Abstract
Lignins, along with condensed tannins (CTs) and salicylate-derived phenolic glycosides, constitute potentially large phenylpropanoid carbon sinks in tissues of quaking aspen (Populus tremuloides Michx.). Metabolic commitment to each of these sinks varies during development and adaptation, and depends on l-phenylalanine ammonia-lyase (PAL), an enzyme catalyzing the deamination of l-phenylalanine to initiate phenylpropanoid metabolism. In Populus spp., PAL is encoded by multiple genes whose expression has been associated with lignification in primary and secondary tissues. We now report cloning two differentially expressed PAL cDNAs that exhibit distinct spatial associations with CT and lignin biosynthesis in developing shoot and root tissues of aspen. PtPAL1 was expressed in certain CT-accumulating, non-lignifying cells of stems, leaves, and roots, and the pattern of PtPAL1 expression varied coordinately with that of CT accumulation along the primary to secondary growth transition in stems. PtPAL2 was expressed in heavily lignified structural cells of shoots, but was also expressed in non-lignifying cells of root tips. Evidence of a role for Pt4CL2, encoding 4-coumarate:coenzyme A ligase, in determining CT sink strength was gained from cellular co-expression analysis with PAL1 and CTs, and from experiments in which leaf wounding increased PAL1 and 4CL2 expression as well as the relative allocation of carbon to CT with respect to phenolic glycoside, the dominant phenolic sink in aspen leaves. Leaf wounding also increased PAL2 and lignin pathway gene expression, but to a smaller extent. The absence of PAL2 in most CT-accumulating cells provides in situ support for the idea that PAL isoforms function in specific metabolic milieus.
l-Phe ammonia-lyase (PAL; EC 4.3.1.5) catalyzes the deamination of l-Phe to yield trans-cinnamic acid, the common precursor for biosynthesis of phenolic derivatives like flavonoids, monolignols, and salicylates that are essential for adaptive, vascular, and reproductive plant development (Hahlbrock and Grisebach, 1979; Jones, 1984). In generating trans-cinnamic acid for phenylpropanoid (secondary) metabolism, PAL diverts carbon from primary metabolic pathways driving cell division and expansion. Control of this gateway can occur by environmental and developmental control of PAL transcription (Liang et al., 1989; Shufflebottom et al., 1993; Kumar and Ellis, 2001), by metabolic feedback inhibition of PAL activity (Bolwell et al., 1986), and by expression of multiple protein isoforms. Isoforms of PAL are posttranslationally modified (Bolwell, 1992; Allwood et al., 1999), exhibit differential metabolite sensitivity (Sarma et al., 1998), and preferentially associate with metabolic channeling complexes (Rasmussen and Dixon, 1999). Thus, differential expression of PAL isoforms could provide regulatory flexibility that may be integral to the ability of rapid-growing tree species like quaking aspen (Populus tremuloides Michx.) to coordinate secondary carbon allocation with carbon fixation and nutrient supply. Condensed tannin (CT) and phenolic glycoside (PG) pools vary 3- to 4-fold over the seasonal course of aspen leaf development (for review, see Lindroth and Hwang, 1996), and vary in response to CO2, light, and nutrient conditions in willow (Salix myrsinifolia), a close aspen relative (Julkunen-Titto et al., 1993). These pools can constitute large enough sinks of phenolic carbon (10%–35% leaf dry weight) to affect overall tree growth, and this is manifest as an inverse relation between aspen clonal growth index and CT-PG pool size (for review, see Lindroth and Hwang, 1996). In comparison, lignin deposited in structural sinks comprises a relatively stable 18% to 25% of stem dry weight in aspen and other Populus spp. (see Higuchi, 1997). Whether one or multiple PAL proteins regulate aspen allelochemistry and lignification is not known.
Evidence linking specific PAL genes with distinct developmental or metabolic roles has been reported for a number of species. The two PAL genes of raspberry (Rubus idaeus) exhibit spatiotemporally distinct expression during flower and fruit development (Kumar and Ellis, 2001), whereas differential responsiveness to environmental stimuli was identified for the PAL genes of bean (Phaseolus vulgaris) and parsley (Petroselinum crispum; Cramer et al., 1989; Liang et al., 1989; Lois et al., 1989; Logemann et al., 1995). In the case of parsley, the four protein isoforms exhibited indistinguishable enzyme kinetic properties, weakening arguments for metabolically distinctive PAL proteins (Appert et al., 1994). However, three tomato (Lycopersicon esculentum) PAL isoforms, kinetically distinct by virtue of their differential sensitivity to phenylpropanoid derivatives, have been purified (Sarma et al., 1998). Although kinetic criteria can be used to argue for metabolic specificity of PAL isoforms, other structural characteristics may also contribute. The metabolically significant association of PAL1 but not PAL2 class isoforms with microsomal proteins in tobacco (Nicotiana tabacum; Rasmussen and Dixon, 1999), for example, is not necessarily defined by kinetic properties of those isoforms.
Support for distinct developmental and environmental regulation of PAL isoforms can be drawn from the studies described above, but differentially regulated PAL isoforms in any given species have not been shown to be limited to, or preferentially associated with, specific metabolic activities: lignification, but not flavonoid biosynthesis, or vice versa, for example. Quaking aspen with its large and segregated CT, PG, and lignin sinks affords a system to identify cell-specific metabolic roles for PAL isoforms in situ. Divergent PAL genes (PkPALg1, PkPALg2a/b, and PkPALg4), exhibiting tissue-specific expression in shoot tips or mature stems, have been identified in a hybrid aspen (Populus kitakamiensis; Osakabe et al., 1995a, 1995b, 1996). Two closely related hybrid poplar (Populus trichocarpa × Populus deltoides) PAL genes, PtdPAL1 and PtdPAL2, differing by 20 bp at their 5′-coding end, and sharing 92% coding region nucleotide identity with PkPALg1, were, like PkPALg1 in hybrid aspen, highly expressed in developing stems and leaves (Subramaniam et al., 1993; Gray-Mitsumune et al., 1999). Based on the known expression pattern of PkPALg2a/b (Osakabe et al., 1995a) and on their own PtdPAL1/2 promoter study, Gray-Mitsumune et al. (1999) suggested developmentally separate lignification roles for PkPALg1/PtdPAL1/2 and PkPALg2a/b in primary and secondary tissues, respectively. Here, we report the cloning and characterization of two distinct aspen PAL cDNAs and their in situ hybridization patterns in developing shoot and root tissues. PtPAL1 was expressed in non-lignifying tissues of shoots and roots. PtPAL2 was expressed in heavily lignified structural cells of shoots, but was also expressed in non-lignifying cells of root tips. We suggest cell-specific metabolic roles for the proteins encoded by these PAL genes based on spatiotemporal correlation of their respective transcripts with CT and lignin distribution. The analysis of PAL function in CT-accumulating cells is further considered in the context of 4CL (4-coumarate:CoA ligase) gene expression and PG abundance.
RESULTS
Isolation and Characterization of PAL cDNAs
Two aspen partial PAL cDNA fragments were used as probes to isolate full-length cDNAs from a λZAPII aspen xylem cDNA library (see “Materials and Methods”). The cDNAs were designated PtPAL1 and PtPAL2 based on sequence homology with hybrid aspen clones PkPALg1 and PkPALg2b, respectively (Osakabe et al., 1995a, 1995b). PtPAL1 was 2,413 bp long with an open reading frame of 2,142 bp and a 5′-untranslated sequence of 122 bp, and a 3′-untranslated region of 149 bp, including a 20-nucleotide poly(A+) tail (GenBank accession no. AF480619). PtPAL2 was 2,515 bp long with an open reading frame of 2,133 bp, a 198-bp 5′-untranslated region, and a 3′-untranslated region of 184 bp, including a 19-nucleotide poly(A+) tail (GenBank accession no. AF480620). The PtPAL1 cDNA sequence predicted a polypeptide of 714 amino acids with a calculated molecular mass of 77.6 kD, and a pI of 5.9. The PtPAL2 cDNA encoded a polypeptide of 711 amino acids, with a molecular mass of 77.5 kD and a pI of 5.75. The homology between the coding regions for PtPAL1 and PtPAL2 was 76% at the nucleotide level and 84% at the amino acid level. However, nucleotide homology in the 3′-untranslated regions was only 52%. PtPAL1 exhibits 99% coding region nucleotide identity with the hybrid aspen ortholog PkPALg1 (Osakabe et al., 1995a, 1995b), and 92% coding region nucleotide identity with the hybrid poplar ortholog PtdPAL1/2 (Subramaniam et al., 1993; Gray-Mitsumune et al., 1999). PtPAL2 exhibits 89% coding region homology with hybrid aspen PkPALg2a/b, and 96% homology in a 1.7-kb overlap with the partial PkPALg4 clone (Osakabe et al., 1995a, 1995b). Southern analysis of four genomic DNA restriction digests with the two full-length cDNA probes revealed distinct hybridization patterns, and suggested that the two PAL genes belong to two distinct small gene families in aspen (data not shown).
RNA-Blot Analysis
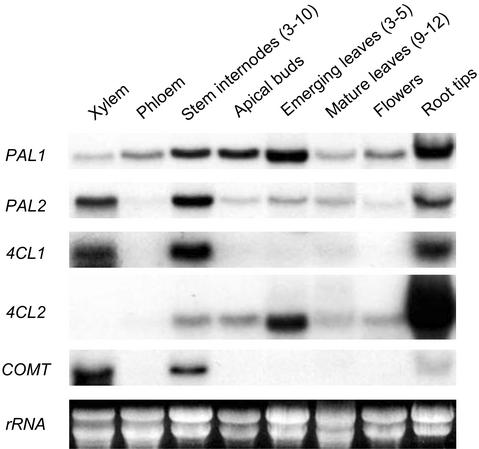
RNA from various tissues was analyzed on northern blots using full-length PtPAL1 and PtPAL2 cDNAs as probes (Fig. 1). PtPAL1 was most abundant in emerging (rapidly expanding) leaves, followed by root tips, apical buds, young stem, female flowers, and developing phloem. Expression was not as strong in mature (fully expanded) leaves or developing xylem. PtPAL2 was most strongly expressed in developing xylem, young stem, and root tips, but was weakly expressed in apical buds, emerging and mature leaves, flowers, and phloem (Fig. 1).
Figure 1.
Northern-blot analysis of PAL, 4CL, and COMT transcript levels in various tissues of aspen. Ten micrograms of total RNA from each tissue was resolved on a 1% (w/v) denaturing agarose gel that was photographed before being blotted and cross-linked onto a nylon membrane. Duplicate blots were hybridized with 32P-labeled full-length PtPAL1 and PtPAL2 cDNAs, stripped between hybridizations, and consecutively probed with 32P-labeled full-length Pt4CL1, Pt4CL2, and PtCOMT cDNAs at high stringency.
The general phenylpropanoid pathway supplied by PAL supports a number of branch pathways through control points regulated by enzymes such as 4CL (for review, see Higuchi, 1997). In aspen, protein isoforms encoded by Pt4CL1 and Pt4CL2 are kinetically distinct and preferentially regulate the lignin and flavonoid biosynthetic branchways, respectively (Hu et al., 1998; Harding et al., 2002). To correlate 4CL1- and 4CL2-modulated branchway activities with PAL expression patterns in various tissues, the two aspen 4CL genes were analyzed along with PAL (Fig. 1). Overall, 4CL1 expression paralleled that of PAL2 with strong expression in stem, xylem, and root tips and weaker expression elsewhere (Fig. 1). 4CL2 expression paralleled that of PAL1, and was strongest in root tips, followed by emerging leaves, apical buds, and stem internodes three through 10. Expression of a lignin pathway gene encoding 5-hydro-xyconiferylaldehyde O-methyltransferase (also known as caffeate O-methyltransferase [COMT]; Bugos et al., 1991; Li et al., 2000) was analyzed to gauge lignification activity in the various tissues represented (Fig. 1). Like 4CL1 and PAL2, COMT was well expressed in young stem, particularly in the xylem. In root tips, however, COMT was detected at a much lower level than 4CL1 and PAL2, a possible indication that 4CL1 and PAL2 expression was not limited to lignifying cells in root tips.
In Situ Hybridization
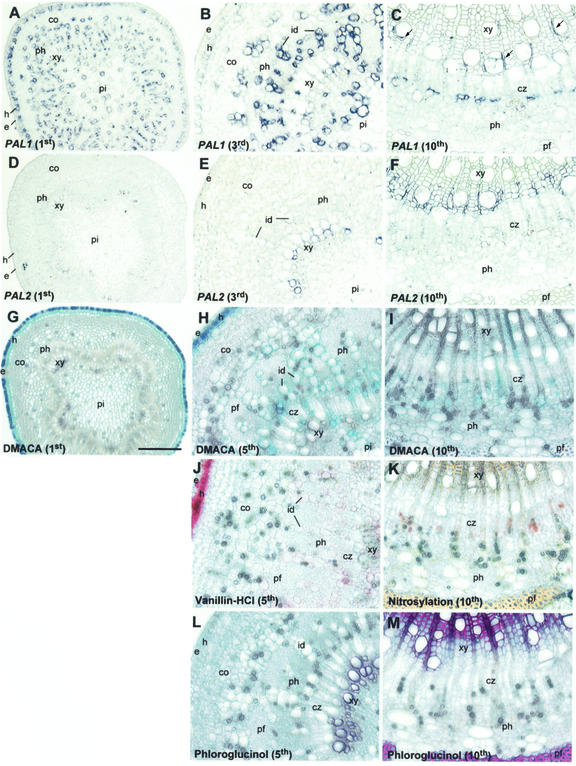
In stems, PtPAL1 first exhibited clear patterns of localized expression at the base of the apical bud and in the first internode (Fig. 2). Expression in radial files of parenchyma cells extended from pith to cortex, and occupied much of the cross-sectional area at the first internode (Fig. 2A). Expression shifted toward radial complexes of proliferative phloem parenchyma, developing idioblasts and nearby cortical cells at the third internode. (Fig. 2B). Expression in pith cells increased as well. Conversely, strong hypodermal and slight epidermal PAL1 expression at the first internode weakened considerably by the third internode (Fig. 2, A and B). In contrast to PtPAL1, expression of PtPAL2 was strictly localized to xylem vessels at these internodes (Fig. 2, D and E). At the 10th internode (Fig. 2C), PtPAL1 expression decreased overall but remained strongest in phloem ray parenchyma near the cambium, followed by ray initials within the cambial zone and xylem ray/axial parenchyma (arrows). This expression pattern was sustained in older internodes (20th and below), with additional expression detected in areas of proliferative primary phloem near the cortex (not shown). PtPAL2 remained highly expressed in developing xylem vessels and fibers undergoing secondary wall thickening at the 10th internode (Fig. 2F). Expression in the cambial zone was weak. PtPAL2 was expressed transiently in phloem fiber cells at the eighth and ninth internodes (not shown).
Figure 2.
In situ localization of PtPAL1 and PtPAL2 mRNAs and histochemical detection of CTs and lignin in aspen stem tissues. Transverse stem sections (10-μm thickness) were hybridized with digoxygenin (DIG)-labeled antisense PAL1 (A–C) or PAL2 (D–F) RNA probes and photographed in bright field. Transverse stem sections (75-μm thickness) were stained with dimethylaminocinnamaldehyde (DMACA; G–I), vanillin-HCl (J), or were nitroso-derivatized (K) for the detection of CTs, or were stained with phloroglucinol for the detection of lignin (L and M). Shown are first internode (A, D, and G), third internode (B and E), fifth internode (H, J, and L), and 10th internode (C, F, I, K, and M). Scale bar = 200 μm (A, D, and G) or 100 μm (all other panels). co, Cortex; cz, cambial zone; e, epidermis; h, hypodermis; id, idioblast, pf, phloem fibers; ph, phloem; pi, pith; xy, xylem.
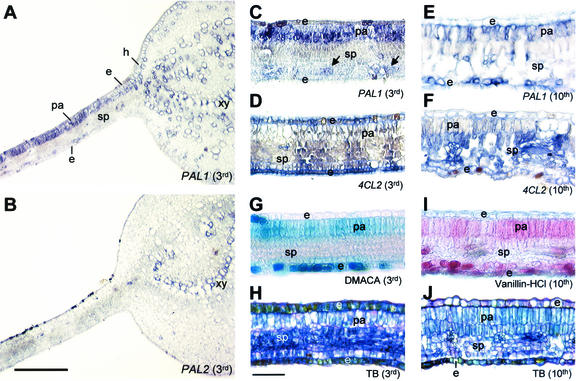
The earliest detectable expression of PtPAL1 in midveins and lamina of preemerged leaves was confined to clusters of cells within a subepidermal layer of the mesophyll (not shown). PtPAL1 became uniformly expressed throughout that layer as it differentiated into palisade parenchyma in the lamina of newly emerged leaf 3 (Fig. 3, A and C). Sporadic expression was detected in the lower spongy mesophyll (Fig. 3C, arrows), with weak expression also evident in the lower epidermis (Fig. 3C). In the midvein, PAL1 was readily detected in hypodermis, scattered clusters of cortical mesophyll cells, and xylem ray parenchyma (Fig. 3A). Expression of PtPAL2 (not to be confused with dark deposits sometimes noted in leaf epidermis) was limited to xylem vessels of the midvein and minor veins of the lamina (Fig. 3B). Those patterns of PAL1 and PAL2 expression were sustained as leaves expanded, though expression tapered off, and was difficult to visualize beyond leaf 10 (not shown).
Figure 3.
In situ localization of PtPAL1, PtPAL2, and Pt4CL2 mRNAs and histochemical detection of CTs in developing aspen leaves. Sections (10-μm thickness) were hybridized with DIG-labeled antisense PAL1 (A, C, and E), PAL2 (B), or 4CL2 (D and F) RNA probes, or stained with toluidine blue (TB) for the detection of total phenolics (H and J). Sections (75-μm thickness) were stained with DMACA (G) or vanillin-HCl (I) for the detection of CTs. Shown are third leaf midvein and lamina (A, B, C, D, G and H) and 10th leaf lamina (E, F, I and J). Scale bar = 200 μm (A and B) or 50 μm (C–J). e, Epidermis; h, hypodermis; pa, palisade; sp, spongy mesophyll; xy, xylem.
Expression of 4CL and PAL1 were analyzed in parallel in leaves three and 10 because 4CL proteins CoA-activate cinnamic acid derivatives for distribution into competing metabolic pathways including those for the synthesis of CTs and lignin. We compared 4CL2 expression with that of PAL1, choosing 4CL2 instead of 4CL1 because it encodes the protein isoform most kinetically suited for non-lignin metabolic activities expected in leaf lamina (Harding et al., 2002) and because it was more strongly expressed than 4CL1 in deveined expanding leaves (Fig. 1). The sites of strongest 4CL2 and PAL1 expression in leaf 3 were largely distinct, although overlap was evident (Fig. 3, C and D). 4CL2 was expressed in epidermal cells and spongy mesophyll cells but was very weakly expressed at best in palisade cells where PAL1 was strongly expressed (Fig. 3, C and D). In a fully expanded leaf at internode 10, PAL1 was detected in the upper palisade layer and in the lower epidermis (Fig. 3E), whereas 4CL2 was more strongly expressed in the second tier of palisade cells, and in spongy mesophyll and lower epidermis (Fig. 3F).
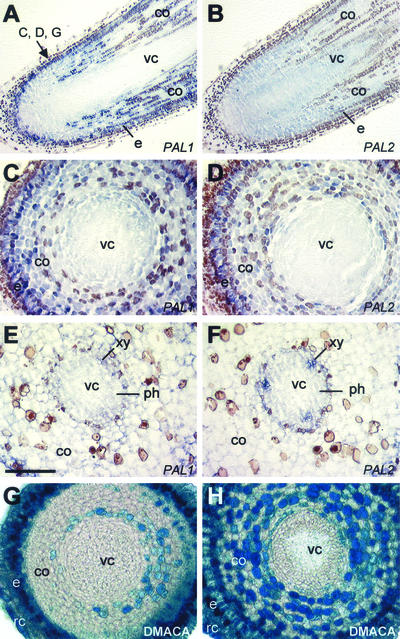
PAL1 and PAL2 were both well expressed in the distal 5 mm of root tips based on RNA-blot analysis (Fig. 1). The cellular distribution patterns of these two transcripts differed, however, primarily due to stronger PAL1 expression in the epidermis and in localized files of dividing cortex cells approximately 300 μm to 1 mm from the root cap (Fig. 4, A and B). Likewise, PAL1 was co-expressed with, but more abundant than, PAL2 in epidermal, exodermal, and many cortical cells shown in cross sections 300 μm from the root tip (Fig. 4, C and D). Cortical and epidermal expression of both genes tapered off with increasing distance from the root tip. In cross sections approximately 2 mm from the tip, PAL2 was primarily expressed in lignifying meta/protoxylem, and less clearly in phloem of the developing stele (Fig. 4F). Expression of PAL1, although detectable in phloem, was weak overall in this region of the root (Fig. 4E). Phloroglucinol staining of root tips was negative except for developing xylem (not shown), and was coupled with weak expression of COMT (Fig. 1), indicating that there were few lignifying cells there and that the high level of PAL2 expression in root tips (Figs. 1 and 4D) was not strictly to support lignin biosynthesis.
Figure 4.
In situ localization of PtPAL1 and PtPAL2 mRNAs and histochemical detection of CTs in aspen root tips. Longitudinal tangential sections (A and B) and transverse sections (C–F) 10 μm in thickness were hybridized with DIG-labeled antisense PAL1 (A, C, and E) or PAL2 (B, D, and F) RNA probes. Transverse sections (75-μm thickness) were stained with DMACA for the detection of CTs (G and H). Arrows in A indicate locations of transverse sections represented in C, D, and G. Transverse sections E, F, and H correspond to a distance approximately 2 mm from root tips. Scale bar = 200 μm (A and B) or 100 μm (C–H). co, Cortex; e, epidermis; ph, phloem; rc, root cap; vc, vascular cylinder; xy, xylem.
Distribution of CTs and PGs
The PGs, salicin, salicortin, tremulacin, and tremuloidin, along with CTs, constitute separate, and potentially quite large allelochemical pools (approximately 10%–35% dry weight) in developing leaves and phloem of aspen (for review, see Lindroth and Hwang, 1996). PGs are biosynthesized from the PAL product, cinnamic acid, after its likely oxidation and hydroxylation to benzoic and salicylic acid (Yalpani et al., 1993; Coquoz et al., 1998; Ribnicky et al., 1998). CTs are biosynthesized from the cinnamic acid derivative, 4-coumaric acid, after its 4CL-dependent entry into the flavonoid pathway. Localization of one or both of these large pools would offer the possibility of correlating expression patterns of specific PAL genes with cell-specific metabolic roles. For comparison of CT localization and in situ gene expression patterns, vibratome sections of stem internodes one, five, and 10 were stained with 4-DMACA (Feucht and Treutter, 1990). CTs were localized within or adjacent to cells with strong PAL1 expression at these internodes. CTs accumulated in epidermal cells of the first internode (Fig. 2G) where PAL1 expression was weak, but accumulated in hypodermal cells at lower internodes (Fig. 2H). PAL1 was more strongly expressed in hypodermal than epidermal cells at these internodes, suggesting that besides PAL1 expression, additional factors, perhaps intercellular transport, need to be considered to explain CT localization. Staining in cortex or primary vascular tissues of the first internode (Fig. 2G) was very weak, but at the fifth internode, files of aqua-blue, CT-staining idioblasts could be discriminated from the gray-blue background of smaller phloem cells (Fig. 2H). Light staining also became evident in clusters of proliferative parenchyma and cortical cells at the outer margin of primary phloem fibers, in ray initials, and sporadically in xylem parenchyma and pith (Fig. 2H). At the 10th internode, staining was strongest in cambial ray initials and in newly developed phloem parenchyma, and was occasionally seen in xylem ray parenchyma (Fig. 2I).
The DMACA localization of CT was verified in adjacent serial sections by vanillin-HCl staining (Gardner, 1975) and a nitroso-derivatization procedure (Reeve, 1951), as illustrated in Figure 2, J and K. Vanillin-HCl produced a pink stain that was identical to the pattern of staining observed using DMACA (Fig. 2, H and J). Nitroso-derivatization produced light reddish-brown staining, consistent with the color produced by purified catechin tannins (Reeve, 1951), in the area of ray initials and newly developed phloem ray parenchyma at the 10th internode (Fig. 2K). The nitroso-derivatization procedure stained lignified cells yellow (Fig. 2K), as verified by phloroglucinol staining (Fig. 2, L and M). Phloroglucinol staining of xylem vessels was readily detectable at the fifth internode (Fig. 2L), whereas staining of phloem fibers, though not detected in primary internodes, was strong at the 10th internode (Fig. 2M).
Localization of CT in newly emerged (third) and fully expanded (10th) leaves (Fig. 3) and root tips (Fig. 4) was also determined by DMACA and vanillin-HCl staining. Light staining was observed in palisade and the lower tier of spongy mesophyll cells, whereas dark staining was detected in the abaxial epidermis in leaf 3 (Fig. 3G). The pattern was similar in leaf 10, although there was a shift in the distribution of darkly stained cells from the abaxial epidermis to the subepidermal layer of spongy mesophyll (Fig. 3I), reminiscent of the CT shift between epidermal and hypodermal cells observed in developing stems. Very little staining was detected in the upper epidermis or midvein of either leaf. PAL1 was expressed to varying degree in all of the CT-containing cells of leaf lamina. Intensely stained CT deposits were observed in PAL1-expressing areas of the lateral root cap, epidermis, and exodermis of the distal most 0.3 to 2 mm of the root tip (Fig. 4, G and H). Staining in cortical cells approximately 300 μm from the tip proper was sporadic (Fig. 4G) but increased in frequency and intensity in a region roughly corresponding to the meristematic and early elongation zones, 500 μm to 2 mm from the root tip (Fig. 4H), before decreasing in more mature root tissues (not shown).
Wounding Experiments
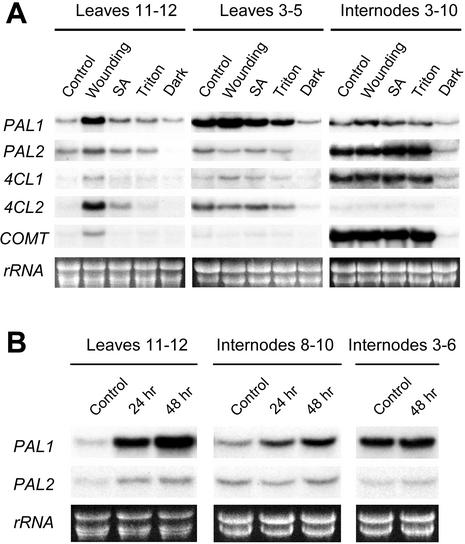
Leaf wounding was used as a strategy to investigate changes in PAL/4CL expression and to monitor correlative changes in CT to PG abundance. Twenty-four hours after wounding, leaves (11 and 12) exhibited stronger local increases in PAL1 than PAL2 expression (Fig. 5A). Steady-state transcript levels of genes encoding downstream lignin pathway (4CL1 and COMT) and non-lignin pathway (4CL2) proteins were low compared with those of both PAL genes in unwounded leaves 11 and 12. Wounding increased 4CL2 expression far more than 4CL1 or COMT expression, indicating, together with the PAL expression data, that non-lignin pathways supported by PAL1-4CL2 were more strongly wound stimulated than lignin-related pathways supported by PAL2-4CL1-COMT in leaf lamina. Along with the increases in PAL1 and 4CL2 expression, PG and CT levels increased 9% and 28%, respectively, in wounded leaves 24 h after treatment (Table I). This short-term leaf wound response is consistent with the interpretation that increased PAL1 and 4CL2 expression led to a general increase in phenylpropanoid metabolism, and favored CT synthesis over that of PG and lignin biosynthesis. CT comprised approximately 10% of the net increase in the CT + PG pool after wounding, versus 3% before wounding. The increased abundance of CT relative to PG upon wounding is consistent with an interpretation of increased CT sink strength in cells with elevated 4CL2 expression. Lignification also may have increased in xylem of leaf veins where PAL2 (Fig. 3B) and 4CL1 (Harding et al., 2002) are localized and COMT is expected to be localized. There was little evidence of a systemic wounding effect on the expression of these genes, but a very slight increase of the PAL1 signal from pooled stem internodes and upper leaves (3–5) was noted on this blot (Fig. 5A). After more severe wounding experiments as described by Parsons et al. (1989), a systemic induction of PAL1 was detected, but only in stem internodes just above the wounded leaves (Fig. 5B). Spraying leaves with 1 mm salicylic acid did not lead to significant local or systemic effects on PAL, 4CL, or COMT expression (Fig. 5A). Dark treatment reduced expression of all genes analyzed, with the exception of PAL1 in leaves 11 and 12 (Fig. 5A).
Figure 5.
Local and systemic wounding effects on PAL transcript and CT levels in aspen. A, PAL, 4CL, and COMT transcript levels in leaves and stems of aspen that were wounded, sprayed with 1 mm salicylic acid in 0.01% (v/v) Triton X-100 (SA) or with 0.01% (v/v) Triton X-100 as a control for SA treatment (Triton) on leaves 11 and 12, or were dark acclimated for 72 h. Wounding and spray treatments were repeated after 2 h and tissues harvested 24 h after the initial treatment for RNA analysis as described in Figure 2. B, Effects of a more severe 24- to 48-h wounding treatment on PAL1 and PAL2 transcript levels in injured leaves (11th and 12th) and uninjured nearby (eighth–10th) or distant (third–sixth) stem internodes.
Table I.
PG and CT concentration in leaf lamina before and 24 h after wounding
| Treatment | PGs | CTsa |
|---|---|---|
| mg g dry wt−1 | ||
| Control | 96 ± 3.0 | 3.2 ± 0.4 |
| Wounded | 105 ± 10.0 | 4.1 ± 0.1 |
PG and CT levels from leaves eight through 10 of control or wounded plants were analyzed as described in “Materials and Methods.” Values are means ± sd (n = 3 individual plants). The experiment was repeated with another aspen clone with equivalent results.
CT levels before and after wounding differed significantly (Student's t test, P < 0.01).
DISCUSSION
Two full-length PAL cDNAs, PtPAL1 and PtPAL2, were isolated from xylem tissue of quaking aspen. Throughout stem development, PAL1 was best expressed in non-lignifying cells exhibiting CT accumulation. PAL2 was expressed in lignifying structural and conducting elements of xylem and phloem, and its expression decreased after lignification of those cells was complete. We also analyzed 4CL expression and obtained quantitative data on CT and PG pool size in leaves where the co-expression of PAL1 and 4CL2 varied between cell layers, potentially affecting the balance of CT and PG in those cell layers. The present study advances previous reports alluding to kinetically adapted PAL proteins (Sarma et al., 1998) by correlating differentially localized PAL mRNAs with distinct metabolic roles in situ. Our data is generally consistent with the interpretation that PAL1 and 4CL2 are associated with CT accumulation. However, developmental variation in metabolite export to adjacent cell layers from PAL1/4CL2co-expressing cells may also have affected CT localization as suggested; for example, in outer cell layers of young stem.
Distinct Roles for PtPAL1 and PtPAL2 in Developing Stem Internodes
By correlating the expression pattern of PAL1 with the distribution of CTs in developing stems, we have identified a specific metabolic role that is distinct from lignification for PAL1 in vascular tissues. PAL1 expression anticipated, or was synchronous with, CT deposition in epidermis/hypodermis, phloem idioblasts, and developing ray parenchyma of phloem and xylem of young stem. To elaborate, CTs initially became abundant in the epidermis/hypodermis between the first and fifth internodes after the early PAL1 expression maximum reached in hypodermis at the first internode (Fig. 2, A, B, G, H, and J). Next, localized PAL1 expression increased in certain radial files of phloem parenchyma cells between the first and third internodes (Fig. 2, A and B), preceding CT accumulation in those cells as they developed into idioblasts (Fig. 2, H and J). PAL1 expression and CT staining subsequently decreased in idioblasts between the fifth and 10th internodes. At the 10th internode, PAL1 expression and CT accumulation shifted to ray initials as well as to phloem and xylem ray parenchyma adjacent to the vascular cambium (Fig. 2, C, I, and K). Developmental lags in the appearance of CTs after changes in PAL1 expression are consistent with time required for synthesis of active PAL protein, a 12-h process in kinetin-treated tobacco suspension cells (Nagai et al., 1994), and product accumulation. Slow or undetected CT turnover, such as that noted in the hypodermis after reduced PAL1 expression, is consistent with reports on CT metabolic stability (Kleiner et al., 1999, and refs. therein). Based on expression pattern, PAL2 appeared to be less associated with CT metabolism than PAL1, but rather was important in lignifying cells (Fig. 2, F and M). This assessment was strengthened by the results from wounding experiments in which increases in PAL1-4CL2 and CT abundance were correlated, whereas PAL2-4CL1 expression increased less markedly in wounded leaves (Fig. 5; Table I).
Similar to stem epidermis/hypodermis, the distribution of root tip cells expressing PAL1 paralleled the distribution of cells producing intense DMACA staining. Interestingly, PAL2 was expressed in many cortical cells in the distal most 300 μm of the root tip (Fig. 4, B and D) that were both lignin and CT negative (Fig. 4G). A key lignin-associated gene, COMT, was only weakly expressed in root tips (Fig. 1), and phloroglucinol staining for lignin was negative for cortical and epidermal cells of root tips (not shown). In this context, the strong expression of PAL2 in roots may support synthesis of certain hydroxycinnamate derivatives important for suberin synthesis or for non-lignin/CT-related activities associated with root protection. The intense CT staining approximately 2 mm from the root tip (Fig. 4H), where PAL expression had decreased (Fig. 4, E and F), can be ascribed to metabolic stability of CTs (Kleiner et al., 1999).
PAL expression has been most specifically identified with lignification in developing vasculature of stems and leaves of poplar species. In hybrid aspen, PAL protein was only detected in lignifying secondary vascular tissues (Osakabe et al., 1996). In hybrid poplar, activity of the PtdPAL1/2 (a PtPAL1 ortholog) promoter was high in primary xylem, phloem, and interfasicular cambium of primary internodes, but became limited to parenchyma cells near phloem fibers and xylem vessels in older internodes (Gray-Mitsumune et al., 1999). The authors hypothesized that PtdPAL1/2 and PkPAL2g (a PtPAL2 ortholog expressed in secondary xylem of hybrid aspen; Osakabe et al., 1996) represent distinctly regulated PAL genes that respond to separate lignification signals specific to primary and secondary tissues (Gray-Mitsumune et al., 1999). It was acknowledged that PAL is also likely to be associated with the large accumulation of phenolics considered to be present in developing shoot tissues of poplar, but previous studies have stopped short of identifying those associations specifically. Rather than suggesting two signals for lignification, one for PtPAL1 in primary tissues and another for PtPAL2 in secondary tissues, we concluded from our data that aspen PtPAL1 expressed in various tissues, including developing phloem or xylem, was connected with the biosynthesis of CTs and other non-lignin metabolites to be discussed below. On the other hand, PtPAL2 expression in shoots was always confined to phloem fiber and xylem cells destined to form heavily lignified secondary walls.
Co-Expression with 4CL2 Modulates the Metabolic Role of PAL1
We observed a correlation between PAL1 and CT localization in developing shoot and root tissues, but across cell types, the intensity of PAL1 expression did not closely correlate with the intensity of CT staining. This could indicate that CTs form an important phenolic reserve in PAL1-expressing cells, but that competing PAL-dependent phenolic pools are also maintained in many of these cells. Because leaves as well as phloem and bark of aspen stems accumulate variable, and often very large, reserves of CTs and PGs (for review, see Lindroth and Hwang, 1996), we reasoned that PG synthesis could alter the correlation between CT abundance and PAL1 expression. The PAL product cinnamic acid is hydroxylated to p-coumaric acid, which is activated by 4CL for biosynthesis of flavonoid-derived CTs. In contrast, conversion of cinnamic acid into precursors for salicylate-derived PGs is not likely to require 4CL because neither aspen 4CL isoform isolated so far recognizes cinnamic acid as a substrate (Harding et al., 2002). Although there are reports of cinnamic acid utilization by tobacco and poplar 4CL isoforms, no kinetic data were presented in the tobacco case (Lee and Douglas, 1996). Kinetic data were reported for the poplar isoform, but the Km was approximately 13-fold higher and the Vmax/Km, approximately 44-fold lower for cinnamic than for p-coumaric acid (Allina et al., 1998). Moreover, because hydroxycinnamate mixtures were not used in those studies, competitive inhibition effects of p-coumaric acid that would further suppress cinnamate utilization (Harding et al., 2002) were not considered. Assuming, therefore, that 4CL2 is required for flavonoid CT but not for PG biosynthesis, and assuming that PGs form a major competing sink in young stem as they do in leaf tissue (Table I), CT sink strength would be relatively weak in cells with low 4CL2, despite strong PAL1 expression. In accordance, stem idioblast cells stained weakly for CTs compared with stem epidermis/hypodermis (Fig. 2), whereas leaf lower epidermis and spongy mesophyll stained strongly for CTs compared with palisade cells (Fig. 3).
In the upper cell layers of young leaves, PAL1 and 4CL2 were strongly expressed, but tended to segregate into the palisade and epidermal layers, respectively (Fig. 3, C–F). Interestingly, CTs were detected in PAL1-expressing palisade cells, and not in the 4CL2-expressing epidermal cells. This is an exception to our general model that CTs accumulate in or adjacent to PAL1/4CL2 co-expressing cells. Therefore, TB was used to dissect the relationship between PAL1 and 4CL2 expression and phenolic product accumulation in the leaf cells (Fig. 3, H and J). TB stains phenolics turquoise-blue to green (O'Brien et al., 1964). In leaf three, TB-staining phenolics were abundant in the upper epidermis (Fig. 3H), despite the absence of PAL1 transcripts there (Fig. 3C). Conversely, CT was absent in the upper epidermis (Fig. 3G) despite expression of 4CL2 (Fig. 3D). It is possible that an additional, unidentified PAL was expressed in leaf epidermal cells to account for the abundant phenolics deposits there. However, the strong nucleotide homology of our two aspen PAL probes with all PAL genes reported in Populus spp. to date (see “Results”) argues against this possibility. Alternatively, intercellular transport of phenolics from the palisade could account for the abundant phenolics in the upper epidermal layers in the absence of PAL1 expression. The exported phenolics clearly were not converted into CTs in the upper epidermis, but could have given rise to 4CL2-promoted synthesis of various flavonoid derivatives that may be more important than CTs for protection from UV light in young leaves. At the same time, flavonoid CT precursors may have been exported back to the palisade cells where we detected their accumulation. The concept of intercellular transport of phenylpropanoid derivatives was first reported to explain the segregation of mesophyllar flavonoid-synthesizing enzymes from epidermal flavonoid products in oat (Avena sativa) leaf (Knogge and Weissenbock, 1986). In that work, PAL activity was detected in the epidermis of younger leaf cells, suggesting trafficking of metabolic precursors from epidermis to mesophyll and of products from mesophyll to epidermis during oat leaf development. We suggest intercellular phenolic trafficking and segregation of 4CL2 and PAL1 may add regulatory flexibility specific to upper cell layers of aspen leaves. In general, our data provide examples of how variable expression of PAL1 and 4CL2, along with intercellular transport, determine the dynamics of CT and PG distribution in non-lignifying leaf, stem, and root cells of quaking aspen.
Limiting effects of 4CL on flavonoid synthesis were reported in PAL overexpressing tobacco (Howles et al., 1996). In the transgenic leaves, chlorogenic acid levels increased with increasing PAL activity, whereas levels of the flavonoid rutin remained unaffected. At the same time, glucoside derivatives of the 4CL substrate 4-coumaric acid increased, suggesting metabolic constriction at 4CL (Howles et al., 1996). Data from our leaf wounding experiments (Fig. 5; Table I) also support the idea of a limiting effect of 4CL on flavonoid biosynthesis. Strong wound up-regulated expression of PAL1 and 4CL2 correlated with a small net shift in carbon allocation from PG to CT during the wound response. In other experiments, the CT to PG ratio in leaves of aspen with severe 4CL down-regulation (Hu et al., 1999) was about 20% lower than in control plants (not shown), consistent with the idea that 4CL and, perhaps, the ratio of 4CL to PAL is important for governing the allocation of carbon between these two pools.
Shifts in the allocation of carbon between CT and PG pools have also been reported in response to changes in nutrient and CO2 regime. Poor nutrient regime and low-light intensity at ambient CO2 levels consistently increased CT to PG ratios in three clones of willow (Julkunen-Titto et al., 1993), a phylogenetic neighbor to quaking aspen. In Salix spp., CT and PG allelochemical pools comprise 10% to 20% of leaf dry weight. Thus, a shift toward CT biosynthesis under nutrient poor conditions may represent a significant cost-saving metabolic adjustment in favor of the more stable, albeit less potent, allelochemical, CTs. PG biosynthesis, turnover, steady-state level, and, thus, metabolic expense, are several times greater than that of CT in sink and source leaves of aspen (Kleiner et al., 1999). The shift we observed toward CT biosynthesis during wounding could represent one of many adjustments regulated by complex Glc/Suc signaling and sink source relations in plants (Ehness et al., 1997), including Populus spp. (Davis et al., 1991, 1993). The expression and roles of individual PAL and 4CL isoforms in the context of sink-source signaling and environmental constraints, including wounding, warrant interest, particularly in light of current interest in global climate and forestry issues.
MATERIALS AND METHODS
Plant Materials
Apical buds, young (rapidly expanding) leaves, mature (fully expanded) leaves, young stems, developing xylem and phloem, and root tips (distal 5 mm) were collected from greenhouse-grown quaking aspen (Populus tremuloides Michx.) plants. Intact, nearly mature female flower tissues were kindly provided by Dr. David Dixon and Dr. Gopi Podila (Michigan Technological University). Tissues were immediately frozen and stored in liquid nitrogen until used for RNA or DNA isolation.
Cloning of PAL cDNAs
Two degenerate primers (PALS, 5′-GTYACTACTGGTTTTGGTGC; and PALAS, 5′-GCATYAATGGATAGGTWGCACT) flanking an approximately 1,400-bp fragment were designed based on the conserved regions of poplar PAL cDNAs available in GenBank. Total RNA (5 μg) isolated from developing xylem according to Bugos et al. (1995) was reverse transcribed using Superscript II (Invitrogen-Life Technologies, Carlsbad, CA), and subsequently PCR amplified using 2 μm each PALS and oligo-dT20 primers, 20 μm dNTPs, and 2.5 units of Taq DNA polymerase (Fisher Scientific, Chicago). PCR parameters were: 94°C for 3 min, followed by 30 cycles of 94°C for 45 s, 50°C for 30 s, and 72°C for 2 min, and a single 5-min extension at 72°C. Reverse transcription (RT)-PCR product was subjected to nested PCR under the same conditions using PALS and PALAS as primers. The PCR products were blunt-ended and cloned into the EcoRV site of pBluescript SK− (Stratagene, La Jolla, CA). An approximately 1.4-kb PAL cDNA fragment, most similar to the hybrid aspen PkPALg2b and designated PtPAL2-1.4, was obtained and used to screen 5 × 105 plaque-forming units of an aspen xylem λgt22 unidirectional cDNA library (Ge and Chiang, 1996). Several positive clones were obtained after three rounds of plaque purification and one partial-length clone was obtained and fully sequenced. This partial clone exhibited 99% identity with hybrid aspen PkPALg1 and was designated PtPAL1-0.9.
A second aspen xylem cDNA library constructed in the λZAPII (Stratagene) vector (kindly provided by Dr. Laigeng Li and Dr. Vincent Chiang [Michigan Technological University]) was screened using PtPAL1-0.9 or PtPAL2-1.4 as probes. Full-length cDNAs, designated PtPAL1 and PtPAL2, were sequenced on both strands using the ABI Prism BigDye Terminator Cycle Sequencing Kit and ABI Prism 310 Genetic analyzer (Perkin-Elmer Applied Biosystems, Foster City, CA). Sequence analysis was performed using the GCG software package (Genetics Computer Group, Madison, WI).
Nucleic Acid Extraction, Blotting, and Hybridization
Genomic DNA was extracted from young aspen leaves, digested with EcoRI, BamHI, HindIII, or XbaI, and blotted onto nylon membrane as described (Tsai et al., 1994). Total RNA was isolated from various aspen tissues according to Chang et al. (1993) and resolved on a denaturing formaldehyde-agarose gel (Sambrook et al., 1989) in the presence of ethidium bromide to allow visualization of RNA after electrophoresis. DNA- and RNA-blot hybridizations were performed according to Tsai et al. (1998) using full-length PtPAL1 and PtPAL2 as probes.
In Situ Hybridization
In situ hybridization was performed using semithin (10-μm) sections of leaf, stem, and root tissues embedded in paraffin according to Harding et al. (2002). DIG-UTP-labeled RNA transcripts were generated by in vitro transcription of PtPAL1-0.9 and PtPAL2-1.4 in sense or antisense orientation using T3 or T7 RNA polymerase (Roche Applied Science, Indianapolis). After hybridization, washing, and blocking, DIG-labeled RNA transcripts reacting with alkaline phosphatase-conjugated anti-DIG Fab fragment (1:750 [v/v]; Roche Applied Science) were colorimetrically detected using nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate. The gene specificity of PtPAL1-0.9 and PtPAL2-1.4 probes was confirmed by DNA dot-blot analysis against the full-length PtPAL1 and PtPAL2 cDNAs (not shown).
Histochemical Staining
Sections (75 μm thick) were sliced from fresh tissues using a vibratome (Ted Pella, Redding, CA), and placed into cold fixative containing 0.5% (w/v) glutaraldehyde in10 mm MOPS (pH 7.0). CTs, specifically flavan-3-ols, or catechin tannins, were detected using 0.5% (w/v) 4-DMACA in butanol-H2SO4 (Feucht and Treutter, 1990). For Vanillin-HCl staining, fresh sections were treated for 1 to 2 min in ethanolic vanillin (20% [w/v]), followed with 1 volume of concentrated HCl. Control reactions performed in 6 n HCl/ethanol for the detection of anthocyanins were negative (not shown). Nitrosylation was performed by the sequential addition to fresh sections of 10% (w/v) NaNO2, 20% (w/v) urea, and 10% (v/v) acetic acid (1:1:1 [v/v]), followed with 2 volumes of 2 n NaOH 3 min later. Lignin was detected by mounting sections in a solution of 2% (w/v) phloroglucinol in ethanol:1 m HCl (1:1 [v/v]). Paraffin-embedded sections were stained for 1 min in a solution of 0.05% (w/v) TB, pH 5.5, rinsed with distilled water, air dried, deparaffinized, and mounted in Permount (Fisher Scientific) for the detection of green- and blue-green-staining phenolic derivatives (O'Brien et al., 1964). Images were recorded using an E-400 microscope (Nikon, Tokyo) equipped with a digital imaging system
PG and CT Measurements
Young, expanded leaves eight to 10 internodes below the terminal apical bud of three individual wounded (see below) or unwounded plants were deribbed and placed into liquid nitrogen for subsequent freeze drying. Leaves were freeze dried in vacuum flasks on dry ice, and dried powders were analyzed in the laboratory of Richard Lindroth (Department of Entomology, University of Wisconsin, Madison) for PGs (Lindroth et al., 1993) and CTs (Porter et al., 1986) using purified aspen CT as the standard.
Wounding, Salicylic Acid, and Dark Treatments
Greenhouse-grown plants approximately 1 m in height were used in the wounding experiments. Leaves 11 and 12 were pinched 15 times around their perimeters with pliers, and the treatment was repeated after 2 h. Leaves and stem sections were harvested just before and 24 h after the initial wounding for RNA-blot analysis. Salicylic acid was applied as a thorough spray of 1 mm Na-salicylate (pH 7.0) in 0.01% (v/v) Triton X-100 to leaves 11 and 12 and repeated after 2 h. Controls for the salicylic acid experiment received a spray of 0.01% (v/v) Triton X-100. Two plants were used for each treatment and tissues were harvested and pooled for RNA-blot analysis. Dark treatments were conducted by placing plants in a ventilated, darkened growth chamber at 20°C for 72 h. In a follow-up wounding experiment, 20 leaves (9–29) were wounded with pliers and wounding was repeated 24 h later. For RNA analysis, two plants were harvested just before wounding, followed by harvests 24 h and again 48 h after the initial wounding. The salicylic acid and both wounding experiments were repeated once.
ACKNOWLEDGMENTS
We thank Dr. Vincent Chiang (Michigan Technological University) for providing aspen xylem cDNA libraries and 4CL and COMT cDNA clones, and for his support of the work. We also thank Dr. Chandrashekhar Joshi (Michigan Technological University) for critical review of the manuscript and Dr. James Mauseth (University of Texas, Austin) for valuable assistance with anatomical interpretation.
Footnotes
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.006262.
LITERATURE CITED
- Allina SM, Aviva P-H, Theilmann DA, Ellis BE, Douglas CJ. 4-Coumarate:coenzyme A ligase in hybrid poplar. Plant Physiol. 1998;116:743–754. doi: 10.1104/pp.116.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allwood EG, Davies DR, Gerrish C, Ellis BE, Bolwell GP. Phosphorylation of phenylalanine ammonia-lyase: evidence for a novel protein kinase and identification of the phosphorylated residue. FEBS Lett. 1999;457:47–52. doi: 10.1016/s0014-5793(99)00998-9. [DOI] [PubMed] [Google Scholar]
- Appert C, Logemann E, Hahlbrock K, Schmid J, Amrhein N. Structural and catalytic properties of the four phenylalanine ammonia-lyase isoenzymes from parsley (Petroselinum crispum Nym.) Eur J Biochem. 1994;225:491–499. doi: 10.1111/j.1432-1033.1994.00491.x. [DOI] [PubMed] [Google Scholar]
- Bolwell GP. A role for phosphorylation in the down-regulation of phenylalanine ammonia-lyase in suspension-cultured cells of French bean. Phytochemistry. 1992;31:4081–4086. [Google Scholar]
- Bolwell GP, Cramer CL, Lamb CJ, Schuch W, Dixon RA. l- Phenylalanine ammonia-lyase from Phaseolus vulgaris: modulation of the levels of active enzyme by trans-cinnamic acid. Planta. 1986;169:97–107. doi: 10.1007/BF01369780. [DOI] [PubMed] [Google Scholar]
- Bugos RC, Chiang VL, Campbell WH. cDNA cloning, sequence analysis and seasonal expression of lignin-bispecific caffeic acid/5-hydroxyferulic acid O-methyltransferase of aspen. Plant Mol Biol. 1991;17:1203–1215. doi: 10.1007/BF00028736. [DOI] [PubMed] [Google Scholar]
- Bugos RC, Chiang VL, Zhang XH, Campbell ER, Podila GK, Campbell WH. RNA isolation from plant tissues recalcitrant to extraction in guanidine. BioTechniques. 1995;19:734–737. [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep. 1993;11:113–116. [Google Scholar]
- Coquoz J-L, Buchala A, Metraux J-P. The biosynthesis of salicylic acid in potato plants. Plant Physiol. 1998;117:1095–1101. doi: 10.1104/pp.117.3.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer CL, Edwards K, Dron M, Liang X, Dildine SL, Bolwell GP, Dixon RA, Lamb CJ, Schuch W. Phenylalanine ammonia-lyase gene organization and structure. Plant Mol Biol. 1989;12:367–383. doi: 10.1007/BF00017577. [DOI] [PubMed] [Google Scholar]
- Davis JM, Egelkrout EE, Coleman GD, Chen THH, Haissig BE, Riemenschneider DE, Gordon MP. A family of wound-induced genes in Populus shares common features with genes encoding vegetative storage proteins. Plant Mol Biol. 1993;23:135–143. doi: 10.1007/BF00021426. [DOI] [PubMed] [Google Scholar]
- Davis JM, Gordon MP, Smit BA. Assimilate movement dictates remote sites of wound-induced gene expression in poplar leaves. Proc Natl Acad Sci USA. 1991;88:2393–2396. doi: 10.1073/pnas.88.6.2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehness R, Ecker M, Godt DE, Roitsch T. Glucose and stress independently regulate source and sink metabolism and defense mechanisms via signal transduction pathways involving protein phosphorylation. Plant Cell. 1997;9:1825–1841. doi: 10.1105/tpc.9.10.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feucht W, Treutter D. Flavan-3-ols in trichomes, pistils and phelloderm of some tree species. Ann Bot. 1990;65:225–230. [Google Scholar]
- Gardner RO. Vanillin-hydrochloric acid as a histochemical test for tannin. Stain Technol. 1975;50:315–317. doi: 10.3109/10520297509117081. [DOI] [PubMed] [Google Scholar]
- Ge L, Chiang VL. A full length cDNA encoding trans-cinnamate 4-hydroxylase from developing xylem of Populus tremuloides (accession no. U47293) (PGR96-075) Plant Physiol. 1996;112:861. [Google Scholar]
- Gray-Mitsumune M, Molitor EK, Cukovic D, Carslon JE, Douglas CJ. Developmentally regulated patterns of expression directed by poplar PAL promoters in transgenic tobacco and poplar. Plant Mol Biol. 1999;39:657–669. doi: 10.1023/a:1006148715050. [DOI] [PubMed] [Google Scholar]
- Hahlbrock K, Grisebach H. Enzymic controls in the biosynthesis of lignin and flavonoids. Ann Rev Plant Physiol. 1979;30:105–130. [Google Scholar]
- Harding SA, Leshkevich J, Chiang VL, Tsai C-J. Differential substrate inhibition couples kinetically distinct 4-coumarate:CoA ligases with spatially distinct metabolic roles in quaking aspen. Plant Physiol. 2002;128:428–438. doi: 10.1104/pp.010603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi T. Biochemistry and Molecular Biology of Wood. New York: Springer-Verlag; 1997. pp. 93–99. [Google Scholar]
- Howles PA, Sewalt VJH, Paiva NL, Elkind Y, Pate NJ, Lamb CJ, Dixon RA. Overexpression of l-phenylalanine ammonia-lyase in transgenic tobacco plants reveals control points for flux into phenylpropanoid biosynthesis. Plant Physiol. 1996;112:1617–1624. doi: 10.1104/pp.112.4.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W-J, Harding SA, Lung J, Popko JL, Ralph J, Stokke DD, Tsai C-J, Chiang VL. Repression of lignin biosynthesis promotes cellulose accumulation and growth in transgenic trees. Nat Biotechnol. 1999;17:808–812. doi: 10.1038/11758. [DOI] [PubMed] [Google Scholar]
- Hu W-J, Kawaoka A, Tsai C-J, Lung J, Osakabe K, Ebinuma H, Chiang VL. Compartmentalized expression of two structurally and functionally distinct 4-coumarate:CoA ligase genes in aspen (Populus tremuloides) Proc Natl Acad Sci USA. 1998;95:5407–5412. doi: 10.1073/pnas.95.9.5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones HD. Phenylalanine ammonia-lyase: regulation of its induction, and its role in plant development. Phytochemistry. 1984;23:1349–1359. [Google Scholar]
- Julkunen-Titto R, Tahvanainen J, Silvola J. Increased CO2 and nutrient status changes affect phytomass and the production of plant defensive secondary chemicals in Salix myrsinifolia (Salisb.) Oecologia. 1993;95:495–498. doi: 10.1007/BF00317433. [DOI] [PubMed] [Google Scholar]
- Kleiner KW, Raffa KF, Dickson RE. Partitioning of 14C-labeled photosynthate to allelochemicals and primary metabolites in source and sink leaves of aspen: evidence for secondary metabolite turnover. Oecologia. 1999;119:408–418. doi: 10.1007/s004420050802. [DOI] [PubMed] [Google Scholar]
- Knogge W, Weissenbock G. Tissue-distribution of secondary phenolic biosynthesis in developing primary leaves of Avena sativa L. Planta. 1986;167:196–205. doi: 10.1007/BF00391415. [DOI] [PubMed] [Google Scholar]
- Kumar A, Ellis BE. The phenylalanine ammonia-lyase gene family in raspberry. Structure, expression, and evolution. Plant Physiol. 2001;127:230–239. doi: 10.1104/pp.127.1.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Douglas CJ. Two divergent members of a tobacco 4-coumarate:coenzyme A ligase (4CL) gene family. cDNA structure, gene inheritance and expression, and properties of recombinant proteins. Plant Physiol. 1996;112:193–205. doi: 10.1104/pp.112.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Popko JL, Umezawa T, Chiang VL. 5-Hydroxy-coniferyl aldehyde modulates enzymatic methylation for syringyl monolignol formation: a new view of monolignol biosynthesis in angiosperms. J Biol Chem. 2000;275:6537–6545. doi: 10.1074/jbc.275.9.6537. [DOI] [PubMed] [Google Scholar]
- Liang X, Dron M, Cramer CL, Dixon RA, Lamb CJ. Differential regulation of phenylalanine ammonia-lyase genes during plant development and by environmental cues. J Biol Chem. 1989;264:14486–14492. [PubMed] [Google Scholar]
- Lindroth RL, Hwang S-Y. Diversity, redundancy, and multiplicity in chemical defense systems of aspen. In: Romeo JT, Saunders JA, Barbosa P, editors. Phytochemical Diversity and Redundancy in Ecological Interactions. New York: Plenum Press; 1996. pp. 25–51. [Google Scholar]
- Lindroth RL, Kinney YY, Platz CL. Responses of deciduous trees to elevated atmospheric CO2: productivity, phytochemistry and insect performance. Ecology. 1993;74:763–777. [Google Scholar]
- Logemann E, Parniske M, Hahlbrock K. Modes of expression and common structural features of the complete phenylalanine ammonia-lyase gene family in parsley. Proc Natl Acad Sci USA. 1995;92:5905–5909. doi: 10.1073/pnas.92.13.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois R, Dietrich A, Hahlbrock K, Schultz W. A phenylalanine ammonia-lyase gene from parsley: structure, regulation and identification of elicitor and light responsive cis-acting elements. EMBO J. 1989;8:1641–1648. doi: 10.1002/j.1460-2075.1989.tb03554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai N, Kitauchi F, Okamoto K, Kanda T, Shimosaka M, Okazaki M. A transient increase of phenylalanine ammonia-lyase transcript in kinetin-treated tobacco cells. Biosci Biotechnol Biochem. 1994;58:558–559. doi: 10.1271/bbb.58.558. [DOI] [PubMed] [Google Scholar]
- O'Brien TP, Feder N, McCully ME. Polychromatic staining of plant cell walls by toluidine blue O. Protoplasma. 1964;59:366–373. [Google Scholar]
- Osakabe Y, Nanto K, Kitamura H, Kawai S, Kondo Y, Fujii T, Takabe K, Katayama Y, Morohoshi N. Immunocytochemical localization of phenylalanine ammonia-lyase in tissues of Populus kitakamiensis. Planta. 1996;200:13–19. doi: 10.1007/BF00196643. [DOI] [PubMed] [Google Scholar]
- Osakabe Y, Ohtsubo Y, Kawai S, Katayama Y, Morohoshi N. Structure and tissue-specific expression of genes for phenylalanine ammonia-lyase from a hybrid aspen, Populus kitakamiensis. Plant Sci. 1995a;105:217–226. [Google Scholar]
- Osakabe Y, Osakabe K, Kawai S, Katayama Y, Morohoshi N. Characterization of the structure and determination of mRNA levels of phenylalanine ammonia-lyase gene family from Populus kitakamiensis. Plant Mol Biol. 1995b;28:1133–1141. doi: 10.1007/BF00032674. [DOI] [PubMed] [Google Scholar]
- Parsons TJ, Bradshaw HD, Jr, Gordon MP. Systemic accumulation of specific mRNAs in response to wounding in poplar trees. Proc Natl Acad Sci USA. 1989;86:7895–7899. doi: 10.1073/pnas.86.20.7895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter LJ, Hrstich LN, Chan BG. The conversion of procyanidins and prodelphinidins to cyanidin and delphinidin. Phytochemistry. 1986;25:223–230. [Google Scholar]
- Rasmussen S, Dixon RA. Transgene-mediated and elicitor-induced perturbation of metabolic channeling at the entry point into the phenylpropanoid pathway. Plant Cell. 1999;11:1537–1551. doi: 10.1105/tpc.11.8.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve RM. Histological tests for polyphenols in plant tissues. Stain Technol. 1951;26:91–96. doi: 10.3109/10520295109113187. [DOI] [PubMed] [Google Scholar]
- Ribnicky DM, Shulaev V, Raskin I. Intermediates of salicylic acid biosynthesis in tobacco. Plant Physiol. 1998;118:565–572. doi: 10.1104/pp.118.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sarma AD, Sreelakshimi Y, Sharma R. Differential expression and properties of phenylalanine ammonia-lyase isoforms in tomato leaves. Phytochemistry. 1998;49:2233–2243. doi: 10.1016/s0031-9422(98)00336-7. [DOI] [PubMed] [Google Scholar]
- Shufflebottom D, Edwards K, Schuch W, Bevan M. Transcription of two members of a gene family encoding phenylalanine ammonia-lyase leads to remarkably different cell specificities and induction patterns. Plant J. 1993;3:835–845. doi: 10.1111/j.1365-313x.1993.00835.x. [DOI] [PubMed] [Google Scholar]
- Subramaniam R, Reinold S, Molitor EK, Douglas CJ. Structure, inheritance, and expression of hybrid poplar (Populus trichocarpa × Populus deltoides) phenylalanine ammonia-lyase genes. Plant Physiol. 1993;102:71–83. doi: 10.1104/pp.102.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C-J, Podila GK, Chiang VL. Agrobacterium-mediated transformation of quaking aspen (Populus tremuloides) and regeneration of transgenic plants. Plant Cell Rep. 1994;14:94–97. doi: 10.1007/BF00233768. [DOI] [PubMed] [Google Scholar]
- Tsai C-J, Popko JL, Mielke MR, Hu W-J, Podila GK, Chiang VL. Suppression of O-methyltransferase gene by homologous sense transgene in quaking aspen causes red-brown wood phenotypes. Plant Physiol. 1998;117:101–112. doi: 10.1104/pp.117.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalpani N, Leon J, Lawton MA, Raskin I. Pathway of salicylic acid biosynthesis in healthy and virus-inoculated tobacco. Plant Physiol. 1993;103:315–321. doi: 10.1104/pp.103.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]