Abstract
A screen for Arabidopsis mutants that were insensitive to methyl jasmonate (MeJA) in an assay for seedling root growth yielded only alleles of previously isolated mutants jar1 and coi1, with one exception. Mapping of the locus and morphological characterization of the new mutant suggested it might be allelic to axr1, which had not previously been reported to show resistance to MeJA. The F1 from a cross of the new mutant with axr1-3 did not show complementation, confirming that these are the same genes. The new allele is called axr1-24. In addition to MeJA and indole-3-acetic acid (IAA), axr1-24 had decreased sensitivity to 1-aminocyclopropane-1-carboxylic acid, 6-benzylamino-purine, epi-brassinolide, and abscisic acid. Both axr1-24 and the previously characterized axr1-3 allele were shown to be susceptible to the opportunistic pathogen Pythium irregulare, a trait found in other jasmonate response mutants, including jar1-1. The double mutant jar1-1/axr1-3 was more resistant to inhibition of root growth by MeJA and was more susceptible to P. irregulare infection than either single mutant, suggesting these genes might act in independent response pathways. In contrast, resistance to IAA in the double mutant was not different from axr1-3. Northern-blot analysis showed that IAA induced the jasmonate-responsive lipoxygenase 2, AOS, and AtVSP gene transcripts and induction was strongly impaired in axr1-3. However, transcript induction by MeJA was only minimally affected in axr1-3. This study demonstrates that in addition to auxin signaling, the AXR1 locus is involved in MeJA response, providing a mechanistic link between jasmonate and auxin-signaling pathways.
Plant hormones control a diverse array of plant responses affecting growth and development, defense against microorganisms and insects, and protection from abiotic stresses (Davies, 1995). These plant signals interact with each other in both complementary and antagonistic ways to accomplish their signaling roles. Many hormone response mutants have been isolated, and characterization of some of these has revealed further evidence for interactions among plant hormones at the level of signal transduction.
Jasmonate plays a critical role in plant reproductive development (McConn and Browse, 1996; Sanders et al., 2000; Stintzi and Browse, 2000), in protecting plants from pathogens and insects (Farmer and Ryan, 1990; Penninckx et al., 1996; McConn et al., 1997; Staswick et al., 1998), and in limiting damage from abiotic agents (Overmyer et al., 2000; Rao et al., 2000). The emerging evidence indicates that jasmonate signaling involves a complex interaction between several cyclopentanone derivatives of linolenic acid metabolism, including jasmonic acid (JA), methyl jasmonate (MeJA; Seo et al., 2001), and the JA precursor 12-oxo-phytodienoic acid (Mueller, 1997; Stintzi et al., 2001). Related products of other synthetic pathways are probably involved as well (Weber et al., 1997).
Three Arabidopsis loci have been identified in screens for resistance to MeJA or the bacterial phytotoxin coronatine, which mimics jasmonate activity. Mutations in JAR1, COI1, and JIN1 lead to reduced sensitivity in the inhibition of root growth by MeJA and various other jasmonate-associated defects, but not to insensitivity to several other tested hormones (Staswick et al., 1992; Feys et al., 1994; Berger et al., 1996). Mutation of a mitogen-activated protein kinase (mpk4) also shows defects in some jasmonate responses (Peterson et al., 2000), although this may by a downstream component that is involved in response to other signals as well. Mutations in some of these genes impair only portions of the jasmonate response. For example, whereas JA is required for male fertility and coi1-1 is male sterile (McConn and Browse, 1996; Stintzi and Browse, 2000), all known jar1 alleles are fertile (Staswick et al., 2002). We recently demonstrated that JAR1 does not encode a signal transduction component, but rather, an enzyme that biochemically modifies JA (Staswick et al., 2002). The inhibition of root growth by MeJA may integrate many of the subprograms that are modulated by jasmonates (Berger et al., 1996), thus additional loci affecting jasmonate response may await discovery.
Auxin is key a hormone that controls plant growth and development, and is involved in cell division and elongation. Identification of the defective genes from several auxin response mutants has led to a model for auxin response involving an ubiquitin-proteasome pathway that includes an SCF-type E3-ubiquitin ligase complex (Gray and Estelle, 2000). Interestingly, COI1 encodes an F-box protein that is related to the TIR1 component of the auxin-signaling SCF complex. This suggests that jasmonate and auxin use a similar signaling mechanism. However, coi1 is not altered in its response to auxin (Feys et al., 1994), suggesting that these are separate signaling pathways. This study was initiated to isolate and characterize new mutants that affect response to jasmonate. The results revealed that jasmonate and auxin act through a common signaling intermediate that also affects response to other plant hormones.
RESULTS
Isolation of a New MeJA-Insensitive Mutant
A screen of about 200,000 M2 seedlings representing around 50,000 M1 parents for resistance of root growth to inhibitory concentrations of MeJA yielded only alleles of the previously isolated mutant loci jar1 and coi1, with one exception. The exception was crossed to wild type (Ler). Analysis of 270 of the resulting F2 progeny showed a χ2 value of 0.11 (P = 0.73) for a 3:1 segregation ratio (MeJA sensitive:MeJA resistant), indicating that this was a recessive single-gene mutation.
The new mutant also had a phenotype distinct from that of other jasmonate response mutants. Plants were shorter, had crinkled leaves, and exhibited partial male sterility. A detailed analysis the phenotype is presented in Table I. This phenotype contrasts with the jasmonate response mutants jar1, coi1, and jin1 that all appear indistinguishable from wild type, except that coi1-1 is male sterile. All 52 of the F2 MeJA-resistant seedlings that survived transfer to soil and grew to maturity exhibited the aberrant phenotype, whereas MeJA-sensitive plants did not. This indicated that a single gene was involved in both jasmonate response and the distinct phenotype. Thus, the new mutant appeared to define a novel locus that is associated with response to MeJA.
Table I.
Morphology of wild-type and axr1-24 plants
| Parameter | Wild Type | axr1-24 |
|---|---|---|
| Height (cm) | 41.2 ± 3.7a | 27.2 ± 3.9 |
| No. of inflorescences | 4.8 ± 1.0 | 6.1 ± 1.1 |
| No. of branches | 13.1 ± 3.9 | 23.1 ± 11.6 |
| Distance between siliques (cm) | 1.0 ± 0.1 | 0.6 ± 0.1 |
| No. of siliques | 154 ± 73 | 22 ± 30 |
| Length of siliques (cm) | 1.5 ± 0.1 | 0.6 ± 0.1 |
| Etiolated seedling hypocotyl length (cm) | 12.9 ± 1.8 | 12.8 ± 1.9 |
| No. of pollen grains/flower | 2,389 ± 719 | 680 ± 565 |
Mean ± sd (n = 12 plants).
The MeJA-Insensitive Mutant Is Allelic to axr1
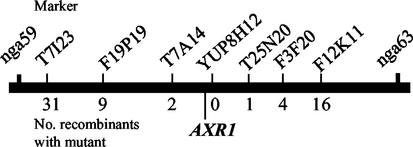
The phenotype of our new mutant was strikingly similar to that of the previously described axr1-3 allele of the auxin response mutant (Lincoln et al., 1990). Furthermore, the locus was mapped to an interval of about 134 kb on chromosome 1 that is flanked by SSLP markers T7A14 and T25N20 (Fig. 1). AXR1 is located within this interval, suggesting these might be the same genes. The F1 cross of our mutant with axr1-3 showed noncomplementation, producing only mutant plants. These results confirmed that these are the same loci and our new mutant is hereafter called axr1-24.
Figure 1.
Mapping of the MeJA resistance locus. Molecular markers used are indicated above the chromosome I interval that is depicted. Numbers below denote the number of recombinants between the mutant locus and the respective marker. The relative position of AXR1 is shown.
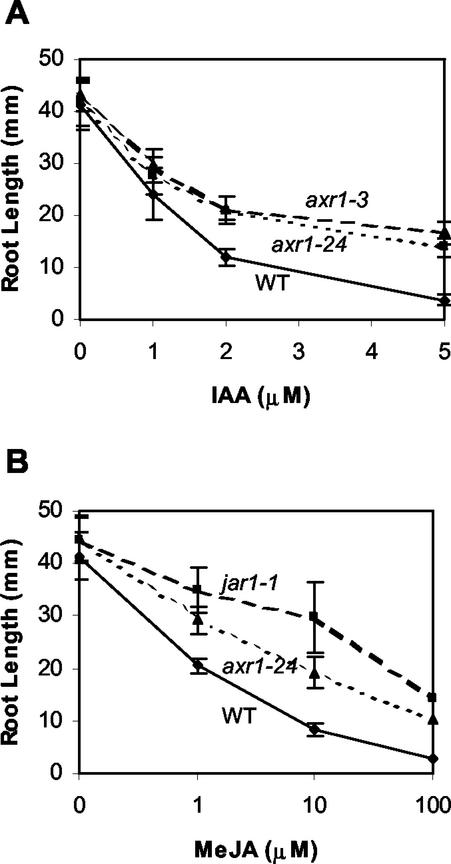
Several alleles of axr1 have been isolated, including axr1-3, which displays only partial loss of gene function (Lincoln et al., 1990). We sequenced two independent cDNA clones of axr1-24 but found no evidence of a mutation that would alter protein translation. This suggests that the mutation in axr1-24 might be in a noncoding region that affects gene expression. The level of resistance to auxin (50% inhibition of root growth) for axr1-24 was essentially the same as in axr1-3 (Fig. 2A). Comparison of the phenotype of axr1-24 (Table I) with the published results for axr1-3 (Lincoln et al., 1990) also indicated that these alleles were similar in the severity of their defect. Together, these results suggest that the phenotype of axr1-24 is attributable to a partial loss of gene function.
Figure 2.
Dose response curve for root growth inhibition on IAA and MeJA. Wild-type (wt) and mutant (jar1-1, axr1-24, and axr1-3) seedling root length was measured after 10 d growth at 21 C. Error bars indicate sd (n = 20). A, Growth on IAA. B, Growth on MeJA.
axr1-24 Is Defective in Its Response to Several Plant Hormones
Previous studies documented that AXR1 confers sensitivity to ethylene and cytokinin as well as auxin (Timpte et al., 1995), but a role in MeJA response had not previously been reported. The MeJA dose response of axr1-24 in primary root growth was tested over a range of concentrations and compared with wild type and jar1-1 (Fig. 2B). axr1-24 had a level of resistance that was less than that seen in jar1-1 over all concentrations tested. At about 10 μm in axr1-24, 50% inhibition of growth occurred, whereas the concentration for 50% inhibition was 5- to 10-fold higher in jar1-1.
To further investigate the hormone insensitivity of axr1-24, seedling root growth was tested on a range of concentrations of 6-benzylamino-purine (BA), epi-brassinolide (BR), and the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC). The response to abscisic acid (ABA) was also examined in a seed germination assay.
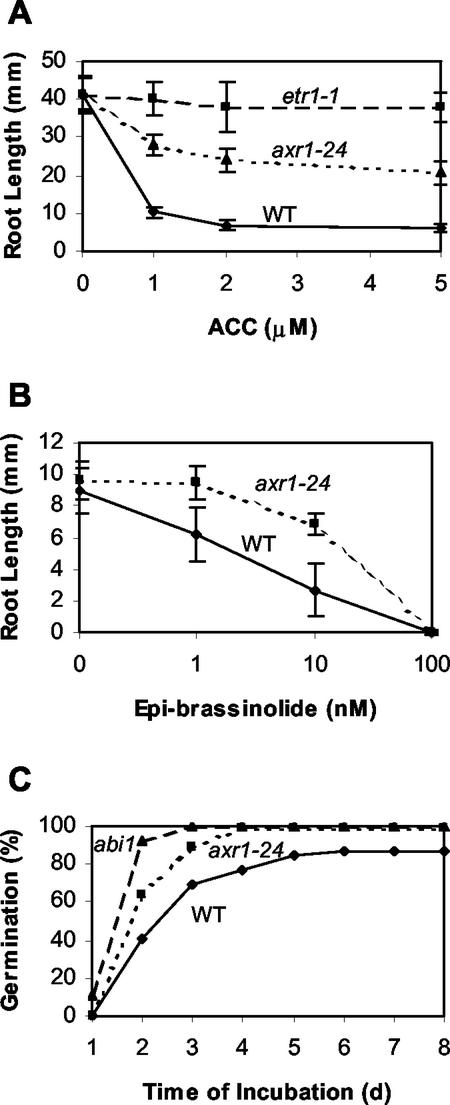
ACC inhibited root growth in axr1-24 to a level intermediate between that of wild type and the ethylene-insensitive mutant etr1-1 (Fig. 3A). Root elongation was inhibited 8%, 50.4%, and 85.3% for etr1, axr1-24, and wild type, respectively, at 5 μm ACC. No difference among the genotypes was observed on control medium.
Figure 3.
Insensitivity of mutants to hormones. Root length determined as in Figure 2. A, Inhibition of root growth by the ethylene precursor ACC. B, Inhibition of root growth by BR. C, Seed germination on 0.5 μm ABA.
Brassinolides are powerful inhibitors of root growth and development. Prolonged growth in the presence of BR caused root curling, so measurements were taken after 5 d rather than 10 d of growth. axr1-24 was resistant to both 1 and 10 nm BR, whereas strong inhibition was observed for both mutant and wild type at 100 nm (Fig. 3B).
In the assay for germination in the presence of ABA, axr1-24 was more resistant than wild type at 0.5 μm ABA (Fig. 3C), although inhibition was similar to wild type at concentrations above 5 μm (data not shown). axr1-24 was less resistant to ABA than the well-characterized mutant abi1, which germinates even at 10 μm ABA.
The effect of cytokinins on root elongation in axr1-24 was also examined. axr1-24 was more resistant in its response to BA than wild type. Total inhibition of root elongation was 66.4% and 73.7% for axr1-24 and wild type, respectively, at 1.5 μm BA. Although the magnitude of difference was small compared with the other hormones tested, an analysis of variance indicated the root length was significantly different (P < 0.05).
axr1-24 Is Resistant to Other Inhibitors of Root Growth
The results indicated that axr1-24 has an altered sensitivity to all tested plant hormones. To further explore the specificity of this locus in sensitivity to other chemicals, we tested root growth response in the presence of various compounds that inhibit root growth.
axr1-24 was more resistant than wild type (P < 0.01) at 30 and 50 μm salicylic acid (SA), although the differences were small compared with most of the plant hormones described earlier (Table II). axr1-24 was also resistant to the inhibitory effects of both ferulic and gallic acids. On the other hand, no difference from wild type was observed over a range of concentrations for gentisic acid, arachidonate, linolenate, juglone, and patulin (data not shown).
Table II.
Resistance of axr1-24 seedling root growth to various organic acids
| Acid | Concentration | Seedling Root Length
|
|
|---|---|---|---|
| Wild type | axr1-24 | ||
| μm | mma | ||
| Salicylic | 0 | 39.5 ± 3.4 | 38.7 ± 2.6 |
| 30 | 14.9a ± 0.9 | 18.8b ± 1.1 | |
| 50 | 13.3a ± 1.0 | 16.9b ± 1.4 | |
| Ferulic | 0 | 46.4 ± 1.7 | 47.6 ± 1.1 |
| 15 | 24.1a ± 1.18 | 31.0b ± 1.4 | |
| 25 | 19.5a ± 0.9 | 27.5b ± 1.4 | |
| Gallic | 0 | 49.7 ± 1.8 | 51.7 ± 1.5 |
| 15 | 26.7a ± 1.5 | 32.0b ± 2.1 | |
| 25 | 22.1a ± 1.6 | 29.0b ± 1.3 | |
Means with different letters (a and b) indicates significant difference between genotypes for each concentration (P = 0.01).
Mean ± sd (n = 20).
axr1-3 Is More Susceptible to the Fungus Pythium irregulare
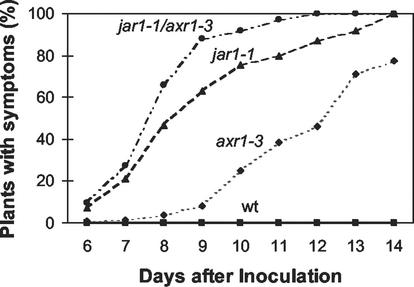
A primary function of the jasmonates is in plant stress response, including defense against the opportunistic fungal pathogen P. irregulare (Staswick et al., 1998; Vijayan et al., 1998). Having determined that the locus responsible for defects in MeJA response in our mutant is in fact AXR1, we next tested whether the well-characterized axr1-3 was defective in this jasmonate-requiring pathogen defense response. Inoculation of 5-week-old plants at the root zone with an isolate of P. irregulare caused the symptoms of wilting and tissue collapse reported previously for jar1-1 but not seen in wild type (Staswick et al., 1998). However, symptoms appeared later in axr1-3 than in jar1-1 (Fig. 4), suggesting a weaker defect in this defense-signaling response compared with jar1-1. One-half of the jar1 plants displayed symptoms by 8 to 9 d after inoculation, whereas axr1-3 did not show a similar level of symptoms until 12 to 13 d. Similar results were also found for axr1-24 (data not shown). This is consistent with the lower level of insensitivity to MeJA observed in the root growth assay for axr1-24 compared with jar1-1 (Fig. 1). Two-week-old plants showed symptoms earlier, but the timing of symptom appearance in axr1-3 was still later than for jar1 (data not shown). These results demonstrate that in addition to its involvement in MeJA-suppressed root growth, AXR1 is necessary for resistance to P. irregulare in Arabidopsis.
Figure 4.
Infection of mutant and wild-type seedlings with P. irregulare. One hundred and fifty-three seedlings were used per genotype. The percentage of plants with symptoms at the times indicated is shown for each genotype. Wild type did not show symptoms during the duration of the experiment.
The Effect of JAR1 and AXR1 Is Additive
Our results showed that AXR1 provides a link between jasmonate and auxin signaling. To investigate whether JAR1 and AXR1 are involved in the same or in distinct response pathways, a double mutant homozygous for both jar1-1 and axr1-3 was constructed. The double mutant was phenotypically similar to axr1-3 in all aspects except flowering time, which was about 1 week earlier than in either parent (data not shown). The roots of jar1-1/axr1-3 were about 3.4 times longer than wild type on 50 μm MeJA, and nearly the same as the length of wild type on control media (Table III). The effect of the lesions in jar1 and axr1 appears to be additive, because the roots of the double mutant grew to about twice the length of either single mutant on 50 μm MeJA. Although root elongation of the double mutant was also slightly greater than wild type on control medium, the difference was small compared with that seen after growth on MeJA-containing medium. Response to 2 μm indole-3-acetic acid (IAA) was the same as for axr1-3 and axr1-3/jar1-1 (Table III), which is consistent with the fact that JAR1 is not known to influence auxin response.
Table III.
Resistance to IAA and MeJA in jar1-1, axr1-3, and jar1-1/axr1-3
| Genotype | Root Length
|
||
|---|---|---|---|
| Control | IAA (2 μm) | MeJA (50 μm) | |
| mma | |||
| wt | 29.1a ± 0.9 | 9.7a ± 0.8 | 6.3a ± 0.4 |
| jar1 | 29.1a ± 1.1 | 9.4a ± 0.8 | 13.5c ± 1.2 |
| axr1 | 29.9a ± 1.6 | 20.3b ± 1.5 | 10.1b ± 1.5 |
| jar1/axr1 | 35.4b ± 1.6 | 19.7b ± 1.6 | 27.9d ± 2.1 |
Means with different letters (a, b, c, and d) between genotypes are significantly different (P = 0.01).
Mean ± sd (n = 20).
To determine whether the pathogen response of the double mutant was also altered, its response to P. irregulare infection was compared with the single mutants. Figure 4 shows that symptom appearance was earlier in jar1-1/axr1-3 than in jar1 and axr1-3, although by 14 d essentially all plants were affected in both jar1-1 and jar1-1/axr1-3. In contrast, about 22% of axr1 plants appeared healthy 14 d after inoculation (Fig. 4).
Expression of Early Auxin and Jasmonate-Responsive Genes in axr1-24
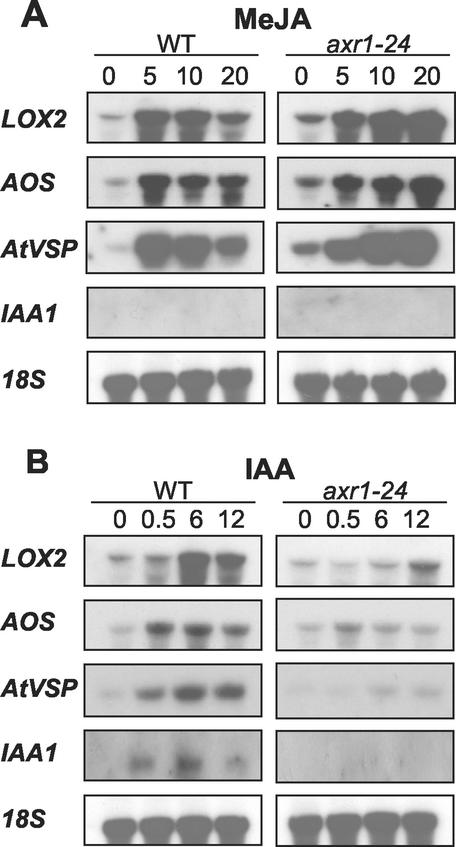
The link between auxin and jasmonate response through AXR1 prompted an examination of MeJA- and auxin-induced gene expression in wild type and in axr1-24 seedlings. Jasmonate-inducible AOS, lipoxygenase 2 (LOX2), and AtVSP transcripts were elevated by MeJA treatment in wild type with a maximum level reached by about 5 h (Fig. 5A). This was followed by a decline in transcript abundance, which is consistent with the transient elevation previously reported for AOS, AtVSP, and several other jasmonate-inducible transcripts in Arabidopsis (Titarenko et al., 1997; Laudert and Weiler, 1998). These transcripts were also elevated to a similar level in axr1-24, although the time for maximal induction was somewhat delayed relative to wild type. MeJA treatment did not raise the mRNA level for the auxin-inducible gene IAA1 in either genotype. Surprisingly, LOX2, AOS, and AtVSP were also induced by IAA, although the maximum transcript level was somewhat less than when induced by MeJA. Induction of these genes by IAA was suppressed in axr1-24, as was also the case for IAA1.
Figure 5.
Transcript level for LOX2, AOS, AtVSP, and IAA1 in light-grown wild-type (WT) and axr1-24 seedlings. Numbers above lanes indicate time (h) after treatment was initiated. Hybridization probe for each row of samples is indicated to the left. A, Total RNA isolated from volatile MeJA-treated seedlings. B, RNA isolated after spraying with 1 μm IAA.
DISCUSSION
This report describes the identification and characterization of a novel Arabidopsis mutant that was defective in its response to MeJA but with additional characteristics that were atypical of those previously identified in jasmonate response mutants. The new mutant had phenotypic abnormalities in plant growth and development and was insensitive to multiple plant hormones. This contrasts with other jasmonate response mutants that, with the exception of male sterility in coi1 (Feys et al., 1994), appear phenotypically normal and are resistant only to jasmonates. Except for male sterility, mutants defective in JA biosynthesis are also developmentally normal, indicating that JA does not play a major role in most aspects of growth and development in Arabidopsis (McConn and Browse, 1996; Sanders et al., 2000).
The discrepancy between results for other jasmonate response mutants and those reported here was resolved by the finding that the new mutant was allelic to the previously characterized auxin response mutant axr1. This was surprising because, although known to modestly affect ethylene and cytokinin response (Timpte et al., 1995), axr1 was not previously documented to alter response to jasmonates. The fact that JA is essential for pollen maturation and anther dehiscence in Arabidopsis suggests that the partial male sterility observed in axr1 alleles may reflect an impairment in development that is mediated by JA, rather than by auxin. In contrast, most other phenotypic abnormalities associated with axr1 alleles are likely not related to jasmonate signaling.
Previous studies have identified numerous genes that act in concert with AXR1 in a ubiquitin-like proteasome pathway that mediates auxin signaling in a manner that is still not fully understood (Ruegger et al., 1998; Yeh et al., 2000). AXR1 and ECR1 form a heterodimeric enzyme that activates the ubiquitin-like RUB protein (del Pozo et al., 2002). Activated RUB is then conjugated to cullins (del Pozo et al., 1998), which are in turn components of a ubiquitin E3 ligase complex called SCF. Along with cullin, the SCF complex consists of SKP1, RBX1, and an F-box protein. SCF is involved in the transfer of ubiquitin from ubiquitin ligase to target proteins in a variety of signaling paths, the ubiquitination specificity being determined by unique SCF components. In the case of auxin signaling, the F-box protein is TIR1, which is closely related to the jasmonate response factor encoded by COI1 (Xie et al., 1998). This suggests that jasmonate signaling also involves an SCF-mediated ubiquitination pathway. However, coi1 is male sterile and does not show altered sensitivity to auxin (Feys et al., 1994), whereas tir1 is resistant to auxin and fertile (Ruegger et al., 1998), suggesting that COI1 and TIR1 function in largely independent proteasome-signaling pathways.
Our present results demonstrate that the auxin and jasmonate proteasome pathways are directly connected through AXR1, which acts upstream of TIR1/COI1. RUB modification of cullins is also important for SCF function in species other than Arabidopsis. Therefore, we hypothesize that altered jasmonate signaling in axr1 is attributable to impaired RUB activation by AXR1/ECR1, which leads to defects in RUB modification of a cullin family member of the SCF-COI1 complex. Mutants affecting other proteins of the auxin proteasome-signaling pathway could be used to test this hypothesis, and the results would further clarify the relationship between auxin and jasmonate response.
An unresolved question is why the effect of an AXR1 mutation on jasmonate response is relatively weak compared with the auxin-related phenotype. As mentioned earlier, it is possible that partial male sterility observed in axr1 is attributable to defects in JA signaling. However, inhibition of root growth and gene induction by MeJA was minimal compared with jar1-1, and pathogen susceptibility was also less than in jar1-1. It should be noted that insensitivity of jar1 alleles to MeJA is itself weak compared with coi1 (Staswick et al., 2002).
The minimal effect on JA responses may simply reflect the fact that like axr1-3, axr1-24 is apparently a weak allele. The phenotype of these two alleles was similar, and we did not find evidence for a mutation in the AXR1-coding sequence from axr1-24, suggesting that (possibly minor) changes in AXR1 gene expression are involved. Another explanation could be that there is gene redundancy for AXR1 function in jasmonate response. Recent evidence suggests that a closely related Arabidopsis gene, called AXL1, may partially complement the function of AXR1 (del Pozo et al., 2002). The extent of complementation might vary in different tissues, possibly explaining why even severe axr1 alleles exhibit no defects in embryogenesis. AXR1 may similarly be relatively less important than AXL1 in RUB activation of cullins that are involved in SCF complexes associated with jasmonate response.
That AXR1 is necessary for defense against the opportunistic pathogen P. irregulare suggests that AXR1 plays an important role in jasmonate-mediated responses. The mechanism of resistance to this microorganism is not known, but it has been suggested that a disruption in the induction of jasmonate-regulated defense genes may lead to increased susceptibility in mutants that are impaired in jasmonate response pathways (Staswick et al., 1998; Vijayan et al., 1998). Although jasmonate-induced transcripts were elevated by MeJA in axr1-24 (Fig. 5), subtle differences in the timing of induction might account for the low level of pathogen susceptibility we observed. We cannot rule out the possibility that auxin also plays a role in resistance to P. irregulare. Analysis of other auxin-signaling mutants, such as tir1 and ask1, as well as auxin biosynthetic mutants should help to answer this question.
The induction of two genes involved in JA biosynthesis by IAA was unexpected. The fact that LOX2 and AOS induction by IAA was suppressed in axr1-24 may indicate another link between jasmonate and auxin signaling, in this case at the level of jasmonate synthesis. It has been proposed that there is positive feedback regulation on the expression of LOX2 and AOS because JA elevates expression of both genes, and their expression is suppressed in JA response mutants (Berger et al., 1995; Mueller, 1997; Laudert and Weiler, 1998). This might provide a mechanism for the amplification of the JA signal at the level of jasmonate biosynthesis. Our results indicate that auxin might also have a role in regulating JA level.
Cross-resistance of mutants to multiple hormones is well documented (Wilson et al., 1990; Hobbie and Estelle, 1994) and suggests that the action of hormones is coordinated by common intermediates or modulators. But the physiological role of loci involved in multihormone response is less clear. Our results suggest that earlier evidence for the involvement of AXR1 in response to other hormones may be functionally important. In addition to jasmonates, other signals critical for plant disease resistance include ethylene and SA. In some cases ethylene complements the role of JA, whereas SA usually acts in defense pathways distinct from, or even antagonistic to, those mediated by jasmonate and ethylene (Penninckx et al., 1996; Bowling et al., 1997). Although not explicitly tested here, the fact that axr1-24 is impaired in its response to ACC and SA raises the interesting possibility that AXR1 may help to integrate diverse defense pathways that are mediated by jasmonates, ethylene, and SA.
As for MeJA response, the effect of axr1-24 on response to other hormones was modest compared with auxin, which is consistent with earlier results for certain other axr1 alleles (Timpte et al., 1994; Nagpal et al., 2000; Rahman et al., 2001). The degree of insensitivity to ACC and ABA in axr1-24 was less than that found in the respective hormone response mutants used as positive controls in this study. Furthermore, decreased sensitivity in axr1-24 was seen even for some compounds not recognized to have specific hormone-like activity (Table II). In some of these responses, the effect of an AXR1 mutation may be indirect, rather than the direct result of a signaling defect. Previous studies found no difference between wild type and axr1 for GA-stimulated hypocotyl elongation (Collett et al., 2000), leading to the conclusion that auxin, ethylene, and GA independently control hypocotyl elongation. However, recent evidence suggests that the GA-response gene SLY1 is related to F-box proteins, raising the possibility that GA signaling also involves a ubiquitin-proteasome pathway with a potential role for AXR1 or a related protein (McGinnis et al., 2002).
In summary, we have provided new evidence that jasmonate signaling involves a ubiquitin-proteasome pathway. Furthermore, our results demonstrate that this pathway is dependent on a component of the RUB-activating enzyme, AXR1, which is shared with the auxin proteasome-signaling pathway. This result provides important new insight into the mechanistic basis for interactions between auxin and jasmonate signaling in plants.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis seeds were sown in Redi Earth (W.R. Grace, Cambridge, MA) in 8- × 8- × 8-cm plastic pots. Pots were covered with plastic film for 3 d and subsequently watered from above. Plants were grown at 21°C under continuous fluorescent illumination (approximately 100 μE m−2 s−1). All morphological measurements were made on 9-week-old plants as described (Lincoln et al., 1990). Pollen grains were counted with a hemocytometer, and the development of stamens and pistil were examined as described previously (Estelle and Somerville, 1987).
Seedling root growth was assayed on Murashige and Skoog basal-salt agar plates as described previously (Staswick et al., 1998). All hormones and compounds were diluted in ethanol and filter sterilized, except that SA was dissolved in ddH2O, BA in 1 n NaOH, and juglone and patulin in ethyl acetate. Each was added to sterile medium at the concentrations indicated for each experiment. The plates were chilled for 2 d at 4°C before being placed vertically in a continuously illuminated incubator at 21°C. The root length of 20 seedlings for each genotype was measured 10 d later, and each experiment was repeated at least four times. Germination on ABA was assayed on Murashige and Skoog agar medium at 21°C as described previously (Staswick et al., 1992).
Mutant Screening and Gene Mapping
Ethyl methanesulfonate-mutagenized-M2 seeds (Columbia) were purchased from Lehle Seeds (Round Rock, TX) and screened for MeJA insensitivity as described earlier (Staswick et al., 1992). Gene mapping was done by crossing the homozygous recessive mutant to Landsberg (NW151 and CS3078), and recessive MeJA-resistant F2 seedlings were rescued to soil. DNA from individual plants was assayed to assess the allelic status for each of several SSLP markers. An initial analysis of 133 plants placed the mutant locus between markers nga59 and nga63 on chromosome 1. Higher resolution mapping was done with 408 plants using new SSLP markers that were developed from the Cereon Genomics (Cambridge, MA) database of SNPs (http://www.Arabidopsis.org/cereon/index.html). Sequence intervals that included about 500 bp flanking the polymorphic sites were used to create primers using Seq Web, v1.2. Genomic DNA was isolated according to the methods of Yu and Pauls (1994) and was amplified using conditions previously described Bell and Ecker (1994).
Assay for Infection by Pythium irregulare
P. irregulare was grown and inoculated to soil containing 5-week-old wild-type and mutant plants using the same inoculation technique as described previously (Staswick et al., 1998) except that one agar plug was used for each seedling. Each pot contained nine seedlings, and 17 pots were used for each genotype. After inoculation, plants were returned to the growth chamber and monitored daily for symptoms of loss of turgor and tissue collapse.
RNA Isolation and Assay
Surface-sterilized seeds were germinated on sterilized 3MM gel-blot paper supported by a glass plate placed in a nearly vertical orientation in Magenta-boxes that contained 50 mL of liquid Murashige and Skoog basal-salt medium. Seedlings were grown 2 weeks under the same conditions as for root assays and then treated either by spraying with 1 μm IAA or with MeJA volatilized from 3MM paper taped to the inside of the container lid. Boxes were returned to the incubator and tissue samples were collected at the time intervals indicated and frozen at −80°C. Total RNA was isolated, and hybridizations were at 42°C and washes at 0.1× SSC at 62 C as described before (Staswick et al., 1998).
All probes for hybridization were generated with [32P]dCTP by random primer labeling using gel-purified DNA. The IAA1 probe was generated using PCR primers CGGAGCACAAGAAGAAC AAC (forward) and ATGGAACATCACCGACCAAC (reverse) based on the sequence of accession no. L15448. The probes for Arabidopsis allene oxide synthase (AOS) and vegetative storage protein (AtVSP) were obtained from mRNA by reverse transcriptase-PCR using primers based on the published sequences (accession no. AB007647; Staswick, 1999). The LOX2 probe was generated by PCR from the cloned Arabidopsis cDNA (Bell and Mullet, 1993). Equal loading of RNA was verified by ethidium bromide staining and by rehybridizing the blots with an 18S rDNA probe.
Generation of jar1/axr1 Double Mutant
axr1 homozygotes were identified among F2 progeny of a cross between axr1-3 and jar1-1 by their distinct abnormal development compared with jar1-1 (Lincoln et al., 1990; Staswick et al., 1992). The genotype of the JAR1 locus in putative double mutants was established with a CAPS marker for the mutant allele (P.E. Staswick, unpublished data). The double mutant was tested on both IAA- and MeJA-containing medium as described previously.
ACKNOWLEDGMENTS
Lines NW151, axr1-3, etr1, and abi1 were obtained from the Arabidopsis Biological Resource Center. The LOX2 cDNA clone was kindly provided by J. Mullet. The technical assistance of Martha Rowe is greatly appreciated.
Footnotes
This work was supported by the Nebraska Research Initiative and by the University of Nebraska Center for Biotechnology. This paper is a contribution of the University of Nebraska Agricultural Research Division (Lincoln). This is journal series no. 13,647.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.005272.
LITERATURE CITED
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Bell E, Mullet JE. Characterization of an Arabidopsis lipoxygenase gene responsive to methyl jasmonate and wounding. Plant Physiol. 1993;103:1133–1137. doi: 10.1104/pp.103.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S, Bell E, Mullet JE. Two methyl jasmonate-insensitive mutants show altered expression of AtVsp in response to methyl jasmonate and wounding. Plant Physiol. 1996;111:525–531. doi: 10.1104/pp.111.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S, Bell E, Sadka A, Mullet JE. Arabidopsis thaliana Atvsp is homologous to soybean VspA and VspB, genes encoding vegetative storage protein acid phosphatases, and is regulated similarly by methyl jasmonate, wounding, sugars, light and phosphate. Plant Mol Biol. 1995;27:933–942. doi: 10.1007/BF00037021. [DOI] [PubMed] [Google Scholar]
- Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X. The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell. 1997;9:1573–1584. doi: 10.1105/tpc.9.9.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collett CE, Harberd NP, Leyser O. Hormonal interactions in the control of Arabidopsis hypocotyl elongation. Plant Physiol. 2000;124:553–562. doi: 10.1104/pp.124.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies PJ. The plant hormones: their nature, occurrence and functions. In: Davies PJ, editor. Plant Hormones: Physiology, Biochemistry and Molecular Biology. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1–12. [Google Scholar]
- del Pozo JC, Dharmasiri S, Hellmann H, Walker L, Gray WM, Estelle M. AXR1-ECR1-dependent conjugation of RUB1 to the Arabidopsis cullin AtCUL1 is required for auxin response. Plant Cell. 2002;14:421–433. doi: 10.1105/tpc.010282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Timpte C, Tan S, Callis J, Estelle M. The ubiquitin-related protein RUB1 and auxin response in Arabidopsis. Science. 1998;280:1760–1763. doi: 10.1126/science.280.5370.1760. [DOI] [PubMed] [Google Scholar]
- Estelle MA, Somerville C. Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Mol Gen Genet. 1987;206:200–206. [Google Scholar]
- Farmer EE, Ryan CA. Interplant communication: airborne methyl jasmonate induces synthesis of proteinase inhibitors in plant leaves. Proc Natl Acad Sci USA. 1990;87:7713–7716. doi: 10.1073/pnas.87.19.7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJF, Benedetti CE, Penfold CN, Turner JG. Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistance to a bacterial pathogen. Plant Cell. 1994;6:751–759. doi: 10.1105/tpc.6.5.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Estelle I. Function of the ubiquitin-proteasome pathway in auxin response. Trends Biochem Sci. 2000;25:133–138. doi: 10.1016/s0968-0004(00)01544-9. [DOI] [PubMed] [Google Scholar]
- Hobbie L, Estelle M. Genetic approaches to auxin action. Plant Cell Environ. 1994;17:525–540. doi: 10.1111/j.1365-3040.1994.tb00147.x. [DOI] [PubMed] [Google Scholar]
- Laudert D, Weiler EW. Allene oxide synthase: a major control point in Arabidopsis thaliana octadecanoid signaling. Plant J. 1998;15:675–684. doi: 10.1046/j.1365-313x.1998.00245.x. [DOI] [PubMed] [Google Scholar]
- Lincoln C, Britton JH, Estelle M. Growth and development of the axr1 mutants of Arabidopsis. Plant Cell. 1990;2:1071–1080. doi: 10.1105/tpc.2.11.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn M, Browse J. The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell. 1996;8:403–406. doi: 10.1105/tpc.8.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn M, Creelman RA, Bell E, Mullet JE, Browse J. Jasmonate is essential for insect defense in Arabidopsis. Proc Natl Acad Sci USA. 1997;94:5473–5477. doi: 10.1073/pnas.94.10.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis KM, Soule JM, Zale J, Steber CM. Control of GA signaling by SLY1, a putative F-box subunit of an E3 SCF ubiquitin ligase (abstract no. 483). American Society of Plant Biologists 2002 Meeting Abstracts, http://abstracts.aspb.org/pb2002/public/P57/0361.html. 2002. p. 118. [Google Scholar]
- Mueller MJ. Enzymes involved in jasmonic acid biosynthesis. Physiol Plant. 1997;100:653–663. [Google Scholar]
- Nagpal P, Walker LM, Young JC, Sonawala A, Timpte C, Estelle M, Reed JW. AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 2000;123:563–574. doi: 10.1104/pp.123.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmyer K, Tuominen H, Kettunen R, Betz C, Langebartels C, Sandermann H, Jr, Kangasjarvi J. Ozone-sensitive Arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signaling pathways in regulating superoxide-dependent cell death. Plant Cell. 2000;12:1849–1862. doi: 10.1105/tpc.12.10.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, Johansen B, Nielsen HB, Lacy M, Austin MJ, Parker JE et al. Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell. 2000;103:1111–1120. doi: 10.1016/s0092-8674(00)00213-0. [DOI] [PubMed] [Google Scholar]
- Penninckx IA, Eggermont K, Terras FR, Thomma BP, De Samblanx GW, Buchala A, Metraux JP, Manners JM, Broekaert WF. Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell. 1996;8:2309–2323. doi: 10.1105/tpc.8.12.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A, Amakawa T, Goto N, Tsurumi S. Auxin is a positive regulator for ethylene-mediated response in the growth of Arabidopsis roots. Plant Cell Physiology. 2001;42:301–307. doi: 10.1093/pcp/pce035. [DOI] [PubMed] [Google Scholar]
- Rao MV, Lee H, Creelman RA, Mullet JE, Davis KR. Jasmonic acid signaling modulates ozone-induced hypersensitive cell death. Plant Cell. 2000;12:1633–1646. doi: 10.1105/tpc.12.9.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev. 1998;12:198–207. doi: 10.1101/gad.12.2.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders PM, Lee PY, Biesgen C, Boone JD, Beals TP, Weiler EW, Goldberg RB. The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell. 2000;12:1041–1061. doi: 10.1105/tpc.12.7.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Song JT, Cheong JJ, Lee YH, Lee YW, Hwang I, Lee JS, Choi YD. Jasmonic acid carboxyl methyltransferase: a key enzyme for jasmonate-regulated plant responses. Proc Natl Acad Sci USA. 2001;98:4788–4793. doi: 10.1073/pnas.081557298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE. Sequence of an allene oxide synthase cDNA from Arabidopsis thaliana (accession no. AF172727) Plant Physiol. 1999;121:312. [Google Scholar]
- Staswick PE, Su W, Howell S. Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc Natl Acad Sci USA. 1992;89:6837–6840. doi: 10.1073/pnas.89.15.6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I, Rowe M. The jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell. 2002;14:1405–1415. doi: 10.1105/tpc.000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Yuen GY, Lehman CC. Jasmonate signaling mutants of Arabidopsis are susceptible to the soil fungus Pythium irregulare. Plant J. 1998;15:747–754. doi: 10.1046/j.1365-313x.1998.00265.x. [DOI] [PubMed] [Google Scholar]
- Stintzi A, Browse J. The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci USA. 2000;97:10625–10630. doi: 10.1073/pnas.190264497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A, Weber H, Reymond P, Browse J, Farmer EE. Plant defense in the absence of jasmonic acid: the role of cyclopentenones. Proc Natl Acad Sci USA. 2001;98:12837–12842. doi: 10.1073/pnas.211311098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpte C, Lincoln C, Pickett FB, Turner J, Estelle M. The AXR1 and AUX1 genes of Arabidopsis function in separate auxin-response pathways. Plant J. 1995;8:561–569. doi: 10.1046/j.1365-313x.1995.8040561.x. [DOI] [PubMed] [Google Scholar]
- Timpte C, Wilson AK, Estelle M. The axr2-1 mutation of Arabidopsis thaliana is a gain-of-function mutation that disrupts an early step in auxin response. Genetics. 1994;138:1239–1249. doi: 10.1093/genetics/138.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titarenko E, Rojo E, Leon J, Sanchez-Serrano JJ. Jasmonic acid-dependent and -independent signaling pathways control wound-induced gene activation in Arabidopsis thaliana. Plant Physiol. 1997;115:817–826. doi: 10.1104/pp.115.2.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayan P, Shockey J, Levesque CA, Cook RJ, Browse J. A role for jasmonate in plant defense of Arabidopsis. Proc Natl Acad Sci USA. 1998;95:7209–7214. doi: 10.1073/pnas.95.12.7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Vick BA, Farmer EE. Dinor-oxo-phytodienoic acid: a new hexadecanoid signal in the jasmonate family. Proc Natl Acad Sci USA. 1997;94:10473–10478. doi: 10.1073/pnas.94.19.10473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AK, Pickett FB, Turner JC, Estelle M. A dominant mutation in Arabidopsis confers resistance to auxin, ethylene and abscisic acid. Mol Gen Genet. 1990;222:377–383. doi: 10.1007/BF00633843. [DOI] [PubMed] [Google Scholar]
- Xie D-X, Feys BF, Nieto-Rostro M, Turner JG. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science. 1998;280:1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- Yeh ET, Gong L, Kamitani T. Ubiquitin-like proteins: new wine in new bottles. Gene. 2000;248:1–14. doi: 10.1016/s0378-1119(00)00139-6. [DOI] [PubMed] [Google Scholar]
- Yu K, Pauls KP. Optimization of DNA-extraction and PCR procedures for random amplified polymorphic DNA (RAPD) analysis in plants. In: Griffin HG, Griffin AM, editors. PCR Technology: Current Innovations. London: CRC Press; 1994. pp. 193–200. [Google Scholar]