Abstract
The Arabidopsis TCH4 gene is up-regulated in expression by diverse environmental and hormonal stimuli. Because TCH4 encodes a xyloglucan endotransglucosylase/hydrolase, this change in expression may reflect a recruitment of cell wall-modifying activity in response to environmental stress and growth. How diverse stimuli lead to the common response of TCH4 expression regulation is not known. Here, we show that induction of expression by the diverse stimuli of touch, darkness, cold, heat, and brassinosteroids (BRs) is conferred to reporter genes by the same 102-bp 5′-untranscribed TCH4 region; this result is consistent with the idea that shared regulatory elements are employed by diverse stimuli. Distal regions influence magnitude and kinetics of expression and likely harbor regulatory elements that are redundant with those located more proximal to the transcriptional start site. Substitution of the proximal regulatory region sequences in the context of distal elements does not disrupt inducible expression. TCH4 expression induction is transcriptional, at least in part because 5′-untranscribed sequences are sufficient to confer this regulation. However, 5′-untranslated sequences are necessary and sufficient to confer the marked transience of TCH4 expression, most likely through an effect on mRNA stability. Perception of BR is not necessary for TCH4::GUS induction by environmental stimuli because regulation is intact in the BR-insensitive mutant, bri1-2. The full response to auxin, however, requires the functioning of BRI1. Developmental expression of TCH4 is unlikely to be meditated by BR because TCH4::GUS is expressed in BR perception and biosynthetic mutants bri1-2 and det2-1, respectively.
Plants are sensitive to a number of abiotic environmental stimuli including light, wind, and temperature. Changes in these environmental conditions often result in rapid and dramatic alterations in plant gene expression, and these molecular responses likely aid plants in acclimating to or withstanding the potential stresses of the environment.
There are sets of genes that change their expression level in response to light stimuli (Ma et al., 2001), others that show elevated expression in extreme heat (Sung et al., 2001), and others that are induced in expression by cold (Thomashow, 1999). The existence of distinct gene sets that respond to different stimuli suggests that specific receptors and signal transduction pathways are utilized in response to alterations in light and different temperature extremes to drive distinct gene expression changes.
In addition to genes whose expression is regulated in response to a single stimulus, there are genes that are induced in expression by multiple, diverse stimuli. For example, the TCH4 gene of Arabidopsis was originally discovered because of its dramatic response to the seemingly innocuous stimulus of touch (Braam and Davis, 1990). TCH4 encodes a xyloglucan endotransglucosylase/hydrolase (XTH, formerly abbreviated XET; Xu et al., 1995; Campbell and Braam, 1998). TCH4 is also up-regulated by darkness, heat shock, and cold shock (Braam and Davis, 1990; Braam, 1992; Polisensky and Braam, 1996). In addition, TCH4 expression is elevated by brassinosteroids (BRs) and auxin (indole-3-acetic acid [IAA]; Xu et al., 1995). Because TCH4 expression is strongly influenced by environmental and hormonal stimuli and the encoded protein acts on a major component of the plant cell wall, we hypothesize that TCH4 plays a role in cell wall modifications in response to environmental stress and during morphogenesis (Xu et al., 1995, 1996; Campbell and Braam, 1999). How these diverse stimuli lead to the common molecular response of TCH4 regulation of expression is unknown.
One possibility is that the TCH4 regulatory region may contain separate cis-acting elements, with each responding to activation of a separable signal transduction pathway. For example, the TCH4 locus, being elevated in expression by both heat and cold, would harbor the heat shock element and dehydration-responsive element, cis-elements defined to drive expression by heat and cold, respectively, in Arabidopsis (Baker et al., 1994; Yamaguchi-Shinozaki and Shinozaki, 1994; Schöffl et al., 1998). Alternatively, genes such as TCH4 may be controlled by a single cis-element that is responsive to multiple stimuli. In this scenario, various signal transduction pathways may converge at some point before the induction of TCH4 transcriptional activity. For example, various environmental stimuli could cause increases in an endogenous growth regulator, such as BRs, which could serve to mediate gene expression changes. Furthermore, there may exist multiple signal transduction pathways with various degrees of shared and separate components. One way to distinguish between these possible scenarios is to identify the region responsible for TCH4 expression regulation and determine if separate regulatory cis-elements exist. In addition, mutants defective in hormone biosynthesis and/or response can be used to investigate the potential roles of hormones in mediating gene expression responses to environmental stimuli.
We tested the transcriptional activity of sequences found within the TCH4 locus to localize regions controlling induction of TCH4 expression. We found that both transcriptional and posttranscriptional mechanisms are involved in TCH4 gene regulation. In addition, using BR biosynthesis and response mutants, we assessed the role of BR as a mediator of TCH4 regulation.
RESULTS
The Regulatory Region of TCH4
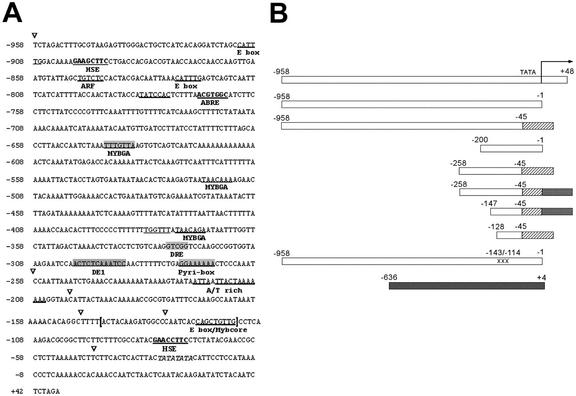
Approximately 1 kb of 5′-non-coding sequences, including the 5′-untranslated region (UTR), of the TCH4 locus were shown to confer upon a reporter gene the regulatory properties of TCH4 (Xu et al., 1995). To begin identifying the DNA sequences responsible for TCH4 regulation, we compared the TCH4 5′ sequences with known regulatory motifs (Fig. 1A). At position −299/−288 relative to the transcriptional start site (+1), 10 of 12 bases match a reverse complement DE1 element (GGATTTTACAGT) shown to be sufficient for darkness inducibility of expression (Inaba et al., 2000). There are three E box motifs (CANNTG) at positions −912/−907, −826/−821, and −122/−117. E boxes in eukaryotic genomes act as binding sites of basic helix-loop-helix transcription factors (Massari and Murre, 2000). The E box at position −122/−117 is overlapping with CTGTTG, a reverse complement of an MYB core motif (YAACNG). There are three additional MYB-related motifs [YAACA(A/G)A], found at −642/−636, −520/−514, and −376/−371. These sequences are similar to those important for the functioning of the GA-responsive element (Lovegrove and Hooley, 2000). GA-responsive elements can be found associated with a pyrimidine box (also called box 2) and a TATCCAC motif, elements thought to enhance regulation (Lovegrove and Hooley, 2000). Interestingly, sequences related to all three of these components are present in the upstream region of TCH4 but are in a different relative placement and spacing from the transcriptional start site than would be expected based on analyses of GA-regulated genes (Gubler and Jacobsen, 1992; Lanahan et al., 1992). A pyrimidine box is present at −275/−268 and a TATCCAC box resides at −794/−788. At −848/−843, there is an auxin response factor-binding site consensus sequence (TGTCTC) that is found in early auxin response genes (Ulmasov et al., 1997). An ACGT-containing abscisic acid (ABA) response element (Busk and Pages, 1998) resides at −771/−765. However, full ABA inducibility requires the presence of multiple copies of ABA response element or a coupling element (Shen and Ho, 1995; Shen et al., 1996; Hobo et al., 1999). Located at −328/−324 is an inverted copy CCGAC, a core sequence of the dehydration-responsive element (Baker et al., 1994; Yamaguchi-Shinozaki and Shinozaki, 1994) found in many cold- and drought-responsive genes. Two potential heat shock elements (Schöffl et al., 1998), each consisting of two copies of alternating units of nGAAn, are located at −899/−892 and −81/−74. However, because efficient binding of heat shock factor requires at least three tandem nGAAn units (Barros et al., 1992), it is unclear whether these sites would be sufficient to confer heat shock regulation.
Figure 1.
5′ TCH4 genomic sequence and schematic representation of TCH4 regions generated and tested for transcriptional regulatory activity. A, Genomic sequence of TCH4 upstream region sufficient to confer up-regulation of expression. The inverted triangles indicate the 5′ positions of TCH4 regions generated and tested for transcriptional regulatory activity (see B). Sequences related to defined regulatory motifs in the sense orientation are underlined, whereas inverted motifs are shaded in gray (motifs are defined in text). The putative TATA box is indicated in italics. Numbering refers to distance from transcriptional start site designated as +1. B, White rectangles represent TCH4 sequences between −958 and +48 and subregions fused to the GUS and LUC reporter genes. The numbering refers to distance relative to the transcriptional start site (+1). The arrow indicates the start position and direction of transcription. For some constructs, the endogenous promoter was replaced with −90- or −46-bp cauliflower mosaic virus (CaMV) 35S promoter (hatched, size indicated by rectangle length); some constructs also have a 143-bp region of a 5′-UTR from tobacco etch virus (TEV) (cross hatched). The penultimate rectangle from the bottom represents a construct containing TCH4 sequences from −958 to −1 that has substituted nucleotides between −143 and −114. The black rectangle at the bottom represents the sequences present from the UBQ10 locus for UBQ10::LUC. These representations of transgenes are not drawn to scale.
To test the functional relevance of these sequences and identify potential cis-regulatory regions, we generated subregions (Fig. 1B), fused them to the reporter genes encoding β-glucuronidase (GUS) and firefly luciferase (LUC), introduced them into Arabidopsis plants, and assayed gene activity in response to environmental and hormonal stimuli.
Role of TCH4 5′-UTR in the Transience of Induced Gene Expression
TCH4 induction of expression is remarkably transient in nature (Braam and Davis, 1990), indicating that if TCH4 is regulated by transcriptional induction, transcription initiation must cease rapidly and the TCH4 transcripts must be unstable. Previously, we showed that GUS transgenes containing −958 to +48 TCH4 sequences, which include the 5′-UTR, are up-regulated in expression with similar magnitude and kinetics as endogenous TCH4 (Xu et al., 1995). This result indicates that the TCH4 −958 to +48 sequences are sufficient to confer transient expression to a reporter gene. Because the only TCH4 sequences present in the transcript are the 48-base 5′-UTR, it is likely that this UTR is sufficient to result in mRNA instability. We further tested this possibility by analyzing the mRNA accumulation kinetics from a GUS transgene fused to only the TCH4 upstream −958 to −1 sequences. Removal of the 48-base UTR does not affect the inducibility of the transgene but results in a prolonged accumulation of GUS mRNAs relative to that of the endogenous TCH4 transcripts in plants stimulated by touch (Fig. 2A), darkness (Fig. 2B), or heat (Fig. 3A, III). These results are consistent with a role for the 5′-UTR of TCH4 in mRNA instability. Furthermore, these data indicate that only 5′-untranscribed sequences, and not sequences in the mRNA, are required for conferring touch, darkness, and heat inducibility to transgenes. Therefore, TCH4 is most likely increased in expression by transcriptional up-regulation and not by posttranscriptional mechanisms acting through the mRNA.
Figure 2.
The loss of 48 bp of the 5′-UTR of TCH4 affects the transient accumulation of mRNA after touch or darkness. A and B, Total RNA was isolated from plants after no stimulus (0) or after touch (A) or darkness (B). Plants were harvested at the indicated times. Four micrograms of total RNA was size fractionated on gels, blotted onto nylon membranes, and hybridized sequentially with the probes shown at right. The prime indicates minutes; “h” stands for hour(s). TUB4 (Tubulin) is shown as a loading and transfer control. The transgene assayed is represented by the rectangle at the bottom (not drawn to scale); see Fig. 1 legend for details.
Figure 3.
TCH4 sequences between −258 and −45 are sufficient to confer up-regulation of expression in response to heat, touch, and darkness. A, Plants harboring transgenes with TCH4 sequences between −258 and −45 (I), −958 and −45 (II) and the −90 CaMV 35S minimal promoter fused to GUS, and −958 and −1 (III) fused to GUS were unstimulated (0) or placed at 35°C. B and C, Plants harboring transgenes composed of TCH4 sequences between −258 and −45 and the 90 bp of the CaMV 35S promoter (“I”) or TCH4 sequences between −258 and −45, the 46 bp of the CaMV 35S promoter, and the TEV leader (“IV”) were left unstimulated (0) or stimulated with touch (B) or darkness (C). A through C, Plants were harvested at the indicated times. Eight micrograms of total RNA was size fractionated on gels, blotted to nylon filters, and hybridized sequentially to probes listed on the right. The lower band in the GUS panel in B and C is frequently observed and is nonspecific. The prime indicates minutes; h indicates hour(s). TUB4 is shown as a loading and transfer control. The transgenes assayed are represented by rectangles at the bottom of the figure (not drawn to scale); see Fig. 1 legend for details.
Figure 3 illustrates data relevant to identifying the 5′-regulatory sequences of TCH4 (to be discussed in the next section) and cis-elements affecting mRNA accumulation kinetics (addressed here). Surprisingly, transcripts with 5′-untranslated sequences from the TEV, which we added to the transgenes in efforts to enhance translation (Carrington and Freed, 1990; Nunberg et al., 1994), also accumulate only transiently. Figure 3, B and C, compare the accumulation kinetics of GUS mRNA derived from transgenes harboring the −258 to −45 TCH4 region and a CaMV 35S minimal promoter region with (transgene IV) or without (transgene I) the TEV 5′-UTR. For historical reasons, the transgenes without the TEV 5′-UTR have 90 bp of the CaMV 35S promoter, whereas transgenes with the TEV 5′-UTR have 46 bp of the CaMV 35S promoter. The −90/+8 CaMV 35S region was originally thought to serve as a minimal promoter; subsequently, it was determined that only the −46/−1 region is required for minimal promoter activity and lacks an element between −83 and −63 that can confer root-specific expression (Benfey et al., 1989; Lam et al., 1989). In response to touch (Fig. 3B) and darkness (Fig. 3C), TCH4 expression is transiently up-regulated with a prominent decrease in transcript accumulation by 2 h. Transcripts derived from the TCH4::GUS transgenes that harbor the TEV 5′-UTR have accumulation kinetics closely comparable with that of TCH4 transcripts (Fig. 3, BIV and CIV). The response of the TCH4::GUS transgenes lacking a TCH4 or TEV 5′-UTR is more prolonged with an abundance of GUS transcripts present up to 2 h poststimulation (Fig. 3, AI, BI, and CI). There are two possible explanations for this result. First, the TEV 5′-UTR may result in GUS mRNA instability. Alternatively, because the TCH4::GUS transgenes differ not only in the presence of the TEV 5′-UTR but also in the length of the CaMV 35S minimal promoter region, it is also possible that transcriptional initiation fails to shut off efficiently in the context of the −90 CaMV 35S promoter. Overall, these results indicate that the TCH4 and, most likely, the TEV UTRs, can confer mRNA instability to the GUS mRNA. The TCH4 5′-UTR, therefore, most likely plays a role in the transient nature of TCH4 induction of expression.
The Sequences between −258 and −45 Are Sufficient to Confer Response to Heat, Touch, and Darkness
We tested transgenic plants harboring subregions of the TCH4 sequences (Fig. 1B) fused to reporter genes for the ability to confer up-regulation of expression in response to stimuli known to lead to an increase in expression of TCH4. Subjecting plants to 35°C results in an increase in TCH4 mRNA within 20 min (Fig. 3A, TCH4). The level of TCH4 expression induction by heat can vary among plants (Fig. 3A, TCH4); therefore, we compared reporter gene expression levels with those of the native TCH4 gene. The GUS gene driven by the −258 to −45 region of the TCH4 locus and including the −90/+8 CaMV promoter is up-regulated in expression by high temperature, similar to TCH4; however, there is a delay in GUS mRNA accumulation compared with that of the endogenous TCH4 mRNA (Fig. 3A). This transgene is identical to that used for assessment of touch and darkness inducibility shown in Figure 3, B, I; and C, I. The alteration in accumulation kinetics of this transgene's transcripts is also seen after touch and darkness stimuli, as discussed below. GUS mRNAs derived from TCH4::GUS transgenes harboring additional TCH4 sequences (−958 to −45 and −958 to −1) accumulate to much higher levels than those derived from the −258/−45 TCH4::GUS transgenes (Fig. 3, A, II and III, GUS). Similar reductions in magnitude are seen for cold inducibility of TCH4::GUS transgenes harboring −258/−45 TCH4 sequences as compared with those with additional distal sequences (data not shown). With respect to BR induction, the kinetics and magnitude of response to BR treatment of transgenic plants harboring a −958/−1 TCH4::GUS construct are nearly identical to that of the endogenous TCH4, whereas a −200/−1 TCH4::GUS transgene shows dramatic reduction in expression levels while still retaining BR inducibility (data not shown). These results suggest that sequences between −958 and −258 affect the magnitude of mRNA accumulation, perhaps acting as quantitative enhancers or due to the presence of redundant functional motifs. Therefore, although there are regulatory elements within the −258 to −45 region, these 213 bp do not represent the complete TCH4 control region.
The −258 to −45 region of TCH4 is also sufficient to confer touch and darkness inducibility of expression upon the GUS reporter gene (Fig. 3, B and C, respectively). The transgene with the −46-bp CaMV 35S promoter and the TEV 5′-UTR has expression kinetics that closely reflect that of the endogenous TCH4 (Fig. 3, BIV and CIV); whereas transcripts derived from the transgene with the −90-bp CaMV 35S promoter and lacking a 5′-UTR region are delayed in both up- and down-regulation (Fig. 3, BI and CI). The observed delay in accumulation of mRNA is unlikely to be due to loss of transcriptional regulatory sequences because the same TCH4 region is sufficient to up-regulate expression with the rapid kinetics of the endogenous TCH4 when in the context of the −46-bp CaMV minimal promoter and the TEV 5′-UTR. Therefore, the delay in kinetics may be due to the combination of the −90-bp CaMV 35S promoter region and the short −258 to −45 TCH4 regulatory region in transgene I; the longer minimal promoter may impact the functioning of these TCH4 regulatory sequences, perhaps by placing them too far from the transcriptional start site.
Assessment of the regulatory activity of subregions of the −258 to −45 sequences using GUS reporter gene fusions and northern analysis was not feasible because the amount of GUS mRNA generated by these gene fusions was below the level of detection (data not shown). Therefore, we employed firefly LUC as a more sensitive reporter and monitored expression as real-time activity displayed as luminescence. LUC activity can be detected at very low levels and because LUC activity, unlike GUS, has a relatively short half-life (Millar et al., 1992), it is a good tool for reporting rapid and relatively transient changes in gene expression.
To verify that LUC activity reflects gene expression, we monitored luminescence emission over time of transgenics harboring the −258 to −45 TCH4::LUC transgene. As shown in Figure 4A, the touched transgenics (individual plants represented by red traces in top panel, right portion of bottom panel) emit higher levels of light than the untouched control population (blue traces in top panel, left portion of bottom panel). There is basal LUC activity in unstimulated transgenic plants, primarily detected from the shoot apex (Fig. 4A, left, bottom). Similarly, TCH4::GUS activity is found in young expanding leaves in the shoot apex (Xu et al., 1995). In comparison, touched transgenics show LUC activity primarily at sites directly mechano-stimulated such as leaves and petioles, while retaining expression at the shoot apex (Fig. 4A, right, bottom). Independent transgenics show similar responses (data not shown). The touch-induced increases in luminescence are due to the function of the TCH4 regulatory sequences, because a similar LUC transgene driven by the UBQ10 regulatory region (Sun and Callis, 1997) shows no significant change in activity over time after comparable stimulation (Fig. 4C). UBQ10 expression has been reported to be relatively constitutive (Sun and Callis, 1997). This is an important control because luminescence from transgenics with constitutively expressed LUC genes have been seen to increase after wounding under some conditions, apparently due to increased uptake of the luciferin substrate (Nass and Scheel, 2001; D.H. Polisensky, E. Iliev, and J. Braam, unpublished results). We found that the use of young plants, sodium citrate (pH 5.6) as a solvent for luciferin, and a 30-min absorption time improves the uptake of luciferin and results in no significant changes in UBQ10::LUC activity (Fig. 4C; D.H. Polisensky, E. Iliev, and J. Braam, unpublished data).
Figure 4.
Inducible in vivo LUC activity conferred by the −258 to −45 and −147 to −45 TCH4 sequences in response to touch stimulation. Plants harboring −258 to −45 TCH4::LUC (A), −147 to −45 TCH4::LUC (B), or UBQ10::LUC (C) transgenes, were sprayed with 1 mm luciferin and 50 mm sodium citrate, pH 5.6, and allowed to absorb the substrate for 30 min. The plants were then placed in a low-light imager and an initial luminescence reading was obtained before stimulation. Control plants were left undisturbed, whereas others were touch stimulated by gently bending them back and forth 20 times. Fifteen-minute (A and B) or 5-min (C) luminescence readings were collected, and the data were extracted and analyzed with Excel (Microsoft, Redmond, WA). The graphs at the top represent the response profiles of individual plants. Traces of control plants are shown in blue; touch-stimulated plants are shown in red. Bottom, Computer-generated false-color overlays of the light emission at 45 min poststimulation. The color bar inset represents the dynamic range of light levels, where minimum light is indicated with dark blue and maximum light is shown in magenta. The transgenes assayed are represented by rectangles below each panel (not drawn to scale); see Figure 1 legend for details.
The TCH4 Upstream Region between −147 and −45 Is Sufficient to Confer Responses to Touch, Temperature Shocks, Darkness, and 24-Epibrassinolide (24-epiBL)
To further define the potential cis-regulatory element(s) conferring TCH4 up-regulation of expression in response to environmental and hormonal stimuli, we tested subregions of the −258 to −45 sequences of TCH4. Figure 4B illustrates that the −147 to −45 TCH4 sequences confer touch-inducible expression to LUC (Fig. 4B). Similar to that of the −258 to −45 TCH4::LUC transgene, the −147 to −45 TCH4::LUC expression in touched plants is observed in leaves, petioles, and shoot apex, whereas unstimulated plants show active LUC primarily in the shoot apex. However, the basal and induced expression levels conferred by the shorter 102 bp of the −147 to −45 TCH4 region are lower (approximately 10-fold reduction) than that conferred by the −258 to −45 region. In addition, activity peaks at approximately 60 min when regulation is conferred by the shorter regulatory region, whereas the −258 to −45 TCH4::LUC transgenics show a maximal response at approximately 30 min. Independent transformants harboring the −147/−45 transgene show similar expression behaviors with respect to magnitude and kinetics of induction (data not shown). These results indicate that the TCH4 sequences between −147 and −45 are sufficient to confer touch-induced up-regulation of expression; however, additional sequences, residing between −258 and −147, play a role in controlling the magnitude of basal and induced expression in addition to enabling the rapidity of enhanced transcription initiation. We find that this 102-bp region between −147 and −45 may be approaching the minimal length for assaying regulatory activity conferred upon reporter genes. When we removed 19 additional bases to generate a −128 to −45 TCH4::GUS reporter, we were unable to detect activity even under induced conditions in multiple independent transgenics (data not shown).
To test whether the −147 to −45 TCH4 genomic sequences are sufficient to confer full regulatory properties of TCH4 to a reporter gene, we subjected transgenic plants to other inducing stimuli, allowed the newly synthesized LUC to accumulate for 1 to 3 h (as indicated), and then applied luciferin and monitored levels of LUC activity. The luminescence data were collected and binned according to relative levels, and the numbers of individual plants per bin are reported in Figure 5 (bar graphs in top panels). False-color representations of luminescence overlaid on photographs of plants are shown in the lower panels of Figure 5. Similar results were obtained with an additional independent transgenic (data not shown).
Figure 5.
TCH4 sequences between −147 and −45 are sufficient to confer up-regulation of expression in response to cold shock, heat shift, darkness, and 24-epiBL. Plants harboring −147 to −45 TCH4::LUC were grown under constant light on agar plates for 10 to 12 d (A–C) or in soil for 14 d (D). A, Control plants remained at room temperature (23°C), whereas others were placed for 10 min on ice (cold); then all plants were left to recover for 2 h at room temperature. B, Control plants remained at room temperature (23°C), and others were placed in 37°C incubator for 1 h (heat). C, Control plants were left undisturbed, whereas others were exposed to darkness for 2 h. D, Control plants were misted with 0.01% (v/v) Triton X-100, whereas others were misted with 10 μm 24-epiBL and 0.01% (v/v) Triton X-100 and incubated for 3 h. After treatments, plants were sprayed with 1 mm luciferin and 50 mm sodium citrate, pH 5.6, and placed in a low-light imager. The data acquisition was delayed for 5 min to avoid chlorophyll phosphorescence. Fifteen-minute luminescent readings were collected and the data were extracted and analyzed with Microsoft Excel. Top, Relative light units (cts/pix s) of individual control and stimulated plants were binned and the distribution plots. Computer-generated false-color overlays of the light emission at 30 min after substrate addition are shown at the bottom of each panel. Minimum light is indicated with dark blue and maximum light is shown in magenta.
The −147 to −45 TCH4::LUC transgenic plants emit increased luminescence in response to a 10-min cold treatment (Fig. 5A), 1 h at 37°C (Fig. 5B), 2 h of darkness (Fig. 5C), and 3 h after application of 24-epiBL, a commonly used form of synthetic brassinolide (Fig. 5D). There is inherent variation in luminescence from control and stimulated plants; however, in all cases, the differences in luminescence between control and stimulated plants are apparent. The induced responses are strongest for darkness and heat shock. Similar assays for auxin (IAA) induction of the −147 to −45 TCH4::LUC expression showed no detectable response in multiple transgenics (data not shown). Transgenic plants with the UBQ10 regulatory region driving LUC were used as controls for these experiments to assess whether any of the treatments affect LUC activity and/or luminescence production. UBQ10::LUC-generated luminescence levels after treatments with touch, darkness, or 24-epiBL application used here were comparable with that of untreated plants (Fig. 4C, additional data not shown). After the cold and heat treatments administered in these experiments, slight decreases in luminescence from UBQ10::LUC plants are observed (data not shown). Reductions in luminescence after transgenic plant exposure to temperature extremes may reflect reduced photosynthetic rates and ATP availability. Overall, the data in Figures 4B and 5 indicate that the TCH4 genomic sequences between −147 and −45 harbor cis-regulatory element(s) sufficient to confer up-regulation of expression in response to mechanical stimulation, cold shock, heat shock, darkness, and 24-epiBL.
We compared sequences between −147 and −45 to potential regulatory regions of other touch-inducible genes, including TCH1 (Braam and Davis, 1990), TCH2 (Braam and Davis, 1990; Khan et al., 1997), TCH3 (Sistrunk et al., 1994), and CBF1 and 2 (Gilmour et al., 1998) and found potential sequence similarities with the TCH4 region −143/−114. To test the necessity of these sequences for gene regulation, we generated a −958/−1 TCH4::GUS transgene in which the sequences between −143 and −114 were altered by interchanging purine and pyrimidine residues. In the context of the −958/−1 region, the sequences between −143 and −114 are nonessential for up-regulation because this transgene still showed touch, darkness, auxin (IAA), and 24-epiBL inducibility of expression (data not shown).
Induced Expression of TCH4 in Response to Cold, Heat, Touch, Darkness, and Auxin Is Conserved in bri1-2
One scenario to explain how diverse stimuli lead to the common response of TCH4 up-regulation of expression is that all the inducing stimuli lead to an increase in an endogenous hormone. Therefore, we tested the hypothesis that mechanical stimuli, darkness, temperature shifts, and application of exogenous IAA all result in increases in endogenous BR that act to up-regulate TCH4 expression. BRI1 encodes a Ser/Thr receptor kinase that perceives BR via its extracellular domain (He et al., 2000). The bri1-2 mutant is insensitive to BR and develops as a severe dwarf (Clouse et al., 1996; Kauschmann et al., 1996). Figure 6A illustrates that TCH4 mRNAs accumulate significantly within 2 h after treatment of wild-type plants with 0.1 to 10 μm 24-epiBL, similar to that reported previously (Xu et al., 1995). In contrast, bri1-2 shows insensitivity to 24-epiBL; TCH4 expression is not enhanced by application of 24-epiBL, except possibly at high concentrations of 24-epiBL where a weak response is observed (Fig. 6A). The TCH4 expression response to IAA is maintained in bri1-2, although the magnitude of the response is reduced compared with wild type (Fig. 6A). Figure 6B shows that TCH4 mRNAs also accumulate in bri1-2 in response to temperature extremes, mechanical perturbation, and darkness. These results indicate that the signal transduction pathways utilized by Arabidopsis to induce TCH4 expression in response to these exogenous stimuli do not require the perception of BR. Therefore, the signal transduction pathway used for BR activation of TCH4 expression must be distinct from the signaling pathways employed by other stimuli, or alternatively, the signaling pathways activated by these diverse inducing stimuli converge at some point downstream of BR perception.
Figure 6.
TCH4 up-regulation of expression in bri1-2 plants in response to environmental and hormonal stimuli. Wild-type and bri1-2 (A) or bri1-2 (B) plants were grown in liquid media, shaking gently and continuously at 60 rpm for approximately 2 weeks. A, Control plants (C) were treated with solvent alone (0.001% [v/v] ethanol); others were treated with increasing concentrations of 24-epiBL or auxin (IAA) as indicated in 0.001% (v/v) ethanol. The concentrations of 24-epiBL and IAA shown are in micromolar. Plants were collected after 2 h and immediately frozen in liquid nitrogen. Total RNA was extracted and 4 μg of RNA was size fractionated on 1% (w/v) formaldehyde gels, blotted to nylon filters, and hybridized sequentially to probes listed on the right. The prime indicates min; “h” indicates hour(s). TUB4 is shown as a loading and transfer control. B, “Cold,” Control plants (C) were left undisturbed, whereas others were shaken gently in 0°C water bath for 2.5 min, quickly brought back to room temperature by gently shaking in a 23°C water bath, and returned back to room temperature shaker for the indicated times. “Heat,” Control plants (C) were left undisturbed, whereas others were transferred to a 35°C water bath and collected at the indicated times. “Touch,” Plants were removed from the shaker and left undisturbed a day before the experiment. Control plants (C) were untreated, whereas others were shaken for 10 s and collected at the indicated times. “Darkness,” Control plants (C) were left undisturbed, whereas others were covered to minimize light and collected at the indicated times.
TCH4::GUS Expression in bri1-2 and det2-1
The availability of bri1-2 and BR biosynthesis mutants such as det2-1 (Chory et al., 1991; Fujioka et al., 1997) enables an investigation into the potential role of endogenous BR in regulating developmental expression of TCH4. TCH4 expression, assessed by TCH4::reporter gene fusions (Xu et al., 1995) and immunolocalization of XTHs (Antosiewicz et al., 1997), correlates with growth and cell expansion and with predicted presence of mechanical stress. One possibility is that TCH4 expression at these sites may be regulated by endogenous BR. That is, BR may mediate gene regulation during cell expansion or in response to mechanical stress. For example, TCH4::GUS expression is high in etiolated hypocotyls, but low in hypocotyls of photomorphogenetic seedlings. Because hypocotyl elongation in the dark requires BR (Azpiroz et al., 1998), TCH4::GUS expression detected in etiolated hypocotyls could be a consequence of BR regulation. The components of the TCH4::GUS expression pattern that are dependent on BR should be lost or at least reduced in the bri1-2 and det2-1 mutants. As shown in Figure 7, TCH4::GUS expression is high in young leaves of light-grown plants and the hypocotyl of etiolated seedlings. Surprisingly, in bri1-2 and det2-1, TCH4::GUS expression remains strong in the hypocotyls and leaves of light-grown plants (Fig. 7, Light) and in dark-grown plants (Fig. 7, Dark) even though expansion of these organs is strongly inhibited. The intense blue staining in the mutants may be a consequence of more concentrated accumulation of X-Gluc precipitate because the mutant cells fail to expand normally (Chory et al., 1991; Clouse et al., 1996; Kauschmann et al., 1996). These results indicate that neither the presence nor perception of BR are required for TCH4::GUS expression in these organs. Therefore, we conclude that other response pathways most likely function to regulate TCH4 expression during morphogenesis.
Figure 7.
TCH4::GUS expression in bri1-2 and det2-1 mutants. 5-Bromo-4-chloro-3-indolyl β-d-glucuronide (X-Gluc) staining of wild-type, bri1-2, and det2-1 plants harboring the −958 to +48 TCH4::GUS transgene. Seedlings were grown for 8 d under 24 h of light (light, top) or in the dark (dark, bottom) for 3 d. The bar in each panel = 1 mm.
DISCUSSION
TCH4 is an unusual gene in that its expression is up-regulated by a variety of seemingly unrelated stimuli, including mechanical perturbations, such as touch, temperature extremes, darkness, and the growth-promoting hormones BR and IAA. Induced TCH4 expression is also remarkably transient. In this report, we investigated which regions of the TCH4 locus contribute to its regulatory behaviors.
Regulation of Transient Gene Expression Induction
By northern analyses, one can demonstrate that TCH4 transcripts accumulate very rapidly after stimulation of plants; for example, 10 to 30 min after touch stimulation, TCH4 mRNA levels peak (Braam and Davis, 1990; Figs. 2A and 3B). Subsequently, there is also a rapid decline in transcripts with an apparent return to basal levels of mRNA by 1 to 3 h after stimulation (Braam and Davis, 1990; Figs. 2A and 3B). Two mechanisms probably account for this rapid disappearance of transcripts. Most likely, transcription initiation is quickly inhibited to halt the production of nascent transcripts. In addition, the TCH4 mRNAs must be unstable. As yet, we do not know whether TCH4 mRNA stability is decreased in a regulated manner or, alternatively, the mRNAs are always unstable. In the latter case, the transient accumulation of TCH4 mRNAs in stimulated plants would be due solely to changes in transcriptional activity. GUS transgenes composed of both 5′-untranscribed TCH4 sequences and transcribed but untranslated TCH4 sequences show expression kinetics that resemble those of the endogenous TCH4 gene (Xu et al., 1995). Because the rapid decay of transcripts after stimulation requires that the mRNAs are unstable, the TCH4-derived sequences, +1 to +48 of the UTR, present in the reporter mRNA are most likely responsible for the transcript instability. When the TCH4 UTR is removed, the GUS transcripts remain abundant at least 2 h, a time when the endogenous TCH4-derived transcripts are near basal levels (Figs. 2 and 3, A; 3, B, I; and C, I). This prolonged response of transgenes lacking the TCH4 UTR is seen consistently with transgenes driven by the −958/−1, the −958/−45, and the −258/−45 untranscribed regions of the TCH4 (Figs. 2 and 3A). However, with the latter two constructs, it is also possible that transcription initiation may continue for a longer duration, which could contribute to the prolonged accumulation of transcripts. Sequences from −45 to −1 of TCH4 were replaced with a −90 to +8 region of the CaMV 35S regulatory region that has been used as a minimal promoter. However, this −90 region harbors additional regulatory sequences in addition to the TATA and CAT boxes (Benfey et al., 1989; Lam et al., 1989); therefore, there may be sequences that also influence the arrest of transcription initiation. Surprisingly, transgenes driven by the comparable transcriptional regulatory regions of TCH4 in which the TEV 5′-UTR was added (to enhance translation initiation) show transcript accumulation kinetics nearly indistinguishable from the native TCH4 (Fig. 3, B, IV; and C, IV), suggesting that the TEV 5′-UTR may also confer RNA instability; such an effect of the TEV 5′-UTR on RNA stability has not been reported previously.
Transcriptional Regulation of TCH4 Induction of Expression
Sequences found upstream of the TCH4 transcribed region are sufficient to confer touch, darkness, 24-epiBL, cold shock, and heat shock inducibility of expression upon reporter genes (Figs. 2–5). This indicates that TCH4 regulation of expression most likely takes place, at least in part, through regulation of the rate of transcription initiation. BRU1 is an XTH-encoding gene from soybean (Glycine max) that is also regulated in expression by BR; however, in contrast to TCH4, BRU1 is thought to be regulated through a posttranscriptional mechanism based on nuclear run-on assays (Zurek and Clouse, 1994).
Sequences Sufficient to Confer Sensitivity to Diverse Stimuli
The 5′-upstream region of TCH4 has numerous motifs that might be predicted to function in the complex regulation of TCH4 expression. Surprisingly, we found that a single, relatively short 102-bp region between −147 and −45 is sufficient to confer upon a reporter gene the ability to be up-regulated in expression in response to touch, darkness, cold shock, heat shock, and 24-epiBL (Figs. 4 and 5). Because GUS activity could not be detected in transgenics harboring −128/−45 TCH4::GUS transgenes (data not shown), sequences between −147 and −128 must be important for TCH4 expression regulation. However, sequences between −143 and −114, in the context of approximately 1 kb of TCH4 upstream sequences, are not necessary for the regulated expression. It is likely, therefore, that there are redundant functional elements within the 1-kb region upstream of the TCH4 transcriptional start site. Consistent with the idea of redundant functional elements, longer 5′ regions generally conferred regulatory behavior that more closely reflected that of the native gene. Sequences between −958 and −258 and −258 and −147 enhance the magnitude of the expression induction and the kinetics of up-regulation (Figs. 3A and 4; additional data not shown). Whether sequences in these regions can also act alone to confer inducible expression is not yet known. However, within these distal sequences, there are additional E boxes and Myb motifs related to those found between −143 and −114 (Fig. 1A). The finding that inducible expression by multiple, diverse stimuli can be conferred by the same subregion is consistent with the possibility that there is a common cis-element that can serve to control at least most of the complex regulation of TCH4 expression. The inability to define separable regions able to confer only a subset of inducible properties indicates that the signal transduction pathways activated by the diverse stimuli that lead to inducible TCH4 expression most likely share at least some common elements. Precise identification of the regulatory sequences that drive TCH4 expression characteristics will require examination of the effects of combinations of site-specific mutations of the 5′-upstream sequences of TCH4.
The Role of BR in TCH4 Expression Regulation
BR has been implicated as a hormone that can influence stress responses in plants. For example, BR treatment improves the cold tolerance of maize (Zea mays) and cucumber (Cucumis sativus) seedlings (Khripach et al., 1999). Exogenous BR also leads to the up-regulation of TCH4 expression; expression of TCH4 is also regulated by diverse abiotic stresses (Xu et al., 1995). Therefore, we tested the possibility that TCH4 induction of expression is mediated through activation of BR signaling. Inducible expression of TCH4 by touch, darkness, cold, and heat remains robust in bri1-2, a BR-insensitive mutant (Fig. 6B); therefore, BR perception is not required for TCH4 regulation of expression by these environmental stimuli. Although steady-state TCH4 mRNA in unstimulated wild-type and bri1-2 plants is comparable (Fig. 6A), TCH4 up-regulation of expression in response to 24-epiBL is abolished in bri1-2 plants, except at very high concentrations. Previous studies (Kauschmann et al., 1996) observed a reduction in steady-state TCH4 mRNA in bri1-2 compared with wild type. No up-regulation of TCH4 mRNA expression in bri1-2 was observed when grown on media supplemented with 0.5 μm 24-epiBL for 13 d (Kauschmann et al., 1996); surprisingly, however, no up-regulation of TCH4 expression was evident in wild-type plants grown on 24-epiBL-supplemented media as well (Kauschmann et al., 1996). It is possible that 24-epiBL induction of TCH4 expression occurs transiently; therefore, the differences between the results described by Kauschmann et al. (1996) and our results (Xu et al., 1995; Fig. 6A) could be attributed to different growth conditions and the duration of 24-epiBL exposure.
IAA induction of TCH4 expression is weaker in bri1-2 than in wild type (Fig. 6A); this result indicates that IAA may act in a synergistic manner with BR in some aspects, including the regulation of TCH4 expression. In addition, we have found that bri1-2 has reduced expression of TCH3, a calmodulin-related gene (D.H. Polisensky and J. Braam, unpublished data). This reduction in TCH3 expression in bri1-2 is likely not directly related to BR insensitivity because TCH3 is not up-regulated in expression by exogenous BR (D.H. Polisensky and J. Braam, unpublished data). TCH3 is, however, up-regulated in expression by IAA (Antosiewicz et al., 1995), and TCH3 expression is returned to wild-type levels in the bri1-2 mutant when supplemented with exogenous IAA (D.H. Polisensky and J. Braam, unpublished data). These results are consistent with the possibility that IAA levels are reduced in plants that are incapable of sensing BR, strengthening the hypothesis that BR and IAA regulation may occur in a synergistic manner. Links between BR and IAA have been reported; for example, the addition of 24-epiBL restores wild-type sensitivity to auxin in sax1 plants (Ephritikhine et al., 1999). However, BR and auxin have also been shown to act independently in many systems (Clouse and Sasse, 1998).
BR is also not required for the expression of TCH4 during morphogenesis; there is strong TCH4::GUS expression in developing bri1-2 and det2-1 seedlings. TCH4::GUS expression in wild type strongly correlates with cell growth and expansion (Xu et al., 1995; Fig. 7, wild type). Because TCH4 expression is not lost in the bri1-2 and det2-1 mutants, it is unlikely that TCH4 developmental regulation is a result of BR signaling or a consequence of the process of cell expansion. However, because TCH4 encodes a cell wall-modifying activity, one possibility is that properties of the wall and, in the case of bri1-2 and det2-1, deviations from wall homeostasis are sensed and transmitted through a signaling pathway that impacts TCH4 expression.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis plants were grown at 22°C to 24°C in 65% to 75% humidity under constant 90 μmol m−2 s−1 light. For northern analyses, plants were grown for 12 to 14 d in liquid Murashige and Skoog media (Murashige and Skoog, 1962). Plants for LUC analyses were grown in 1-inch pots at a density of one plant per pot or on agar plates containing 0.5× Murashige and Skoog media supplemented with 1% (w/v) Suc. Plants for GUS assays were grown on filter paper on agar-containing plates with 0.5× Murashige and Skoog with 0.2% (w/v) Suc for 4 to 10 d under constant light or for 3 to 4 d covered (darkness).
Generation of TCH4::GUS and TCH4::LUC Reporter Constructs
For construction of −958 to −1 TCH4::GUS plasmid, the region from −958 to −1 was isolated by PCR using the forward primer (5′-TCTAGACTTTGCGTAAG-3′) and reverse primer (5′-TTTGAGGGTTTATGGAGG-3′) and cloned into pCRII vector (Invitrogen, Carlsbad, CA). The recombinant plasmid was cut with XbaI, and ligated into XbaI-cut pBI101 vector (CLONTECH, Palo Alto, CA) in front of the GUS gene. For construction of −958 to −45 TCH4::GUS and −258 to −45 TCH4::GUS with −90/+8 CaMV 35S promoter, the region from −958 to −45 was PCR amplified using the forward primer (5′-TCTAGACTTTGCGTAAG-3′) and the reverse primer (5′-AAGATTTTTAAGAG-3′) and the region from −258 to −45 was PCR amplified using the forward 5′-CCAATTAAATCTGAAACC-3′ and reverse primer 5′-AAGATTTTTAAGAG-3′ and cloned into pCRII. The TCH4 regions between HindIII and EcoRV were ligated into a pBI221- (CLONTECH) derived plasmid with sequences between BamHI and EcoRI deleted; this cloning step resulted in fusion between TCH4 sequences and the −90/+8 CaMV 35S promoter. The hybrid regulatory regions, flanked by HindIII and XbaI sites, were inserted at the HindIII and XbaI sites of pBI101, forming the −958 to −45 TCH4::GUS and −258 to −45 TCH4::GUS plasmids. The construct containing TCH4 region from −128 to −45 linked with the −90/+8 CaMV 35S promoter was made by nested deletion from the 5′ end of the TCH4 region in the recombinant plasmid, −258/−45 TCH4::GUS. The exact site of the deletion was determined by sequencing analysis.
For construction of fusion genes with the −258 to −45 region and smaller TCH4::GUS and TCH4::LUC fusions with the −46-bp CaMV 35S promoter, promoter fragments were PCR amplified as follows: −258 to −45 using the forward primer (5′-CCCAAGCTTCCAATTAAATCT-3′) and the reverse primer (5′-GAAGATCTAAGATTTTTAAGA-3′), and −147 to −45 using the forward primer (5′-CCCAAGCTTCTTTTACTACAA-3′) and the reverse primer (5′-GAAGATCTAAGATTTTTAAGA-3′). The primer-introduced HindIII and BglII sites are underlined. The PCR products were cloned into pCRII and sequenced. For TCH4::LUC constructs, the TCH4 regions were excised as HindIII-BglII fragments, then subcloned into pKS 35S-TEV, a pBluescript KSII-based vector that has a −46-bp CaMV 35S minimal promoter and 143-bp TEV UTR inserted at the BamHI site. The fragments were further subcloned as HindIII-BamHI fragments from pKS 35STEV into pCR 35S-TEV, a pCRII-based vector that harbors a PCR-amplified −46-bp CaMV 35S minimal promoter and 143-bp TEV UTR. Direct cloning of the HindIII-BglII TCH4 fragments into pCR 35S-TEV was not possible due to the presence of an additional BglII site. The TCH4 sequences, along with the −46-bp CaMV 35S minimal promoter and TEV UTR, were subcloned as HindIII-XhoI into pKAJ201, a pBI101-based vector into which the GUS gene was replaced with SalI-SacI fragment from pJD300 (Luehrsen et al., 1992) containing the LUC gene. For TCH4::GUS constructs, the PCR-amplified TCH4 sequences were subcloned from pCRII as HindIII-BglII fragments into pBI 35S-TEV GUS, a pBI 101-based vector in which a PCR-amplified −46-bp CaMV 35S minimal promoter and 143-bp TEV UTR were inserted in front of GUS.
To generate a TCH4 region with mutated sequence between −143 and −114, a two-step PCR procedure was followed starting with the plasmid pBITG as the template. pBITG consists of bases −958 to −1 of TCH4 upstream sequences (nucleotides 40, 185 through 39, and 227 of GenBank accession no. AB011482), the GUS gene, and the nopaline synthase terminator of pBI101, all cloned into the binary vector BIN19 (Frisch et al., 1995). First step PCR utilized the 5′ primer lsp5 (5′-CTCAAAGCTTGCATGCCTGCAGGTCGAC-3′) coupled with a 54-mer (jmb2, 5′-ACCACTAGTTGTCGGTTTAACGAGGTGCTGAAAAGCCTGTGTTTTATTTATTGG-3′) consisting of 24 bases complementary to wild-type TCH4 promoter sequence, and 30 bases mutated by exchange of pyrimidines and purines. The second PCR reaction contained the 3′ primer, ls3p (5′-CTCAGGATCCTCTAGATGCATGCTCGAT-3′) and a 34-mer (jmb1, 5′-GACAACTAGTGGTCCTCAAAGACGCGGCTTCTTC-3′) consisting of 13 bases of overlap with jmb2 and 21 bases complementary to wild-type TCH4 sequence. PCR conditions were 94°C for 3 min, followed by 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 2.5 min. After a final extension of 10 min at 72°C, PCR products were purified by phenol/chloroform extraction and ethanol precipitation and digested with SpeI. The two fragments were then ligated and further digested with SalI/BamHI. The mutated TCH4 was cloned into SalI/BamHI-digested pBITG from which the wild-type TCH4 region had been removed. The resulting construct was identical to pBITG with the exception of 30 bp within the TCH4 region, which was verified by sequencing to confirm an exchange of Gs for Ts, Ts for Gs, Cs for As, and As for Cs in the region between −143 and −114.
The constructs were moved into Arabidopsis via Agrobacterium tumefaciens-mediated vacuum infiltration (Bechtold et al., 1993). Independent homozygous T3 lines were obtained by selfing followed by selection on kanamycin and DNA-blot hybridization analyses. Two to five independent TCH4::reporter lines per construct were examined and found to have comparable inducible expression behavior.
RNA Analysis
For RNA analyses, treated and control plants were harvested at the indicated time points and then immediately frozen in liquid nitrogen. Total RNA was purified (Verwoerd et al., 1989), electrophoresed on formaldehyde gels, blotted overnight onto nylon membranes (Micron Separations, Westborough, MA), and hybridized with hexamer-labeled DNA fragments (Feinberg and Vogelstein, 1983). The probes used were described by Xu et al. (1995).
LUC Data Acquisition and Analysis
In vivo LUC analyses were performed with a NightOWL low light imager (Perkin-Elmer Applied Biosystems, Foster City, CA). Plants were finely misted from a 15-cm distance with 1 mm luciferin (Biosynth AG, Staad, Switzerland) and 50 mm sodium citrate, pH 5.6, and placed in the NightOWL after 30 min. Multiple images were acquired over 5- to 15-min intervals as noted. Computer-generated representations of luminescence emissions were overlaid with photographs acquired before and/or after the completion of time courses. Data extraction and analysis were performed with WinLight software (Perkin-Elmer Applied Biosystems) and exported into Excel spreadsheets (Microsoft). Control plants were similarly and simultaneously treated (with the exclusion of the stimulus) and concurrently viewed.
Histochemical Analysis
Plant assays of GUS activity were performed as described by Gallagher (1992). In brief, the plants were fixed in 2% (w/v) paraformaldehyde, 100 mm sodium phosphate, pH 7.0, and 1 mm EDTA for 20 to 25 min on ice. After washing twice with 100 mm sodium phosphate, pH 7.0, the plants were incubated in 2 mm X-Gluc (Molecular Probes, Eugene, OR), 50 mm sodium phosphate (pH 7.0), and 0.01% (v/v) Triton X-100 overnight at 37°C. After the reaction was stopped with a water wash, the tissues were cleared with several washes of 70% (v/v) ethanol. The tissues were mounted on microscope slides in 50% (v/v) glycerol, and the slides were fitted into a Pathscan Enabler (Meyer Instruments, Houston) and scanned with a 35-mm film scanner (Nikon, Tokyo).
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial purposes. No restrictions or conditions will be placed on the use of any materials described in this paper that would limit their use in noncommercial research purposes.
ACKNOWLEDGMENTS
We thank Judy Callis (University of California, Davis) for the use of the UBQ10::LUC transgenic plants and the Braam lab members for comments on the manuscript.
Footnotes
This work was supported by the National Science Foundation (grant no. IBN9982654 to J.B.), by the Department of Energy (grant no. DE–FG03–99ER20331 to J.B.), in part by the National Institutes of Health (Biotechnology Training Grant no. T32–Gm08362 to E.A.I.), by the National Science Foundation (Integrative Biology and Neuroscience; Integrative Plant Biology, to S.D.C.'s laboratory), by the U.S. Department of Agriculture/National Research Initiative (Plant Growth and Development, to S.D.C.'s laboratory), and by the North Carolina Agricultural Research Service (to S.D.C.'s laboratory).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.008680.
LITERATURE CITED
- Antosiewicz DM, Polisensky DH, Braam J. Cellular localization of the Ca2+ binding TCH3 protein of Arabidopsis. Plant J. 1995;8:623–636. doi: 10.1046/j.1365-313x.1995.08050623.x. [DOI] [PubMed] [Google Scholar]
- Antosiewicz DM, Purugganan MM, Polisensky DH, Braam J. Cellular localization of the Arabidopsis TCH4 XET during development and after wind stimulation. Plant Physiol. 1997;115:1319–1328. doi: 10.1104/pp.115.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpiroz R, Wu Y, LoCascio JC, Feldmann KA. An Arabidopsis brassinosteroid-dependent mutant is blocked in cell elongation. Plant Cell. 1998;10:219–230. doi: 10.1105/tpc.10.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SS, Wilhelm KS, Thomashow MF. The 5′ region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought-, and ABA-regulated gene expression. Plant Mol Biol. 1994;24:701–713. doi: 10.1007/BF00029852. [DOI] [PubMed] [Google Scholar]
- Barros MD, Czarnecka E, Gurley WB. Mutational analysis of a plant heat shock element. Plant Mol Biol. 1992;19:665–675. doi: 10.1007/BF00026792. [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris Life Sci. 1993;316:1194–1199. [Google Scholar]
- Benfey PN, Ren L, Chua NH. The CaMV 35S enhancer contains at least two domains which can confer different developmental and tissue-specific expression patterns. EMBO J. 1989;8:2195–2202. doi: 10.1002/j.1460-2075.1989.tb08342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braam J. Regulated expression of the calmodulin-related TCH genes in cultured Arabidopsis cells: induction by calcium and heat shock. Proc Natl Acad Sci USA. 1992;89:3213–3216. doi: 10.1073/pnas.89.8.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braam J, Davis RW. Rain-, wind- and touch-induced expression of calmodulin and calmodulin-related genes in Arabidopsis. Cell. 1990;60:357–364. doi: 10.1016/0092-8674(90)90587-5. [DOI] [PubMed] [Google Scholar]
- Busk PK, Pages M. Regulation of abscisic acid-induced transcription. Plant Mol Biol. 1998;37:425–435. doi: 10.1023/a:1006058700720. [DOI] [PubMed] [Google Scholar]
- Campbell P, Braam J. Co- and/or post-translational modifications are critical for TCH4 XET activity. Plant J. 1998;15:553–561. doi: 10.1046/j.1365-313x.1998.00239.x. [DOI] [PubMed] [Google Scholar]
- Campbell P, Braam J. Xyloglucan endotransglycosylases: diversity of genes, enzymes and potential wall-modifying functions. Trends Plant Sci. 1999;4:361–366. doi: 10.1016/s1360-1385(99)01468-5. [DOI] [PubMed] [Google Scholar]
- Carrington JC, Freed DD. Cap-independent enhancement of translation by a plant potyvirus 5′ nontranslated region. J Virol. 1990;64:1590–1597. doi: 10.1128/jvi.64.4.1590-1597.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Nagpal P, Peto CA. Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell. 1991;3:445–460. doi: 10.1105/tpc.3.5.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Sasse JM. Brassinosteroids: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:427–451. doi: 10.1146/annurev.arplant.49.1.427. [DOI] [PubMed] [Google Scholar]
- Ephritikhine G, Fellner M, Vannini C, Lapous D, Barbier-Brygoo H. The sax1 dwarf mutant of Arabidopsis thaliana shows altered sensitivity of growth responses to abscisic acid, auxin, gibberellins and ethylene and is partially rescued by exogenous brassinosteroid. Plant J. 1999;18:303–314. doi: 10.1046/j.1365-313x.1999.00454.x. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Frisch DA, Harris-Haller L, Yokubaitis NT, Thomas TL, Hardin SH, Hall TC. Complete sequence of the binary vector Bin19. Plant Mol Biol. 1995;27:405–409. doi: 10.1007/BF00020193. [DOI] [PubMed] [Google Scholar]
- Fujioka S, Li J, Choi YH, Seto H, Takatsuto S, Noguchi T, Watanabe T, Kuriyama H, Yokota T, Chory J. The Arabidopsis deetiolated2 mutant is blocked early in brassinosteroid biosynthesis. Plant Cell. 1997;9:1951–1962. doi: 10.1105/tpc.9.11.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher SR. GUS Protocols: Using the GUS Gene as a Reporter of Gene Expression. San Diego: Academic Press; 1992. [Google Scholar]
- Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 1998;16:433–442. doi: 10.1046/j.1365-313x.1998.00310.x. [DOI] [PubMed] [Google Scholar]
- Gubler F, Jacobsen JV. Gibberellin-responsive elements in the promoter of a barley high-pI alpha-amylase gene. Plant Cell. 1992;4:1435–1441. doi: 10.1105/tpc.4.11.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Wang ZY, Li J, Zhu Q, Lamb C, Ronald P, Chory J. Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science. 2000;288:2360–2363. doi: 10.1126/science.288.5475.2360. [DOI] [PubMed] [Google Scholar]
- Hobo T, Asada M, Kowyama Y, Hattori T. ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent. Plant J. 1999;19:679–689. doi: 10.1046/j.1365-313x.1999.00565.x. [DOI] [PubMed] [Google Scholar]
- Inaba T, Nagano Y, Reid JB, Sasaki Y. DE1, a 12-base pair cis-regulatory element sufficient to confer dark-inducible and light on-regulated expression to a minimal promoter in pea. J Biol Chem. 2000;275:19723–19727. doi: 10.1074/jbc.M001337200. [DOI] [PubMed] [Google Scholar]
- Kauschmann A, Jessop A, Koncz C, Szekeres M, Willmitzer L, Altmann T. Genetic evidence for an essential role of brassinosteroids in plant development. Plant J. 1996;9:701–713. [Google Scholar]
- Khan AR, Johnson KA, Braam J, James MNG. Comparative modeling of the three-dimensional structure of the calmodulin-related TCH2 protein from Arabidopsis. Proteins. 1997;27:144–153. doi: 10.1002/(sici)1097-0134(199701)27:1<144::aid-prot14>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Khripach VA, Zhabinskii VN, de Groot AE. Brassinosteroids. A New Class of Plant Hormones. San Diego: Academic Press; 1999. [Google Scholar]
- Lam E, Benfey P, Gilmartin PM, Fang R-X, Chua NH. Site specific mutations alter in vitro factor binding and change promoter expression pattern in transgenic plants. Proc Natl Acad Sci USA. 1989;86:7890–7894. doi: 10.1073/pnas.86.20.7890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanahan MB, Ho TH, Rogers SW, Rogers JC. A gibberellin response complex in cereal alpha-amylase gene promoters. Plant Cell. 1992;4:203–211. doi: 10.1105/tpc.4.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovegrove A, Hooley R. Gibberellin and abscisic acid signalling in aleurone. Trends Plant Sci. 2000;5:102–110. doi: 10.1016/s1360-1385(00)01571-5. [DOI] [PubMed] [Google Scholar]
- Luehrsen KR, de Wet JR, Walbot V. Transient expression analysis in plants using firefly luciferase reporter gene. Methods Enzymol. 1992;216:397–414. doi: 10.1016/0076-6879(92)16037-k. [DOI] [PubMed] [Google Scholar]
- Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H, Deng XW. Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell. 2001;13:2589–2607. doi: 10.1105/tpc.010229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ, Short SR, Chua N-H, Kay SA. A novel circadian phenotype based on firefly luciferase expression in transgenic plants. Plant Cell. 1992;4:1075–1087. doi: 10.1105/tpc.4.9.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Nass N, Scheel D. Enhanced luciferin entry causes rapid wound-induced light emission in plants expressing high levels of luciferase. Planta. 2001;212:149–154. doi: 10.1007/s004250000389. [DOI] [PubMed] [Google Scholar]
- Nunberg AN, Li Z, Bogue MA, Vivekananda J, Reddy AS, Thomas TL. Developmental and hormonal regulation of sunflower helianthinin genes: proximal promoter sequences confer regionalized seed expression. Plant Cell. 1994;6:473–486. doi: 10.1105/tpc.6.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polisensky DH, Braam J. Cold-shock regulation of the Arabidopsis TCH genes and the effects of modulating intracellular calcium levels. Plant Physiol. 1996;111:1271–1279. doi: 10.1104/pp.111.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöffl F, Prändl R, Reindl A. Regulation of the heat-shock response. Plant Physiol. 1998;17:1135–1141. doi: 10.1104/pp.117.4.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Ho THD. Functional dissection of an abscisic acid (ABA)-inducible gene reveals two independent ABA-responsive complexes each containing a G-box and a novel cis-acting element. Plant Cell. 1995;7:295–307. doi: 10.1105/tpc.7.3.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Zhang P, Ho THD. Modular nature of abscisic acid (ABA) response complexes: composite promoter units that are necessary and sufficient for ABA induction of gene expression in barley. Plant Cell. 1996;8:1107–1119. doi: 10.1105/tpc.8.7.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sistrunk ML, Antosiewicz DM, Purugganan MM, Braam J. Arabidopsis TCH3 encodes a novel Ca2+ binding protein and shows environmentally induced and tissue-specific regulation. Plant Cell. 1994;6:1553–1565. doi: 10.1105/tpc.6.11.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C-W, Callis J. Independent modulation of Arabidopsis thaliana polyubiquitin mRNAs in different organs and in response to environmental changes. Plant J. 1997;11:1017–1027. doi: 10.1046/j.1365-313x.1997.11051017.x. [DOI] [PubMed] [Google Scholar]
- Sung DY, Vierling E, Guy CL. Comprehensive expression profile analysis of the Arabidopsis Hsp70 gene family. Plant Physiol. 2001;126:789–800. doi: 10.1104/pp.126.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mole Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwoerd TC, Dekker BM, Hoekema A. A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 1989;17:2362. doi: 10.1093/nar/17.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Campbell P, Vargheese AK, Braam J. The Arabidopsis XET-related gene family: environmental and hormonal regulation of expression. Plant J. 1996;9:879–889. doi: 10.1046/j.1365-313x.1996.9060879.x. [DOI] [PubMed] [Google Scholar]
- Xu W, Purugganan MM, Polisensky DH, Antosiewicz DM, Fry SC, Braam J. Arabidopsis TCH4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. Plant Cell. 1995;7:1555–1567. doi: 10.1105/tpc.7.10.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature or high-salt stress. Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurek DM, Clouse SD. Molecular cloning and characterization of a brassinosteroid-regulated gene from elongating soybean epicotyls. Plant Physiol. 1994;104:161–170. doi: 10.1104/pp.104.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]