Abstract
Proline (Pro) is one of the most widely distributed osmolytes in water-stressed plants. We previously isolated from Arabidopsis a gene encoding Pro dehydrogenase (ProDH), a mitochondrial enzyme involved in the first step of the conversion of Pro to glutamic acid. The ProDH gene in Arabidopsis is up-regulated by rehydration after dehydration but is down-regulated by dehydration. ProDH is also induced by l-Pro and hypoosmolarity. The induction of ProDH expression under rehydration seems to be caused by both accumulated Pro and hypoosmolarity. We analyzed a DNA region that is located 5′ to the transcription start site (a promoter region) of ProDH to identify cis-acting elements involved in l-Pro-induced and hypoosmolarity-induced expression in transgenic tobacco (Nicotiana tabacum) and Arabidopsis plants. We found that a 9-bp sequence, ACTCATCCT, in the ProDH promoter is necessary for the efficient expression of ProDH in response to l-Pro and hypoosmolarity. Moreover, ACTCAT is a core cis-acting element, which we have called Pro- or hypoosmolarity-responsive element (PRE), that is necessary for l-Pro-responsive and hypoosmolarity-responsive expression of ProDH. Microarray and RNA gel-blot analyses showed that 21 l-Pro-inducible genes have the PRE sequences in their promoter regions. These results indicate that the PRE sequence play an important role in the l-Pro-responsive gene expression.
Plant growth and productivity are greatly affected by osmotic or water stress caused by drought or high salinity. Plants respond and adapt to osmotic stress to survive. Many plants accumulate compatible osmolytes, such as Pro, Gly betaine, or sugar alcohols, when they are subjected to osmotic stress (Hellebust, 1976; Delauney and Verma, 1993). Among these, Pro is the most diversely used osmolyte accumulated under osmotic stress conditions in plants (Singh et al., 1972; McCue and Hanson, 1990; Delauney and Verma, 1993). Pro is accumulated not only in plants but also in eubacteria, protozoa, marine invertebrates, and algae (Measures, 1975; McCue and Hanson, 1990; Delauney and Verma, 1993). Pro has some significant roles as a sink of energy or reducing power (Walton et al., 1991), as a source of carbon and nitrogen compounds (Ahmad and Hellebust, 1988; Peng et al., 1996), as a hydroxyl radical scavenger (Smirnoff and Cumbes, 1989), and in protection of plasma membrane integrity (Mansour, 1998) in plants under osmotic stress.
There are two routes for Pro biosynthesis: Pro is produced from Glu through the Glu pathway and from Orn through the Orn pathway. Glu is the primary precursor rather than Orn for Pro biosynthesis in osmotically stressed plants (Hu et al., 1992; Delauney et al., 1993; Roosens et al., 1998). The accumulation of Pro in dehydrated plants is caused by both the activation of Pro biosynthesis and the inactivation of Pro metabolism (Yoshiba et al., 1995; Kiyosue et al., 1996; Peng et al., 1996; Verbruggen et al., 1996; Nakashima et al., 1998; Yoshiba et al., 1999). A decrease in the level of accumulated Pro in rehydrated plants is conversely caused by both the inactivation of biosynthesis and the activation of metabolism. In higher plants, l-Pro is synthesized from l-Glu via Δ1-pyrroline-5-carboxylate (P5C) by two enzymes, P5C synthetase and P5C reductase. l-Pro is metabolized to l-Glu via P5C by two enzymes, Pro dehydrogenase (ProDH) and P5C dehydrogenase (Yoshiba et al., 1997; Strizhov et al., 1997). P5C synthetase and ProDH catalyze the first step and the rate-limiting step, respectively. The accumulation of Pro in transgenic Arabidopsis plants that overexpressed antisense ProDH cDNA improved stress tolerance to freezing and high salinity, which indicates the important role of ProDH in Pro metabolism (Nanjo et al., 1999a).
In a previous report, we showed that the expression of the ProDH gene is repressed by dehydration, but is induced by rehydration after dehydration for 10 h (Kiyosue et al., 1996; Peng et al., 1996; Verbruggen et al., 1996). A high level of transcripts of ProDH was also detected when plants were incubated in medium containing l-Pro (Kiyosue et al., 1996; Peng et al., 1996; Verbruggen et al., 1996; Nakashima et al., 1998) or under hypoosmotic conditions (Nakashima et al., 1998). Therefore, the induction of ProDH expression under rehydration seems to be caused not only by accumulated Pro but also by hypoosmolarity. The elucidation of ProDH expression may be helpful in understanding the molecular process of the recovery of plants from osmotic stress.
We analyzed the expression of the β-glucuronidase (GUS) reporter gene, driven by the 1.4-kb ProDH promoter, in transgenic Arabidopsis plants and found that the gene expression of ProDH is controlled by its promoter region (Nakashima et al., 1998). The 1.4-kb ProDH promoter region contains cis-acting elements involved in l-Pro- and hypoosmolarity-inducible expression of ProDH. To understand this expression of ProDH in detail, we analyzed the cis-acting elements by using transgenic tobacco (Nicotiana tabacum) and Arabidopsis plants containing a fused gene consisting of deleted or base-substituted DNA fragments of the ProDH promoter and the luciferase (LUC) or GUS reporter genes. We determined a novel cis-acting element involved in l-Pro- and hypoosmolarity-responsive expression of ProDH.
RESULTS
5′-Deletion Analysis of the ProDH Promoter Regions Involved in l-Pro- and Hypoosmolarity-Inducible Expression
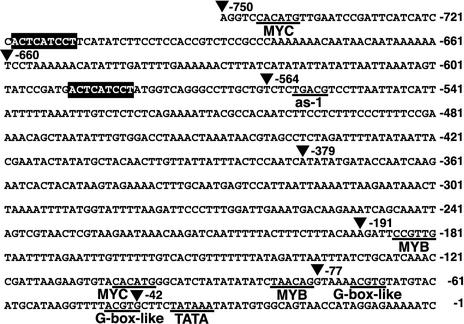
The nucleotide sequence of the 750-bp ProDH promoter region is shown in Figure 1. To examine the cis-acting elements involved in the l-Pro- and hypoosmolarity-inducible expression, we prepared transcriptional fusion genes consisting of the 5′-deleted ProDH promoter regions fused to the LUC reporter gene and introduced them into tobacco and Arabidopsis plants by Agrobacterium tumefaciens infection. We prepared six constructs containing fragments deleted to positions −750, −564, −379, −191, −77, and −42 in the promoter region—namely, 5D1, 5D2, 5D3, 5D4, 5D5, and 5D6 respectively—and made stable transformants (Fig. 2A). Three-week-old T2 transgenic tobacco plants were transferred from germination medium (GM) agar plates and placed in hydroponic conditions for 24 h in DW or 0.09 m l-Pro solution. Incubation in DW or l-Pro solution increased the LUC activity of the T2 transgenic tobacco plants containing the 5D1 construct, but did not increase the activity of the 5D2, 5D3, 5D4, 5D5, or 5D6 transformants (Fig. 2B). We got similar results when we used 3-week-old T2 transgenic Arabidopsis plants (Fig. 2C). These results indicate that at least one cis-acting element for the l-Pro- and hypoosmolarity-inducible expression of ProDH is localized in the region between positions −750 and −564 of the ProDH promoter.
Figure 1.
Nucleotide sequence of the promoter region of ProDH. Underlines show the putative TATA box (TATAA), G-box-like sequence (ACGTG), MYB recognition site (PyAACNPu), MYC recognition site (CANNTG), and as-1 sequence (TGACG). ▪, The direct repeat sequence ACTCATCCT. ▴, Start points of 5′-deleted derivatives. The nucleotide sequence was analyzed with the Genetyx software system (Software Development, Tokyo).
Figure 2.
5′-Deletion analysis of the ProDH promoter for the l-Pro- and hypoosmolarity-responsive induction of the LUC reporter gene in transgenic tobacco and Arabidopsis plants. A, Schematics of the 5′-terminal deletions of the ProDH promoter fused to the LUC reporter gene. Arrows indicate 9-bp direct repeat sequences. B, LUC activity in transgenic tobacco plants containing 5′-terminal deletions of the ProDH promoter fused to the LUC gene. T2 seedlings of tobacco were incubated in distilled water (DW) or in 0.09 m l-Pro for 24 h. LUC activity was measured in three plants of seven independent transformant lines for each construct. Multiplication factors of induction of LUC activity (ratio of after treatment to before treatment) by DW and l-Pro treatments are shown at the right. Bars indicate se. C, LUC activity in transgenic Arabidopsis plants containing 5′-terminal deletions of the ProDH promoter fused to LUC. T2 seedlings of Arabidopsis were incubated in DW or 0.09 m l-Pro for 24 h. LUC activity was measured in seven independent transformant lines for each construct. We measured three plants for each line and showed average values.
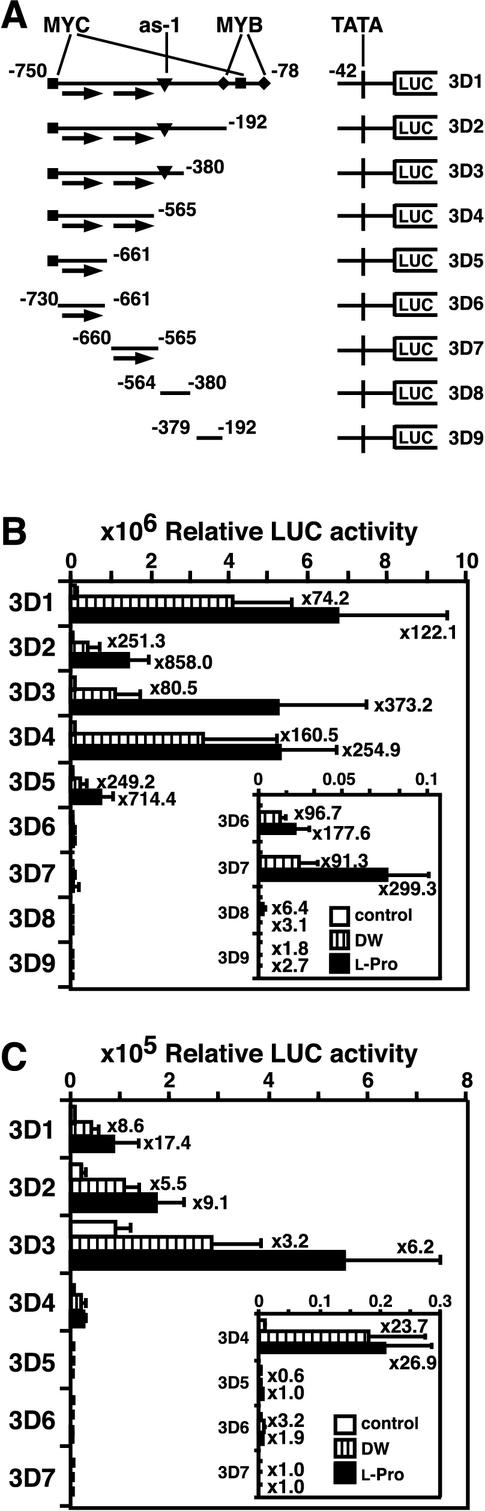
3′-Deletion Analysis of the ProDH Promoter Regions Involved in l-Pro- and Hypoosmolarity-Inducible Expression
We constructed 3′-deleted fragments of the ProDH promoter fused to the LUC reporter gene and introduced the chimeric genes into tobacco and Arabidopsis plants by A. tumefaciens infection. Five chimeric gene fusions contained the ProDH minimal TATA box sequence (5D6 fragment) and the ProDH promoter regions −750 to −78, −750 to −192, −750 to −380, −750 to −565, and −750 to −661 (namely, 3D1, 3D2, 3D3, 3D4, and 3D5, respectively; Fig. 3A). All of the T1 transgenic tobacco plants containing these constructs showed the l-Pro- and hypoosmolarity-inducible expression of the LUC gene (Fig. 3B). The 3D6 construct, which lacked 20-bp fragments (including a MYC recognition site) from the 5′ end of the 3D5 fragment, also functioned in the induction of LUC gene expression (Fig. 3B). The 3D1, 3D2, 3D3, and 3D4 constructs contain a couple of 9-bp direct repeat sequences (ACTCATCCT) in the region between −720 and −712 (first repeat) and between −591 and −583 (second repeat). The 3D5 and 3D6 constructs contained only the first repeat sequence. Then we made construct 3D7, which had only the second repeat sequence in the ProDH promoter. The 3D7 construct also functioned in the l-Pro- and hypoosmolarity-inducible expression of LUC. In contrast, constructs 3D8 and 3D9, which had no direct repeat sequence, did not function in either l-Pro- or hypoosmolarity-inducible expression of LUC. These results suggest that at least one of the 9-bp direct repeat sequences is necessary for l-Pro- and hypoosmolarity-inducible expression in the ProDH promoter in the T1 tobacco plants.
Figure 3.
3′-Deletion analysis of the ProDH promoter for the l-Pro- and hypoosmolarity-responsive induction of the LUC reporter gene in transgenic tobacco and Arabidopsis plants. A, Schematics of the 3′-terminal deletions of promoters fused to the LUC reporter gene. Arrows indicate 9-bp direct repeat sequences. B, LUC activity in transgenic tobacco plants containing 3′-terminal deletions of the ProDH promoter fused to LUC. T1 leaves of tobacco were incubated in DW or in 0.09 m l-Pro for 24 h. LUC activity was measured in 20 independently obtained transgenic plants for each construct. Multiplication factors of induction of LUC activity by DW and l-Pro treatments are shown on the right. Bars indicate se. C, LUC activity in transgenic Arabidopsis plants containing 3′-terminal deletions of the ProDH promoter fused to the LUC reporter gene. T2 seedlings of Arabidopsis were incubated in DW or 0.09 m l-Pro for 24 h. LUC activity was measured in seven independent transformant lines for each construct. We measured three plants for each line and showed average values.
On the other hand, the 3-week-old T2 transgenic Arabidopsis seedlings with the 3D4 construct, containing a couple of direct repeat sequences, showed significant induction of LUC activity by l-Pro or hypoosmolarity, whereas the transgenic Arabidopsis seedlings with the 3D5, 3D6, and 3D7 constructs containing one of the direct repeat sequences did not (Fig. 3C). These results suggest that two direct repeat sequences (ACTCATCCT) may be necessary for l-Pro- and hypoosmolarity-inducible expression of ProDH in Arabidopsis plants.
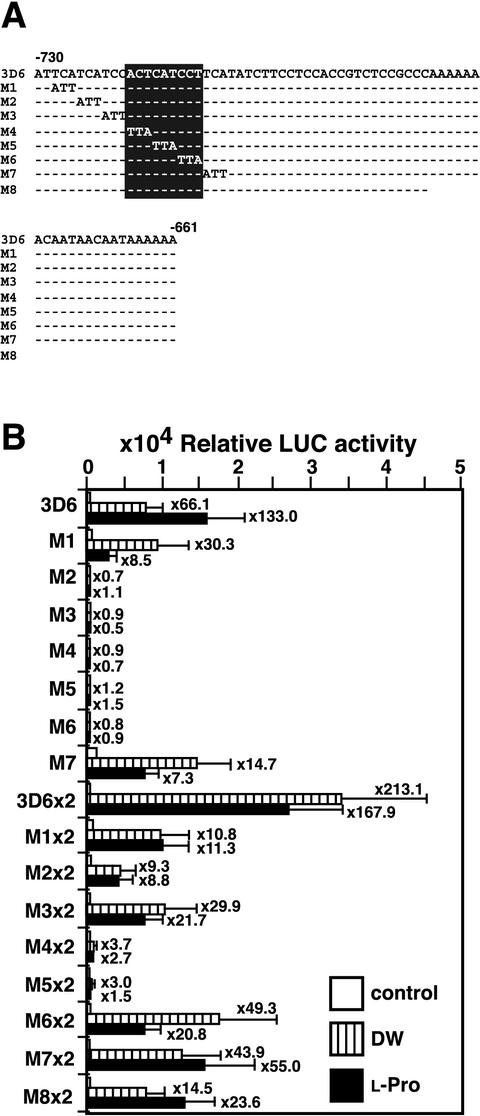
ACTCAT Sequence Is Required for l-Pro- and Hypoosmolarity-Responsive Expression of ProDH
Seven base-substituted 3D6 fragments were fused to the 5D6 minimal TATA promoter and LUC and were introduced into tobacco plants (Fig. 4A). We observed l-Pro- and hypoosmolarity-inducible expression of LUC in transgenic tobacco plants containing the M1 and M7 constructs but not in plants containing the M2, M3, M4, M5, or M6 constructs (Fig. 4B). We then prepared transgenic tobacco plants containing the construct with a tandemly repeated dimer of the mutated 3D6 fragments fused to the 5D6 minimal TATA promoter and LUC. The leaves of the T1 transgenic tobacco plants containing the M1×2, M2×2, M3×2, M6×2, and M7×2 constructs showed l-Pro- and hypoosmolarity-inducible expression of LUC activity, but the leaves of plants containing the M4×2 and M5×2 constructs did not (Fig. 4B). We deleted 23-bp sequences from the 3′ end of the 3D6 fragment, and made the M8 construct by using the deleted fragment fused to the 5D6 minimal TATA promoter and LUC (Fig. 4A). The leaves of the T1 transgenic tobacco plants containing the M8 construct also showed l-Pro- and hypoosmolarity-inducible expression of LUC activity (Fig. 4B). These results indicate that the 9-bp ACTCATCCT sequence and the 5′-flanking sequence are necessary for the efficient expression of the ProDH gene in response to l-Pro and hypoosmolarity and that ACTCAT in the 9-bp sequence is the core element.
Figure 4.
Base substitution analysis of the 70-bp region from positions −730 to −661 of the ProDH promoter for the l-Pro- and hypoosmolarity-responsive induction of LUC in transgenic tobacco plants. A, Upper strand sequences of the 70-bp fragment of the ProDH promoter and its mutated sequences (M1–M8). Each fragment containing each mutation was ligated to the −42 ProDH minimal TATA promoter-LUC construct. Dashes indicate the sequence of the 3D6 construct. B, Effect of base substitutions in the direct repeat sequence for l-Pro- and hypoosmolarity-responsive expression of ProDH. T1 leaves of tobacco were incubated in DW or 0.09 m l-Pro for 24 h. LUC activity was measured in 12 to 26 leaves of independent transformant lines for each construct and is shown as average values. Bars indicate se.
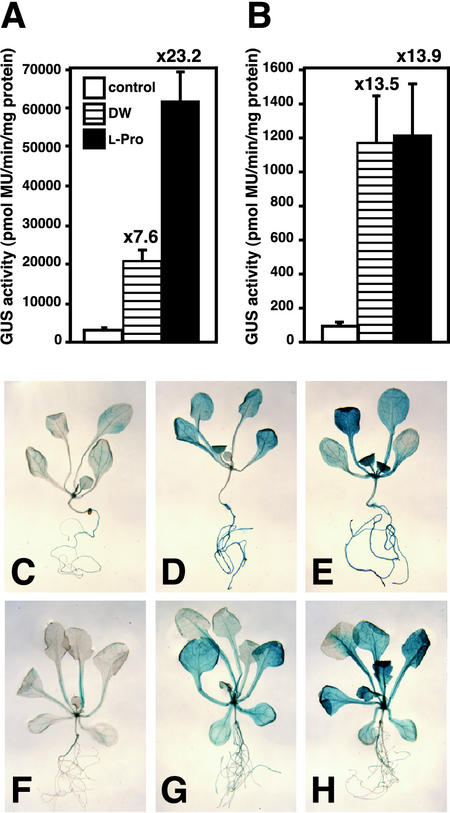
Expression of GUS Activity in Transgenic Arabidopsis Containing the ProDH Promoter Construct with One Direct Repeat Sequence
To examine whether the 3D5 fragment, containing one direct repeat sequence (ACTCATCCT), is sufficient for l-Pro- and hypoosmolarity-inducible expression of ProDH in Arabidopsis plants, we replaced the LUC reporter gene of the 3D5 construct with the GUS reporter gene (3D5-GUS construct). We also analyzed another construct as a positive control: the 1.4-kb promoter-GUS construct, with a chimeric gene consisting of the 1.4-kb ProDH promoter region and the GUS reporter gene. We examined GUS expression in T2 transgenic Arabidopsis plants containing the 1.4-kb promoter-GUS or 3D5-GUS construct (Fig. 5, A and B). Incubation in DW or l-Pro solution increased GUS activity in Arabidopsis plants containing the 1.4-kb promoter-GUS construct (Fig. 5A) and in plants containing the 3D5-GUS construct (Fig. 5B). Two-week-old transgenic Arabidopsis plants containing the 1.4-kb promoter-GUS construct treated with DW or l-Pro showed strong GUS staining in whole Arabidopsis plants (Fig. 5, D and E). On the other hand, 17-d-old 3D5-GUS transgenic plants treated with DW or l-Pro showed strong GUS staining in leaves and stems but weak staining in roots (Fig. 5, G and H).
Figure 5.
GUS activity and histochemical analysis in transgenic Arabidopsis plants containing the 1.4-kb ProDH promoter or the 3D5 fragment fused to the GUS reporter gene after treatment with DW or l-Pro. A, GUS activity in transgenic Arabidopsis containing the 1.4-kb promoter-GUS fusion gene before and after treatment with DW or 0.09 m l-Pro for 24 h. GUS activity was measured in two independent transformant lines for each construct. We measured five plants for each line and showed average values. B, GUS activity in transgenic Arabidopsis containing the 3D5 fragment fused to the GUS reporter gene instead of the LUC reporter gene before and after treatment with DW or 0.09 m l-Pro for 24 h. GUS activity was measured in three independent transformant lines for each construct. We measured five plants for each line and showed average values. C through E, Control plants containing the 1.4-kb promoter-GUS fusion gene grown for 2 weeks and stained for 4 h. F through H, 3D5 constructs containing the GUS reporter gene instead of the LUC reporter gene grown for 14 d and stained overnight. C and F, Before treatment; D and G, after treatment with DW for 24 h; E and H, after treatment with 0.09 m l-Pro for 24 h.
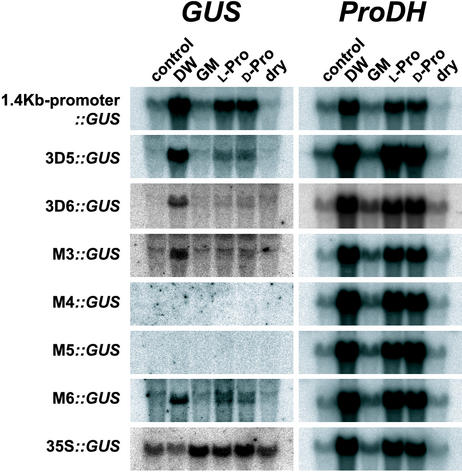
We also carried out RNA gel-blot analysis to analyze the expression of the GUS reporter gene in the 3-week-old T3 transgenic Arabidopsis plants containing the 1.4-kb promoter-GUS, 3D5-GUS, 3D6-GUS, M3-GUS, M4-GUS, M5-GUS, M6-GUS, or 35S-GUS constructs treated with DW, GM, l-Pro, d-Pro, and dehydration (Fig. 6). The 35S-GUS construct was expressed constitutively. The 1.4-kb promoter-GUS, 3D5-GUS, and 3D6-GUS constructs responded to DW, l-Pro, and d-Pro treatments but did not respond to dehydration. The M4-GUS and M5-GUS constructs did not respond to any treatments, despite the M3-GUS and M6-GUS constructs responding to DW, l-Pro, and d-Pro treatments.
Figure 6.
RNA gel-blot analysis in transgenic Arabidopsis plants containing the 1.4-kb ProDH promoter, 3D5, 3D6, M3, M4, M5, or M6 fragments fused to the GUS reporter gene after treatment with DW, GM, 0.09 m l-Pro, or 0.09 m d-Pro for 2 h or dehydration for 2 h. The 35S-GUS construct contains the CaMV 35S-promoter and the GUS reporter gene.
Some Arabidopsis Genes Induced by l-Pro Have ACTCAT Sequence in Their Promoter Regions
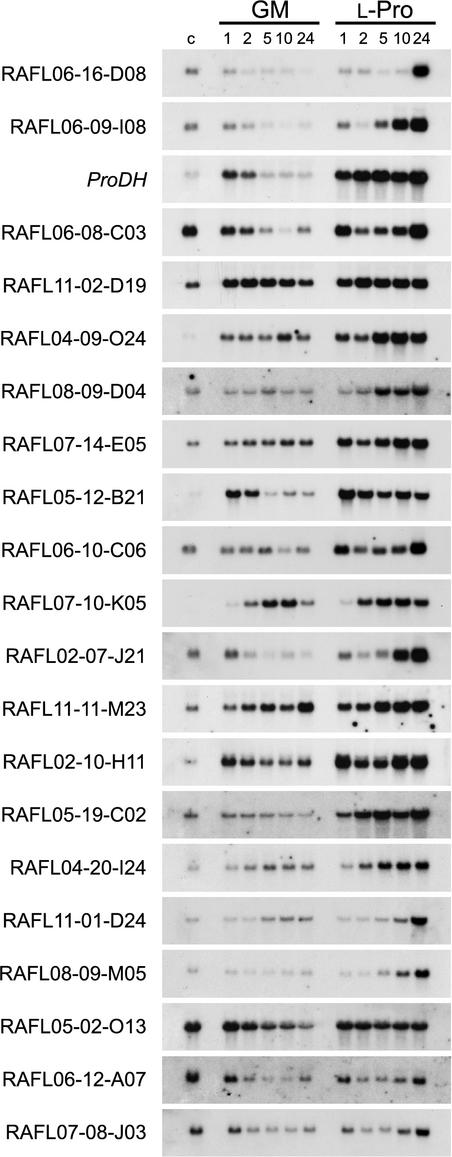
To know whether the ACTCAT sequence is the common cis-acting element in the l-Pro-inducible genes, we surveyed the promoter regions of the other l-Pro-inducible genes. Fifty genes including ProDH were found to be induced by l-Pro treatment using a cDNA microarray containing 7,000 Arabidopsis full-length cDNAs (Seki et al., 2001a, 2001b). mRNAs prepared from the l-Pro- and GM-treated whole plants were used for the generation of Cy3-labeled and Cy5-labeled cDNA probes, respectively. These cDNA probes were hybridized with the cDNA microarray, and the expression profiles of the 7,000 genes were analyzed. We regarded genes with an expression ratio (l-Pro-treated/GM-treated) greater than 1.8 times as putative l-Pro-inducible genes. Fifty l-Pro-inducible genes were identified by cDNA microarray analysis. Computer analysis showed that 27 genes including the ProDH gene have the ACTCAT sequence in their promoter regions (Table I). We subsequently examined l-Pro-inducible expression of each of the 27 genes by RNA gel-blot analysis and found that 21 genes among the 27 genes show l-Pro-inducible expression (Fig. 7). One of these 21 genes was induced by l-Pro slowly after 24 h of the treatment (Fig. 7, RAFL06–16-D08). Six genes among the 27 genes were not induced by l-Pro.
Table I.
l-Pro-inducible genes verified by microarray and RNA gel-blot analysis
| Gene Name | Accession No.b | Mips Codec | Ratiod | ACTCATe | ATGAGT | TATA | Descriptionf |
|---|---|---|---|---|---|---|---|
| RAFL06-16-D08 | AV824224 | At2g25510 | 7.0 | −167 to −162 | −308 to −313 | −30 | Unknown protein |
| RAFL06-09-I08 | AV823852 | At3g22240 | 6.0 | −427 to −422 | −33 | Unknown protein | |
| −521 to −516 | |||||||
| ProDH | AB008810 | At3g30775 | 5.9 | −591 to −586 | −327 to −332 | −37 | Pro dehydrogenase protein |
| −719 to −714 | ProDH | ||||||
| RAFL06-08-C03 | AV823737 | At1g31580 | 5.3 | −927 to −922 | −524 to −519 | −27 | Putative protein |
| −1,031 to −1,026 | |||||||
| RAFL11-02-D19 | AV832077 | At2g45180 | 4.7 | −1,486 to −1,491 | −39 | Putative Pro-rich protein | |
| RAFL04-09-O24 | AV821416 | At4g30270 | 4.6 | −1,021 to −1,016 | −34 | Meri-5 protein | |
| RAFL08-09-D04 | AV826095 | At2g39570 | 4.6 | −775 to −770 | −520 to −525 | −29 | Unknown protein |
| RAFL07-14-E05 | AV825755 | At1g02920 | 4.5 | −707 to −702 | −505 to −510 | −35 | Glutathione S-transferase |
| −1,315 to −1,310 | −1,441 to −1,446 | protein | |||||
| RAFL05-12-B21 | AV822819 | At2g41100 | 4.0 | −1,113 to −1,108 | −1,321 to −1,326 | −35 | Calmodulin-related protein |
| RAFL06-10-C06 | AV823897 | At5g42530 | 3.8 | −190 to −185 | −34 | Unknown protein | |
| RAFL07-10-K05 | AV825528 | At4g14130 | 3.2 | −87 to −92 | −31 | Probable xyloglucan endotransglycosylase-related protein XTR-7 | |
| −180 to −185 | |||||||
| −364 to −369 | |||||||
| RAFL02-07-J21 | AV821161 | At3g22240 | 3.0 | −427 to −422 | −33 | Unknown protein | |
| −521 to −516 | |||||||
| RAFL11-11-M23 | AV832330 | At4g12470 | 2.9 | −304 to −299 | −683 to −688 | −31 | pEARLI 1 protein homolog |
| −1,012 to −1,007 | |||||||
| RAFL02-10-H11 | AV821217 | At3g50480 | 2.7 | −893 to −898 | −39 | Hypothetical protein | |
| −1,257 to −1,262 | |||||||
| RAFL05-19-C02 | AV823433 | At3g57520 | 2.7 | −165 to −170 | −39 | Imbibition protein homolog | |
| RAFL04-20-I24 | AV822062 | At2g44790 | 2.6 | −271 to −266 | −47 | Phytocyanin protein | |
| RAFL11-01-D24 | AV832036 | At3g01420 | 2.0 | −140 to −135 | −34 | Feebly-like protein | |
| RAFL08-09-M05 | AV826133 | At3g22600 | 2.0 | −210 to −205 | −28 | Unknown protein | |
| −1,044 to −1,039 | |||||||
| RAFL05-02-O13 | AV822197 | At1g78830 | 1.9 | −1,443 to −1,438 | −30 | Hypothetical protein | |
| RAFL06-12-A07 | AV824014 | At1g72930 | 1.8 | −210 to −205 | −495 to −500 | −31 | Hypothetical protein |
| RAFL07-08-J03 | AV825348 | At2g41090 | 1.8 | −599 to −594 | −692 to −697 | −34 | Calmodulin-like calcium- |
| −701 to −696 | binding protein CABP-22 |
Twenty-one genes among 50 l-Pro-inducible genes have the ACTCAT sequence in their promoter regionsa.
Upstream sequences (1,500 bp) of the 5′ termini of the cDNA clones are used as promoter regions.
Accession nos. for cDNAs used in this study.
Mips entry codes for cDNAs used in this study.

Nos. indicate the nucleotide exists upstream of the 5′ terminus of the putative transcription start site as described by Seki et al. (2001b).
Descriptions are cited from Mips database.
Figure 7.
RNA gel-blot analysis of l-Pro-inducible genes verified by using the full-length cDNA microarray. RNA samples from Arabidopsis plants transferred from GM plates to GM solution (GM) or GM plates to 0.09 m l-Pro solution (l-Pro) for 1, 2, 5, 10, and 24 h and untreated plants (control) were hybridized with PCR-amplified or sfiI-digested DNA fragments as probes.
DISCUSSION
ProDH is induced not only by rehydration after dehydration and Pro (Kiyosue et al., 1996; Peng et al., 1996), but also by hypoosmolarity (Nakashima et al., 1998). We previously showed by using the GUS reporter gene that the 1.4-kb ProDH promoter region contains cis-acting elements for l-Pro- and hypoosmolarity-inducible expression of ProDH (Nakashima et al., 1998). In this report, we chose the LUC reporter gene for the analysis of the promoter region of ProDH. Using transgenic tobacco plants, we carried out 5′- and 3′-deletion analysis of the ProDH promoter region, and showed that the 90-bp promoter region from positions −750 to −661 (3D5 fragment) functions in l-Pro- and hypoosmolarity-inducible gene expression (Figs. 2 and 3). However, this fragment responded to neither l-Pro- nor hypoosmolarity in transgenic Arabidopsis (Fig. 3). We thought that the LUC activity in response to l-Pro and hypoosmolarity was too low to detect in Arabidopsis plants containing the 3D5 construct. Because we thought that the GUS enzyme is stable and accumulates in the plant tissues, we used the GUS reporter gene instead of LUC. The GUS activity was induced in response to l-Pro and hypoosmolarity in Arabidopsis plants containing the 3D5-GUS construct (Fig. 5). Moreover, we observed l-Pro- and hypoosmolarity-induced accumulation of the GUS transcript in the 1.4-kb promoter-GUS or 3D5-GUS transgenic Arabidopsis (Fig. 6). These results indicate that the 3D5 fragment and the 1.4-kb ProDH promoter respond to DW, l-Pro, and d-Pro but not to GM and dehydration at transcriptional level and support that the 3D5 fragment contains a cis-acting element for positive transcriptional regulation by Pro and hypoosmolarity.
We also measured the LUC activities of T2 transgenic tobacco plants containing the 3D5-LUC, 3D6-LUC, or 3D7-LUC constructs after rehydration by DW (data not shown). These transgenic tobacco plants showed the increase of LUC activities after rehydration. We have shown that the ProDH gene is induced by rehydration in Arabidopsis plants using RNA gel-blot analysis in our previous reports (Kiyosue et al., 1996; Nakashima et al., 1998). Our present data based on transgenic studies indicate that the 3D5 fragment of the ProDH promoter is sufficient to respond to rehydration.
The 3D5 fragment contains a 9-bp ACTCATCCT sequence. The 3D6 and 3D7 fragments contain the first and second ACTCATCCT sequences, respectively (Fig. 3). These fragments also functioned in the l-Pro- and hypoosmolarity-inducible expression, whereas 3D8 and 3D9, with no ACTCATCCT sequence, did not. These results suggest that the ACTCATCCT sequence may function as a cis-acting factor involved in l-Pro- and hypoosmolarity-inducible expression of ProDH. Then, we showed, by using base-substituted DNA fragments, that the ACTCATCCT sequence and the 5′-flanking sequence are required for l-Pro- and hypoosmolarity-responsive expression of ProDH, and that the ACTCAT sequence is the core element in tobacco plants (Fig. 4B). Furthermore, we carried out RNA gel-blot analysis to show that the ACTCAT sequence is also core element in Arabidopsis plants using T3 transgenic Arabidopsis plants containing the 1.4-kb promoter-GUS, 3D5-GUS, 3D6-GUS, M3-GUS, M4-GUS, M5-GUS, or M6-GUS constructs (Fig. 6). The M4-GUS and M5-GUS constructs that contain mutations in the ACTCAT sequence did not responded to any treatments, in spite of the M3-GUS and M6-GUS constructs responded to DW, l-Pro and d-Pro treatments. These suggest that the ACTCAT sequence functions as a cis-acting element for Pro- and hypoosmolarity-responsive expression of ProDH in Arabidopsis plants. Some transcription factors may bind to the ACTCAT sequence and positively regulate ProDH gene expression at high concentrations of Pro or strong hypoosmolarity. We designated the ACTCAT sequence Pro- or hypoosmolarity-responsive element (PRE).
We found that the ACTCAT sequence has high homology with the TGACTC sequence, which is a binding site for GCN4 protein (Struhl, 1982; Donahue et al., 1983; Hinnebusch et al., 1985). The TGACTC sequence occurs as multiple copies in the promoter regions of genes subject to general amino acids control system in yeast and functions as a cis-acting element involved in derepression by the general control mechanism (Struhl, 1982; Donahue et al., 1983; Hinnebusch et al., 1985). The GCN4 protein binds to the TGACTC sequence and functions as a positive transcription factor (Arndt and Fink, 1986). These reports lead to the possibility that GCN4-like proteins might be positive activators for l-Pro- and hypoosmolarity-inducible expression of ProDH by binding to the ACTCAT sequence in the ProDH promoter.
We sought another transcriptional activator, qa-1F, which binds to a 16-bp sequence containing a strand complementary to ACTCAT (Geever et al., 1989). Neurospora crassa, a filamentous fungus, contains a qa gene cluster that controls quinate-shikimate metabolism (Baum et al., 1987). The transcription levels of the qa genes are coordinately controlled by the positive and negative regulators qa-1F and qa-1S, respectively (Geever et al., 1983, 1989). qa-1F encodes an activator protein required for its own mRNA synthesis, called autoregulation, and for the synthesis of other qa mRNAs, including qa-1S (Geever et al., 1983, 1989). A qa-like transcription factor may be involved in the ACTCAT transcription system in the l-Pro- or hypoosmolarity-inducible ProDH expression.
When we stained transgenic Arabidopsis plants containing the 1.4-kb promoter-GUS construct treated by hypoosmolarity or l-Pro, we observed strong GUS staining in whole plants (Fig. 5, D and E). In contrast, in transgenic Arabidopsis plants containing the 3D5-GUS construct, GUS staining was strong in leaves and stems but weak in roots (Fig. 5, G and H). We previously reported that GUS activity in roots increased when transgenic Arabidopsis seeds containing the 1.4-kb promoter-GUS construct were germinated in water (Nakashima et al., 1998). However, GUS activity was not increased in the germination of transgenic Arabidopsis seeds containing the 3D5-GUS construct (data not shown). The 1.4-kb promoter region of ProDH has an as-1 sequence (TGACG, −560 to −556) that is involved in gene expression in roots (Katagiri et al., 1989). An as-1 cis-acting element or other elements may be involved in ProDH expression in roots of Arabidopsis.
Arabidopsis plants exposed to drought or high-salt stress accumulate a high content of Pro (Yoshiba et al., 1995; Kiyosue et al., 1996; Nanjo et al., 1999b). In these cases, ProDH expression is repressed even if Pro is accumulated in stressed Arabidopsis plants (Kiyosue et al., 1996). We showed that the GUS transcript in transgenic Arabidopsis plants containing the 3D5-GUS construct did not increase after 2 h of dehydration (Fig. 6). We also observed that the GUS activity in the transgenics did not increase after 5 h of dehydration or high-salinity treatment with 250 mm NaCl solution (data not shown). These results suggest that the 90-bp region between −750 and −661 of the ProDH promoter also contains negative regulatory elements for l-Pro-inducible expression of the gene under water stress. This region may contain binding sites for negative transcription factors that inhibit l-Pro-inducible expression, in spite of the accumulation of Pro under water stress. There seem to be several transcription factors that bind to the ACTCAT sequence. Some may inactivate ProDH expression under dehydration and others may activate ProDH expression after release from water stress.
To elucidate whether the promoter region of the other l-Pro-inducible genes have the ACTCAT sequence, we used an Arabidopsis full-length cDNA microarray (Table I; Fig. 7). We found that 27 l-Pro-induced genes have the ACTCAT sequence in their promoter regions, and we showed that 19 genes among the 27 genes have similar induction patterns of that of ProDH by RNA gel-blot analysis. These results suggest that the ACTCAT sequence is conserved in many l-Pro-inducible promoters and plays a key role in l-Pro-inducible expression. Two l-Pro-inducible genes have the ACTCAT sequence encode cell wall-associated proteins (RAFL07–10-K05, RAFL04-09-O24, Table I). Pro and Hyp are components of cell wall as Pro-rich proteins and Hyp-rich glycoproteins, respectively (Nanjo et al., 1999b), which suggest that these two may be controlled by l-Pro treatment. However, some l-Pro-inducible genes do not have the ACTCAT sequences in their promoter region, which suggests the existence of other cis-acting elements for l-Pro-inducible gene expression.
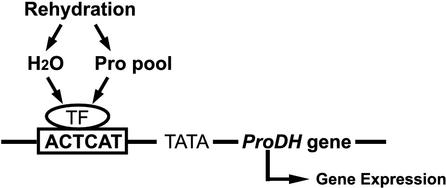
We propose a model for l-Pro- and hypoosmolarity-inducible expression of ProDH based on our results (Fig. 8). During rehydration, plant cells are exposed to high concentration of l-Pro accumulated during dehydration and hypoosmolarity. The ProDH promoter contains the ACTCAT sequence, which may be involved in ProDH expression via l-Pro and hypoosmolarity. To determine the relationship between l-Pro content and DW treatment in Arabidopsis, we measured the l-Pro content in Arabidopsis plants after incubation in DW (data not shown). There was no significant change in the l-Pro content within 2 h after incubation in DW, and but it decreased within 5 h. This l-Pro decrease may be caused by the induction of ProDH expression under hypoosmosis. These results show that DW treatment does not increase the l-Pro content, and that the DW and l-Pro treatments independently affect the induction of ProDH expression (Fig. 8). A high concentration of l-Pro and hypoosmolarity caused by the absorption of water independently regulate the expression of ProDH through the cis-acting element, PRE, with ACTCAT motif under rehydration.
Figure 8.
A model for the induction of ProDH under rehydration. ProDH expression is up-regulated by rehydration after dehydration, through l-Pro accumulation and hypoosmolarity. The transcription factor (TF) may be activated by Pro signaling or hypoosmolarity and bind to the ACTCAT sequence. The expression of ProDH may then be induced.
Because we determined a novel cis-acting element for the l-Pro- and hypoosmolarity-responsive expression of ProDH, we tried to identify DNA-binding proteins that bind to PRE using nuclear extract from l-Pro- or hypoosmolarity-treated Arabidopsis plants (data not shown). However, we could not detect any PRE-binding proteins in the nuclear extract. PRE-binding proteins might be unstable in the nuclear extract. To isolate cDNA encoding the PRE-binding proteins, we are now screening Arabidopsis cDNA library prepared from rehydrated plants using a yeast one-hybrid system. Identification of the transcription factors that bind to PRE will help us understand the molecular mechanisms of rehydration process.
MATERIALS AND METHODS
Transgenic Plants
The 5′-deleted fragments of the ProDH promoter region were synthesized by the PCR using 5′ primers containing a HindIII site at the 5′ end and 3′ primers containing a BamHI site at the 5′ end. The PCR products were cloned into the HindIII and BamHI sites of the pBluescript II vector (Stratagene, La Jolla, CA), and the resulting plasmids were confirmed by sequencing and then digested with HindIII and BamHI. They were ligated into the HindIII and BamHI sites of the promoterless vector pBI101.1 containing the LUC reporter gene instead of the GUS reporter gene. The 3′-deleted or base-substituted fragments of the ProDH promoter region were synthesized by PCR by using 5′ primers containing a HindIII site at the 5′ end and 3′ primers containing a SalI site at the 5′ end. The PCR products were cloned into the HindIII and SalI sites of the pBluescript II vector, and the resulting plasmids were digested with HindIII and SalI. They were ligated into the HindIII and SalI sites of the promoterless GUS or LUC expression vector pBI101.1 containing the minimal promoter region of ProDH (−42 to +122). Tobacco (Nicotiana tabacum cv SR1) and Arabidopsis (Columbia ecotype) were transformed as previously described (Valvekens et al., 1988; Benfey et al., 1989).
Plant Growth and Treatments
T1 transgenic tobacco plants or T2 transgenic tobacco seedlings were grown on Murashige and Skoog agar medium containing 100 μm kanamycin at 25°C under continuous light (2,000 lux). Leaves or whole plants with four to five leaves were removed from the medium and frozen in liquid nitrogen immediately (control) or transferred to plates containing DW for hypoosmolarity or 0.09 m l-Pro and incubated under dim light (100 lux) for 24 h. T2 or T3 seedlings of transgenic Arabidopsis were grown on GM agar plates containing 20 μm kanamycin at 22°C between 3 and 4 weeks under continuous light (2000 lux) as previously described (Nakashima et al., 1998). Unbolted whole plants were pulled out from the agar medium and were frozen in liquid nitrogen immediately (control) or were transferred to plates containing DW, GM solution, GM with 0.09 m l-Pro instead of 0.09 m Suc (l-Pro), or GM with 0.09 m d-Pro instead of 0.09 m Suc (d-Pro) or were subjected to dehydration stress and incubated under dim light (100 lux) for 2 or 24 h.
Assay of LUC Activity
LUC activity was assayed in leaf extracts of T1 transgenic tobacco plants or whole-plant extracts of 2- to 3-week-old T2 transgenic tobacco or Arabidopsis plants subjected to several treatments for 24 h with the Pica Gene LUC assay kit (Toyo-Ink, Tokyo) according to the manufacturer's instructions. Protein concentration of the extracts was determined by the Bradford method (Bio-Rad, Hercules, CA). We measured light intensity of the extract containing 5 μg of protein for 30 s and represented it as relative LUC activities.
Assay of GUS Activity and Histochemistry
For analysis of GUS activity in young seedlings, the transgenic Arabidopsis plants subjected to several treatments for 24 h were grown on GM agar plates for 2 to 3 weeks under continuous light (3,000 lux; Valvekens et al., 1988). GUS activity was assayed in extracts of the seedlings by fluorometric determination of the production of 4-methyl umbelliferone from the glucuronide precursor using a standard protocol (Jefferson et al., 1986). GUS activity was histochemically localized by prefixing 14-d-old whole transgenic plants in 0.3% (v/v) formaldehyde in 50 mm sodium phosphate (pH 7) for 10 min (Hatton et al., 1995), incubating them in 1 mm 5-bromo-4-chloro-3-indolyl glucuronide at 37°C for 4 h or overnight, fixing, and then dehydrating in a 50% to 100% (v/v) ethanol series (Nakashima et al., 1998).
RNA Gel-Blot Analysis
Total RNA was isolated from unbolted whole Arabidopsis plants subjected to several treatments for 2 h as previously described (Kiyosue et al., 1993). Fragments of the coding region of GUS or the ProDH cDNA were labeled by the random-primer method with [α-32P]dCTP (Amersham Biosciences AB, Uppsala) using the random-primed DNA-labeling kit (Roche diagnostics, Mannheim, Germany). The labeled fragments were hybridized with RNA according to standard protocols (Kiyosue et al., 1994).
Arabidopsis Full-Length cDNA Microarray Analysis
Arabidopsis (Columbia ecotype) plants were grown on GM agar at 22°C for 3 weeks under continuous light (2,000 lux) as previously described (Nakashima et al., 1998). Unbolted whole plants were transferred to plates containing GM or 0.09 m l-Pro and incubated under dim light (100 lux) for 24 h. Total RNA was isolated from l-Pro-treated or GM-treated whole plants using TRIZOL Reagent (Invitrogen, Carlsbad, CA). One milligram of total RNA was used for isolation of mRNA by MACS mRNA Isolation Kit (Miltenyi Biotec, Bergisch Glabach, Germany). Three micrograms of mRNA was employed on microarray analysis (Seki et al., 2001a). mRNA samples from l-Pro-treated plants for 24 h were fluorescently labeled with Cy3-dUTP, and samples from GM-treated plants for 24 h were labeled with Cy5-dUTP. After hybridization with the full-length cDNA microarray and scanning, relative expression ratios were calculated.
ACKNOWLEDGMENTS
We thank Atsuko Iuchi, Kyoko Murai, Ekuko Ohgawara, Fumie Saito, Mie Yamamoto, and Satomi Yoshida of Japan International Research Center for Agricultural Sciences for their excellent technical assistance.
Footnotes
This work was supported in part by the Program for the Promotion of Basic Research Activities for Innovative Biosciences. R.S. was supported by the Cooperative System for Supporting Priority Research of the Japan Science and Technology Corporation.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.009993.
LITERATURE CITED
- Ahmad I, Hellebust JA. The relationship between inorganic nitrogen metabolism and proline accumulation in osmoregulatory responses of two euryhaline microalgae. Plant Physiol. 1988;88:348–354. doi: 10.1104/pp.88.2.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt K, Fink GR. GCN4 protein, a positive transcription factor in yeast, binds general control promoters at all 5′ TGACTC 3′ sequences. Proc Natl Acad Sci USA. 1986;83:8516–8520. doi: 10.1073/pnas.83.22.8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum JA, Geever R, Giles NH. Expression of qa-1F activator protein: identification of upstream binding sites in the qa gene cluster and localization of the DNA-binding domain. Mol Cell Biol. 1987;7:1256–1266. doi: 10.1128/mcb.7.3.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey PN, Ren L, Chua N-H. The CaMV 35S enhancer contains at least two domains which can confer different developmental and tissue-specific expression patterns. EMBO J. 1989;8:2195–2202. doi: 10.1002/j.1460-2075.1989.tb08342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delauney AJ, Hu C-AA, Kishor PBK, Verma DPS. Cloning of ornithine-δ-aminotransferase cDNA from Vigna aconitifolia by trans-complementation in Escherichia coli and regulation of proline biosynthesis. J Biol Chem. 1993;268:18673–18678. [PubMed] [Google Scholar]
- Delauney AJ, Verma DPS. Proline biosynthesis and osmoregulation in plants. Plant J. 1993;4:215–223. [Google Scholar]
- Donahue TF, Daves RS, Lucchini G, Fink GR. A short nucleotide sequence required for regulation of HIS4 by the general control system of yeast. Cell. 1983;32:89–98. doi: 10.1016/0092-8674(83)90499-3. [DOI] [PubMed] [Google Scholar]
- Geever RF, Case ME, Tyler BM, Buxton F, Giles NH. Point mutations and DNA rearrangements 5′ to the inducible qa-2 gene of Neurospora allow activator protein-independent transcription. Proc Natl Acad Sci USA. 1983;80:7298–7302. doi: 10.1073/pnas.80.23.7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geever RF, Huiet L, Baum JA, Tyler BM, Patel VB, Rutledge BJ, Case ME, Giles NH. DNA sequence, organization and regulation of the qa gene cluster of Neurospora crassa. J Mol Biol. 1989;207:15–34. doi: 10.1016/0022-2836(89)90438-5. [DOI] [PubMed] [Google Scholar]
- Hatton D, Sablowski R, Yung M-H, Smith C, Schuch W, Bevan M. Two classes of cis sequences contribute to tissue-specific expression of a PAL2 promoter in transgenic tobacco. Plant J. 1995;7:859–876. doi: 10.1046/j.1365-313x.1995.07060859.x. [DOI] [PubMed] [Google Scholar]
- Hellebust JA. Osmoregulation. Annu Rev Plant Physiol. 1976;27:485–505. [Google Scholar]
- Hinnebusch AG, Lucchini G, Fink GR. A synthetic HIS4 regulatory element confers general amino acid control on the cytochrome c gene (CYC1) of yeast. Proc Natl Acad Sci USA. 1985;82:498–502. doi: 10.1073/pnas.82.2.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C-AA, Delauney AJ, Verma DPS. A bifunctional enzyme (Δ1-pyrroline-5-carboxylate synthetase) catalyzes the first two steps in proline biosynthesis in plants. Proc Natl Acad Sci USA. 1992;89:9325–9358. doi: 10.1073/pnas.89.19.9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Burgess SM, Hirsh D. β-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc Natl Acad Sci USA. 1986;83:8447–8451. doi: 10.1073/pnas.83.22.8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri F, Lam E, Chua N-H. Two tobacco DNA-binding proteins with homology to the nuclear factor CREB. Nature. 1989;340:727–730. doi: 10.1038/340727a0. [DOI] [PubMed] [Google Scholar]
- Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K. Characterization of cDNA for a dehydration-inducible gene that encodes a Clp A, B-like protein in Arabidopsis thaliana L. Biochem Biophys Res Commun. 1993;196:1214–1220. doi: 10.1006/bbrc.1993.2381. [DOI] [PubMed] [Google Scholar]
- Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K. Cloning of cDNA for genes that are early responsive to dehydration-stress (ERDs) in Arabidopsis thaliana L.: identification of three ERDs as HSP cognate genes. Plant Mol Biol. 1994;25:791–798. doi: 10.1007/BF00028874. [DOI] [PubMed] [Google Scholar]
- Kiyosue T, Yoshiba Y, Yamaguchi-Shinozaki K, Shinozaki K. A nuclear gene encoding mitochondrial proline dehydrogenase, an enzyme involved in proline metabolism, is upregulated by proline but downregulated by dehydration in Arabidopsis. Plant Cell. 1996;8:1323–1335. doi: 10.1105/tpc.8.8.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour MMF. Protection of plasma membrane of onion epidermal cells by glycinebetaine and proline against NaCl stress. Plant Physiol Biochem. 1998;36:767–772. [Google Scholar]
- McCue KF, Hanson AD. Drought and salt tolerance: towards understanding and application. Trends Biotechnol. 1990;8:358–362. [Google Scholar]
- Measures JC. Role of amino acids in osmoregulation of non-halophilic bacteria. Nature. 1975;257:398–400. doi: 10.1038/257398a0. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Satoh R, Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K. A gene encoding proline dehydrogenase is not only induced by proline and hypoosmolarity, but is also developmentally regulated in the reproductive organs of Arabidopsis. Plant Physiol. 1998;118:1233–1241. doi: 10.1104/pp.118.4.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanjo T, Kobayashi M, Yoshiba Y, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K. Antisense suppression of proline degradation improves tolerance to freezing and salinity in Arabidopsis thaliana. FEBS Lett. 1999a;461:205–210. doi: 10.1016/s0014-5793(99)01451-9. [DOI] [PubMed] [Google Scholar]
- Nanjo T, Kobayashi M, Yoshiba Y, Sanada Y, Wada K, Tsukaya H, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K. Biological functions of proline in morphogenesis and osmotolerance revealed in antisense transgenic Arabidopsis thaliana. Plant J. 1999b;18:185–193. doi: 10.1046/j.1365-313x.1999.00438.x. [DOI] [PubMed] [Google Scholar]
- Peng Z, Lu Q, Verma DPS. Reciprocal regulation of Δ1-pyrroline-5-carboxylate synthetase and proline dehydrogenase genes controls proline levels during and after osmotic stress in plants. Mol Gen Genet. 1996;253:334–341. doi: 10.1007/pl00008600. [DOI] [PubMed] [Google Scholar]
- Roosens NHCJ, Thu TT, Iskandar HM, Jacobs M. Isolation of the ornithine-δ-aminotransferase cDNA and effect of salt stress on its expression in Arabidopsis thaliana. Plant Physiol. 1998;117:263–271. doi: 10.1104/pp.117.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K. Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell. 2001a;13:61–72. doi: 10.1105/tpc.13.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Yamaguchi-Shinozaki K, Carninci P, Kawai J, Hayashizaki Y, Shinozaki K. Arabidopsis encyclopedia using full-length cDNAs and its application. Plant Physiol Biochem. 2001b;39:211–220. [Google Scholar]
- Singh NK, Aspinall D, Paleg LG. Proline accumulation and varietal adaptability to drought in barley: potential metabolic measure of drought resistance. Nature (New Biol) 1972;236:188–190. doi: 10.1038/newbio236188a0. [DOI] [PubMed] [Google Scholar]
- Smirnoff N, Cumbes QJ. Hydroxyl radical scavenging activity of compatible solutes. Phytochemistry. 1989;28:1057–1060. [Google Scholar]
- Strizhov N, Ábrahám E, Ökrész L, Blickling S, Zilberstein A, Schell J, Koncz C, Szabados L. Differential expression of two P5CS genes controlling proline accumulation during salt-stress requires ABA and is regulated by ABA1, ABI1 and AXR2 in Arabidopsis. Plant J. 1997;12:557–569. doi: 10.1046/j.1365-313x.1997.00557.x. [DOI] [PubMed] [Google Scholar]
- Struhl K. Regulatory sites for his3 gene expression in yeast. Nature. 1982;300:285–286. doi: 10.1038/300284a0. [DOI] [PubMed] [Google Scholar]
- Valvekens D, Montagu MV, Lijsebettens MV. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbruggen N, Hua X-J, May M, Montagu MV. Environmental and developmental signals modulate proline homeostasis: evidence for a negative transcriptional regulator. Proc Natl Acad Sci USA. 1996;93:8787–8791. doi: 10.1073/pnas.93.16.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton EF, Clark CJ, Boldingh HL. Effects of hydrogen cyanamide on amino acid profiles in kiwifruit buds during budbreak. Plant Physiol. 1991;97:1256–1259. doi: 10.1104/pp.97.3.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiba Y, Kiyosue T, Katagiri T, Ueda H, Mizoguchi T, Yamaguchi-Shinozaki K, Wada K, Harada Y, Shinozaki K. Correlation between the induction of a gene for Δ1-pyrroline-5-carboxylate synthetase and the accumulation of proline in Arabidopsis thaliana under osmotic stress. Plant J. 1995;7:751–760. doi: 10.1046/j.1365-313x.1995.07050751.x. [DOI] [PubMed] [Google Scholar]
- Yoshiba Y, Kiyosue T, Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K. Regulation of levels of proline as an osmolyte in plants under water stress. Plant Cell Physiol. 1997;38:1095–1102. doi: 10.1093/oxfordjournals.pcp.a029093. [DOI] [PubMed] [Google Scholar]
- Yoshiba Y, Nanjo T, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Stress-responsive and developmental regulation of Δ1-pyrroline-5-carboxylate synthetase 1 (P5CS1) gene expression in Arabidopsis thaliana. Biochem Biophys Res Commun. 1999;261:766–772. doi: 10.1006/bbrc.1999.1112. [DOI] [PubMed] [Google Scholar]