Abstract
A DNA cassette containing an Arabidopsis C repeat/dehydration-responsive element binding factor 1 (CBF1) cDNA and a nos terminator, driven by a cauliflower mosaic virus 35S promoter, was transformed into the tomato (Lycopersicon esculentum) genome. These transgenic tomato plants were more resistant to water deficit stress than the wild-type plants. The transgenic plants exhibited growth retardation by showing dwarf phenotype, and the fruit and seed numbers and fresh weight of the transgenic tomato plants were apparently less than those of the wild-type plants. Exogenous gibberellic acid treatment reversed the growth retardation and enhanced growth of transgenic tomato plants, but did not affect the level of water deficit resistance. The stomata of the transgenic CBF1 tomato plants closed more rapidly than the wild type after water deficit treatment with or without gibberellic acid pretreatment. The transgenic tomato plants contained higher levels of Pro than those of the wild-type plants under normal or water deficit conditions. Subtractive hybridization was used to isolate the responsive genes to heterologous CBF1 in transgenic tomato plants and the CAT1 (CATALASE1) was characterized. Catalase activity increased, and hydrogen peroxide concentration decreased in transgenic tomato plants compared with the wild-type plants with or without water deficit stress. These results indicated that the heterologous Arabidopsis CBF1 can confer water deficit resistance in transgenic tomato plants.
Many environmental stresses, such as heat, salinity, low temperature, and drought, and developmental processes, such as seed maturation, cause water deficit in plants (Ingram and Bartels, 1996). To understand water deficit stress at the molecular level, many genes have been isolated, such as rd (responsive to dehydration), erd (early responsive to dehydration), and Lea (late embryogenesis abundant; Shinozaki and Yamaguchi-Shinozaki, 2000). The accumulation of LEA protein occurs during seed maturation, desiccation, and increases in vegetative tissue when plants are exposed to water deficit (Ingram and Bartels, 1996). Overexpression of a barley (Hordeum vulgare) group 3 LEA protein gene, HVA1, enhances tolerance of water deficit and salt stress in transgenic rice (Oryza sativa; Xu et al., 1996). Arabidopsis RD29A (COR78) responds to water deficit and low-temperature stresses (Horvath et al., 1993; Yamaguchi-Shinozaki and Shinozaki, 1993). Study of the promoter RD29A has lead to the characterization of a 9-bp element, TACCGACAT, referred to as dehydration-responsive element (DRE), that is also found in the promoter regions of many water deficit and cold responsive genes, such as RD17, ERD10, KIN1, COR15a, and COR6.6 (Yamaguchi-Shinozaki and Shinozaki, 1994; Wang et al., 1995; Thomashow, 1999). The DRE element contains a 5-bp core sequence of CCGAC, also known as C repeat (CRT), that plays an important role in regulating gene expression in response to low temperature, water deficit, and high salinity (Baker et al., 1994; Yamaguchi-Shinozaki and Shinozaki, 1994). Proteins that bind to the DRE/CRT element and mediate transcription were isolated by the yeast (Saccharomyces cerevisiae) one-hybrid system and named DRE-binding proteins (DREBs)/CRT-binding factors (CBFs; Stockinger et al., 1997; Liu et al., 1998). DREBs/CBFs are encoded by multigene families. Among them, the DREB1A and DREB2A respond to low temperature and water deficit stresses, respectively (Liu et al., 1998).
CBF1 (DREB1B), a homolog of DREB1A, is a transcriptional activator that binds to the CRT/DRE element, in the promoter region of cold-regulated (COR) genes that respond to both low temperature and water deficit (Stockinger et al., 1997; Gilmour et al., 1998). Overexpression of the CBF1 gene in Arabidopsis plants induces expression of COR genes and increases freezing tolerance in the absence of cold acclimation (Jaglo-Ottosen et al., 1998), suggestive of the role of a master switch of the CBF regulation (Thomashow, 2001). Besides its effect in Arabidopsis, overexpression of CBF1 in canola oilseed rape (Brassica napus) also activates COR homologous genes and enhances freezing tolerance, indicating that the function of CBF1 may be highly conserved in plants (Jaglo et al., 2001). Once ice crystals form in the extracellular spaces of plant cells, water moves out of the cell resulting in water deficit. Therefore, the mechanisms of freezing and water deficit tolerance may be similar to each other (Thomashow, 2001). The freezing, salt, and drought tolerance capabilities of transgenic Arabidopsis have also been achieved by the expression of DREB1A (CBF3) cDNA, driven by a 35S promoter or stress-inducible RD29A promoter (Liu et al., 1998; Kasuga et al., 1999). Therefore, overexpression of CBF1 might also improve water deficit tolerance in Arabidopsis plants.
The existence of a CBF1-like expressed sequence tag (EST) in tomato (Lycopersicon esculentum; Jaglo et al., 2001) suggests that a pathways may exist in tomato that is similar to the Arabidopsis signal transduction. The objective of this experiment was to determine whether overexpression of CBF1 in tomato enhanced water deficit tolerance, as is the case in Arabidopsis expressing DREB1A (CBF3). In this study, we present evidence that the transgenic tomato plants expressing CBF1 are more resistant to water deficit than the wild-type plants.
RESULTS
Overexpression of Heterologous Arabidopsis CBF1 in Transgenic Tomato Plants
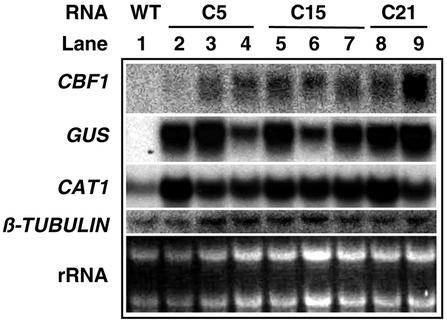
A DNA cassette consisting of an Arabidopsis CBF1 cDNA driven by a cauliflower mosaic virus 35S promoter and a nos terminator was ligated into pCAMBIA2301, which contains β-glucuronidase (GUS) and NPTII reporter genes, to form pJLM1. This plasmid was transferred into the tomato genome by Agrobacterium tumefaciens-mediated transformation. After kanamycin selection, the putative transgenic tomato plants were assayed by GUS staining and Southern-blot analyses to identify the transgenic plants (data not shown). We obtained 22 unique transgenic tomato lines. Northern-blot analysis was performed to reveal the mRNA levels in transgenic T1 plants, three independent T1 lines from C5 (Fig. 1, lanes 2–4), three independent T1 lines from C15 (Fig. 1, lanes 5–7), and two independent T1 lines from C21 (Fig. 1, lanes 8 and 9). The heterologous CBF1 and GUS transcripts accumulated only in transgenic T1 plants. Interestingly, one transgenic T1 plant did not show high expression of Arabidopsis CBF1, but GUS transcripts were detected (Fig. 1, lane 2). This phenomenon is probably due to segregation of T1 seeds. Transgenic T1 plants expressing heterologous CBF1 were evaluated for resistance to water deficit stress. As shown in Figure 1, levels of mRNA of β-TUBULIN and rRNA were used as internal control.
Figure 1.
Northern-blot analysis of transgenic tomato plants. Total RNA (10 μg) was extracted from the wild-type (WT; lane 1) and transgenic T1 plants overexpressing CBF1 (lane 2–9). Probes used were 32P-labeled Arabidopsis CBF1 cDNA, the GUS reporter gene from pCAMBIA2301, tomato CAT1, and β-TUBULIN.
Transgenic Tomato Plants Were More Resistant to Water Deficit Than the Wild-Type Plants
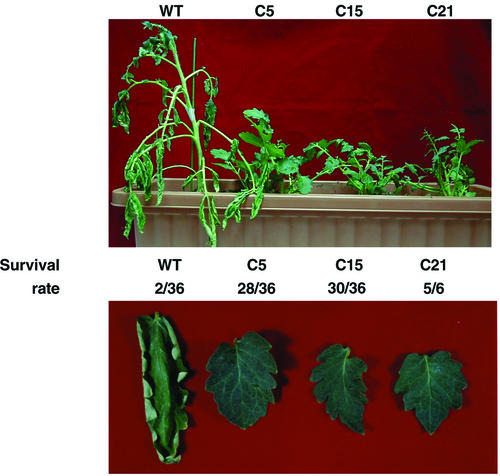
To evaluate the capacity for water deficit resistance, transgenic tomatoes and wild-type plants grown in the same pot with peat moss were not watered for 21 d. It was observed that the leaves of wild-type plant became wilted and curled, whereas the transgenics were not (Fig. 2). To examine the survival rates of the wild-type and transgenic plants under conditions of water deficit, the treatment (water deficit) was extended to 4 weeks. The wild-type plants were sick after 28 d without watering, and did not recover during the 7-d period after rewatering. Compared with the survival rate of wild-type plants, transgenic tomatoes were apparently more resistant to water deficit after 4 weeks of water deprivation (Fig. 2). Less than 6% of the wild-type plants survived after 4 weeks of water deficit treatment, whereas 77.8%, 83.3%, and 83.3% of the transgenic tomato lines C5, C15, and C21 survived the treatment. These results suggest that overexpression of CBF1 can significantly improve water deficit resistance in tomato, similar to the results obtained from transgenic Arabidopsis plants overexpressing DREB1A (CBF3; Kasuga et al., 1999).
Figure 2.
Transgenic tomato plants exhibited more resistance to water deficit stress than wild-type plants. Wild-type and three transgenic T1 plants (WT, C5, C15, and C21) were grown at 24°C without watering for 21 d. Leaves of the wild-type plant significantly curled and wilted. For the survival rate test, wild-type (WT) and three T1 transgenic plants (C5, C15, and C21) were grown at 24°C without watering for 28 d. Numbers of plants alive per total number of tested plants are indicated in the middle of the photograph.
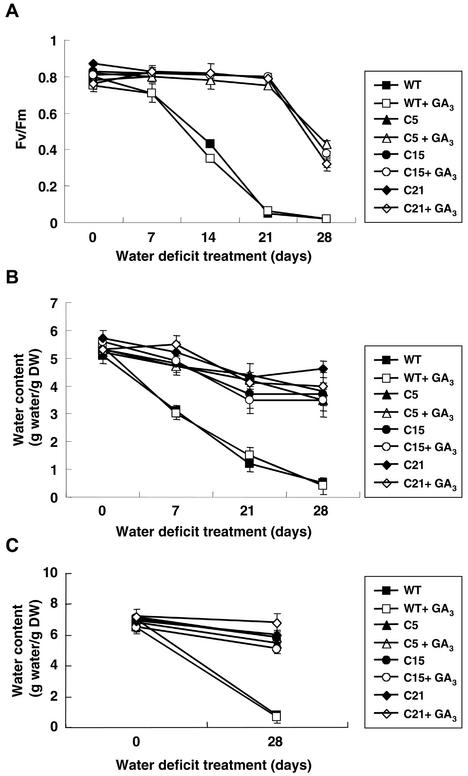
The resistance to water deficit stress was revealed by chlorophyll fluorescence maximum photochemical efficiency of PSII in the dark-adapted state (Fv/Fm) values, measured at d 0, 7, 14, and 28 during water deficit treatment. PS II integrity was significantly more stable in the transgenic plants as compared with the wild-type plants during water deficit stress (Fig. 3A). Fv/Fm decreased in wild-type plants after 21 d without watering and did not recover after rewatering. The transgenic plants, however, maintained an Fv/Fm value of about 50% of their well-watered control even after 28 d without watering (Fig. 3A), and recovered almost completely after rewatering (data not shown).
Figure 3.
Improved resistance of CBF1 transgenic tomato T1 plants to water deficit stress not affected by gibberellic acid (GA3) treatment. GA3-treated and non-treated tomato plants were deprived of water for various times. Fv/Fm values (A) and water content of leaves (B) were measured on d 0, 7, 21, and 28. The water content of the roots (C) of water deficit-stressed transgenic tomato and wild-type plants was measured on d 0 and 28. Each value is the mean ± sd (n = 5 individual plants).
To test the ability of maintaining water in the tissue, water contents of leaves (Fig. 3B) and roots (Fig. 3C) of water deficit-stressed transgenic tomato and wild-type plants were measured at various time points. Water content of transgenic plants remained high during water deficit treatments. In contrast, a marked reduction in water content was observed in the wild-type plants.
Water Deficit Resistance Was Not Affected by Applying GA3 in Transgenic Tomato Plants
All the transgenic tomato plants were shorter than the wild-type plants due to their short internodes. Previously, internode length has been reported to have a positive correlation with GA content (Ross et al., 1989). Recently, we found that the internode length of transgenic plants could be recovered by applying GA3 exogenously (Hsieh et al., 2002). It was of interest to know whether the resistance to water deficit stress of transgenic tomato plants would be affected after GA3 treatment. GA3-treated wild-type and transgenic tomato plants were subjected to the water deficit stress as previously described. Fv/Fm values and the water content of leaves and roots showed that transgenic tomato plants were still more resistant to water deficit stress than GA3-treated wild-type plants (Fig. 3). It is also worth noting that GA3 treatment of both wild-type and transgenic tomato plants seem to have little or no impact on water deficit resistance. There seems to be a correlation of GA content with internode length, but water deficit resistance seems to be independent of it. Hence, the ability to resist water deficit stress was not affected by GA3 in transgenic tomato plants. These results suggest that the CBF1-mediated improvement of water deficit resistance in transgenic tomato plants was probably not due to a morphological change.
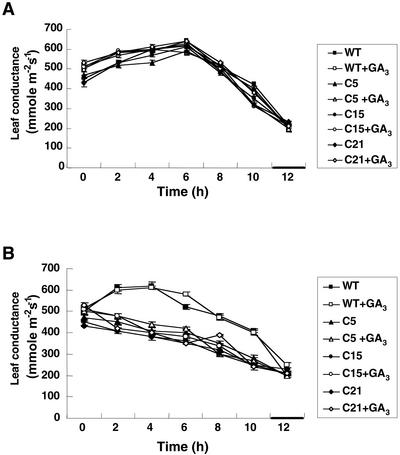
To identify the possible relationship between stomatal movement and water imbalance, changes in leaf conductance were determined and are shown in Figure 4. Leaf conductance in the wild-type plants followed a typical diurnal pattern. In all transgenic tomato and wild-type plants, the stomatal opening increased rapidly after the start of the light period, reached a maximal level at about 6 h, and then decreased (Fig. 4A). The stomata of the transgenic CBF1 tomato plants closed rapidly after water deficit treatment with or without GA3 pretreatment as compared with wild-type plants, which showed a similar pattern during regular watering (Fig. 4, A and B). The effects of CBF1 expression apparently result in retained water, thus negating tissue damage; therefore, the phenotype of transgenic plants appeared normal under water deficit conditions.
Figure 4.
Transgenic tomato T2 plants rapidly close stomata compared with wild-type plants under water deficit condition. Transgenic tomato and wild-type plants were grown at 24°C with regular watering (A) or no watering for 7 d (B). Horizontal bars in the axis of abscissas represent the dark period. The 2-h dark period extended from 10 to 12 h. Each value is the mean ± sd (n = 15 individual plants).
More Pro Was Detected in Transgenic Tomato Plants
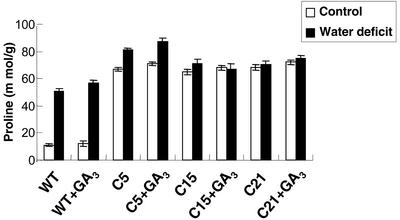
Many plants respond to water deficit by accumulating high concentrations of compatible solutes or osmolytes, such as Pro, mannitol, Fru, Gly betaine, and trehalose (Bajaj et al., 1999; Hoekstra et al., 2001). Because elevated Pro levels occur in transgenic Arabidopsis that overexpressed DREB1A (CBF3; Gilmour et al., 2000), we also measured Pro content in transgenic tomato plants under normal and water deficit conditions. The Pro content in transgenic tomato was higher than in the wild-type plants under both normal and water deficit conditions (Fig. 5). However, there is no further elevation in response to the water stress conditions in transgenic tomato plants, indicating that overexpression of CBF1 under non-stress conditions protects the plants from subsequent stress. GA3 treatment did not affect the Pro content in wild-type and transgenic tomato plants (Fig. 5). These results may suggest that transgenic tomato plants possess an inherent resistance to water deficit conditions, which is much higher than wild-type plants, consistent with results of Fv/Fm value and water content (Fig. 3).
Figure 5.
Transgenic tomato plants contain more Pro than wild-type plants. Wild-type and transgenic T1 plants with or without GA3 treatment were grown at 24°C with daily watering (control) or without watering for 28 d (water deficit). Pro content was measured. Each value is the mean ± sd (n = 5 individual plants).
Exogenous GA3 Treatment Reversed Growth Retardation of Transgenic Tomato Plants without Affecting Water Deficit Resistance
The dwarf phenotype was only observed in transgenic tomato plants that overexpressed heterologous CBF1, not in transgenic tomato plants that only overexpressed the GUS reporter gene (data not shown). Hence, the dwarfism was a result of overexpression CBF1, not the transformation procedure itself. The transgenic tomato plants were not only shorter than wild-type plants, fruit and seed numbers and fresh weights were also less than those of wild-type plants under normal growth conditions (Table I). After exogenous GA3 treatment, fruit, seed number, and fresh weight increased, suggesting GA3 improved the growth of the transgenic tomato plants apparently. However, the seed number of transgenic plants treated with GA3 did not reach the same level as that of the wild-type plants. After water deficit treatment, the fresh weights of transgenic tomato plants were higher than the wild-type plants that had wilted after treatment. No difference in fruit and seed number of transgenic tomato plants without GA3 treatment under normal or water deficit treatment was observed (Table I). Moreover, there was no significant reduction in fruit, seed number, and fresh weights of transgenic plants treated with GA3 under water deficit conditions. However, the production and fresh weight of wild-type plants was severely impaired after water deficit treatment, indicating that wild-type plants were not resistant to water deficit as compared with transgenic tomato plants. Exogenous GA3 treatment showed the same results with non-GA3 treatment, suggesting that the water deficit resistance was not affected in transgenic tomato plants, which can be observed from the fact that there were no change in fruit, seed number, and fresh weight (Table I). Because the phenomenon of chilling treatment was similar to water deficit treatment, we also calculated the fruit, seed number, and fresh weight after chilling treatment. Results of chilling treatment were similar to water deficit treatment, indicating that the transgenic tomato plants were more resistant to chilling and water deficit stress than the wild-type plants.
Table I.
The effects of various treatments on the growth characteristics of transgenic tomato and wild-type plants
| WT | C5 | C15 | C21 | WT+GA3 | C5+GA3 | C15+GA3 | C21+GA3 | |
|---|---|---|---|---|---|---|---|---|
| Control | 22.6 ± 4.3 | 6.0 ± 1.6 | 7.2 ± 1.6 | 1.6 ± 1.1 | 26.6 ± 4.1 | 24.8 ± 3.6 | 22.4 ± 3.2 | 17.4 ± 5.8 |
| 49.7 ± 9.5 | 8.4 ± 2.7 | 6.8 ± 1.3 | 2.4 ± 0.9 | 43.7 ± 9.2 | 25.4 ± 3.0 | 22.6 ± 2.6 | 29.6 ± 14.8 | |
| 131.4 ± 5.1 | 80.6 ± 5.1 | 106.8 ± 9.2 | 85.0 ± 3.9 | 147.4 ± 7.1 | 133.4 ± 13.8 | 138.8 ± 13.6 | 127.6 ± 8.7 | |
| Chilling | 1.1 ± 0.9 | 6.6 ± 0.9 | 8.2 ± 0.8 | 2.4 ± 0.5 | 2.3 ± 0.6 | 21.8 ± 2.4 | 24.4 ± 4.5 | 15.2 ± 2.2 |
| 1.5 ± 1.1 | 9.0 ± 1.5 | 9.0 ± 1.5 | 2.0 ± 0.8 | 1.1 ± 0.8 | 27.0 ± 8.6 | 24.8 ± 7.8 | 10.6 ± 2.8 | |
| 32.5 ± 6.7 | 84.2 ± 5.4 | 120.0 ± 17.3 | 84.4 ± 7.1 | 31.5 ± 5.8 | 123.0 ± 21.4 | 130.2 ± 8.5 | 131.2 ± 6.9 | |
| Water deficit | 6.2 ± 2.2 | 8.8 ± 1.2 | 8.2 ± 0.8 | 2.6 ± 1.8 | 2.2 ± 0.5 | 23.6 ± 2.4 | 21.4 ± 2.1 | 15.6 ± 3.9 |
| 3.5 ± 1.1 | 7.6 ± 2.4 | 9.0 ± 1.5 | 1.8 ± 0.9 | 4.1 ± 1.7 | 37.0 ± 8.2 | 37.7 ± 6.5 | 27.7 ± 13.3 | |
| 39.8 ± 27.1 | 101.2 ± 11.6 | 120.0 ± 17.3 | 88.4 ± 6.1 | 41.2 ± 26.7 | 123.4 ± 15.9 | 129.8 ± 14.4 | 128.8 ± 14.7 |
The order of data shown in each treatment is: fruit no. per plant, seed no. per fruit, and fresh wt (g) per plant. Each value is the mean ± sd (n = 5 individual plants). Chilling treatment is incubated at 0°C for 7 d, then returned to 24°C. Water deficit treatment is without watering for 4 weeks. The measured plants are 3 months old. The stress treatment time is also included in the growth period.
Enhancement of Catalase Activity, and Reduction of Hydrogen Peroxide (H2O2) Concentration in Transgenic Tomato Plants
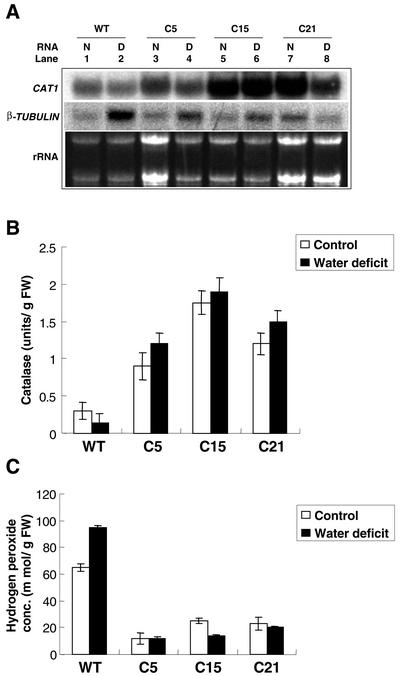
Transgenic tomato plants were analyzed at the molecular level using subtractive hybridization techniques against wild-type plants. Subtractive hybridization experiments were performed to identify any up-regulated genes belonging to the transgenic tomato plant (data not shown). No known COR homologous genes were isolated from the subtractive hybridization experiment, and Arabidopsis COR genes, such as COR47, KIN1, and COR15a, did not cross hybridize with any tomato RNA even in low-stringency hybridization condition (data not shown). Using tomato dehydrin TAS14, which is also responsive to stresses (Godoy et al., 1990, 1994; Parra et al., 1996), as a probe, we failed to detect any mRNA transcripts expressed in any transgenic tomato plants (data not shown). The CAT1 gene, however, was one of the numbers of up-regulated genes we isolated. Northern-blot analysis indicated that transcriptional levels of CAT1 were higher in transgenic tomatoes than in wild-type plants under normal (Figs. 1 and 6A) or water deficit (Fig. 6A) conditions. The catalase and H2O2 concentrations of plants grown under normal conditions with or without watering for 28 d were measured. Catalase activity of transgenic tomato plants was higher than that of wild-type plants under normal or water deficit conditions (Fig. 6B). H2O2 concentrations were lower in transgenic tomato than in wild-type plants under normal or stressed conditions (Fig. 6C). Our results indicate that CAT1 expression and catalase activity increased and H2O2 concentration reduced in transgenic tomato plants.
Figure 6.
Transgenic tomatoes exhibit increased catalase activity but a reduction in H2O2 concentrations under normal and water deficit conditions. Ten micrograms of RNA was extracted from wild-type (WT) and three transgenic plants (C5, C15, and C21) grown under control conditions (N) or without watering for 12 d (D), were used to for northern-blot analysis. Probes used were the 32P-labeled tomato CAT1 gene and β-TUBULIN (A). Plants were grown at 24°C with regular watering (control) or without watering for 28 d (water deficit), catalase activity (B), and H2O2 concentration (C) were measured.
DISCUSSION
CBF genes are considered as “master switches” that activate expression of COR genes, increasing freezing tolerance in transgenic Arabidopsis plants in the absence of cold stimulation (Thomashow et al., 2001). Overexpression of DREB1A (CBF3) not only increases freezing tolerance, but also salt loading and drought tolerance in transgenic Arabidopsis (Kasuga et al., 1999). In this study, we found that overexpression of Arabidopsis CBF1 increases water deficit resistance in transgenic tomato plants. Results of survival rate, Fv/Fm, and water content show that transgenic tomato plants are more resistant to water deficit stress than wild-type plants (Figs. 2 and 3). Results of stomatal movement and Pro content imply that transgenic plants have the ability to cope with water deficit conditions better than wild-type plants. These results suggest that heterologous CBF1 could improve environmental stress resistance in agriculturally important crop plants.
CBF1, however, also severely reduced growth in tomato (Table I). Similar effects were found in Arabidopsis plants overexpressing DREB1A (CBF3; Kasuga et al., 1999), suggesting heterologous CBF1 also affects developmental processes in transgenic tomato plants. The transgenic tomato plants showed a decrease in fruit, seed number, and fresh weight as compared with wild-type plants under normal conditions. This phenomenon could be reversed by GA3 treatment (Table I). However, seed number of transgenic plants treated with GA3 could not be improved as compared with wild-type plants (Table I). Furthermore, there is no obvious difference of fruit, seed production, and fresh weights of transgenic tomato plants between normal or water deficit conditions regardless of GA3 treatment (Table I). These results suggested that overexpression of the CBF1 protein interferes with production in transgenic plants. The increased resistance could have been due to less water evaporation from the dwarf phenotype of the transgenic tomato plants. However, when the growth retardation was reversed by exogenous application of GA3, the water deficit resistance of the transgenic tomato plants was not lost (Fig. 3). These results indicate that the dwarf phenotype may not be a major factor determining resistance to water deficit. As shown in Figure 4, the stomata of the transgenic CBF1 tomato plants closed rapidly after water deficit treatment as compared with wild-type plants. These results indicate that CBF1 might have an effect on the apparent retention of water to avoid damage resultant from water deficit.
Previously, inhibition of growth under non-stressed conditions could be prevented by replacing the constitutive 35S promoter with the RD29A stress-inducible promoter in transgenic Arabidopsis plants expressing DREB1A (CBF3; Kasuga et al., 1999; Smirnoff and Bryant, 1999). However, the stress tolerance of transgenic Arabidopsis plants was not affected by replacing this RD29A stress-inducible promoter. We also conducted experiments replacing the cauliflower mosaic virus 35S promoter, used to drive the CBF1 gene in transgenic tomatoes, with the barley ABRC1 and Arabidopsis COR15A stress-inducible promoter (J. Lee, P.-T. Yang, J.-F. Wu, Y. Charng, T.H.D Ho, and M.-T. Chan, unpublished data). It was found that no plant growth retardation was derived from swapping the promoter. Moreover, water deficit resistance was also not affected in transgenic ARBC1-CBF1 tomato plants.
Overexpression of CBF1 can activate expression of COR genes in Arabidopsis plants (Jaglo-Ottosen et al., 1998) and these induced COR genes may play an important role in freezing tolerance. It was expected that tomato endogenous COR homologs may also exist and be induced by the overexpression of a CBF1 transcriptional factor. Surprisingly, the use of known Arabidopsis COR genes as probes, such as KIN1, COR15a, COR47, and RD29A, did not lead to cross hybridization of any RNA transcripts from transgenic tomato plants, even under low-stringency hybridization condition (data not shown). Moreover, we did not detect any mRNA transcript in transgenic tomato plants using tomato dehydrin TAS14, one of the LEA genes, as a probe (data not shown). Although we have obtained many tomato EST clones and unknown cDNA fragments from subtractive hybridization experiments, we have not obtained any known Arabidopsis COR- or RD-like genes from our subtractive library (T.-H. Hsieh and M.-T. Chan, unpublished data). These results may be due to the low homology between Arabidopsis COR or RD gene probes and tomato endogenous homologs, or unknown tomato COR-like, RD-like, or LEA genes induced by heterologous CBF1. More evidence is needed to confirm or reject this hypothesis.
Antioxidant enzymes, such as glutathione reductase and superoxide dismutase activity, increase in response to water deficit stress (Ingram and Bartels, 1996), and overexpression of antioxidant genes improves tolerance to pathogens, paraquat, and osmotic stresses (e.g. chilling, salinity, and drought; Bray et al., 2000). CAT1 is one of the responsive genes we isolated from subtractive hybridization. Both mRNA level and catalase activity increased in transgenic tomato as compared with wild-type plants (Figs. 1 and 6, A and B). The H2O2 concentration was reduced in transgenic tomatoes as compared with wild-type plants (Fig. 6C). Our results support the hypothesis that activation of antioxidant genes converge the resistance of transgenic tomato plants to water deficit. The up-regulation of CAT1 might be a consequence of the overexpression of CBF1. More evidence is needed to determine if heterologous CBF1 activates these genes directly or indirectly.
Overexpression of Δ1-pyrroline-5-carboxylate synthase (P5CS) in transgenic tobacco plants increases Pro content by 10- to 18-fold as compared with wild-type plants, resulting in better growth under water deficit conditions (Kavi et al., 1995). The mRNA transcripts of P5CS2 and Pro content were highly increased in transgenic Arabidopsis plants expressing CBF3 (Gilmour et al., 2000). Pro concentrations were higher in transgenic tomato plants than in wild-type plants (Fig. 5), similar to the overexpression results of DREB1A (CBF3) in Arabidopsis (Gilmour et al., 2000). This study implies that the tomato P5CS gene(s) may also be induced in transgenic tomato plants. Therefore, we did detect a low-stringency hybridized band in the northern-blot analysis using the tomato EST440219 clone (accession no. BF112629), which is similar to the P5CS gene, as a probe. However, there was no significant difference in expression between wild-type and transgenic tomato plants (data not shown). These results suggest that other P5CS genes may be up-regulated in transgenic CBF1 tomato plants. We had many unknown cDNA fragments and EST clones from subtractive hybridization (data not shown). In future experiments, we will isolate the tomato P5CS genes and other genes responsive to heterologous CBF1 that are important in transgenic tomato plants with water deficit resistance. Characterization of these responsive genes will contribute to an understanding of stress resistance, and help to decipher the stress signal transduction pathways in tomato plants.
It is interesting that the transcriptional activator similar to CBF1 does exist in different plant species (Jaglo et al., 2001), indicating that stress signal transduction pathways may be conserved in various plant species. Overexpression of DREB1A (CBF3) increases tolerance to freezing and water deficit stresses (Kasuga et al., 1999), suggesting that different stress signal transduction pathways might cross talk between each other. Recent findings indicate that there is cross talk between two stress-signaling pathways in Arabidopsis (Shinozaki and Yamaguchi-Shinozaki, 2000). Recently, we also observed that transgenic CBF1 tomato plants have enhanced chilling tolerance as compared with wild-type plants (Hsieh et al., 2002). These results in combination with the results in this study confirm that the enhancement of stress tolerance phenomenon in transgenic tomato plants might be conferred by multiple defense systems. These activated defense systems may also protect transgenic plants from other stress conditions. We believe that a similar approach might be applicable to other important crops, such as rice, maize (Zea mays), wheat (Triticum aestivum), and barley, to improve tolerance against stress conditions. This may be accomplished by transferring several (e.g. three–four) key regulatory genes, rather than a large number of stress-related genes under inducible promoters. Overall, the engineering of stress-tolerant crops by incorporating (a) master switch gene(s) like CBF1 may be an efficient approach to minimize stress damage.
MATERIALS AND METHODS
Plant Materials
Seeds of tomato (Lycopersicon esculentum L. Miller cv CL5915-93D4-1-0-3) were provided by the Asian Vegetable Research and Development Center (Tainan, Taiwan, Republic of China). Seeds were soaked in water at 32°C for 1 h, surface sterilized for 10 min with 1% (v/v) NaOCl, washed twice with sterile water for 5 min, and subsequently germinated on Murashige and Skoog medium under a 16-h photoperiod at 26°C.
DNA Construction and Agrobacterium tumefaciens-Mediated Tomato Transformation
A CBF1 gene was isolated by reverse transcriptase-PCR from 3-week-old Arabidopsis leaves as described previously (Chan and Yu, 1998). The transformation procedure followed was as described previously (Hsieh et al., 2002).
Identification of Transgenic Tomato Plants
Total RNA was isolated using a Triazole (Life Technologies/Gibco-BRL, Cleveland) solution, the DNA- and RNA-blot analyses were performed according to Chan et al. (1994). The GUS DNA, excised from the BamHI-SacI restriction fragment of plasmid pBI221 (CLONTECH Laboratories, Palo Alto, CA), and the CBF1 gene, isolated from pT7Blue-CBF1 (Hsieh et al., 2002), were used as probes. Tomato β-TUBULIN cDNA fragment was isolated by reverse transcriptase-PCR from 3-month-old tomato plant leaves. The 5′ primer (5′-CCCGGGCACACTTGATCCCATTCGT-3′, SmaI site underlined) and the 3′ primer (5′-CCCGGGCATTCTGTCTGGGTACTCT-3′, SmaI site underlined) were chosen to amplify the 539-bp β-TUBULIN partial cDNA fragment. The PCR fragments were cloned into pT7Blue(R) and the DNA sequences were determined by a PRISM 373 automatic DNA sequencing system (ABI, Sunnyvale, CA). CAT1 (accession no. M93719) isolated from subtractive hybridization was also used as a probe. All fragments were labeled with [α-32P]dCTP using the random primer method (Feinberg and Vogelstein, 1983). Tomato seeds produced from transgenic tomato plants were collected and selection procedures were performed as described previously (Hsieh et al., 2002).
Analysis of Transgenic Plants under Water Deficit Conditions
Wild-type and transgenic tomato plants were grown under similar conditions, in pots with peat moss and watered every alternate day. Day temperature was maintained at 26°C ± 2°C and night temperature at 22°C ± 2°C. Relative humidity was maintained at 50% ± 10%. Plants were grown under 16/8 h light (about 120 μmol m−2 s−1). Survival rate of water deficit treatment was defined as healthy plant number divided by total plant number. Pictures were taken to record the phenotypes. The water deficit treatment time was included in the growth period. After 3 months, these plants were harvested, weighed for fresh weight, and the fruit and seed numbers calculated.
For water deficit treatment, wild-type and transgenic T1 plants were grown at 24°C and without water supply for various time periods (0, 7, 14, 21, and 28 d). For GA3 treatment, 3-week-old wild-type and transgenic T1 plants were sprayed with 5 μL L−1 GA3 three times in a week. Three leaves or five roots were detached from each plant and weighed for fresh weight, with sampling and measurements repeated five times. Detached leaves or rootswere then dried at 65°C for 2 d to determine dry weight. The water content was calculated based on the following equation:
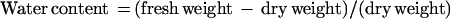 |
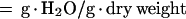 |
Chlorophyll fluorescence values were measured using a pulse-activated modulation fluorimeter (Walz, Effeltrich, Germany) according to the method described by Oberschall et al. (2000).
Leaf conductance measurements were taken from the third and fourth leaves of intact transgenic CBF1 T2 and wild-type plants which were growing under normal (control) and water deficit (7 d) conditions. The results were measured at an interval of a 10 h of light and 2 h of dark. Leaf conductance was measured with an LI-1600 steady-state porometer (LI-COR, Lincoln, NE).
Pro Content, Catalase Activity, and H2O2 Concentration Analyses
Leaves detached from plants were extracted using 3-sulfosalicylic acid, and the supernatant collected after centrifugation. Ninhydrin and acetic acid was added to the supernatant and incubated at 100°C for 60 min. It was snap chilled on ice to terminate the reaction, and then toluene was added and the absorbance at A520 measured.
Wild-type and transgenic T1 plants were grown at 24°C without water for 28 d as described previously (water deficit treatment). The leaf catalase activity was measured according to Pinhero et al. (1997). The H2O2 concentration was analyzed as described by O'Kane et al. (1996).
Subtractive Hybridization
Poly(A+) RNA (0.7 μg) was extracted from leaves of wild-type and transgenic tomato plants that were grown in normal conditions, and used to perform subtractive hybridization according to the CLONTECH PCR select cDNA subtraction kit manual. After PCR amplification, the PCR products were cloned into the pT7Blue(R) vector (Novagen, Madison, WI). DNA sequences were determined by an ABI PRISM 373 automatic DNA sequencing system.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Tuan-Hua David Ho (Washington University, St. Louis), Albert H. Markhart (University of Minnesota, St. Paul), and Ning-Sun Yang and Miss Fang-Fei Yeh (Institute of BioAgricultural Sciences, Academia Sinica, Taipei, Taiwan, Republic of China) for their critical suggestion of this manuscript. We also thank Dr. Virginia Walbot (Stanford University, CA) for providing pJD301 plasmid DNA as our intermediate vector for cloning CBF1. We are grateful to The Institute of Molecular Biology and The Institute of Botany, Academia Sinica; The Asian Vegetable Research and Development Center; and The Department of Agronomy, National Taiwan University, for their technical assistance, and for providing the tissue culture facility, greenhouse, and chemical analysis equipment.
Footnotes
This work was supported by Academia Sinica (grant) and by the National Science Council of the Republic of China (grant no. NSC–90–2311–B–001–071).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.006783.
LITERATURE CITED
- Bajaj S, Targolli J, Liu LF, Ho THD, Wu R. Transgenic approaches to increase dehydration-stress tolerance in plants. Mol Breed. 1999;5:493–503. [Google Scholar]
- Baker SS, Wilhelm KS, Thomashow MF. The 5′-region of Arabidopsis thaliana COR15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol Biol. 1994;24:701–713. doi: 10.1007/BF00029852. [DOI] [PubMed] [Google Scholar]
- Bray EA, Bailey-Serres J, Weretilnyk E. Response to abiotic stresses. In: Buchanan BB, Gruissem W, Jones RL, editors. Biochemistry and Molecular Biology of Plants. Rockville, MD: American Society of Plant Physiologists Press; 2000. pp. 1158–1203. [Google Scholar]
- Chan MT, Chao YC, Yu SM. Novel gene expression for plant cells based on induction of α-amylase promoter by carbohydrate starvation. J Biol Chem. 1994;269:17635–17641. [PubMed] [Google Scholar]
- Chan MT, Yu SM. The 3′ untranslated region of a rice α-amylase gene mediated sugar-dependent abundance of mRNA. Plant J. 1998;15:685–695. doi: 10.1046/j.1365-313x.1998.00244.x. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 2000;124:1854–1865. doi: 10.1104/pp.124.4.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 1998;16:433–442. doi: 10.1046/j.1365-313x.1998.00310.x. [DOI] [PubMed] [Google Scholar]
- Godoy JA, Lunar R, Torres-Schumann S, Moreno J, Rodrigo RM, Pintor-Toro JA. Expression, tissue distribution and subcellular localization of dehydrin TAS14 in salt-stressed tomato plants. Plant Mol Biol. 1994;26:1921–1934. doi: 10.1007/BF00019503. [DOI] [PubMed] [Google Scholar]
- Godoy JA, Pardo JM, Pintor-Toro JA. A tomato cDNA inducible by salt stress and abscisic acid: nucleotide sequence and expression pattern. Plant Mol Biol. 1990;15:695–705. doi: 10.1007/BF00016120. [DOI] [PubMed] [Google Scholar]
- Hoekstra FA, Golovina EA, Buitink J. Mechanisms of plant desiccation tolerance. Trends Plant Sci. 2001;6:1360–1385. doi: 10.1016/s1360-1385(01)02052-0. [DOI] [PubMed] [Google Scholar]
- Horvath DP, McLarney BK, Thomashow MF. Regulation of Arabidopsis thaliana L. (Heyn) COR78 in response to cold. Plant Physiol. 1993;103:1047–1053. doi: 10.1104/pp.103.4.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TH, Lee JT, Yang PT, Chiu LH, Charng Yy, Wang YC, Chan MT. Heterologous expression of the Arabidopsis CBF1 gene confers elevated tolerance to chilling and oxidative stresses in transgenic tomato. Plant Physiol. 2002;129:1086–1094. doi: 10.1104/pp.003442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram J, Bartels D. The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:377–403. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- Jaglo KR, Kleff S, Amundsen KL, Zhang X, Haake V, Zang JZ, Deits T, Thomashow MF. Components of the Arabidopsis C-repeat/Dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol. 2001;127:910–917. [PMC free article] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Tomashow MF. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science. 1998;280:104–106. doi: 10.1126/science.280.5360.104. [DOI] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol. 1999;17:287–291. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- Kavi KPB, Hong Z, Miao G-H, Hu C-A, Verma DPS. Overexpression of Δ1-pyrroline-5-carboxylate synthase increases proline production and confers osmotolerance in transgenic plants. Plant Physiol. 1995;108:1387–1394. doi: 10.1104/pp.108.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transcription pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obershall A, Deak M, Torok K, Sass L, Vass I, Kovacs I, Feher A, Dudits D, Horvath GV. A novel aldose/aldehyde reductase protects transgenic plants against lipid peroxidation under chemical and drought stresses. Plant J. 2000;24:437–446. doi: 10.1046/j.1365-313x.2000.00885.x. [DOI] [PubMed] [Google Scholar]
- O'Kane D, Gill V, Boyd P, Burdon RH. Chilling, oxidative stress and antioxidant responses in Arabidopsis thaliana callus. Planta. 1996;198:366–370. doi: 10.1007/BF00620053. [DOI] [PubMed] [Google Scholar]
- Parra MM, Del Pozo O, Luna R, Godoy JA, Pintor-Toro JA. Structure of the dehydrin tas14 gene of tomato and its developmental and environmental regulation in transgenic tobacco. Plant Mol Biol. 1996;32:453–460. doi: 10.1007/BF00019097. [DOI] [PubMed] [Google Scholar]
- Pinhero RG, Rao MV, Paliyath G, Murr DP, Fletcher RA. Changes in activities of antioxidant enzymes and their relationship to genetic and paclobutrazol-induced chilling tolerance of maize seedlings. Plant Physiol. 1997;114:695–704. doi: 10.1104/pp.114.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JJ, Reid JB, Gaskin P, Macmillan J. Internode length in Pisum. Estimation of GA1 level in genotypes Le, le and led. Physiol Plant. 1989;76:173–176. [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Molecular responses to dehydration and cold: differences and cross-talk between two stress signal pathways. Curr Opin Plant Biol. 2000;3:217–223. [PubMed] [Google Scholar]
- Smirnoff N, Bryant JA. DREB takes the stress out of growing up. Nat Biotechnol. 1999;17:229–230. doi: 10.1038/6968. [DOI] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to cold and water deficit. Proc Natl Acad Sci USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- Thomashow MF. So what's new in the field of plant cold acclimation? Lots! Plant Physiol. 2001;125:89–93. doi: 10.1104/pp.125.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF, Gilmour SJ, Stockinger EJ, Jaglo-Ottosen KR, Zarka DG. Role of the Arabidopsis CBF transcriptional activators in cold acclimation. Physiol Plant. 2001;112:171–175. [Google Scholar]
- Wang H, Datla R, Georges F, Loewen M, Cuter AJ. Promoters from kin1 and cor6.6, two homologous Arabidopsis thaliana genes: transcriptional regulation and gene expression induced by low temperature, ABA osomoticum and dehydration. Plant Mol Biol. 1995;28:605–617. doi: 10.1007/BF00021187. [DOI] [PubMed] [Google Scholar]
- Xu D, Duan X, Wang B, Hong B, Ho THD, Wu R. Expression of a late embryogenesis abundant protein gene, HVA1, from barely confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiol. 1996;110:249–257. doi: 10.1104/pp.110.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Characterization of the expression of a desiccation -responsive rd29A gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol Gen Genet. 1993;236:331–340. doi: 10.1007/BF00277130. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.