Abstract
Plants take up large amounts of K+ from the soil solution and distribute it to the cells of all organs, where it fulfills important physiological functions. Transport of K+ from the soil solution to its final destination is mediated by channels and transporters. To better understand K+ movements in plants, we intended to characterize the function of the large KT-HAK-KUP family of transporters in rice (Oryza sativa cv Nipponbare). By searching in databases and cDNA cloning, we have identified 17 genes (OsHAK1–17) encoding transporters of this family and obtained evidence of the existence of other two genes. Phylogenetic analysis of the encoded transporters reveals a great diversity among them, and three distant transporters, OsHAK1, OsHAK7, and OsHAK10, were expressed in yeast (Saccharomyces cerevisiae) and bacterial mutants to determine their functions. The three transporters mediate K+ influxes or effluxes, depending on the conditions of the experiment. A comparative kinetic analysis of HAK-mediated K+ influx in yeast and in roots of K+-starved rice seedlings demonstrated the involvement of HAK transporters in root K+ uptake. We discuss that all HAK transporters may mediate K+ transport, but probably not only in the plasma membrane. Transient expression of the OsHAK10-green fluorescent protein fusion protein in living onion epidermal cells targeted this protein to the tonoplast.
K is the most abundant cation in living cells, thus making the highest contribution to maintaining the cellular electroneutrality and osmotic equilibrium. Because K+ was selected for these functions very early in the evolution of life, the cellular processes evolved in a K+-rich medium, and many of them became dependent on K+. Many of these requirements were established before the Cambrian explosion of plants, when they conquered the rocks emerging from the sea. Although these rocks offered a poor nutritional environment, plants evolved on them, developing complex mechanisms of K+ uptake, as well as more cellular and physiological functions for K+, when plants gained complexity. At present, soils are less K+ deficient than in the Cambrian era, but plants are more complex and K+ has to be moved to cells that are physically very distant from the K+ source. As a result, transmembrane movements of K+ are crucial for the physiology of contemporary plants (Kochian and Lucas, 1988; Rodríguez-Navarro, 2000).
Transmembrane K+ movements in plants are mediated by several types of channels (Mäser et al., 2001), and by transporters that belong to two families, KcsA-TRK and Kup-HAK (Rodríguez-Navarro, 2000; Mäser et al., 2001). In the Arabidopsis genome, there are 15 genes encoding two different types of rather divergent K+ channels, with 2P/4TM (two P loops and four transmembrane domains) and 1P/6TM structures (one P loop and six transmembrane domains; Mäser et al., 2001). The functions of channels are diverse, mediating inward and outward K+ movements in plasma membrane and tonoplast (Zimmermann and Sentenac, 1999; Ivashikina et al., 2001; Mäser et al., 2001; Reintanz et al., 2002; Schönknecht et al., 2002).
Transporters of the KcsA-TRK family in plants, which are named HKT, exist in low numbers in the same plant. There is only one HKT gene in Arabidopsis (Uozumi et al., 2000), and probably two genes in rice (Oryza sativa cv Nipponbare; Horie et al., 2001) and Eucalyptus sp. (Fairbairn et al., 2000). In contrast, plant transporters of the Kup-HAK family, which are called KT, HAK, and KUP, are almost as numerous as channels. In the Arabidopsis genome, there are 13 genes encoding transporters of this type (Mäser et al., 2001). Two distant members keep a low level of identity, around 40% (Senn et al., 2001), and transporters of this family are expressed in all or in most plant tissues (Rubio et al., 2000). This suggests that KT-HAK-KUP transporters play diverse and important functions in the plant, a notion that is corroborated by the defects exhibited by two Arabidopsis mutants in transporters of this family. In one of them, a T-DNA insertion into the AtAKT3-KUP4 gene produces tiny root hairs (Rigas et al., 2001); in the other, one amino acid change in the AtKT2-KUP2 transporter causes a short hypocotyl and small leaves (Elumalai et al., 2002).
A remarkable characteristic of Kup-HAK transporters is that the range of K+ concentrations at which they are active overlaps with other types of transporters in bacteria, fungi, and plants. In Escherichia coli, the Kup transporter exhibits low affinity as other K+ transporters (Bakker, 1993), and in fungi, HAK transporters exhibit high affinity (Bañuelos et al., 1995) and seem to be redundant with other high-affinity K+ transporters (Haro et al., 1999; Bañuelos et al., 2000). In plants, KT-HAK-KUP transporters have been associated with high-affinity K+ uptake in roots (Santa-María et al., 1997; Fu and Luan, 1998; Kim et al., 1998; Rubio et al., 2000), but recent findings using akt1 and akt2 Arabidopsis mutants suggest that high-affinity K+ uptake in Arabidopsis roots is mediated by channels (Hirsch et al., 1998; Spalding et al., 1999; Dennison et al., 2001). Moreover, there is increasing evidence suggesting that many KT-HAK-KUP transporters are low-affinity transporters (Quintero and Blatt, 1997; Senn et al., 2001), which apparently overlap their K+ concentration range of activities with K+ channels. This apparent duplication of transporters with similar affinities for K+ could be explained if transporters of the Kup-HAK family mediated active K+-H+ cotransport (Rodríguez-Navarro, 2000; Zakharyan and Trchounian, 2001), and the other transporters were uniporters allowing only electrochemical equilibrium. This hypothesis would give a good explanation for the extensive existence of Kup-HAK family of transporters in so many different organisms, but needs to be proved. Therefore, a reasonable understanding of K+ uptake in many organisms requires a more extensive research of the functions of the Kup-HAK transporters. Moreover, in plants, the problem is more complex than in bacteria or fungi, because in addition to K+ uptake from the soil solution, the cation has to be distributed to many cells by mechanisms that are not well understood at the molecular level.
A plausible way to solve the questions posed at present about the plant KT-HAK-KUP transporters could be a systematic functional identification of all them in a specific plant. This is possible in Arabidopsis because the entire genome has been sequenced and many knockout lines are available. Although this line of research in Arabidopsis is absolutely necessary, Arabidopsis is not the best model for plants with agronomic applications, and of the plants that can be taken as models in agronomic research, rice is the most interesting for many reasons: (a) The sequencing of the rice genome almost finished with the concourse of private and public efforts; (b) its genome has a high degree of colinearity with those of wheat (Triticum aestivum), barley (Hordeum vulgare), and maize (Zea mays); and (c) rice is one of the most important human foods (Cantrell and Reeves, 2002).
The aim of the present work was to obtain a complete inventory of the rice HAK genes, after a mixed approach of search in public and private databases, and cDNA cloning. In addition, the cDNAs corresponding to three selected genes were functionally characterized by expressing them in yeast (Saccharomyces cerevisiae), bacteria, and in epidermal root onion cells.
RESULTS
Inventory of HAK Transporters
After a systematic search in public databases and in Monsanto Rice Genome Sequence Database (MRGSD; Pharmacia Rice-research.org Program, St. Louis), using as queries the barley HvHAK1 and HvHAK2 transporters, we identified 14 genes in rice that could encode HAK transporters (OsHAK2, OsHAK3, OsHAK5–15, and OsHAK17). We also identified several identical expressed sequence tag (EST) sequences with high homology to HvHAK1, which corresponded to partial-length cDNAs prepared from leaf, callus, panicle, and young roots. We isolated this cDNA, as described below, and named it OsHAK1. The cDNA OsHAK4 (Rubio et al., 2000) was not further investigated.
In parallel with searches in databases, we performed a systematic reverse transcriptase (RT)-PCR cDNA cloning from rice seedlings grown at 3 mm K+ or in the absence of added K+, obtaining more than 100 HAK cDNA fragments. By restriction analysis, we selected 35 clones for sequencing, from which all but one corresponded to previously identified genes. The new cDNA clone was named OsHAK16. Table I summarizes these results and gives the reference sequence for each gene or cDNA.
Table I.
Inventory of rice cv Nipponbare HAK transporters
| Name | Source and Reference Sequence Accession No. | Notes and Redundant Sequences |
|---|---|---|
| OsHAK1 | This work, AJ427970 | 2.3-kb Incomplete cDNA; ESTs: AU062610, AU164229, and AU062578 |
| OsHAK2 | RGPa, BAB64197 | g.s. and c.t. |
| OsHAK3 | MRGSDb, AJ427974 | g.s. Redundant to AC011806, AP003281, and AP003236, new translation |
| OsHAK4 | AF129485 | 2.3-kb Incomplete cDNA; EST: C71725 |
| OsHAK5 | RGP, BAB67929 | g.s. and c.t. |
| OsHAK6 | RGP, BAB67945 | g.s. and c.t. |
| OsHAK7 | MRGSD, AJ427976 | g.s., Full-length cDNA, AJ427971 |
| OsHAK8 | MRGSD, AJ427977 | Incomplete g.s. and c.t. |
| OsHAK9 | MRGSD, AJ427978 | g.s. and c.t. |
| OsHAK10 | MRGSD, AJ427979 | g.s., Full-length cDNA, AJ427972; EST: C72058 |
| OsHAK11 | MRGSD, AJ427980 | g.s. and c.t. |
| OsHAK12 | MRGSD, AJ427981 | g.s. and c.t. |
| OsHAK13 | MRGSD, AJ427982 | Incomplete g.s. and c.t. |
| OsHAK14 | MRGSD, AJ427983 | Incomplete g.s. and c.t. |
| OsHAK15 | MRGSD, AJ427984 | g.s. and c.t. |
| OsHAK16 | This work, AJ427973 | 2.2-kb Incomplete cDNA |
| OsHAK17 | MRGSD, AJ427975 | g.s. and c.t. |
g.s., Genomic sequence; c.t., conceptual translation.
RGP, Rice Genome Research Program (http://rpd.affrc.go.jp/).
MRGSD (http://www.rice-research.org/).
Finally, we used the 17 identified coding sequences as queries for an exhaustive database search of rice cv Nipponbare sequences. This search produced two additional EST sequences, AU101149 and AU029476, which may encode HAK transporters different from those previously identified, and two short genomic sequences in MRGSD, which we could not identify in any of our cDNA preparations. The two EST sequences probably correspond to two additional genes, but the two short genomic sequences may be remains of pseudogenes. A certain number of rice cv Nipponbare EST sequences in the databases were different from any of the 17 query sequences in just a few bases, which were considered sequencing discrepancies and not new genes. A few EST sequences from non-stated cultivars did not greatly differ from the rice cv Nipponbare sequences, but the difference was more than that expected from sequencing mistakes. This suggests the existence of allelic differences among different cultivars and, with a high probability, between allelic HAK genes in japonica and indica rice subspecies.
When the gene sequences were aligned using as reference the known cDNA sequences, or eventually the homology of the translated sequences, we were able to determine that most of the OsHAK genes shared seven putative introns situated in identical positions (Fig. 1). Five genes lacked some of these introns (HAK3, HAK6, HAK10, HAK14, and HAK17), and most of the 14 genes presented additional introns in non-conserved positions either upstream or downstream of the conserved ones. Analyses of the gene sequences did not reveal defects that could suggest that any of them is not transcribed or is translated into a nonfunctional protein. However, at the moment, the possibility that any of the genes is not functional cannot be ruled out.
Figure 1.
Schematic representation of 14 rice HAK genes. The black bars represent the open reading frames and the positions and lengths of introns are indicated by cuts and gray bars. Interruption of the black bars with two parallel lines denotes that the sequence is incomplete.
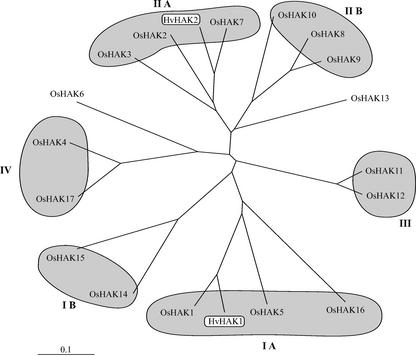
Conceptual translations of the OsHAK cDNAs and genes presented all the characteristics of typical HAK transporters (Rodríguez-Navarro, 2000). The phylogenetic tree of the OsHAK transporters (Fig. 2) could be divided into the four clusters previously described (Rubio et al., 2000). However, restricting the clusters to sequences with an approximate minimum of 60% identity, cluster I and cluster II divided into two subgroups each: IA (OsHAK1, OsHAK5, and OsHAK16), IB (OsHAK14 and OsHAK15), IIA (OsHAK2, OsHAK3, and OsHAK7), and IIB (OsHAK8, OsHAK9, and OsHAK10). Cluster III included OsHAK11 and OsHAK12, and cluster IV included OsHAK4 and OsHAK17. The sequence divergence of OsHAK6 and OsHAK13 segregated these transporters from all clusters.
Figure 2.
Phylogenetic tree of rice cv Nipponbare HAK transporters and barley transporters HvHAK1 and HvHAK2. Alignments of the sequences were performed with the ClustalX program. The scale bar corresponds to a distance of 10 changes per 100 amino acid positions. Accession numbers: HvHAK1, T04379; and HvHAK2, AAF36491; other accession numbers are given in Table I.
OsHAK1 Is Expressed Mainly in Roots, Whereas OsHAK7 and OsHAK10 Are Expressed in Shoots and Roots
During the process of cDNA cloning described above, OsHAK1 cDNA fragments were repeatedly cloned from the roots of K+-starved seedlings, less frequently from the roots of normal plants, and never from the shoots, regardless of the conditions in which the seedlings had been grown. These results suggested that although OsHAK1 was expressed in the whole plant (see Table I EST references), maximum expression was in roots and up-regulated in conditions of K+ deficiency, which is the expression pattern of its closest homolog in barley, HvHAK1 (Santa-María et al., 1997). To investigate this possibility, we studied the expression of the OsHAK1 transcripts in shoots and roots during a period of K+ starvation. In a parallel control experiment, we made similar determinations for OsHAK7, because fragments of this cDNA were systematically cloned from roots and shoots. Moreover, OsHAK7 is highly homologous to the barley HvHAK2 transporter, which has been taken as an archetype of cluster II HAK transporters (Senn et al., 2001). We also included OsHAK10 in the expression studies as a representative of cluster IIB, and because it is expressed in the rice panicle at flowering stage (see EST accession no. C72058), and might be a specific transporter of a few types of cells.
As expected from the cloning results, a real-time RT-PCR approach demonstrated that OsHAK1 was expressed almost exclusively in roots, whereas OsHAK7 was expressed in roots and shoots. Both transcripts were found in control plants grown at 3 mm K+ and moderately enhanced with K+ starvation (Table II summarizes the results obtained with seedlings grown in the dark, but older seedlings grown as described in “Materials and Methods” produced nonsignificantly different results). In the conditions of our experiments (PCR efficiency), we were able to calculate that the expression levels of the OsHAK1 and OsHAK7 mRNAs in the roots of K+-starved seedlings were approximately 5- and 3-fold higher, respectively, than the expression of the OsRAC1 (actin) mRNA. In view of the limited effect of external conditions on the expressions of the OsHAK1 and OsHAK7 transcripts, the expression of OsHAK10 was investigated by regular PCR. The expression of OsHAK10 was very similar to that of OsHAK7, both in shoots and roots, and almost insensitive to many external conditions, such as K+ starvation, Na+ stress, high-pH stress, and high-Ca2+ stress.
Table II.
Relative abundance of OsHAK1 and OsHAK7 mRNA transcripts in rice seedlings grown at 3 mm and when these seedlings were exposed to a K+-free medium for 16 h
| Transcript | Roots
|
Shoots
|
||
|---|---|---|---|---|
| 0 h | 16 h | 0 h | 16 h | |
| OsHAK1 | 0.35 ± 0.09 | 0.64 ± 0.12 | 0.05 ± 0.02 | 0.07 ± 0.04 |
| OsHAK7 | 0.22 ± 0.11 | 0.81 ± 0.14 | 0.20 ± 0.08 | 0.77 ± 0.15 |
The mRNA contents of these plants were referred to the corresponding contents of seedlings grown permanently in a K+-free medium. The results, obtained by real-time RT-PCR in three independent experiments, are presented as the means of the three experiments ± sd.
Functional Expression of OsHAK1-1 in Yeast Mutants
For the functional study, we selected three transporters from clusters IA, IIA, and IIB (OsHAK1, OsHAK7, and OsHAK10, respectively) and their K+ transport capacities were initially investigated through heterologous expression in a yeast mutant deficient in the TRK systems for K+ uptake. To obtain the full-length cDNAs encoding the selected transporters, we followed a RT-PCR method that was satisfactory for OsHAK7 and OsHAK10. However, for unknown reasons, the reverse transcription of OsHAK1 and other HAK transporters belonging to cluster I stopped 50 to 100 bp before the translation initiation codon. For the barley HvHAK1, which also belongs to cluster I, it was extremely difficult to obtain the full-length cDNA (Santa-María et al., 1997), and in rice it was impossible for OsHAK1 and OsHAK16. A comparison of translated sequences of the barley HvHAK1 and Phragmites australis PceHAK1A transporters with our longest OsHAK1 cDNA clone indicated that the rice clone probably lacked 48 bp (Fig. 3). Considering the high degree of similarity between these three transporters, we constructed a chimeric clone, denoted OsHAK1-1, in which the first 48 bp encoding the 16 amino acids of the barley HvHAK1 transporter were added to our incomplete OsHAK1 cDNA.
Figure 3.
Alignments of the N-terminal sequences of barley HvHAK1, P. australis PceHAK1A, and OsHAK1 transporters. Accession numbers: HvHAK1, T04379; and PceHAK1A, AB055631.
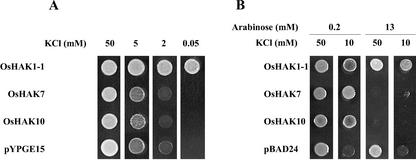
When OsHAK1-1, OsHAK7, and OsHAK10 cDNAs were transformed into the trk1 trk2 yeast mutant, OsHAK1-1 suppressed the K+ dependence of the mutant completely. Cells expressing OsHAK1-1 showed high-affinity K+ uptake (Rb+ Km of 6 μm), and grew rapidly at very low K+ concentrations (Fig. 4A). On the contrary, OsHAK7 and OsHAK10 failed to show any detectable effect on the yeast mutant.
Figure 4.
Growth of OsHAK1-1-, OsHAK7-, and OsHAK10-transformed yeast (A) and bacterial (B) mutants at different K+ concentrations. The yeast trk1 trk2 mutant was transformed with the empty plasmid pYPGE15 or with the plasmid containing the tested cDNAs under the control the PGK1 promoter. The E. coli strain TKW4205, deficient in the K+ transport systems Kdp, TrkA, and Kup, was transformed with the empty plasmid pBAD24 or with the plasmid containing the tested cDNAs under the control of an Ara-responsive promoter (Guzman et al., 1995). Testing growth media contained the K+ concentrations (A) or the K+ and Ara concentrations (B) recorded in each case.
Functional Expression of OsHAK1-1, OsHAK7, and OsHAK10 in E. coli Mutants
Because the OsHAK7 and OsHAK10 cDNAs failed to suppress the K+ transport defect of yeast mutants, we tested these cDNAs in an E. coli K+ transport mutant using an expression vector with an Ara-responsive promoter (Guzman et al., 1995), and followed a similar approach with OsHAK1-1, for comparison. At pH 5.5, which is the most convenient for this type of experiment (Senn et al., 2001), the three clones improved the growth of the transformant bacteria at low K+ and low Ara (10–200 μm), which promotes a moderate transgene expression (Fig. 4B). At high Ara concentration (13 mm), OsHAK7 and OsHAK10, but not OsHAK1-1, were toxic at all K+ concentrations tested (Fig. 4B). Toxicity of overexpressed HAK transporters in bacteria occurs with some of them, but the reasons have not been investigated. The barley transporter HvHAK2 is toxic at high Ara, but only at high K+ (Senn et al., 2001), and of the two HAK transporters studied in Cymodocea nodosa, one is toxic but not the other (Garciadeblas et al., 2002). In K+-starved cells of the E. coli mutant, expression of the OsHAK7 and OsHAK10 transporters at high Ara concentrations elicited K+ or Rb+ uptake at higher rates than the endogenous uptake of the bacterial mutant. Kinetic analysis of the uptake rates at pH 5.5 indicated that both transporters exhibited K+ and Rb+Kms of 5 to 10 mm (not shown). In similar experiments, OsHAK1-1 did not mediate sufficiently rapid K+ or Rb+ uptake, and the kinetic of uptake could not be determined.
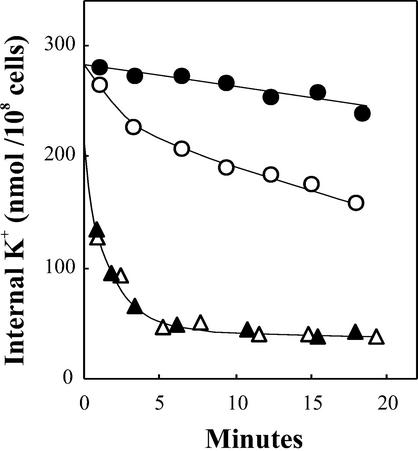
It has been found recently that HAK transporters are reversible in E. coli and mediate rapid K+ losses, which can be tested at pH 5.5, low external K+, and in the presence of 10 mm propionic acid to inhibit the endogenous K+ efflux (Garciadeblas et al., 2002). In these conditions, when the expression of the transgenes was fully induced at 13 mm Ara, both OsHAK7 and OsHAK10 mediated very rapid K+ losses. In contrast, OsHAK1-1 mediated a much slower K+ loss (Fig. 5), which was consistent with the low uptake rates found for this transporter in E. coli.
Figure 5.
K+ loss in the E. coli strain TKW4205 transformed with plasmid pBAD24 containing the OsHAK1-1, OsHAK7, or OsHAK10 cDNAs. Time courses of the K+ contents of cells induced in 13 mm Ara and suspended in K+-free minimal medium, pH 5.5, supplemented with 4.9% (w/v) sorbitol, and 10 mm propionic acid. Black circles, Cells transformed with pBAD24; white circles, cells transformed with OsHAK1-1; black triangles, cells transformed with OsHAK7; white triangles, cells transformed with OsHAK10.
OsHAK10 Locates to the Tonoplast in Onion Epidermal Cells
Phenotypic suppression by the plant transporter OsHAK1-1 of the trk1 trk2 yeast mutant indicated a plasma membrane localization of the transporter in yeast cells, possibly mimicking the location in the plant. In contrast, the discrepancy between yeast and bacterial cells regarding the functionality of OsHAK7 and OsHAK10 suggested that these transporters might be targeted to a yeast membrane other than the plasma membrane, and that they could be K+ transporters of internal membranes of the plant cells. This was an attractive possibility because the high number of rice HAK transporters could be explained by a certain diversity of functions.
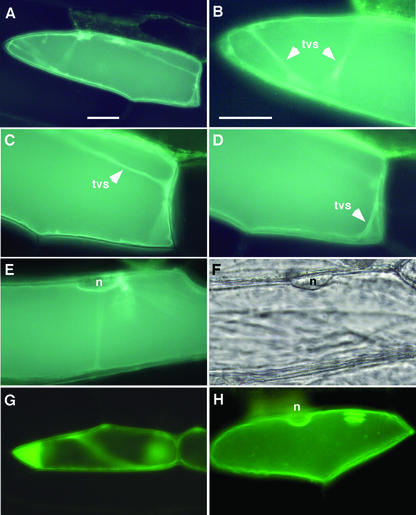
As a first approach to investigate the location of OsHAK7 and OsHAK10, we carried out transient expressions of their green fluorescent protein (GFP) fusion proteins in living onion epidermal cells (Scott et al., 1999). The results depicted in Figure 6 indicate that the OsHAK10:GFP protein located to the tonoplast. Onion epidermal cells have a large vacuole, and as a consequence, the GFP fluorescence contoured the cells, except in places where the tonoplast was separated from the cell periphery by the nuclei. Membrane-lining transvacuolar strands of cytoplasm spanning the cell body also produced fluorescence that upon close-up observation clearly depicted two vacuolar membranes. This fluoresce distribution was identical to those shown by the tonoplast transporters AtNHX1 (Apse et al., 1999) and AtNHX2 (Yokoi et al., 2002), although the fluorescence intensity of the OsHAK10:GFP protein was lower, probably reflecting its lower expression level (experiments with AtNHX1 were carried out in parallel with those of OsHAK10 and are shown in Fig. 6; the results with AtNHX2 have been reported previously by Yokoi et al., 2002). Similar experiments with a OsHAK7:GFP fusion produced inconclusive results because of the low fluorescence intensity in labeled cells, which could be due to poor expression or posttranslational processing of the recombinant protein.
Figure 6.
Tonoplast localization of HAK10:GFP. Transient expressions in onion epidermal cells of a OsHAK10:GFP translational fusion protein was visualized by epifluorescence microscopy. A, GFP fluorescence concentrated to the tonoplast. The large vacuole of onion epidermal cells occupies most of the cell volume. B through D, Captions of different focal planes of the cell imaged in A, showing transvacuolar strands of cytoplasm (tvs). Note that the transvacuolar strands seen in C and D are lined by two tonoplast membranes. E, The tonoplast follows the cell contour except in the perinuclear region (n) where the vacuole detaches from the cell surface. F, Clear field caption of the cell shown in E depicting the perinuclear region (n). As controls, transient expression assays in onion epidermal cells of GFP and AtNHX1:GFP were carried out in parallel. G, In the control cell expressing GFP, the fluoresce concentrated in the cytoplasm and was manifestly absent in the vacuole. H, In the control cell expressing AtNHX1:GFP, the fluorescence concentrated to the tonoplast showing the perinuclear region (n). Bar = 25 μm.
An HAK Transporter Mediates High-Affinity K+ Uptake in Rice Roots
The expression pattern of the OsHAK1 transcript in rice seedlings and the kinetic characteristics of OsHAK1-1-mediated Rb+ influx in yeast mutants suggested that OsHAK1 contributes to the high-affinity K+ transporter of rice roots. To assess the role of HAK transporters in K+ uptake by rice roots, we set up a kinetic study to compare K+ uptake in rice roots and in yeast expressing OsHAK1-1.
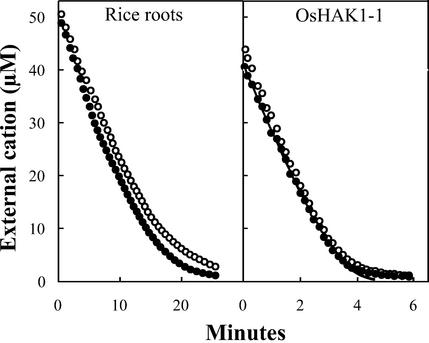
When rice roots of K+-starved seedlings were exposed to a low K+ concentration, the concentration dependence of the rate of depletion followed a Michaelis-Menten equation exactly until K+ reached a concentration of approximately 4 μm, and similar results were obtained with the yeast mutant expressing OsHAK1-1 (Fig. 7). Similar experiments using Rb+, which reached low internal concentrations in short-term experiments and practically fulfilled the condition of zero Rb+ inside, produced kinetics that were almost identical to the K+ kinetics (Fig. 7; deviations are discussed in “Materials and Methods”). Moreover, the kinetics of Rb+ influx in yeast expressing OsHAK1-1 was repeated by recording influxes (Rodríguez-Navarro and Ramos, 1984) and we obtained the same values for both Km and Vmax. All this indicated that the experimental approach was correct.
Figure 7.
Uptake of K+ or Rb+ by roots of K+-starved rice seedlings and by the yeast trk1 trk2 mutant expressing OsHAK1-1. Depletion in the external medium of K+ (white circles) or Rb+ (black circles) was followed with a selective electrode, whose response was calibrated at intervals in the same experiment by atomic emission spectrophotometry. The line drawn in the right panel corresponds to an exact Michaelis-Menten relationship between rate and Rb+ concentration. The Kms calculated for the parts of the curves that follow the Michaelis-Menten equation are: in rice roots, 11 μm K+ or Rb+, and in yeast, 6 μm K+ or Rb+. The corresponding Vmaxs are: 3.2 nmol mg−1 min−1 in roots and 10 nmol mg−1 min−1 in yeast (dry weight in both cases).
The actual Km of K+ uptake by rice roots showed a significant variability depending on the K+ concentration of the external medium that the seedlings maintained at the moment of their use. The lower the K+ concentration, the greater the affinity for K+ (lower Km) that the roots exhibited. Our standard plants were in steady state with 0.1 to 0.2 μm K+ in the external medium. In these plants, the values of the K+, Rb+, and Cs+ Kms varied a little among plants grown in different containers, but were almost identical in repetitions with plants from the same container. In any case, the conclusion of the experiments was clear: The K+ transporter of rice roots did not discriminate between K+ and Rb+, and very little between K+ and Cs+. In the yeast mutant expressing OsHAK1-1, the values and the relationships among the K+, Rb+, and Cs+ Kms were very similar to those in rice (Table III).
Table III.
Kinetic constants for alkali cation influxes in rice roots and in the yeast mutant expressing OsHAK1-1
| Cation | Rice Roots
|
OsHAK1-1 in Yeast
|
|
|---|---|---|---|
| Km | Km | Ki | |
| μm | μm | mm | |
| K+ | 11–18a | 6 ± 1 | – |
| Rb+ | 11–18a | 6 ± 1 | – |
| Cs+ | 30–50a | 11 ± 2 | – |
| Na+ | n.d. | 6,000 ± 500 | 5/11b |
Dependent on the batch of seedlings; see text.
Eleven micromolar on Rb+ influx and 5 mm on K+ uptake using the K+ depletion method, as described “Materials and Methods.”
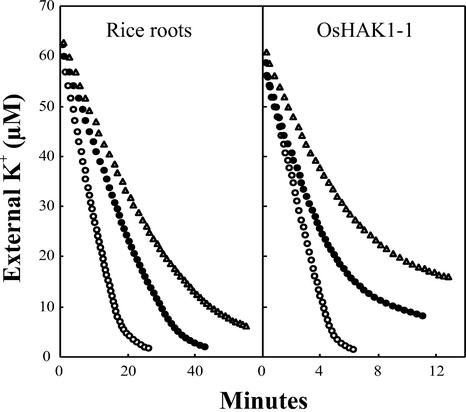
For further comparison, we determined the inhibition of K+ uptake by NH4+ in rice roots and in the yeast mutant expressing OsHAK1-1. The inhibition on the Vmax was similar in yeast and rice roots (approximately 50% inhibition at 0.5 mm NH4+) and the inhibition on the Km only a little stronger in yeast (Fig. 8). Altogether, the effects were not very different; for example, the rate of K+ uptake at 30 μm K+ was inhibited 3-fold by 0.5 mm NH4+ both in yeast and roots. On considering the differences, it should be observed that NH4+ may have important non-comparable indirect effects in yeast and rice roots because the cation can be transported in both systems with different effects on the membrane potential.
Figure 8.
Inhibition by NH4+ of the depletion of the external K+ by roots of K+-starved rice seedlings and by the yeast trk1 trk2 mutant expressing OsHAK1-1. Conditions as in Figure 7. NH4+ concentrations: control, white circles; 250 μm, black circles; and 500 μm, triangles.
Na+ could not be used for the same purpose as NH4+ because Na+ uptake in rice roots was complex, as described for barley roots (Rains and Epstein, 1967a, 1967b), and probably depolarized the plasma membrane as well. The kinetics of Na+ influx mediated by OsHAK1-1 in yeast was simple (5 mm Na+Km and a Vmax 2-fold lower than the Rb+ Vmax) and not related to the kinetics of Na+ uptake in seedling roots. This indicated that Na+ uptake in rice roots was not dominated by HAK transporters.
DISCUSSION
We have identified 17 HAK genes in rice cv Nipponbare, and the existence of two additional genes, although not confirmed, should be taken into consideration. These genes can encode rather divergent proteins, which form a phylogenetic tree with the previously described clusters (Rubio et al., 2000). As in Arabidopsis, pairs of distant transporters keep low amino acid sequence identity, around 40%, and clusters I and II group most of the rice transporters (Fig. 2). This high number of transporters in rice and their sequence divergence give further support to the notion that suggests that KT-HAK-KUP transporters play distinct functions in different types of membranes (Senn et al., 2001). This conclusion is also supported by the abundance of the transcripts of three transporters studied in this report, OsHAK1-1, OsHAK7, and OsHAK10, which were rather abundant in roots, and slightly affected by the external conditions. OsHAK7 and OsHAK10 are similarly expressed in seedling roots and shoots, and OsHAK10 is also expressed in the panicle at flowering stage (see EST accession no. C72058). These results are unlikely to correspond to transporters that are expressed in a few specialized cells.
Experiments of K+ transport either in yeast or in bacteria (Figs. 4, 5, and 7) demonstrated that OsHAK1-1, OsHAK7, and OsHAK10 are K+ transporters. Pending experimental confirmation, it is likely that the other transporters of the rice HAK family are also K+ transporters. Difficulties in obtaining functional expressions in yeast or bacteria have led to proposals of K+ uptake activity of several Arabidopsis KT-KUP transporters with weak experimental support. We would like to emphasize that growth improvement of yeast or E. coli mutants in K+-limiting minimal media by expressing a gene or cDNA does not formally demonstrate that K+ uptake has been enhanced and that the gene or cDNA encodes a K+ transporter. That conclusion could be misleading because the growth rate of genetically manipulated strains of fungi and bacteria in a minimal medium with or without limiting K+ can be enhanced by improving many metabolic restrictions and not only by enhancing K+ uptake.
A relevant question in the study of HAK transporters is their individual cellular location, which may not be the same for all of them. The kinetic characteristics of the high-affinity K+ uptake of barley (Santa-María et al., 1997) and rice (see below) roots strongly suggest that some HAK transporters are involved in this uptake, and that this function has to take place in the plasma membrane. On the other hand, tonoplast localization of some HAK transporters is suggested by the analysis of the cellular K+ distribution in K+-starved barley roots, in which a K+-H+ symporter, the putative functional mechanism of HAK transporters (Rodríguez-Navarro, 2000), is needed to explain the release of K+ from the vacuole to the cytoplasm when the vacuolar concentration is low (Walker et al., 1996). Although the formal demonstration of the cellular location of every HAK transporter requires the use of antibodies, the targeting of the OsHAK10-GFP fusion protein to the tonoplast of living onion epidermal cells (Fig. 6) strongly suggests that this transporter locates to the tonoplast of rice cells.
The kinetic characteristics of a transporter are key data for formulating hypotheses of its physiological functions. However, even in those transporters that have been kinetically characterized in yeast or bacteria, their kinetic characteristics in plant cells are almost impossible to assess while we ignore the membrane in which each transporter is expressed. Two factors, pH and membrane potential, can affect the kinetics of an HAK transporter (Blatt et al., 1987), and neither of them is constant for all cells and membranes in a plant. Our test conditions in bacteria at pH 5.5, at which the membrane potential may be around −60 mV in E. coli (Ramos and Kaback, 1977; Kashket, 1982), probably mimic the functional conditions of a tonoplast transporter, but may be very different from those prevailing for a plasma membrane transporter. In contrast, expression in the plasma membrane of yeast cells may situate the transporter in conditions very similar to those prevailing in a plasma membrane transporter of root epidermal or cortex cells, but different from those prevailing in other cells.
According to this, the low affinity exhibited by OsHAK10 for K+ and Rb+ when expressed in bacteria probably mimics its affinity for K+ in functional conditions in the tonoplast of plant cells, and has a value that can be expected for a transporter permanently exposed to high K+. The most likely function of a tonoplast HAK K+ transporter would be to mediate the transfer of K+ from the vacuole to the cytoplasm, coupling the movement of K+ to the movement of H+, which changes the energetics of the process with reference to K+ channels. As explained above, this coupling is required for the vacuolar loss of K+ in K+-starved barley roots because the vacuolar K+ concentration is low (Walker et al., 1996) but, to our knowledge, a similar requirement has not been proposed for cells in normal conditions, in which OsHAK10 has an extensive expression. Because in the absence of energetic restrictions for a K+ channel, a K+-H+ symport is equivalent to the coupling of a K+ channel and a K+/H+ antiporter (two K+ move outward through the channel and one returns through the antiporter), the function of a tonoplast HAK transporter cannot be understood with our current knowledge of tonoplast physiology. However, one possibility to be considered is that, in some conditions, the tonoplast electrical potential is depolarized to increase the transfer of anions (e.g. malate) from the vacuole to the cytoplasm through anion channels (Barbier-Brygoo et al., 2000) and that this made the concomitant transfer of K+ through channels impossible.
The most frequently proposed function for KT-HAK-KUP transporters is root K+ uptake. Although the results obtained with akt1 and akt2 mutants of Arabidopsis (Hirsch et al., 1998; Spalding et al., 1999; Dennison et al., 2001) suggest that K+ channels may dominate root K+ uptake, and that the involvement of transporters in this function may be secondary, this is not the case for rice. To our knowledge, no report has demonstrated that Arabidopsis can deplete external K+ down to 0.1 to 0.2 μm, as we have found for rice. At these external K+ concentrations, assuming that the cytoplasmic K+ concentration in epidermal and cortex root cells is 50 mm (Walker et al., 1996, 1998), the K+ equilibrium potential across the plasma membrane is at least −320 mV, a value much more negative than the most negative membrane potential value recorded in plants. This suggests that a K+ channel cannot mediate K+ uptake (Rodríguez-Navarro, 2000) and that a K+ transporter capable of mediating active K+ uptake exists in rice roots, which also exhibits a low capacity to discriminate among K+, Rb+, and Cs+ (Table III). At least some HAK transporters fulfill the two characteristics because they can be K+-H+ symporters (Rodríguez-Navarro, 2000) and do not discriminate significantly among K+, Rb+, and Cs+ (Rubio et al., 2000; Table III). In addition to the general kinetic similarities, the NH4+ sensitivity of K+ uptake in roots was very similar to that found for OsHAK1-1 in yeast. Although we could not clone the complete OsHAK1 cDNA, it is unlikely that all the coincidences between K+ uptake in roots and yeast expressing OsHAK1-1 are due to the 48 bp with which we completed the OsHAK1 cDNA (Fig. 3). More likely, OsHAK1-1 mimics functionally OsHAK1, and OsHAK1, or a closely related transporter dominates root K+ uptake.
OsHAK1 is expressed in young roots but also in leaf, panicle, callus, and seedling shoots. If OsHAK1 is the root high-affinity K+ transporter, its function in leaf or panicle is not clear and two different causes may explain its presence in these organs: Either some cells are exposed to very low K+ concentrations or some cells are depolarized and, in these cells, OsHAK1 exhibits a much lower affinity for K+ than in root cells.
In conclusion, one or perhaps several HAK transporters mediate K+ uptake by roots in rice, whereas other HAK transporters locate to the tonoplast with the probable function of mediating the release of K+ from the vacuoles. However, the high number of HAK transporters in rice and their extensive expression in many parts of the plant makes it difficult to identify at this moment particular functions with specific transporters. Moreover, there may be functions of HAK transporters that cannot be predicted at this moment.
MATERIALS AND METHODS
Plant Seedlings
Rice (Oryza sativa cv Nipponbare) seeds were surface sterilized and germinated in filter paper wetted with a 1.0 mm CaSO4 solution. Then the seeds were transferred to 5- to 10-L plastic containers with a 5 mm CaCl2 solution, in which they were supported by cheesecloth stapled to floating frames. Seedlings were grown in the dark for 7 to 10 d at 28°C. The presence of root-associated bacteria or fungi was checked systematically with microbiological counts to rule out misleading results. In typical batches of plants, the number of bacteria was low and fungi were almost absent. Excised roots were cut approximately 5 mm below the seeds.
Strains, Media, and Growth Conditions
The Escherichia coli strain DH5α was routinely used for plasmidic DNA propagation. The bacterial strain TKW4205 (thi rha lacZ nagA recA Sr::Tn10 ΔkdpABC5 trkA405 kup1) deficient in the three K+ uptake systems (Kdp, TrkA, and Kup) was used for complementation assays (Senn et al., 2001). The yeast (Saccharomyces cerevisiae) strain WΔ3 (Mat a ade2 ura3 trp1 trk1Δ::LEU2 trk2Δ::HIS3; Haro et al., 1999) deficient in the endogenous K+ uptake systems TRK1 and TRK2 was used for functional complementation assays in yeast. Bacterial TKW4205 strain and derivatives were grown in Luria-Bertani medium supplemented with 30 mm K+. For bacterial growth tests at low K+, serial dilution drops of strains grown in LB supplemented with 30 mm K+ were inoculated on a solid medium containing 5 mm PO4H3, 0.4 mm MgSO4, 6 μm FeSO4, 1 mm citric acid, 1 mg L−1 thiamine, 0.2% (w/v) glycerol, 8 mm Asn, 10 mm MES, 20 μm CaCl2, brought to pH 5.5 with Arg, and supplemented with the indicated K+ and Ara concentrations (Senn et al., 2001). Yeast strains were grown in synthetic dextrose medium (Sherman, 1991) supplemented with 50 mm K+. For yeast growth experiments at low K+ concentrations, serial dilution drops of strains were inoculated on Arg phosphate medium (Rodríguez-Navarro and Ramos, 1984) supplemented with the indicated K+ concentrations.
Recombinant DNA Techniques
Manipulation of nucleic acids was performed by standard protocols or, when appropriate, according to the manufacturer's instructions. PCRs were performed in a thermocycler (Perkin-Elmer Applied Biosystems, Foster City, CA), using the Expand-High-Fidelity PCR System (Roche Molecular Biochemicals, Summerville, NJ). Some of the PCR fragments were first cloned into the PCR2.1-Topo vector using the TOPO TA Cloning Kit (Invitrogen, Carlsbad, CA). For yeast expressions, the cDNAs were cloned into vector pYPGE15 and transformed into the trk1 trk2 yeast mutant, as described previously (Santa-María et al., 1997). For expression in the E. coli mutant TKW4205, the cDNAs were cloned into vector pBAD24, following the procedure described previously (Senn et al., 2001). DNA sequencing was performed in an automated ABI PRISM 377 DNA sequencer (Perkin-Elmer Applied Biosystems). Total rice RNA was prepared using the RNeasy Plant Kit (Qiagen USA, Valencia, CA). PCR amplifications of HAK fragments were carried out on double-stranded cDNA synthesized from total RNA by using the cDNA Synthesis System Kit (Roche) and degenerate primers deduced from HAK conserved regions as described by Rubio et al. (2000). The full-length cDNAs were obtained by using the 5′/3′RACE Kit (Roche) according to the manufacturer's instructions.
Protein Alignments and Phylogenetic Tree Generations
Protein sequence alignments and phylogenetic trees were obtained by using the ClustalX program (Thompson, 1997). Almost identical trees were generated using the complete sequences or the sequences spanning from the first to the last putative transmembrane fragments (Rodríguez-Navarro, 2000), which were the sequences used to generate the tree shown in Figure 2. Because the OsHAK4, OsHAK8, and OsHAK16 were incomplete, their phylogenetic positions might vary slightly when the full-length sequences were used.
Real-Time PCR Assays
For the experiments reported in Table II, seedlings were grown as described above, in the absence of K+ or in the presence of 3 mm KCl. PCR assays were repeated three times with mRNA obtained from the same set of plants; two repetitions were carried out using the same cDNA, which was prepared using the cDNA Synthesis System Kit (Roche), as described above. For the third repetition, the cDNA was prepared using the First-strand cDNA Synthesis Kit (Amersham-Pharmacia Biotech, Uppsala). In another experiments plants were grown in the mineral medium described previously (Yeo et al., 1999) supplemented with KCl up to 3 mm K+. In this medium, plants were grown for 15 d in a 14-h photoperiod of 400 μmol m−2 s−1 and 27°C/20°C (light and dark, respectively). The K+-free medium was a modification of the 3 mm K+ medium that contained the following differential elements: 1 mm (NH)4PO4H, 0.75 mm Ca(NO3)2, 0.25 mm CaCl2, 0.5 mm MgSO4, and 1.14 mm MgCl2. In these experiments, the cDNA was prepared using the First-strand cDNA Synthesis Kit (Amersham-Pharmacia Biotech). Real-time PCRs were performed using an ABI Prism 7700 Sequence Detection System (Perkin-Elmer Applied Biosystems) and SYBR Green PCR Master Mix (Perkin-Elmer Applied Biosystems). The PCR amplification was carried out with an initial step at 95°C for 10 min followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. In every series of PCR reactions, there were two negative controls to exclude possible external or genomic DNA contamination, one in which the PCR reaction was performed without template cDNA and another which contained an aliquot of the original RNA sample. All reaction mixtures were analyzed by agarose gel electrophoresis to confirm that only the expected PCR product was produced. To normalize the results, the OsRAC1 cDNA (encoding rice actin) was amplified in all the experiments in parallel with the cDNAs under analysis. Quantification of the PCR reaction was carried out in the exponential phase of the reaction, where yield is proportional to the input cDNA. Serial dilutions of the cDNA prepared from roots of K+-starved seedlings were used to construct standard regression lines, which relate the OsHAK cDNA contents and the threshold cycle (a measure of the amplified cDNA). Then, the contents of the OsHAK1 and OsHAK7 cDNAs in every sample were determined by interpolation in the standard line, and the results were expressed relative to the corresponding OsHAK cDNA contents of the roots of K+-starved seedlings. The primers used were: OsRAC1, forward primer 5′-CCTCGCACCAAGCAGCATGAAGA-3′ and reverse primer 5′-CGACTCATCATACTCTCCCTTTG-3′; OsHAK1, forward primer 5′-CAAGAGGATCGCGGTGAACTACA-3′ and reverse primer 5′-CTTGAGCAGCTGATCGTTTGGA-3′; and OsHAK7, forward primer 5′-GAACTCCAACTTCCTCAAGACG-3′ and reverse primer 5′-AGATCATGCCGACTTCGACGAG-3′.
Cation Uptake Experiments in Roots and Yeast Cells
To generate enough kinetic data for our study, it was necessary to develop a rapid and reliable method to measure K+ influx, which is not simple if 42K+ is not available, and the use of 86Rb+ to label K+ is avoided (Rodríguez-Navarro, 2000). Therefore, we improved a rapid method for the analysis of the depletion of K+ in a vessel with a high density of roots (Huang et al., 1992), using a K+-selective electrode and a simple calculation program. In this type of experiment, if the concentration dependence of the rate of K+ uptake follows a Michaelis-Menten equation, the time course of the depletion can be used to obtain the Km and Vmax. The problem of this procedure is that a Michaelis-Menten equation explains the rate of an enzyme-catalyzed reaction when the backward reaction is negligible; otherwise, the rate would also depend on the product concentration (internal K+ in our case). Although recording initial rates of reaction (initial rates of uptake in this case) is sufficient to fulfill the requirement; in some cases, as those described here, the product is present but the backward reaction is insignificant.
Although the cation depletion experiments described here were carried out with excised roots, control experiments with whole seedlings produced identical results, as has been reported previously (Huang et al., 1992). Excised roots (80–270 mg dry weight) were directly immersed in a 100-mL thermostatic vessel maintained at 28°C. A K+ electrode (Mettler-Toledo GmbH, Urdorf, Switzerland) was used to monitor K+ or Rb+ concentrations. In every experiment, the response of the electrode was controlled by taking samples at regular intervals and analyzing the K+ or Rb+ concentrations by atomic emission spectrophotometry. The mV readings of the ionometer were transferred to an Excel sheet (Microsoft, Redmond, WA) and transformed in micromolar values using the individually determined response equation of the electrode. In Cs+ and Na+ uptake experiments, the concentrations were monitored exclusively by atomic emission spectrophotometry.
To calculate the Km and Vmax from the cation depletion curves (Fig. 7), we integrated the following equation (C+ represents an unspecified cation, V is the volume of the vessel, and p the weight of roots or cells):
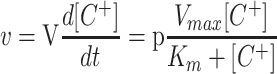 |
1 |
obtaining
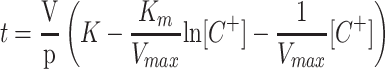 |
2 |
By constructing a table with the experimental values of t and [C+], the Km and Vmax values were obtained by fitting the data to Equation 2 using the Solver function of the Microsoft Excel Program. Because the rates of uptake (depletion) deviated from a Michaelis-Menten equation at K+, Rb+, or Cs+ concentrations below 4 μm (see Fig. 7), approximately, in the table constructed with the t and [C+] values, the pair of values corresponding to the lowest concentrations were deleted before applying the fitting program. The concentration limit at which the deviation was appreciable was experimentally calculated in each experiment by using sets of data extended down to different micromolar concentrations. The deviation from Michaelis-Menten kinetics at low cation concentrations can be explained by two factors. In the cases of Rb+ and Cs+, the deviation can be explained by the accumulation in the testing medium of K+ lost from the root or yeast cells. In the case of K+, the deviation can be explained because we measured net uptake, and the difference between net uptake and influx becomes more important when the influx is very low. Repeated experiments carried out with seedlings from the same container produced almost identical results, regardless of the amount of roots used, from 80 to 270 mg dry weight, except for the proportional variation of the Vmax. The most significant source of variation of the Kms among different containers was the K+ concentration in the medium in which the seedlings were growing, as described in “Results.” In yeast cells, the same procedure was applied in parallel with traditional experiments of Rb+ and Na+ influx in which the initial rates of cation uptake were plotted as a function of the cation concentration (Rodríguez-Navarro and Ramos, 1984). Satisfactorily, for Rb+, both procedures produced identical results.
K+ and Rb+ Fluxes in Bacteria
K+ and Rb+ uptake assays in bacteria were carried out as described elsewhere (Senn et al., 2001). In brief, cells were grown at 37°C in LB medium supplemented with 100 μg mL−1 ampicilin and 30 mm K+ up to an absorbance of 1.0. Ara was then added to a final concentration of 13 mm and cells were incubated for 15 min. Then the cells were centrifuged, transferred, and kept for 30 min in K+-free minimal medium (Senn et al., 2001) supplemented with 13 mm Ara. After this K+ starving period, the cells were transferred to fresh K+-free minimal medium to which Rb+ or K+ was added to assay the initial rates uptake. For K+ efflux experiments, after incubation for 15 min in LB with 13 mm Ara, the cells were centrifuged and suspended in K+-free minimal medium, pH 5.5, supplemented with 4.9% (w/v) sorbitol, 10 mm propionic acid, and 13 mm Ara (the minimal medium contains 22 mm glycerol). In this medium, the K+ loss of the cells was recorded during 10 to 20 min (Garciadeblas et al., 2002). In uptake and efflux assays, cell samples were taken at intervals, filtered through 0.45-μm pore membrane filters (Millipore, Bedford, MA), and washed with the same minimal medium supplemented with 10 mm MgCl2. Cells were acid extracted overnight in a 0.1 m HCl solution and the Rb+ or K+ concentrations in the supernatant were determined by atomic emission spectrophotometry. Reported experiments of K+ efflux were repeated at least three times. Only a representative experiment is presented because the results showed a low variability, similar to the differences found between cells transformed with OsHAK7 or OsHAK10 in Figure 5.
Localization of HAK10:GFP
Intracellular localization of OsHAK10 was determined by monitoring the transient expression of a OsHAK10:GFP translational fusion product in onion (Allium cepa) epidermal cells after DNA particle bombardment. The coding region of GFP (Sheen et al., 1995) was fused, in-frame, to the 3′ terminus of full-length OsHAK10 cDNA and this chimeric gene was cloned into the vector pGreen-35S (John Innes Centre, Norwich, UK, http://www.pgreen.ac.uk). For control experiments, identical constructs containing the AtNXH1:GFP and GFP cDNAs were prepared. The OsHAK10:GFP, AtNXH1:GFP, and GFP constructs were coated onto gold particles (1 μm) and delivered into onion cells with a Biolistic PDS-1000-He apparatus (Bio-Rad Laboratories, Hercules, CA). The bombardment parameters were: rupture disc bursting pressure, 900 psi; distance to macro-carrier, 8 mm; distance to stopping screen, 6 mm; and distance to target tissue, 6 cm. Onion epidermal cells were placed into Murashige and Skoog medium with 2% (w/v) Suc before bombardment and incubated in the dark at 28°C for 24 to 36 h after particle delivery. GFP fluorescence was visualized under an Axioskop microscope (Zeiss, Jena, Germany) equipped with a fluorescein isothiocyanate filter set (band pass 450–490 nm, farb teiler [color splitter] 510 nm, and long pass 515 nm).
Database Searches
BLAST searches for rice sequences, using the sequences of the barley (Hordeum vulgare) transporters HvHAK1 and HvHAK2, were carried out in the two databases (http://www.rice-research.org/ and http://www.ncbi.nlm.nih.gov/). The former is a working draft of the rice genome sequence.
ACKNOWLEDGMENTS
We thank Manuel Aguilar for the multiplication of rice cv Nipponbare seeds, Pharmacia rice-research.org program for allowing us the use of its database, and the National Institute of Agrobiological Resources/Society for Technoinnovation of Agriculture, Forestry and Fisheries Institute for providing several cDNA clones and rice cv Nipponbare seeds.
Footnotes
This work was supported by the Consejería de Educación y Cultura de la Comunidad de Madrid (Programa de Grupos Estratégicos) and by the Ministerio de Ciencia y Tecnología (grant no. BIO2000–0938).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.007781.
LITERATURE CITED
- Apse MP, Aharon GS, Snedden WA, Blumwald E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science. 1999;285:1256–1258. doi: 10.1126/science.285.5431.1256. [DOI] [PubMed] [Google Scholar]
- Bakker EP. Low-affinity K+uptake systems. In: Bakker EP, editor. Alkali Cation Transport Systems in Prokaryotes. Boca Raton, FL: CRC Press; 1993. pp. 253–276. [Google Scholar]
- Bañuelos MA, Klein RD, Alexander-Bowman SJ, Rodríguez-Navarro A. A potassium transporter of the yeast Schwanniomyces occidentalis homologous to the Kup system of Escherichia colihas a high concentrative capacity. EMBO J. 1995;14:3021–3027. doi: 10.1002/j.1460-2075.1995.tb07304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bañuelos MA, Madrid R, Rodríguez-Navarro A. Individual functions of the HAK and TRK potassium transporters of Schwanniomyces occidentalis. Mol Microbiol. 2000;37:671–679. doi: 10.1046/j.1365-2958.2000.02040.x. [DOI] [PubMed] [Google Scholar]
- Barbier-Brygoo H, Vinauger M, Colcombet J, Ephritikhine G, Frachisse J-M, Maurel C. Anion channels in higher plants: functional characterization, molecular structure and physiological role. Biochim Biophys Acta. 2000;1465:199–218. doi: 10.1016/s0005-2736(00)00139-5. [DOI] [PubMed] [Google Scholar]
- Blatt MR, Rodríguez-Navarro A, Slayman CL. Potassium-proton symport in Neurospora: kinetic control by pH and membrane potential. J Membr Biol. 1987;98:169–189. doi: 10.1007/BF01872129. [DOI] [PubMed] [Google Scholar]
- Cantrell RP, Reeves TG. The cereal of the world's poor takes center stage. Science. 2002;296:53. doi: 10.1126/science.1070721. [DOI] [PubMed] [Google Scholar]
- Dennison KL, Robertson WR, Lewis BD, Hirsch RE, Sussman MR, Spalding EP. Functions of AKT1 and AKT2 potassium channels determined by studies of single and double mutants of Arabidopsis. Plant Physiol. 2001;127:1012–1019. [PMC free article] [PubMed] [Google Scholar]
- Elumalai RP, Nagpal P, Reed JW. A mutation in the Arabidopsis KT2/KUP2potassium transporter gene affects shoot cell expansion. Plant Cell. 2002;14:119–131. doi: 10.1105/tpc.010322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbairn DJ, Liu W, Schachtman DP, Gomez-Gallego S, Day SR, Teasdale RD. Characterisation of two distinct HKT1-like potassium transporters from Eucalyptus camaldulensis. Plant Mol Biol. 2000;43:515–525. doi: 10.1023/a:1006496402463. [DOI] [PubMed] [Google Scholar]
- Fu H-H, Luan S. AtKUP1: a dual affinity K+transporter from Arabidopsis. Plant Cell. 1998;10:63–73. doi: 10.1105/tpc.10.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garciadeblas B, Benito B, Rodríguez-Navarro A (2002) Molecular cloning and functional expression in bacteria of the potassium transporters CnHAK1 and CnHAK2 of the seagrass Cymodocea nodosa. Plant Mol Biol (in press) [DOI] [PubMed]
- Guzman L-M, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBADpromoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haro R, Sainz L, Rubio F, Rodríguez-Navarro A. Cloning of two genes encoding potassium transporters in Neurospora crassa and expression of the corresponding cDNAs in Saccharomyces cerevisiae. Mol Microbiol. 1999;31:511–520. doi: 10.1046/j.1365-2958.1999.01192.x. [DOI] [PubMed] [Google Scholar]
- Hirsch RE, Lewis BD, Spalding EP, Sussman MR. A role for the AKT1 potassium channel in plant nutrition. Science. 1998;280:918–921. doi: 10.1126/science.280.5365.918. [DOI] [PubMed] [Google Scholar]
- Horie T, Yoshida K, Nakayama H, Yamada K, Oiki S, Shinmyo A. Two types of HKT transporters with different properties of Na+ and K+ transport in Oryza sativa. Plant J. 2001;27:115–128. doi: 10.1046/j.1365-313x.2001.01077.x. [DOI] [PubMed] [Google Scholar]
- Huang ZZ, Yan X, Jalil A, Norlyn JD, Epstein E. Short-term experiments on ion transport by seedlings and excised roots. Plant Physiol. 1992;100:1914–1920. doi: 10.1104/pp.100.4.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashikina N, Becker D, Ache P, Meyerhoff O, Felle HH, Hedrich R. K+ channel profile and electrical properties of Arabidopsisroot hairs. FEBS Lett. 2001;508:463–469. doi: 10.1016/s0014-5793(01)03114-3. [DOI] [PubMed] [Google Scholar]
- Kashket ER. Stiochiometry of the H+-ATPase of growing and resting aerobic Escherichia coli. Biochemistry. 1982;21:5534–5538. doi: 10.1021/bi00265a024. [DOI] [PubMed] [Google Scholar]
- Kim EJ, Kwak JM, Uozumi N, Schroeder JI. AtKUP1: an Arabidopsis gene encoding high-affinity potassium transport activity. Plant Cell. 1998;10:51–62. doi: 10.1105/tpc.10.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochian LV, Lucas WJ. Potassium transport in roots. Adv Bot Res. 1988;15:93–178. [Google Scholar]
- Mäser P, Thomine S, Schroeder JI, Ward JM, Hirschi K, Sze H, Talke IN, Amtmann A, Maathuis FJM, Sanders D et al. Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 2001;126:1646–1667. doi: 10.1104/pp.126.4.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero J, Blatt MR. A new family of K+ transporters from Arabidopsisthat are conserved across phyla. FEBS Lett. 1997;415:206–211. doi: 10.1016/s0014-5793(97)01125-3. [DOI] [PubMed] [Google Scholar]
- Rains DW, Epstein E. Sodium absorption by barley roots: the role of the dual mechanisms of alkali cation transport. Plant Physiol. 1967a;42:314–318. doi: 10.1104/pp.42.3.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rains DW, Epstein E. Sodium absorption by barley roots: its mediation by mechanism 2 of alkali cation transport. Plant Physiol. 1967b;42:319–323. doi: 10.1104/pp.42.3.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos S, Kaback HR. The relationship between the elctrochemical proton gradient and active transport in Escherichia colimembrane vesicles. Biochemistry. 1977;16:854–859. doi: 10.1021/bi00624a007. [DOI] [PubMed] [Google Scholar]
- Reintanz B, Szyroki A, Ivashikina N, Ache P, Godde M, Becker D, Palme K, Hedrich R. AtKC1, a silent Arabidopsis potassium channel α-subunit modulates root hair K+influx. Proc Natl Acad Sci USA. 2002;99:4079–4084. doi: 10.1073/pnas.052677799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigas S, Debrosses G, Haralampidis K, Vicente-Agullo F, Feldmann KA, Grabov A, Dolan L, Hatzopoulos P. TRH1encodes a potassium transporter required for tip growth in Arabidopsis root hairs. Plant Cell. 2001;13:139–151. doi: 10.1105/tpc.13.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Navarro A. Potassium transport in fungi and plants. Biochim Biophys Acta. 2000;1469:1–30. doi: 10.1016/s0304-4157(99)00013-1. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Navarro A, Ramos J. Dual system for potassium transport in Saccharomyces cerevisiae. J Bacteriol. 1984;159:940–945. doi: 10.1128/jb.159.3.940-945.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio F, Santa-María GE, Rodríguez-Navarro A. Cloning of Arabidopsisand barley cDNAs encoding HAK potassium transporters in root and shoot cells. Physiol Plant. 2000;109:34–43. [Google Scholar]
- Santa-María GE, Rubio F, Dubcovsky J, Rodríguez-Navarro A. The HAK1gene of barley is a member of a large gene family and encodes a high-affinity potassium transporter. Plant Cell. 1997;9:2281–2289. doi: 10.1105/tpc.9.12.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönknecht G, Spoormaker P, Steinmeyer R, Brüggerman L, Ache P, Dutta R, Reintanz B, Godde M, Hedrich R, Palme K. KCO1 is a component of the slow-vacuolar (SV) ion channel. FEBS Lett. 2002;511:28–32. doi: 10.1016/s0014-5793(01)03273-2. [DOI] [PubMed] [Google Scholar]
- Scott A, Wyatt S, Tsou PL, Robertson D, Allen NS. Model system for plant cell biology: GFP imaging in living onion epidermal cells. Biotechniques. 1999;26:1128–1132. doi: 10.2144/99266st04. [DOI] [PubMed] [Google Scholar]
- Senn ME, Rubio F, Bañuelos MA, Rodríguez-Navarro A. Comparative functional features of plant potassium HvHAK1 and HvHAK2 transporters. J Biol Chem. 2001;276:44563–44569. doi: 10.1074/jbc.M108129200. [DOI] [PubMed] [Google Scholar]
- Sheen J, Hwang S, Niwa Y, Kobayashi H, Galbraith DW. Green-fluorescent protein as a new vital marker in plant cells. Plant J. 1995;8:777–784. doi: 10.1046/j.1365-313x.1995.08050777.x. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Spalding EP, Hirsch RE, Lewis DR, Qi Z, Sussman MR, Lewis BD. Potassium uptake supporting plant growth in the absence of AKT1 channel activity. Inhibition by ammonium and stimulation by sodium. J Gen Physiol. 1999;113:909–918. doi: 10.1085/jgp.113.6.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uozumi N, Kim EJ, Rubio F, Yamaguchi T, Muto S, Tsuboi A, Bakker EP, Nakamura T, Schroeder JL. The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol. 2000;122:1249–1259. doi: 10.1104/pp.122.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DJ, Black CR, Miller AJ. The role of cytosolic potassium and pH in the growth of barley roots. Plant Physiol. 1998;118:957–964. doi: 10.1104/pp.118.3.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DJ, Leigh RA, Miller AJ. Potassium homeostasis in vacuolate plant cells. Proc Natl Acad Sci USA. 1996;93:10510–10514. doi: 10.1073/pnas.93.19.10510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo AR, Flowers SA, Rao G, Welfare K, Senanayake N, Flowers TJ. Silicon reduces sodium uptake in rice (Oryza sativaL.) in saline conditions and this is accounted for by a reduction in the transpirational bypass flow. Plant Cell Environ. 1999;22:559–565. [Google Scholar]
- Yokoi S, Quintero FJ, Cubero B, Ruiz MT, Bressan RA, Hasegawa PM, Pardo JM. Differential expression and function of Arabidopsis thaliana NHX Na+/H+antiporters in the salt stress response. Plant J. 2002;30:529–539. doi: 10.1046/j.1365-313x.2002.01309.x. [DOI] [PubMed] [Google Scholar]
- Zakharyan E, Trchounian A. K+ influx by Kup in Escherichia coli is accompanied by a decrease in H+efflux. FEMS Microbiol Lett. 2001;204:61–64. doi: 10.1111/j.1574-6968.2001.tb10863.x. [DOI] [PubMed] [Google Scholar]
- Zimmermann S, Sentenac H. Plant ion channels: from molecular structures to physiological functions. Curr Opin Plant Biol. 1999;2:477–482. doi: 10.1016/s1369-5266(99)00020-5. [DOI] [PubMed] [Google Scholar]