Abstract
The Arabidopsis genome contains 14 genes encoding the serine protease DegP. Products of four of these genes are located in the chloroplast: three in the thylakoid lumen and one on the stromal side of the membrane. We expressed the gene encoding DegP1 as a His-tagged fusion protein in Escherichia coli, purified the protein by affinity chromatography, and characterized it biochemically. Size-exclusion chromatography suggested that DegP1 eluted from the column as a mixture of monomers and hexamers. Proteolytic activity was characterized using β-casein as a model substrate. DegP1 demonstrated concentration-dependent activity, a pH optimum of 6.0 and increasing activity at elevated temperatures. DegP1 was capable of degrading two lumenal proteins, plastocyanin and OE33, suggesting a role as a general-purpose protease in the thylakoid lumen. The results of this work are discussed in the context of the recent elucidation of the structure of the E. coli homolog and the possible physiological role of the protease in the chloroplast lumen.
Intracellular proteolysis plays two major roles in all organisms. As a component of the protein quality control system, together with molecular chaperones, proteases ensure protein quality by either remodeling denatured or damaged ones, or degrading them to free amino acids (Wickner et al., 1999). The second role is fine-tuning of proper levels of regulatory proteins (Gottesman, 1996; Hershko and Ciechanover, 1998). As a semiautonomous organelle, the chloroplast is expected to have these two functions fulfilled by organelle-located proteases (Adam, 2000). The chloroplast contains a proteolytic machinery composed of different families of proteases, distributed in the different compartments of the organelle. The ATP-dependent Ser protease Clp is located mostly in the stroma, where it can degrade both soluble and membrane-bound substrates (e.g. Shanklin et al., 1995; Halperin and Adam, 1996; Majeran et al., 2000). It is composed of the proteolytic subunit ClpP and the regulatory ATPase ClpC, which form oligomeric structures (Sokolenko et al., 1998; Peltier et al., 2001), both encoded by multiple genes (Adam et al., 2001; Zheng et al., 2002). Another ATP-dependent protease is FtsH, which harbors both its proteolytic and ATPase domains in the same polypeptide. This is a metalloprotease that is bound to the thylakoid membrane, exposing its functional domains to the stromal side of the membrane (Lindahl et al., 1996). It has been implicated in the degradation of unassembled thylakoid membrane proteins (Ostersetzer and Adam, 1997), and degradation of the D1 protein of photosystem II reaction center after photoinhibition (Lindahl et al., 2000; Bailey et al., 2002). This protease is also encoded by multiple enzymes (Adam et al., 2001), and one of its isomers, FtsH2, is apparently involved in chloroplast development as well (Chen et al., 2000; Takechi et al., 2000). SppA was recently identified as another thylakoid membrane-bound Ser protease facing the stroma (Lensch et al., 2001). In addition to these proteases, several processing and amino peptidases, capable of limited proteolysis, are found in chloroplasts (for review, see Sokolenko et al., 2002).
The innermost compartment of the chloroplast, the thylakoid lumen, contains more than 80 proteins (Peltier et al., 2002; Schubert et al., 2002). Proteolytic degradation of only a few of these has been previously documented. In Chlamydomonas reinhardtii, the copper-containing electron carrier plastocyanin (PC) is rapidly degraded in the lumen when the alga is grown in a copper-deficient medium (Merchant and Bogorad, 1986; Li and Merchant, 1995). Further support for the existence of a proteolytic machinery in the lumen comes from the demonstration of the short half-life of truncated forms of the 23-kD subunit of the oxygen-evolving complex (Roffey and Theg, 1996). Although the identity of the protease(s) involved is still unknown, the existence of one family of proteases in the lumen is now established. The Ser protease DegP1 has been identified as a protease that is tightly associated with the lumenal side of thylakoid membranes, and its level appears to increase transiently in response to exposure of plants to heat (Itzhaki et al., 1998). This protease is encoded in Arabidopsis by a family of genes; the products of at least four of them are targeted to chloroplasts (Adam et al., 2001). Two isomers, highly similar to DegP1, DegP5, and DegP8, were recently identified together with DegP1 in a proteomic study of lumenal proteins (Schubert et al., 2002). Another isomer, somewhat less conserved, designated DegP2, was found associated with the stromal side of the thylakoid membrane and was implicated in the initial cleavage of the D1 protein of photosystem II reaction center (Haussuhl et al., 2001).
As a first step toward understanding the physiological functions of thylakoid proteases, we overexpressed DegP1 in Escherichia coli and characterized its proteolytic activity. Here, we demonstrate that recombinant DegP1 is proteolytically active toward both model and lumenal substrates, and we characterize its activity with respect to enzyme concentration, pH, and temperature.
RESULTS
Expression and Purification of Plant DegP1 from E. coli
In an initial attempt to express plant DegP1 in E. coli, we subcloned the cDNA encoding the mature protein into the expression vector pT7–7. Upon induction with isopropylthio-β-galactoside (IPTG), the protein was expressed. However, this system was very inefficient because only small amounts of the protein could be observed. Attempts to express DegP1 as a fusion protein with an intein and a chitin-binding domain also failed. In this case, only a mutant of the cDNA, in which the active Ser residue was replaced by a Gly, was expressed correctly, whereas the wild-type fusion protein was much smaller, probably as result of self-degradation (data not shown).
A third attempt to overexpress the protein was made with the pET-15b vector. The 5′ end of the DegP1 cDNA fragment was ligated to the 3′ end of the His-tag-encoding domain of the vector, to yield mature DegP1 with a His-tag at its N terminus. Two hours after induction of expression with IPTG, a band of approximately 37 kD was observed, whose level decreased 2 h later (Fig. 1A). Because this was the only extra band that appeared after induction and its size was in the range of the expected protein, we suspected that the overexpressed protein had undergone degradation. To test this possibility, we took samples from the culture at different times after induction and subjected them to immunoblot analysis, using an antibody against the His tag. As shown in Figure 1B, a His-tagged protein accumulated 1 h after induction with IPTG, but its level gradually decreased with time, suggesting that this protein is unstable in the bacteria.
Figure 1.
Expression of DegP1 in E. coli. A, BL21-DE3 cells, transformed with pET-15b-DegP1, were induced with 0.5 mm IPTG. Samples were taken at the times indicated, normalized to cell density, and subjected to SDS-PAGE on a 12% (w/v) acrylamide gel. The location of an induced protein, suspected to be DegP1, is indicated by an arrow. B, Immunoblot of a similar gel with an anti-poly-His monoclonal antibody.
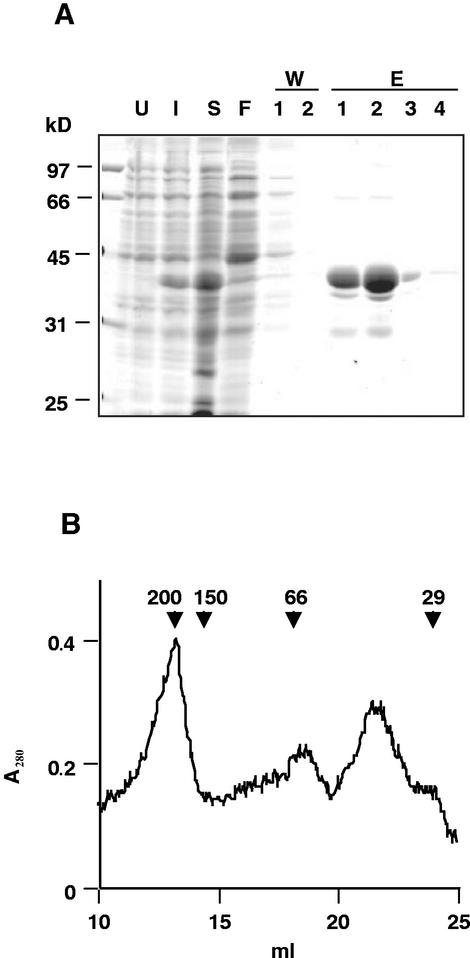
After sonication of the bacterial cells, followed by centrifugation, DegP1 was found equally distributed between the soluble and insoluble fractions (data not shown). The soluble fraction was subjected to affinity purification on a nickel column. As shown in Figure 2A, most proteins did not bind to the column (lane F). After two washes with a buffer containing 20 mm imidazole, no more proteins come off the column (lane W2). DegP1 was eluted from the column with a buffer containing 250 mm imidazole. Most of the protein was found in the first two eluted fractions, with the remainder coming out in the third and fourth fractions (lanes E1–E4). The first two eluted fractions contained three additional minor bands, two smaller and one larger than DegP1. Immunoblot analysis with an antibody against DegP revealed cross-reaction with all four bands (data not shown), suggesting that the smaller bands may represent degradation products of DegP1, whereas the larger one may be an oligomeric form.
Figure 2.
Purification of DegP1. A, BL21-DE3 cells, transformed with pET-15b-DegP1 (U), were induced with 0.5 mm IPTG for 1 h (I). Cells were harvested, sonicated, and centrifuged to obtain a soluble fraction (S). This fraction was mixed with nickel-nitrilotriacetic acid (Ni-NTA) agarose and loaded onto a column, and the flow-through liquid was collected (F). The column was then washed twice (W1 and W2), and eluted four times (E1–E4) with 250 mm imidazole. Samples were resolved by SDS-PAGE on a 12% (w/v) acrylamide gel followed by Coomassie Blue staining. B, The pooled fractions E1–E4 were concentrated and loaded onto a Sephacryl S-200 column, and the A280 of the eluted protein was monitored. Relative migration of standard proteins and their molecular mass in kilodaltons is indicated by arrowheads.
To estimate the native size of the affinity-purified His-tagged enzyme, we performed size-exclusion chromatography on a Sephacryl S-200 column. The elution profile presented in Figure 2B shows two main peaks with calculated molecular masses of 200 and 40 kD, respectively. Given the mass of DegP1, 35 kD, and the 2.5-kD N-terminal tag, this suggested that the enzyme is found in a mixture of monomeric and hexameric forms.
Recombinant DegP1 Is Proteolytically Active
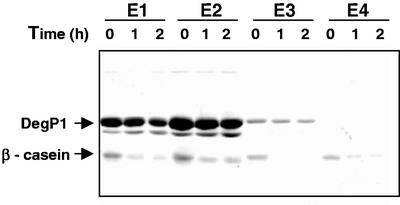
To test whether the recombinant DegP1, containing a His tag at its N terminus, was proteolytically active, we incubated it with the model substrate β-casein. Equal volumes of the four eluted fractions, containing different amounts of DegP1, were incubated with equal amounts of substrate. From the results presented in Figure 3, it appears that DegP1 in all four eluted fractions can degrade the protein substrate. However, it should be noted that degradation in the more dilute fractions (E3 and E4) was more efficient. Thus, all four fractions were combined and used for further characterization of the enzyme. Interestingly, when fractions from the size-exclusion chromatography were assayed for proteolytic activity, both the oligomeric and monomeric fractions were active (data not shown). Given the purity of the affinity-purified enzyme, further characterization was done with this preparation.
Figure 3.
β-Casein degradation by purified DegP1 fractions. Fractions eluted from the Ni-NTA agarose column (5 μL each) were incubated with 1 μg of β-casein for the times indicated. At the end of the incubation, reaction mixtures were subjected to SDS-PAGE on a 12% (w/v) acrylamide gel. The locations of DegP1 and β-casein are indicated on the left.
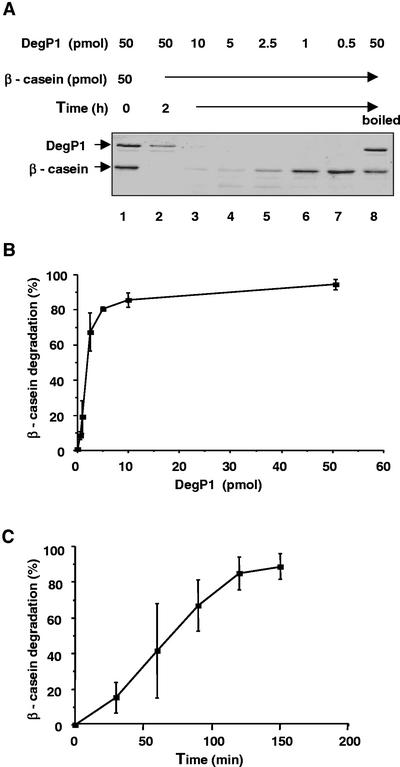
When 50 pmol of β-casein was incubated with 0.5 to 50 pmol of DegP1, we observed concentration-dependent degradation of the substrate (Fig. 4, A and B). Interestingly, it appears that DegP1 itself undergoes self-degradation during the incubation (Fig. 4A, compare lanes 1 and 2). During a 2-h incubation, approximately 2 pmol of the enzyme was sufficient for degradation of 50% of the substrate (Fig. 4B). The rate of degradation progressed linearly with time for 2 h, and then started to level off even before the substrate had been completely degraded (Fig. 4C). We also tested whether the presence of the His tag has any effect on proteolytic activity. For that purpose, the His tag was cleaved from the fusion protein with thrombin and the remaining DegP1 was purified. Removal of the tag did not increase the activity of the protein (data not shown). Thus, we continued the characterization with the His-tagged enzyme.
Figure 4.
Concentration and time dependence of β-casein degradation by DegP1. A, β-Casein (50 pmol) was incubated with 0.5 to 50 pmol of DegP1 for 2 h, after which the reaction mixtures were subjected to SDS-PAGE. A reaction containing 50 pmol of preboiled DegP1 is also included in the experiment. B, The experiment presented in A was repeated three times and the β-casein bands were quantified. Means ± se are presented. C, β-Casein (50 pmol) was incubated with 5 pmol of DegP1 for the indicated times, and the remaining β-casein was quantified. Means ± se of three experiments are presented.
Effect of pH and Temperature on Degradation Rate
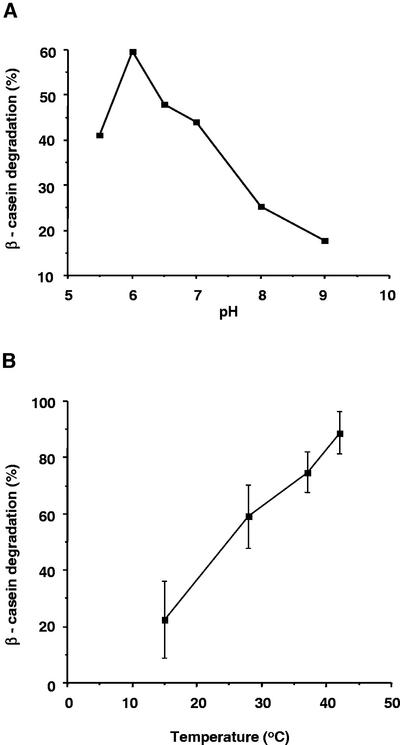
β-Casein degradation by DegP1 was tested at different pH values in the range of 5.5 to 9.0. As shown in Figure 5A, degradation was most efficient at pH values between 5.5 and 7.0, with highest activity observed at pH 6.0. At slightly basic pHs, degradation rate dropped dramatically. DegP1 is a resident of the thylakoid lumen, and optimal activity at pH 6.0 is consistent with the slightly acidic pH found in this compartment.
Figure 5.
pH and temperature dependence of β-casein degradation by DegP1. A, β-Casein (50 pmol) was incubated with 5 pmol of DegP1 in buffers with pH values ranging from 5.5 to 9.0. Percentage of β-casein degradation is presented. B, β-Casein degradation assays were carried out at different temperatures. Means ± se of three experiments are presented.
Because bacterial DegP is essential for survival at elevated temperatures (Lipinska et al., 1990) and because levels of plant DegP rise in response to exposure to heat (Itzhaki et al., 1998), we tested the activity of DegP1 at different temperatures. Within the range of 15°C to 42°C, degradation of β-casein rose gradually (Fig. 5B). Degradation at 55°C was even higher (data not shown). At this stage, it is not clear whether the enzyme itself operates better at higher temperatures or whether heat induces unfolding of the substrate, thereby making it more accessible to the protease. However, it should be noted that even after boiling the enzyme for 5 min, it remained partially active (Fig. 4A, compare lane 8 with lanes 1 and 7).
Degradation of Lumenal Proteins by DegP1
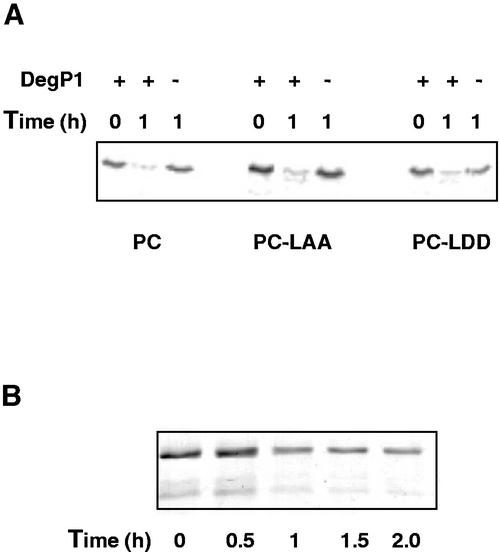
To test whether DegP1 is capable of degrading residents of the lumen compartment, we incubated the enzyme with in vitro synthesized PC. Degradation of three variants of the protein were tested: the wild-type PC and PC containing either a normal (ANDENYALAA) or mutated (ANDENYALDD) SsrA tag (Keiler et al., 1996) at its C terminus. The results in Figure 6A show that wild-type PC and the two C-terminal-extended proteins are equally susceptible to DegP, whereas in the absence of the protease, the proteins are stable. Although the C terminus of PC appears to play no role in its degradation, these results demonstrate that DegP can indeed degrade a lumenal protein.
Figure 6.
Degradation of PC and OE33 by DegP1. A, In vitro synthesized PC and its derivatives were incubated for 1 h with or without DegP1, followed by SDS-PAGE and imaging of the radioactive protein by phosphor imager. B, Overexpressed OE33 was incubated with DegP1, followed by SDS-PAGE and staining.
To further test the ability of DegP to degrade lumenal proteins, we incubated it with the 33-kD subunit of the oxygen-evolving complex (OE33) that had been overexpressed in E. coli. Here again, the protein was degraded, although degradation appeared to slow down after 1 h of incubation (Fig. 6B). Why degradation did not proceed further is not known, but it is conceivable that different forms of the overexpressed protein have differential stability. However, the ability of DegP1 to degrade both PC and OE33 suggests that it may be involved in complete degradation of lumenal proteins in vivo.
DISCUSSION
A common feature of all our attempts to express plant DegP1 in E. coli was its instability in the bacterium (Fig. 1B). Although the amount of protein accumulated 1 h after induction was sufficient for further purification and characterization, the reason for the observed instability remains unclear. Circumstantial evidence suggests that the recombinant protein is prone to self-degradation. Under conditions in which DegP1, fused to an intein and a chitin-binding domain, did not accumulate, a protein containing a single amino acid substitution at the active site did (data not shown). Some degree of self-degradation was also evident in the in vitro activity assay of the purified protein (Fig. 4A, compare lanes 1 and 2). This observation is consistent with partial cleavage of the overexpressed bacterial protein (Lipinska et al., 1990) and its self-degradation observed at elevated temperature (Kim et al., 1999). Whether plant DegP1 is unstable in vivo as well is not yet known, but it should be noted that upon continuous exposure of pea (Pisum sativum) seedlings to elevated temperatures, we observed a transient increase in the level of the protein, followed by a decrease to the basal level within 16 h (Itzhaki et al., 1998). Whether this has any functional significance remains to be determined.
DegP1 appears to be well suited for activity in the thylakoid lumen, because its pH optimum determined here was slightly acidic (Fig. 5A). Its increased activity at elevated temperatures (Fig. 5B), a characteristic of the E. coli enzyme as well (Skorko-Glonek et al., 1995; Kim et al., 1999; Spiess et al., 1999), is also consistent with its proposed role in response to heat. Why this enzyme, unlike most others, becomes increasingly active with increasing temperature was a mystery until very recently. Elucidation of the three-dimensional structure of the bacterial enzyme (Krojer et al., 2002) offers a clue to this question. At low temperature, access to the active site is blocked because of a twist in one of the loops that compose this site. Thermal motion of this loop may give the substrates access to the active site, and catalysis ensues (Krojer et al., 2002). Whether the structure of DegP1 is identical to that of the E. coli enzyme is not yet known, but the high similarity between the amino acid sequences in the region of the active site suggests similar structures. Thus, the reason for thermal activation of both enzymes may be the same.
In addition to DegP1, two other DegP homologs, DegP5 and DegP8, have been identified in the lumen (Schubert et al., 2002). DegP1 and DegP8 are very similar, having calculated molecular masses of 35.2 and 37.2 kD, respectively, whereas DegP5 is shorter, with a molecular mass of 23.5 kD. The main difference between these proteins and their bacterial homolog is the number of PDZ domains. PDZ domains have been implicated in protein-protein interactions, including protease-substrate recognition, in different biological systems (Saras and Heldin, 1996; Levchenko et al., 1997; Ponting, 1997; Beebe et al., 2000; Sheng and Sala, 2001). Whereas E. coli DegP contains two PDZ domains in tandems in its C terminus, DegP1 and DegP8 contain only the first domain, and DegP5 has none. This may have implications for substrate recognition and specificity of the three thylakoid lumen enzymes.
The finding that DegP1 forms hexamers is consistent with the oligomeric form of E. coli DegP. Although it was previously demonstrated that the bacterial enzyme forms a six-member ring-like structure (Kim et al., 1999), the crystal structure shows a staggered association of two trimeric rings (Krojer et al., 2002). Similar discrepancy between previous molecular and biochemical work and the crystal structure relates to the role of PDZ domains in oligomerization. Deletion of these domains from the bacterial enzyme results in smaller oligomers, either dimers or trimers, leading to the conclusion that the PDZ domains are important for the hexameric structure (Sassoon et al., 1999). In contrast, the crystal structure shows that the PDZ domains are oriented toward the perimeter of the ring, and do not contribute to the packing (Krojer et al., 2002). Thus, the high similarity between the protease domains of the three lumenal enzymes suggests that they may interact. The question of whether they form hetero-hexamers in vivo or whether the formation of homo-oligomers is favored remains to be determined experimentally.
To test the activity of DegP1 on lumenal proteins, we used PC and OE33 as substrates. Both could be degraded by the protease, suggesting that their degradation in vivo may be mediated by DegP1. In addition, we tested the effect of the SsrA tag on PC degradation. Proteins tagged with SsrA peptide have been previously shown to be rapidly degraded both in vitro and in vivo. Tagged λcI repressor was degraded by both E. coli Clp protease (Gottesman et al., 1998) and FtsH (Herman et al., 1998). Thus, the identity and conformation of the C termini of protein substrates appear to be important for their recognition and degradation by proteases. However, the presence of wild-type versus mutated tags did not affect PC degradation by DegP1, and thus it seems that the C terminus of PC does not play a role in its degradation. It was recently shown that that the C terminus of the short-lived bacterial ς32 is similarly not involved in its degradation by FtsH (Tomoyasu et al., 2001). This differential behavior suggests that different substrates interact differently with the same proteases and that multiple domains on the surface of the protease may be involved with substrate recognition and binding.
Here, we demonstrate the capability of DegP1 to degrade both model and lumenal protein substrates. Its lumenal location and membrane association suggest that DegP is involved in the degradation of integral membrane proteins as well. The current understanding of the mechanisms involved in the degradation of integral membrane proteins that span the membrane several times is very limited. It is plausible that enzymes from both sides of the membrane cooperate in the degradation of such substrates, as has been shown in mitochondria for a model substrate that spans the membrane only once (Leonhard et al., 2000). In the context of thylakoids, DegP could cleave lumenal loops, thus facilitating translocation and degradation of trans-membrane helices by ATP-dependent proteases such as FtsH on the stromal side of the membrane. The purification and characterization of DegP1 reported here will allow us to deal experimentally with this question and with the question of degradation of heat-denatured proteins in the thylakoid lumen.
MATERIALS AND METHODS
Subcloning, Overexpression, and Purification of DegP1
A fragment encoding the mature portion of DegP1 was generated by PCR, using the cDNA in pSPORT (Itzhaki et al., 1998) as a template. A codon for Met and the NdeI restriction site was added by using the oligonucleotide 5′-GGAATTCATATGTTTGTAGTTAGT-3′ as one primer in the reaction and SP6 as the second primer. The PCR product was cut with NdeI and BamHI and inserted into the corresponding sites in the plasmids pT7–7 and pET-15b. Escherichia coli BL21-DE3 cells were transformed with the vectors containing the DegP1 insert. Cultures were grown at 37°C to an OD600 of 0.3 to 0.4, and expression of DegP1 was induced by the addition of 0.5 mm IPTG. Cultures were then further grown for 1 to 4 h, and the cells were harvested by centrifugation at 5,000g for 5 min and assayed for expression by resuspension in 4× solubilization buffer followed by SDS-PAGE. To confirm the identity of the expressed protein, we excised the suspected band from the gel and subjected it to digestion with trypsin, followed by electrospray ionization-mass spectrometry analysis. The amino acid sequences of seven peptides were obtained, and all of them matched the sequence of DegP1. These peptides gave 24.3% coverage of the mature protein sequence, and given the fact that they originated from different regions of the protein, including its C terminus, the identification of the expressed protein as DegP1 was beyond doubt.
For purification of DegP1, cells from a 500-mL culture were harvested 1 h after induction with IPTG and resuspended in 10 mL of a buffer containing 50 mm NaH2PO4 pH 8.0, 300 mm NaCl, and 20% (w/v) glycerol (buffer A) containing 10 mm imidazole. lysozyme was added to a final concentration of 1 mg mL−1, and the solution was sonicated in a ice bath three times for 3 min each with 1-min intervening cooling periods. The sonicated cells were centrifuged at 5,000g for 5 min, the supernatant was mixed with a 1/4 volume of Ni-NTA agarose (Qiagen USA, Valencia, CA), and the mixture was gently agitated for 1 h at 4°C. The mixture was then loaded onto a column and washed twice with 4 mL of buffer A containing 20 mm imidazole, followed by four elutions with 0.5 mL of buffer A containing 250 mm imidazole. For assessment of the size of DegP1, fractions E1 to E4 were pooled, concentrated by lyophilization, and loaded onto a Sephacryl S-200 column. The column was then eluted with 10 mm Tris-HCl, pH 8.0, 150 mm NaCl, and 1 mm EDTA, and absorbance of the eluant at 280 nm was monitored.
Proteolytic Degradation Assays
To assay proteolytic activity of DegP1, a standard reaction mixture included 5 pmol of DegP1, 50 mm MES, pH 6.0, and 50 pmol β-casein (Sigma-Aldrich, St. Louis) in a total volume of 10 μL. The mixture was incubated for 1 h at 37°C and subjected to SDS-PAGE (see below) after the addition of 10 μL of 4× solubilization buffer. Where indicated, different amounts of DegP1 (0.5–50 pmol) were added, or the duration of incubation (30–150 min), pH (5.5–9.0), or incubation temperature (15°C–42°C) was changed. Degradation of OE33 was assayed in the same reaction mixture, except that β-casein was replaced with 15 pmol of OE33 that had been overexpressed in E. coli as previously described (Betts et al., 1994). Degradation of PC and its derivatives was measured in the same manner, except that the substrate in the reaction was in vitro synthesized protein (TNT coupled wheat germ extract system, Promega, Madison, WI), equivalent to 50,000 cpm.
Miscellaneous
SDS-PAGE was performed on 12% or 15% (w/v) acrylamide gels as described previously (Adam and Hoffman, 1993). Gels were stained with Coomassie Blue R-250, and degradation of β-casein was quantified using image analysis software (Image Master 2D Elite v3.01, Amersham Biosciences AB, Uppsala). For visualization and quantification of radioactive PC and its derivatives, gels were analyzed using a phosphor imager (Bas 1000, Fuji Photo Film, Tokyo). Western blot was performed with a monoclonal anti-poly His antibody (Sigma-Aldrich) at a 1:3,000 dilution, which was visualized using a chemiluminescence kit (Pierce Chemical, Rockford, IL).
ACKNOWLEDGMENTS
We thank Prof. Arie Admon and members of his group at The Protein Center, Faculty of Biology, The Technion (Haifa, Israel) for the electrospray ionization-mass spectrometry analysis.
Footnotes
This work was supported by The Israel Science Foundation (grant no. 122/00 to Z.A.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.007922.
LITERATURE CITED
- Adam Z. Chloroplast proteases: possible regulators of gene expression? Biochimie. 2000;82:647–654. doi: 10.1016/s0300-9084(00)00612-x. [DOI] [PubMed] [Google Scholar]
- Adam Z, Adamska I, Nakabayashi K, Ostersetzer O, Haussuhl K, Manuell A, Vallon O, Rodermel SR, Shinozaki K, Clarke AK. Chloroplast and mitochondrial proteases in Arabidopsis: a proposed nomenclature. Plant Physiol. 2001;125:1912–1918. doi: 10.1104/pp.125.4.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam Z, Hoffman NE. Biogenesis of a photosystem I light harvesting complex: evidence for a membrane intermediate. Plant Physiol. 1993;102:35–43. doi: 10.1104/pp.102.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S, Thompson E, Nixon PJ, Horton P, Mullineaux CW, Robinson C, Mann NH. A critical role for the Var2 FtsH homologue of Arabidopsis thaliana in the photosystem II repair cycle in vivo. J Biol Chem. 2002;277:2006–2011. doi: 10.1074/jbc.M105878200. [DOI] [PubMed] [Google Scholar]
- Beebe KD, Shin J, Peng J, Chaudhury C, Khera J, Pei D. Substrate recognition through a PDZ domain in tail-specific protease. Biochemistry. 2000;39:3149–3155. doi: 10.1021/bi992709s. [DOI] [PubMed] [Google Scholar]
- Betts SD, Hachigian TM, Pichersky E, Yocum CF. Reconstitution of the spinach oxygen-evolving complex with recombinant Arabidopsis manganese-stabilizing protein. Plant Mol Biol. 1994;26:117–130. doi: 10.1007/BF00039525. [DOI] [PubMed] [Google Scholar]
- Chen M, Choi Y, Voytas DF, Rodermel S. Mutations in the Arabidopsis VAR2 locus cause leaf variegation due to the loss of a chloroplast FtsH protease. Plant J. 2000;22:303–313. doi: 10.1046/j.1365-313x.2000.00738.x. [DOI] [PubMed] [Google Scholar]
- Gottesman S. Proteases and their targets in Escherichia coli. Annu Rev Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- Gottesman S, Roche E, Zhou Y, Sauer RT. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halperin T, Adam Z. Degradation of mistargeted OEE33 in the chloroplast stroma. Plant Mol Biol. 1996;30:925–933. doi: 10.1007/BF00020804. [DOI] [PubMed] [Google Scholar]
- Haussuhl K, Andersson B, Adamska I. A chloroplast DegP2 protease performs the primary cleavage of the photodamaged D1 protein in plant photosystem II. EMBO J. 2001;20:713–722. doi: 10.1093/emboj/20.4.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman C, Thevenet D, Bouloc P, Walker GC, D'Ari R. Degradation of carboxy-terminal-tagged cytoplasmic proteins by the Escherichia coli protease hflB. Genes Dev. 1998;12:1348–1355. doi: 10.1101/gad.12.9.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Itzhaki H, Naveh L, Lindahl M, Cook M, Adam Z. Identification and characterization of DegP, a serine protease associated with the luminal side of the thylakoid membrane. J Biol Chem. 1998;273:7094–7098. doi: 10.1074/jbc.273.12.7094. [DOI] [PubMed] [Google Scholar]
- Keiler KC, Waller PRH, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- Kim KI, Park SC, Kang SH, Cheong GW, Chung CH. Selective degradation of unfolded proteins by the self-compartmentalizing HtrA protease, a periplasmic heat shock protein in Escherichia coli. J Mol Biol. 1999;294:1363–1374. doi: 10.1006/jmbi.1999.3320. [DOI] [PubMed] [Google Scholar]
- Krojer T, Garrido-Franco M, Huber R, Ehrmann M, Clausen T. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature. 2002;416:455–459. doi: 10.1038/416455a. [DOI] [PubMed] [Google Scholar]
- Lensch M, Herrmann RG, Sokolenko A. Identification and characterization of SppA, a novel light-inducible chloroplast protease complex associated with thylakoid membranes. J Biol Chem. 2001;276:33645–33651. doi: 10.1074/jbc.M100506200. [DOI] [PubMed] [Google Scholar]
- Leonhard K, Guiard B, Pellecchia G, Tzagoloff A, Neupert W, Langer T. Membrane protein degradation by AAA proteases in mitochondria: extraction of substrates from either membrane surface. Mol Cell. 2000;5:629–638. doi: 10.1016/s1097-2765(00)80242-7. [DOI] [PubMed] [Google Scholar]
- Levchenko I, Smith CK, Walsh NP, Sauer RT, Baker TA. PDZ-like domains mediate binding specificity in the Clp/Hsp100 family of chaperones and protease regulatory subunits. Cell. 1997;91:939–947. doi: 10.1016/s0092-8674(00)80485-7. [DOI] [PubMed] [Google Scholar]
- Li HH, Merchant S. Degradation of plastocyanin in copper-deficient C. reinhardtii: evidence for a protease-susceptible conformation of the apoprotein and regulated proteolysis. J Biol Chem. 1995;270:23504–23510. doi: 10.1074/jbc.270.40.23504. [DOI] [PubMed] [Google Scholar]
- Lindahl M, Spetea C, Hundal T, Oppenheim AB, Adam Z, Andersson B. The thylakoid FtsH protease plays a role in the light-induced turnover of the photosystem II D1 protein. Plant Cell. 2000;12:419–431. doi: 10.1105/tpc.12.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl M, Tabak S, Cseke L, Pichersky E, Andersson B, Adam Z. Identification, characterization, and molecular cloning of a homologue of the bacterial FtsH protease in chloroplasts of higher plants. J Biol Chem. 1996;271:29329–29334. doi: 10.1074/jbc.271.46.29329. [DOI] [PubMed] [Google Scholar]
- Lipinska B, Zylicz M, Georgopoulos C. The HtrA (DegP) protein, essential for Escherichia coli survival at high temperatures, is an essential endopeptidase. J Bacteriol. 1990;172:1791–1797. doi: 10.1128/jb.172.4.1791-1797.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W, Wollman F-A, Vallon O. Evidence for a role of ClpP in the degradation of the chloroplast cytochrome b6f complex. Plant Cell. 2000;12:137–149. doi: 10.1105/tpc.12.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S, Bogorad L. Rapid degradation of apoplastocyanin in Cu(II)-deficient cells of Chlamydomonas reinhardtii. J Biol Chem. 1986;261:15850–15853. [PubMed] [Google Scholar]
- Ostersetzer O, Adam Z. Light-stimulated degradation of an unassembled Rieske FeS protein by a thylakoid-bound protease: the possible role of the FtsH protease. Plant Cell. 1997;9:957–965. doi: 10.1105/tpc.9.6.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier J-B, Emanuelsson O, Kalume DE, Ytterberg J, Friso G, Rudella A, Liberles DA, Soderberg L, Roepstorff P, von Heijne G et al. Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell. 2002;14:211–236. doi: 10.1105/tpc.010304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier J-B, Ytterberg J, Liberles DA, Roepstorff P, van Wijk KJ. Identification of a 350 kDa ClpP protease complex with 10 different Clp isoforms in chloroplasts of Arabidopsis thaliana. J Biol Chem. 2001;276:16318–16327. doi: 10.1074/jbc.M010503200. [DOI] [PubMed] [Google Scholar]
- Ponting CP. Evidence for PDZ domains in bacteria, yeast, and plants. Protein Sci. 1997;6:464–468. doi: 10.1002/pro.5560060225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roffey RA, Theg SM. Analysis of the import of carboxyl-terminal truncations of the 23-kilodalton subunit of the oxygen-evolving complex suggests that its structure is an important determinant for thylakoid transport. Plant Physiol. 1996;111:1329–1338. doi: 10.1104/pp.111.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saras J, Heldin CH. PDZ domains bind carboxy-terminal sequences of target proteins. Trends Biochem Sci. 1996;21:455–458. doi: 10.1016/s0968-0004(96)30044-3. [DOI] [PubMed] [Google Scholar]
- Sassoon N, Arie JP, Betton JM. PDZ domains determine the native oligomeric structure of the DegP (HtrA) protease. Mol Microbiol. 1999;33:583–589. doi: 10.1046/j.1365-2958.1999.01505.x. [DOI] [PubMed] [Google Scholar]
- Schubert M, Petersson UA, Haas BJ, Funk C, Schroder WP, Kieselbach T. Proteome map of the chloroplast lumen of Arabidopsis thaliana. J Biol Chem. 2002;277:8354–8365. doi: 10.1074/jbc.M108575200. [DOI] [PubMed] [Google Scholar]
- Shanklin J, Dewitt ND, Flanagan JM. The stroma of higher plant plastids contain ClpP and ClpC, functional homologs of Escherichia coli ClpP and ClpA: an archetypal two-component ATP-dependent protease. Plant Cell. 1995;7:1713–1722. doi: 10.1105/tpc.7.10.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, Sala C. PDZ domains and the organization of supramolecular complexes. Annu Rev Neurosci. 2001;24:1–29. doi: 10.1146/annurev.neuro.24.1.1. [DOI] [PubMed] [Google Scholar]
- Skorko-Glonek J, Krzewski K, Lipinska B, Bertoli E, Tanfani F. Comparison of the structure of wild-type HtrA heat shock protease and mutant HtrA proteins: a Fourier transform infrared spectroscopic study. J Biol Chem. 1995;270:11140–11146. doi: 10.1074/jbc.270.19.11140. [DOI] [PubMed] [Google Scholar]
- Sokolenko A, Lerbs-Mache S, Altschmied L, Herrmann RG. Clp protease complexes and their diversity in chloroplasts. Planta. 1998;207:286–295. doi: 10.1007/s004250050485. [DOI] [PubMed] [Google Scholar]
- Sokolenko A, Pojidaeva E, Zinchenko V, Panichkin V, Glaser VM, Shestakov SV, Herrmann RG. The gene complement for proteolysis in the cyanobacterium Synechocystis sp. PCC 6803 and Arabidopsis thaliana chloroplasts. Curr Genet. 2002;41:291–310. doi: 10.1007/s00294-002-0309-8. [DOI] [PubMed] [Google Scholar]
- Spiess C, Beil A, Ehrmann M. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell. 1999;97:339–347. doi: 10.1016/s0092-8674(00)80743-6. [DOI] [PubMed] [Google Scholar]
- Takechi K, Sodmergen, Murata M, Motoyoshi F, Sakamoto W. The YELLOW VARIEGATED (VAR2) locus encodes a homologue of FtsH, an ATP-dependent protease in Arabidopsis. Plant Cell Physiol. 2000;41:1334–1346. doi: 10.1093/pcp/pcd067. [DOI] [PubMed] [Google Scholar]
- Tomoyasu T, Arsene F, Ogura T, Bukau B. The C terminus of ς32 is not essential for degradation by FtsH. J Bacteriol. 2001;183:5911–5917. doi: 10.1128/JB.183.20.5911-5917.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner S, Maurizi MR, Gottesman S. Posttranslational quality control: folding, refolding, and degrading proteins. Science. 1999;286:1888–1893. doi: 10.1126/science.286.5446.1888. [DOI] [PubMed] [Google Scholar]
- Zheng B, Halperin T, Hruskova-Heidingsfeldova O, Adam Z, Clarke AK. Characterization of chloroplast Clp proteins in Arabidopsis: localization, tissue specificity and stress responses. Physiol Plant. 2002;114:92–101. doi: 10.1034/j.1399-3054.2002.1140113.x. [DOI] [PubMed] [Google Scholar]