve when highly similar genes belonging to the same gene family are studied. Dornelas et al. (2000) previously reported on the cellular expression profile of five of the SHAGGY-like genes in embryos and in floral meristems. They showed that AtSK11 and AtSK12 genes displayed identical expression pattern, whereas AtSK21, AtSK22, and AtSK23 presented similar but different expression patterns within the organs tested, thereby increasing even more the complexity, at the functional level, of the SHAGGY-like gene family. Second, the roles filled by these genes in plants should be assessed by a study of the phosphorylation state of the AtSK isoenzymes, which requires antibodies to be produced for each of them. Finally, the analysis of null or negative-dominant mutants, none of which has been described so far, is an unavoidable goal to elucidate the functions fulfilled by each member of the SHAGGY-like gene family.
MATERIALS AND METHODS
Plant Material
Arabidopsis ecotype Columbia seeds were sown in soil in a growth chamber (17°C, 65% [v/v] hygrometry, 100% light HQI, and 0% [w/v] sodium) under a 16-h photoperiod. Seedlings were harvested 17 d after sowing (two cotyledons + four leaves stage). Roots, rosette leaves, cauline leaves, inflorescence stems, flower buds, open flowers, and 0.5-cm-long green siliques were harvested from a pool of 50 plants, 60 d after sowing. While harvesting the inflorescence stems, mainly primary inflorescence stems were taken and stem nodes were avoided. To test the effect of abiotic treatments on gene expression, 50 Columbia seeds were incubated in 250 mL of Murashige and Skoog (catalogue no. M0222, Duchefa, Haarlem, The Netherlands) liquid medium, complemented with 20 g L−1 Suc, pH 5.7 (MS20) in a growth chamber at 25°C. After 15 d, the seedlings were transferred to different environments to make them undergo several types of abiotic stimuli, after a kinetics at time 0, 4, and 8 h. Concerning the temperature stimuli, the flasks were put in an ice box, or in a 40°C water bath. The dehydration test was performed by taking out the seedlings from the flask and laying them on Whatman 3MM (Whatman, Clifton, NJ), according to Iuchi et al. (2000). The salt and the osmotic stimuli were carried out by transferring the seedlings into MS20 + 150 mm NaCl or 25% (w/v) PEG 6000, respectively. Wounding was performed by lacerating all the leaves of plants grown for 49 d in short-day conditions in greenhouses at 19.5°C day/17.5°C night temperature, with 60% (v/v) hygrometry and a light intensity of 300 μE m−2 s−1. Ten plantlets were harvested 30 and 60 min after wounding. Darkness was applied to 46-d-old plantlets grown in short-day conditions (see above). Ten plantlets were then transferred under total darkness in the same temperature and hygrometry conditions as the control plantlets. The 10 plantlets were harvested after 50 h or 10 d in the dark, as the corresponding 10 control plants grown under light. All the plant materials were frozen in liquid nitrogen immediately after harvesting. All these processes were repeated at least twice.
RNA Extraction and cDNA Preparation
Total RNA has been extracted from the frozen materials using the Plant RNeasy extraction kit (Qiagen USA, Valencia, CA). To eliminate the residual genomic DNA present in the preparation, the RNA was treated by an RNAse-free DNAse I according to the manufacturer's instructions (Qiagen USA). Total RNA was then quantified with a spectrophotometer, and loaded on a denaturing agarose gel to check their concentration and their integrity. Five micrograms of total RNA was reverse transcribed using the Superscript II RT kit (catalogue no. 18089-011, Life Technologies/Gibco-BRL, Cleveland) according to the manufacturer's instructions. cDNA was diluted at a concentration depending on the level of expression of the studied gene (for example, 10 ng of equivalent total RNA was tested in the PCR experiment for the AtSK genes). cDNA was aliquoted and kept at 4°C during the whole experiment to avoid discrepancy in the data due to freezing-thawing cycle repetitions.
Real-Time Quantitative RT-PCR
Gene and Oligonucleotide Designation
The PCR amplification has been performed with oligonucleotides specific for 21 genes. The genes, as well as the sequence of their specific oligonucleotides, are presented in Table II. For the amplification of the ACTIN genes, two oligonucleotides were designed so that the two genes ACTIN 2 and ACTIN 8 were amplified simultaneously. The position of these oligonucleotides has been chosen so that the size of the PCR product ranges between 50 and 150 bp. The suitability of the oligonucleotide sequences in term of efficiency of annealing has been tested, first by using the Primer Express 1.0 (Perkin-Elmer Applied Biosystems, Foster City, CA) and the Oligo 4.0 (W. Rychlik) softwares, and second, by determining the rate of amplification at each cycle for each couple of primers.
Table II.
Genes and oligonucleotides used in the real-time RT-PCR experiments
| Sequence of the 5′/3′ Oligonucleotides, Respectively | Gene (GenBank Accession No.) | Gene Reference |
|---|---|---|
| 5′-CTCTTAATGTAGCATGAACACAACAAAC-3′/5′-ATGAACCAACCAACCATAGTAATAACAC-3′ | AtSK11 (AJ000732) | Dornelas et al. (1998) |
| 5′-AAACTAGAGCAAAGCAGTCGAGATATTC-3′/5′-TTTCTCTACTCATACAACAAACAAAGGG-3′ | AtSK12 (Y12710) | Dornelas et al. (1998) |
| 5′-CTTATACCTGACCACGCCCG-3′/5′-AGATGAAAGCTAATGATAATCCTGAGAAG-3′ | AtSK13 (AL163792) | – |
| 5′-GTACCATTACACGAGCCACAAGG-3′/5′-TTTTTGTTTTCGCAGATTTGGG-3′ | AtSK21 (X94939) | Dornelas et al. (1999) |
| 5′-TGTGCATGTCTGAAGAGAAAGAGG-3′/5′-CGGGTATAGAAGCTGATAATAGTCGTG-3′ | AtSK22 (X99696) | Dornelas et al. (1999) |
| 5′-AAAATGCCGCCTTATGGAATG-3′/5′-TGGGACACAATCATCGCCTC-3′ | AtSK23 (X94938) | Dornelas et al. (1999) |
| 5′-TTGGGTTAATGGCTGCTTCTCTAC-3′/5′-TGTGGTTGCAGAGGTCAGTGAG-3′ | AtSK31 (AJ224338) | Tavares et al. (2000) |
| 5′-TGCGACAGCGTCTAATTCCAG-3′/5′-GCAGAAGTGGTTCAAAATTGATGTAC-3′ | AtSK32 (AF058919) | Tavares et al. (2000) |
| 5′-GGAGCAGCCAGTTTTCAGCC-3′/5′-CGTGAAACAAACAAGTGGTCTGTAAC-3′ | AtSK41 (X79279) | Jonak et al. (1995) |
| 5′-CCTATCGGGTGTTCTCTTCTTTTCTC-3′/5′-TCATTGCTTCTTTTTCCAGTGATTTC-3′ | AtSK42 (AAG31189) | – |
| 5′-TGTGAGTCCACCAAAACGATAATATC-3′/5′-GGAAGGGAAGAAGAACAGGCAC-3′ | AtMAP3Kε1 (AJ224982) | Jovannic et al. (2001) |
| 5′-TCTGATAGAAGAACGACGTGATGG-3′/5′-TGGAAGATGATATACACTTAGCTGGG-3′ | AtMAP3Kε2 (AC013483) | – |
| 5′-AAACTCACAGAAGACAGAACATAAGAATG-3′/5′-GATAAAACTAAAAAGGCCAAAATAAGGTAG-3′ | AtMAP4Kα1 (AAF69529) | – |
| 5′-GGTCATCTGCGACTGGGAGTAC-3′/5′-CATGGTCATTCTGAAAAGGGTG-3′ | AtMAP4Kα2 (BAB02151) | – |
| 5′-GGTAACATTGTGCTCAGTGGTGG-3′/5′-AACGACCTTAATCTTCATGCTGC-3′ | ACTIN 2 (U41998) and ACTIN 8 (U42007) | An et al. (1996) |
| 5′-AGAGGATTGGTCTATTTGCGGAG-3′/5′-GACGGGCTTCCTTAGACCTTTTAG-3′ | MEDEA (AF060485) | Grossniklaus et al. (1998) |
| 5′-AGAAAGAGCTTGCACATATGGAGAG-3′/5′-GGCCATGAAAACCCTAGAAGATAATC-3′ | SUPERMAN (U38946) | Sakai et al. (1995) |
| 5′-AAGCGACTCTACTGTTGGGAATG-3′/5′-AAACTAAGAACGAGGCGTCCAAG-3′ | SCARECROW (U62798) | Di Laurenzio et al. (1996) |
| 5′-GAGAAGATCGAAGGAGAGTACAGAGAAC-3′/5′-GGGGCGGTCTAATCTGCAAC-3′ | ASYMMETRIC LEAVES 1 (AF175996) | Byrne et al. (2000) |
| 5′-ATCACTTGGCTCCACTGTTGTTC-3′/5′-ACAAAACACACATAAACATCCAAAGTG-3′ | RD29A (D13044) | Yamaguchi-Shinozaki and Shinozaki (1993) |
| 5′-CAAGTCCAGTGCATCAGTTTCG-3′/5′-AAGATGGGGGATAAAGTTTTGAGG-3′ | Ats1A (X13611) | Krebbers et al. (1988) |
Amplification
The cDNA was amplified using the SYBR-GreenR PCR Master kit containing a Hot Start Taq polymerase (catalogue no. 430 9155, Perkin-Elmer Applied Biosystems) on the GeneAmp 9600 thermocycler (Perkin-Elmer Applied Biosystems). To determine the absolute number of specific cDNA molecules present in the samples, a standard was needed. Because more than 20 genes were tested, and to improve the comparison between specific gene amplifications, we used Arabidopsis genomic DNA (gDNA) as a reference matrix. For each experiment, a range of six dilutions of gDNA from Arabidopsis ecotype Columbia was tested in the same conditions as the cDNA samples. Knowing accurately the size and the mass of the genome of Arabidopsis ecotype Columbia, we were able to determine the number of genomes present in this dilution range, which, thus, can be used as a reference for the calculation of the number of cDNA molecules present in each sample tested. Therefore, the data were expressed as a number of Arabidopsis ge per nanogram of total RNA. At the end of the PCR cycles, the data were analyzed with the GeneAmp 5700 SDS software (Perkin-Elmer Applied Biosystems). To check the specificity of annealing of the oligonucleotides, a dissociation kinetics was performed by the machine at the end of the experiment. In addition, each amplified product was sequenced. Despite a treatment of the RNA with DNase I before cDNA amplification (see above), the contamination by gDNA was checked by amplification of gDNA using primers annealing on introns. The number of genome copies ranged from 0 to 20 ng of total RNA−1 (data not shown). Despite that these values are negligible compared with the >2,000 copies ng RNA−1 amplified for most of the genes tested in this study, we nevertheless took them into account for the calculation of the final value obtained for the cDNA samples.
In one experiment, at least four values, corresponding to the absolute transcript levels, were produced for each sample. The experiments were repeated at least twice independently, and the data were averaged. Depending on the samples, the sds were between 5% and 30% of the average value (data not shown). To standardize the data, the ratio of the absolute transcript level of each gene to the absolute transcript level of ACTIN 2/8 was calculated for each sample. Due to this ratio calculation, the sds were not displayed in Figures 3 and 4.
ACKNOWLEDGMENTS
We are grateful to Annaich Mingam and Alain Lecharny for their helpful discussions about the real-time RT-PCR technology. We are also grateful to Jean-Paul Bares and Gilles Santé for the maintenance of the plant facilities.
Footnotes
This work was supported by the “Pluriformation Genome” (contribution toward the purchasing of the real-time PCR apparatus).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.009175.
LITERATURE CITED
- Amador V, Monte E, Garcia-Martinez JL, Prat S. Gibberellins signal nuclear import of PHOR1 a photoperiod-responsive protein with homology to Drosophilaarmadillo. Cell. 2001;106:343–354. doi: 10.1016/s0092-8674(01)00445-7. [DOI] [PubMed] [Google Scholar]
- An YQ, McDowell JM, Huang S, McKinney EC, Chambliss S, Meagher RB. Strong, constitutive expression of the Arabidopsis ACT2/ACT8actin subclass in vegetative tissues. Plant J. 1996;10:107–121. doi: 10.1046/j.1365-313x.1996.10010107.x. [DOI] [PubMed] [Google Scholar]
- Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA. ASYMMETRIC LEAVES 1 mediates leaf patterning and stem cell function in Arabidopsis. Nature. 2000;408:967–971. doi: 10.1038/35050091. [DOI] [PubMed] [Google Scholar]
- Chikano H, Ogawa M, Ikeda Y, Koizumi N, Kusano T, Sano H. Two novel genes encoding SNF-1 related protein kinases from Arabidopsis thaliana: differential accumulation of AtSR1 and AtSR2transcripts in response to cytokinins and sugars and phosphorylation of sucrose synthase by AtSR2. Mol Gen Genet. 2001;264:674–681. doi: 10.1007/s004380000354. [DOI] [PubMed] [Google Scholar]
- Dajani R, Fraser E, Roe SM, Young N, Good V, Dale TC, Pearl LH. Crystal structure of glycogen synthase kinase 3β: structural basis for phosphate-primed substrate specificity and autoinhibition. Cell. 2001;105:721–732. doi: 10.1016/s0092-8674(01)00374-9. [DOI] [PubMed] [Google Scholar]
- Dan I, Watanabe NM, Kusumi A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 2001;11:220–230. doi: 10.1016/s0962-8924(01)01980-8. [DOI] [PubMed] [Google Scholar]
- Davletova S, Meszaros T, Miskolczi P, Oberschall A, Torok K, Magyar Z, Dudits D, Deak M. Auxin and heat shock activation of a novel member of the calmodulin like domain protein kinase gene family in cultured alfalfa cells. J Exp Bot. 2001;52:215–221. [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsisroot. Cell. 1996;86:423–433. doi: 10.1016/s0092-8674(00)80115-4. [DOI] [PubMed] [Google Scholar]
- Dornelas MC, Lejeune B, Dron M, Kreis M. The ArabidopsisSHAGGY-related protein kinase (ASK) gene family: structure organization and evolution. Gene. 1998;212:249–257. doi: 10.1016/s0378-1119(98)00147-4. [DOI] [PubMed] [Google Scholar]
- Dornelas MC, Van Lammeren AA, Kreis M. Arabidopsis thalianaSHAGGY-related protein kinases (AtSK11 and 12) function in perianth and gynoecium development. Plant J. 2000;21:419–429. doi: 10.1046/j.1365-313x.2000.00691.x. [DOI] [PubMed] [Google Scholar]
- Dornelas MC, Wittich P, von Recklinghausen I, van Lammeren A, Kreis M. Characterization of three novel members of the ArabidopsisSHAGGY-related protein kinase (ASK) multigene family. Plant Mol Biol. 1999;39:137–147. doi: 10.1023/a:1006102812280. [DOI] [PubMed] [Google Scholar]
- Emily-Fenouil F, Ghiglione C, Lhomond G, Lepage T, Gache C. GSK3beta/shaggy mediates patterning along the animal-vegetal axis of the sea urchin embryo. Development. 1998;125:2489–2498. doi: 10.1242/dev.125.13.2489. [DOI] [PubMed] [Google Scholar]
- Gallagher TF, Ellis RJ. Light-stimulated transcription of genes for two chloroplast polypeptides in isolated pea leaf nuclei. EMBO J. 1982;1:1493–1498. doi: 10.1002/j.1460-2075.1982.tb01345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner O, Day PJ, Bosshard PP, Imeri F, Altwegg M, Nadal D. Quantitative detection of Streptococcus pneumoniaein nasopharyngeal secretions by real-time PCR. J Clin Microbiol. 2001;39:3129–3134. doi: 10.1128/JCM.39.9.3129-3134.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus U, Vielle-Calzada JP, Hoeppner MA, Gagliano WB. Maternal control of embryogenesis by MEDEA, a Polycomb group gene in Arabidopsis. Science. 1998;280:446–450. doi: 10.1126/science.280.5362.446. [DOI] [PubMed] [Google Scholar]
- Hajouj T, Michelis R, Gepstein S. Cloning and characterization of a receptor-like protein kinase gene associated with senescence. Plant Physiol. 2000;124:1305–1314. doi: 10.1104/pp.124.3.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Saint-Jeannet JP, Woodgett JR, Varmus HE, Dawid IB. Glycogen synthase kinase-3 and dorsoventral patterning in Xenopusembryos. Nature. 1995;374:617–622. doi: 10.1038/374617a0. [DOI] [PubMed] [Google Scholar]
- Heitzler P, Simpson P. The choice of cell fate in the epidermis of Drosophila. Cell. 1991;64:1083–1092. doi: 10.1016/0092-8674(91)90263-x. [DOI] [PubMed] [Google Scholar]
- Hernandez M, Rio A, Esteve T, Prat S, Pla M. A rapeseed-specific gene acetyl-coa carboxylase can be used as a reference for qualitative and real-time quantitative PCR detection of transgenes from mixed food samples. J Agric Food Chem. 2001;49:3622–3627. doi: 10.1021/jf010173n. [DOI] [PubMed] [Google Scholar]
- Huang S, An YQ, McDowell JM, McKinney EC, Meagher RB. The Arabidopsis ACT11 actingene is strongly expressed in tissues of the emerging inflorescence pollen and developing ovules. Plant Mol Biol. 1997;33:125–139. doi: 10.1023/a:1005741514764. [DOI] [PubMed] [Google Scholar]
- Hunter T, Plowman GD. The protein kinases of budding yeast: six score and more. Trends Biochem Sci. 1997;22:18–22. doi: 10.1016/s0968-0004(96)10068-2. [DOI] [PubMed] [Google Scholar]
- Ichimura K, Mizoguchi T, Irie K, Morris P, Giraudat J, Matsumoto K, Shinozaki K. Isolation of ATMEKK1 (a MAP kinase kinase kinase)-interacting proteins and analysis of a MAP kinase cascade in Arabidopsis. Biochem Biophys Res Commun. 1998;253:532–543. doi: 10.1006/bbrc.1998.9796. [DOI] [PubMed] [Google Scholar]
- Ingham DJ, Beer S, Money S, Hansen G. Quantitative real-time PCR assay for determining transgene copy number in transformed plants. Biotechniques. 2001;31:136–140. doi: 10.2144/01311rr04. [DOI] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K. A stress-inducible gene for 9-cis-epoxycarotenoid dioxygenase involved in abscisic acid biosynthesis under water stress in drought-tolerant cowpea. Plant Physiol. 2000;123:553–562. doi: 10.1104/pp.123.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonak C, Beisteiner D, Beyerly J, Hirt H. Wound-induced expression and activation of WIG, a novel glycogen synthase kinase 3. Plant Cell. 2000;12:1467–1475. doi: 10.1105/tpc.12.8.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonak C, Heberle-Bors E, Hirt H. Inflorescence-specific expression of AtK-1 a novel Arabidopsis thalianahomologue of shaggy/glycogen synthase kinase-3. Plant Mol Biol. 1995;27:217–221. doi: 10.1007/BF00019194. [DOI] [PubMed] [Google Scholar]
- Jouannic S, Champion A, Segui-Simarro JM, Salimova E, Picaud A, Tregear J, Testillano P, Risueno MC, Simanis V, Kreis M et al. The protein kinases AtMAP3Kepsilon1 and BnMAP3Kepsilon1 are functional homologues of S. pombecdc7p and may be involved in cell division. Plant J. 2001;26:637–649. doi: 10.1046/j.1365-313x.2001.01065.x. [DOI] [PubMed] [Google Scholar]
- Kariyazono H, Ohno T, Ihara K, Igarashi H, Joh-o K, Ishikawa S, Hara T. Rapid detection of the 22q11.2 deletion with quantitative real-rime PCR. Mol Cell Probe. 2001;5:71–73. doi: 10.1006/mcpr.2000.0340. [DOI] [PubMed] [Google Scholar]
- Kim L, Kimmel AR. GSK3, a master switch regulating cell-fate specification and tumorigenesis. Curr Opin Genet Dev. 2000;10:508–514. doi: 10.1016/s0959-437x(00)00120-9. [DOI] [PubMed] [Google Scholar]
- Krebbers E, Seurinck J, Herdies L, Cashmore AR, Timko MP. Four genes in two diverged subfamilies encode the ribulose-1,5-bisphosphate carboxylase small subunit polypeptides of Arabidopsis thaliana. Plant Mol Biol. 1988;11:745–759. doi: 10.1007/BF00019515. [DOI] [PubMed] [Google Scholar]
- Lammers PJ, Jun J, Abubaker J, Arreola R, Gopalan A, Bago B, Hernandez-Sebastia C, Allen JW, Douds DD, Pfeffer PE et al. The glyoxylate cycle in an arbuscular mycorrhizal fungus. Carbon flux and gene expression. Plant Physiol. 2001;127:1287–1298. [PMC free article] [PubMed] [Google Scholar]
- Leprince A, Jouannic S, Hamal A, Kreis M, Henry Y. Molecular characterization of plant cDNAs BnMAP4Kalpha1 and BnMAP4Kalpha2belonging to the GCK/SPS1 subfamily of MAP kinase kinase kinase kinase. Biochim Biophys Acta. 1999;1444:1–13. doi: 10.1016/s0167-4781(98)00246-2. [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science. 2002;295:1299–1301. doi: 10.1126/science.1065769. [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH, Vafeados D, Chory J. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 2001;127:14–22. doi: 10.1104/pp.127.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth AM, Kvarnheden A, von Arnold S. Isolation of a PSTAIRE CDC2 cDNA from Pinus contortaand its expression during adventitious root development. Plant Physiol Biochem. 2001;39:107–114. [Google Scholar]
- Marcote MJ, Carbonell J. Transient expression of a pea MAP kinase gene induced by gibberellic acid and 6-benzyladenine in unpollinated pea ovaries. Plant Mol Biol. 2000;44:177–186. doi: 10.1023/a:1006434330381. [DOI] [PubMed] [Google Scholar]
- Martinek S, Inonog S, Manoukian AS, Young MW. A role for the segment polarity gene shaggy/GSK-3 in the Drosophilacircadian clock. Cell. 2001;105:769–779. doi: 10.1016/s0092-8674(01)00383-x. [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Irie K, Hirayama T, Hayashida N, Yamaguchi-Shinozaki K, Matsumoto K, Shinozaki K. A gene encoding a mitogen-activated protein kinase kinase kinase is induced simultaneously with genes for a mitogen-activated protein kinase and an S6 ribosomal protein kinase by touch cold and water stress in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1996;93:765–769. doi: 10.1073/pnas.93.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli G, Nagy F, Fraley RT, Rogers SG, Chua NH. A short conserved sequence is involved in the light-inducibility of a gene encoding ribulose 1,5-bisphosphate carboxylase small subunit of pea. Nature. 1985;315:200–204. [Google Scholar]
- Moreno J, Spreitzer RJ. C172S substitution in the chloroplast-encoded large subunit affects stability and stress-induced turnover of ribulose-15-bisphosphate carboxylase/oxygenase. J Biol Chem. 1999;274:26789–26793. doi: 10.1074/jbc.274.38.26789. [DOI] [PubMed] [Google Scholar]
- Morris PC, Guerrier D, Leung J, Giraudat J. Cloning and characterization of MEK1, an Arabidopsisgene encoding a homologue of MAP kinase kinase. Plant Mol Biol. 1997;35:1057–1064. doi: 10.1023/a:1005963222768. [DOI] [PubMed] [Google Scholar]
- Murillo I, Jaeck E, Cordero MJ, San Segundo B. Transcriptional activation of a maize calcium-dependent protein kinase gene in response to fungal elicitors and infection. Plant Mol Biol. 2001;45:145–158. doi: 10.1023/a:1006430707075. [DOI] [PubMed] [Google Scholar]
- Oreña SJ, Torchia AJ, Garofalo RS. Inhibition of glycogen-synthase kinase 3 stimulates glycogen synthase and glucose transport by distinct mechanisms in 3T3–L1 adipocytes. J Biol Chem. 2000;275:15765–15772. doi: 10.1074/jbc.M910002199. [DOI] [PubMed] [Google Scholar]
- Pay A, Jonak C, Bögre L, Meskiene I, Mairinger T, Szalay A, Heberle-Bors A, Hirt E. The MSK family of alfalfa protein kinase genes encodes homologues of shaggy/glycogen synthase kinase-3 and shows differential expression patterns in plant organs and development. Plant J. 1993;3:847–856. doi: 10.1111/j.1365-313x.1993.00847.x. [DOI] [PubMed] [Google Scholar]
- Perez-Perez JM, Ponce MR, Micol JL. The UCU1 Arabidopsisgene encodes a SHAGGY/GSK3-like kinase required for cell expansion along the proximodistal axis. Dev Biol. 2002;242:161–173. doi: 10.1006/dbio.2001.0543. [DOI] [PubMed] [Google Scholar]
- Piao HL, Lim JH, Kim SJ, Cheong GW, Hwang I. Constitutive over-expression of AtGSK1 induces NaCl stress responses in the absence of NaCl stress and results in enhanced NaCl tolerance in Arabidopsis. Plant J. 2001;27:305–314. doi: 10.1046/j.1365-313x.2001.01099.x. [DOI] [PubMed] [Google Scholar]
- Piao HL, Pih KT, Lim JH, Kang SG, Jin JB, Kim SH, Hwang I. An ArabidopsisGSK3/shaggy-like gene that complements yeast salt stress-sensitive mutants is induced by NaCl and abscisic acid. Plant Physiol. 1999;119:1527–1534. doi: 10.1104/pp.119.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raitt DC, Posas F, Saito H. Yeast Cdc42 GTPase and Ste20 PAK-like kinase regulate Sho1-dependent activation of the Hog1 MAPK pathway. EMBO J. 2000;19:4623–4631. doi: 10.1093/emboj/19.17.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reintanz B, Szyroki A, Ivashikina N, Ache P, Godde M, Becker D, Palme K, Hedrich R. AtKC1, a silent Arabidopsispotassium channel alpha-subunit modulates root hair K+ influx. Proc Natl Acad Sci USA. 2002;99:4079–4084. doi: 10.1073/pnas.052677799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruel L, Stambolic V, Ali A, Manoukian AS, Woodgett JR. Regulation of the protein kinase activity of Shaggy (Zeste-white3) by components of the wingless pathway in Drosophilacells and embryos. J Biol Chem. 1999;274:21790–21796. doi: 10.1074/jbc.274.31.21790. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei N. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sakai H, Medrano LJ, Meyerowitz EM. Role of SUPERMAN in maintaining Arabidopsisfloral whorl boundaries. Nature. 1995;378:199–203. doi: 10.1038/378199a0. [DOI] [PubMed] [Google Scholar]
- Shin HJ, Lee DE, Shin DH, Kim KU, Kim HY, Ohashi Y, Han O, Baik MG, Back K. Molecular cloning and cultivar specific expression of MAP kinases from Capsicum annuum. Mol Cell. 2001;11:48–54. [PubMed] [Google Scholar]
- Siegfried E, Chou TB, Perrimon N. wingless signaling acts through zeste-white 3 the Drosophilahomolog of glycogen synthase kinase-3 to regulate engrailed and establish cell fate. Cell. 1992;71:1167–1179. doi: 10.1016/s0092-8674(05)80065-0. [DOI] [PubMed] [Google Scholar]
- Sivakumar P, Sharmila P, Saradhi PP. Proline alleviates salt-stress-induced enhancement in ribulose-15-bisphosphate oxygenase activity. Biochem Biophys Res Commun. 2000;279:512–515. doi: 10.1006/bbrc.2000.4005. [DOI] [PubMed] [Google Scholar]
- Tavares R, Aubourg S, Lecharny A, Kreis M. Organization and structural evolution of four multigene families in Arabidopsis thaliana: AtLCAD AtLGT AtMYST and AtHD-GL2. Plant Mol Biol. 2000;42:703–717. doi: 10.1023/a:1006368316413. [DOI] [PubMed] [Google Scholar]
- Tichtinsky G, Tavares R, Takvorian A, Schwebel-Dugue N, Twell D, Kreis M. An evolutionary conserved group of plant GSK-3/shaggy-like protein kinase genes preferentially expressed in developing pollen. Biochim Biophys Acta. 1998;1442:261–273. doi: 10.1016/s0167-4781(98)00187-0. [DOI] [PubMed] [Google Scholar]
- Wang QM, Fiol CJ, Depaoli-Roach AA, Roach PJ. Glycogen synthase kinase-3beta is a dual specificity kinase differentially regulated by tyrosine and serine/threonine phosphorylation. J Biol Chem. 1994;269:14566–14574. [PubMed] [Google Scholar]
- Webster MT, Rozycka M, Sara E, Davis E, Smalley M, Young N, Dale TC, Wooster R. Sequence variants of the axin gene in breast, colon and other cancers: an analysis of mutations that interfere with GSK3 binding. Gene Chrom Can. 2001;28:443–453. [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thalianaand analysis of its promoter in transgenic plants. Mol Gen Genet. 1993;236:331–340. doi: 10.1007/BF00277130. [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Nishitani K. A comprehensive expression analysis of all the members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell-wall construction in specific organs of Arabidopsis. Plant Cell Physiol. 2001;42:1025–1033. doi: 10.1093/pcp/pce154. [DOI] [PubMed] [Google Scholar]
- Zumbrunn J, Kinoshita K, Hyman AA, Näthke IS. Binding of the adenomatous polyposis coli protein to microtubules increases microtubule stability and is regulated by GSK3 beta phosphorylation. Curr Biol. 2001;11:44–49. doi: 10.1016/s0960-9822(01)00002-1. [DOI] [PubMed] [Google Scholar]