Abstract
We have isolated a new recessive dwarf mutant of rice (Oryza sativa L. cv Nipponbare). Under normal growth conditions, the mutant has very short leaf sheaths; has short, curled, and frizzled leaf blades; has few tillers; and is sterile. Longitudinal sections of the leaf sheaths revealed that the cell length along the longitudinal axis is reduced, which explains the short leaf sheaths. Transverse sections of the leaf blades revealed enlargement of the motor cells along the dorsal-ventral axis, which explains the curled and frizzled leaf blades. In addition, the number of crown roots was smaller and the growth of branch roots was weaker than those in the wild-type plant. Because exogenously supplied brassinolide considerably restored the normal phenotypes, we designated the mutant brassinosteroid-dependent 1 (brd1). Further, under darkness, brd1 showed constitutive photomorphogenesis. Quantitative analyses of endogenous sterols and brassinosteroids (BRs) indicated that BR-6-oxidase, a BR biosynthesis enzyme, would be defective. In fact, a 0.2-kb deletion was detected in the genomic region of OsBR6ox (a rice BR-6-oxidase gene) in the brd1 mutant. These results indicate that BRs are involved in many morphological and physiological processes in rice, including the elongation and unrolling of leaves, development of tillers, skotomorphogenesis, root differentiation, and reproductive growth, and that the defect of BR-6-oxidase caused the brd1 phenotype.
Brassinosteroids (BRs) have various physiological and morphological effects on plants and are involved in the elongation of stems, unrolling of leaves, responses to environmental stress (Bishop and Yokota, 2001), and differentiation of tracheary elements (Yamamoto et al., 1997, 2001). Numerous mutants unable to synthesize BRs have been isolated, including de-etiolated2-1 to -8 (Chory et al., 1991; Li et al., 1996), constitutive photomorphogenesis and dwarfism (Szekeres et al., 1996), dwarf4-1 to -4 (Azpiroz et al., 1998; Choe et al., 1998), diminuto (Takahashi et al., 1995; Klahre et al., 1998), sterol1-2 and -3 (Choe et al., 1999), dwarf5-1 to -6 (Choe et al., 2000) in Arabidopsis, dwarf (Bishop et al., 1996, 1999) in tomato (Lycopersicon esculentum), and lkb (Nomura et al., 1997, 1999; Schultz et al., 2001) in pea (Pisum sativum). The causative genes have been cloned by using such mutants, and this has clarified the physiological effects of BRs (Bishop and Yokota, 2001). These BR-defective mutants usually exhibit a constitutive photomorphogenesis—a de-etiolated phenotype with less hypocotyl elongation than that shown by wild-type plants, even in the dark (Chory et al., 1991; Kauschmann et al., 1996)—which means that BRs are essential for skotomorphogenesis. Treatment with brassinazole (brz) or Brz2001, both of which are specific inhibitors of brassinolide (BL) biosynthesis, is effective to repress endogenous BL functions in dicots, such as Arabidopsis (Asami et al., 2000; Sekimata et al., 2001). It was recently reported that brz binds to the DWARF4 protein (Asami et al., 2001).
To identify components of the BR signaling pathway, researchers have used a genetic approach: identifying mutants that are insensitive to exogenously applied BRs. The Arabidopsis mutants brassinosteroid insensitive 1 (bri1; Clouse et al., 1996) and brassinosteroid insensitive 2 (Li et al., 2001b) were identified. BRI1, encoding a Leu-rich repeat receptor-like protein kinase (Li and Chory, 1997), is a plasma membrane protein and functions as a Ser/Thr protein kinase in vitro (Friedrichsen et al., 2000; Oh et al., 2000). Further, the treatment of Arabidopsis seedlings with BL activates autophosphorylation of the BRI1 protein (Wang et al., 2001). These results suggest that BRI1 is a BL receptor. Activation tagging of a weak bri1 allele (bri1-5) resulted in the identification of BRI1 SUPPRESSOR (BRs1), which was predicted to encode a secreted carboxypeptidase. Overexpression of BRs1 protein can suppress the bri1 allele, strongly suggesting that BRs1 is involved in BR signaling (Li et al., 2001a). It was recently shown that BRASSINOSTEROID INSENSITIVE 2 encodes a GSK3/SHAGGY-like kinase and acts as a negative regulator to control BR signaling in plants (Li and Nam, 2002).
In contrast to the many studies performed in Arabidopsis and some other dicots, only a few reports have been published on the physiological effects of BRs on the growth and development of monocots. The most well-known effect is that exogenous BR, alone or in combination with auxin, enhances the bending of the lamina joint of rice (Oryza sativa; Maeda, 1965). Because brz and Brz2001 do not have marked effects on rice (Sekimata et al., 2001), it is important to isolate and analyze BR biosynthetic mutants of rice to understand the function of BR in rice and other monocots. OsBRI1, with extensive sequence similarity to the Arabidopsis BRI1 gene, which encodes a putative BR receptor kinase, was recently cloned (Yamamuro et al., 2000). Through analyses of a loss-of-function mutant of OsBRI1, it was found that the gene functioned in internode elongation, bending of the lamina joint, and skotomorphogenesis (Yamamuro et al., 2000).
To our knowledge, this is the first report of a BR biosynthesis mutant of rice. We characterized its morphological and physiological features, measured the contents of sterols and BRs, and found a defect in the rice BR-6-oxidase (OsBR6ox) gene.
RESULTS
Isolation of a Recessive Dwarf Mutant of Rice
The mutant used in this study was originally identified as a rice cv Nipponbare dwarf mutant in transgenic lines produced in our laboratory. The phenotype of the mature plant was extreme dwarfism. The phenotype was not observed in the T0 (M1) generation but appeared at T1 (M2). The mutant phenotype segregated as a monogenic recessive mutation; T1 (M2) plants segregated at 115 (wild type):34 (mutant). Southern-blot analyses of the T0 and T1 plants revealed that the copy number of T-DNA at T0 was 1 and that linkage between the dwarf phenotype and the T-DNA insertion was not detectable in the T1 (M2; data not shown). As a result, some T1 plants either with or without the T-DNA insertion showed the dwarf phenotype. We later designated the dwarf mutation that was not derived from the T-DNA insertion as brassinosteroid dependent 1 (brd1), and we have used the line without the T-DNA insertion in this study. T2 plants from a brd1 heterozygote without the T-DNA insertion segregated at 137 (wild type):45 (mutant), which showed that the recessive brd1 mutation is stably inherited by the next generation. It is suggested that abortive integration of the T-DNA caused the mutation. As an alternative, the cell culture and regeneration process used in transgenic rice production by the method of Hiei et al. (1994) might cause the mutation.
Mutant Phenotypes of Leaves and Panicles
Figure 1, A through C, illustrates the mutant phenotypes grown under white light. Compared with the wild-type plant, the mutant has extremely short leaf sheaths and short leaf blades. In addition, the leaf blades of the mutant are curled and frizzled. The mutant plant has less tillering than the wild type and does not usually have panicles, although very rarely short panicles with some small and sterile seeds were observed in the greenhouse (Fig. 1, D–F). An examination of the seeds, by removing lemmas and paleas, revealed that the albumen had not developed in the mutant seeds.
Figure 1.
Morphology of brd1 plant. A, Three-week-old brd1 seedling grown in the growth chamber. Bar = 1 cm. B, Eighty-day-old wild-type (WT) and brd1 plants grown in soil in the growth chamber. Bar = 10 cm. C, Close-up of brd1 in B. D, Six-month-old brd1 plant with a short panicle grown in the greenhouse in soil at 28°C. E, Wild-type and brd1 panicles. Bar = 1 cm. F, Seeds of wild-type and brd1 plants. Bar = 5 mm.
The Mutant Phenotypes Are Rescued by BRs
Many Arabidopsis dwarf mutants revert to normal phenotype with addition of BL (Li et al., 1996; Szekeres et al., 1996; Azpiroz et al., 1998; Klahre et al., 1998; Choe et al., 1999). However, no rice dwarf mutants have been reported to recover. We cultured plants in solution to clarify the effects of BL on mutant rice. Different concentrations of BL were added to the solution. Figure 2 shows the results. At 1 nm, BL was not able to remarkably change either the length of the leaf sheaths and blades or the curled and frizzled appearance after 30 d. However, at 10 and 100 nm, it was able to rescue both of the mutant phenotypes. The leaf sheaths of the mutant grown in 100 nm BL were slightly longer than those grown in 10 nm BL, but old leaf blades grown in 100 nm BL bent, which indicated that the higher concentration is superfluous. A lamina inclination assay showed that exogenous BL bends leaf blades of wild-type rice (Maeda, 1965). Leaf sheaths of the mutant in 1,000 nm BL extended for about 2 weeks after addition, but the growth gradually deteriorated.
Figure 2.
Phenotypic restoration of brd1 plant by BL. Seedlings were germinated and grown on one-half-strength Murashige and Skoog medium containing both 3% (w/v) Suc and 0.4% (w/v) Gelrite. When the fourth leaf emerged (about 10 d after sowing), plants were transplanted into Kimura's B solution (Sato et al., 1996) with or without BL. The photograph was taken after an additional 30 d of growth.
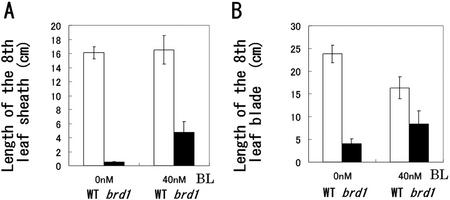
To measure the restoration of the leaf length by exogenously supplied BL, we cultured plants in the presence or absence of 40 nm BL, an intermediate concentration between 10 and 100 nm, because the leaf sheaths of the mutant grown in the solution culture containing 40 nm BL were longer than those grown in the solution culture containing 10 nm BL after an additional 80 d. The lengths of the eighth leaf sheath and the eighth leaf blade were measured as representative values. The length of wild-type leaf sheaths did not increase with the addition of 40 nm BL, but that of mutant leaf sheaths increased about nine times after 30 d (Fig. 3A). The length of wild-type leaf blades decreased to about two-thirds on addition of 40 nm BL, but that of mutant leaf blades approximately doubled (Fig. 3B). The values obtained by dividing the length of the eighth leaf blade with that of the eighth leaf sheath are 1.5 (wild type, 0 nm), 8.0 (mutant, 0 nm), 1.0 (wild type, 40 nm), and 1.8 (mutant, 40 nm). These results indicate that the addition of 40 nm BL also restored the normal ratio of leaf blade to leaf sheath in the mutant. Application of 100 nm castasterone (CS), another BR, also restored the mutant phenotypes (data not shown). We designated this rice mutant brd1, because the phenotypes are restored by BRs.
Figure 3.
Length of the eighth leaf of wild-type (WT) and brd1 plants grown with or without BL. A, Length of the eighth leaf sheath grown with or without BL. B, Length of the eighth leaf blade grown with or without BL. Seedlings were germinated and grown on one-half-strength Murashige and Skoog medium containing both 3% (w/v) Suc and 0.4% (w/v) Gelrite. When the fourth leaf emerged (about 10 d after sowing), plants were transplanted to Kimura's B solution culture medium with or without 40 nm BL. The lengths of the leaves were measured after an additional 30 d of growth. The results are presented as mean values ± sd from five to seven plants.
Histological Observation
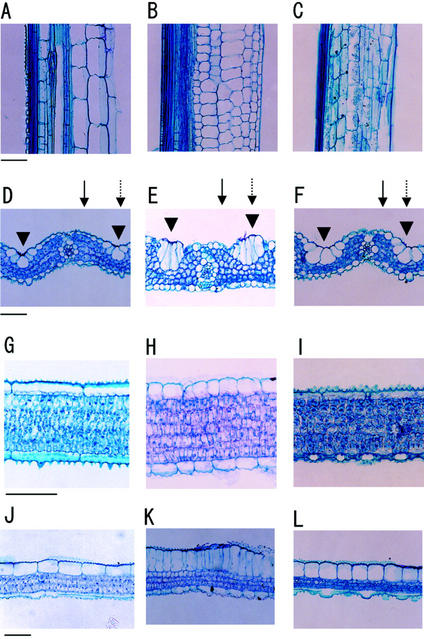
We examined leaf histology to determine the cause of the morphological change mentioned above. We examined the eighth leaf (L8) at the L11 emergence stage. Figure 4, A and B, indicates the cell morphology of the leaf sheaths in longitudinal sections of wild-type and mutant plants. In the wild type, cells were longitudinally elongated, whereas in the mutant they were shorter (by >75%). Figure 4, D and E, indicates the cell morphology of the L8 blades in transverse sections of wild-type and mutant plants. The larger size of the motor cells of the mutant plant might cause the curly phenotype of the leaf blades, because the motor cells play an important role in leaf rolling (Hoshikawa, 1989). Figure 4, G, H, J, and K, indicates the cell morphology of the L8 blades in longitudinal sections of both plants. Mutant mesophyll cells in the leaf blades were packed more tightly than their wild-type counterparts, and the leaf blades of mutant plants had smaller intercellular spaces than their wild-type counterparts. In addition, both epidermal cells and motor cells along the longitudinal axis of the mutant leaf blades were much shorter (25%–50% of the size of wild-type cells). As a result, mutant epidermal and motor cells are more expanded in the dorsal-ventral axis of the leaf blade than wild-type cells, thereby increasing the thickness of the leaf blades. All of these phenotypes of the mutant cells were restored by 40 nm BL (Fig. 4, C, F, I, and L).
Figure 4.
Light microscopy of wild-type and brd1 leaves sectioned longitudinally and transversely. A through C, Longitudinal sections of the central region of the eighth leaf sheath of wild-type (A), brd1 (B), and brd1 in the presence of 40 nm BL (C). D through F, Transverse sections of the central region of the eighth leaf blade of wild-type (D), brd1 (E), and brd1 in the presence of 40 nm BL (F). Arrows indicate the cutting point shown in G to I. Broken arrows indicate the cutting point shown in J to L. Arrowheads indicate motor cells. G through L, Longitudinal sections of the central region of the eighth leaf blade of wild-type (G and J), brd1 (H and K), and brd1 in the presence of 40 nm BL (I and L). Seedlings were germinated and grown on one-half-strength Murashige and Skoog medium containing both 3% (w/v) Suc and 0.4% (w/v) Gelrite. When the fourth leaf emerged (about 10 d after sowing), plants were transplanted to Kimura's B solution culture medium with or without 40 nm BL. When the 11th leaf blade emerged, the eighth leaf was examined. Bar = 50 μm.
Brd1 Exhibits Constitutive Photomorphogenesis in Darkness
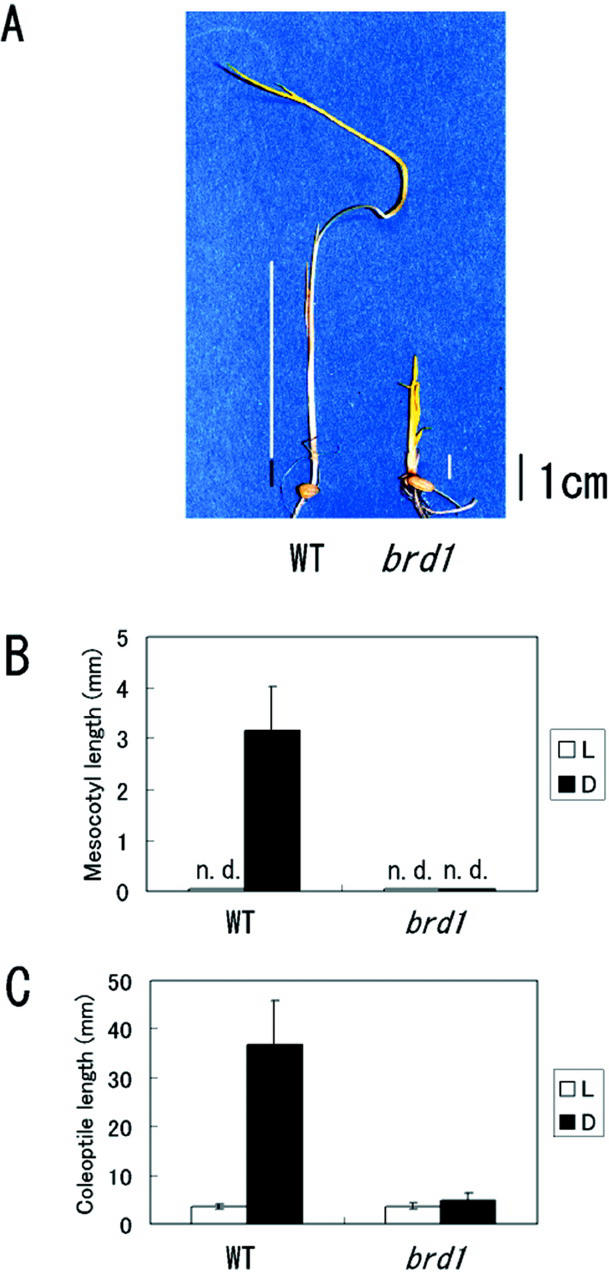
Many Arabidopsis mutants, tomato extreme dwarf mutant, and rice d61 mutant, which are defective in BR biosynthesis or BR signal transduction, exhibit a constitutive photomorphogenic phenotype in the dark (Bishop et al., 1996; Clouse and Sasse, 1998; Schumacher and Chory, 2000; Yamamuro et al., 2000). To test whether brd1 also shows such a phenotype, seedlings were grown in the dark. The elongation of coleoptiles and mesocotyls is a typical rice response to constant darkness (Takano et al., 2001). Figure 5A shows wild-type and mutant rice seedlings grown in the dark. Wild-type rice plants germinated and grown in the dark showed an obvious elongation of the mesocotyl and coleoptile compared with those grown under light. No such elongation occurred in the mutant (Fig. 5, B and C). In addition, an extension of the second internode in wild-type plants grown in the dark was detected as reported by Yamamuro et al. (2000), whereas it was not detected in brd1 (data not shown).
Figure 5.
Photomorphogenic reaction. A, Two-week-old wild-type (WT) and brd1 seedlings grown in the dark. Black bar indicates the mesocotyl length; white bars indicate coleoptile lengths. B and C, WT and brd1 seedlings were grown under white light (L) or in the dark (D) for 2 weeks. The lengths of mesocotyls (B) and coleoptiles (C) of these seedlings were measured individually. WT and brd1 seedlings were germinated and grown on one-half-strength Murashige and Skoog medium containing 3% (w/v) Suc and 0.4% (w/v) Gelrite. The results are presented as mean values ± sd from seven to 10 plants. n.d., The corresponding tissue was not detected.
Root Phenotypes
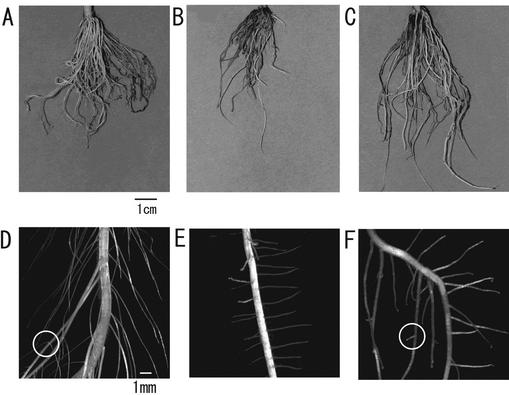
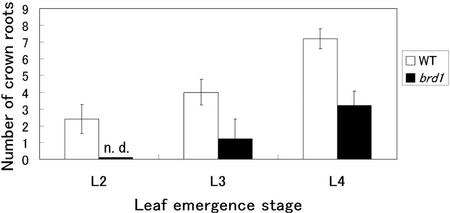
Exogenously supplied BRs inhibit Arabidopsis root growth (Clouse et al., 1993) and many Arabidopsis severe mutants in BR biosynthesis have shorter roots than wild-type plants (Chory et al., 1991; Takahashi et al., 1995; Azpiroz et al., 1998). However, little is known about the function of endogenous BRs in root development. brd1 in solution culture did not have shorter roots, at least not 70 d after sowing. However, because the number of crown roots in the mutant seemed to be small (see Fig. 7, A and B), we compared the number with that of the wild type. Figure 6 indicates that at the L2 emergence stage, two or three crown roots were elongated in the wild-type plant but none was elongated in the mutant. At the L4 stage, about seven crown roots were similarly elongated in the wild type compared with three in the mutant. These results suggest that BRs promotes the development of crown roots in rice.
Figure 7.
Root morphology. A through C, Photographs of roots. D through F, Roots observed under stereomicroscope. A and D, Wild type; B and E, brd1; C and F, brd1 in the presence of 1 nm BL. Seedlings were germinated and grown on one-half-strength Murashige and Skoog medium containing 3% (w/v) Suc and 0.4% (w/v) Gelrite. When the fourth leaf emerged (about 10 d after sowing), plants were transplanted into Kimura's B solution with or without BL. The photograph was taken after an additional 60 d of growth. Bar = 1 cm in A through C and 1 mm in D through F. Circles indicate secondary branched roots.
Figure 6.
Number of crown roots at different leaf emergence stages. The number of crown roots which extended over 2 mm long was counted. Seedlings were grown in the light on one-half-strength Murashige and Skoog medium containing 3% (w/v) Suc and 0.4% (w/v) Gelrite. The results are presented as mean values ± sd from 10 plants. WT, Wild-type. n.d., Crown roots were not detected.
Figure 7 shows the root morphology of wild-type plants, brd1 mutant plants, and mutant plants grown in the presence of 1 nm BL. The number of crown roots in the mutant was smaller than in the wild type but was partly rescued by 1 nm BL (Fig. 7, A–C). We further observed the branched roots by stereomicroscopy. Two types of primary branched roots (thick and thin) grow on the wild-type crown, and secondary branched roots grow on the thick primary branched roots (Hoshikawa, 1989). Wild-type roots had thick and long primary branched roots with secondary branches (Fig. 7D). However, only thin and short primary branched roots were found on the mutant crown (Fig. 7E). Secondary branched roots were found on some long primary branched roots of the mutant grown in the presence of 1 nm BL, which also indicates that the mutant root phenotype was partly rescued by BL. The mutant grown in the presence of 10 nm BL also extended secondary branched roots, but BL at both 100 and 1,000 nm inhibited the extension of crown roots, and no secondary branched roots were found (data not shown). Our results suggest that the endogenous level of BL is related to the development of crown roots, thick primary branched roots, and secondary branched roots, and to the extension of the thin primary branched roots.
Quantitative Analyses of Sterols and BRs
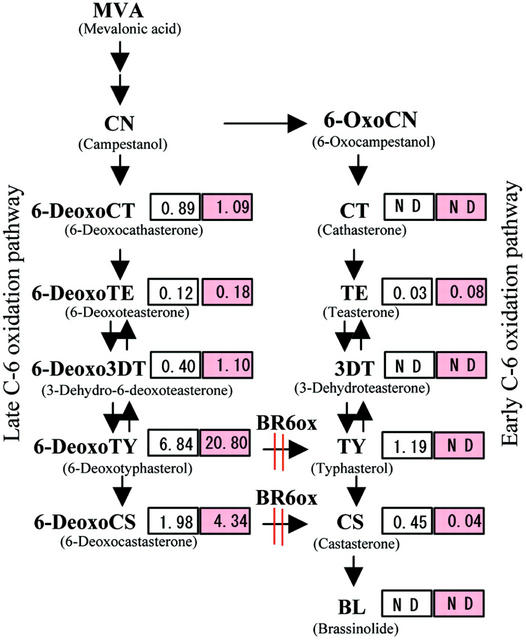
To identify the metabolic block in sterol and BR biosynthesis, we used gas chromatography-selected ion monitoring (GC-SIM) to chemically analyze brd1 and wild-type plant material. Analysis of the content of both campesterol (CR) and campestanol (CN) preceding the BR biosynthetic pathway did not show large differences between brd1 (84.1 μg g−1 fresh weight CR and 2.9 μg g−1 fresh weight CN) and wild-type (63.0 μg g−1 fresh weight CR and 1.4 μg g−1 fresh weight CN) plants. In contrast, analysis of the content of BRs of brd1 showed a decrease of CS and typhasterol (TY), and an increase of 6-deoxocastasterone (6-DeoxoCS), 6-deoxotyphasterol (6-DeoxoTY), and 3-dehydro-6-deoxoteasterone (6-Deoxo3DT) compared with wild-type plants (Fig. 8). Shimada et al. (2001) reported that BR-6-oxidases from Arabidopsis and tomato catalyze multiple C-6 oxidations in BR biosynthesis. This suggests that rice BR-6-oxidase would be defective in brd1.
Figure 8.
BL biosynthetic pathway and BR content. BR amounts, in nanograms per gram fresh weight, of wild-type and brd1 plants are shown in white and red boxes, respectively. ND, Not detected. BR6ox, BR-6-oxidase.
Molecular Characterization of the brd1 Mutation
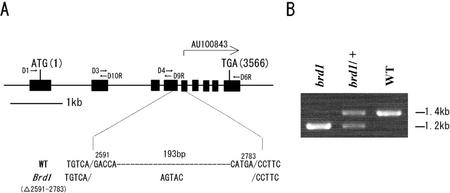
We searched rice databases for homologs of the tomato Dwarf gene (Bishop et al., 1996), which encodes a BR-6-oxidase, and of the Arabidopsis BR6ox gene (Shimada et al., 2001), and we found one expressed sequence tag (EST) clone (GenBank/EMBL/DNA data bank of Japan accession no. AU100843) with the highest sequence similarity. When the sequences were compared, it was apparent that the EST was partial. Therefore, we compared the sequence of AU100843 with the draft sequence of the rice genome (Yu et al., 2002) and found that only 1 contig (18,223) was homologous to the sequence of AU100843 (E value, 0.0). Comparison of the genomic sequence with the sequences of AU100843, tomato Dwarf, and Arabidopsis BR6ox allowed us to deduce that the rice homolog, designated OsBR6ox, consists of nine exons (Fig. 9A). Because only 1 contig was homologous to the sequence of AU100843, we consider OsBR6ox to be a single-copy gene in the rice genome.
Figure 9.
A, Schematic representation of OsBR6ox and the location of the deletion in the brd1 mutant. The closed boxes indicate exons. The approximate locations of primer sequences are shown by arrows. AU100843 is an EST clone of rice. B, PCR analysis of OsBR6ox in brd1, brd1/+, and wild-type plants. An ethidium bromide-stained agarose gel shows PCR products generated by using primers D4 and D6R, which amplify the genomic DNA containing exons 4 to 9.
PCR analysis of OsBR6ox in brd1, brd1/+, and wild-type plants in the T2 generation showed that a deletion of about 200 bp occurred in the genomic region from exon 4 to exon 9 in brd1 when we used primers D4 and D6R (Fig. 9B). In total, we analyzed 30 T2 plants. PCR products from eight brd1 plants showed only a 1.2-kb band. PCR products from seven wild-type plants showed only a 1.4-kb band. PCR products from 15 brd1/+ plants that showed the wild-type phenotype but produced both wild-type and mutant plants in the T3 generation showed both 1.2- and 1.4-kb bands. These results indicate that the genotypes of all 30 plants were perfectly linked with the PCR polymorphism and strongly suggest that the 0.2-kb deletion in OsBR6ox caused the brd1 mutation. Sequence analysis of the genomic region from exon 4 to exon 9 in brd1 showed that both a 193-bp deletion and a 5-bp (AGTAC) insertion occurred between exons 4 and 5 (Fig. 9A). The deletion and the insertion also caused a frameshift, causing a loss-of-function mutation of OsBR6ox.
DISCUSSION
Although Yamamuro et al. (2000) have characterized the BR-insensitive mutant d61 of rice, to our knowledge, no BR-biosynthetic mutant of rice has been published. Yamamuro et al. stated that the main functions of BRs in rice are internode elongation, bending of lamina joints, and skotomorphogenesis. Although the phenotypes of leaf sheaths of both d61, a loss-of-function mutant of OsBRI1, and transgenic rice carrying the antisense strand of OsBRI1 (Yamamuro et al., 2000) suggested a relationship between BRs and the elongation of leaf sheaths, direct evidence that BRs elongate the leaf sheaths had not been obtained.
By using the brd1 mutant, we have shown that BRs play an essential role in the elongation of leaf sheaths and leaf blades (Figs. 2 and 3). One reason for the extremely short leaf sheaths of brd1 is the prevention of longitudinal cell elongation (Fig. 4, A and B). Cells of the eighth leaf sheath are several times larger in the wild-type plant than in the brd1 mutant. But the length of the mutant sheaths is one-thirtieth that of the wild-type sheaths (Fig. 3A). Prevention of cell elongation cannot entirely explain such short sheaths, and we suggest that cell division is also prevented in the development of leaf sheaths in brd1. The recent report that BRs control the proliferation of leaf cells of Arabidopsis (Nakaya et al., 2002) reinforces our suggestion.
Motor cells are characteristic of leaves of gramineous plants. When leaf moisture decreases, turgor pressure is lost, and the cells shrink along the transverse axis. As a result, the leaf rolls up, with the upper surface inside. The rolling of rice leaves when they lose moisture is attributable to the action of these motor cells (Hoshikawa, 1989). In the mutant leaf blades, motor cells and epidermal cells were tightly pressed along the longitudinal axis (Fig. 4, H and K) and elongated along the dorsal-ventral axis compared with those in the wild-type leaf blades. Although different from the rolling in the dry condition, the abnormal cell morphology of the mutant motor cells might cause the curled and frizzled leaf blades.
One reason for the short leaf blades of brd1 is the prevention of longitudinal cell elongation of motor cells and epidermal cells (Fig. 4, H and K). Although the leaf blades of mutant plants had smaller intercellular spaces than their wild-type counterparts, the reduction in cell size of mesophyll cells was not as remarkable as that of motor and epidermal cells (Fig. 4, G, H, J, and K). These results suggest that cell division is also prevented in the development of leaf blades of brd1 and that the effect of BL on cell extension depends on cell type more strongly for the epidermal and motor cells than for the mesophyll cells in leaf blades.
Exogenously supplied BL decreased the ratio of leaf blade to leaf sheath in rice (Fig. 3), which suggests that endogenous BRs might also control leaf proportions in rice. However, exogenously added BL (40 nm) could not restore the reproductive development of brd1, which suggests that the endogenous levels of BRs were not high enough to start or maintain reproductive growth. As an alternative, related steroids that are essential for reproductive development might also be defective in brd1.
The addition of 5 μm gibberellin A3 (GA3) induced the elongation of leaf blades and sheaths in wild-type rice. In the mutant, 5 μm GA3 did not have a large effect on the elongation of leaf sheaths or on the curling of leaf blades, but induced the elongation of leaf blades (data not shown). These results suggest that in the wild type, BRs are less involved in the extension of leaf blades by GA3 but contributes to the extension of the leaf sheaths by GA3. In the mutant, GA3 was not involved in leaf expansion, but BRs were involved. The addition of 1, 10, or 100 μm 3-indoleacetic acid did not induce any remarkable effects on mutant rice, at least not 2 weeks after the addition (data not shown).
In the BR-defective constitutive photomorphogenesis and dwarfism mutant of Arabidopsis and brz-treated cress (Lepidium sativum) plants, a predominant differentiation of phloem and partial inhibition of the development of xylem was detected, which indicates that BRs function in xylem development (Szekeres et al., 1996; Nagata et al., 2001). We examined phloem and xylem of wild-type and brd1 rice plants (Fig. 4, D and E), but we did not detect any obvious differences. Because the developmental mechanism of phloem and xylem is different in dicots and monocots, BRs might not contribute to the development of phloem and xylem in monocots.
In tomato, conversion of 6-DeoxoCS to CS by the DWARF enzyme is the only major C-6 oxidation pathway (Bishop et al., 1999). In Arabidopsis, in addition to the C-6 oxidation of 6-DeoxoCS to CS, another C-6 oxidation of 6-DeoxoTY to TY is also present (Noguchi et al., 2000). Both steps are catalyzed by the product of AtBR6ox, a homolog of the tomato Dwarf gene (Shimada et al., 2001). Molecular characterization of OsBR6ox, a homolog of Dwarf and AtBR6ox, showed that the gene was defective in the brd1 mutant (Fig. 9). The content of BRs in the mutant suggests that there are at least two C-6 oxidation pathways catalyzed by OsBR6ox in rice, as in Arabidopsis, because TY was not detected in the mutant, but it was detected in the wild type (Fig. 8). The level of 6-Deoxo3DT also increased in brd1, maybe as a result of reversible conversion between 6-DeoxoTY and 6-Deoxo3DT (Noguchi et al., 2000). The results in Figure 8 also suggest that the early C-6 oxidation pathway would be a minor pathway in rice, because the level of TE was very low, and neither CT nor 3DT was detected.
The deduced amino acid sequence of OsBR6ox consists of 470 amino acid residues, has characteristics of cytochrome P450s, and is similar to that of tomato DWARF and AtBR6ox, with 63% and 58% sequence identity, respectively (Fig. 10). As a result of the frameshift in OsBR6ox in brd1, however, the carboxy-terminal region of the open reading frame (ORF; from Asp-300 to Tyr-470) is disrupted. Because the essential heme-binding consensus sequence of cytochrome P450, FxxGxxxCxG (lowercase x indicates variable amino acid residues; Nelson et al., 1996), is located in the carboxy-terminal region (Fig. 10), BR-6-oxidase in brd1 would be nonfunctional. Because rice cv 93-11 was used for the draft sequence (Yu et al., 2002) but we used rice cv Nipponbare, we sequenced the genomic region of rice cv Nipponbare corresponding to the deduced ORF of OsBR6ox and confirmed that the deduced amino acid sequence of OsBR6ox is perfectly conserved between rice cvs Nipponbare and 93-11, except for the loss of Ser-165 in rice cv Nipponbare by a 3-bp deletion (data not shown). All of these data strongly suggest that the 193-bp deletion and the 5-bp insertion in OsBR6ox lead to reductions of TY and CS, which causes the brd1-conferred phenotype.
Figure 10.
Multiple sequence alignment of OsBR6ox with known sequences for BR-6-oxidases. The GenBank/EMBL/DNA data bank of Japan accession numbers are AB035868 for the Arabidopsis BR6ox gene and U54770 for the tomato Dwarf gene. The amino acid sequence of OsBR6ox was deduced from the DNA sequence of contig 18,223 from the Web site at http://btn.genomics.org.cn/rice (Yu et al., 2002). The heme-binding signature sequence of cytochrome P450 is underlined. As a result of the frameshiftings deletion in OsBR6ox in brd1, the carboxy-terminal region of the ORF (from Asp-300 to Tyr-470) is disrupted. Reverse contrast characters highlight identical amino acid residues. Gaps introduced to improve alignment are shown by hyphens.
Brd1 should be useful to help identify the BR signaling cascade in monocots in near future because signaling is thought to be arrested under normal conditions but restored with application of BL.
MATERIALS AND METHODS
Growth Conditions
Dehusked seeds of wild-type (Oryza sativa L. cv Nipponbare) and mutant rice were surface-sterilized and sown on one-half-strength Murashige and Skoog medium containing 3% (w/v) Suc and 0.4% (w/v) Gelrite (Wako Pure Chemicals, Osaka). Seedlings were transplanted into soil or solution culture medium about 10 d after sowing. Kimura's B medium (Sato et al., 1996) with or without BL was used for solution culture. The medium was changed twice a week. Unless specified, plants were grown in a growth chamber at 28°C under long-day conditions (14 h light [60–70 μmol m−2 s−1]/10 h dark).
Chemicals
BL, CS, and 3-indoleacetic acid were purchased from Wako Pure Chemicals. Gibberellin A3 was purchased from Sigma-Aldrich (St. Louis). Murashige and Skoog salt was purchased from Wako Pure Chemicals.
Histological Observations
For histological examination, leaves were fixed in formalin:acetic acid:70% (v/v) ethanol (1:1:18, v/v). Excised leaves were dehydrated in a graded ethanol/tert-butanol series, embedded in paraffin, and sectioned to 10 μm on a rotary microtome (HM 335E, Carl Zeiss Co., Oberkochen, Germany). Paraffin sections were stained with toluidine blue O (0.01%, w/v) and observed under a light microscope.
Extraction and Purification of Sterols
Mature shoots (wild type, 2.83 g fresh weight; brd1, 0.35 g fresh weight) grown in solution culture were harvested and extracted with methanol:chloroform (5:1, v:v). The extract was evaporated to dryness and partitioned between ethyl acetate and 0.5 m K2HPO4 buffer. A portion (100 mg fresh weight equivalent) of the ethyl acetate phase was spiked with [2H6]campestanol as an internal standard, evaporated to dryness, and saponified with 1 n sodium hydroxide in methanol at 80°C for 1.5 h. The hydrolysate was evaporated to dryness and partitioned between chloroform and water. The chloroform phase was evaporated to dryness, dissolved in chloroform, and passed through a short column of silica gel (Wakogel C-300, Wako Pure Chemicals). The eluate was subjected to GC-SIM analysis.
GC-SIM Analysis of Sterols
The sterol sample was trimethylsilylated with N-methyl-N-trimethylsilyl-trifluoroacetamide at room temperature for 5 min and then subjected to GC-SIM under the same conditions as described by Nomura et al. (2001), except that the column was a DB-5 column (0.25 mm i.d. × 15 m, 0.25-μm film thickness; J&W Scientific, Folsom, CA). The levels of sterols were determined from calibration curves constructed from the ratios of the M+ peak area of [2H6]campestanol-trimethylsilyl ether to the M+ peak areas of the trimethylsilyl ether derivatives of authentic sterols.
Extraction and Purification of BRs
Mature shoots (wild type, 15 g fresh weight; brd1, 15 g fresh weight) grown in the greenhouse in soil under long-day condition (16 h light/8 h dark) were harvested and extracted with methanol. The extracts were spiked with 2H6-labeled BRs as internal standards before reduction to an aqueous residue. The aqueous residue was partitioned between ethyl acetate and 0.5 m K2HPO4 buffer. The ethyl acetate phase was evaporated to dryness and partitioned between hexane and 80% (v/v) methanol. The 80% (v/v) methanol phase was evaporated to dryness, and the residual solid was purified on a column of charcoal (chromatography grade, Wako Pure Chemicals), which was eluted with methanol:water (6:4 and 8:2, v/v), methanol, methanol:chloroform (9:1, 7:3, 5:5, 3:7, and 1:9, v/v), and chloroform. To monitor the biological activity of BRs, sample aliquots were assayed in a rice lamina inclination test (Yokota et al., 1996). The methanol:chloroform (7:3 and 5:5, v/v) fractions were combined and chromatographed on a Sephadex LH-20 column (bed volume, 500 mL; Amersham Biosciences Inc., Piscataway, NJ) with a methanol:chloroform (4:1, v/v) mixture as the mobile phase. Successive 10-mL fractions were collected. Fractions 37 to 40 were combined, dissolved in methanol, and passed through columns of diethylaminopropyl silica (Bondesil, Varian, Palo Alto, CA) and octadecyl silica (ODS-SS-1020-T, Senshu Science, Tokyo). Reverse-phase HPLC was carried out using the same conditions as described by Nomura et al. (2001), and the BR fractions were collected for analysis by GC-SIM.
GC-SIM Analysis of BRs
BRs were converted to methaneboronates, methaneboronate-trimethylsilyl ethers, or trimethylsilyl ethers for GC (Nomura et al., 2001). GC-SIM analyses were carried out in the electron ionization mode on a 6890A/5973 N MSD instrument (Agilent Technologies, Palo Alto, CA) fitted with a DB-5 column (0.25 mm i.d. × 15 m, 0.25-μm film thickness; J&W Scientific). The carrier gas was He at a flow rate of 2 mL min−1, the injection port temperature was 280°C, and the samples were introduced by splitless injection. The column oven temperature was programmed at 170°C for 0.5 min before being elevated to 280°C at 74°C min−1 and then to 300°C at 3°C min−1. The contents of BRs were calculated from the peak area ratios of 2H6 and 2H0 M+ ions or 2H6 and 2H0 fragment ions.
DNA Analysis
Rice genomic DNA was isolated from leaf blades by using a cetyl-trimethyl-ammonium bromide procedure (Murray and Thompson, 1980). For the brd1 mutant analyses, the 5′ to 3′ sequences of primers used were as follows: D1, CAGCACAAGCAAGCAGCTTG; D3, CCAAGATCGACGCCTTCATG; D4, GCAAGGAAGAAGCTTGTT; D6R, GGACCAAAAGAATACAGGAG; D9R, GCAACAAGCTTCTTCCTT; and D10R, CGCATGAAGGCGTCGATCTT. Genomic DNA isolated from the mutant and wild-type plants was subjected to PCR, using these primer sets. The DNA fragment amplified by using the primers D4 and D6R were purified with a QIAquick PCR Purification Kit (Qiagen Inc., Chatsworth, CA) and sequenced. The sequence was confirmed by sequencing independently amplified fragments at least twice to eliminate PCR misincorporation.
ACKNOWLEDGMENTS
We thank Dr. Suguru Takatsuto (Joetsu University, Joetsu-shi, Japan) for deuterated steroids, Ko Murakami for technical advice and help with the histological examination, and Yumiko Yamada for technical assistance. We also thank Dr. Toshifumi Nagata for his many suggestions, and Tomiko Senba, Hiromi Satoh, Lois Ishizaki, Toshiko Shibata, Chikako Tomita, and Noriko Nakajima for their help.
Footnotes
This work was supported by the Ministry of Agriculture, Forestry, and Fisheries of Japan (rice genome project no. MP–1202), by the Human Frontiers Science Program (grant no. RG00162–2000 to T.Y.), and by the Japan Society for the Promotion of Science (Grant-in-Aid for Scientific Research no. 11460057 to T.Y.). T.N. is a research fellow of the Japan Society for the Promotion of Science since 1998.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.007179.
LITERATURE CITED
- Asami T, Min YK, Nagata N, Yamagishi K, Takatsuto S, Fujioka S, Murofushi N, Yamaguchi I, Yoshida S. Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol. 2000;123:93–100. doi: 10.1104/pp.123.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asami T, Mizutani M, Fujioka S, Goda H, Min YK, Shimada Y, Nakano T, Takatsuto S, Matsuyama T, Nagata N et al. Selective interaction of triazole derivatives with DWF4, a cytochrome P450 monooxygenase of the brassinosteroid biosynthetic pathway, correlates with brassinosteroid deficiency in planta. J Biol Chem. 2001;276:25687–25691. doi: 10.1074/jbc.M103524200. [DOI] [PubMed] [Google Scholar]
- Azpiroz R, Wu Y, LoCascio JC, Feldmann KA. An Arabidopsis brassinosteroid-dependent mutant is blocked in cell elongation. Plant Cell. 1998;10:219–230. doi: 10.1105/tpc.10.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop GJ, Harrison K, Jones J. The tomato Dwarf gene isolated by heterologous transposon tagging encodes the first member of a new cytochrome P450 family. Plant Cell. 1996;8:959–969. doi: 10.1105/tpc.8.6.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop GJ, Nomura T, Yokota T, Harrison K, Noguchi T, Fujioka S, Takatsuto S, Jones JD, Kamiya Y. The tomato DWARF enzyme catalyzes C-6 oxidation in brassinosteroid biosynthesis. Proc Natl Acad Sci USA. 1999;96:1761–1766. doi: 10.1073/pnas.96.4.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop GJ, Yokota T. Plants steroid hormones, brassinosteroids: current highlights of molecular aspects on their synthesis/metabolism, transport, perception and response. Plant Cell Physiol. 2001;42:114–120. doi: 10.1093/pcp/pce018. [DOI] [PubMed] [Google Scholar]
- Choe S, Dilkes BP, Fujioka S, Takatsuto S, Sakurai A, Feldmann KA. The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22α-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell. 1998;10:231–243. doi: 10.1105/tpc.10.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Noguchi T, Fujioka S, Takatsuto S, Tissier CP, Gregory BD, Ross AS, Tanaka A, Yoshida S, Tax FE et al. The Arabidopsis dwf7/ste1 mutant is defective in the Δ7 sterol C-5 desaturation step leading to brassinosteroid biosynthesis. Plant Cell. 1999;11:207–221. [PMC free article] [PubMed] [Google Scholar]
- Choe S, Tanaka A, Noguchi T, Fujioka S, Takatsuto S, Ross AS, Tax FE, Yoshida S, Feldmann KA. Lesions in the sterol “delta”7 reductase gene of Arabidopsis cause dwarfism due to a block in brassinosteroid biosynthesis. Plant J. 2000;21:431–443. doi: 10.1046/j.1365-313x.2000.00693.x. [DOI] [PubMed] [Google Scholar]
- Chory J, Nagpal P, Peto CA. Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell. 1991;3:445–459. doi: 10.1105/tpc.3.5.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Hall AF, Langford M, McMorris TC, Baker ME. Physiological and molecular effects of brassinosteroids on Arabidopsis thaliana. J Plant Growth Regul. 1993;12:61–66. [Google Scholar]
- Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Sasse JM. Brassinosteroids: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:427–451. doi: 10.1146/annurev.arplant.49.1.427. [DOI] [PubMed] [Google Scholar]
- Friedrichsen DM, Joazeiro CA, Li J, Hunter T, Chory J. Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 2000;123:1247–1256. doi: 10.1104/pp.123.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- Hoshikawa K. The Growing Rice Plant. Tokyo: Nobunkyo; 1989. [Google Scholar]
- Kauschmann A, Jessop A, Koncz C, Szekeres M, Willmitzer L, Altmann T. Genetic evidence for an essential role of brassinosteroids in plant development. Plant J. 1996;9:701–713. [Google Scholar]
- Klahre U, Noguchi T, Fujioka S, Takatsuto S, Yokota T, Nomura T, Yoshida S, Chua NH. The Arabidopsis DIMINUTO/DWARF1 gene encodes a protein involved in steroid synthesis. Plant Cell. 1998;10:1677–1690. doi: 10.1105/tpc.10.10.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- Li J, Lease KA, Tax FE, Walker JC. BRs1, a serine carboxypeptidase, regulates BRI1 signaling in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2001a;98:5916–5921. doi: 10.1073/pnas.091065998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J. A role for brassinosteroids in light-dependent development of Arabidopsis. Science. 1996;272:398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH. Regulation of brassinosteroid signaling by a GSK/SHAGGY-like kinase. Science. 2002;295:1299–1301. doi: 10.1126/science.1065769. [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH, Vafeados D, Chory J. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 2001b;127:14–22. doi: 10.1104/pp.127.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda E. Rate of lamina inclination in excised rice leaves. Physiol Plant. 1965;18:813–827. [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata N, Asami T, Yoshida S. Brassinazole, an inhibitor of brassinosteroid biosynthesis, inhibits development of secondary xylem in cress plants (Lepidium sativum) Plant Cell Physiol. 2001;42:1006–1011. doi: 10.1093/pcp/pce122. [DOI] [PubMed] [Google Scholar]
- Nakaya M, Tsukaya H, Murakami N, Masahiro K. Brassinosteroids control the proliferation of leaf cells of Arabidopsis thaliana. Plant Cell Physiol. 2002;43:239–244. doi: 10.1093/pcp/pcf024. [DOI] [PubMed] [Google Scholar]
- Nelson DR, Koymans L, Kamataki T, Stegeman JJ, Feyereisen R, Waxman DJ, Waterman MR, Gotoh O, Coon MJ, Estabrook RW. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics. 1996;6:1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Tax FE, Feldmann KA. Biosynthetic pathways of brassinolide in Arabidopsis. Plant Physiol. 2000;124:201–209. doi: 10.1104/pp.124.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Kitasaka Y, Takatsuto S, Reid JB, Fukami M, Yokota T. Brassinosteroid/sterol synthesis and plant growth as affected by lka and lkb mutations of pea. Plant Physiol. 1999;119:1517–1526. doi: 10.1104/pp.119.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Nakayama M, Reid JB, Takeuchi Y, Yokota T. Blockage of brassinosteroid biosynthesis and sensitivity causes dwarfism in garden pea. Plant Physiol. 1997;113:31–37. doi: 10.1104/pp.113.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Sato T, Bishop GJ, Kamiya Y, Takatsuto S, Yokota T. Accumulation of 6-deoxocathasterone and 6-deoxocastasterone in Arabidopsis, pea and tomato is suggestive of common rate-limiting steps in brassinosteroid biosynthesis. Phytochemistry. 2001;57:171–178. doi: 10.1016/s0031-9422(00)00440-4. [DOI] [PubMed] [Google Scholar]
- Oh MH, Ray WK, Huber SC, Asara JM, Gage DA, Clouse SD. Recombinant brassinosteroid insensitive 1 receptor-like kinase autophosphorylates on serine and threonine residues and phosphorylates a conserved peptide motif in vitro. Plant Physiol. 2000;124:751–766. doi: 10.1104/pp.124.2.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Imiya Y, Ida S, Ichii M. Characterization of molybdenum cofactor mutant of rice, Oryza sativa L. Plant Sci. 1996;119:39–47. [Google Scholar]
- Schultz L, Huub L, Kerckhoffs J, Klahre U, Yokota T, Reid JB. Molecular characterization on the brassinosteroid-deficient lkb mutant of pea. Plant Mol Biol. 2001;47:491–498. doi: 10.1023/a:1011894812794. [DOI] [PubMed] [Google Scholar]
- Schumacher K, Chory J. Brassinosteroid signal transduction: still casting the actors. Curr Opin Plant Biol. 2000;3:79–84. doi: 10.1016/s1369-5266(99)00038-2. [DOI] [PubMed] [Google Scholar]
- Sekimata K, Kimura T, Kaneko I, Nakano T, Yoneyama K, Takeuchi Y, Yoshida S, Asami T. A specific brassinosteroid biosynthesis inhibitor, Brz2001: evaluation of its effects on Arabidopsis, cress, tobacco, and rice. Planta. 2001;213:716–721. doi: 10.1007/s004250100546. [DOI] [PubMed] [Google Scholar]
- Shimada Y, Fujioka S, Miyauchi N, Kushiro M, Takatsuto S, Nomura T, Yokota T, Kamiya Y, Bishop GJ, Yoshida S. Brassinosteroid-6-oxidases from Arabidopsis and tomato catalyze multiple C-6 oxidations in brassinosteroid biosynthesis. Plant Physiol. 2001;126:770–779. doi: 10.1104/pp.126.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres M, Nemeth K, Koncz Kalman Z, Mathur J, Kauschmann A, Altmann T, Redei GP, Nagy F, Schell J et al. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Gasch A, Nishizawa N, Chua N-H. The DIMINUTO gene of Arabidopsis is involved in regulating cell elongation. Genes Dev. 1995;9:97–107. doi: 10.1101/gad.9.1.97. [DOI] [PubMed] [Google Scholar]
- Takano M, Kanegae H, Shinomura T, Miyao A, Hirochika H, Furuya M. Isolation and characterization of rice phytochrome A mutants. Plant Cell. 2001;13:521–534. doi: 10.1105/tpc.13.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z-Y, Hideharu S, Fujioka S, Yoshida S, Chory J. BRI1 is a critical component of a plasma membrane receptor for plant steroids. Nature. 2001;410:380–383. doi: 10.1038/35066597. [DOI] [PubMed] [Google Scholar]
- Yamamoto R, Demura T, Fukuda H. Brassinosteroids induce entry into the final stage of tracheary element differentiation in cultured Zinnia cells. Plant Cell Physiol. 1997;38:980–983. doi: 10.1093/oxfordjournals.pcp.a029262. [DOI] [PubMed] [Google Scholar]
- Yamamoto R, Fujioka S, Demura T, Takatsuto S, Yoshida S, Fukuda H. Brassinosteroid levels increase drastically prior to morphogenesis of tracheary elements. Plant Physiol. 2001;125:556–563. doi: 10.1104/pp.125.2.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M. Loss of function of a rice brassinosteroid insensitive 1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell. 2000;12:1591–1606. doi: 10.1105/tpc.12.9.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota T, Matsuoka T, Koarai T, Nakayama M. 2-Deoxybrassinolide, a brassinosteroid from Pisum sativum seed. Phytochemistry. 1996;42:509–511. [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]