Abstract
LeCTR1 was initially isolated by both differential display reverse transcriptase-polymerase chain reaction screening for tomato (Lycopersicon esculentum) fruit ethylene-inducible genes and through homology with the Arabidopsis CTR1 cDNA. LeCTR1 shares strong nucleotide sequence homology with Arabidopsis CTR1, a gene acting downstream of the ethylene receptor and showing similarity to the Raf family of serine/threonine protein kinases. The length of the LeCTR1 transcribed region from ATG to stop codon (12,000 bp) is more than twice that of Arabidopsis CTR1 (4,700 bp). Structural analysis reveals perfect conservation of both the number and position of introns and exons in LeCTR1 and Arabidopsis CTR1. The introns in LeCTR1 are much longer, however. To address whether this structural conservation is indicative of functional conservation of the corresponding proteins, we expressed LeCTR1 in the Arabidopsis ctr1-1 (constitutive triple response 1) mutant under the direction of the 35S promoter. Our data clearly show that ectopic expression of LeCTR1 in the Arabidopsis ctr1-1 mutant can restore normal ethylene signaling. The recovery of normal ethylene sensitivity upon heterologous expression of LeCTR1 was also confirmed by restored glucose sensitivity absent in the Arabidopsis ctr1-1 mutant. Expression studies confirm ethylene responsiveness of LeCTR1 in various tissues, including ripening fruit, and may suggest the evolution of alternate regulatory mechanisms in tomato versus Arabidopsis.
The plant hormone ethylene is involved in a variety of developmental and physiological processes in plants, including senescence, fruit ripening, and abscission (Abeles et al., 1992; Lelièvre et al., 1997; Giovannoni, 2001). It also plays an important role in physiological responses to environmental stresses such as water deficit, mechanical wounding, and pathogen attack (Abeles et al., 1992). The unraveling of the molecular basis of the ethylene perception and signal transduction pathway has been enhanced by the use of Arabidopsis mutants altered in the seedling triple response (Guzmàn and Ecker, 1990). The triple response is exhibited by seedlings treated with ethylene and results in: (a) inhibition of root elongation, (b) shortening and radial swelling of the hypocotyl, and (c) exaggerated curvature of the apical hook. Numerous loci have been identified and many corresponding genes cloned, representing various steps in ethylene signaling from receptors through transcription factors (Ecker, 1995; Johnson and Ecker, 1998; Stepanova and Ecker, 2000). The ETR1 gene was the first to be cloned (Chang et al., 1993) and was shown to encode a functionally active ethylene receptor (Schaller and Bleecker, 1995). Subsequently, it has been demonstrated that ETR1 belongs to a multigene family whose five members are differentially regulated (Schaller and Bleecker, 1995; Hua and Meyerowitz, 1998). Despite significant divergence at the structural and primary sequence level, all members of the ethylene receptor family are functionally active (Hua and Meyerowitz, 1998).
In contrast to the ethylene insensitivity phenotype conferred by the ethylene response mutation (etr), disruption of the ctr1 locus confers constitutive ethylene response in the absence of the hormone. Epistatic studies revealed that the CTR1 gene product acts downstream of the ethylene receptors (Kieber et al., 1993). Furthermore, the regulatory domain of CTR1 was found to associate with ETR1 and ethylene response sensor (ERS1) in yeast two-hybrid and in vitro protein association assays (Clark et al., 1998), raising the possibility that ethylene receptors directly regulate CTR1 activity.
In the tomato (Lycopersicon esculentum), initial inroads into ethylene perception were made via cloning of the Never-ripe gene, which proved to be a tomato ethylene receptor most like the ERS receptor of Arabidopsis (Hua et al., 1995; Wilkinson et al., 1995). Subsequent studies regarding ethylene perception and signal transduction have focused on the isolation and characterization of the receptor gene family and a family of putative transcription factors related to the Arabidopsis EIN3 gene. In recent years, five ETR1 homologs have been identified in tomato (Payton et al., 1996; Zhou et al., 1996; Tieman and Klee, 1999) and heterologous expression and complementation studies have been employed on a subset of ETR gene family members to demonstrate ethylene receptor activity (Wilkinson et al., 1995; Tieman et al., 2000).
Although two tomato sequences showing significant sequence homology with Arabidopsis CTR1 have been reported, data addressing their functional significance are lacking. The Arabidopsis CTR1 gene is reported to be constitutively expressed (Kieber et al., 1993). Constitutive expression was not the case for the tomato LeCTR1 gene that we have shown previously to be regulated by ethylene and during fruit ripening (Giovannoni et al., 1998; Zegzouti et al., 1999). Isolation of LeCTR1 and its regulation by ethylene and induction during ripening suggested the possibility that this step in the ethylene-signaling network may be a target for differential regulation in species displaying aspects of development critically dependent upon ethylene. Before addressing this question, however, it was first necessary to establish LeCTR1 function.
Here, we show that the genomic structure of LeCTR1 is highly conserved with the Arabidopsis CTR1 gene and is capable of furnishing CTR1 function when expressed in the ctr1-1 mutant of Arabidopsis. We also demonstrate that LeCTR1 mRNA accumulates during fruit ripening and upon ethylene treatment not only in fruit but also in additional non-fruit tissues. Together, these results may suggest regulatory modification of a necessary component of ethylene signal transduction in tomato as compared with Arabidopsis.
RESULTS AND DISCUSSION
LeCTR1 Encodes a Putative Raf Kinase Protein
The ethylene-inducible LeCTR1 differential display-derived fragment, initially called ER50 (Zegzouti et al., 1999), was extended by primer extension to obtain a full coding sequence of the cDNA clone. Accurate LeCTR1 sequence was confirmed by RACE-PCR of a partial cDNA isolated by screening a ripe fruit cDNA library with the Arabidopsis CTR1 cDNA as probe, and followed by reverse transcriptase (RT)-PCR and sequencing of the resulting full-length cDNA (Giovannoni et al., 1998). Translation of the longest open reading frame of the resulting cDNA sequence predicts a protein with a molecular mass of 92 kD and no obvious membrane-spanning domains.
The predicted LeCTR1 protein shares significant homology with different members of the Raf family of Ser/Thr protein kinases of the class mitogen-activated protein kinase kinase kinase from both animals (e.g. Rattus norvegicus C-Raf-1) and plants. Database searches revealed that LeCTR1 shows the strongest homology with AtCTR1, a negative regulator of ethylene signaling in Arabidopsis (Kieber et al., 1993). A similar level of homology was also found with other plant Raf kinases, like the AtEDR1 (Enhanced Disease Resistance 1), an Arabidopsis gene shown to act as a negative regulator of defense responses (Frye et al., 2001) and LeCTR2, another tomato CTR-like protein (LeCTR2/TCTR2, GenBank accession no. AJ005077). Within the kinase domain of LeCTR1, AtCTR1, AtEDR1, and LeCTR2, there is perfect conservation of 11 subdomains typical of the catalytic site of Ser/Thr protein kinases (Hanks et al., 1988; Hanks and Quinn, 1991). These domains include two signature patterns. The first, spanning amino acids 562 to 569, corresponds to a typical ATP-binding site motif (GxGxxGxV; Schenk and Snaar-Jagalska, 1999) present in all protein kinases (Fig. 1). The second sequence signature (HRDLKxxN) located at amino acid 678 to 685 represents the consensus sequence for Ser/Thr protein kinases (Schenk and Snaar-Jagalska, 1999) and is well conserved in AtCTR1 and both tomato homologs. The overall comparison of the full sequences indicate that LeCTR1 shares between 58% and 62% identity at the protein sequence level with AtCTR1, AtEDR1, and LeCTR2. The kinase domain of LeCTR1 exhibits more identity with AtCTR1 (82%) than with LeCTR2 (59%) and AtEDR1 (62%), whereas the kinase domain of LeCTR2 and AtEDR1 exhibits 85% identity (Frye et al., 2001). In the N-terminal region, LeCTR1 also shows higher identity with AtCTR1 (55%) than with LeCTR2 and AtEDR1 (36%). However, the amino-terminal region lacks significant homology to the amino-terminal portion of Raf, suggesting that LeCTR1 and AtCTR1 may be regulated by different factors and/or by a distinct mechanism (Kieber, 1997). In summary, the LeCTR1 sequence displays greater similarity to AtCTR1 than LeCTR2, suggesting LeCTR1 is more likely to bear AtCTR1 function as a negative regulator of the ethylene transduction pathway in tomato.
Figure 1.
Sequence comparison between tomato LeCTR1 (AAl87456) and LeCTR2 (AJ005077), AtCTR1 (CAB82938), AtEDR1 (AAG31143), and R. norvegicus C-Raf (P11345) proteins. Identities between proteins are indicated by shaded squares. The kinase catalytic domain is located in the C-terminal side, including the 11 subdomains (roman numerals). The sequence consensus for the ATP-binding site and Ser/Thr protein kinase are also indicated (… … and - - - - - -, respectively). Arabic numbers indicate the starting of each exon for LeCTR1 and AtCTR1.(Figure continues on facing page.)
Isolation and Structural Analysis of the Tomato LeCTR1 Genomic Clone
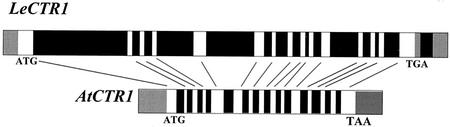
The LeCTR1 genomic clone has been obtained by PCR amplification of tomato genomic DNA. Comparison of genomic and cDNA sequences allowed the delineation of intron and exon positions. Both AtCTR1 and LeCTR1 have 15 exons and 14 introns with the position and size of the exons perfectly conserved between the two species (Fig. 2). Despite this structural conservation, the LeCTR1 introns are typically larger than those of AtCTR1. As a consequence, the overall size of the LeCTR1 transcribed region from ATG to stop codon is more than twice that of AtCTR1 (12,006 bp versus 4,701 bp, respectively). Such high conservation of genomic structure might be indicative of conserved function.
Figure 2.
Comparison of the genomic structure of the tomato LeCTR1 (AY079048) and the Arabidopsis AtCTR1 gene (L08790). Black portions represent the introns, white portions represent the exons, and gray portions represent the untranslated region. Arrows indicate that each exon of LeCTR1 gene correspond to its homolog in the AtCTR1 gene.
Reversion of the Arabidopsis ctr1-1 Mutant Phenotype by Complementation with LeCTR1
Expression of LeCTR1 in the Arabidopsis ctr1-1 Mutant Restores the Wild-Type Phenotype
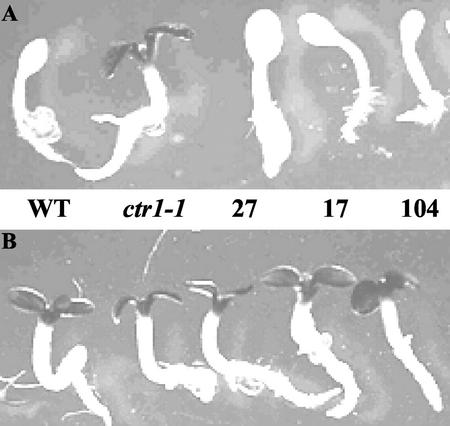
To assess the functional significance of LeCTR1, we attempted complementation of the ctr1-1 mutant of Arabidopsis (Columbia ecotype) using a sense construct of the LeCTR1 cDNA driven by the 35S promoter. Figure 3 shows the typical seedling phenotype of the ctr1-1 mutant. Light-grown ctr1-1 seedlings display a considerable delay in the opening of the apical hook and in cotyledon expansion, a greater darkening of the cotyledons, and significant reduction of root elongation. Dark-grown seedlings displayed a constitutive triple response (Kieber et al., 1993). Seventeen transgenic lines corresponding to independent transformation events harboring the 35S:LeCTR1 sense construct were generated. Three transgenic lines presenting different levels of recovery of the wild-type phenotype were selected for detailed molecular and physiological analysis.
Figure 3.
Phenotypes of the transgenic LeCTR1-overexpressing lines (27, 17, and 104) compared with that of Arabidopsis wild type and the ctr1-1 mutant. A, Adult plants at the rosette stage grown in the greenhouse. B, Adult plants at the flowering stage grown in the greenhouse. C, Four-day-old seedlings grown in the light. D, Four-day-old etiolated seedlings.
Light-grown transgenic lines displayed variable degrees of complementation of different aspects of the mutant phenotype (Fig. 3). For instance, adult plants from all three transformed lines displayed a wild-type phenotype in terms of rosette size and inflorescence development (Fig. 3, A and B). The cotyledon shape, color, and timing of development were completely identical between the complemented lines and wild-type control, whereas root length of transgenic plants was highly variable, ranging from the wild-type to ctr1-1 mutant phenotypes (Fig. 3C). Specifically, line 27 grown in the light developed normal roots similar in size to those of wild-type plants, whereas under identical growth conditions, line 104 has short roots only slightly longer than those of ctr1-1. Line 17 has roots with intermediate elongation. Moreover, etiolated seedlings of all LeCTR1-overexpressing lines displayed a gradual recovery of the hypocotyl elongation rate compared with the ctr1-1 mutant (Fig. 3D).
Recovery of Normal Ethylene Response of the ctr1-1 Mutant Complemented with LeCTR1
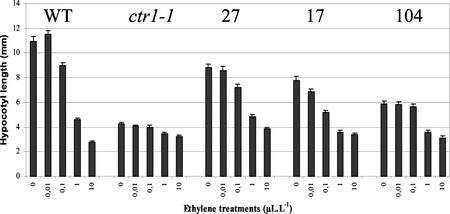
To more fully assess the degree to which LeCTR1 complementation restores the capacity of the ctr1-1 mutant to respond to ethylene, we established a dose response curve of hypocotyl length in response to exogenous ethylene treatment (Fig. 4). After 3 d of growth in the absence of ethylene, hypocotyl length reached 80%, 72%, and 54% of that of wild-type size in lines 27, 17, and 104, respectively, compared with 33% in the non-complemented ctr1-1 controls. When complemented seedlings were treated with ethylene, hypocotyl elongation decreased in correlation with increasing ethylene concentration. The minimal threshold ethylene concentration yielding scorable alteration of hypocotyl length was 0.1 μL L−1, whereas 1 μL L−1 resulted in identical inhibition of hypocotyl elongation in both wild-type and transgenic ctr1-1 lines expressing LeCTR1 (Fig. 4).
Figure 4.
Ethylene response of the transgenic LeCTR1-overexpressing lines (27, 17, 104) compared with that of wild type and the ctr1-1 mutant. Etiolated seedlings untreated or treated with increasing ethylene concentration (0.01, 0.1, 1, and 10 μL L−1) were grown for 3 d before monitoring hypocotyl length. Each histogram represents the mean of 30 measurements and the vertical bars indicate the confidence interval.
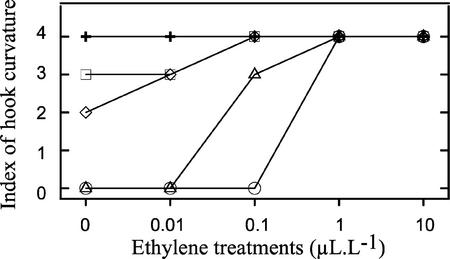
Analysis of apical hook curvature revealed that all three complemented lines responded to ethylene, though, consistent with the hypocotyl phenotype, the degree of response varied among the three lines. Specifically, lines 17 and 104 displayed a 90° (index 2) and 180° (index 3) apical curvature, respectively, even in the absence of ethylene treatment (Fig. 5), suggesting partial complementation for these two lines. Line 27 displayed a fully open hook in the absence of ethylene but exhibited hook formation at 0.1 μL L−1 ethylene as compared with a requirement of 1 μL L−1 for the wild-type control (Fig. 5). These observations show that expression of LeCTR1 in the ctr1-1 mutant was capable of restoring seedling responsiveness to ethylene both in the hypocotyl and in the hook. Although the hypocotyl response was triggered by the same ethylene concentration in wild-type and complemented mutant lines, hook curvature required less ethylene in the LeCTR1-expressing lines, suggesting differential sensitivity to the hormone in various complemented tissues.
Figure 5.
Effect of ethylene on the apical hook curvature of the transgenic LeCTR1-complemented lines (27, 17, and 104) compared with that of wild type and the ctr1-1 mutant. ○, Wild type; +, ctr1-1 mutant; Δ, line 27; ⋄, line 17; □, line 104. The level of apical curvature was estimated visually for 30 seedlings using a scale ranging from 0 to 4 (0, no apical hook; 1, 90° curvature; 2, 180° curvature; 3, beginning of hook formation; and 4, full hook). The experiment was repeated twice.
LeCTR1 Expression Restores Glc Sensitivity to the ctr1-1 Mutant
Wild-type seedlings of Arabidopsis undergo growth arrest when cultivated in light in the presence of 6% (w/v) Glc. Exogenous ethylene allows seedlings to overcome Glc-induced inhibition of growth (Zhou et al., 1998). The ctr1-1 mutant is capable of normal seedling growth in the presence of 6% (w/v) Glc, even in the absence of ethylene (Fig. 6A), presumably due to constitutive activation of ethylene signaling. Complementation of ctr1-1 with LeCTR1 resulted in recovery of Glc-induced growth inhibition similar to that shown by the wild type (Fig. 6A). When seedlings were supplemented with 10 μL L−1 of ethylene, all lines including wild-type control, overcame Glc-induced growth inhibition (Fig. 6B). The reversion of the ctr1-1 phenotype relative to Glc tolerance was total and included all growth arrest-associated symptoms that had been previously described (Zhou et al., 1998). Specifically, ctr1-1 seedlings expressing LeCTR1 demonstrated ethylene reversible inhibition of: (a) expansion and greening of cotyledons, (b) abnormal development, and (c) root elongation (Fig. 6). These data further demonstrate the ability of LeCTR1 to complement for the loss of CTR1 function in the Arabidopsis ctr1-1 mutant.
Figure 6.
Effect of Glc on the development of the transgenic LeCTR1-complemented lines (27, 17, and 104) compared with wild type and the ctr1-1 mutant. For each line, 50 seedlings are grown on Murashige and Skoog medium containing 6% (w/v) Glc during 10 d in the light. Wild type, ctr1-1 mutant, or LeCTR1-complemented lines grown in a sealed box with air (A) or in the presence of 10 μL L−1 ethylene (B). The experiment was repeated three times.
Molecular Analysis of the Transformed Lines
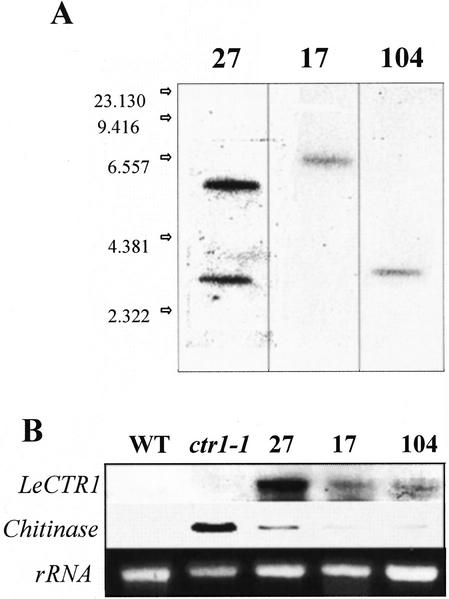
The incorporation of the transgene in the three transformed lines selected for this study was confirmed by Southern-blot analysis. Figure 7A shows that line 27 contained two copies of the transgene, whereas lines 17 and 104 contained only one. As a consequence, homozygous progenies were easily selected for lines 17 and 104, whereas for line 27, because of the presence of multiple insertions, it was difficult to ascertain whether the progenies were homozygous for all the insertions. Therefore, it was assumed that a mixed population of plants was used in the case of line 27. Northern-blot analysis clearly indicated the presence of transgene-derived transcripts in the transformed lines, but not in the wild type nor in the ctr1-1 mutant. Moreover, LeCTR1 transcripts accumulated to higher levels in line 27 as compared with the two other transgenic lines. The level of transgene expression correlated positively with the number of T-DNA insertions consistent with the apparent gene dose effect described for hypocotyl elongation. Line 27, which displayed the highest level of LeCTR1 transcript accumulation, also showed the greatest degree of complementation (Fig. 7B).
Figure 7.
Molecular analysis of the transgenic LeCTR1-complemented lines (27, 17, and 104) compared with wild type and the ctr1-1 mutant. A, Southern analysis of the transgenic lines with a NPTII probe. Numbers indicate the fragment size in kilobase pairs. Line 27 contained two insertions, whereas line 17 and 104 contained only one copy of the transgene. B, Northern-blot analysis of LeCTR1 and basic chitinase transcript accumulation. Equal loading of the gel with the RNA samples is checked by ethidium bromide staining (bottom).
To investigate the effect of LeCTR1 at the level of ethylene-responsive gene expression, we analyzed the accumulation of an ethylene-inducible basic chitinase (Samac et al., 1990). Chitinase gene expression is ethylene dependent and has been shown to be constitutively expressed in the ctr1-1 mutant (Kieber et al., 1993). As expected, in the absence of ethylene, the chitinase transcripts were undetectable in wild-type lines, while accumulating to substantial levels in the ctr1-1 mutant (Fig. 7B). LeCTR1-complemented ctr1-1 lines exhibited a dramatic reduction of chitinase expression as compared with the untransformed mutant, indicating reversion toward the wild-type phenotype. However, there was no clear correlation between transgene copy number and the level of chitinase gene expression.
LeCTR1 Gene Expression Is Regulated by Ethylene in Tomato
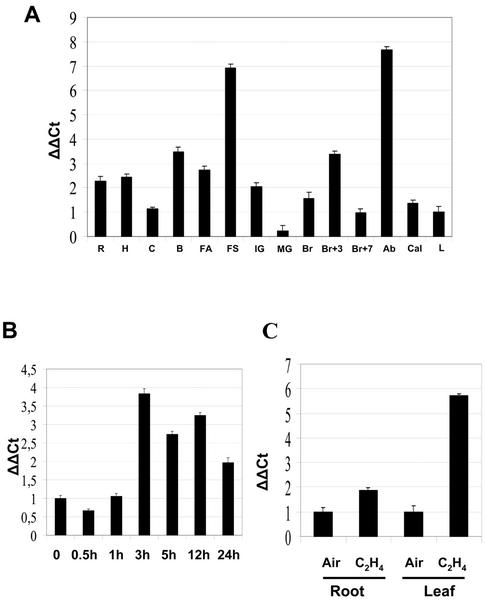
A variety of tissues were harvested for RNA extraction and analyzed for levels of LeCTR1 mRNA expression using real-time quantitative PCR (Fig. 8A). The LeCTR1 message was detected at varying levels in all tissues examined. The LeCTR1 message increased, coincident with the onset of fruit ripening. LeCTR1 transcript levels were relatively low in mature green fruit and increased during the breaker and 3 d post-breaker stages, followed by a decline during later fruit development.
Figure 8.
Ethylene-dependent and tissue-specific expression of LeCTR1 in tomato. The levels of LeCTR1 transcripts were assessed by real-time quantitative PCR. The experiments were carried out in triplicate. A, LeCTR1 mRNA accumulation was monitored in the root (R), hypocotyl (H), cotyledon (C), unopened buds (B), flower at anthesis (FA), senescent flowers (FS), young fruit 7 DPA (IG), mature green fruit (MG), breaker fruit (Br), breaker + 3 (Br+3), breaker + 7 (Br+7), abscission zone (Ab), callus (Cal), and leaf (L). ΔΔCt on the y axis refers to the fold difference in LeCTR1 expression relative to the leaf. B, Ethylene responsiveness of LeCTR1 in mature green fruit treated with 20 μL L−1 ethylene. ΔΔCt on the y axis refers to the fold difference in LeCTR1 expression relative to the control. C, LeCTR1 ethylene regulation in root and leaves. Ethylene treatment was performed as in B. ΔΔCt on the y axis refers to the fold difference in LeCTR1 expression relative to air-treated root and leaf, respectively.
It has been shown previously that LeCTR1 expression is inducible in mature green fruit treated with ethylene (Zegzouti et al., 1999). To more fully characterize the dynamics of ethylene responsiveness during fruit development and particularly at the onset of ripening, LeCTR1 gene responsiveness to ethylene was examined in mature green fruit (Fig. 8B). LeCTR1 responded relatively rapidly to ethylene treatment, demonstrating a 4-fold induction within 3 h of treatment. Modest levels of induction were maintained throughout the 24-h experimental time course and analysis of expression in ripening fruit indicated this induction is likely to persist throughout the later stages of ripening and senescence (Fig. 8A).
The highest levels of LeCTR1 observed during flower development occur in association with senescence, which is also marked by considerable ethylene biosynthesis. Figure 8C shows that 4-week-old tomato plants demonstrated increased LeCTR1 expression in response to ethylene in leaves (6-fold) and roots (2-fold). These results suggest that LeCTR1 is ethylene inducible in a range of tomato tissues, in contrast to the relative constitutive expression reported for its Arabidopsis counterpart, AtCTR1 (Kieber et al., 1993). The fact that substantial induction of LeCTR1 occurs during stages of significant ethylene-mediated developmental modification (e.g. ripening, senescence, and abscission) may reflect evolutionary modification of ethylene signal transduction pathway regulation to meet the signaling needs of tissues greatly influenced by ethylene action.
CONCLUSION
Although a number of CTR1-like sequences from tomato and other plant species are available in the gene databases, there is no experimental evidence regarding their putative involvement in the ethylene signal transduction pathway. The isolation of the LeCTR1 genomic clone showed that its structural organization was very well conserved when compared with the Arabidopsis CTR1 gene. The availability of a well-characterized Arabidopsis ctr1-1 mutant offered a unique opportunity to investigate the predicted function of LeCTR1 through heterologous expression. By converting the constitutive triple-response phenotype of the Arabidopsis ctr1-1 mutant to a largely normal ethylene-responsive phenotype, we demonstrated that LeCTR1 encodes a functional ethylene signal transduction component capable of interacting with upstream and downstream partners of the Arabidopsis CTR1 protein. These data strongly support that the ethylene response pathways in tomato and Arabidopsis are composed of conserved components, yet there are potential differences.
In contrast with what has been previously reported in Arabidopsis for CTR1 (Kieber et al., 1993), we have shown that LeCTR1 is ethylene inducible. In accordance, LeCTR1 mRNA accumulation was up-regulated during tomato fruit ripening and in additional tissues synthesizing large amounts of ethylene. However, even though the data reported so far indicated that the expression of AtCTR1 was not affected significantly by ethylene (Kieber et al., 1993), it must be stressed here that there is only limited information on ethylene inducibility of this gene in Arabidopsis. The discrepancy observed between the two species for the expression of the CTR gene might also arise from the fact that the Arabidopsis CTR protein is encoded by a single gene, whereas in the tomato, at least two CTR isoforms exist that are encoded by a small multigene family (L. Adams and J. Giovannoni, unpublished data).
In tomato, it has been shown that the five gene members encoding the ethylene receptors are differentially regulated at the transcriptional level (Lashbrook et al., 1998). Interestingly, the tomato ethylene receptor, NR (LeETR3), shows similar induction to LeCTR1 during the course of fruit ripening in concert with increased ethylene evolution, which could be an effective method of regulating ethylene responsiveness of various plant tissues (Tieman et al., 2000). The observation that a second component of ethylene signaling in tomato, LeCTR1, demonstrates similar regulation suggests that multiple targets may exist for modulation of ethylene responsiveness in at least some plant species. It is striking that a negative regulator of ethylene response is induced during fruit ripening when just the opposite might be logically anticipated. This expression pattern may simply represent the ethylene-inducible nature of LeCTR1 in a tissue producing large amounts of ethylene, though said induction may have little physiological relevance. The fact that tomato fruit produce ethylene in concentrations greatly exceeding those required to induce ripening would support this possibility (Oeller et al., 1991). Alternatively, induction of LeCTR1 during ripening may represent a damping mechanism to prevent ripening and subsequent senescence from proceeding too rapidly. Analysis of LeCTR1 expression in tomato cultivars demonstrating variable fruit ethylene evolution rates and/or ripening times could clarify the role of LeCTR1 induction during fruit ripening.
MATERIALS AND METHODS
Plant Material
Arabidopsis (ecotype Columbia) plants were grown under standard greenhouse conditions. Agrobacterium tumefaciens-mediated transformation was carried out using the pGA643 binary vector according to Bird et al. (1988). The sense construct was generated by cloning the full LeCTR1 open reading frame under the transcriptional control of the cauliflower mosaic virus 35S promoter and the nopaline synthase terminator. The transformation protocol for in planta transformation of Arabidopsis was as described by Clough and Bent (1998). A. tumefaciens strain C58 carrying the binary plasmid pGA643 was grown to stationary phase in LennoxL-Broth medium at 28°C, 250 rpm. Cells were harvested by centrifugation for 20 min at room temperature at 5,500g and then resuspended in inoculation medium containing 5% (w/v) Suc and 0.05% (v/v) Silwet L-77 (OSI Specialties, Inc., Danbury, CT). Plants were inverted into this suspension such that all aboveground tissues were submerged, and plants were then removed after 3 to 5 s of gentle agitation. Plants were left in a low-light or dark location overnight and returned to the greenhouse the next day. Plants were grown for further 3 to 5 weeks until siliques were brown and dry. The selection of putative transformants was done on a 70 mg L−1 kanamycin-containing agar medium.
Isolation of the Genomic Clone
Genomic DNA was extracted from 1 g of ground tomato (Lycopersicon esculentum) leaves. The powder was mixed with 5 mL of extraction buffer (2% [w/v] hexadecyl-trimethyl-ammonium bromide, 1.4 m NaCl, 20 mm EDTA, and 100 mm Tris-HCl, pH 8) and warmed at 65°C for 10 min. After a phenol/chloroform/isoamylalcohol and chloroform extraction, DNA was precipitated with 1 volume of isopropanol for 20 min on ice. After centrifugation (5 min for 2,000g), the pellet was resuspended in 10 mL of wash buffer (76% [v/v] ethanol and 10 mm ammonium acetate). After centrifugation (10 min for 2,000g), the DNA was resuspended in 200 μL of sterile water. An RNAse treatment was done at 37°C for 10 min. Several primers were chosen based on the cDNA sequence and polymerization chain reactions were performed on the genomic DNA (primer 1, GAGCTGAAAATTGCTAATGGCAG, with primer 2, CCATCCCATAAATCCAATAAAATCC; primer 3, GGATTTTATTGGATTTATGGGATGG, with primer 4, CTTCTCCAGCAGGAGCTGCACCCC; primer 5, GGGGTGCAGCTCCTGCTGGAGAAG, with primer 6, CGCTTAAAACTCCAGGCTTACC; primer 7, GGTAAGCCTGGAGTTTTAAGCG, with primer 8, CTGTATCTGGTCGTGTCATCGGTG; primer 9, CACCGATGACACGACCAGATACAG, with primer 10, TGAGAGCAACTGCATGTCTGTGTG; and primer 11, CTCCTCTACCTCCACCAGGT, with primer 12, CATACAGTTATACAAGAATCCTGGGC). The fragments were cloned and sequenced. Comparative analysis between the genomic clone and cDNA sequences allowed the delimitation of introns and exons.
Southern Analysis
The genomic DNA was extracted as described above. DNA (5 μg) was digested with EcoRV enzyme. Digested DNA was separated in an agarose gel and blotted on a nylon membrane as described by Sambrook et al. (1989). A probe corresponding to the NPTII gene was labeled with [32P]dCTP using a random primer kit (Ready-to-Go, Amersham Biosciences, Piscataway, NJ). Blots were hybridized with a fragment of NPTII gene in a buffer containing 0.3 volumes of 1 m sodium phosphate buffer (pH 7.2), 0.7 volumes of 10% (w/v) SDS, and 1:500 (v/v) volumes of 0.5 m EGTA, pH 8. Washes were carried out as described by Sambrook et al. (1989).
Northern Analysis
Leaves from Arabidopsis plants were collected at the rosette stage and total RNA was obtained from 0.5 g of leaf tissue ground in liquid nitrogen and extracted with phenol as previously described (Verwoerd et al., 1989), except that the extraction buffer was 100 mm Tris-HCl (pH 8.0), 100 mm LiCl, 10 mm EDTA, and 1% (w/v) SDS. Total RNA (8 μg) was fractionated on a 1.2% (w/v) agarose gel containing formaldehyde in MOPS buffer (Sambrook et al., 1989) and then transferred onto GeneScreen membranes (PerkinElmer Life Sciences, Boston) following the manufacturer's instructions. Probes were labeled with [32P]dCTP using a random primer kit (Ready-to-Go, Amersham Biosciences). Blots were hybridized, as described for the Southern analysis, with a fragment of the basic chitinase cDNA (Samac et al., 1990) or with a fragment of the LeCTR1. To check for equal loading, a reverse picture of the ethidium bromide-stained gel was used. Washes were carried out under high stringency (Sambrook et al., 1989).
Ethylene Treatment
Sterilized seeds were put on Murashige and Skoog agar medium plates and placed at 4°C for 4 d. Ethylene treatment was carried out in sealed boxes. For air control, contaminating ethylene was removed using KMnO4. The different concentrations of ethylene applied were checked by gas chromatography and adjusted to 0.01, 0.1, 1, and 10 μL L−1. Hypocotyl measurements were made on 30 seedlings grown in darkness during 3 d with or without ethylene. The experiment was repeated three times and the level of apical curvature was estimated visually using a scale ranging from 0 to 4 (0, no apical hook; 1, 90° curvature; 2, 180° curvature; 3, beginning of hook formation; and 4, full hook). At a given ethylene concentration, the level of apical curvature was homogenous, i.e. >90% of the seedlings exhibited the same phenotype.
Mature green fruit were placed in a sealed chamber and gassed with 20 μL L−1 ethylene for 0 to 24 h. Four-week-old tomato plants were placed in a sealed chamber and gassed with or without 20 μL L−1 ethylene for 8 h.
Glc Sensitivity
Sterilized seeds were put on Murashige and Skoog agar medium containing 6% (w/v) filter-sterilized Glc. After 4 d at 4°C, plates were placed in light in a sealed box containing either air or 10 μL L−1 ethylene. Every 2 d, the boxes were opened to allow a renewal of the atmosphere and put back either with air or 10 μL L−1 ethylene. The experiment was stopped after 10 d of culture.
Quantitative RT-PCR
To obtain total RNA, the same protocol for northern analysis was used. The pellet was allowed to air dry and was resuspended in diethyl pyrocarbonate water. After quantification, 10 μg of RNA was treated with DNAse I (Promega, Madison, WI) and cleaned up with a phenol-chloroform extraction.
Real-time quantitative PCR was performed using 250 ng of total RNA for LeCTR1 and 2.5 pg for 18S in a 20-μL reaction volume using Taq-Man One-Step RT-PCR Master Mix reagents kit (PE-Applied Biosystems, Foster City, CA) on an ABI PRISM 7900HT sequence-detection system. PRIMER EXPRESS software (PE-Applied Biosystems) was used to design gene-specific primers and Taq-Man probes: LeCTR1 forward primer, CATCCTCTTTCTTACTGTGAGAAAATTTAGA; LeCTR1 reverse primer, CATTTCCCTGTATAAAAACGTTCAGTT; LeCTR1 Taq-Man probe, VIC-CCAACTGCCATTAGCAATTTTCAGCTCAA-TAMRA; 18S forward primer, CGGAGAGGGAGCCTGAGAA; 18S reverse primer, CCCGTGTTAGGATTGGGTAATTT; and 18S Taq-Man probe, 6FAM-CGGCTACCACATCCAAGGAAGGCA-TAMRA. For LeCTR1, optimal primer concentration was 900 nm and optimal probe concentration was 250 nm. Optimal primer and probe concentrations for 18S were 300 and 125 nm, respectively. RT-PCR conditions were as follows: 48°C for 30 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Samples were run in triplicate on each 384-well plate and were repeated on at least two plates for each experiment. For each sample, a Ct (threshold cycle) value was calculated from the amplification curves by selecting the optimal ΔRn (emission of reporter dye over starting background fluorescence) in the exponential portion of the amplification plot.
Relative fold differences were calculated based on the comparative Ct method using the 18S as an internal standard. To demonstrate that the efficiencies of the LeCTR1 and 18S primers and probes were approximately equal, the absolute value of the slope of log input amount versus delta Ct was calculated for both LeCTR1 and 18S and was determined to be <0.1. To determine relative fold differences for each sample in each experiment, the Ct value for LeCTR1 was normalized to the Ct value for 18S and was calculated relative to a calibrator (leaf for Fig. 8A and control tissues for Fig. 8, B and C) using the formula 2−ΔΔCt.
ACKNOWLEDGMENT
We are grateful to Simone Albert for excellent technical assistance in Arabidopsis genetic transformation and analysis of plant growth.
Footnotes
This work was supported in part by the European Union (grant no. FAIR CT–95 0225), by the Institut National de la Recherche Agronomique (Action Transversale Structurante Tomate, grant no. 2001–2003), by the “Midi-Pyrénées” Regional Council (grant nos. 99009080 and 01002710 to M.B.), and by the National Science Foundation (grant nos. IBN–9604115 and DBI–9872617 to J.J.G.). This research forms part of the requirement for the degree of PhD for J.L.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.009415.
LITERATURE CITED
- Abeles FB, Morgan PW, Salveit ME. Ethylene in Plant Biology. Ed 2. New York: Academic Press; 1992. [Google Scholar]
- Bird CR, Smith CJS, Ray JA, Moureau P, Bevan MW, Bird AS, Hughes S, Morris PC, Grierson D, Schuch W. The tomato polygalacturonase gene and ripening-specific expression in transgenic plants. Plant Mol Biol. 1988;11:651–662. doi: 10.1007/BF00017465. [DOI] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerovitz EM. Arabidopsis ethylene response gene ETR1: similarity of product two-component regulators. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- Clark KL, Larsen PB, Wang X, Chang C. Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc Natl Acad Sci USA. 1998;95:5401–5406. doi: 10.1073/pnas.95.9.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Ecker JR. The ethylene signal transduction pathway in plants. Science. 1995;268:667–675. doi: 10.1126/science.7732375. [DOI] [PubMed] [Google Scholar]
- Frye CA, Tang D, Innes RW. Negative regulation of defense responses in plant by conserved MAPKK kinase. Proc Natl Acad Sci USA. 2001;98:373–378. doi: 10.1073/pnas.98.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni JJ. Molecular biology of fruit maturation and ripening. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:725–749. doi: 10.1146/annurev.arplant.52.1.725. [DOI] [PubMed] [Google Scholar]
- Giovannoni JJ, Kannan P, Lee S, Yen H-C. Genetic approaches to manipulation of fruit development and quality in tomato. In: Cockshull KE, Gray D, Seymour GB, Thomas B, editors. Genetic and Environmental Manipulation of Horticultural Crops. New York: CABI Publishing; 1998. pp. 1–15. [Google Scholar]
- Guzmàn P, Ecker JR. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell. 1990;2:513–523. doi: 10.1105/tpc.2.6.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK, Quinn AM. Protein kinase catalytic domain sequence database: identification of conserved features of primary structure and classification of family members. Methods Enzymol. 1991;200:38–62. doi: 10.1016/0076-6879(91)00126-h. [DOI] [PubMed] [Google Scholar]
- Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- Hua J, Chang C, Sun Q, Meyerowitz EM. Ethylene insensitivity conferred by Arabidopsis ERS gene. Science. 1995;269:1712–1714. doi: 10.1126/science.7569898. [DOI] [PubMed] [Google Scholar]
- Hua J, Meyerowitz EM. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell. 1998;94:261–271. doi: 10.1016/s0092-8674(00)81425-7. [DOI] [PubMed] [Google Scholar]
- Johnson PR, Ecker JR. The ethylene gas signal transduction pathway: a molecular perspective. Annu Rev Genet. 1998;32:227–254. doi: 10.1146/annurev.genet.32.1.227. [DOI] [PubMed] [Google Scholar]
- Kieber JJ. The ethylene response pathway in Arabidopsis. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:277–296. doi: 10.1146/annurev.arplant.48.1.277. [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldman KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Lashbrook CC, Tieman DM, Klee HJ. Differential regulation of the tomato ETR gene family throughout plant development. Plant J. 1998;15:243–252. doi: 10.1046/j.1365-313x.1998.00202.x. [DOI] [PubMed] [Google Scholar]
- Lelièvre J-M, Latché A, Jones B, Bouzayen M, Pech J-C. Ethylene and fruit ripening. Physiol Plant. 1997;101:727–739. [Google Scholar]
- Oeller PW, Wong LM, Taylor LP, Pike DA, Theologis A. Reversible inhibition of tomato fruit senescence by antisense 1-aminocyclopropane-1-carboxylate synthase. Science. 1991;254:427–439. doi: 10.1126/science.1925603. [DOI] [PubMed] [Google Scholar]
- Payton S, Fray RG, Brown S, Grierson D. Ethylene receptor expression is regulated during fruit ripening, flower senescence and abscission. Plant Mol Biol. 1996;31:1227–1231. doi: 10.1007/BF00040839. [DOI] [PubMed] [Google Scholar]
- Samac DA, Hironaka CM, Yallaly PE, Shah DM. Isolation and characterization of the genes encoding basic and acidic chitinase in Arabidopsis thaliana. Plant Physiol. 1990;93:907–914. doi: 10.1104/pp.93.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schaller GE, Bleecker AB. Ethylene-binding site generated in yeast expressing the Arabidopsis ETR1 gene. Science. 1995;270:1809–1811. doi: 10.1126/science.270.5243.1809. [DOI] [PubMed] [Google Scholar]
- Schenk PW, Snaar-Jagalska BE. Signal perception and transduction: the role of protein kinase. Biochem Biophys Acta. 1999;1449:1–24. doi: 10.1016/s0167-4889(98)00178-5. [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Ecker JR. Ethylene signaling: from mutants to molecules. Curr Opin Plant Biol. 2000;3:353–360. doi: 10.1016/s1369-5266(00)00096-0. [DOI] [PubMed] [Google Scholar]
- Tieman DM, Klee HJ. Differential expression of two novel members of the tomato ethylene-receptor family. Plant Physiol. 1999;120:165–172. doi: 10.1104/pp.120.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman DM, Taylor MG, Ciardi JA, Klee HJ. The tomato ethylene receptors NR and LeETR4 are negative regulators of ethylene response and exhibit functional compensation within a multigene family. Proc Natl Acad Sci USA. 2000;97:5663–5668. doi: 10.1073/pnas.090550597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwoerd TC, Dekker BMM, Hoekema A. A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 1989;17:2362. doi: 10.1093/nar/17.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson JQ, Lanahan MB, Yen HC, Giovanoni JJ, Klee HJ. An ethylene inducible component of signal transduction encoded by Never-ripe. Science. 1995;257:967–971. doi: 10.1126/science.270.5243.1807. [DOI] [PubMed] [Google Scholar]
- Zegzouti H, Jones B, Frasse P, Marty C, Maître B, Latché A, Pech J-C, Bouzayen M. Ethylene-regulated gene expression in tomato fruit: characterization of novel ethylene-response and ripening-related genes isolated by differential-display. Plant J. 1999;18:589–600. doi: 10.1046/j.1365-313x.1999.00483.x. [DOI] [PubMed] [Google Scholar]
- Zhou D, Kaliatzis P, Mattoo AK, Tucker M. The mRNA for an ETR1 homologue in tomato is constitutively expressed in vegetative and reproductive tissues. Plant Mol Biol. 1996;30:1331–1338. doi: 10.1007/BF00019564. [DOI] [PubMed] [Google Scholar]
- Zhou L, Jank J-C, Jones TL, Sheen J. Glucose and ethylene signal transduction cross talk revealed by an Arabidopsis glucose-insensitive mutant. Proc Natl Acad Sci USA. 1998;95:10294–10299. doi: 10.1073/pnas.95.17.10294. [DOI] [PMC free article] [PubMed] [Google Scholar]