Abstract
The protein content of tomato (Lycopersicon esculentum) xylem sap was found to change dramatically upon infection with the vascular wilt fungus Fusarium oxysporum. Peptide mass fingerprinting and mass spectrometric sequencing were used to identify the most abundant proteins appearing during compatible or incompatible interactions. A new member of the PR-5 family was identified that accumulated early in both types of interaction. Other pathogenesis-related proteins appeared in compatible interactions only, concomitantly with disease development. This study demonstrates the feasibility of using proteomics for the identification of known and novel proteins in xylem sap, and provides insights into plant-pathogen interactions in vascular wilt diseases.
In land plants, xylem sap plays an essential role in the supply of water and mineral salts to the aerial tissues (De Boer and Volkov, 2003). It also constitutes an environment in which microorganisms can thrive, be it endophyte or pathogen. Through colonization of xylem vessels, pathogenic fungi or bacteria cause vessel clogging leading to wilting of the plant. Verticillium spp. and Fusarium oxysporum are xylem-colonizing fungi that cause important diseases in crops (Tjamos and Beckman, 1989). Other vascular wilt fungi are responsible for tree diseases such as the devastating Dutch elm disease (Hubbes, 1999; http://www.dutchelmdisease.org). As far as molecular analysis of fungal wilt diseases is concerned, the interaction between tomato (Lycopersicon esculentum) and the host-specific Fusarium oxysporum f. sp. lycopersici is currently one of the best-studied model systems.
Resistance and susceptibility of tomato toward F. oxysporum is at least partly determined by interactions occurring within xylem vessels. In an incompatible interaction, the fungus is apparently contained within the vessel it has invaded, whereas in a compatible interaction, it invades neighboring parenchyma tissue and spreads laterally to other vessels, eventually colonizing the entire vascular system (Gao et al., 1995; Mes et al., 2000). Furthermore, the only dominant resistance gene against F. oxysporum that has been cloned was shown to be expressed specifically in xylem parenchyma cells that are in contact with vessels (Simons et al., 1998; Mes et al., 2000). It is therefore plausible that in an incompatible interaction, recognition of a fungal component takes place by these cells as soon as the fungus enters the vessel, leading to effective defense responses.
One of the responses to pathogen attack commonly observed is the production of so-called pathogenesis-related (PR) proteins, many of which have antimicrobial activity (Kitajima and Sato, 1999; Van Loon and Van Strien, 1999). The vast majority of studies related to antimicrobial defense of plants deals with leaf pathogens; little is known about proteins secreted in xylem sap after invasion by pathogens. In the case of citrus trees affected by citrus blight, increased levels of several peroxidases (Nemec, 1995) and an expansin (Ceccardi et al., 1998) were associated with disease development. In rice (Oryza sativa), a peroxidase accumulates in xylem vessels in response to invasion by Xanthomonas oryzae (Young et al., 1995).
To obtain a more comprehensive overview of the response of a plant to xylem invasion, we initiated an analysis of the changes in xylem sap protein content of tomato upon infection with F. oxysporum. Individual proteins that accumulated upon infection were identified using mass spectrometry (MS). Our results demonstrate that fungal colonization of xylem vessels triggers a response that partly overlaps with the response to leaf colonization, but which also has unique features such as the accumulation of a basic glucanase and a novel member of the PR-5 family.
RESULTS
Changes in Xylem Sap Protein Content after Infection with F. oxysporum
Before looking at the consequences of F. oxysporum infection, the protein content of xylem sap obtained from healthy plants was investigated. Xylem sap was collected from stems of 5-week-old tomato plants that were cut off below the second true leaf (see “Materials and Methods”). The first 3 mL of sap generally contained between 30 and 70 μg mL−1 protein. When sap yield was higher (up to 10 mL), overall protein concentration was in the range of 20 to 30 μg mL−1. This may be attributable to the experimental setup: Cutting the stem leads to an increase in sap stream, which may cause dilution of xylem sap constituents (Liang and Zhang, 1997). SDS-PAGE and silver staining of sap proteins revealed the presence of a prominent 10-kD species and many minor bands in the 20- to 60-kD range. Identical protein patterns were observed in mock-inoculated plants (Fig. 1, lanes C).
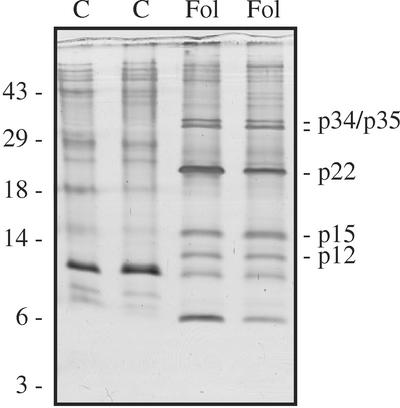
Figure 1.
F. oxysporum infection causes accumulation of disease-related proteins in tomato xylem sap. Five-week-old GCR161 plants were either mock-inoculated (C) or inoculated with the compatible race 2 isolate Fol007 (Fol). After 3 weeks, when F. oxysporum-inoculated plants showed severe disease symptoms, sap was collected from individual plants, concentrated and analyzed with SDS-PAGE on a Tris-Tricine gel. Proteins were visualized by silver staining. Lanes represent sap from different plants. Molecular masses of marker proteins are indicated on the left (in kD). The most abundant disease-related proteins are indicated on the right, designated according to their estimated sizes.
To investigate whether disease-related proteins accumulate in xylem sap after colonization by F. oxysporum, 5-week-old plants were root-inoculated with the compatible race 2 isolate Fol007. Sap was collected at 3 weeks after infection, at which time the plants showed severe disease symptoms. As shown in Figure 1, at least five disease-related proteins in the range of 10 to 40 kD appeared in this interaction (lanes Fol; the band at 6 kD was not always clearly observed, see Fig. 2). At higher molecular masses we did not observe consistent differences between protein patterns of F. oxysporum-infected and mock-infected plants. Having established that new proteins accumulate in xylem sap during F. oxysporum colonization, we proceeded to investigate the timing of appearance of these proteins in compatible and incompatible interactions. Very little difference with control plants was seen in infected plants at 4 d after inoculation (not shown). After 1 week, however, the 22-kD protein appeared in both compatible and incompatible interactions (Fig. 2). At later stages of infection, disease-related proteins of 12, 15, 34, and 35 kD accumulated only in compatible interactions. The level of a 10-kD protein, present in uninfected plants, conversely decreased during compatible interactions. The timing of these events coincided with visible disease symptoms.
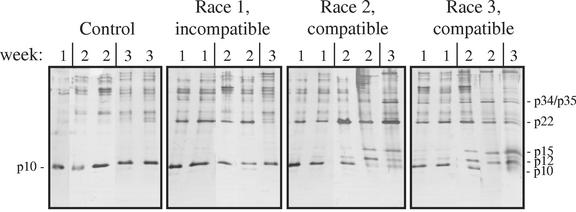
Figure 2.
Time-dependent accumulation of disease-related proteins in compatible and incompatible interactions. GCR161 plants were mock-inoculated (Control) or inoculated with the incompatible race 1 isolate Fol004, the compatible race 2 isolate Fol007, or the compatible race 3 isolate Fol029. Sap was collected at 1, 2, or 3 weeks after inoculation and analyzed as described in Figure 1. The disease-related proteins p12, p15, p22, p34, and p35 are indicated, as is the p10 protein present in healthy plants.
When the F. oxysporum isolate used for the incompatible interaction (Fol004) was used to infect the susceptible plant line C32, severe disease symptoms ensued, and disease-related xylem sap proteins appeared that were indistinguishable from the ones shown in Figure 2 (results not shown). Thus, the differences observed between the compatible and incompatible interactions cannot be ascribed to different fungal races producing different proteins in planta.
Identification of Xylem Sap Proteins
To investigate whether the disease-related proteins in xylem sap are identical to proteins already identified in other tomato-pathogen interactions or still unknown proteins secreted by either plant or fungus, we used MS to obtain sequence information. Proteins were digested in gel with trypsin and a mass spectrum of the resulting peptides (a peptide mass fingerprint) was acquired with a matrix-assisted laser-desorption ionization time of flight (MALDI-TOF) mass spectrometer. The list of apparent peptide masses was then used to screen databases for correspondence to predicted tryptic digests of known proteins. When enough material could be obtained, individual peptides were selected for sequence analysis with tandem MS, either to confirm a putative identity or to obtain “sequence tags” allowing additional database searches. The 12-kD band contained a protein of fungal origin; its characterization will be reported elsewhere. The identification of the remaining proteins is described below.
p15 Contains Isoforms of PR-1
The peptide mass fingerprint of p15 corresponds to PR-1a and PR-1b, two secreted isoforms of PR protein 1 sharing 97% sequence identity (Table I). Sequence information was obtained with tandem MS (MS/MS) of four peptides. These peptides fully match the PR-1b sequence and cover 47% of the mature, 135-amino acid protein. Two of these peptides match PR-1a as well (Table I). The two putative PR-1a-specific peptides unfortunately could not be analyzed with MS/MS because of low abundance or poor ionization. However, the presence of the expected masses in the peptide mass fingerprint and the previous demonstration that PR-1a and PR-1b accumulate together in tomato leaves infected with Cladosporium fulvum (Joosten et al., 1990) makes it highly likely that p15 contains both PR-1 isoforms. Interestingly, both the overall mass and the MS/MS spectrum of the amino-terminal peptide (which is the same for PR-1a and PR-1b) confirm the previous identification of pyro-Glu at the N terminus resulting from cyclization of the N-terminal Gln (Lucas et al., 1985). Although such modifications decrease the chance of finding correct matches in databases with peptide mass lists, this example demonstrates that the information retrieved from mass spectra goes beyond primary sequence information.
Table I.
Predicted tryptic peptides of PR-1a and/or PR-1b that were detected by MALDI-TOF mass spectrometry of p15
| MH+ Iona | Position in PR-1ab | Position in PR-1bb | Sequence | Confirmation with MS/MS? |
|---|---|---|---|---|
| 923.43 | 33–40 | 33–40 | AQNYANSR | |
| 1,088.56 | 93–100 | HYTQVVWR | Yes | |
| 1,404.70 | 91–100 | CRHYTQVVWRc | ||
| 1,436.66 | 90–100 | MCGHYTQVVWR | ||
| 1,710.78 | 1–15 | 1–15 | [pGlu]NSPQDYLAVHNDARd | Yes |
| 1,726.82 | 41–57 | 41–57 | AGDCNLIHSGAGENLAK | |
| 1,758.86 | 16–32 | 16–32 | AQVGVGPMSWDANLASR | Yes |
| 2,698.30 | 66–89 | AAVQLWVSERPSYNYATNQCVGGK | Yes | |
| 2,726.30 | 66–89 | AAVQLWVSERPDYNYATNQCVGGK |
m/z value of single-protonated peptides, with Cys modified by iodoacetamide.
Position of tryptic peptide in mature protein.
One missed cleavage.
pGlu, Pyroglutamate.
p34 and p35 Are β-1,3-Glucanases
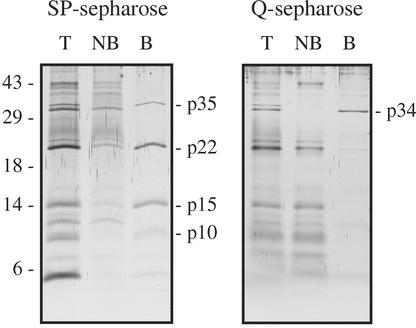
Peptide mass fingerprints of p34 and p35 unambiguously identified these proteins as previously described β-1,3-glucanases (PR-2 proteins; Tables II and III). p34 is identical to the 35-kD acidic glucanase PR-2a, which accumulates in leaf apoplast during infection by the leaf mold fungus C. fulvum (Joosten and De Wit, 1989; Van Kan et al., 1992). Its acidic nature is supported by its ability to bind positively charged (Q) Sepharose (Fig. 3). It is the only acidic PR protein detected in xylem sap because all other disease-related proteins bind to negatively charged (SP) Sepharose, implying a basic nature (Fig. 3).
Table II.
Predicted peptides of the translation product of PR-Q'ba that were detected by MALDI-TOF mass spectrometry of p35
| MH+ Ionb | Positionc | Sequence |
|---|---|---|
| 923.40 | 1–8 | [pGlu]TGVCYGRd |
| 932.52 | 156–163 | GYVDPIIR |
| 1,007.53 | 257–264 | TYNNNLIR |
| 1,088.56 | 298–306 | HFGLFTPNR |
| 1,551.83 | 33–45 | IYDPHQPTLQALR |
| 1,597.87 | 114–129 | NIQNAISGAGLGNQIK |
| 1,753.86 | 9–25 | NGNGLPSPADVVALCNRe |
| 1,783.91 | 211–226 | NLFDALLDATYSALEK |
| 1,936.98 | 190–207 | LDYALFTSPGVVVNDNGRe |
| 1,980.99 | 130–147 | VSTAIETELTTDTYPPSR |
| 2,250.23 | 273–291 | RPSKPIEAYIFALFNENLK |
| 2,836.45 | 88–113 | YIAVGNEVSPLNGNAQYVPFVINAMRe |
| 2,902.42 | 227–256 | AGGSSLDIVVSESGWPSAGAGQLTSIDNAR |
| 3,616.85 | 46–79 | GSNIELILGVPNPDLQNIASSQANANAWVQNNVR |
TC94164 of the TIGR Lycopersicon Gene Index.
m/z value of single-protonated peptides, with Cys modified by iodoacetamide.
Position of tryptic peptide in predicted mature protein.
Conversion of Gln to pyroglutamate (pGlu) assumed.
Underlined Asn mostly deamidated (see text).
Table III.
Predicted peptides of acidic glucanase PR-2aa that were detected by MALDI-TOF mass spectrometry of p34
| MH+ Ionb | Positionc | Sequence |
|---|---|---|
| 1,289.65 | 89–100 | YIAVGNEVDPGR |
| 1,363.77 | 157–168 | SFINPIIGFLSR |
| 1,426.75 | 10–22 | IANNLPSDQDVIK |
| 1,555.82 | 34–46 | IYFPETNVFNALK |
| 1,749.82 | 274–287 | TIETYLFAMFDENR |
| 1,867.00 | 131–147 | VSTATYLGLLTNTYPPR |
| 3,212.66 | 169–196 | HNLPLLANIYPYFGHADDNVPLPYALFK |
As encoded by TC94480 of the TIGR Lycopersicon Gene Index.
m/z value of single-protonated peptides, with Cys modified by iodoacetamide.
Position of tryptic peptide in predicted mature protein.
Figure 3.
Most disease-related proteins bind to negatively charged Sepharose. After pH adjustment, xylem sap of tomato collected 3 weeks after infection with a compatible race of F. oxysporum (Fol007) was incubated with either SP- or Q-Sepharose with affinity for basic or acidic proteins, respectively. Proteins were washed off the Sepharose beads with 250 mm NaCl. Proteins were separated in Tris-Tricine gels and silver-stained. T, Total sap protein; NB, non-bound fraction; B, bound fraction.
p35 corresponds to the translation product of PR-Q'b, an mRNA that accumulates in tomato leaves upon infection by citrus exocortis viroid (Domingo et al., 1994). Interestingly, this protein does not accumulate in tomato leaf apoplast after colonization by C. fulvum (Joosten and De Wit, 1989), indicating that the xylem and leaf (mesophyll) apoplastic spaces, although both extracellular compartments, show differential responses to pathogen challenge. However, a basic glucanase does accumulate in total leaf homogenates after C. fulvum infection. Published peptide sequences of this basic glucanase (Van Kan et al., 1992) match the primary structure of the PR-Q'b translation product and hence p35. Its presence in leaf homogenate most likely reflects its accumulation in leaf xylem (see “Discussion”).
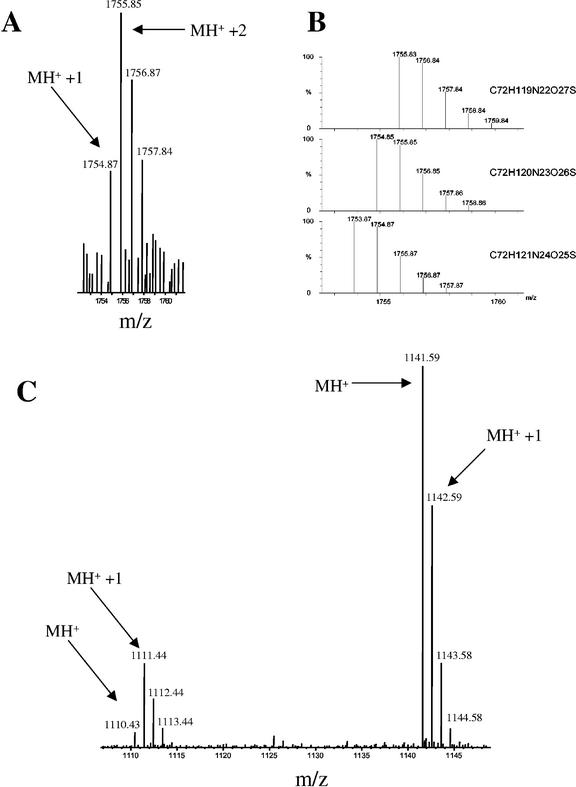
Some details of the mass spectrum of p35 are worth mentioning. One is the assignment of one peak (at m/z = 923.4) to the N-terminal peptide assuming modification of the N-terminal Gln to pyro-Glu, as in the PR-1 proteins described above. Such a modification is consistent with the predicted cleavage site of the signal peptide and the resistance of the mature N terminus to Edman degradation (Van Kan et al., 1992). Another remarkable observation is that four Asn residues in three p35-derived peptides appeared to be mostly deamidated, causing a 1-D increase in mass (Table II; Fig. 4). Such a modification is known to occur in vitro or in vivo especially when an Asn is followed by a Gly residue (Meinwald et al., 1986; Emslie et al., 2000) and apparently occurs in all four NG-pairs present in p35 (Table II). Taking into account these posttranslational modifications, all peptide peaks larger than 900 D in the MALDI-TOF spectrum were accounted for.
Figure 4.
Evidence for Asn deamidation. Shown are two examples of peptides whose molar mass distribution corresponds to partial deamidation of Asn residues. A, This cluster of peaks in a MALDI-TOF spectrum of p35 corresponds to the tryptic peptide NGNGLPSPADVVALCNR (predicted m/z of 12C monoisotopic peptide: 1,753.86 D) with partial deamidation of Asn residues in both Asn-Gly pairs. At this peptide molar mass size, the 12C monoisotopic peptide peak (MH+) should be at least equal to the one with one 13C atom (MH+ + 1), as shown in B. The higher abundance of the MH+ + 1 and MH+ + 2 peaks and the presence of two Asn-Gly pairs in the peptide suggests deamidation of Asn leading to 1 D mass increase per Asn. B, Predicted isotope distributions of the peptide described in A (lower trace) and with one (middle trace) or two (upper trace) Asn residues deamidated. Chemical formulae used for the calculations are shown next to the traces. From this peak shape, it was concluded that one and in some cases two Asn residues were deamidated. C, In this MALDI-TOF spectrum of p22, the cluster of peaks at the left corresponds to TNCNFNGAGR of PR-5x (predicted m/z of 12C monoisotopic peptide: 1,110.48 D). At this peptide molar mass size, the 12C monoisotopic peptide peak (MH+) should be higher than the one with one 13C atom (MH+ + 1), as seen for the larger peptide on the right (predicted m/z: 1,141.61). The higher abundance of the MH+ + 1 peak and the presence of an Asn-Gly pair in the peptide strongly suggests deamidation of Asn.
P22 Is a New PR-5 Isoform
Database searches with tryptic peptide masses of p22 yielded two PR-5 proteins as candidates: Two peptides of the peptide mass fingerprint match the vacuolar PR-5 protein NP24 (King et al., 1988), and four peptides match the highly related AP24 (=p23/NP24-II/TPM1; Rodrigo et al., 1991; Woloshuk et al., 1991; Ruiz-Medrano et al., 1992; Table IV). However, four prominent peptides in the spectrum could not be matched, and many predicted peptides from both NP24 and AP24 were not seen in the spectrum, casting some doubt on the identification. For p22, sequence tags obtained with MS/MS proved to be important for the identification of the real coding sequence, which differed from the ones in the database. Four peptides of p22 were sequenced with MS/MS, one of which was the carboxyl-terminal peptide (not visible in the MALDI-TOF spectrum because of interference of matrix material; see Fig. 5). Two of the internal peptides are identical with predicted peptides of NP24 and AP24 (T2 and T5 in Fig. 5). However, the third internal peptide (T16) differed at two positions from both proteins. Furthermore, the length of the carboxyl-terminal peptide (T17) did not match the expected C terminus of either NP24 or AP24, based on the experimentally determined C terminus of the highly similar tobacco (Nicotiana tabacum) AP24 resulting from cleavage of the vacuolar targeting sequence (Melchers et al., 1993). In fact, the C-terminal sequence of p22 is similar to that of secreted PR-5 proteins in tobacco (Cornelissen et al., 1986; Pierpoint and Tatham, 1987) and Arabidopsis (Uknes et al., 1992). Together with the extracellular location of p22, these observations made us suspect that p22 was in fact a new, secreted isoform of PR-5.
Table IV.
Predicted peptides of PR-5x that were detected by MALDI-TOF mass spectrometry of p22
| MH+ Iona | Positionb | No.c | Sequence | Also Present in Isoforms: | Confirmation with MS/MS? |
|---|---|---|---|---|---|
| 1,110.48 | 49–58 | T9 | TNCNFNGAGRd | ||
| 1,141.61 | 29–38 | T5 | GQTWVINAPR | AP24, NP24 | Yes |
| 1,525.82 | 26–38 | T4–5 | LDRGQTWVINAPR | AP24 | |
| 1,681.93 | 25–38 | T3–5 | RLDRGQTWVINAPR | AP24 | |
| 1,920.90 | 7–24 | T2 | NNCPYTVWAASTPIGGGR | AP24, NP24 | Yes |
| 2,206.99 | 120–138 | T11 | CHAIHCTANINGECPSPLRd | ||
| 2,998.26 | 175–200 | T16 | CPNAYSYPQDDPTSLFTCPSGSTNYR | Yes | |
| 3,103.26 | 139–166 | T12 | VPGGCNNPCTTFGGQQYCCTQGPCGPTK |
m/z value of single-protonated peptides, with Cys modified by iodoacetamide.
Position of tryptic peptide in predicted mature protein.
Predicted tryptic peptides are numbered from the amino terminus.
Underlined Asn mostly deamidated (see text).
Figure 5.
The p22 band contains a novel PR-5 protein. The vacuolar PR-5 proteins NP24 (accession no. P12670) and AP24 (the full translation product of The Institute for Genomic Research [TIGR] tentative consensus sequence TC52651 is shown) are aligned with the putative translation product (PR-5x) of the newly identified cDNA. Asterisks below the alignment indicate divergence in sequence between the proteins. White triangles above the aligned sequences indicate carboxyl-terminal amino acids of peptides predicted for a trypsin digest of any of the proteins in the alignment. Predicted tryptic peptides of PR-5x are numbered (T1–17). Peptides corresponding to MS/MS sequences are in bold. Peptides whose mass corresponds to peaks in the peptide mass fingerprint are underlined (including peptides T3–5 and T4–5 with missed cleavages; see Table IV). Cleavable N- and C-terminal signal sequences are in lowercase. Mature N termini were confirmed experimentally for NP24 (King et al., 1988) and AP24 (=P23; Rodrigo et al., 1991; Woloshuk et al., 1991); mature C termini are predicted by comparison with tobacco AP24 (Melchers et al., 1993).
The cDNA for Xylem Sap PR-5
No gene or cDNA encoding a PR-5 isoform with a C terminus as found in the 22-kD xylem sap protein was present in sequence databases. However, in a screen for F. oxysporum-infection-specific cDNA-amplified fragment-length polymorphism (AFLP) fragments, one cDNA fragment was found that could encode the C-terminal part of p22 (M. Haring, S. de la Fuente van Bentem, and B.J.C. Cornelissen, unpublished data). To establish whether this fragment was part of the coding sequence for p22, we set out to isolate the remaining part of the cDNA to compare the sequence with the mass spectrometric data. Using a cDNA library made from roots and hypocotyls of a compatible F. oxysporum-tomato interaction (see “Materials and Methods”), we amplified the full coding sequence with primers specific for the AFLP fragment. The new sequence is very closely related to the AP24 and NP24 coding sequences but is clearly different and encodes a new PR-5 isoform (Fig. 6). Henceforth, we refer to this protein as PR-5x (PR-5 of xylem sap). PR-5x matches the same two MS/MS sequence tags as the vacuolar isoforms. However, in contrast to the vacuolar proteins, the third internal peptide matches as well, as does the C-terminal peptide (Fig. 5). Moreover, three additional peptides in the peptide mass fingerprint that did not match with NP24 or AP24 correspond to peptides that are unique for PR-5x (Table IV; Fig. 5, T9, T11, and T12). We are therefore confident that the identified cDNA encodes the 22-kD protein found in xylem sap of infected tomato plants. As in p35, all Asn residues that are followed by a Gly notably appeared to be mostly deamidated (Table IV; Fig. 4).
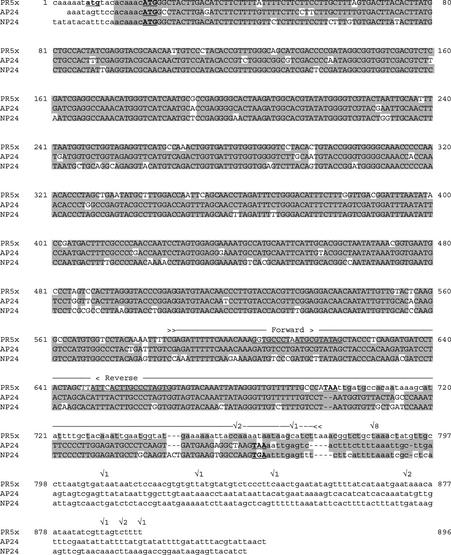
Figure 6.
The coding sequence of PR-5x is highly similar to coding sequences of vacuolar PR-5 proteins. PR-5x encoding cDNA (GenBank accession no. AY093595) is aligned with coding sequences for the vacuolar PR-5 proteins NP24 (accession no. AF093743; Jia et al., 2000) and AP24 (Ruiz-Medrano et al., 1992; Rodrigo et al., 1993); the sequence shown here is TC52651 from the TIGR Lycopersicon Gene Index, which includes the start codon. The line above PR-5x cDNA between arrow heads (≫–≪) indicates the sequence of a tomato-F. oxysporum-interaction-specific cDNA-AFLP fragment. Stop codons and potential start codons are bold and underlined. Coding sequences are in uppercase. Poly(A) addition sites, based on the 3′ ends of PR-5x cDNA clones, are indicated (✓), with numbers corresponding to the number of cDNAs ending there. Underlined sequences in PR-5x correspond to forward and reverse primers used for cloning of overlapping fragments of the PR-5x cDNA.
Relationship of PR-5x to Other PR-5 Proteins
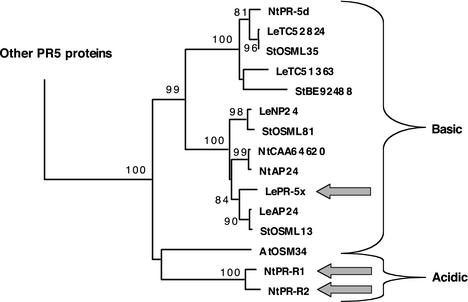
As mentioned above, PR-5x is very similar in sequence to NP24 and especially AP24 (91% and 93% identity, respectively). This is reflected in a phylogenetic tree of PR-5 proteins (Fig. 7). The three proteins form a separate cluster together with two proteins from tobacco and two from potato (Solanum tuberosum). Interestingly, this group is part of a larger cluster with additional Solanaceae proteins and only one ortholog from Arabidopsis (AtOSM34) from among the 22 sequences found in the Arabidopsis genome (Fig. 7). Strong diversification has apparently occurred in this particular group of PR-5 proteins in Solanaceae. The similarity of PR-5x to NP24 and AP24 coding sequences suddenly disappears beyond the stop codon of PR-5x, suggesting that PR-5x was relatively recently derived from the ancestor of AP24 through a recombination event near the 3′ end of the coding sequence (Fig. 6).
Figure 7.
PR-5x belongs to a subgroup of PR-5 proteins that diversified in Solanaceae. All available full-length (predicted) protein sequences of PR-5 proteins of Arabidopsis (At), tomato (Le), tobacco (Nt), and potato (St), were aligned. Additions to the core PR-5 consensus sequence at N termini (signal sequences) and C termini (vacuolar targeting sequences or other extensions) were trimmed. This alignment was used to construct a phylogenetic tree. Only the clade containing PR-5x is shown. Published sequences are referred to by protein names: NP24 (accession no. P12670) and AP24 (accession no. CAA50059; complete sequence derived from TC52651 of the TIGR Lycopersicon Gene Index) from tomato; OSML13 (accession no. P50701), OSML35 (accession no. P50703), and OSML81 (accession no. P50702) from potato; AP24 (“osmotin”, accession no. P14170), PR-5d (accession no. BAA11180), PR-R1 (“major isoform”, accession no. P13046), and PR-R2 (“minor isoform”, accession no. P07052) from tobacco and AtOSM34 (accession no. CAA61411) from Arabidopsis. Remaining sequences are either derived from tentative consensus (TC) sequences in TIGR databases (Quackenbush et al., 2001) or database accessions. All sequence names are preceded by species abbreviations. Bootstrap percentages are provided for branches receiving 70% or more support. Branch length reflects the extent of sequence divergence. Thick arrows indicate secreted isoforms. All other proteins are known or predicted to be vacuolar (based on C-terminal propeptides). Only the two PR-R isoforms of tobacco are acidic.
Immunodetection of a PR-3 Isoform
One class of PR proteins that was not abundantly present in xylem sap of F. oxysporum-infected tomato is PR-3 (chitinase). This was somewhat surprising because PR-3 proteins are commonly found in tomato-pathogen interactions. To see whether a PR-3 protein is present in xylem sap after F. oxysporum infection, immunoblotting with an antibody against tobacco chitinase was performed. In sap of infected plants, a protein of 29 kD was detected, whereas in mock-infected plants, a protein of around 34 kD reacted with the antibody (Fig. 8). In silver-stained gels, no protein could be detected that clearly corresponded to either of these proteins. It appears, therefore, that a PR-3 isoform is present in sap of F. oxysporum-infected plants, but at much lower levels than the PR-1, PR-2, and PR-5 proteins.
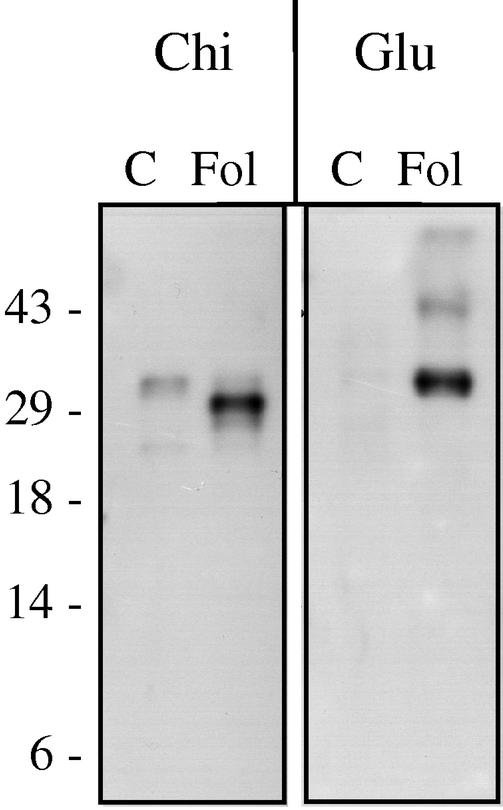
Figure 8.
Immunodetection of PR-2 and PR-3 isoforms in tomato xylem sap. Five-week-old C32 plants were either mock-inoculated (C) or inoculated with the compatible race 2 isolate Fol007 (Fol). Three weeks after infection, xylem sap proteins were isolated, separated in a Tris-Tricine gel, and blotted for immunodetection with antibodies raised against tobacco chitinase (Chi) or glucanase (Glu). Molecular masses (in kD) of marker proteins are indicated on the left.
DISCUSSION
In this study, we show that a small set of proteins reproducibly accumulates in tomato xylem sap upon infection by F. oxysporum, a root-invading, xylem-colonizing fungus. The limited number of these proteins, together with the observation that all proteins identified are known or predicted to be secretory proteins, makes it very unlikely that these proteins originate from leakage of cell content caused by damage occurring during infection or sap collection.
Secretion of specific proteins in response to pathogen attack or chemical stress has been investigated in a variety of plant species, but nearly always in leaf apoplast. In the case of tomato, PR proteins have been detected in leaf intercellular washing fluid after challenge with Phytophthora infestans and C. fulvum (De Wit et al., 1986; Christ and Mösinger, 1989; Joosten and De Wit, 1989). To our knowledge, the only report in which a PR protein in xylem sap of pathogen-infected vessels was identified concerns a peroxidase that accumulates in X. oryzae-infected rice (Young et al., 1995). Although xylem is part of the apoplast, it is separated from the apoplast of the cortex by the endodermis (Kuhn et al., 2000; Sattelmacher, 2001). The findings presented here show, on the one hand, that leaf apoplastic PR proteins (PR-1a, PR-1b, and PR-2a) can also be secreted into xylem sap in response to infection. On the other hand, the response to a xylem-invading pathogen has unique aspects, such as the accumulation of a basic glucanase and of PR-5x, a newly identified member of the PR-5 family.
Proteins belonging to the PR-1 family accumulate in high amounts during virtually all plant-microbe interactions investigated (Van Loon and Van Strien, 1999). With immunogold labeling, PR-1 has previously been detected in secondary thickenings of xylem vessels of tomato roots after infection with F. oxysporum radicis-lycopersici (Benhamou et al., 1991). A similar observation was done in leaves after treatment with 3-abscisic acid or tobacco necrosis virus (Jeun, 2000). β-1,3-Glucanases (PR-2 proteins) are also commonly secreted upon pathogen attack (Stintzi et al., 1993; Simmons, 1994). In the case of fungal vascular wilt diseases, glucanases accumulate in roots and stems of tomato infected with Verticillium albo-atrum (Pegg and Young, 1981, 1982; Young and Pegg, 1981) and in muskmelon (Cucumis melo) infected with F. oxysporum f. sp. melonis (Netzer and Kritzman, 1979). However, in these studies, it was not clear which isoform was involved, nor was it clear whether the enzymes were intra- or extracellularly located. Later studies revealed the presence of extracellular glucanases in xylem tissue of plants affected by wilt disease. In eggplant (Solanum melongena), a glucanase was found to accumulate in secondary cell walls of xylem vessels upon infection with V. albo-atrum (Benhamou et al., 1989), whereas in carnation (Dianthus caryophyllus), the intercellular fluid of vascular cylinders contained glucanases after infection with F. oxysporum f. sp. dianthi (Van Pelt-Heerschap and Smit-Bakker, 1999).
In the present study, the identities of an acidic glucanase and a basic glucanase in xylem sap were established. The acidic isoform was found previously in the apoplast of tomato leaves infected with C. fulvum (Joosten and De Wit, 1989; Van Kan et al., 1992). On the basis of published peptide sequences, the basic isoform found here is likely the same as the basic glucanase that was found to accumulate in C. fulvum-infected leaves (Van Kan et al., 1992). Its mRNA-level rises in leaves infected with citrus exocortis viroid, suggesting that the protein not only accumulates during fungal colonization but also upon viroid challenge (Domingo et al., 1994). The protein predicted from the cDNA sequence has no carboxyl-terminal propeptide for vacuolar targeting, consistent with its presence in xylem sap. However, because this protein was found only in total leaf homogenate of C. fulvum-infected leaves and not in the intercellular washing fluid, it was initially thought to be intracellularly localized (Van Kan et al., 1992). The method used to obtain intercellular washing fluid from leaves probably does not extract xylem sap. We conclude that the basic extracellular glucanase is secreted in vascular tissue but not in leaf mesophyll.
Another dominant PR protein that accumulates in leaf apoplast in the tomato-C. fulvum interaction is a 26-kD chitinase (Joosten and De Wit, 1989). This protein did not accumulate in xylem sap of F. oxysporum-infected plants to levels comparable with the other PR proteins, again showing that the two compartments respond differently to fungal invasion. However, anti-chitinase antibodies did react with a 29-kD protein in xylem sap of infected tomato (Fig. 8). A protein of this size was sometimes observed in silver-stained gels as a very faint band, suggesting that it is present in low amounts compared with the other PR proteins identified in this study. Because of this low abundance, we could not determine whether this protein is in fact the same as the 26-kD chitinase accumulating during C. fulvum infection. In healthy plants, an apparently different protein of around 34 kD was detected with the anti-chitinase antibody. The constitutive presence of chitinases in healthy plants has been demonstrated in several plants and tissues, including xylem sap of cucumber (Cucumis sativus; Masuda et al., 2001).
The best illustration of compartmental specificity in responses to fungal infection is the accumulation of PR-5x, which may constitute a specific reaction to pathogen challenge in roots and/or vascular tissues. This protein is clearly not present in SDS-PAGE gels of leaf apoplastic proteins after infection with C. fulvum (Joosten and De Wit, 1989). Within the family of PR-5 proteins, PR-5x is not closely related to secreted, acidic PR-5-proteins of other plant species like tobacco PR-R (Cornelissen et al., 1986; Payne et al., 1988) or Arabidopsis PR-5 (Uknes et al., 1992). In contrast, it is very closely related to basic, vacuolar PR-5 proteins (also referred to as osmotins; Fig. 7). PR-5x is of particular interest for two additional reasons: It accumulates in xylem sap relatively early after infection, and it is the only protein produced in high amounts in an incompatible interaction.
Apart from PR-5x, the other PR proteins observed in this study accumulate only in compatible interactions, concomitantly with the appearance of disease symptoms. Several explanations for this come to mind. One is that the majority of PR proteins observed in this study are produced only in aerial tissue. These would then only accumulate once the fungus grows into the stem, which occurs only sporadically in incompatible interactions (Gao et al., 1995; Mes et al., 2000). The production of these proteins may alternatively depend on a more advanced stage of infection, at which the fungus starts to invade xylem parenchyma cells (Beckman and Roberts, 1995; Mes et al., 2000). Such a scenario resembles compatible interactions of tomato with C. fulvum (De Wit and van der Meer, 1986). Another explanation for the absence of most disease-related proteins in an incompatible interaction is that their production is local, occurring only in invaded vessels. This would lead to low overall levels in xylem sap in incompatible interactions, where F. oxysporum is restricted to a limited number of vessels. PR-5x would then be exceptional in that it is produced systemically, being also secreted into uninfected vessels. Whichever explanation will hold true, investigation of the way in which PR-5x gene expression is induced is of special interest in the tomato-F. oxysporum interaction, and possibly in interactions with other root- and/or xylem-invading pathogens.
The fact that F. oxysporum colonization proceeds despite accumulation of PR-1, PR-2, and PR-5 proteins implies that F. oxysporum can withstand or avoid the potential antifungal activity that these proteins may have. It is well-established that β-1,3-glucanases (PR-2 proteins) can inhibit fungal growth by degrading cell walls, usually in concert with chitinases (Mauch et al., 1988; Sela-Buurlage et al., 1993; Stintzi et al., 1993). Until now, anti-microbial activity of PR-1 proteins from tomato and tobacco has only been shown against oomycetes in vitro (Niderman et al., 1995) and in PR-1-overproducing tobacco plants (Alexander et al., 1993). The antifungal activity of PR-5 proteins has been established more extensively. Some have been shown to be active in vitro against F. oxysporum. These include AP24 and NP24, which are very similar to PR-5x (Rodrigo et al., 1993; Abad et al., 1996; Hu and Reddy, 1997). If the disease-related xylem sap proteins identified here can inhibit growth of F. oxysporum, they may play a role in delaying colonization. However, it may also be that in a compatible interaction these proteins are produced too late and are not in close contact with the potentially sensitive hyphal tips of F. oxysporum.
This study shows that peptide mass fingerprinting, especially in combination with MS/MS, is an excellent tool to identify proteins in tomato xylem sap, for several reasons. First, because MS analysis only requires small amounts of protein, the low amount of protein present in xylem sap is sufficient. Second, MS/MS yields multiple internal sequence tags from a single sample, without the need for peptide purification. This proved to be especially important to distinguish between highly similar proteins like PR-1a/PR-1b and NP24/AP24/PR-5x. Moreover, multiple sequence tags of a novel protein can be used to identify the corresponding (partial) cDNA in databases for which as yet no complete tryptic digest database is available. Finally, evidence for posttranslational modifications can be obtained, as exemplified in this study by Asn deamidation and modification of N-terminal Gln to pyro-Glu.
In a compatible interaction between F. oxysporum and tomato, xylem sap proteins can be of either plant or fungal origin. In addition to plant defense-related proteins, pathogen-derived proteins are of interest because they are good candidates for virulence and/or avirulence factors. Apart from p12, which contains at least one protein of fungal origin (M. Rep, unpublished data), all the proteins analyzed in this initial survey were clearly of plant origin. Because there may be additional fungal proteins among the less abundant disease-related proteins, we are now applying two-dimensional PAGE to allow more extensive coverage of the proteome of xylem sap of infected tomato.
MATERIALS AND METHODS
Plant Material, Fungal Isolates, and Infections
Fusarium oxysporum f. sp. lycopersici (Fol) isolates Fol004 (race 1), Fol007 (race 2), and Fol029 (race 3) were described before (Mes et al., 1999). Spores were collected from 5-d-old cultures in potato (Solanum tuberosum) dextrose broth and used for root-inoculation of 5-week-old tomato (Lycopersicon esculentum) plants at a spore density of 0.5 × 107 mL−1. Tomato lines used were GCR161 (resistant to Fol race 1) and C32 (susceptible to all Fol races; Kroon and Elgersma, 1993). Disease symptoms in compatible interactions started to appear around 10 d after infection. These include epinasty, yellowing, and eventually browning and abscission of leaves, beginning with the lowest leaves and continuing upward, and the appearance of adventitious roots on the stem, also starting at the base of the stem and proceeding upward. These symptoms were accompanied by browning of vascular bundles in the stem.
Isolation of Xylem Sap and Analysis of Protein Content
Xylem sap was collected from 5- to 8-week-old tomato plants according to the method described by Satoh et al. (1992). In short, stems were cut off below the second true leaf, the first droplet appearing on the cut surface was removed with blotting paper, and the plant was placed in a horizontal position. Sap dripping from the cut surface was collected in tubes placed on ice for a period of 3 to 6 h, generally yielding between 2 and 10 mL of sap. In plants inoculated with a compatible race of F. oxysporum, fungal spores were present in the first 50 μL of sap collected at the cut surface as early as 11 d after infection. Xylem sap was concentrated by freeze drying, and the protein concentration was measured with the bicinchoninic acid method (Sigma-Aldrich, St. Louis). Volumes were adjusted so that each sample contained 1 μg μL−1 protein. It should be noted that this is an overestimation because polysaccharides are also detected with the bicinchoninic acid method. SDS-PAGE was done with Hoefer Mighty Small SE250 minigel equipment (Amersham Biosciences AB, Uppsala) using the Tris/Tricine buffer system and 20% acrylamide (Schagger and von Jagow, 1987). Silver staining was used to visualize proteins. For immunodetection, proteins were blotted on polyvinylidene difluoride membranes. Antibody detection was done with the ECL western blotting system (Amersham Biosciences AB). Antibodies against tobacco (Nicotiana tabacum) chitinase (FB154) and glucanase (FB150) were kindly provided by Syngenta (Basel).
Q/SP Sepharose-Binding
Xylem sap was adjusted to a pH of 7.5 (for SP-Sepharose binding) or 8.5 (for Q-Sepharose binding) by addition of 0.1 volume of 300 mm Tris buffer. One milliliter of sap was then incubated with 50 μL of SP Sepharose Fast Flow or Q Sepharose Fast Flow (Amersham Biosciences AB) for 1 h at room temperature. The beads were collected by centrifugation. Proteins in the supernatant (unbound fraction) were precipitated overnight at 20°C after addition of 4 volumes of ethanol:0.1 m NaAc (19:1, v/v) and dissolved in 100 μL of water. Proteins that bound to the beads were eluted by incubation with 100 μL of Tris buffer containing 250 mm NaCl for 1 h at room temperature.
MS
Protein bands of interest were cut from the stained gel. For MS analysis, the gel slices were S-alkylated with iodoacetamide and vacuum dried using a speedvac. The in-gel digestion with trypsin (sequencing grade, Roche Diagnostics, Indianapolis) and extraction of the peptides after the overnight incubation were done according to Shevchenko et al. (1996). The collected eluates were either dried overnight in a speedvac or directly concentrated and washed on a μC18 ZipTip (Millipore, Bedford, MA). The peptides were eluted or redissolved (speedvac) in 5 to 10 μL of 60% (v/v) acetonitrile/1% (v/v) formic acid. The peptide solutions were mixed 1:1 (v/v) with a solution containing 52 mm α-cyano-4-hydroxycinnamic acid (Sigma-Aldrich) in 49% (v/v) ethanol/49% (v/v) acetonitrile/2% (v/v) trifluoroacetic acid and 1 mm ammonium acetate. Before dissolving, the α-cyano-4-hydroxycinnamic acid was washed briefly with acetone. The mixture was spotted on a target plate and allowed to dry at room temperature. Reflectron MALDI-TOF spectra were acquired on either a TofSpec 2E or a MALDI-TOF mass spectrometer (both Micromass, Wythenshawe, UK). The resulting peptide spectra were used to search the ABCC Non-Redundant Protein Database release 20010401 (Advanced Biomedical Computing Center, Frederick, MD; http://www-fbsc.ncifcrf.gov/) and an in-house translated version of the Lycopersicon Gene Index database (v7.1, release date: August 15, 2001) with MassLynx ProteinProbe (Micromass). When enough material was available MS/MS analysis was performed to obtain sequence information. If necessary, the peptide solutions were desalted using a μC18 ZipTip (Millipore) and eluted in 3 to 5 μL of 60% (v/v) acetonitrile/1% (v/v) formic acid. A gold-plated nanospray needle (Protana [Odense, Denmark] or New Objective [Woburn, MA]) was filled with 2 to 5 μL of the peptide mixture and analyzed on a Micromass Q-TOF mass spectrometer using nano electrospray ionization. Ions were selected from the survey spectra for low-energy collision-induced dissociation experiments using argon as a collision gas. The resulting MS/MS spectra were analyzed with MassLynx Pepseq and Biolynx software. Additional database searching was done with the generated sequence tag.
cDNA Isolation and Sequencing
On the basis of the sequence of a cDNA-AFLP fragment, primers were designed (PR-5x-F, GACCATGGGGTGCCCTAATGCGTATAG; and PR-5x-R, TACTCGAGCACTAGGGCAAGTGAATAA) that specifically anneal to PR-5x cDNA (indicated in Fig. 6) and contain a NcoI and a XhoI restriction site, respectively. PR-5x cDNA sequences were isolated using these primers and a cDNA library as follows. cDNAs derived from F. oxysporum-infected tomato root and stem tissue were directionally cloned into a pACT-2 vector (BD Biosciences Clontech, Palo Alto, CA; J. Vossen, unpublished data). 5′ Fragments of PR-5x were PCR-amplified with the pACT-F (TAATACCACTACAATGGATG) and PR-5x-R primers using the proof reading Pfu polymerase (Stratagene, La Jolla, CA). 3′ Fragments were amplified with the pACT-R (GTGCACGATGCACAGTTG) and PR-5x-F primers. The fragments were cloned, and their sequences were determined with a Gene ReadIR 4200 (LI-COR, Lincoln, NE). All 5′ fragments had the same nucleotide sequence. The 3′ fragments did not differ in sequence but there were differences in length, presumably caused by alternative poly(A) addition sites (see Fig. 6). Sixty-three basepairs of the sequences at the end of 5′ fragments and at the beginning of the 3′ fragments overlapped, indicating that they were derived from mRNAs from the same gene. The complete sequence was deposited at GenBank (accession no. AY093595).
Protein Sequence Alignment and Tree Construction
Sequence alignments and phylogenetic tree construction was done with MacVector (Oxford Molecular Group, Oxford).
ACKNOWLEDGMENTS
We thank Maarten Stuiver and Els van Deventer (Syngenta) for kindly providing us with antibodies against tobacco PR proteins. Peter Sterk is thanked for his help with translation of the TIGR Lycopersicon Gene Index. Frank Takken and Michel Haring are gratefully acknowledged for critical reading of the manuscript.
Footnotes
This work was supported in part by the Council for Medical Sciences of the Netherlands Organization for Scientific Research.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.007427.
LITERATURE CITED
- Abad L, D'Urzo MP, Liu D, Narasimhan ML, Reuveni M, Zhu JK, Niu X, Singh NK, Hasegawa PM, Bressan RA. Antifungal activity of tobacco osmotin has specificity and involves plasma membrane permeabilization. Plant Sci. 1996;118:11–23. [Google Scholar]
- Alexander D, Goodman RM, Gut-Rella M, Glascock C, Weymann K, Friedrich L, Maddox D, Ahl-Goy P, Luntz T, Ward E et al. Increased tolerance to two oomycete pathogens in transgenic tobacco expressing pathogenesis-related protein 1a. Proc Natl Acad Sci USA. 1993;90:7327–7331. doi: 10.1073/pnas.90.15.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman CH, Roberts EM. On the nature and genetic basis for resistance and tolerance to fungal wilt diseases of plants. Adv Bot Res. 1995;21:35–77. [Google Scholar]
- Benhamou N, Grenier J, Asselin A. Immunogold localization of pathogenesis-related protein P14 in tomato root cells infected with Fusarium oxysporum f. sp. radicis-lycopersici. Physiol Mol Plant Pathol. 1991;38:237–253. [Google Scholar]
- Benhamou N, Grenier J, Asselin A, Legrand M. Immunogold localization of beta-1,3-glucanases in two plants infected by vascular wilt fungi. Plant Cell. 1989;1:1209–1221. doi: 10.1105/tpc.1.12.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccardi TL, Barthe GA, Derrick KS. A novel protein associated with citrus blight has sequence similarities to expansin. Plant Mol Biol. 1998;38:775–783. doi: 10.1023/a:1006039016393. [DOI] [PubMed] [Google Scholar]
- Christ U, Mösinger E. Pathogenesis-related proteins of tomato: I. Induction by Phytophthora infestansand other biotic and abiotic inducers and correlations with resistance. Physiol Mol Plant Pathol. 1989;35:53–65. [Google Scholar]
- Cornelissen BJ, Hooft van Huijsduijnen RA, Bol JF. A tobacco mosaic virus-induced tobacco protein is homologous to the sweet-tasting protein thaumatin. Nature. 1986;321:531–532. doi: 10.1038/321531a0. [DOI] [PubMed] [Google Scholar]
- De Boer AH, Volkov V (2003) Logistics of water and salt transport through the plant: structure and functioning of the xylem. Plant Cell Environ (in press)
- De Wit PJ, Buurlage MB, Hammond KE. The occurrence of host-, pathogen-, and interaction-specific proteins in the apoplast of Cladosporium fulvum (syn. Fulvia fulva) infected tomato leaves. Physiol Mol Plant Pathol. 1986;29:159–172. [Google Scholar]
- De Wit PJ, van der Meer FE. Accumulation of the pathogenesis-related tomato leaf protein p14 as an early indicator of incompatibility in the interaction between Cladosporium fulvum (Syn. Fulvia fulva) and tomato. Physiol Mol Plant Pathol. 1986;28:203–214. [Google Scholar]
- Domingo C, Conejero V, Vera P. Genes encoding acidic and basic class III beta-1,3-glucanases are expressed in tomato plants upon viroid infection. Plant Mol Biol. 1994;24:725–732. doi: 10.1007/BF00029854. [DOI] [PubMed] [Google Scholar]
- Emslie KR, Molloy MP, Barardi CRM, Jardine D, Wilkins MR, Bellamy AR, Williams KL. Serotype classification and characterization of the rotavirus SA11 VP6 protein using mass spectrometry and two-dimensional gel electrophoresis. Funct Integr Genomics. 2000;1:12–24. doi: 10.1007/s101420000002. [DOI] [PubMed] [Google Scholar]
- Gao H, Beckman CH, Mueller WC. The nature of tolerance to Fusarium oxysporum f. sp. lycopersiciin polygenically field-resistant marglobe tomato plants. Physiol Mol Plant Pathol. 1995;46:401–412. [Google Scholar]
- Hu X, Reddy AS. Cloning and expression of a PR5-like protein from Arabidopsis: inhibition of fungal growth by bacterially expressed protein. Plant Mol Biol. 1997;34:949–959. doi: 10.1023/a:1005893119263. [DOI] [PubMed] [Google Scholar]
- Hubbes M. The American elm and Dutch elm disease. For Chron. 1999;75:265–273. [Google Scholar]
- Jeun YC. Immunolocalization of PR-protein P14 in leaves of tomato. J Plant Dis Prot. 2000;107:352–367. [Google Scholar]
- Jia Y, Martin GB. Rapid transcript accumulation of pathogenesis-related genes during an incompatible interaction in bacterial speck disease resistant tomato plants. Plant Mol Biol. 1999;40:455–465. doi: 10.1023/a:1006213324555. [DOI] [PubMed] [Google Scholar]
- Joosten MH, Bergmans CJB, Meulenhoff EJS, Cornelissen BJC, De Wit PJ. Purification and serological characterization of three basic 15-kilodalton pathogenesis-related proteins from tomato. Plant Physiol. 1990;94:585–591. doi: 10.1104/pp.94.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten MH, De Wit PJ. Identification of several pathogenesis-related proteins in tomato leaves inoculated with Cladosporium fulvum (syn. Fulvia Fulva) as 1,3-β-glucanases and chitinases. Plant Physiol. 1989;89:945–951. doi: 10.1104/pp.89.3.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King GJ, Turner VA, Hussey CE, Jr, Syrkin Wurtele E, Lee M. Isolation and characterization of a tomato cDNA clone which codes for a salt-induced protein. Plant Mol Biol. 1988;10:401–412. doi: 10.1007/BF00014946. [DOI] [PubMed] [Google Scholar]
- Kitajima S, Sato F. Plant pathogenesis-related proteins: molecular mechanisms of gene expression an protein function. J Biochem. 1999;125:1–8. doi: 10.1093/oxfordjournals.jbchem.a022244. [DOI] [PubMed] [Google Scholar]
- Kroon BAM, Elgersma DM. Interactions between race 2 of Fusarium oxysporum f. sp. lycopersiciand near-isogenic resistant and susceptible lines of intact plants or callus of tomato. J Phytopathol. 1993;137:1–9. [Google Scholar]
- Kuhn AJ, Schröder WH, Bauch J. The kinetics of calcium and magnesium entry into mycorrhizal spruce roots. Planta. 2000;210:488–496. doi: 10.1007/PL00008156. [DOI] [PubMed] [Google Scholar]
- Liang J, Zhang J. Collection of xylem sap at flow rate similar to in vivo transpiration flux. Plant Cell Physiol. 1997;38:1375–1381. [Google Scholar]
- Lucas J, Henriquez AC, Lottspiech F, Henschen A, Sänger HL. Amino acid sequence of the “pathogenesis-related” leaf protein p14 from viroid-infected tomato reveals a new type of structurally unfamiliar proteins. EMBO J. 1985;4:2745–2749. doi: 10.1002/j.1460-2075.1985.tb03998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda S, Kamada H, Satoh S. Chitinase in cucumber xylem sap. Biosci Biotechnol Biochem. 2001;65:1883–1885. doi: 10.1271/bbb.65.1883. [DOI] [PubMed] [Google Scholar]
- Mauch F, Mauch-Mani B, Boller T. Antifungal hydrolases in pea tissue: II. Inhibition of fungal growth by combinations of chitinase and β-1,3-glucanase. Plant Physiol. 1988;88:936–942. doi: 10.1104/pp.88.3.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinwald YC, Stimson ER, Scheraga HA. Deamidation of the asparaginyl-glycyl sequence. Int J Pept Protein Res. 1986;28:79–84. doi: 10.1111/j.1399-3011.1986.tb03231.x. [DOI] [PubMed] [Google Scholar]
- Melchers LS, Sela-Buurlage MB, Vloemans SA, Woloshuk CP, Van Roekel JS, Pen J, van den Elzen PJ, Cornelissen BJ. Extracellular targeting of the vacuolar tobacco proteins AP24, chitinase and beta-1,3-glucanase in transgenic plants. Plant Mol Biol. 1993;21:583–593. doi: 10.1007/BF00014542. [DOI] [PubMed] [Google Scholar]
- Mes JJ, van Doorn AA, Wijbrandi J, Simons G, Cornelissen BJC, Haring MA. Expression of the Fusarium resistance gene I-2colocalizes with the site of fungal containment. Plant J. 2000;23:183–194. doi: 10.1046/j.1365-313x.2000.00765.x. [DOI] [PubMed] [Google Scholar]
- Mes JJ, Weststeijn EA, Herlaar F, Lambalk JJM, Wijbrandi J, Haring MA, Cornelissen BJC. Biological and molecular characterization of Fusarium oxysporum f. sp. lycopersicidivides race 1 isolates into separate virulence groups. Phytopathology. 1999;89:156–160. doi: 10.1094/PHYTO.1999.89.2.156. [DOI] [PubMed] [Google Scholar]
- Nemec S. Stress-related compounds in xylem fluid of blight-diseased citrus containing Fusarium solaninaphtazarin toxins and their effects on the host. Can J Microbiol. 1995;41:515–524. [Google Scholar]
- Netzer D, Kritzman G. Beta-(1,3) Glucanase activity and quantity of fungus in relation to Fusariumwilt in resistant and susceptible near-isogenic lines of muskmelon. Physiol Plant Pathol. 1979;14:47–55. [Google Scholar]
- Niderman T, Genetet I, Bruyere T, Gees R, Stintzi A, Legrand M, Fritig B, Mosinger E. Pathogenesis-related PR-1 proteins are antifungal: isolation and characterization of three 14-kilodalton proteins of tomato and of a basic PR-1 of tobacco with inhibitory activity against Phytophthora infestans. Plant Physiol. 1995;108:17–27. doi: 10.1104/pp.108.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne G, W. M, Williams S, Desai N, Parks TD, Dincher S, Carnes M, Ryals J. Isolation and nucleotide sequence of a novel cDNA clone encoding the major form of pathogenesis-related protein R. Plant Mol Biol. 1988;11:223–224. doi: 10.1007/BF00015674. [DOI] [PubMed] [Google Scholar]
- Pegg GF, Young DH. Changes in glycosidase activity and their relationship to fungal colonization during infection of tomato by Verticillium albo-atrum. Physiol Mol Plant Pathol. 1981;19:371–382. [Google Scholar]
- Pegg GF, Young DH. Purification and characterization of chitinase enzymes from healthy and Verticillium albo-atrum-infected tomato plants, and from V. albo-atrum. Physiol Mol Plant Pathol. 1982;21:389–409. [Google Scholar]
- Pierpoint WS, Tatham AS. Identification of the virus-induced protein of tobacco leaves that resembles the sweet-protein thaumatin. Physiol Mol Plant Pathol. 1987;31:291–298. [Google Scholar]
- Quackenbush J, Cho J, Lee D, Liang F, Holt I, Karamycheva S, Parvizi B, Pertea G, Sultana R, White J. The TIGR gene indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res. 2001;29:159–164. doi: 10.1093/nar/29.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo I, Vera P, Frank R, Conejero V. Identification of the viroid-induced tomato pathogenesis-related (PR) protein P23 as the thaumatin-like tomato protein NP24 associated with osmotic stress. Plant Mol Biol. 1991;16:931–934. doi: 10.1007/BF00015088. [DOI] [PubMed] [Google Scholar]
- Rodrigo I, Vera P, Tornero P, Hernandez-Yago J, Conejero V. cDNA cloning of viroid-induced tomato pathogenesis-related protein P23: characterization as a vacuolar antifungal factor. Plant Physiol. 1993;102:939–945. doi: 10.1104/pp.102.3.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Medrano R, Jimenez-Moraila B, Herrera-Estrella L, Rivera-Bustamante RF. Nucleotide sequence of an osmotin-like cDNA induced in tomato during viroid infection. Plant Mol Biol. 1992;20:1199–1202. doi: 10.1007/BF00028909. [DOI] [PubMed] [Google Scholar]
- Satoh S, Iizuka C, Kikuchi A, Nakamura N, Fujii T. Proteins and carbohydrates in xylem sap from squash root. Plant Cell Physiol. 1992;33:841–847. [Google Scholar]
- Sattelmacher B. The apoplast and its significance for plant mineral nutrition. New Phytol. 2001;149:167–192. doi: 10.1046/j.1469-8137.2001.00034.x. [DOI] [PubMed] [Google Scholar]
- Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Sela-Buurlage MB, Ponstein AS, Bres-Vloemans SA, Melchers LS, van den Elzen PJM, Cornelissen BJC. Only specific tobacco (Nicotiana tabacum) chitinases and β-1,3-glucanases exhibit antifungal activity. Plant Physiol. 1993;101:857–863. doi: 10.1104/pp.101.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Simmons CR. The physiology and molecular biology of plant 1,3-beta-d-glucanases and 1,3;1,4-beta-d-glucanases. Crit Rev Plant Sci. 1994;13:325–387. [Google Scholar]
- Simons G, Groenendijk J, Wijbrandi J, Reijans M, Groenen J, Diergaarde P, van der Lee T, Bleeker M, Onstenk J, de Both M et al. Dissection of the Fusarium I2gene cluster in tomato reveals six homologs and one active gene copy. Plant Cell. 1998;10:1055–1068. doi: 10.1105/tpc.10.6.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A, Heitz T, Prasad V, Wiedemann-Merdinoglu S, Kauffmann S, Geoffroy P, Legrand M, Fritig B. Plant “pathogenesis-related” proteins and their role in defense against pathogens. Biochimie. 1993;75:687–706. doi: 10.1016/0300-9084(93)90100-7. [DOI] [PubMed] [Google Scholar]
- Tjamos EC, Beckman CH. NATO ASI Series H: Cell Biology. Vol. 28. Berlin: Springer-Verlag; 1989. Vascular wilt diseases of plants: basic studies and control. [Google Scholar]
- Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J. Acquired resistance in Arabidopsis. Plant Cell. 1992;4:645–656. doi: 10.1105/tpc.4.6.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kan JA, Joosten MH, Wagemakers CA, Van den Berg-Velthuis GC, De Wit PJ. Differential accumulation of mRNAs encoding extracellular and intracellular PR proteins in tomato induced by virulent and avirulent races of Cladosporium fulvum. Plant Mol Biol. 1992;20:513–527. doi: 10.1007/BF00040610. [DOI] [PubMed] [Google Scholar]
- Van Loon LC, Van Strien EA. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol Mol Plant Pathol. 1999;55:85–97. [Google Scholar]
- Van Pelt-Heerschap H, Smit-Bakker O. Analysis of defense-related proteins in stem tissue of carnation inoculated with a virulent and avirulent race of Fusarium oxysporum f.sp. dianthi. Eur J Plant Pathol. 1999;105:681–691. [Google Scholar]
- Woloshuk CP, Meulenhoff JS, Sela-Buurlage M, van den Elzen PJ, Cornelissen BJ. Pathogen-induced proteins with inhibitory activity toward Phytophthora infestans. Plant Cell. 1991;3:619–628. doi: 10.1105/tpc.3.6.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DH, Pegg GF. Purification and characterization of 1,3-beta-glucan hydrolases from healthy and Verticillium albo-atrum-infected tomato plants. Physiol Plant Pathol. 1981;19:391–417. [Google Scholar]
- Young SA, Guo A, Guikema JA, White FF, Leach JE. Rice cationic peroxidase accumulates in xylem vessels during incompatible interactions with Xanthomonas oryzae pv oryzae. Plant Physiol. 1995;107:1333–1341. doi: 10.1104/pp.107.4.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]