Abstract
The importance of maternal cells in controlling early embryogenesis is well understood in animal development, yet in plants the precise role of maternal cells in embryogenesis is unclear. We demonstrated previously that maternal activity of the SIN1 (SHORT INTEGUMENTS1) gene of Arabidopsis is essential for embryo pattern formation and viability, and that its postembryonic activity is required for several processes in reproductive development, including flowering time control and ovule morphogenesis. Here, we report the cloning of SIN1, and demonstrate its identity to the CAF (CARPEL FACTORY) gene important for normal flower morphogenesis and to the SUS1 (SUSPENSOR1) gene essential for embryogenesis. SIN1/SUS1/CAF has sequence similarity to the Drosophila melanogaster gene Dicer, which encodes a multidomain ribonuclease specific for double-stranded RNA, first identified by its role in RNA silencing. The Dicer protein is essential for temporal control of development in animals, through the processing of small RNA hairpins that in turn inhibit the translation of target mRNAs. Structural modeling of the wild-type and sin1 mutant proteins indicates that the RNA helicase domain of SIN1/SUS1/CAF is important for function. The mRNA was detected in floral meristems, ovules, and early embryos, consistent with the mutant phenotypes. A 3.3-kb region 5′ of the SIN1/SUS1/CAF gene shows asymmetric parent-of-origin activity in the embryo: It confers transcriptional activation of a reporter gene in early embryos only when transmitted through the maternal gamete. These results suggest that maternal SIN1/SUS1/CAF functions early in Arabidopsis development, presumably through posttranscriptional regulation of specific mRNA molecules.
Molecular details behind genetic regulation in the early development of plants are beginning to emerge (for a recent review, see Chaudhury et al., 2001; Baroux et al., 2002). In Arabidopsis, only two genes have been identified whose activities are required in the maternal sporophyte (or female somatic cells) for normal pattern formation during embryo development (Ray et al., 1996b; Prigge and Wagner, 2000). Of these, SERRATE encodes a zinc finger protein presumably involved in chromatin structure modulation, and is required maternally for normal cotyledon initiation (Prigge and Wagner, 2000). At least one wild-type allele of SIN1 (SHORT INTEGUMENTS1), the only other gene identified in this class, is required in the maternal sporophyte for normal patterning in the embryo (Ray et al., 1996b). Plant strains homozygous for the hypomorphic sin1-2 mutant allele show a strong maternal effect on embryo development, producing developmentally arrested embryos with loss of apical, basal, or radial symmetry elements regardless of embryo genotype. Most of these form defective plants with no differentiation of shoot or root apical meristems (Ray et al., 1996b).
The exact role of the mother plant in regulating embryogenesis is the subject of some debate. The discussion so far has focused on the role of gene products contributed by the maternal gametophyte (Vielle-Calzada et al., 2001; Weijers et al., 2001). Many genes of this class are preferentially transcribed from the maternally contributed genome in the zygote, but, unlike the genetically defined expression of SIN1 in the female sporophyte, their expression appears to be required in the female gametophyte (Springer et al., 2000; Vielle-Calzada et al., 2000). This preferential transcription of some genes from the maternally contributed genome does not reflect complete transcriptional inactivation of the paternal genome, sometimes called imprinting when it occurs in animal systems, because at least some paternal genes are transcriptionally active early in embryogenesis (Weijers et al., 2001) but appears to affect the majority of genes (Vielle-Calzada et al., 2000, 2001; Baroux et al., 2002).
Mutant phenotype analysis shows a broader role for SIN1 than just maternal sporophytic control of embryo pattern formation. Originally isolated in the Landsberg erecta (La-er) strain, homozygous sin1-1 and sin1-2 mutations cause female sterility and ovule defects due to uncoordinated growth of integuments and over-proliferation of chalazal nucellus (Robinson-Beers et al., 1992; Lang et al., 1994; Ray et al., 1996a). The severity of integument defects in sin1 ovules is enhanced by the simultaneous presence of the erecta mutation (Lang et al., 1994; Ray et al., 1996a). Homozygous sin1 plants derived from embryos rescued by a heterozygous maternal sporophyte are also late flowering and produce an excess of vegetative leaves and lateral inflorescence axes (Ray et al., 1996a). This suggests a role of SIN1 in controlling meristem fate determination. The range of mutant phenotypes suggests that sin1 mutants are retarded in their ability to progress through developmental stages. This is reminiscent of the serrate phenotype, which also has transitional delays (Clarke et al., 1999).
We demonstrate here that SIN1 (At1g01040) is identical to previously identified CAF (CARPEL FACTORY) and SUSPENSOR1 genes (Schwartz et al., 1994; Jacobsen et al., 1999). The caf-1 mutation, due to a T-DNA insertion near the 3′ end of the gene (Jacobsen et al., 1999), causes pleiotropic defects throughout the plant. These include disruption of cell fate and cell division pattern in the floral meristem, producing excess stamens and carpels, but with no reported change in flowering time. In contrast, sin1 alleles were not previously known to affect floral organ number. Unlike homozygous caf or sin1, the homozygous sus1 embryos are inviable, as are the majority of developmentally arrested offspring derived from a homozygous sin1-2 mother plant. We provide new data that resolve some of these apparent differences among the mutant alleles of the same gene. The cloned SIN1/SUS1/CAF gene is homologous to the Drosophila melanogaster gene Dicer (a multidomain ribonuclease) essential for RNA silencing (Bernstein et al., 2001). Although little is known about the function of Dicer-like genes in plants, this gene family is required for the production of 21- to 25-nucleotide (nt) double-stranded RNA (dsRNA) fragments from larger dsRNA targets in animals (Bernstein et al., 2001; Grishok et al., 2001; Hutvágner et al., 2001; Knight and Bass, 2001). Here, we demonstrate the functional importance of an RNA helicase domain of a Dicer-like protein, provide evidence for preferential activation of the gene's maternally transmitted regulatory region in Arabidopsis embryos, and propose a model for its mechanism of action in plants.
RESULTS
SIN1, a Dicer Homolog, Is an Essential Gene
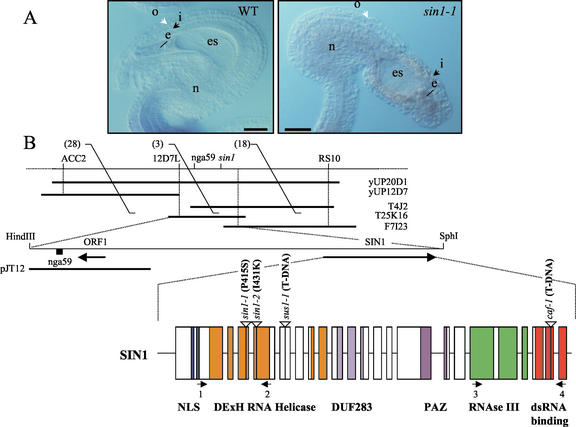
Homozygous sin1 mutants exhibit defects in flowering time (Ray et al., 1996a; Table I), delayed transition between developmental states, and failure to coordinate integument formation in ovules (Fig. 1A). These pleiotropic effects suggest that SIN1 plays a key role in several developmental processes. We cloned the SIN1 gene (Golden, 1999) by mapping first to an overlapping series of BACs, sequencing the 25,992-bp overlap region on the top of chromosome I (gi6684981), predicting the coding sequence, and sequencing mutant alleles within the predicted gene (Fig. 1B). The original genomic sequence around SIN1 was derived from BACs of the Col ecotype and was subsequently verified by resequencing 8.2 kb of Col and La-er chromosomal DNA. The same region was also sequenced from the sin1 mutants, with no nucleotide changes over the entire 8.2-kb region except for a single C to T transition in sin1-1 and a single T to A transversion in sin1-2. The isolated gene (gi11559646) is identical to the CAF gene (gi6102609), except for two single base sequencing differences whose identity we confirmed by resequencing the caf mutant allele (Jacobsen et al., 1999).
Table I.
Effects of sin1 and caf mutations on flowering time and leaf no.
| Strain | Segregant Phenotype | n | Average Bolting Time | Average No. of Leaves | Significant Difference between Segregants |
|---|---|---|---|---|---|
| Columbia-glabrous1(Co-gl) | Wild type | 41 | 27.4 ± 1.4 | 9.1 ± 1.0 | – |
| sin1-1/+ (Co-gl) | Sin1+ | 32 | 28.1 ± 1.4 | 9.2 ± 0.9 | – |
| Sin1− | 10 | 45.0 ± 2.9 | 18.8 ± 2.7 | d > 99% | |
| sin1-2/+ (Co-gl) | Sin1+ | 33 | 28.4 ± 1.6 | 9.3 ± 1.0 | – |
| Sin1− | 15 | 41.0 ± 2.3 | 18.7 ± 2.0 | d > 99% | |
| sin1-1/+ (La-er) | Sin1+ | 38 | 27.9 ± 1.6 | 8.3 ± 0.8 | – |
| Sin1− | 15 | 42.5 ± 5.9 | 14.6 ± 2.8 | d > 99% | |
| sin1-2/+ (La-er) | Sin1+ | 26 | 26.6 ± 2.0 | 7.7 ± 0.9 | – |
| Sin1− | 9 | 36.2 ± 3.6 | 12.3 ± 2.2 | d > 99% | |
| caf-1/+ (La-er) | Caf+ | 42 | 27.8 ± 2.0 | 8.4 ± 0.8 | – |
| Caf− | 7 | 35.0 ± 2.1 | 11.1 ± 1.7 | d > 99% |
Regardless of the genetic background, sin1 and caf mutants exhibit a delay in flowering (as assayed by the no. of days it took the plants to bolt), as well as an increase in the production of leaves. Data were analyzed using the Fisher-Behrens procedure (for means with unequal population variances), with the d value significant at the 99% level when comparing the mutants with their wild-type segregants, for both flowering time and leaf no. (Campbell, 1989). The data are presented as mean ± sd.
Figure 1.
SIN1 gene structure and its mutant phenotype in the ovule. A, In wild-type ovules, the outer integument cell layers entirely cover the inner integument that encloses the embryo sac that contains the egg. Most mutant ovules show uncoordinated growth of both the inner and outer integuments and the nucellus, such that the embryo sac, with the egg, is extruded. o, Outer integuments; i, inner integuments; es, embryo sac; n, nucellus. B, Map of the chromosomal region overlapping SIN1. RS10, nga59, 12D7L, and ACC2 are DNA sequence markers. Numbers within parentheses are numbers of crossovers between La-er and Columbia (Col) chromosomes. yUP20D1 and yUP12D7 are yeast (Saccharomyces cerevisiae) artificial chromosome clones; T4J2, T25K16, and F7I23 are bacterial artificial chromosomes (BACs); and pJT12 is a plasmid subclone of Arabidopsis chromosome I. The positions of the only two identified open reading frames (ORFs) in this region (ORF1 and SIN1) are represented by large arrows. The lower diagram shows intron-exon boundaries of SIN1 (exons are boxed), with different protein-coding domains color coded. The two primer sets (1 and 2, 3 and 4) used for reverse transcriptase (RT)-PCR (Fig. 2A) are represented by arrows corresponding to the primers location in the SIN1 gene.
A 6.2-kb cDNA (accession no. AF292940) corresponding to the SIN1 mRNA was isolated from a flower- and seed-enriched cDNA library (Lou et al., 1999) using probes generated from PCR-amplified genomic regions from the predicted SIN1 exons 1 through 5 and 10 through 14 (Golden, 1999). The cDNA has a 5,727-bp ORF, a 378-bp 5′-untranslated region (UTR), a 74-bp 3′-UTR, and nine adenines at the 3′ end, likely to be from the poly(A+) tail. The cDNA sequence confirmed the presence of 19 introns and 20 exons. The C to T mutation in sin1-1 and T to A in sin1-2 are in exons 3 and 4, respectively. The non-complementing mutation, sus1-1, which confers embryo lethality (Errampalli et al., 1991; Castle et al., 1993), contains 17 kb of a tandem array of T-DNA insertions in the fifth exon. The identity between SIN1 and SUS1 confirmed that the gene is essential for viability.
Transcription of SIN1/SUS1/CAF
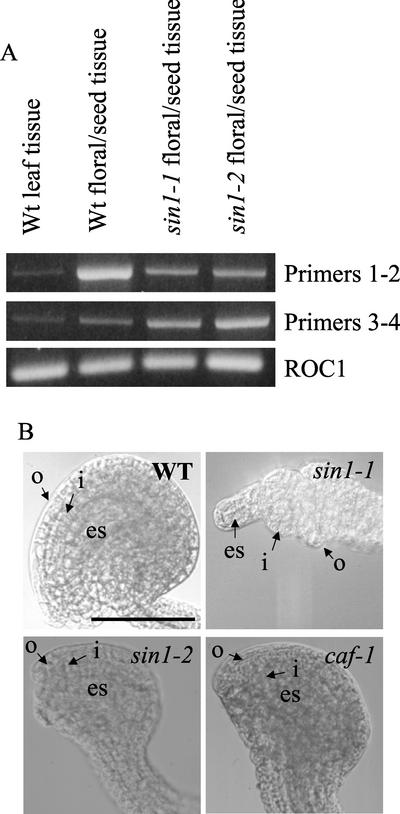
Hybridization with either the 6.2-kb cDNA or a 310-nt probe corresponding to the 5′ terminus of the SIN1/SUS1/CAF ORF to total RNA extracted from flowers and seeds detected a major 6.2-kb and a minor 2.6-kb species only, as previously published (Jacobsen et al., 1999). Jacobsen et al. (1999) showed by RNA-blot analysis that exons 3 and 4 (where the sin1 point mutations map) were not present in the 2.6-kb transcript. This suggests that if the sin1 point mutations affect message stability, then they would alter the steady-state mRNA level of the 6.2-kb transcript and not the 2.6-kb transcript. To investigate the level of the 6.2-kb SIN1/SUS1/CAF message in sin1 mutant plants, RT-mediated PCR analysis of RNA was performed with two sets of primers (Fig. 2A). Primer set 1-2 amplified RNA sequences encoding parts of exon 1 through sequences in exon 4, whereas primer set 3-4 amplified RNA sequences from exon 17 to the end of exon 19. The two RT-PCR signals were barely visible from wild-type leaf RNA, but higher levels were detected with RNA from wild-type flowers and seeds. Both SIN1/SUS1/CAF-specific PCR products were detected in RNA from flowers and seeds of sin1-1 and sin1-2 point mutants, suggesting that Sin1− phenotypes are due to functionally aberrant proteins and not due to a significant reduction in SIN1/SUS1/CAF RNA concentration. This is consistent with previous observations for caf-1 (Jacobsen et al., 1999).
Figure 2.
Analysis of sin1 and caf mutants. A, RT-PCR analysis of SIN1/SUS1/CAF mRNA expression. The top row represents signal from RT-PCR with a primer set that amplifies +457 to +1,763 (1 and 2), and the middle row is signal with a set that amplifies +4,545 to +6,138 (3 and 4). The bottom row is signal from the constitutively expressed mRNA of the ROC1 gene. SIN1/SUS1/CAF mRNA is expressed at a higher level in floral tissue than in leaves, and it is not significantly lower in the sin1 point mutations. The relative levels of the two RT-PCR-amplified bands from mutant and wild-type flowers were not reproducibly different. B, Ovule morphology affected by the sin1 and caf mutant alleles in La-er. o, Outer integuments; i, inner integuments; es, embryo sac. Bar = 100 microns.
Previous studies on localization of SIN1/SUS1/CAF message by in situ hybridization indicated that the transcript is present in the vegetative shoot apical meristem (SAM), the inflorescence meristem, and the developing floral meristem, but the expression pattern at other times and places in development was not reported (Jacobsen et al., 1999). In an effort to identify the cytological domains of expression within reproductive tissues, we carried out in situ hybridization analysis in developing flowers and siliques (Fig. 3). Antisense probes generated from the 6.2-kb full-length cDNA revealed gene-specific signals within the inflorescence meristem and the central region of developing flowers including immature stamen and carpel, in the stigmatic transmitting tissue, and in early ovule primordia (Fig. 3, A, C, and I). In immature ovules, SIN1/SUS1/CAF-specific transcript is localized within the integument initials, nucellus, and in the megaspore mother cell (Fig. 3E). In mature ovules until the time of fertilization, no message is detectable in the ovule except in the funiculus and chalazal nucellus. After fertilization, SIN1/SUS1/CAF-specific mRNA is detectable in the embryo proper, and not in the suspensor, up to the globular stage (Fig. 3G), and undetectable beyond this stage (data not shown). The SIN1/SUS1/CAF-specific transcript (detectable above background hybridization) appears confined to the funiculus (Fig. 3J) and the chalaza in sporophytically derived tissues of the seed. These RNA localization results are consistent with the domains affected by sin1, sus1, and caf mutations, namely ovule integuments, inflorescence meristem, the embryo, and the floral meristem, respectively.
Figure 3.
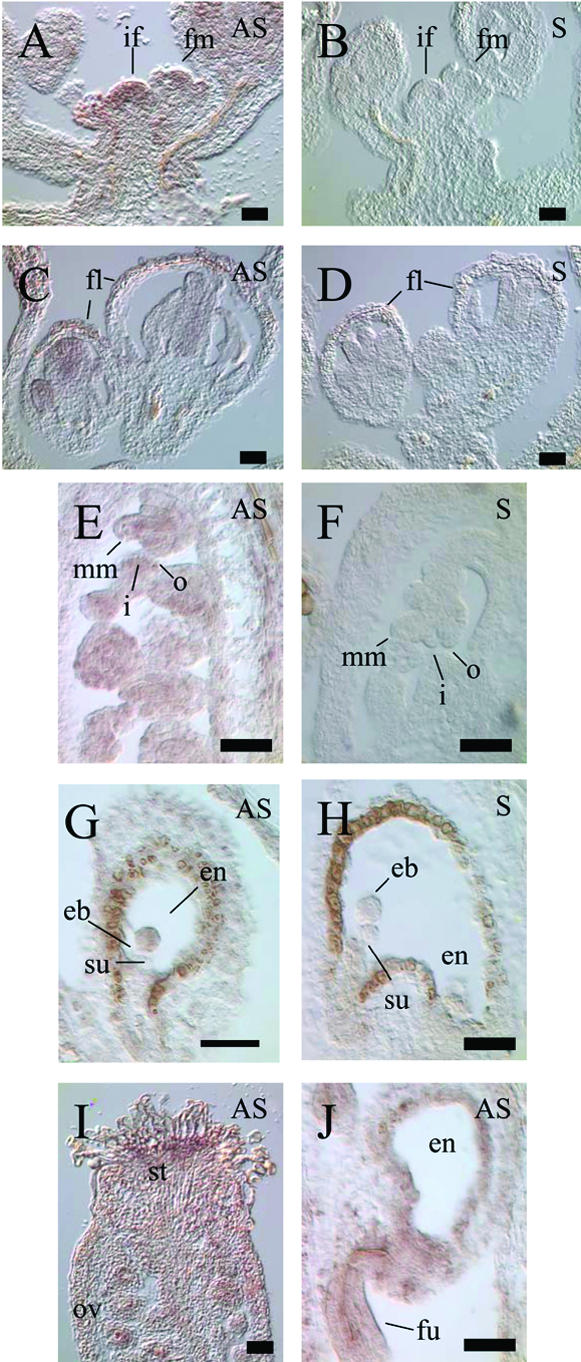
Localization of SIN1/SUS1/CAF transcript by in situ hybridization. A and B, Inflorescence and early floral meristems, showing strong expression in the inflorescence meristem and in the center of the floral meristem. C and D, Developing flowers, with expression in the center of the flower. E and F, RNA accumulated in all cell types of the early ovule. G and H, Expression in early embryo is located in the embryo proper, and is not detected in the suspensor or the endosperm. I, Stigmatic tissue shows strong expression, and message is detected in immature ovules. J, Strong expression is coincident with vascular elements of the funiculus in seeds with heart stage embryos. Sections hybridized with full-length cDNA antisense probe (A, C, E, G, I, and J) are indicated by AS, whereas sections hybridized with the sense probe (B, D, F, H, and J) are indicated by S. if, Inflorescence meristem; fm, floral meristem; fl, developing flowers; mm, megaspore mother cell; o, outer integuments; i, inner integuments; eb, embryo; en, endosperm; su, suspensor; st, stigmatic tissue; ov, ovules; fu, funiculus. All bars = 100 microns.
The Genomic Region Upstream of SIN1/SUS1/CAF Is Active throughout Development
To study transcriptional regulation of SIN1/SUS1/CAF in more detail, we fused a 3.3-kb 5′-upstream region adjacent to the SIN1/SUS1/CAF ORF (including 38 bp of the 5′-UTR) to the β-glucuronidase (GUS) reporter gene (Fig. 4A, pSP2). Of the 12 independent T2 plant lines that were originally isolated, four that segregated the T-DNA as single insert locus were verified by DNA-blot analysis to contain single T-DNA inserts. Of these lines, two (labeled pSP2-1 and pSP2-8 in Table II) with the most intense staining patterns were made homozygous for the T-DNA insert by self-crosses, and were used in further experiments.
Figure 4.
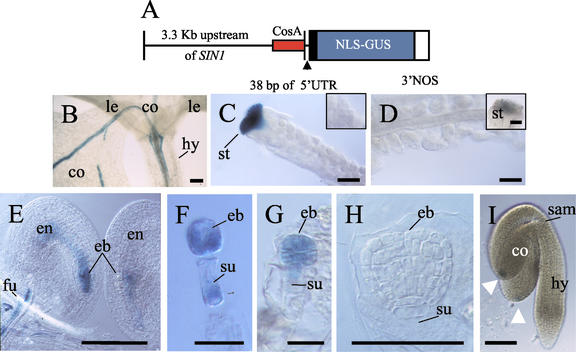
Expression pattern of the SIN1/SUS1/CAF upstream genomic region, assayed by promoter GUS fusion. A, The SIN1/SUS1/CAF putative promoter construct pSP2, containing 3.3 kb of sequence upstream of the SIN1/SUS1/CAF gene (with 38 bp of the 5′-UTR), fused to the GUS reporter gene. The minimal promoter region from the cosmid CosA (which rescues the caf-1 mutant phenotype) is identified in red (Jacobsen et al., 1999). B, Two-leaf stage seedling (10 d post-germination) showing GUS activity in developing veins of the cotyledons and some expression in the hypocotyl. C, GUS expression is only detectable in the stigmatic tissue, and undetected in the initiating integuments at floral stage 11 (inset). D, Mature ovules at floral stage 13 show no detectable activity, and a reduction in GUS activity is seen in the stigma (inset). E and F, Post-fertilization ovules showing staining in the zygote and the endosperm. GUS activity is also present in the funiculus, which was observed throughout seed development. F, GUS expression in a dissected two-cell embryo, with activity in both the embryo and suspensor. G, GUS expression in octant embryo, but now absent from the suspensor. H, No detectable GUS activity in late globular or later embryonic stages (data not shown). I, Linear to cotyledon stage embryo showing GUS expression at the tip of the cotyledon (white arrowheads). co, Cotyledon; le, leaf; hy, hypocotyl; st, stigma; eb, embryo; en, endosperm; fu, funiculus; su, suspensor; sam, SAM. All bars = 100 microns, except F and G, which = 50 microns.
Table II.
Expression of GUS reporter gene in reciprocal crosses
| Recipient Pistil | Pollen Donor | Expression in
Early Embryo
|
Late Expression in Mature Seeds
|
||
|---|---|---|---|---|---|
| GUS+ | GUS− | GUS+ cotyledons | GUS− cotyledons | ||
| pSP2-1 | Co-gl | 35 | 2 | 86 | 31 |
| Co-gl | pSP2-1 | 0 | 63 | 69 | 133 |
| pSP2-8 | Co-gl | 67 | 16 | 73 | 69 |
| Co-gl | pSP2-8 | 0 | 78 | 28 | 28 |
The SIN1/SUS1/CAF putative promoter GUS fusion pSP2 is transcriptionally active post-fertilization only if contributed from the maternal gamete. This silencing of the paternal allele is released by the time pSP2 activity reappears, at the late walking stick stage of embryogenesis. The pollinated pistils were dissected either 3 d postpollination, at a pistil length of 2.5 to 3.0 mm, to score for GUS activity in early embryogenesis, or 10 d postpollination to score for activity in late embryogenesis. The GUS fusion lines are labeled pSP2-1 and pSP2-8, whereas the wild-type control is Co-gl.
At the seedling stage, GUS activity was detected in the veins and in the central vasculature, especially in phloem cells, the cotyledons, the hypocotyl, and roots (Fig. 4B). GUS activity, completely absent during ovule development until fertilization, was rapidly induced in the embryo sac and in vascular cells of the funiculus after fertilization (in Fig. 4, compare C and D with E). GUS activity was detected in the two-cell embryo (Fig. 4F) and persisted until the octant stage of the embryo (Fig. 4G), then declined to an undetectable level past the globular stage (Fig. 4H; data not shown). GUS activity in the vascular tissue of the funiculus was detectable throughout seed development (data not shown). In embryos at the linear to cotyledon stages of development, GUS activity was consistently detected only in a few cells at the tips of the cotyledons and never in the embryo shoot or root apical meristem (Fig. 4I). The most dynamic GUS expression was detected in the stigma of flowers until just before pollination, which then ended abruptly (compare Fig. 4C with the inset in D). Compared with the mRNA localization data from in situ hybridization experiments, the lack of GUS activity in the meristem and in the early ovule integuments indicates that pSP2 reporter construct does not completely recapitulate the endogenous expression pattern. However, the expression patterns in stigmatic tissue, in early embryos, and in the vasculature of the funiculus exactly parallels what was observed by RNA localization.
Only the Maternal SIN1/SUS1/CAF Upstream Genomic Region Activates Transcription in Early Embryos
The initial characterization of self-crossed heterozygous pSP2 lines showed that approximately one-half of the fertilized ovules lacked detectable GUS activity (data not shown). Therefore, we set out to test whether the asymmetric parental requirement of SIN1/SUS1/CAF activity reported earlier could, in part, be the result of preferential transcriptional activation of the SIN1/SUS1/CAF allele that is transmitted through the female gametophyte. In other words, does activation of the SIN1/SUS1/CAF upstream region in the embryo show a disparity depending on the parent-of-origin? To address this question, reciprocal crosses were conducted between flowers of homozygous pSP2 lines and wild-type flowers, and resulting embryos were assayed for GUS activity. Results in Table II show that GUS activity was detected in the post-fertilization embryo sac only when the GUS transgene was transmitted through the female gametophyte (102 Gus+ ovules/120 ovules tested), but not when transmitted through the pollen (0 Gus+ ovules/141 ovules tested). The effect of maternal inheritance was most evident during the initial activation of GUS expression in the embryo sac and in early embryos up to the early globular stage. Late GUS activity in cotyledons of mature seeds was independent of the asymmetric parental induction of the reporter gene seen early in development. These results suggest that the early transcription of SIN1/SUS1/CAF in the post-fertilization embryo is primarily from the maternally transmitted allele. Sequencing of three different ecotypes revealed no DNA sequence polymorphism in the SIN1/SUS1/CAF ORF; therefore, preferential inheritance through the female gametophyte could not be directly confirmed by sequence polymorphism-guided RT-PCR.
Phenotypes of sin1, sus1, and caf Alleles Overlap
We were initially struck by the phenotypic differences reported between the sin1 mutations and the caf-1 allele. Based on mutant phenotypes, SIN1 was thought to regulate ovule and embryo morphogenesis, and control flowering time (Robinson-Beers et al., 1992; Lang et al., 1994; Ray et al., 1996a, 1996b), whereas CAF was thought to regulate cell division in the floral meristem (Jacobsen et al., 1999). Closer examination revealed that caf-1 also affects flowering time and ovule development. Because caf and sin1 mutations were originally isolated in different genetic backgrounds, we directly compared the phenotypic effects of the caf-1 allele introgressed into La-er with those of two sin1 alleles originally isolated in La-er. These studies revealed that the caf-1 mutation delays flowering as much as the hypomorphic sin1-2 allele (Table I). In addition, both caf-1 and sin1-2 in the La-er background show approximately the same frequency of petal/stamen and stamen/carpel mosaicism; however, 94% of caf-1 carpels remained unfused compared with only 8% of sin1-2 carpels (Table III). Ovules of both mutants were abnormal, being smaller and having less curvature than wild-type ovules, but with comparable expansion of inner and outer integuments (Fig. 2B). Because these phenotypic effects of caf-1 are milder than those of sus1 or sin1-1, the deletion of one of the two dsRNA-binding domains (see later) in caf-1 is not sufficient to cause a null phenotype.
Table III.
Effects of sin1 and caf mutations on floral architecture
| Strain/Segregant Phenotype | Se | Se/Pe | Pe | Pe/St | St | St/Ca | Ca | Unfused Carpels |
|---|---|---|---|---|---|---|---|---|
| % | ||||||||
| Co-gl | ||||||||
| Sin1+ | 4.0 ± 0.0 | 0 | 4.0 ± 0.0 | 0 | 5.8 ± 0.4 | 0 | 2.0 ± 0.0 | 0 |
| sin1-1/+ (Co-gl) | ||||||||
| Sin1+ | 4.0 ± 0.1 | 0 | 4.0 ± 0.0 | 0 | 5.9 ± 0.4 | 0 | 2.0 ± 0.0 | 0 |
| Sin1− | 4.1 ± 0.3* | 0 | 3.8 ± 0.4** | 0.1 ± 0.2 (6%) | 6.0 ± 0.5 | 0 | 2.0 ± 0.0 | 0 |
| sin1-2/+ (Co-gl) | ||||||||
| Sin1+ | 4.0 ± 0.1 | 0 | 4.0 ± 0.0 | 0 | 5.9 ± 0.4 | 0 | 2.0 ± 0.0 | 0 |
| Sin1− | 4.2 ± 0.5** | 0.1 ± 0.3 (6%) | 4.0 ± 0.3 | 0.1 ± 0.4** (14%) | 5.9 ± 0.5 | 0 | 2.0 ± 0.0 | 0 |
| sin1-1/+ (La-er) | ||||||||
| Sin1+ | 4.0 ± 0.0 | 0 | 4.0 ± 0.0 | 0 | 5.7 ± 0.6 | 0 | 2.0 ± 0.0 | 0 |
| Sin1− | 4.0 ± 0.1 | 0 | 4.0 ± 0.1 | 0 | 5.8 ± 0.4 | 0 | 2.0 ± 0.0 | 0 |
| sin1-2/+ (La-er) | ||||||||
| Sin1+ | 4.0 ± 0.0 | 0 | 4.0 ± 0.0 | 0 | 5.9 ± 0.4 | 0 | 2.0 ± 0.1 | 0 |
| Sin1− | 4.0 ± 0.0 | 0 | 4.0 ± 0.0 | 0.0 ± 0.1 (2%) | 6.2 ± 0.9* | 0.2 ± 0.5* (12%) | 2.5 ± 1.0** | 8 |
| caf-1/+ (La-er) | ||||||||
| Caf+ | 4.0 ± 0.0 | 0 | 4.0 ± 0.0 | 0.0 ± 0.1 (2%) | 5.4 ± 0.6 | 0 | 2.0 ± 0.0 | 0 |
| Caf− | 3.9 ± 0.2 | 0.0 ± 0.1 (2%) | 3.9 ± 0.3** | 0.1 ± 0.4 (12%) | 5.4 ± 0.9 | 0.1 ± 0.2 (6%) | 1.9 ± 0.3* | 94 |
Aberrant floral organ no. and morphology were observed in both sin1 and caf mutants, regardless of genetic background (either La-er or Co-gl). For each phenotypic class, n = 50 flowers and the data are presented as the mean organ number ± sd. se, Sepals; se/pe, sepaloid petals; pe, petals; pe/st, petaloid stamen; st, stamen; st/ca, stamenoid carpels; ca, carpels. The frequency at which morphologically abnormal floral organs arose is noted. Data were analyzed using the Fisher-Behrens procedure, with an asterisk representing the d value significant at the 95% level, and two asterisks representing the d value significant at the 99% level (Campbell, 1989).
The Structural Basis of SIN1/SUS1/CAF Function
The predicted SIN1/SUS1/CAF protein contains 1,909 amino acid residues (Golden, 1999; Jacobsen et al., 1999; gi6102610), and has multiple conserved motifs, including a bipartite nuclear localization signal (NLS), a DExH box RNA helicase C motif (Gorbalenya and Koonin, 1993), two regions that are conserved among Dicer-like proteins (DUF283, a domain of unknown function, http://www.cgr.ki.se/Pfam/DQL_sel_domains.html; and a Piwi-Argonaute-Zwille (PAZ) domain, Cerutti et al., 2000; A. Mushegian and A. Ray, unpublished data), two RNase III motifs (Mian, 1997), and two dsRNA-binding domains (Ramos et al., 2000; Fig. 1B). These domains, except for the N-terminal approximately 225 amino acid residues that contains a region of highly charged amino acids and the NLS, are shared among Dicer and its homologs in Caenorhabditis elegans and humans (Bernstein et al., 2001; Grishok et al., 2001; Knight and Bass, 2001). The Dicer protein was originally purified based on its catalytic ability to degrade long dsRNA molecules into short approximately 22-nt products that are associated with both posttranscriptional gene silencing and RNAi (Bernstein et al., 2001). Although the essential roles of dsRNA-binding and RNAse III domains in Dicer function are clear, the role of the RNA helicase domain in the process of RNA silencing is less well understood, partially due to the lack of weak loss-of-function alleles in other Dicer family members.
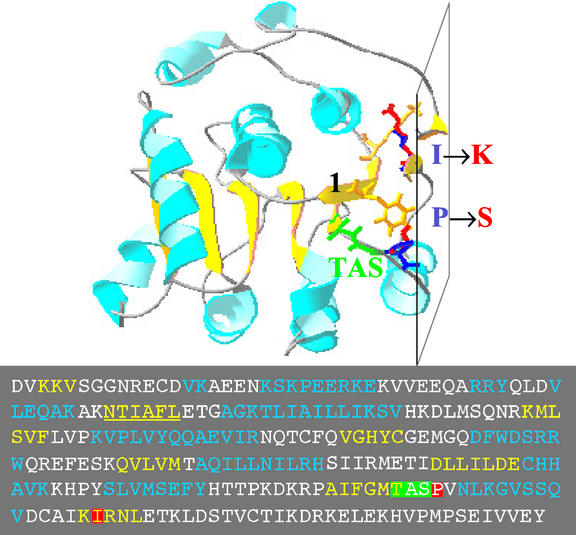
Perturbations of different structural domains of the SIN1/SUS1/CAF protein result in discrete mutant phenotypes. The previously described caf-1 mutation is predicted to delete 75 amino acid residues from the C terminus of the SIN1/SUS1/CAF protein, generating a protein that lacks one of two dsRNA-binding domains (Jacobsen et al., 1999). In contrast, the large T-DNA insertion in exon 5 of sus1-1 results in a more significant disruption near the N terminus of the protein. This large lesion probably causes a null mutation because it phenocopies the sus1-3 mutation, which contains a deletion of greater than 20 kb that removes the 5′ end of the gene. Additional sus1 knockouts generated from a large-scale insertional mutagenesis project (McElver et al., 2001) have also been described recently (http://www.seedgenes.org). The sin1-1 and sin1-2 alleles both map to the C-terminal region of the helicase domain, having P415S and I431K substitutions, respectively. Neither amino acid residue had previously been described as crucial for helicase function. Amino acid sequence alignment and homology modeling, using the yeast translation initiation factor 4A (yIF4A) helicase as the template, suggests a similar structural basis for the effects of both point mutations in sin1: Both amino acid substitutions map on the same face of the predicted helicase domain (Fig. 5). The P415S substitution is adjacent to a predicted α-helix, which follows the TAS signature (apparently corresponding to the helix E and the conserved SAT motif of yIF4A, respectively), and the I431K change is in the last β-strand of the helicase structural core. The side chains of both affected residues point inside the core, and the mutated residues are predicted to produce side chain clashes with residues within the conserved β-strand 1. The α-helix and the β-strand harboring the mutations are on the same plane, which forms a nearly flat outer surface of the molecule, thought to interact with the RNA substrate (Pause and Sonenberg, 1992). The mutations are predicted to distort the face of this plane, thus either impairing the ability of the helicase domain to bind to its RNA substrate or the efficiency with which the nearby TAS motif mediates RNA unwinding. Replacement of a Pro residue at the beginning of an α-helix by Ser in P415S substitution may cause repacking of the α-helix, which is expected to cause a more drastic perturbation of the helicase-RNA interaction plane than that caused by the I431K replacement. This expectation is consistent with genetic evidence that sin1-1 is stronger than sin1-2. Perturbation of this interaction surface may reduce, but not eliminate, the activity of the SIN1/SUS1/CAF helicase on some substrate RNAs, thus explaining the hypomorphic nature of the two missense alleles.
Figure 5.
Structural model of the SIN1/SUS1/CAF helicase domain. Yeast translation initiation factor yIF4A was used as a template for alignment of the SIN1/SUS1/CAF helicase domain residues (inset shows the amino acid sequence, with mutated residues boxed). α-helices and β-strands are light-blue and yellow ribbons, respectively. The peptide backbone of the TAS motif is in green. The side chains of residues in the mutated positions are shown; wild-type residues are dark blue and mutated residues are red. The side chains of those residues in the first β-strand (underlined in the inset) that clash with the mutated residues are in yellow. The plane, formed by an α-helix and two β-strands, predicted to be involved in RNA binding and unwinding, is indicated.
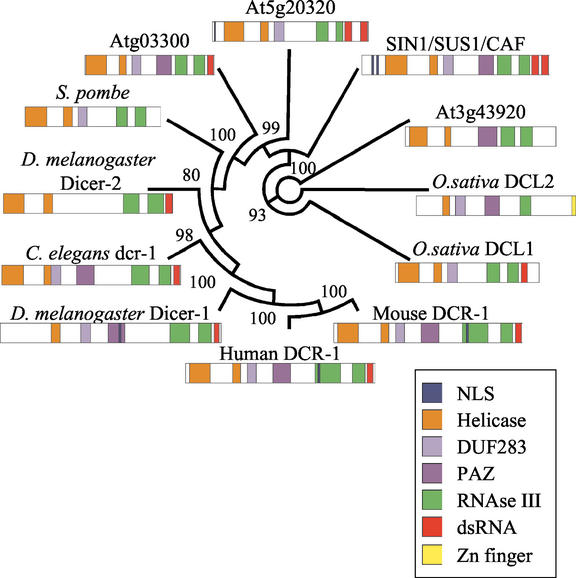
Three additional Dicer family members in Arabidopsis were recognized by BLAST searches using as query the entire SIN1/SUS1/CAF protein sequence (Fig. 6). Of these, T15B3.60 (At3g43920) and T17B22.28 (At3g03300) are on chromosome III and F5024.210 (At5g20320) is on chromosome V. T17B22.28 and F5024.210 have a domain architecture (determined by CDART and PFAM searches) that is very similar to that of SIN1/SUS1/CAF. Unlike all other Dicer family members, T15B3.8 and the Dicer homolog found in S. pombe lack a dsRNA-binding domain, which may indicate a somewhat different biochemical role for these proteins. Finally, F5024.210, unlike all other Dicer homologs in Arabidopsis, does not appear to have a PAZ domain. Phylogenetic analysis suggests that the known members of the Dicer family diverged early in eukaryotic evolution (Fig. 6). On the whole, Arabidopsis Dicer-like proteins are more similar to each other than to Dicer-like proteins in organisms from other kingdoms. Several other proteins with sequence similarity to SIN1/SUS1/CAF were identified in Arabidopsis, but because they lacked either the RNA helicase or the RNAse III domain, which are thought to be integral for Dicer function, they were not included in this analysis.
Figure 6.
Phylogenetic analysis of known Dicer-like proteins. A neighbor-joining circle tree, constructed by using full-length protein sequences of SIN1/SUS1/CAF and all known Dicer homologs. The predicted domain structures for Dicer-like proteins were identified by Pfam and Prosite searches. Sequence accessions are: human Dicer-1 (gi14748177), Schizosaccharomyces pombe CAB37423 (gi2130449), Arabidopsis F5024.210 and At5g20320 (gi15241323), Arabidopsis T15B3.8 and At3g43920 (gi7594553), SIN1/SUS1/CAF and At1g01040 (gi11559646), Arabidopsis T17B22.1 and Atg03300 (gi6714410), C. elegans dcr-1 (gi630692), D. melanogaster Dicer-1 (gi17738129), D. melanogaster Dicer-2 (gi16215719), Oryza sativa DCL1 (gi18087887), Mus musculus mDCR-1 (gi20385913), and O. sativa DCL2 (gi:20804934). Analysis using only the conserved helicase or RNase III domains produced identical tree structures.
DISCUSSION
The results presented here raise the possibility that dsRNA processing is important for plant development, and that epigenetic control of transcriptional activation at least partly regulates this process in the early embryo. We have demonstrated that sin1 and sus1 mutations both affect the same gene (now understood to be a Dicer homolog) as the previously described caf-1 mutation. The RNA helicase activity of SIN1/SUS1/CAF is important for function, which is consistent with biochemical models of Dicer's activity. The gene is essential for viability. SIN1/SUS1/CAF RNA is found in reproductive tissues and in very early embryos. Using a promoter fusion construct, we demonstrate that this expression in the early embryo is from the maternally contributed genome. These findings attest to a novel mechanism of gene regulation during development, which appears to be conserved among higher eukaryotes.
sin1 and caf-1 Alleles Have Similar Phenotypes
Previously published accounts reveal little apparent similarity between the phenotypes of sin1 and caf-1 mutations. Closer examination in more uniform genetic backgrounds described here did, however, identify phenotypic similarities in flowering time delay, floral morphology, and ovule formation. A role for SIN1/SUS1/CAF in the control of cell division was suggested on the basis of microscopic observation of cell number in caf mutant plants, which is consistent with previous observations of increased cell number in sin1 SAM and ovule nucellus (Lang et al., 1994; Ray et al., 1996a, 1996b; Jacobsen et al., 1999). Furthermore, caf-1 in La-er causes a similar delay in SAM fate transition, as do the sin1 alleles in La-er. RNA localization via in situ hybridization largely confirmed expression in those same organs and tissues that are affected in the mutant plants. Assuming that SIN1/SUS1/CAF functions like Dicer, we attribute the slight phenotypic differences between the two alleles as due either to differences in the requirements of the target dsRNA molecules for helicase or RNAse III activity or to differential interactions of the two mutant proteins with overlapping (but nonidentical) target dsRNA populations in different organs or in distinct spatio-temporal domains.
SIN1/SUS1/CAF mRNA Localization Domains Overlap with Its 5′-UTR Activity Domains
Utilizing the reporter construct pSP2, which is a fusion between the GUS gene and a 3.3-kb region upstream of the SIN1/SUS1/CAF ORF that contains only 38 bp of the 378-bp 5′-UTR, we investigated how SIN1/SUS1/CAF transcriptional activity was regulated throughout development. RNA localization and GUS expression data together suggest roles for the SIN1/SUS1/CAF gene in developmental processes that had not been elucidated through mutant analysis. For example, we are uncertain of the significance of strong promoter activity and the accumulation of SIN1/SUS1/CAF transcript in the stigma immediately before pollination; neither sin1 nor caf-1 is known to be defective in pollination and pollen tube growth. Like its role in other aspects of development, SIN1/SUS1/CAF could regulate the timing of stigma maturation. Alternatively, the burst of SIN1/SUS1/CAF transcription may underlie an RNA silencing-related defensive mechanism against opportunistic dsRNA viral infiltration through tracts of pollen tubes.
After fertilization, the SIN1/SUS1/CAF promoter region is activated in vascular (sporophytic) cells of the ovule funiculus, which is correlated with the presence of SIN1/SUS1/CAF RNA in this region. These observations raise the intriguing possibility that SIN1/SUS1/CAF RNA expressed in this part of the sporophyte may be required for normal embryo development. This presumed effect-at-a-distance of funicular RNA expression was observed before. For example, mutant analysis indicated that the activity of the predicted transcription factor DAG1, whose mRNA is also localized to the vasculature of the funiculus, is required sporophytically in seed coat cells for maintaining seed dormancy (Papi et al., 2000).
Although pSP2 reporter expression did recapitulate most aspects of the RNA localization pattern, there was no detectable GUS activity in either the developing ovule or the SAM. Therefore, the reporter construct is probably missing key strands for transcription regulation. Alternatively, the GUS mRNA that is fused to the 38-nt 5′-UTR of SIN1/SUS1/CAF may not be translated, or is unstable, in these cellular domains.
The Role of SIN1/SUS1/CAF in Early Development. Sporophytic, Gametophytic, or Zygotic?
In early Arabidopsis embryo, paternal alleles of many genes are selectively silenced, whereas their maternal alleles are activated through transcriptional (Vielle-Calzada et al., 2000) and possibly posttranscriptional (Springer et al., 2000) mechanisms in the gametophyte. Due to their pattern of inheritance, mutations in these genes are thought to exert a gametophytic maternal effect on the development of the embryo, in which gene expression in the embryo is controlled by events occurring in the maternal gametophyte. Our previous work characterizing the SIN1 maternal effect did not indicate a similar mechanism, suggesting instead that its role in patterning the embryo is primarily mediated through the gene's expression in the maternal sporophyte (Ray et al., 1996b). Specifically, a sin1/+ heterozygous embryo is as affected as a sin1/sin1 homozygous embryo when either genotype is borne on a sin1/sin1 homozygous mother. In contrast, a self-fertilized sin1/+ heterozygous mother will produce sin1/sin1 homozygous embryos that progress though embryogenesis normally. Nonetheless, post-zygotic expression of SIN1/SUS1/CAF transcript must be essential for embryo viability because the homozygous sus1 deletion is embryo lethal even when such embryos are borne on a hemizygous sus1/+ sporophyte. Here, we have investigated the source of the post-zygotic message by in situ RNA localization, and have also shown that a 5′-upstream region of SIN1/SUS1/CAF is transcriptionally active in early embryos only when it is maternally transmitted. This pattern of inheritance of gene expression is reminiscent of genes that exert a gametophytic maternal effect mentioned above, suggesting an additional maternal source of SIN1/SUS1/CAF activity. Furthermore, the developmental arrest of sus1/sus1 embryos on a hemizygous sus1/+ sporophyte indicates a requirement of post-zygotic expression. Alternatively, it may indicate a dosage-sensitive sporophytic maternal effect, in which a hemizygous sporophyte cannot rescue a homozygous null mutant embryo. Thus, there may be at least three components of SIN1/SUS1/CAF expression in the ovule. First, there is an early sporophytic expression in ovule integuments and nucellus, which is correlated with integument morphogenesis. The second is an early zygotic component that is presumably essential for embryo viability, which appears to be expressed off the maternally transmitted allele. Third, there is a maternal sporophytic component that is required for embryo morphogenesis, which may act at distance from the funiculus to the embryo.
Given the expression pattern and the molecular nature of the predicted protein product, it is likely that SIN1/SUS1/CAF's role in early embryogenesis is to down-regulate the activity of RNA targets required for early embryogenesis, but whose continued activity has detrimental effects on later embryo development. This would mean that the embryo-lethal phenotype of sus1 alleles (as well as the patterning defects of the maternal effect sin1 alleles) is due to the embryo's inability to proceed beyond the globular stage. Although we cannot distinguish whether the mRNAs that SIN1/SUS1/CAF potentially targets are zygotic or uniparental in origin, it is an intriguing coincidence that known genes whose paternally transmitted alleles are silent in early embryos become active in early globular stages (Springer et al., 2000; Vielle-Calzada et al., 2000). Thus, symmetric transcriptional activation of most genes that show early asymmetric inheritance of expressivity occurs after SIN1/SUS1/CAF transcripts disappear in the embryo.
Molecular Mechanisms of SIN1/SUS1/CAF Activity and Its Role in Development
The conservation of sequence and domain architecture in Dicer-like proteins, and their involvement in reproduction and development across plant and animal kingdoms, suggest a fundamental function for these proteins in a conserved cellular process (for recent review, see Matzke et al., 2001; Vance and Vaucheret, 2001). All of the conserved protein modules defining the Dicer family implicate RNA as the substrate for these proteins, and, specifically, suggest a role in binding, cleavage, and subsequent unwinding of dsRNA. Effects of the mutant alleles of SIN1/SUS1/CAF are entirely consistent with such a function. The sin1-1, sin1-2, and caf-1 alleles are the only reported reduction-of-function alleles in any Dicer gene family member. The sin1 point mutations mapped here establish the critical requirement of a functional RNA helicase domain for Dicer-like proteins. Several pathways of RNA silencing are known, but so far only one of these seems to be used in the context of development (Grishok et al., 2001; Hutvágner et al., 2001). In animals, several developmentally important dsRNAs have been identified, which form short hairpin structures, generally less than 100 nt in length (Lee et al., 1993; Reinhart et al., 2000). Dicer cleaves the hairpins, which then inhibit initiation of translation of their target mRNAs through binding of the target RNA's 3′-UTR (Olsen and Ambros, 1999; Grishok et al., 2001; Hutvágner et al., 2001). To date, no regulatory dsRNA hairpin has been identified in plants (see “Note Added in Proof”), but even in animals their demonstration has been difficult due to small mutational target size and refractoriness to single base changes (Lee et al., 1993). Our work so far does not exclude the involvement of other Dicer-like proteins of Arabidopsis in RNA silencing pathways; in fact, putative Arabidopsis proteins F5024.210 and T15B3.8 appear to be more closely related to animal Dicer than is SIN1/SUS1/CAF. It is possible that one or both of these proteins, or even T17B22.1, a more distant paralogue, participate in RNA silencing.
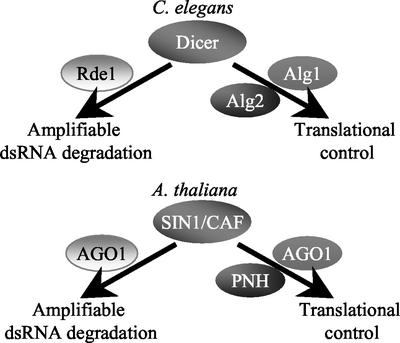
Recent work with the C. elegans Dicer gene (dcr-1) showed that it is required for correctly transitioning between developmental stages, contributed to the zygote maternally, and that it genetically interacts with a pair of highly homologous ARGONAUTE gene family members, alg1 (argonaute-like1) and alg2 (argonaute-like2) (Grishok et al., 2001; Knight and Bass, 2001; S. Schauer, K. Gentile, F. Hagen, and A. Ray, unpublished data). Specifically, alg1 alg2 double mutant worms are dcr-1 null phenocopies (Grishok et al., 2001). These characteristics of C. elegans dcr-1 have parallels with those observed for SIN1/SUS1/CAF in Arabidopsis. SIN1/SUS1/CAF has a role in the correct timing of development, not only in embryogenesis but also in mature plants for flowering time. We have shown here that in very early stages of embryogenesis, SIN1/SUS1/CAF gene product is expressed from the maternally contributed allele. Null alleles of SIN1/SUS1/CAF phenocopy a double mutant in a pair of highly homologous ARGONAUTE gene family members in Arabidopsis: AGO1 (ARGONAUTE1) and PNH (ZWILLE/PINHEAD; Bohmert et al., 1998; Moussian et al., 1998; Lynn et al., 1999). As in sus1 mutant embryos, the ago1 pnh double mutant embryos arrest for cell differentiation at the globular stage, but growth and cell division continue in post-globular stages (Schwartz et al., 1994; Lynn et al., 1999). Given these similarities, we speculate that in Arabidopsis, SIN1/SUS1/CAF genetically interacts with AGO and PNH to silence developmentally important target mRNAs (Fig. 7). In D. melanogaster, Dicer physically interacts with ARGONAUTE2, presumably through the PAZ domain shared by both proteins (Hammond et al., 2001); likewise, there may be physical interaction in Arabidopsis between SIN1/SUS1/CAF and AGO or PNH through their common PAZ domains. This hypothesis is consistent with the known expression pattern of these genes (Bohmert et al., 1998; Lynn et al., 1999; this work). Because the ARGONAUTE family includes the eukaryotic translation initiation factor, eIF2C, it is likely that in plants, these three proteins together may control the translation initiation of their target mRNAs, as has been demonstrated in C. elegans (Olsen and Ambros, 1999). The occurrence of selective translational regulation of mRNA in early plant embryo has been proposed before (Springer et al., 2000) but remains to be characterized.
Figure 7.
Models of SIN1/SUS1/CAF function in vivo. Based on parallel genetic evidence in C. elegans (Olsen and Ambros, 1999; Grishok et al., 2001; Ruvkun, 2001), we propose that in Arabidopsis, SIN1/SUS1/CAF interacts with AGO1 and/or PNH to regulate developmentally important genes through control of translation initiation. In addition, we propose that SIN1/SUS1/CAF mediates degradation of dsRNA in conjunction with AGO1 (Fagard et al., 2000).
Plants may use small dsRNA hairpins (or their cleaved products) as developmental regulators over long distances in much the same way as 21- to 25-nt dsRNA fragments of RNA viral genomes induce systemic signaling for defense against viral pathogens (Jorgensen et al., 1998). Movement of a target RNA from the maternal sporophyte into the developing embryo in a SIN1/SUS1/CAF-dependent manner could also explain the role of the sporophyte in embryogenesis. Recent studies have implicated dsRNA molecules in transcriptional repression of transgenic promoter sequences in plants (Mette et al., 1999, 2000, 2001). Whether this mechanism of gene regulation is used in a developmental context is not yet known. In summary, results presented here suggest that at least one multidomain protein that is presumably involved in RNA silencing is crucial for regulating several aspects of plant development. Its precise mechanism of action, its target molecules, and other participating proteins in this process remain to be elucidated.
MATERIALS AND METHODS
Plant Strains
The sin1-1 and sin1-2 mutations in Arabidopsis were originally isolated in the La-er background (Robinson-Beers et al., 1992; Ray et al., 1996a). As the ER gene interacts with the SIN1/SUS1/CAF gene during integument formation (Lang et al., 1994), these mutant alleles were introgressed in Col lines carrying the gl1 (glaborus1) mutation by repeated crosses. Analysis of molecular markers confirmed the introgressions (S.E. Schauer, unpublished data). The caf-1 allele was originally isolated in Wassilewskija and introgressed into La-er (Jacobsen et al., 1999). All Col lines used in these experiments were homozygous for gl1, with the exception being the pSP2 lines, which were Gl1+. All plant growth conditions were as previously noted (Ray et al., 1996a).
Cloning of SIN1
Chromosome Walking
Pollen from sin1-1 (La-er) was crossed to wild-type Col stigma and F1 were self-crossed. DNA samples from F3 families originating from individual sin1-1/+ heterozygous F2 descendants were analyzed for recombination. Sampling over 700 F2 chromosomes yielded no crossover between sin1 and nga59. sin1 was mapped to the Arabidopsis yeast (Saccharomyces cerevisiae) artificial chromosome clone yUP20D1 against the markers RS10 (18 recombinants) and 12D7LE, the left end rescued from yUP12D7 (three recombinants). A BAC alignment was created over sin1 using nga59 as the anchor point, and the mutation was mapped on the BACs T4J2 and F7I23. A 29.9-kb contig of DNA over sin1 was established from partially overlapping BACs F7I23, T4J2, and T25K16, and the contig was sequenced (GenBank accession no. gi6684981). Analysis of the sequenced region with GENSCAN (Burge, 1998), trained on Arabidopsis splice sites, revealed two putative genes: a leftward complex gene with multiple introns, and a rightward single ORF1 that encodes a 358-amino acid-long protein with highly conserved (67% similarity over a 95-residue stretch) sequence to RAV1 and RAV2, which are members of an AP2-like DNA-binding protein family (Altschul et al., 1997). Screening over 105 cDNA clones of a seed-enriched library (Lou et al., 1999) did not yield an ORF1 cDNA, although other members of the RAV family were isolated. Sequencing of ORF1 DNA from sin1-1 and sin1-2 mutants failed to reveal any mutation. Attempts to complement sin1 with an 18.5-kb subclone (pJT12) overlapping ORF1 failed, strengthening the likelihood that the complex predicted gene could be SIN1.
Complementation
Due to very close map positions of the previously described suspensor1 (sus1 or embryo lethal76) mutation (Castle et al., 1993; Schwartz et al., 1994) and sin1, we suspected that they could be allelic. Complementation tests were performed by crossing flowers heterozygous for sus1 to sin1 pollen and by scoring the appearance of phenotypically Sin1− plants among the F1 progeny. Sixteen of 36 F1 plants from a sin1-1 × sus1-1/+ cross, 14 of 22 from a sin1-1 × sus1-2/+ cross, and 30 of 55 from a sin1-1 × sus1-3/+ cross were Sin1−, demonstrating allelism. To confirm, pollen from a Sin1− F1 segregant from the sin1-1 × sus1-2 cross was backcrossed to a sin1-1/+ heterozygote, which produced 12 phenotypically wild-type, 17 Sin1−, and nine Sus1− progeny. When pollen from the same strain was backcrossed to genotypically wild-type flowers, the progeny produced five phenotypically wild-type, zero Sin1−, and three Sus1− progeny. These results confirmed that sin1 and sus1 mutations are allelic. The failure of sus1-2 and sus1-3 to complement sin1 may be due to the loss of one of several genes potentially covered by the rearranged region in these alleles, whereas that of sus1-1 could be due to one of the two linked T-DNA insertion mutations. We confirmed the identity of the non-complementing T-DNA insertion locus in sus1-1 by first separating the two linked T-DNA loci by recombination, then testing for non-complementation (Golden, 1999). Sequencing of chromosomal DNA surrounding the deduced non-complementing T-DNA insertion point showed that it disrupts the predicted gene. To identify the sin1-1 and sin1-2 mutations, 8.2 kb of the genomic region (from approximately 450 bp upstream of the predicted ATG to approximately 150 bp downstream of the termination codon) was sequenced from wild-type La-er and Co-gl1, and from sin1-1 and sin1-2 mutants (in La-er), respectively. The PCR and sequencing primers are described by Golden (1999).
cDNA Isolation
The lambda ZAP II (Stratagene, La Jolla, CA) cDNA library (Lou et al., 1999) was screened by hybridization to two probes of 913 and 2,008 bp, respectively, made by PCR amplification of a genomic clone covering the highest probability exons of the predicted gene that was disrupted by T-DNA in sus1-1. The primer pairs were: 5′d[ATGGTGTCGTGGAGGGTTC]3′, 5′d[ACTTGAGGGTCCTGTTTGCAG]3′, 5′d[CACTGAGGTATGATTCTTG]3′, and 5′d[ATCGATGATCTCGTGTCTG]3′.
RNA Analysis
RNA localization via in situ hybridization was performed as described by Vielle-Calzada et al. (2000) with some modifications (a detailed protocol can be obtained from Dr. Ueli Grossniklaus, Institute of Plant Biology, University of Zürich, Zollikerstrasse 107, CH-8008 Zürich, Switzerland; grossnik@botinst.unizh.ch). For RT-PCR, total RNA was isolated from 2 to 4 g of leaves or flowers with Triazol (Invitrogen, Carlsbad, CA). The total RNA was digested with RNAse-free DNase (Boehringer Mannheim/Roche, Indianapolis), extracted with phenol:chloroform:isoamyl alcohol (25:24:1 [v/v]), and precipitated with 8 m LiCl (Sigma, St. Louis). Five micrograms of total RNA was used to make cDNA using SuperScript II RNase H− RT (Invitrogen) following the manufacturer's instructions. The cDNA was precipitated with 8 m LiCl, resuspended in 25 μL of 10 mm Tris-HCl (pH 8.0), 1 mm EDTA (pH 8.0), and digested with RNase H (Invitrogen) and RNase A (Sigma). One microliter was used in the PCR reactions, using the following primers: RT-PCR1-flem2A, 5′d[ATCGATGATCTCGTGTCTG]3′; New-F-emb76, 5′d[CGACGACTATCTCTGAAGGCATAGGC]3′; RT-PCR5-emb-cDNA-rev2, 5′d[TCTAAAATGGGTTGTTAGTCG]3′; emb-cDNA-For, 5′d[GTAATGACTACATCTCGTTGAAG]3′; 5′-ROC1 (cytoplasmic cyclophilin), 5′d[TGGCGTTCCCTAAGGTATACT]3; and 3′-ROC1, 5′d[TTCCCGGCGGTGAAATCT]3′. PCR reactions were performed for 35 cycles with Taq polymerase (Invitrogen) and the products were separated by electrophoresis.
Construction of the pSP2 Reporter Lines
The pSP2 vector was constructed by cloning an approximately 5.0-kb SphI KpnI genomic fragment from p3A1 (see Golden, 1999) into pBJ61 (which has the GUS gene in the +1 reading frame). This construct was digested with ClaI and subsequently recircularized, which deleted 1,291 bp of SIN1/SUS1/CAF genomic sequence, leaving approximately 3.3 kb of genomic sequence (including 38 bp of 5′-UTR) in front of the GUS gene. The predicted start site of the SIN1/SUS1/CAF gene is found 379 bp from the beginning of the cDNA, and is not present in this construct. The fusion protein was cloned into the binary vector, pART27 (Gleave, 1992), which contains the neomycin phosphotransferase gene driven under control of the nopaline synthase promoter, and transformed into Agrobacterium tumefaciens (as described in Golden, 1999). These A. tumefaciens strains were vacuum infiltrated into Arabidopsis and T1 lines were plated out on kanamycin (as described by Golden, 1999). GUS staining was done as described by Vielle-Calzada et al. (2000).
Sequence Analysis
The nonredundant sequence database at National Center for Biotechnology Information (Bethesda, MD) and the PSI-BLAST program were used for database searches (Kagaya et al., 1999). The SMART server (Schultz et al., 1998) and Pfam server (Bateman et al., 1999) were used to search for the conserved domains. Database scans using profiles were performed with Wisetools (Birney et al., 1996). NLS was detected using the PSORT server (Nakai and Horton, 1999). All similarities reported here were supported statistically; typically, related sequences had probabilities of matching by chance below 10−5 upon first time passing the threshold in PSI-BLAST searches, and scores above 5,000 in Wise searches. Whole protein sequences were first aligned using ClustalX (Thompson et al., 1997) with the following settings: the Pairwise parameter Gap Opening 35.00; the Pairwise parameter Gap Extension Penalty 0.75; the Multiple Alignment parameter Gap Opening 15.00; the Multiple Alignment parameter Gap Extension Penalty 0.3; and the Multiple Alignment parameter Delay Divergent Sequence 25% (Hall, 2001). The gap-only columns were removed from the alignment, and the sequences were realigned. After removing the gap-only columns, the final alignment was then used to produce an un-rooted neighbor joining circle tree using Paup* version 4.0b8, which was boot strapped for 1,000 replicates (Swofford, 2000).
Protein Homology Modeling
Helicase sequences that are most closely related to the helicase domain of SIN1/SUS1/CAF were retrieved by the PSI-BLAST iterative searches (Kagaya et al., 1999) of the SWISSPROT database until the sequence of yIF4A from yeast was detected. Multiple related sequences were realigned using the Gibbs sampler option of the MACAW program (Schuler et al., 1991), and this alignment was used as a guide to model the SIN1/SUS1/CAF helicase domain structure onto the known structure of yIF4A (PDB ID 1QVA; Johnson and McKay, 1999) using the ProMod algorithm (Guex et al., 1999).
Note Added in Proof
Recent work (B.J. Reinhart, E.G. Weinstein, M.W. Rhoades, B. Bartel, D.P. Bartel [2002] Genes and Dev 16: 1616–1626) has shown that caf-1 plants fail to process endogenous short RNA hairpins of presumed regulatory function.
ACKNOWLEDGMENTS
We thank Chuck Gasser, Steve Jacobson, and The Arabidopsis Stock Center for plant strains; Sarah Bean, Joanne Lundholm, Delwin Merchant, Jack Tsai, and Kerri Vacher for technical assistance; Terry Delaney, Lynne and Bob Angerer, Bob Fleming, Don Kane, and Vicki Vance for advice and comments on the manuscript; Abdul Chaudhury for cDNA library; Bart Janssen for vectors; and Elliot Meyerowitz for sharing manuscript before publication.
Footnotes
This work was supported by the National Science Foundation (grant nos. IBN–9728239 and IBN–9982414 to A.R.), and by a Searle Scholarship (to U.G., University of Zürich).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.003491.
LITERATURE CITED
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroux C, Spillane C, Grossniklaus U. Genomic imprinting during seed development. In: Dunlap JC, Wu C-t, editors. Advances in Genetics: Homology Effects. San Diego: Academic Press; 2002. pp. 165–214. [DOI] [PubMed] [Google Scholar]
- Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 1999;27:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy A, Hammond S, Hannon G. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Birney E, Thompson JD, Gibson TJ. PairWise and SearchWise: finding the optimal alignment in a simultaneous comparison of a protein profile against all DNA translation frames. Nucleic Acids Res. 1996;24:2730–2739. doi: 10.1093/nar/24.14.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmert K, Camus I, Bellini C, Bouchez D, Caboche M, Benning C. AGO1defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 1998;17:170–180. doi: 10.1093/emboj/17.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge CB. Modeling dependencies in pre-MRNA splicing signals. In: Salzberg S, Searls D, Kasif S, editors. Computational Methods in Molecular Biology. Amsterdam: Elsevier Science; 1998. pp. 127–163. [Google Scholar]
- Campbell RC. Statistics for Biologists. Ed 3. UK: Cambridge University; 1989. [Google Scholar]
- Castle LA, Errampalli D, Atherton TL, Franzmann LH, Yoon ES, Meinke DW. Genetic and molecular characterization of embryonic mutants identified following seed transformation in Arabidopsis. Mol Gen Genet. 1993;241:504–514. doi: 10.1007/BF00279892. [DOI] [PubMed] [Google Scholar]
- Cerutti L, Mian N, Bateman A. Domains in gene silencing and cell differentiation proteins: the novel PAZ domain and redefinition of the piwi domain. Trends Biochem Sci. 2000;25:481–482. doi: 10.1016/s0968-0004(00)01641-8. [DOI] [PubMed] [Google Scholar]
- Chaudhury A, Koltunow A, Payne T, Lou M, Tucker M, Dennis E, Peacock W. Control of early seed development. Annu Rev Cell Dev Biol. 2001;17:677–699. doi: 10.1146/annurev.cellbio.17.1.677. [DOI] [PubMed] [Google Scholar]
- Clarke J, Tack D, Findlay K, Van Montagu M, Van Lijsebettens M. The SERRATE locus controls the formation of the early juvenile leaves and phase length in Arabidopsis. Plant J. 1999;20:493–501. doi: 10.1046/j.1365-313x.1999.00623.x. [DOI] [PubMed] [Google Scholar]
- Errampalli D, Patton D, Castle L, Mickelson L, Hansen K, Schnall J, Feldmann K, Meinke D. Embryonic lethals and T DNA insertional mutagenesis in Arabidopsis. Plant Cell. 1991;3:149–157. doi: 10.1105/tpc.3.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard M, Boutet S, Morel J-B, Bellini C, Vaucheret H. AGO1, QDE-2, and RDE-1 are related proteins for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc Natl Acad Sci USA. 2000;97:11650–11654. doi: 10.1073/pnas.200217597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave AP. A versatile binary vector system with a T-DNA organizational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol. 1992;20:1203–1207. doi: 10.1007/BF00028910. [DOI] [PubMed] [Google Scholar]
- Golden T. Plant development: a view through the SHORT INTEGUMENTS1 gene of Arabidopsis thaliana. PhD thesis. NY: University of Rochester; 1999. [Google Scholar]
- Gorbalenya AE, Koonin EV. Helicases: amino acid sequence comparisons and structure-function relationships. Curr Opin Struct Biol. 1993;3:419–429. [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello C. Genes and mechanisms related to RNA interference regulate the expression of the small temporal RNAs that control C. elegansdevelopmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Guex N, Diemand A, Peitsch MC. Protein modeling for all. Trends Biochem Sci. 1999;2:364–367. doi: 10.1016/s0968-0004(99)01427-9. [DOI] [PubMed] [Google Scholar]
- Hall BG. Phylogenetic Trees Made Easy: A How-To Manual for Molecular Biologists. Sunderland, MA: Sinauer Associates; 2001. [Google Scholar]
- Hammond S, Boettcher S, Caudy A, Kobayashi R, Hannon G. Argonaute2, a link between genetic and biochemical analysis of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- Hutvágner G, McLachlan J, Pasquinelli A, Bálint É, Tuschl T, Zamore P. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Running MP, Meyerowitz EM. Disruption of an RNA helicase/RNAse III gene in Arabidopsiscauses unregulated cell division in floral meristems. Development. 1999;126:5231–5243. doi: 10.1242/dev.126.23.5231. [DOI] [PubMed] [Google Scholar]
- Johnson ER, McKay DB. Crystallographic structure of the amino terminal domain of yeast initiation factor 4A, a representative DEAD-box RNA helicase. RNA. 1999;5:1526–1534. doi: 10.1017/s1355838299991410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen R, Atkinson R, Forster R, Lucas W. An RNA based information superhighway in plants. Science. 1998;279:1486–1487. doi: 10.1126/science.279.5356.1486. [DOI] [PubMed] [Google Scholar]
- Kagaya Y, Ohmiya K, Hattori T. RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 1999;27:470–478. doi: 10.1093/nar/27.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight S, Bass B. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang JD, Ray S, Ray A. sin1, a mutation affecting female fertility in Arabidopsis, interacts with mod1, its recessive modifier. Genetics. 1994;137:1101–1110. doi: 10.1093/genetics/137.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R, Feinbaum R, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lou M, Bilodeau P, Koltunow A, Dennis E, Peacock WJ, Chaudhury AM. Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci. 1999;96:296–301. doi: 10.1073/pnas.96.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn K, Fernandez A, Aida M, Sedbrook J, Tasaka M, Masson P, Barton MK. The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1gene. Development. 1999;126:469–481. doi: 10.1242/dev.126.3.469. [DOI] [PubMed] [Google Scholar]
- Matzke M, Matzke A, Kooter J. RNA: guiding gene silencing. Science. 2001;293:1080–1083. doi: 10.1126/science.1063051. [DOI] [PubMed] [Google Scholar]
- McElver J, Tzafrir I, Aux G, Rogers R, Ashby C, Smith K, Thomas C, Schetter A, Zhou Q, Cushman MA et al. Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics. 2001;159:1751–1763. doi: 10.1093/genetics/159.4.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mette M, Aufsatz W, van der Winden J, Matzke M, Matzke A. Transcriptional silencing and promoter methylation triggered by double stranded RNA. EMBO J. 2000;19:5194–5201. doi: 10.1093/emboj/19.19.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mette M, Matzke A, Matzke M. Resistance of RNA mediated TGS to HC-Pro, a viral suppressor of PTGS, suggests alternative pathways for dsRNA processing. Curr Biol. 2001;11:1119–1123. doi: 10.1016/s0960-9822(01)00315-3. [DOI] [PubMed] [Google Scholar]
- Mette M, van der Winden J, Matzke M, Matzke A. Production of aberrant promoter transcripts contributes to methylation and silencing of unlinked homologous promoters in trans. EMBO J. 1999;18:241–248. doi: 10.1093/emboj/18.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mian IS. Comparative sequence analysis of ribonucleases HII, III, II PH and D. Nucleic Acids Res. 1997;25:3187–3195. doi: 10.1093/nar/25.16.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussian B, Schoof H, Haecker A, Jurgens G, Laux T. Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsisembryogenesis. EMBO J. 1998;17:1799–1809. doi: 10.1093/emboj/17.6.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- Olsen P, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegansby blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- Papi M, Sabatini S, Bouchez D, Camilleri C, Costantino P, Vittorioso P. Identification and disruption of an Arabidopsiszinc finger gene controlling seed germination. Genes Dev. 2000;14:28–33. [PMC free article] [PubMed] [Google Scholar]
- Pause A, Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge M, Wagner D. The Arabidopsis SERRATEgene encodes a zinc-finger protein required for normal shoot development. Plant Cell. 2000;13:1263–1279. doi: 10.1105/tpc.13.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A, Grunert S, Adams J, Micklem DR, Proctor MR, Freund S, Bycroft M, St. Johnston D, Varani G. RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J. 2000;19:997–1009. doi: 10.1093/emboj/19.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, Lang JD, Golden T, Ray S. SHORT INTEGUMENTS1 (SIN1), a gene required for ovule development in Arabidopsis, also controls flowering time. Development. 1996a;122:2631–2638. doi: 10.1242/dev.122.9.2631. [DOI] [PubMed] [Google Scholar]
- Ray S, Golden T, Ray A. Maternal effects of the short integuments1mutation on embryo development. Dev Biol. 1996b;180:365–369. doi: 10.1006/dbio.1996.0309. [DOI] [PubMed] [Google Scholar]
- Reinhart B, Slack F, Basson M, Pasquinelli A, Bettinger J, Rougvie A, Horvitz H, Ruvkun G. The 21 nucleotide let-7 RNA regulated developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Robinson-Beers K, Pruitt RE, Gasser CS. Ovule development in wild type Arabidopsisand two female sterile mutants. Plant Cell. 1992;4:1237–1250. doi: 10.1105/tpc.4.10.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvkun G. Glimpses of a tiny RNA world. Science. 2001;294:797–799. doi: 10.1126/science.1066315. [DOI] [PubMed] [Google Scholar]
- Schuler GD, Altschul SF, Lipman DJ. A workbench for multiple alignment construction and analysis. Proteins. 1991;9:180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz BW, Yeung EC, Meinke DW. Disruption of morphogenesis and transformation of the suspensor in abnormal suspensor mutants of Arabidopsis. Development. 1994;120:3235–3245. doi: 10.1242/dev.120.11.3235. [DOI] [PubMed] [Google Scholar]
- Springer PS, Holding DR, Groover A, Yordan C, Martienssen R. The essential Mcm7 protein PROLIFERA is localized to the nucleus of dividing cells during the G(1) phase and is required maternally for early Arabidopsisdevelopment. Development. 2000;127:1815–1822. doi: 10.1242/dev.127.9.1815. [DOI] [PubMed] [Google Scholar]
- Swofford DL. Paup*. Phylogenetic Analysis Using Parsimony (* and Other Methods) Version 4. Sunderland, MA: Sinauer Associates; 2000. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies of multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance V, Vaucheret H. RNA silencing in plants: defense and counterdefense. Science. 2001;292:2277–2280. doi: 10.1126/science.1061334. [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada J-P, Baskar R, Grossniklaus U. Delayed activation of the paternal genome during seed development. Nature. 2000;404:91–94. doi: 10.1038/35003595. [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada J-P, Baskar R, Grossniklaus U. Early paternal gene activity in Arabidopsis(response) Nature. 2001;414:710. [Google Scholar]
- Weijers D, Geldner N, Offringa R, Jürgens G. Early paternal gene activity in Arabidopsis. Nature. 2001;414:709–710. doi: 10.1038/414709a. [DOI] [PubMed] [Google Scholar]