Abstract
The production of anthocyanins in fruit tissues is highly controlled at the developmental level. We have studied the expression of flavonoid biosynthesis genes during the development of bilberry (Vaccinium myrtillus) fruit in relation to the accumulation of anthocyanins, proanthocyanidins, and flavonols in wild berries and in color mutants of bilberry. The cDNA fragments of five genes from the flavonoid pathway, phenylalanine ammonia-lyase, chalcone synthase, flavanone 3-hydroxylase, dihydroflavonol 4-reductase, and anthocyanidin synthase, were isolated from bilberry using the polymerase chain reaction technique, sequenced, and labeled with a digoxigenin-dUTP label. These homologous probes were used for determining the expression of the flavonoid pathway genes in bilberries. The contents of anthocyanins, proanthocyanidins, and flavonols in ripening bilberries were analyzed with high-performance liquid chromatography-diode array detector and were identified using a mass spectrometry interface. Our results demonstrate a correlation between anthocyanin accumulation and expression of the flavonoid pathway genes during the ripening of berries. At the early stages of berry development, procyanidins and quercetin were the major flavonoids, but the levels decreased dramatically during the progress of ripening. During the later stages of ripening, the content of anthocyanins increased strongly and they were the major flavonoids in the ripe berry. The expression of flavonoid pathway genes in the color mutants of bilberry was reduced. A connection between flavonol and anthocyanin synthesis in bilberry was detected in this study and also in previous data collected from flavonol and anthocyanin analyses from other fruits. In accordance with this, models for the connection between flavonol and anthocyanin syntheses in fruit tissues are presented.
Fruit development from flower to ripe fruit is a complex process that involves modification of cellular compartments, loss of cell wall structure causing softening, and accumulation of carbohydrates (Brady, 1987). The production of secondary metabolites during the ripening process is an essential phenomenon for the contribution of seed dispersal of the plant in the form of accumulation of pigments and flavor compounds. The significance of secondary products in defense against diseases in developing fruits should also be remembered (Harborne, 1997; Mercier, 1997).
Flavonoids are a large group of phenolic secondary metabolites that are widespread among plants and are involved in many plant functions. Anthocyanins, a flavonoid subclass, are the main pigments in flowers and fruits, acting as insect and animal attractants (Bohm, 1998; Harborne and Williams, 2000). Anthocyanins are synthesized via the phenylpropanoid pathway (Fig. 1). Anthocyanin biosynthesis has been extensively studied in several plant species, and, therefore, detailed information of the course of reactions is available. Two classes of genes are required for anthocyanin biosynthesis, the structural genes encoding the enzymes that directly participate in the formation of anthocyanins and other flavonoids, and the regulatory genes that control the transcription of structural genes. The enzyme activities in the various branch pathways are highly regulated. Transcriptional controls play an important role in regulating the overall activity of flavonoid biosynthesis. The pathway is also controlled in response to different developmental and environmental cues (for review, see Koes et al., 1994; Holton and Cornish, 1995; Mol et al., 1998; Weisshaar and Jenkins, 1998; Winkel-Shirley, 2001). There is also evidence that the enzymes involved in flavonoid metabolism might be acting as membrane-associated multienzyme complexes, which have implications on overall efficiency, specificity, and regulation of the pathway (Stafford, 1991; Winkel-Shirley, 1999, 2001).
Figure 1.
A schematic presentation of the anthocyanin biosynthetic pathway, with emphasis on the flavonols, proanthocyanidins, and anthocyanidins found in bilberry. Enzyme abbreviations: C4H, Cinnamate 4-hydroxylase4; 4CL, 4-coumaroyl:CoA ligase; CHI, chalcone isomerase; F3′H, flavonoid 3′ hydroxylase; F3′5′H, flavonoid 3′5′ hydroxylase; LCR, leucoanthocyanidin reductase; UFGT, UDP Glc-flavonoid 3-O -glucosyl transferase; MT, methyltransferase.
Bilberry (Vaccinium myrtillus), or European blueberry, is among the most significant wild berries in northern Europe. Blueberries (Vaccinium) are recognized for their high anthocyanin content, which is believed to provide health benefits. Bilberry, of all blueberries, contains exceptionally high amounts of anthocyanins (Kalt and Dufour, 1997; Prior et al., 1998). The content and composition of anthocyanins in bilberries, as well as in blueberries, has been determined earlier by Suomalainen and Keränen (1961), Martinelli et al. (1986), and Wang et al. (2000). Several studies have also been done on the bioactive properties of bilberries and blueberries (Bomser et al., 1996; Kalt and Dufour, 1997; Youdim et al., 2000), especially concerning the antioxidant activity of their anthocyanin fractions (Prior et al., 1998; Smith et al., 2000; Ehlenfeldt and Prior, 2001). However, to our knowledge, there are no published studies on flavonoid biosynthesis in blueberries with particular regard to the expression of genes in the phenyl propanoid pathway.
The color of bilberries varies normally blue to almost black. As rare variants, bilberry mutants with white or pink fruits have been found in nature (Fig. 2, B and C). In these mutants, the taste and the size of the fruit are the same as in wild-type bilberry, and the color of the berries is the only difference. Therefore, it is assumed that the change in color in these bilberry mutants would be due to the mutation in structural or regulatory genes involved in anthocyanin biosynthesis.
Figure 2.
A, Bilberry from flower to ripe fruit. Six different stages were examined in this study. Stages: 1, flower; 2, small-sized green fruits; 3, middle-sized green fruits; 4, half-expanded, just after coloring began; 5, nearly expanded, half-colored, red fruits; 6, fully colored, blue, ripe fruits. B, The color mutation of bilberry with white berries. C, The color mutation of bilberry with pink berries.
This study describes the isolation of cDNA fragments of five genes involved in flavonoid biosynthesis from bilberry, and the study of expression of the genes during the development of fruit from flower to ripe berry (Fig. 2A), and in parallel, the accumulation of flavonols, proanthocyanidins, and anthocyanins. The color mutation forms of bilberry, with white or pink fruits, were studied as well.
RESULTS
Isolation and Sequence Analysis of the cDNA Fragments of the Structural Genes Involved in Flavonoid Biosynthesis
The sizes of isolated cDNA fragments of the bilberry flavonoid pathway genes, Phe ammonia-lyase (PAL), chalcone synthase (CHS), flavanone 3-hydroxylase (F3H), dihydroflavonol 4-reductase (DFR), and anthocyanidin synthase (ANS), ranged from 171 to 476 bp. The size of the fragment of glyceraldehyde-3-phosphate dehydrogenase (GPD) gene, used as control, was 333 bp (AY123769). For all fragments, similarity to the corresponding genes from other species was detected. In comparison with some related, previously reported sequences, each fragment exhibited 65% to 93% identity to the corresponding genes from other species in nucleotide sequences (Table I). More than one different sequence was found from all the isolated cDNA fragments in sequencing analysis, but the ones showing highest homology with genes from other species were selected for the probes for the gene expression analysis.
Table I.
Comparison of deduced amino acid sequences of structural genes involved in anthocyanin biosynthesis from bilberry with the known related nucleotide sequences
| Gene | Species | Amino Acid Sequences
|
Ref. | |
|---|---|---|---|---|
| Length/range of similarity | Identity | |||
| % | ||||
| PAL | Bilberry | 258 | AY123770 | |
| Raspberry | 1,655–1,867 | 79.5 | AF304366 | |
| Grape | 318–576 | 79.6 | X75967 | |
| Gerbera | 16–244 | 84.0 | Z38099 | |
| Arabidopsis | 1,374–1,622 | 75.2 | AY045919 | |
| CHS | Bilberry | 170 | AY123765 | |
| Raspberry | 1,140–1,301 | 84.0 | AF292367 | |
| Grape | 812–972 | 86.3 | AB015872 | |
| Gerbera | 390–549 | 88.7 | Z38096 | |
| Arabidopsis | 590–759 | 87.0 | AF1122086 | |
| F3H | Bilberry | 476 | AY123766 | |
| Grape | 490–951 | 72.6 | X75965 | |
| Arabidopsis | 1,376–1,747 | 72.6 | U33932 | |
| DFR | Bilberry | 227 | AY123767 | |
| Grape | 566–845 | 74.1 | X75964 | |
| Gerbera | 579–694 | 78.5 | Z17221 | |
| Arabidopsis | 1,371–1,601 | 73.5 | AJ251982 | |
| ANS | Bilberry | 286 | AY123768 | |
| LDOX | Grape | 715–1,000 | 84.9 | X75966 |
| LDOX | Arabidopsis | 660–945 | 77.9 | U70478 |
The sequence analyses were made with the GCG, Inc. bestfit analysis with the gap creation penalty = 20 and gap extension penalty = 3. Ref., Accession nos. of the nucleotide sequences in the DDBJ, EMBL, and GenBank databases.
Expression of the Flavonoid Pathway Genes in Developing Fruits
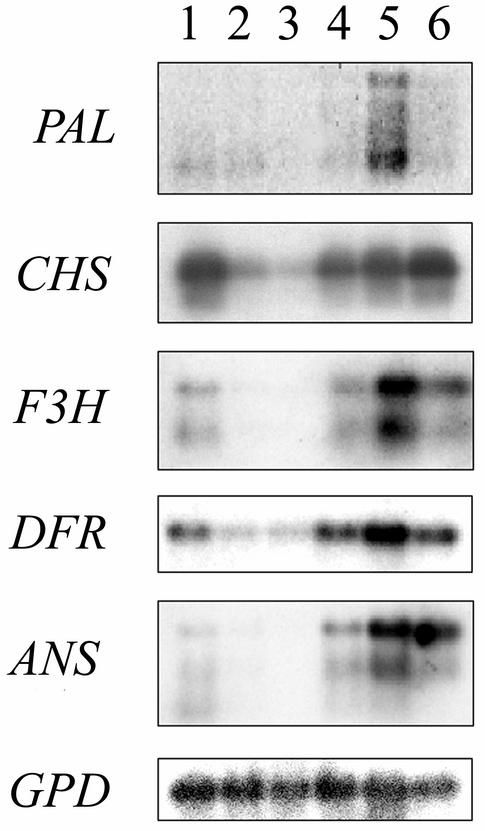
The expression of five flavonoid pathway genes (PAL, CHS, F3H, DFR, and ANS) was investigated in samples taken at six different stages of fruit development of bilberries (Fig. 3). Data indicated that the expression of flavonoid pathway genes occurs in two phases. The genes are highly expressed in flowers and especially at the stages of ripening where the color development in fruits occurs. All of the genes examined were expressed in flowers and in berries. The expression of CHS and DFR was observable throughout the ripening, but was reduced at stages 2 and 3 compared with the later stages of ripening. The expression of flavonoid pathway genes was highest at stage 5 when the berry was still pale inside but the skin was already red. In ripe bilberries, the expression began to decrease again.
Figure 3.
Temporal expression of the anthocyanin pathway genes of bilberry during berry development, probed with bilberry cDNA fragments for PAL, CHS, F3H, DFR, and ANS. Numbers (1–6) indicate the different stages examined from flower to ripe berry. The same membrane was also rehybridized with a GPD probe to show the equal loading of the RNA and cDNA amounts of the samples.
Expression of the Flavonoid Pathway Genes in Color Mutants
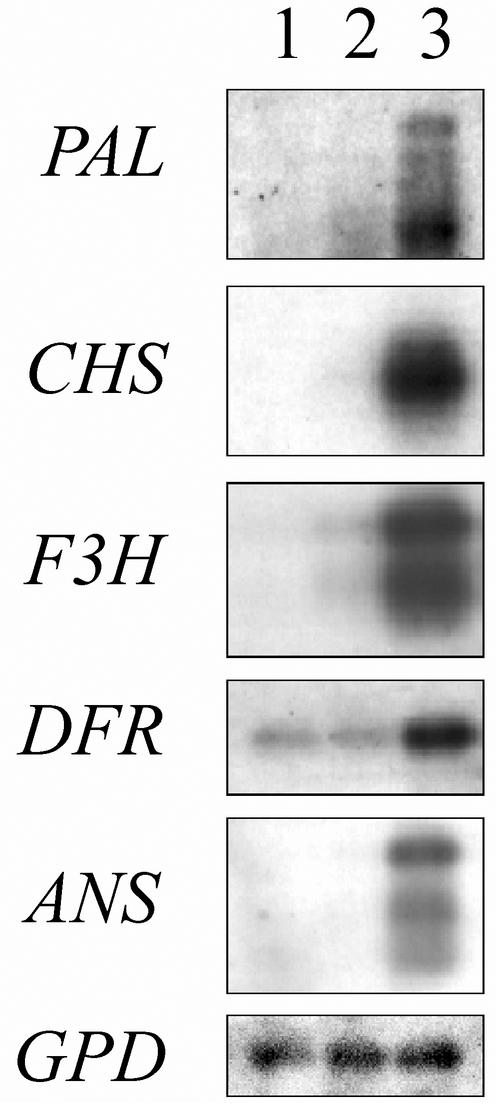
The expression of the five target genes in color mutation forms of bilberry was studied at the ripening stage 5, when the expression in wild-type bilberries was at its maximum. In both of the color mutants (pink and white), the expression of flavonoid biosynthetic genes was reduced compared with the wild-type bilberries (Fig. 4). In the pink mutant, the expression of the flavonoid pathway genes studied, except for ANS, was at a detectable level, whereas in the white mutant, only the expression of PAL and DFR was detected.
Figure 4.
Expression of the anthocyanin pathway genes of bilberry in color mutation forms of bilberry, probed with bilberry cDNA fragments for PAL, CHS, F3H, DFR, and ANS. Five micrograms of RNA translated to cDNA of the white bilberry (lane 1), of the pink bilberry (lane 2), and of the wild bilberry (lane 3) was used. The same membrane was also rehybridized with GPD probe to show the loading of the equal RNA and cDNA amounts of the samples.
Accumulation of Flavonols, Proanthocyanidins, and Anthocyanins
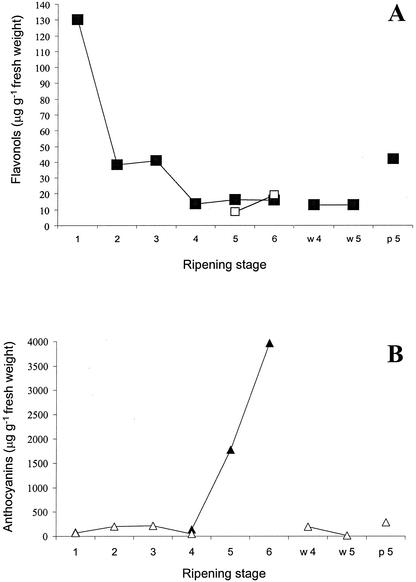
The composition and contents of flavonols, proanthocyanidins, and anthocyanins were determined in all samples of the wild and color mutation forms of bilberries. Water content increased from 78% (w/w) in raw bilberry to 85% (w/w) during the progress of ripening, but this variation did not affect valid comparison of ripening stages on fresh weight basis. The content of the major flavonol of bilberry, quercetin, was highest in the flower of the wild bilberry (130 μg g−1 fresh weight) and at the beginning of berry development (stages 2–3), whereas it decreased at the later stages of ripening (Fig. 5A). At stage 4, another flavonol, myricetin, was distinctly detected and quantified. During the progress of ripening, the content of myricetin doubled and reached the level of quercetin in the ripe berry (stage 6). In the white color mutant, the content of quercetin (13 μg g−1) was at the same level as in the wild bilberries at the same maturity stages (4 and 5), and was somewhat higher (42 μg g−1) in the pink color mutant. Myricetin was not found in the color mutants.
Figure 5.
A, Contents of the flavonols quercetin (▪) and myricetin (□) during development and ripening of bilberries. B, Contents of anthocyanins (▴) and proanthocyanins (▵) during the same stages as in A. Numbers 1 through 6 indicate the different stages of ripening examined. W4 and W5, White mutant at the ripening stages 4 and 5. P5, Pink mutant at the ripening stage 5.
The content of oligo- and polymeric procyanidins (i.e. proanthocyanidins analyzed as cyanidin) was estimated to be as about 50 μg g−1 in the wild-type flowers. Procyanidins were the major flavonoids in the early developmental stages of wild bilberries and in the pink mutants (200, 216, and 286 μg g−1, respectively). The content of procyanidins in the white mutant was found to decrease during ripening from 193 to 12 μg g−1 (Fig. 5B). Also, in the wild-type bilberry, the content of procyanidins decreased to an undetectable level before the synthesis of anthocyanins. Prodelphinidins (i.e. proanthocyanidins analyzed as delphinidin) were not found in flowers but could be detected in small green wild-type fruits (stages 2–3) at the level below the quantification limit (<10 μg g−1). The oligo- and polymeric proanthocyanidins were not quantifiable from stage 5 in the highly pigmented bilberries.
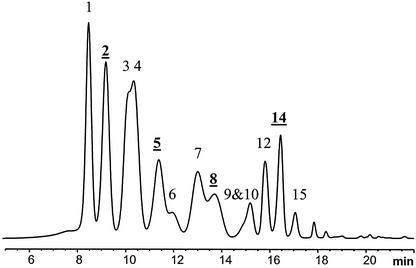
The HPLC-diode array detector (DAD) profile of anthocyanins (anthocyanidin glycosides) in the ripe wild fruits is shown in Figure 6. The profile of anthocyanins in the flower consisted of cyanidin 3-O-β-galactoside, -glucoside, and -arabinoside. The total content of the cyanidin glycosides in the flower samples was 86 μg g−1 (as aglycons; Fig. 5B). Anthocyanins were quantifiable at stage 4 of the wild bilberry (140 μg g−1), after which the content increased strongly, reaching 3,960 μg g−1 in the ripe fruit (Fig. 5B). At the beginning of the anthocyanin biosynthesis (stage 4), seven highly abundant anthocyanins were detected and quantified using HPLC-DAD, whereas at stage 5, 13 anthocyanins could be quantified. With the more sensitive electrospray ionization mass spectrometry (ESI-MS) method, all 15 anthocyanins were detected (Table II). The identification was based on combining the ESI-MS data of anthocyanins with the well-known chromatographic profile of anthocyanins of bilberry (Kader et al., 1996).
Figure 6.
HPLC-DAD profiles of anthocyanins found in flower and ripe bilberry. Standards were available for the anthocyanins with underlined peak numbers. Peak numbers refer to Table II.
Table II.
ESI-MS identification data of anthocyanins found in ripe bilberry
| ESI-MS Identification
|
Sugar | Anthocyaninsc | Rta | ||
|---|---|---|---|---|---|
| Peaksa | MS | MS-MSb | |||
| M+ | M+-sugar | min | |||
| 1 | 465 | 303 | 162 | Delphinidin-3-galactoside | 8.4 |
| 2 | 465 | 303 | 162 | Delphinidin-3-glucoside | 9.1 |
| 3 | 449 | 287 | 162 | Cyanidin-3-galactoside | 10.0 |
| 4 | 435 | 303 | 132 | Delphinidin-3-arabinoside | 10.3 |
| 5 | 449 | 287 | 162 | Cyanidin-3-glucoside | 11.3 |
| 6 | 479 | 317 | 162 | Petunidin-3-galactoside | 11.5 |
| 7 | 419 | 287 | 132 | Cyanidin-3-arabinoside | 12.9 |
| 8 | 479 | 317 | 162 | Petunidin-3-glucoside | 13.7 |
| 9 | 463 | 301 | 162 | Peonidin-3-galactoside | 14.8 |
| 10 | 449 | 317 | 132 | Petunidin-3-arabinoside | 15.1 |
| (11) | 463 | 301 | 162 | Peonidin-3-glucoside | nd |
| 12 | 493 | 331 | 162 | Malvidin-3-galactoside | 15.9 |
| (13) | 433 | nd | Peonidin-3-arabinoside | nd | |
| 14 | 493 | 331 | 162 | Malvidin-3-glucoside | 16.1 |
| 15 | 463 | 331 | 132 | Malvidin-3-arabinoside | 16.4 |
nd, Not determined.
Refer to peak numbers and retention times (Rt) in Figure 6. Peaks in parentheses were not detected with HPLC-DAD.
MS signal of peak 13 was overlapped and not fragmented in MS-MS.
Tentative identification data combined with literature (Kader et al., 1996).
DISCUSSION
The beginning of the fruit development after fertilization involves the cell division and cell expansion phases. The ripening is an aspect of development and is initiated after seed maturation has been completed. Tissue softening and accumulation of pigments occur during the ripening phase (Gillaspy et al., 1993). The development of bilberries from flower to ripe fruit lasts usually 8 to 10 weeks, varying between different years (Sjörs, 1989). In bilberry, the ripening phase lasts 2 to 3 weeks, and the accumulation of anthocyanins is rapid during that period.
To clarify the flavonoid biosynthesis in developing bilberries, cDNA fragments of five structural genes encoding PAL, CHS, F3H, DFR, and ANS were isolated. Fragments were subcloned and five different clones were sequenced from all genes. Therefore, more than one isoform was found from each gene. This suggests that all of the examined flavonoid pathway genes in bilberry would represent multigene families, which is also the case with many other species (Holton and Cornish, 1995). The sequences of the isoforms were in some cases highly similar, which also explains the multiple bands for F3H and ANS as well as for PAL in gene expression analysis. Multiple bands of F3H and ANS genes have also been observed in gene expression analysis of other related studies, for example, with Perilla frutescens (Gong et al., 1997).
Anthocyanin production is limited in most plants to certain tissues, and it occurs during specific stages of development. The visible accumulation of these compounds usually reflects the activity of biosynthetic enzymes functioning in the pathway (Koes et al., 1994). In bilberry flowers, the expression of flavonoid pathway genes agreed with the accumulation of anthocyanins, as three different anthocyanins were determined and expression of all flavonoid biosynthetic genes, including ANS, was detected. The profile of anthocyanins found in flowers in this study was the same as has previously been found in bilberry cell cultures (Madhavi et al., 1998). In bilberry fruits, the anthocyanin synthesis occurs first in the epidermal cell layers, after which the inner cells of the berry also become fully pigmented. Our results show that the mRNA levels encoding PAL, CHS, F3H, DFR, and ANS in developing bilberries increase in concurrence with the accumulation of anthocyanins. The expression of anthocyanin pathway genes was specifically up-regulated at the period when anthocyanin accumulation grows most rapidly. In ripe berries, the expression levels started to drop again. Similar results have been obtained in the developmental studies of pea (Pisum sativum), snapdragon (Antirrhinum majus), and petunia (Petunia hybrida) flowers (Jackson et al., 1992; Quattrocchio et al., 1993; Uimari and Strömmer, 1998). The results obtained in this study provide additional evidence for the correlation between the expression of structural flavonoid pathway genes and anthocyanin production during fruit development. On the other hand, the way the flavonoid pathway genes in bilberry were expressed at the very beginning of the fruit development and again at the end of ripening (Fig. 3) is similar to what has been found in strawberry (Fragaria spp.; Manning, 1998) and in grape berry (Boss et al., 1996; Kobayashi et al., 2001).
The flavonols quercetin and myricetin are found in bilberry (Häkkinen and Auriola, 1998; Häkkinen et al., 1999). In plants, flavonols have been found to possess a protective role as a UV filter, and they also may function as copigments for anthocyanins in fruits and flowers (Koes et al., 1994; Bohm, 1998). In the present study, the amount of quercetin was interestingly highest in flowers and at the beginning of berry development.
The levels of oligo- and polymeric proanthocyanidins were also high at the beginning of berry development, which agrees with the results from gene expression analysis. The expression of DFR, which reduces dihydroflavonols to leucoanthocyanidins (flavan-3, 4-diols), was detected throughout the berry development. Leucoanthocyanidins are the precursors of anthocyanins and proanthocyanidins (Bohm, 1998). The amounts of monomeric (-)-epicatechin and (+)-catechin, dimeric, oligo-, and polymeric proanthocyanidins have also been found to decrease in the progression of ripening in grape (Vitis vinifera; Boss et al., 1996), in bilberry (Morazzoni and Bombardelli, 1996), and in other fruit tissues (Wrangham and Waterman, 1983; Harborne, 1997; Mercier, 1997). According to the present results, the same phenomenon was detectable also in the white bilberry mutant. It has been suggested that the presence of proanthocyanidins in unripe fruits could provide protection against too early feeding, as the taste of proanthocyanidins is astringent (Harborne, 1997). Proanthocyanidins have also been found to protect developing fruit tissues against fungal pathogens (Mercier, 1997).
The color mutants of flowers have been attracting plant scientists from the very beginning of plant study. The detailed knowledge of the course of events of the flavonoid pathway is based on the work done with color mutation forms of maize (Zea mays) and ornamental plants like petunia, snapdragon, and Arabidopsis (Koes et al., 1994; Holton and Cornish, 1995; Mol et al., 1998; Quattrocchio et al., 1999; Winkel-Shirley, 2001). It has been found that the change in flower color may be due to the mutation in structural genes or regulatory genes of the flavonoid pathway. At the molecular level, the regulatory mutants for anthocyanin biosynthesis are characterized by reduced mRNA amounts for several structural genes and lower levels of the corresponding enzymes (Uimari and Strömmer, 1998). Little is known about regulatory genes in fruit tissues. In the present study, the reduction in levels of PAL, CHS, F3H, DFR, and ANS mRNA was observed in color mutation forms of bilberry with white or pink berries. Likewise, the total content of flavonols in the ripe white bilberry mutant was one-half of the amount analyzed in the corresponding wild type. Also notable was the absence of myricetin in the color mutants. Instead, quercetin and proanthocyanidins were detected. We previously made a similar observation in the study of phenolic compounds in berries of black and red currants (Ribes sp.) and their unpigmented green and white variants, respectively (Määttä et al., 2001). The composition of flavonol glycosides was the same, but the contents were lower in unpigmented variants compared with black and red currants. Further identification with ESI-MS revealed that the contents of myricetin glycosides were distinctly lower and the contents of quercetin glycosides were higher in green currants compared with black currants (K. Määttä, unpublished data).
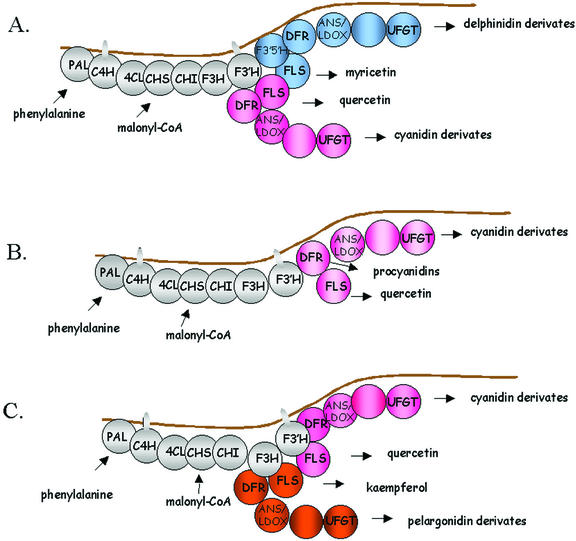
An interesting observation was also the absence of the flavonol and anthocyanidins synthesized directly from dihydrokaempferol, namely kaempferol and pelargonidin in bilberries. Dihydrokaempferol is also the precursor for dihydroquercetin and dihydromyricetin, and flavonol synthase (FLS) catalyzes the dehydrogenation of these 3-hydroxyflavanones to the corresponding flavonols (Fig. 1; Bohm, 1998). In strawberry, flavonols kaempferol and quercetin are found, and pelargonidin is the main anthocyanidin (Häkkinen and Törrönen, 2000; Nyman and Kumpulainen, 2001). To further analyze this observation, and to combine the present information, we assembled a table of flavonol and anthocyanidin contents in different fruits (Table III). The data collected in Table III shows that a similar trend is also observed in other fruits. In accordance with this, in fruit tissues, there appears to be three predominant models for flavonol and anthocyanin synthesis, which are presented in Figure 7. Quercetin from flavonols and cyanidin derivative anthocyanins are found in all fruits, which agrees with the knowledge that cyanidin-derived anthocyanins are regarded to be more primitive in evolution than pelargonidin- or delphinidin-derived anthocyanins (Harborne and Williams, 2000). In addition to quercetin- and cyanidin-derived anthocyanins, myricetin- and delphinidin-derived anthocyanins or kaempferol- and pelargonidin-derived anthocyanins may be produced in different fruits. The connection between myricetin- and delphinidin-derived anthocyanins is explained by the activity of flavonoid 3′5′ hydroxylase, which hydroxylates the 3′ and 5′ position of the dihydrokaempferol or dihydroquercetin and is required for biosynthesis of dihydromyricetin, the precursor of myricetin- and delphinidin-derived anthocyanins. Quercetin was produced also, regardless of anthocyanin production in raw berries and in color mutants of bilberry. Still, the connection between the occurrence of quercetin- and cyanidin-derived anthocyanins is evident when analyzing the data collected from other fruits.
Table III.
Distribution of flavonols and anthocyanins in fruits
| Fruit | Flavonol
|
Anthocyanin Derivative
|
||||
|---|---|---|---|---|---|---|
| Quercetin | Myricetin | Kaempferol | Cyanidin | Delphinidin | Pelargonidin | |
| Family Empetraceae | ||||||
| Crowberry (Empetrum nigrum) | x | x | x | x | ||
| Family Ericaceae | ||||||
| Bilberry | x | x | x | x | ||
| Blueberry (Vaccinium corymbosum) | x | x | x | x | ||
| Bog whortleberry (Vaccinium uliginosum) | x | x | x | x | ||
| Cranberry (Vaccinium oxycoccos) | x | x | x | |||
| Lingonberry (Vaccinium vitis-idaea) | x | x | ||||
| Family Grossulariaceae | ||||||
| Black currant (Ribes nigrum) | x | x | xa | x | x | |
| Red currant (Ribes x pallidum) | x | x | ||||
| Red gooseberry (Ribes uva-crispa) | x | x | x | |||
| Family Rosaceae | ||||||
| Apple (Malus spp.) | x | x | ||||
| Chokeberry (Aronia mitschurinii) | x | x | x | |||
| Nectarine (Prunus persica) | x | x | ||||
| Peach (Prunus persica) | x | x | ||||
| Plum (Prunus domestica) | x | x | x | |||
| Red raspberryb (Rubus idaeus) | x | x | x | x | ||
| Rosehip (Rosa spp.) | x | x | ||||
| Sour cherry (Prunus cerasus) | x | x | x | x | ||
| Strawberry | x | x | x | x | ||
| Sweet cherry (Prunus avium) | x | x | x | x | ||
| Rowanberry (Sorbus aucuparia) | x | x | x | |||
| Family Vitaceae | ||||||
| Grape | x | x | xa | x | x | |
Data were collected from Häkkinen et al. (1999), Kumpulainen et al. (2001), Tomas-Barberan et al. (2001), and Macheix et al. (1990). x, The occurrence of compound.
In black currant and grape, small amounts of kaempferol are found from some varieties whereas quercetin and myricetin are the predominant flavonols.
Kaempferol and pelargonidin derivatives are not found in all raspberry varieties.
Figure 7.
Models for the organization of flavonoid pathway enzymes for production of anthocyanidins and flavonols in fruits, supposedly as macromolecular complexes at the endoplasmic reticulum. A, Model for the production of myricetin and quercetin in connection to delphinidin- and cyanidin-derived anthocyanins (e.g. bilberry, blueberries, grape, and black currant). In grape and black currant, small amounts of kaempferol are found, but quercetin and myricetin are the predominant flavonols. B, Model for the production of quercetin in connection with cyanidin-derived anthocyanins (e.g. lingonberry, apple, rosehip , and flowers and callus cultures of bilberry, etc.). C, Model for the production of kaempferol and quercetin in connection with cyanidin- and pelargonidin-derived anthocyanins (strawberry, raspberry [partially]). Modified from Winkel-Shirley (1999). Enzyme names are abbreviated as in Figure 1.
It appears that in fruits, FLSs and DFRs are specialized in using as substrates dihydromyricetin or dihydrokaempferol alternatively, in addition to dihydroquercetin. In flowers, it has been found that FLSs, or dihydroflavonol 4-reductases specialized to converting certain flavonols, may also be able to use different dihydroflavonols as substrates to a lesser extent (Holton and Cornish, 1995; Bohm, 1998; Johnson et al., 2001). This explains the small amounts of kaempferol found in species where quercetin and myricetin are the predominant flavonols (e.g. black currant), as dihydrokaempferol, being the precursor of dihydroquercetin and dihydromyricetin, is present in all species.
As flavonoids are recognized for their beneficial effects for human health, a lot of research is still needed to clarify the bioactive effects of different flavonoid compounds. For better understanding and progressing with the breeding work of different fruits, an important research target would be to clarify the detailed course of reactions and controlling system involved in the flavonoid biosynthesis of fruit tissues. Another challenging aspect would also be to clarify the effects of environmental factors on flavonoid biosynthesis in fruits. Future research with the flavonoid biosynthesis will involve the study of the gene families and isolation of other structural and regulatory genes of the flavonoid pathway from different fruits.
In conclusion, our results demonstrate the coordinated expression of flavonoid biosynthetic genes in relation to the accumulation of anthocyanins, proanthocyanidins, and flavonols in developing fruits of bilberry. The expression of flavonoid pathway genes in color mutant forms of bilberry was reduced, and no myricetin or anthocyanins were found from them. The connection between flavonol and anthocyanidin synthesis in bilberry was detected in this study and also in data collected from flavonol and anthocyanidin analyses from other fruits. In accordance with this, models for the connection of flavonol and anthocyanin synthesis in fruits are presented.
MATERIALS AND METHODS
Plant Material
The flowers and fruits of wild bilberries (Vaccinium myrtillus), growing in the natural forest stands in Oulu, Finland, were harvested at six different ripening stages (Fig. 2A). The color mutation forms of bilberry, i.e. bilberries with white or pink fruits (Fig. 2, B and C), were obtained from a test field at the Botanical Garden, University of Oulu. Flowers and fruits for the RNA isolation and flavonoid analysis were collected directly into liquid nitrogen and were stored at −70°C until used.
Isolation of Total RNA
Total RNA was isolated from bilberry flowers and berries with the method described for pine (Pinus spp.) trees by Chang et al. (1993), modified by Jaakola et al. (2001a). The quality of the isolated RNA was verified on 1% (w/v) ethidium bromide-stained agarose gel and from the absorbance spectrum at wavelengths from 220 to 300 nm.
PCR Cloning and Sequencing
The cDNA was prepared from 10 μg of bilberry fruit total RNA, which was reverse-transcribed by M-MuLV reverse transcriptase (Invitrogen, Carlsbad, CA) from an anchored oligo-dT primer using standard methods in a reaction volume of 20 μL. Fragments of flavonoid pathway genes and of GPD gene were amplified from the cDNA by the PCR. Partially degenerated primers designed for gerbera were used for amplifying fragments of PAL, CHS, and DFR coding sequences (Helariutta et al., 1993, 1995), and 5′-ACGTGTCGACIGG(T/C) TGIAC(A/C/G/T) GT(T/A/G) ATCCA-3′ and 5′-ACGTCAGCTG(T/C) GA(A/G) GA(T/C) TGGGGIAT(T/C/A) TT-3′ of F3H. For amplifying fragment of the ANS gene coding sequence, partially degenerated primers 5′-T(C/G) CAAA(T/A) GAAGAT(A/C) AACTACTACCC(A/C) A-3′ and 5′-CA(G/A) AA(A/G) ACAGCCCA(A/T) GAAA(C/T) CCTIACC-3′ were designed based on homologies found in genes isolated earlier. The primers 5′-GCTCCCAGCAAGGATGCCCC-3′ and 5′-CGGAAGGCCATTCCAGTCAACT-3′ were designed for amplifying the fragment of GPD gene. Dynazyme DNA-polymerase (Finnzymes, Espoo, Finland) was used for amplification. Conditions for PCR of PAL and CHS fragments were 94°C for 75 s, 55°C for 2 min, and 72°C for 2 min, with 25 cycles. For DFR, F3H, and ANS fragments, PCR was performed using a “touch down” strategy: 10 times (94°C for 75 s; 50°C for 5 min adding 1°C per cycle, slope +22°C, per 10 s; 72°C for 5 min) followed by 31 times (94°C for 75 s; 53°C for 2 min; and 72°C for 5 min). The PCR products were cloned into a pUC19 vector (Sambrook and Russel, 2001). Sequencing reactions were carried out using a BigDye Terminator Cycle Sequencing Kit (PE Applied Biosystems, Warrington, UK). A DNA sequencer (model 377; PE Applied Biosystems) was used for sequencing.
Gene Expression Analysis
As the high anthocyanin content of bilberries caused problems in the conventional northern-blotting procedure using a nonradioactive detection method, gene expression in developing bilberries was studied with the method developed by Jaakola et al. (2001b). The method is based on using cDNA instead of RNA for the blotting step. The RNA samples were translated to cDNA directly after isolation. Equal sample amounts were verified by measuring the amount of RNA with spectrophotometry and in gel before transcription to cDNA. The cDNAs were then separated by electrophoresis, stained with ethidium bromide to further verify the equal sample amounts, and blotted onto nylon membranes by Southern transfer. Probes consisting of PCR-amplified fragments of the flavonoid pathway genes from bilberry were labeled with digoxigenin-dUTP according to the manufacturer's instructions (Roche Diagnostics, Mannheim, Germany). Hybridization was performed overnight at 42°C in Ultrahyb hybridization solution (Ambion, Austin, TX) in a probe concentration of 1 ng mL−1. The membrane was washed twice in 2× SSC and 0.1% (w/v) SDS for 5 min and twice in 0.5× SSC and 0.1% (w/v) SDS for 15 min at 42°C. Detection was performed with the DIG Nucleic Acid Detection kit and CDP-Star (Roche Diagnostics) according to manufacturer's instructions. The membrane was exposed to film (XAR; Kodak, Rochester, NY) for 13 min. The negative was developed and photographed using the Fluor-S Quantity One program (Bio-Rad, Hercules, CA). The same membrane used for the flavonoid gene expression analysis was rehybridized with GPD probe for showing equal loading of the samples.
Analysis of Water Content
To measure the water content, bilberry samples were freeze-dried (Dura-Dry Condenser; FTS Systems, Stone Ridge, TX). The sample sizes of white and pink bilberry mutants were not sufficient for the analysis of water content.
Analysis of Flavonoids
Frozen berries (10 g) were crushed and powdered using a mortar and pestle. Two subsamples of 1 to 2 g were weighed and suspended in 10 mL of acidified methanol (0.6 m of HCl) by heating and mixing for 1 min. After sampling 1 mL of suspension for analysis of anthocyanins, the refluxing of samples was continued. Upon heating in acidic methanol, flavonol glycosides and anthocyanins were deconjugated to aglycons, and proanthocyanidins were converted to anthocyanidins. The refluxing time of 2 h was previously found to be optimal for the highest possible yield of flavonol aglycons (Häkkinen et al., 1999), as well as of anthocyanidins released from proanthocyanidins (Määttä et al., 2001). The samples were filtered through a 0.45-μm Regen cellulose syringe filter (TITAN, Gloucester, UK) prior to analysis.
An HPLC combined to a DAD was used for analysis. The chromatographic conditions (column and gradient systems) were as used previously by Määttä et al. (2001). Anthocyanins were further identified by HPLC with ESI-MS interface. The HPLC-ESI-MS apparatus and ionization conditions were as described by Häkkinen and Auriola (1998).
A mixture of 3-O-β-glucosides of delphinidin, cyanidin, petunidin, pelargonidin, peonidin, and malvidin (5 μm of each) was obtained from Polyphenols (Sandnes, Norway) and was dissolved in 20 mL of methanol for a stock solution. Cyanidin and delphinidin chlorides were purchased from Extrasynthese (Geney, France), quercetin was obtained from Sigma Chemical, and myricetin was purchased from Fluka (Buchs, Switzerland). These standards were dissolved in methanol to a concentration of approximately 1 mg mL−1 and they were stored at −20°C as stock solutions.
Retention times and spectra of the peaks at maximum absorption wavelengths (λmax) in DAD detection were matched with authentic standards for identification of flavonol aglycons as quercetin and myricetin (λmax = 360 nm), anthocyanidins as cyanidin and delphinidin (λmax = 520 nm), and anthocyanins as 3-O-β-glucosides of delphinidin, cyanidin, petunidin, peonidin, and malvidin (λmax = 520 nm). The other sugar derivatives of anthocyanidins were identified in ESI-MS as their flavylium cations (M+) and in sequential MS-MS fragmentation as aglycons (M+-sugar). The major ion in MS ionization was selected to MS-MS fragmentation; therefore, one overlapped minor anthocyanin flavylium cation was not fragmented (Table II). The sugar moieties of anthocyanins found in bilberry were specified according to previous publications (Kader et al., 1996).
Standard curves for quantification were prepared using authentic standards as follows: flavonols 2 to 110 μg mL−1, anthocyanidins 2 to 130 μg mL−1, and anthocyanins 1.5 to 70 μg mL−1 (as aglycon) in methanol. The concentration of acid (0.6 m) in methanolic solutions of anthocyanins and anthocyanidins was adjusted to the same level as in the samples by adding HCl. In the pigmented ripening stages of bilberries (stages 4–6), the content of proanthocyanidins was assessed to be zero, and upon heating in acidic methanol about 80% (w/v) of cyanidin glycosides was found to be deconjugated to cyanidin aglycons. The contents of proanthocyanidins (as cyanidin) in the flower and in the slightly pigmented bilberries (stages 2–3) were estimated by subtracting 80% (w/v) of the content of the aglycon from the total quantified content of cyanidins.
ACKNOWLEDGMENTS
We thank Taina Uusitalo for technical assistance in molecular biological analysis and Dr. Seppo Auriola for ESI-MS analysis. We also thank Prof. Teemu Teeri and his “Gerbera group,” especially Dr. Paula Elomaa, for kindly providing the primers for PAL, CHS, DFR, and F3H genes, and for helpful advice, especially at the beginning of the work.
Footnotes
This work was supported by the Eemil Aaltonen foundation and by the Oscar Öflund foundation. The research is a part of the Cooperation Program of the University of Oulu and Kuusamo Town and was financed by the European Union (to E.S.F.), by the Regional Council of Northern Ostrobothnia, and by Kuusamo Town.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.006957.
LITERATURE CITED
- Bohm B. Introduction of Flavonoids. Singapore: Harwood Academic Publishers; 1998. [Google Scholar]
- Bomser J, Madhavi DL, Singletary K, Smith MAL. In vitro anticancer activity of fruit extracts from Vaccinium species. Planta Med. 1996;62:212–216. doi: 10.1055/s-2006-957862. [DOI] [PubMed] [Google Scholar]
- Boss PK, Davies C, Robinson SP. Analysis and the expression of anthocyanins pathway genes in developing Vitis vinifera L. cv Shiraz grape berries and the implications for pathway regulation. Plant Physiol. 1996;11:1059–1066. doi: 10.1104/pp.111.4.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady CJ. Fruit ripening. Annu Rev Plant Physiol. 1987;38:155–178. [Google Scholar]
- Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep. 1993;11:113–116. [Google Scholar]
- Ehlenfeldt M, Prior R. Oxygen radical absorbance capacity (ORAC) and phenolic and anthocyanin concentrations in fruit and leaf tissues of highbush blueberry. J Agric Food Chem. 2001;49:2222–2227. doi: 10.1021/jf0013656. [DOI] [PubMed] [Google Scholar]
- Gillaspy G, Ben-David H, Gruissem W. Fruits: a developmental perspective. Plant Cell. 1993;5:1449–1451. doi: 10.1105/tpc.5.10.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Yamazaki M, Sugiyama M, Tanaka Y, Saito K. Cloning and molecular analysis of structural genes involved in anthocyanin biosynthesis and expressed in a forma-specific manner in Perilla frutescens. Plant Mol Biol. 1997;35:915–927. doi: 10.1023/a:1005959203396. [DOI] [PubMed] [Google Scholar]
- Häkkinen SH, Auriola S. High-performance liquid chromatography with electrospray ionization mass spectrometry and diode array ultraviolet detection in the identification of flavonol aglycones and glycosides in berries. J Chromatogr. 1998;829:91–100. doi: 10.1016/s0021-9673(98)00756-0. [DOI] [PubMed] [Google Scholar]
- Häkkinen SH, Kärenlampi SO, Heinonen IM, Mykkänen HM, Törrönen AR. Content of the flavonols quercetin, myricetin, and kaempferol in 25 edible berries. J Agric Food Chem. 1999;47:2274–2279. doi: 10.1021/jf9811065. [DOI] [PubMed] [Google Scholar]
- Häkkinen SH, Törrönen AR. Content of flavonols and selected phenolic acids in strawberries and Vaccinium species: influence of cultivar, cultivation site and technique. Food Res Int. 2000;33:517–524. [Google Scholar]
- Harborne J. Tomas-Barberan F, ed, Phytochemistry of Fruit and Vegetables. New York: Oxford University Press; 1997. Phytochemistry of fruits and vegetables: an ecological overview; pp. 353–367. [Google Scholar]
- Harborne J, Williams C. Advances in flavonoid research since 1992. Phytochemistry. 2000;55:481–504. doi: 10.1016/s0031-9422(00)00235-1. [DOI] [PubMed] [Google Scholar]
- Helariutta Y, Elomaa P, Kotilainen M, Griesbach RJ, Schröder J, Teeri TH. Chalcone synthase-like genes active during corolla development are differentially expressed end encode enzymes with different catalytic properties in Gerbera hybrida (Asteraceae) Plant Mol Biol. 1995;28:47–60. doi: 10.1007/BF00042037. [DOI] [PubMed] [Google Scholar]
- Helariutta Y, Elomaa P, Kotilainen M, Seppänen P, Teeri TH. Cloning of cDNA coding for dihydroflavonol-4-reductase (DFR) and characterization of dfr expression in the corollas of Gerbera hybrida var. Regina (Compositae) Plant Mol Biol. 1993;22:183–193. doi: 10.1007/BF00014927. [DOI] [PubMed] [Google Scholar]
- Holton TA, Cornish EC. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell. 1995;7:1071–1083. doi: 10.1105/tpc.7.7.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakola L, Pirttilä AM, Hohtola A. cDNA blotting offers an alternative method for gene expression studies. Plant Mol Biol Rep. 2001b;19:125–128. [Google Scholar]
- Jaakola L, Pirttilä AM, Halonen M, Hohtola A. Isolation of high quality RNA from the bilberry (Vaccinium myrtillus L.) fruit. Mol Biotechnol. 2001a;19:201–203. doi: 10.1385/MB:19:2:201. [DOI] [PubMed] [Google Scholar]
- Jackson D, Roberts K, Martin C. Temporal and spatial control of expression of anthocyanin biosynthetic genes in developing flowers of Anthirrum majus. Plant J. 1992;2:425–434. [Google Scholar]
- Johnson ET, Ryu S, Yi H, Shin B, Cheong H, Choi G. Alteration of a single amino acid changes the substrate specificity of dihydroflavonol 4-reductase. Plant J. 2001;25:325–333. doi: 10.1046/j.1365-313x.2001.00962.x. [DOI] [PubMed] [Google Scholar]
- Kader F, Rovel B, Girardin M, Metche M. Fractionation and identification of the phenolics compounds of highbush blueberries (Vaccinium corymbosum L.) Food Chem. 1996;55:35–40. [Google Scholar]
- Kalt W, Dufour D. Health functionality of blueberries. HortTechnology. 1997;7:216–221. [Google Scholar]
- Kobayashi S, Ishimaru M, Ding CK, Yakushiji H, Goto N. Comparison of UDP-glucose:flavonoid 3-O-glucosyltransferase (UFGT) gene sequences between white grapes (Vitis vinifera) and their sports with red skin. Plant Sci. 2001;160:543–550. doi: 10.1016/s0168-9452(00)00425-8. [DOI] [PubMed] [Google Scholar]
- Koes R, Quattrocchio R, Mol J. The flavonoid biosynthetic pathway in plants: function and evolution. BioEssays. 1994;16:123–132. [Google Scholar]
- Määttä K, Kamal-Eldin A, Törrönen R. Phenolic compounds in berries of black, red, green, and white currants (Ribes sp.) Antioxid Redox Signal. 2001;3:981–993. doi: 10.1089/152308601317203521. [DOI] [PubMed] [Google Scholar]
- Macheix JJ, Fleuriet A, Billot J. Fruit Phenolics. Boca Raton, FL: CRC Press; 1990. [Google Scholar]
- Madhavi DL, Bomser J, Smith MAL, Singletary K. Isolation of bioactive constituents from Vaccinium myrtillus (bilberry) fruits and cell cultures. Plant Sci. 1998;131:95–103. [Google Scholar]
- Manning K. Isolation of a set of ripening-related genes from strawberry: their identification and possible relationship to fruit quality traits. Planta. 1998;205:622–631. doi: 10.1007/s004250050365. [DOI] [PubMed] [Google Scholar]
- Martinelli EM, Baj A, Bombardelli E. Computer-aided evaluation of liquid-chromatographic profiles for anthocyanins in Vaccinium myrtillus fruits. Anal Chim Acta. 1986;191:275–281. [Google Scholar]
- Mercier J. Tomas-Barberan F, ed, Phytochemistry of Fruit and Vegetables. New York: Oxford University Press; 1997. Role of phytoalexins and other antimicrobial compounds from fruits and vegetables in postharvest disease resistance; pp. 221–241. [Google Scholar]
- Mol J, Grotewold E, Koes R. How genes paint flowers and seeds. Trends Plant Sci. 1998;3:212–217. [Google Scholar]
- Morazzoni P, Bombardelli E. Vaccinium myrtillus L. Fitoterapia. 1996;67:3–29. [Google Scholar]
- Nyman A, Kumpulainen J. Determination of anthocyanins in berries and red wine by high-performance liquid chromatography. J Agric Food Chem. 2001;49:4183–4187. doi: 10.1021/jf010572i. [DOI] [PubMed] [Google Scholar]
- Prior RL, Cao G, Martin A, Sofic E, McEwen J, O'Brien C, Lischner N, Ehlenfeldt M, Kalt W, Krewer G et al. Antioxidant capacity as influenced by total phenolic and anthocyanin content, maturity, and variety of Vaccinium species. J Agric Food Chem. 1998;46:2686–2693. [Google Scholar]
- Quattrocchio FM, Wing J, Leppen H, Mol J, Koes R. Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. Plant Cell. 1993;5:1497–1512. doi: 10.1105/tpc.5.11.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio FM, Wing J, van der Woude K, de Vetten N, Mol J, Koes R. Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cell. 1999;11:1433–1444. doi: 10.1105/tpc.11.8.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russel DW. Molecular Cloning: A Laboratory Manual. 3rd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Sjörs H. Blåbär, Vaccinium myrtillus: ett växtporträtt. Svensk Bot Tidskr. 1989;83:411–428. [Google Scholar]
- Smith MAL, Marley KA, Seigler D, Singletary KW, Meline B. Bioactive properties of wild blueberry fruits. J Food Sci. 2000;65:352–356. [Google Scholar]
- Stafford H. Flavonoid evolution an enzymatic approach. Plant Physiol. 1991;96:680–685. doi: 10.1104/pp.96.3.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen H, Keränen A. The first anthocyanins appearing during the ripening of blueberries. Nature. 1961;191:498–499. [Google Scholar]
- Uimari A, Strömmer J. Anthocyanin regulatory mutations in pea: effects on gene expression and complementation by R-like genes of maize. Mol Gen Genet. 1998;257:198–204. doi: 10.1007/s004380050639. [DOI] [PubMed] [Google Scholar]
- Wang J, Kalt W, Sporns P. Comparison between HPLC and MALDI-TOF MS analysis of anthocyanins in highbush blueberries. J Agric Food Chem. 2000;48:3330–3335. doi: 10.1021/jf000101g. [DOI] [PubMed] [Google Scholar]
- Weisshaar B, Jenkins GI. Phenylpropanoid biosynthesis and its regulation. Curr Opin Plant Biol. 1998;1:251–257. doi: 10.1016/s1369-5266(98)80113-1. [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. Evidence for enzyme complexes in the phenylpropanoid and flavonoid pathways. Physiol Plant. 1999;107:142–149. [Google Scholar]
- Winkel-Shirley B. Flavonoid biosynthesis: a colorful model for genetics, biochemistry, cell biology and biotechnology. Plant Physiol. 2001;126:485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrangham RW, Waterman PG. Condensed tannins in fruits eaten by chimpanzees. Biotropica. 1983;15:217–222. [Google Scholar]
- Youdim K, Shukitt-Hale B, Martin A, Wang H, Denisova N, Bickford P, Joseph J. Short-term dietary supplementation of blueberry polyphenolics: beneficial effects on ageing brain performance and peripheral tissue function. Nutr Neurosci. 2000;3:383–397. [Google Scholar]