Abstract
Human genetic variation is an important determinant of the outcome of infection with Mycobacterium tuberculosis. We have conducted a two-stage genome-wide linkage study to search for regions of the human genome containing tuberculosis-susceptibility genes. This approach uses sibpair families that contain two full siblings who have both been affected by clinical tuberculosis. For any chromosomal region containing a major tuberculosis-susceptibility gene, affected sibpairs inherit the same parental alleles more often than expected by chance. In the first round of the screen, 299 highly informative genetic markers, spanning the entire human genome, were typed in 92 sibpairs from The Gambia and South Africa. Seven chromosomal regions that showed provisional evidence of coinheritance with clinical tuberculosis were identified. To identify whether any of these regions contained a potential tuberculosis-susceptibility gene, 22 markers from these regions were genotyped in a second set of 81 sibpairs from the same countries. Markers on chromosomes 15q and Xq showed suggestive evidence of linkage (lod = 2.00 and 1.77, respectively) to tuberculosis. The potential identification of susceptibility loci on both chromosomes 15q and Xq was supported by an independent analysis designated common ancestry using microsatellite mapping. These results indicate that genome-wide linkage analysis can contribute to the mapping and identification of major genes for multifactorial infectious diseases of humans. An X chromosome susceptibility gene may contribute to the excess of males with tuberculosis observed in many different populations.
One-third of the world's population is estimated to be infected with Mycobacterium tuberculosis (1). Half of those exposed to the organism become infected (2), but only 1 in 10 persons who become infected will ever develop clinical disease (3). In only a minority of cases is there an obvious identifiable risk factor such as diabetes, advanced age, alcohol abuse, HIV infection, or corticosteroid usage. In the remainder, a complex interaction of genetic and environmental factors causes the development of clinical tuberculosis. There is substantial evidence from studies on racial variation in susceptibility to tuberculosis (4, 5) and twin studies (6, 7) that host genetic factors are important in determining the susceptibility to infection with M. tuberculosis and the subsequent development of clinical disease.‡‡ In a large case-control study in Gambians, including over 800 subjects, we have shown that genetic variants of the natural resistance-associated macrophage protein (NRAMP1) and vitamin D receptor (VDR) genes are associated with smear-positive pulmonary tuberculosis (8, 9). However, together, these can only account for a small proportion of the overall genetic component suggested by twin studies.
It is now possible to screen the entire human genome for genes exerting a major effect on susceptibility to multifactorial diseases, and several complete screens of non-infectious disease have now been completed (10–19). These studies represent a systematic approach to finding genes that exert a high locus-specific sibling recurrence risk (λs). For example, simulation analyses have shown that there is a 99.9% chance of obtaining a maximum lod score >2.3 for a locus with λs = 2.5 by using markers spaced at approximately 11-centimorgan intervals and 96 affected sibpair families (10). However, whether such an approach would be valuable in common infectious diseases, most of which may be highly polygenic, is unknown.
Segregation analysis of families with multiple cases of tuberculosis in Brazil has suggested that there are one or two major genes determining individual tuberculosis-susceptibility (20), at least in such selected pedigrees. We have conducted a two stage genome-wide search on 136 families, including 173 sibpairs, affected by clinical tuberculosis from The Gambia and South Africa to attempt to localize any genes exerting a major effect on risk of this disease. No marker was strongly linked to disease, indicating that tuberculosis-susceptibility is not a monogenic disease in Africans. Two regions on chromosomes 15q and Xq showed suggestive evidence of linkage to tuberculosis. The presence of susceptibility genes on these chromosomes was supported by an independent analysis that we term common ancestry using microsatellite (CAM) mapping. Collectively, these data and analysis support the localization of novel tuberculosis susceptibility genes in both of these chromosomal regions.
Materials and Methods
Design of Family Studies.
Families with two or more siblings affected by tuberculosis were identified from the records of all nine tuberculosis clinics located throughout The Gambia, from four clinics around Tygerberg Hospital in the Western Cape, South Africa and from Hlabisa Hospital, KwaZulu-Natal. Microsatellites [genetic markers consisting of a (CA)n sequence with marked length variation] were used to screen the entire human genome in a two stage process. In the first round, 288 microsatellite markers were typed in 67 Gambian families including 73 fully independent sibpairs and 16 KwaZulu-Natal families including 19 independent sibpairs. A further 11 markers on chromosomes 15q and Xq were then typed to increase the marker density in these two regions. The second round of the screen using 22 microsatellite markers was conducted on 12 Gambian sibpair families and 41 families from the Western Cape including 69 independent sibpairs (table 1). Both parents of the affected siblings were typed if available. When parental samples were not available, unaffected siblings were recruited to the study to reconstruct parental genotypes. In the first round screen, 33 Gambian families included 2 parents, 20 families only one parent, and 14 families no parents and, in KwaZulu-Natal, 9 families included both parents and 7 families one parent. In the second screen, 6 Gambian families had both parents, 3 families 1 parent, and 3 families no parents available and, in the Western Cape, 12 families had two parents, 17 families 1 parent, and 12 families no parents.
Table 1.
Number of families and sibpairs in two-stage genome screen
| Number of families with: | First screen
|
Second screen
|
||
|---|---|---|---|---|
| The Gambia | South Africa | The Gambia | South Africa | |
| Two affected siblings | 61 | 13 | 12 | 21 |
| Three affected siblings | 6 | 3 | 0 | 14 |
| Four affected siblings | 0 | 0 | 0 | 5 |
| Six affected siblings | 0 | 0 | 0 | 1 |
| Total families | 67 | 16 | 12 | 41 |
| Total sibpairs | 73 | 19 | 12 | 69 |
The number of fully independent sibpairs in each family is equal to the number of affected siblings minus one.
Patient Characteristics.
The Gambia has seven principal ethnic groups that have been shown to be closely genetically related (21). The patients recruited from KwaZulu-Natal were all Zulus, and the patients from the Western Cape belonged to the racial group previously designated Cape Coloureds.
Of 164 affected siblings from The Gambia, 102 had smear-positive pulmonary disease. The other patients were diagnosed by experienced clinicians and were only included in the study if they fulfilled all three of the following criteria: (i) significant symptoms consistent with a diagnosis of tuberculosis (at least two of the following: loss of >10% body weight, chronic cough every day for >4 weeks, prolonged fever or night sweats for >4 weeks, and significant cervical lymphadenopathy); (ii) a known smear-positive tuberculosis contact; and (iii) a chest x-ray consistent with active disease (alveolar infiltrates with or without cavitation) unless extrapulmonary disease was diagnosed. Culture facilities for acid fast bacilli were not available at the time of the study. Fifty-one patients were under 16 years old at the time of diagnosis. The Gambia has a much lower incidence of HIV infection than is found in the majority of sub-Saharan Africa, and less than 2% of the general population are HIV positive. Patients who gave their consent (>95%) were screened for HIV antibodies, and only three affected siblings tested positive.
All affected siblings from the Western Cape had positive sputum smears and/or culture, and all were tested for HIV antibodies. No patients were HIV-positive, a reflection of the low prevalence of HIV in this community (22). Two affected siblings were less than 16 years old. Of 35 affected siblings from KwaZulu-Natal, 15 were sputum smear-positive, and the remainder were diagnosed by experienced clinicians, using the criteria described above. Seven patients were HIV-positive, and thirteen siblings were less than 16 years old.
Genotyping.
Two hundred and ninety-nine highly polymorphic microsatellite markers (mean heterozygosity 0.83 in these populations) covering all 22 autosomes and the X chromosome were typed in the first round screen. The average interval between adjacent markers was <11 centimorgans, and <0.5% of the genome was >20 centimorgans from the nearest marker, according to the Genethon linkage map (23). Microsatellite genotyping was performed by using fluorescence-based semiautomated technology as described by Reed et al. (24). PCR conditions were optimized for each set of fluorescence-labeled primers by using a range of annealing temperatures (50–61°C) and magnesium concentrations (1.0–3.0 mM) on MJ Research machines. The 15-μl total reaction mix contained 50 ng of DNA, 40 ng of each primer, 2.5 mmol of potassium chloride buffer, 100 μM dNTPs, 1.0–3.0 mM MgCl2, and 0.2 units of Taq polymerase. Pooled amplified PCR products were electrophoresed through 6% acrylamide gels on 373A DNA sequencers (Perkin–Elmer). DNA fragment sizing was performed by using the genescan 672 and genotyper software programs (Perkin–Elmer).
Statistical Analysis.
The statistical methodology underlying genome-wide linkage analysis is discussed in ref. 25. For each individual marker, affected sibpair analysis was performed with the sibpair analysis program (16). sibpair analysis provides a likelihood-based test statistic for linkage that is equivalent to the lod score calculated assuming recessive disease and phase unknown matings. When parental genotypes are not available, the likelihood is a sum of terms corresponding to each of the possible parental genotype combinations using genotype frequencies calculated assuming Hardy-Weinberg equilibrium and including information from affected and unaffected offspring. Segregation of alleles to unaffected offspring is assumed to be Mendelian (16). Because it is impossible to be sure that unaffected siblings are resistant to tuberculosis, affected-unaffected sibling pairs were not used in linkage analysis.
Maximum-likelihood multipoint mapping was performed by using mapmaker/sibs (26). This is a multipoint method based on the single-point affected pedigree member lod score method of Risch (27, 28). Only fully independent affected sibpairs were utilized in the analysis, and maximum likelihood lod scores were calculated by using the possible triangle constraints (29). The program uses genotype information for each affected sibpair, with information from parents and unaffected sibs where available, to infer the identity-by-descent distribution for each point along a chromosome (26). Multipoint mapping is more powerful than single marker analysis as more families are informative in the linkage analysis. For the X chromosome, lod scores are calculated separately for brother-brother, brother-sister, and sister-sister pairs and then combined. Any marker that showed a lod score >1 on sibpair analysis or any region that showed a lod >1 on mapmaker/sibs on the first round of the screen was followed up in the second round. Exclusion maps (regions with a very low probability of containing a tuberculosis-susceptibility gene) were produced for a putative disease-susceptibility gene with λs = 2.0, assuming no dominance variance using mapmaker/sibs.
The technique of common ancestry using microsatellite (CAM) mapping was also used to analyze the data. This method is described in further detail elsewhere (W.A., R.B., G. Cooper, and A.V.S.H., unpublished work). Whereas traditional linkage analysis treats both alleles at a locus independently, CAM mapping focuses on the inherited genotype. In essence, CAM mapping is an extended form of homozygosity mapping that looks for an association between disease incidence and regions of high homozygosity/heterozygosity. However, whereas homozygosity mapping treats each marker independently, CAM mapping looks at patterns of relatedness among all markers on the same chromosome. The principle of CAM mapping is as follows. In any population, the expected time to most recent common ancestor (“time depth”) will be the same for every gene. However, recombination, independent assortment of chromosomes, and genetic drift together ensure that time depth varies greatly among genes about this expectation. Similarly, the average time depth for entire chromosomes will vary, both among chromosomes in the same population and for any given chromosome in different subpopulations. Given this variation, genes on the same pair of homologous chromosomes will tend to have more similar time depths than expected by chance, simply by dint of being from the same subpopulation. In other words, time depth is correlated on homologous pairs. Given this time depth correlation, individuals who are homozygous at one locus on a particular chromosome will tend to show lower mean time depth for all other markers on that chromosome. Thus, a chromosome that carries a homozygous disease susceptibility factor can be identified by its lower mean time depth in affected relative to unaffected individuals. In CAM mapping, data from microsatellite-based genome screens are used to compare average time depths in cases and controls. Chromosomes that exhibit a large, disease-status-dependent difference in time depth are considered candidates for the location of susceptibility factors. Furthermore, individual markers were compared to facilitate the localization of susceptibility genes on a chromosome. In any individual at any locus, the genetic distance between parental genomes is calculated as the squared difference in allele length, d (2). These differences are then compared in affected versus unaffected and transmitted versus nontransmitted genotypes by using a simple t test to generate a statistic that reflects the magnitude and direction of the difference. Precise significance levels have yet to be determined, but work to date suggests that values exceeding 1.5 should be treated as large and significant (data not shown).
Results
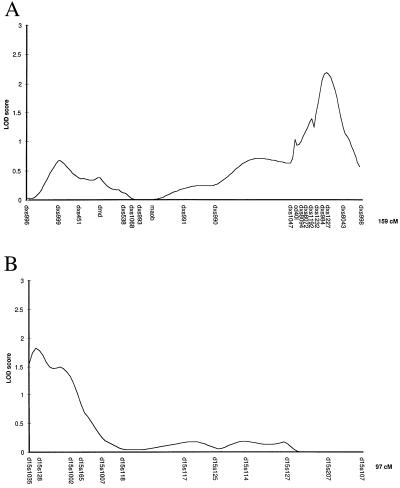
In the first round of the genome screen, conducted on 92 independent sibpairs, seven regions on chromosomes 3, 5, 6, 8, 9, 15, and X showed nominal evidence for linkage (lod >1.0) using mapmaker/sibs and sibpair analysis. Twenty-two markers from these regions were typed in a second round screen on a further 81 independent sibpairs. Two regions, on chromosomes 15q and Xq, showed evidence of linkage to putative tuberculosis-susceptibility genes in the combined analysis (table 2). For chromosomes 15 and X, multipoint lod scores were produced for the combined screens by using mapmaker/sibs (Fig. 1). The maximum multipoint lod scores for these regions are 1.82 and 2.18, respectively. There is a difference between the maximum lod score for the X chromosome produced by the two programs (1.77 at CD40l for sibpair analysis and 2.18 for mapmaker/sibs at DXS1227). This is because mapmaker/sibs analyzes the data for brother-brother, brother-sister, and sister-sister pairs separately and then combines lod scores. lod scores cannot be less than zero, so if one pairing produces allele sharing of less than the expected 0.5, this is ignored. Thus, we prefer the more conservative estimate for the X chromosome produced by sibpair analysis. The apparent lower lod scores for D15S1007 and D15S1002 in the single locus analysis (Table 2) compared with the multipoint analysis (Fig. 1) are caused by low marker information content.
Table 2.
Individual marker linkage analysis for tuberculosis
| Marker | Screen 1
|
Screen 2
|
Combined analysis
|
|||
|---|---|---|---|---|---|---|
| lod | P value | lod | P value | lod | P value | |
| D15S1035 | 1.03 | 0.015 | 0.66 | 0.041 | 1.64 | 0.003 |
| D15S128 | 1.43 | 0.005 | 0.60 | 0.048 | 2.00 | 0.001 |
| D15S1002 | 0.00 | 0.50 | 1.04 | 0.001 | 0.39 | 0.090 |
| D15S165 | 0.017 | 0.39 | 0.72 | 0.035 | 0.51 | 0.063 |
| D15S1007 | 0.24 | 0.14 | 0.24 | 0.15 | 0.50 | 0.065 |
| CD40l | 0.71 | 0.035 | 1.00 | 0.016 | 1.77 | 0.002 |
| DXS8094 | 0.64 | 0.040 | 0.77 | 0.030 | 1.49 | 0.004 |
| DXS8072 | 0.00 | 0.50 | 0.15 | 0.21 | 0.08 | 0.27 |
| DXS1192 | 0.27 | 0.13 | 0.52 | 0.060 | 0.71 | 0.035 |
| DXS1232 | 0.082 | 0.27 | 0.55 | 0.056 | 0.61 | 0.047 |
| DXS984 | 0.17 | 0.19 | 0.87 | 0.022 | 0.98 | 0.017 |
| DXS1227 | 0.00 | 0.50 | 0.17 | 0.19 | 0.086 | 0.27 |
| DXS8043 | 0.029 | 0.36 | 0.077 | 0.28 | 0.11 | 0.24 |
Analyses were performed by using sibpair analysis for each individual marker. Markers are shown in order of position along chromosome.
Figure 1.
Multipoint maximum lod score analysis for chromosomes 15 and X for combined screen 1 and 2 data, calculated by using mapmaker/sibs.
No region of the genome showed strong evidence (e.g., a lod score > 3.0) of linkage to tuberculosis in these African populations. This indicates that tuberculosis-susceptibility in our populations is not a monogenic trait. Multipoint exclusion mapping was performed on data from the first round screen for a putative tuberculosis-susceptibility gene with a λs = 2.0 or 3.0 by using mapmaker/sibs. The majority of the genome could not be excluded for a λs = 2.0, but 78% of the genome could be excluded for a gene with a λs = 3.0 (30). Therefore, a gene exerting a moderate population-wide sibling recurrence risk for tuberculosis could have been missed by this genome screen.
For the markers on chromosomes other than 15 and X, combined analysis of the first and second rounds of the screen produced a lod score of less than 0.7. These regions are unlikely to contain a major tuberculosis-susceptibility gene unless there is significant heterogeneity between tuberculosis-susceptibility in The Gambia and South Africa. None of these markers produced a lod score of more than 1.5 when The Gambian families alone were analyzed (30).
In view of the suggestive evidence of tuberculosis susceptibility genes on chromosomes 15q and Xq provided by linkage analysis, we proceeded to analyze the same dataset by using a form of homozygosity mapping termed common ancestry using microsatellites (CAM) mapping. This independent analysis identified both chromosomes 15 and X as showing a significant excess of homozygosity in the tuberculosis cases compared with controls (data not shown). The concordance between linkage analysis and CAM analysis was equally striking when individual markers were compared. Of all of the microsatellite markers studied across the genome, three of the five highest scores on CAM analysis (Table 3) were for makers lying in the regions of linkage on chromosomes 15q and Xq (Fig. 1; Table 2).
Table 3.
Individual marker CAM mapping analysis for tuberculosis
| Microsatellite | Score | P value |
|---|---|---|
| D15S1035 | 3.63 | 0.0001 |
| D14S63 | 2.69 | 0.004 |
| D1S197 | 2.28 | 0.011 |
| DXS1047 | 2.25 | 0.012 |
| CD40l | 2.21 | 0.014 |
| DXS993 | 1.96 | 0.025 |
| D11S903 | 1.79 | 0.037 |
| D6S314 | 1.75 | 0.040 |
| D8S285 | 1.75 | 0.040 |
| D17S799 | 1.73 | 0.042 |
Results of calculating mean d2 for affected and unaffected individuals, for every marker in turn in a time to most recent common ancestor analysis. The 10 microsatellite markers with the highest scores are shown. Significance was assessed by randomizing the data with respect to affected status 100 times. The score given to each marker is the observed difference divided by the standard deviation of the randomizations, with P values taken directly from the resulting z value (one-tailed, no adjustment for multiple tests).
Discussion
A variety of different approaches have previously been used to identify genes involved in host susceptibility to infectious diseases, including candidate gene studies on tuberculosis (8, 9, 31) and malaria (32) and studies on inbred families with extreme susceptibility to non-tuberculous mycobacteria (33). To date, linkage studies have addressed only candidate genes or small numbers of extended families with measurements of schistosomal parasite burdens rather than disease (34, 35). Here, we report a genome-wide search on a large number of families with an infectious disease. The linkage analysis alone provides suggestive evidence for tuberculosis-susceptibility genes on chromosomes 15q and Xq, and these results are supported by the results of the CAM mapping.
Complex segregation analysis on some Brazilian families with multiple cases of tuberculosis rejected a polygenic model of inheritance and suggested that only one or two genes were important in determining susceptibility to disease (20). In our study, no marker was strongly linked to disease, indicating that there is unlikely to be a single major tuberculosis-susceptibility gene among Africans. Whether this apparent difference relates to differences in tuberculosis-susceptibility between Brazilians and Africans, the analysis of selected pedigrees in the Brazilian study, or some of the inherent limitations of complex segregation analysis (36), requires further investigation. Combining the data from the three African populations could have resulted in a gene exerting a major tuberculosis-susceptibility effect in a single population being overlooked. However, when the Gambian and South African families were evaluated separately, no additional significantly linked loci were identified (30).
In a case-control study in The Gambia, two candidate genes, NRAMP1 and VDR, were found to be associated with tuberculosis (8, 9), but, in the present family study, microsatellites near to these genes were not found to be significantly genetically linked to susceptibility. Although the patients in the case-control study were clinically more homogeneous (all HIV-negative adults with smear-positive pulmonary tuberculosis) than those in the family study, there is no inconsistency between the datasets. NRAMP1 and VDR do not exert a large enough familial clustering effect to produce statistically significant evidence of linkage in this study. The nearest marker to NRAMP1, D2S1471, had a weakly positive lod score of 0.47. This issue has previously been discussed by Risch and Merikangas, who showed that linkage analysis does not usually have the power to identify disease-susceptibility loci conferring genotype relative risks of around 2 (similar to NRAMP1) as unrealistically large numbers of families would be required (37). Conversely, this implies that the putative susceptibility genes mapped to chromosomes 15q and Xq may have substantially larger effects than any tuberculosis susceptibility locus implicated previously.
CAM analysis may be viewed as a more sensitive type of homozygosity mapping. It employs a sensitive index of genetic distance (d2) to identify chromosomes and chromosomal regions that share more recent ancestry and thus can map susceptibility loci that are recessive or (with less power) codominant. Thus, using the same dataset, information in the population structure of the families studied is extracted to provide mapping information that is independent of linkage analysis of the sibling pairs. In this first application of such a combined analysis, a concordance in the results of linkage and CAM mapping is observed, supporting the localization of susceptibility genes on chromosomes 15q and Xq. For example, the highest scoring individual markers from the whole genome using CAM analysis (Table 3), D15S1035, is located in the region of linkage on chromosome 15. The presence of high CAM mapping scores in the regions of linkage argues that the susceptibility genes suggested by linkage analysis are more likely to be recessive, or at least codominant, rather than dominant.
Positional candidate genes located in the region of interest on chromosome 15 include the P protein and the HERC2 genes (38, 39). Several candidate genes that lie in the region of interest on the X chromosome, Xq26, particularly CD40 ligand, also warrant investigation. The microsatellite within the CD40 ligand gene that was typed in this study did not show evidence of transmission disequilibrium to affected offspring. It is of note that there are approximately twice as many male compared with female tuberculosis patients in both The Gambia and South Africa (data not shown; ref. 40). This significant male excess is consistently observed in many different ethnic groups throughout the developing and industrially developed world (41–44). Although this excess of male patients could be attributable to a hormonal effect or to environmental risk factors, it is also consistent with a tuberculosis-susceptibility gene on the X chromosome.
In summary, this study has found evidence against the proposition that tuberculosis susceptibility is largely monogenic, at least in these African populations. Two regions of the genome on chromosomes 15q and Xq showed evidence of tuberculosis susceptibility genes both by linkage and CAM mapping, and further work toward gene identification in these regions is indicated. The characterization of susceptibility genes with effects large enough to be detectable in whole genome screens should provide new insights into the pathogenesis of this major cause of global morbidity and mortality.
Acknowledgments
Ethical approval was provided by the joint Gambian Government/Medical Research Council Ethical Committee and the Ethics Committee of the Faculty of Medicine, University of Stellenbosch. We are very grateful for the help of the Gambian National TB Control Program Directors, Dr. V. Bouchier and Dr. K. Manneh, and The City of Tygerberg Head of Health, Dr. I. Toms, and to Prof. G. M. Lathrop for statistical advice. This work was funded by the Wellcome Trust (Grant ref. 044418/Z/95/Z/139) and the Glaxo Wellcome Action TB Initiative. R.B. is a Wellcome Training Fellow in Tropical Medicine and A.V.S.H. is a Wellcome Trust Principal Research Fellow.
Abbreviations
- CAM mapping
common ancestry using microsatellite mapping
- lod
logarithm of odds
Footnotes
Throughout this paper, we follow conventional genetic terminology by referring to tuberculosis-susceptibility or disease-susceptibility. However, in considering the evolution of host-pathogen interactions, it may be more appropriate to describe genetic resistance to disease
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.140201897.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.140201897
References
- 1.Dolin P J, Raviglione M C, Kochi A. Bull W H O. 1994;72:213–220. [PMC free article] [PubMed] [Google Scholar]
- 2.Rouillon A, Perdrizet S, Parrot R. Tubercle. 1976;57:275–279. doi: 10.1016/s0041-3879(76)80006-2. [DOI] [PubMed] [Google Scholar]
- 3.Murray C J L, Styblo K, Rouillon A. Bull Int Union Tuberc Lung Dis. 1990;65:6–24. [PubMed] [Google Scholar]
- 4.Stead W W, Senner J W, Reddick W T, Lofgren J P. N Engl J Med. 1990;322:422–427. doi: 10.1056/NEJM199002153220702. [DOI] [PubMed] [Google Scholar]
- 5.Stead W W. Ann Intern Med. 1992;116:937–941. doi: 10.7326/0003-4819-116-11-937. [DOI] [PubMed] [Google Scholar]
- 6.Kallmann F J, Reisner D. Am Rev Tuberc. 1942;47:549–574. [Google Scholar]
- 7.Comstock G W. Am Rev Respir Dis. 1978;117:621–624. doi: 10.1164/arrd.1978.117.4.621. [DOI] [PubMed] [Google Scholar]
- 8.Bellamy R, Ruwende C, Corrah T, McAdam K P W J, Whittle H C, Hill A V S. N Engl J Med. 1998;338:640–644. doi: 10.1056/NEJM199803053381002. [DOI] [PubMed] [Google Scholar]
- 9.Bellamy R, Ruwende C, Corrah T, McAdam K P W J, Thursz M, Whittle H C, Hill A V S. J Infect Dis. 1999;179:721–724. doi: 10.1086/314614. [DOI] [PubMed] [Google Scholar]
- 10.Davies J L, Kawaguchi Y, Bennett S T, Copeman J B, Cordell H J, Pritchard L E, Reed P W, Gough S C L, Jenkins S C, Palmer S M, et al. Nature (London) 1994;371:130–136. doi: 10.1038/371130a0. [DOI] [PubMed] [Google Scholar]
- 11.Hanis C L, Boerwinkle E, Chakraborty R, Ellsworth D L, Concannon P, Stirling B, Morrison V A, Wapelhorst B, Spielman R S, Gogolin-Ewens K J, et al. Nat Genet. 1996;13:161–166. doi: 10.1038/ng0696-161. [DOI] [PubMed] [Google Scholar]
- 12.Sawcer S, Jones H B, Feakes R, Gray J, Smaldon N, Chataway J, Robertson N, Clayton D, Goodfellow P N, Compston A. Nat Genet. 1996;13:464–468. doi: 10.1038/ng0896-464. [DOI] [PubMed] [Google Scholar]
- 13.The Multiple Sclerosis Genetics Group. Nat Genet. 1996;13:469–471. doi: 10.1038/ng0896-469. [DOI] [PubMed] [Google Scholar]
- 14.Ebers G C, Kukay K, Bulman D E, Sadovnick A D, Rice G, Anderson C, Armstrong H, Cousin K, Bell R B, Hader W, et al. Nat Genet. 1996;13:472–476. doi: 10.1038/ng0896-472. [DOI] [PubMed] [Google Scholar]
- 15.Daniels S E, Bhattacharya S, James A, Leaves N I, Young A, Hill M R, Faux J A, Ryan G F, Le Soeuf P N, Lathrop G M, et al. Nature (London) 1996;383:247–250. doi: 10.1038/383247a0. [DOI] [PubMed] [Google Scholar]
- 16.Satsangi J, Parkes M, Louis E, Hashimoto L, Kato N, Welsh K, Terwilliger J D, Lathrop G M, Bell J I, Jewell D P. Nat Genet. 1996;14:199–202. doi: 10.1038/ng1096-199. [DOI] [PubMed] [Google Scholar]
- 17.Zhong F, McCombs C C, Olson J M, Elston R C, Stevens F M, McCarthy C F, Michalski J P. Nat Genet. 1996;14:329–333. doi: 10.1038/ng1196-329. [DOI] [PubMed] [Google Scholar]
- 18.Trembath R C, Clough R L, Rosbotham J L, Jones A B, Camp R D R, Frodsham A, Browne J, Barber R, Terwilliger J, Lathrop G M, Barker N W N. Hum Mol Genet. 1997;6:813–820. doi: 10.1093/hmg/6.5.813. [DOI] [PubMed] [Google Scholar]
- 19.The Collaborative Study on the Genetics of Asthma. Nat Genet. 1997;15:389–392. [Google Scholar]
- 20.Shaw M A, Collins A, Peacock C S, Miller E N, Black G F, Sibthorpe D, Lins-Lainson Z, Shaw J J, Ramos F, Silveira F, Blackwell J M. Tubercle Lung Dis. 1997;78:35–45. doi: 10.1016/s0962-8479(97)90014-9. [DOI] [PubMed] [Google Scholar]
- 21.Allsopp C E M, Harding R H, Taylor C, Bunce M, Kwiatkowski D, Anstey N, Brewster D, McMichael A J, Greenwood B M, Hill A V S. Am J Hum Genet. 1992;50:411–421. [PMC free article] [PubMed] [Google Scholar]
- 22.Beyers N, Gie R P, Zietsman H L, Kunneke M, Hauman J, Tatley M, Donald P R. S Afr Med J. 1996;86:40–44. [PubMed] [Google Scholar]
- 23.Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, et al. Nature (London) 1996;380:152–154. doi: 10.1038/380152a0. [DOI] [PubMed] [Google Scholar]
- 24.Reed P W, Davies J L, Copeman J B, Bennett S T, Palmer S M, Pritchard L E, Gough S C L, Kawaguchi Y, Cordell H J, Balfour K M, et al. Nat Genet. 1994;7:390–395. doi: 10.1038/ng0794-390. [DOI] [PubMed] [Google Scholar]
- 25.Weeks D E, Lathrop G M. Trends Genet. 1995;11:513–520. doi: 10.1016/s0168-9525(00)89163-5. [DOI] [PubMed] [Google Scholar]
- 26.Kruglyak L, Lander E S. Am J Hum Genet. 1995;57:439–454. [PMC free article] [PubMed] [Google Scholar]
- 27.Risch N. Am J Hum Genet. 1990;46:229–241. [PMC free article] [PubMed] [Google Scholar]
- 28.Risch N. Am J Hum Genet. 1990;46:242–253. [PMC free article] [PubMed] [Google Scholar]
- 29.Holmans P. Am J Hum Genet. 1993;52:362–374. [PMC free article] [PubMed] [Google Scholar]
- 30.Bellamy R. Ph.D. thesis. United Kingdom: Univ. of Oxford; 1998. [Google Scholar]
- 31.Singh S P, Mehra N K, Dingley H B, Pande J N, Vaidya M C. J Infect Dis. 1983;148:676–681. doi: 10.1093/infdis/148.4.676. [DOI] [PubMed] [Google Scholar]
- 32.Hill A V S, Allsopp C E M, Kwiatkowski D, Anstey N M, Twumasi P, Rowe P A, Bennett S, Brewster D, McMichael A J, Greenwood B M. Nature (London) 1991;352:595–560. doi: 10.1038/352595a0. [DOI] [PubMed] [Google Scholar]
- 33.Newport M J, Huxley C M, Huston S, Hawrylowicz C M, Oostra B A, Williamson R, Levin M. N Engl J Med. 1996;335:1941–1949. doi: 10.1056/NEJM199612263352602. [DOI] [PubMed] [Google Scholar]
- 34.Marquet S, Abel L, Hillaire D, Dessein H, Kalil J, Feingold J, Weissenbach J, Dessein A J. Nat Genet. 1996;14:181–184. doi: 10.1038/ng1096-181. [DOI] [PubMed] [Google Scholar]
- 35.Marquet S, Abel L, Hillaire D, Dessein A. Eur J Hum Genet. 1999;7:88–97. doi: 10.1038/sj.ejhg.5200243. [DOI] [PubMed] [Google Scholar]
- 36.McGuffin P, Huckle P. Am J Hum Genet. 1990;46:994–999. [PMC free article] [PubMed] [Google Scholar]
- 37.Risch N, Merikangas K. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 38.Gardner J M, Nakatsu Y, Gondo Y, Lee S, Lyon M F, Kinf R A, Brilliant M H. Science. 1992;257:1121–1124. doi: 10.1126/science.257.5073.1121. [DOI] [PubMed] [Google Scholar]
- 39.Ji Y, Walkowicz M J, Buiting K, Johnson D K, Tarvin R E, Rinchik E M, Horsthemke B, Stubbs L, Nicholls R D. Hum Mol Genet. 1999;8:533–542. doi: 10.1093/hmg/8.3.533. [DOI] [PubMed] [Google Scholar]
- 40.Kustner H G V. S Afr Med J. 1979;55:460–473. [PubMed] [Google Scholar]
- 41.Rieder H L, Kelly G D, Bloch A B, Cauthen G M, Snider D E. Chest. 1991;100:678–811. doi: 10.1378/chest.100.3.678. [DOI] [PubMed] [Google Scholar]
- 42.Heng B H, Tan K K. Bull Int Union Tuberc Lung Dis. 1991;66:125–128. [PubMed] [Google Scholar]
- 43.Medical Research Council Cardiothoracic Epidemiology Group. Thorax. 1992;47:770–775. doi: 10.1136/thx.47.10.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aktogu S, Yorgancioglu A, Cirak K, Kose T, Dereli S M. Eur Resp J. 1996;9:2031–2035. doi: 10.1183/09031936.96.09102031. [DOI] [PubMed] [Google Scholar]