Abstract
The components of the cellular machinery that accomplish the various complex and dynamic membrane fusion events that occur at the division plane during plant cytokinesis, including assembly of the cell plate, are not fully understood. The most well-characterized component, KNOLLE, a cell plate-specific soluble N-ethylmaleimide-sensitive fusion protein (NSF)-attachment protein receptor (SNARE), is a membrane fusion machine component required for plant cytokinesis. Here, we show the plant ortholog of Cdc48p/p97, AtCDC48, colocalizes at the division plane in dividing Arabidopsis cells with KNOLLE and another SNARE, the plant ortholog of syntaxin 5, SYP31. In contrast to KNOLLE, SYP31 resides in defined punctate membrane structures during interphase and is targeted during cytokinesis to the division plane. In vitro-binding studies demonstrate that AtCDC48 specifically interacts in an ATP-dependent manner with SYP31 but not with KNOLLE. In contrast, we show that KNOLLE assembles in vitro into a large approximately 20S complex in an Sec18p/NSF-dependent manner. These results suggest that there are at least two distinct membrane fusion pathways involving Cdc48p/p97 and Sec18p/NSF that operate at the division plane to mediate plant cytokinesis. Models for the role of AtCDC48 and SYP31 at the division plane will be discussed.
Plant cell division is completed by the highly dynamic process of de novo cell plate construction leading to the separation of two daughter cells. Formation of this unique cytokinetic organelle involves at least three key membrane fusion steps: (a) fusion of secretory vesicles across the division plane to form a membranous tubular-vesicular network, (b) consolidation of the tubular-vesicular network, and (c) fusion of the cell plate leading-edge with the original parental plasma membrane to complete division (Samuels et al., 1995). These distinct stages of cell plate biogenesis likely involve both heterotypic and homotypic membrane fusion events. In addition to the cell plate, the division plane contains an extensive endoplasmic reticulum (ER) network that has been suggested to function in the formation of the cell plate (Hepler, 1982). Assembly of cell plate-associated ER is likely to involve homotypic fusion of ER membrane within the division plane.
Two homologous classes of ATPases associated with various cellular activities (AAA) proteins (Fröhlich, 2001), Sec18p N-ethylmaleimide-sensitive fusion protein (NSF) and Cdc48p/p97 (p97 is also known as VCP), regulate a variety of secretory membrane fusion processes (Acharya et al., 1995; Latterich et al., 1995; Rabouille et al., 1995). Monomers of Sec18p/NSF and Cdc48p/p97 consist of two Mg2+-dependent ATPase domains and an N-terminal substrate/adapter domain that assemble into biologically active ring-shaped oligohexamers (Peters et al., 1990; Hanson et al., 1997). Of these two AAA complexes, the biochemical function of Sec18p/NSF, which along with its cofactor, soluble NSF-attachment protein (α-SNAP) regulates hetero- and homotypic membrane fusion, has been the most well characterized.
Secretory vesicle targeting and fusion is mediated through the pairing of cognate, cytoplasmically oriented integral membrane proteins, SNAP receptors (SNAREs), which reside on the two fusing membrane species (i.e. vesicle (v)-SNAREs pair with target membrane (t)-SNAREs) to yield SNARE complexes (Jahn and Südhof, 1999; Brunger, 2001). Sec18p/NSF functions as a molecular chaperone to disassemble, at the expense of ATP, these SNARE complexes to facilitate another round of secretory membrane targeting and fusion (Littleton et al., 2001; May et al., 2001).
SNAREs are not only involved in the heterotypic fusion of secretory vesicles with their appropriate acceptor compartment but also function in the homotypic fusion of like-like membranes such as vacuoles (Nichols et al., 1997) and ER-membranes (Patel et al., 1998; Roy et al., 2000). In contrast to Sec18p/NSF-dependent homotypic yeast (Saccharomyces cerevisiae) vacuolar fusion, homotypic yeast ER, and animal transitional-ER (t-ER) fusion is dependent on Cdc48p/p97 and its interaction with the t-SNAREs Ufe1p and Sed5p/syntaxin 5, respectively (Latterich et al., 1995; Roy et al., 2000). Interestingly, Ufe1p and Sed5p/syntaxin 5 have dual roles: (a) ER-ER homotypic fusion required for ER biogenesis and maintenance, and (b) heterotypic fusion of early secretory compartment (i.e. ER and Golgi) transport vesicles.
Cdc48p/p97 is likely to modulate SNARE folding and complex integrity. In support, the Cdc48p/p97 archaeal ortholog, VAT, has been demonstrated to mediate the ATP-dependent folding and unfolding of cyclophilin (Golbik et al., 1999). Unlike Sec18p/NSF, however, the chaperone activity of Cdc48p/p97 is required for apparently distinct cellular activities beyond membrane fusion including protein degradation (Ghislain et al., 1996; Hoppe et al., 2000; Dai and Li, 2001; Hitchcock et al., 2001; Rape et al., 2001; Ye et al., 2001; Braun et al., 2002; Jarosch et al., 2002; Rabinovich et al., 2002) and DNA metabolism (Yamada et al., 2000; Zhang et al., 2000a). Specific adapter proteins regulate the recruitment of Cdc48p/p97-chaperone activity to various pathways. Interaction of p97 and Sed5p/syntaxin 5 is mediated by the mammalian cofactor p47 (Kondo et al., 1997). The Ufd1/Npl4 heterodimer functions as an adapter for p97/Cdc48p in ER protein dislocation and degradation in both yeast and mammalian cell systems (Hitchcock et al., 2001; Rape et al., 2001; Ye et al., 2001; Braun et al., 2002; Jarosch et al., 2002; Rabinovich et al., 2002) as well as functioning in the early stages of nuclear envelope fusion in Xenopus sp. (Hetzer et al., 2001). The binding of these adapters to soluble p97 is competitive (Meyer et al., 2000).
Sec18p/NSF and Cdc48p/p97 likely regulate membrane fusion through distinct reaction mechanisms. In support, adapter proteins associate with cytosolic Cdc48p/p97 hexamers (Kondo et al., 1997), whereas Sec18p/NSF binds to α-SNAP only in the presence of SNARE complexes (Wilson et al., 1992). In addition, the rate of ATPase hydrolysis by Cdc48p/p97 is reduced in the presence of p47 (Meyer et al., 1998), whereas NSF ATPase activity is simulated by α-SNAP (Morgan et al., 1994).
The assembly of animal Golgi cisternae from both mitotic Golgi fragments and vesiculated Golgi membranes of ilimaquinone-treated mammalian cells requires the combined action of p97/p47 and NSF/α-SNAP (Acharya et al., 1995; Rabouille et al., 1995) through a common t-SNARE, Sed5p/syntaxin 5 (Rabouille et al., 1998). These studies provide additional evidence that the two ATPases perform distinct biochemical processes required for Golgi maturation. Reminiscent of mammalian Golgi reassembly, peroxisome biogenesis in yeast requires two AAA proteins, Pex1p and Pex6p (Distel et al., 1996), mediating distinct membrane fusion events.
The recent identification and characterization of several division plane-localized Arabidopsis membrane fusion factors have provided some insight into division plane membrane fusion machinery. KNOLLE is a cell plate-associated t-SNARE required for cell plate formation (Lukowitz et al., 1996; Lauber et al., 1997). KNOLLE has recently been shown to interact with (a) SNAP33, a SNAP25 homolog (Heese et al., 2001); (b) NSPN11, a plant-specific SNARE (Zheng et al., 2002); and (c) KEULE (Assaad et al., 2001), a Sec1p homolog. Cells of severely malformed KNOLLE, KEULE, and AtSNAP33 mutant Arabidopsis plants are multinucleated and have incomplete cross walls; these phenotypes are consistent with cytokinesis defects. Suggestive evidence that Cdc48p/p97 may also play a role in cell plate formation comes from immunofluorescence microscopy studies (Feiler et al., 1995).
In this paper, we further characterize the Arabidopsis ortholog of Cdc48p/p97, AtCDC48, and provide evidence that both Cdc48p/p97- and Sec18p/NSF-dependent membrane fusion pathways exist at the plane of cell division during plant cytokinesis. These pathways are likely to mediate the extensive membrane dynamics including cell plate and ER assembly that occur during plant cell division.
RESULTS
Generation and Characterization of AtCDC48 Antibodies
The Arabidopsis genome encodes three CDC48 isoforms: AtCDC48A (At3g09840), AtCDC48B (At3g53230), and AtCDC48C (At5g03340; Initiative, 2000). AtCDC48A represents the original gene isolated by Feiler et al. (1995) and likely represents the most abundant isoform because of the presence of >100 expressed sequence tags (ESTs) and several full-length cDNAs in the Arabidopsis Information Resource (http://www.Arabidopsis.org) and Salk Institute Genomic Analysis Laboratory (http://signal.salk.edu/cgi-bin/tdnaexpress) databases compared with those identified to date for AtCDC48B (eight ESTs) and AtCDC48C (15 ESTs). Amino acid sequence analysis suggests that all isoforms are localized in the cytoplasm.
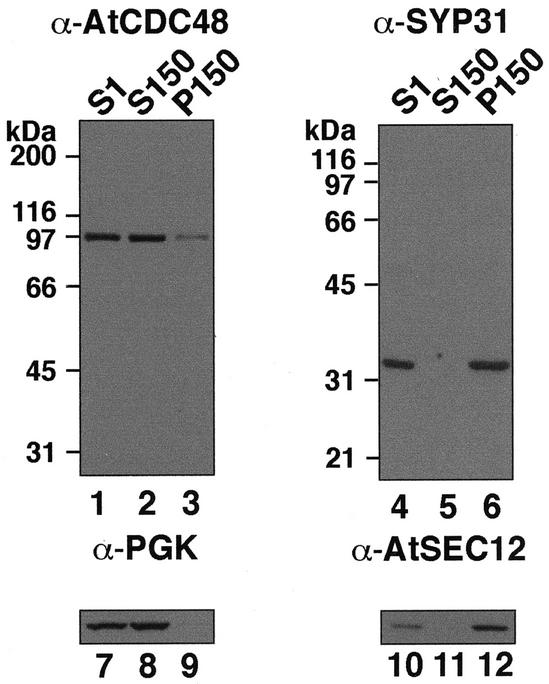
Initial characterization of AtCDC48a by Feiler et al. (1995) was hampered by the use of heterologous peptide-specific antibodies raised against the mammalian AtCDC48 homolog, VCP/p97. To more effectively examine its localization and function in plants, we generated affinity-purified antibodies against the C terminus (amino acids 690–809) of AtCDC48A. Given that AtCDC48B and AtCDC48C share 91% and 95% amino acid sequence identity with full-length AtCDC48A, respectively, these antibodies are expected to detect all three isoforms in Arabidopsis total protein extracts. As shown in Figure 1 the C-terminal AtCDC48 antibodies detect a single 97-kD polypeptide in immunoblots of protein extracts prepared from Arabidopsis suspension-cultured cells (Fig. 1A, lanes 1–3). As reported previously, the mobility of AtCDC48 is slower than the predicted molecular mass of AtCDC48A-C (approximately 89 kD; Feiler et al., 1995). Immunoblot analysis of whole-cell protein extracts prepared from yeast cdc48-1 mutants expressing AtCDC48A in the sense but not antisense orientation further confirmed the specificity of the C-terminal antibodies for AtCDC48 (data not shown).
Figure 1.
Specificity of AtCDC48 and SYP31 antibodies. Arabidopsis subcellular fractions (20 μg; S1, S150, and P150) were resolved by SDS-PAGE and analyzed by immunoblotting with AtCDC48 (lanes 1–3) and SYP31 (lanes 4–6) antibodies. Cytosolic PGK (lanes 7–9) and membrane-associated AtSEC12 (lanes 10–12) were used to confirm the identity and relative purity of each subcellular fraction.
AtCDC48 Is a Peripheral Membrane Protein
The biochemical characterization of CDC48/p97 in plants has not been performed previously. To examine the intracellular distribution of AtCDC48, the fractionation of AtCDC48 was compared with the distribution of the soluble and membrane subcellular markers, phosphoglycerokinase (PGK; cytosol; Kang et al., 2001) and AtSEC12 (ER; Bar-Peled and Raikhel, 1997), respectively, in post-nuclear supernatant (S1) and 150,000g cytosol-free microsomal membrane (P150) and membrane-free cytosolic (S150) fractions (Fig. 1). In contrast to the marker proteins, AtCDC48 was detected in both soluble (S150) and membrane (P150) fractions, suggesting that AtCDC48 is a peripheral membrane protein (Fig. 1A).
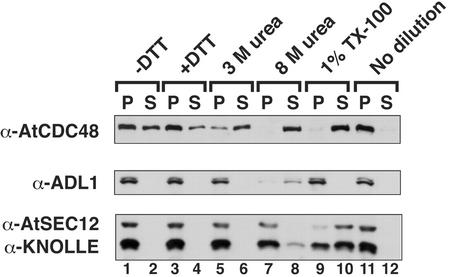
The membrane association of AtCDC48 in the P150 fraction was confirmed by its flotation on Suc density gradients (data not shown) and was further analyzed under a variety of conditions that release integral and nonintegral proteins from membranes to examine the biochemical nature of the association of AtCDC48 with membranes. Figure 2 demonstrates that AtCDC48 was preferentially released into the soluble fraction when the microsomal membranes were diluted into membrane isolation buffer (MIB) containing increasing concentrations of the denaturant urea (Fig. 2A, lanes 5–8). Under the same conditions, the integral membrane marker proteins KNOLLE (Lauber et al., 1997) and AtSEC12 (Bar-Peled and Raikhel, 1997) remained in the particulate fraction. AtSEC12p and KNOLLE were only solubilized upon treatment of the membrane fraction with detergent (Fig. 2A, lanes 9 and 10). Additional experiments indicated that AtCDC48 membrane association is not sensitive to salt (up to 2 m NaCl) and, therefore, is not simply mediated by electrostatic interactions (data not shown). In contrast to the peripheral membrane marker protein ADL1 (Gu and Verma, 1996; Park et al., 1997; Kang et al., 2001), AtCDC48 membrane association was also found to be destabilized in the absence of thiol reductant, DTT (Fig. 2A, lanes 1–4), suggesting that the protein is sensitive to the redox potential of its environment. All subcellular fractionation and biochemical experiment conditions in this manuscript, therefore, contain DTT. Protein recovery in the soluble and particulate fractions was assessed and found to be near 100% except for samples treated with 8 m urea (data not shown). These data strongly support the conclusion that AtCDC48 is a peripheral membrane-associated protein.
Figure 2.
AtCDC48 is peripherally associated with membranes. Microsomal membranes (P150) were diluted and incubated in the absence (lanes 1–2) or presence of 1 mm dithiothreitol (DTT; lanes 3–4), 3 m urea (lanes 5–6), 8 m urea (lanes 7–8), 1% (v/v) TX-100 (lanes 9–10), or no dilution was made (lanes 11–12). Samples were fractionated by differential centrifugation. Protein equivalents (20 μg) of soluble and pelletable material were analyzed by immunoblotting using antibodies directed against AtCDC48, Arabidopsis dynamin-like protein 1 (ADL1; peripheral), AtSEC12 (integral ER), and KNOLLE (integral SNARE). Protein recovery and loading was analyzed by PonceauS staining before immunoblot development.
Soluble AtCDC48 Is Predominantly Associated with Large Oligomers
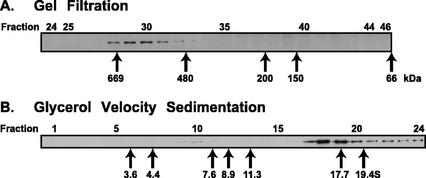
From biochemical and cryoelectron microscopy studies, both Cdc48p/p97 and Sec18p/NSF have been shown to assemble as stacked hexameric rings (Hanson et al., 1997; Peters et al., 1992; Rouiller et al., 2000). Hexameric p97 has also been shown to form higher-ordered hetero-oligomers with its adapter protein p47 (Kondo et al., 1997). To examine the native oligomeric structure of soluble AtCDC48, cytosolic protein extracts were subjected to sizing by gel filtration and velocity sedimentation (Fig. 3). Similar to the reported mass of cytosolic p97/CDC48 from various organisms including pure homohexameric rat p97 (600 kD; Kondo et al., 1997) Xenopus sp. p97 (570 kD; Peters et al., 1990) and the Trypanosoma sp. Cdc48p/p97 ortholog, TbVCP (555 kD; Roggy and Bangs, 1999), native cytosolic AtCDC48 was found to fractionate by Superose 6 gel filtration chromatography with an estimated molecular mass of 640 kD. For comparison, the majority of Arabidopsis cytosolic proteins were found to fractionate with a molecular mass less than 250 kD (data not shown). By glycerol gradient velocity sedimentation analysis, we confirmed the oligomeric nature of AtCDC48. The fractionation profile of AtCDC48 was highly reproducible between separate gradients, and a typical immunoblot of gradient fractions is shown in Figure 3B. The majority of cytosolic AtCDC48 was found to sediment at approximately 17S (fractions 18 and 19), which is much larger than reported for the Xenopus sp. p97 14.5S homohexameric and Trypanosoma sp. TbVCP complexes (Peters et al., 1990; Roggy and Bangs, 1999). One possible explanation for the substantial difference in sedimentation rates between AtCDC48 and these other p97/CDC48 orthologs may be related to the different gradient density medium used in these studies (i.e. glycerol versus Suc), which could affect the hydrodynamic properties of the CDC48p/p97 complex and, hence, its observed sedimentation rate. In addition to the 17S AtCDC48 peak, a significant fraction of cytosolic AtCDC48 was found to sediment further than the 19.4S marker (Fig. 3B) with a small but reproducible peak (fraction 22), suggesting that cytosolic hexameric AtCDC48 exists in higher-ordered complexes similar to the 740-kD rat p97/p47 complex (Kondo et al., 1997). A minor fraction of AtCDC48 was also observed to sediment at approximately 6.7S (fraction 10; approximately 125 kD) which is likely to correspond to monomeric AtCDC48 and/or a subcomplex composed of a single AtCDC48 subunit and a low-molecular mass adapter protein(s). These results suggest that cytosolic AtCDC48 is predominantly associated with homohexameric and high-ordered protein oligomers in actively dividing and expanding Arabidopsis cells.
Figure 3.
Cytosolic AtCDC48 exists as a large heterogeneous protein complex. Arabidopsis cytosol (S150) was fractionated by Superose-6 HR 10/30 gel filtration chromatography (A) and glycerol velocity gradient sedimentation (B). Protein mobility standards (arrows) were run in parallel to the sample. Individual fractions were analyzed by immunoblotting and Coomassie staining to localize AtCDC48 or mass standards, respectively.
Immunolocalization of AtCDC48
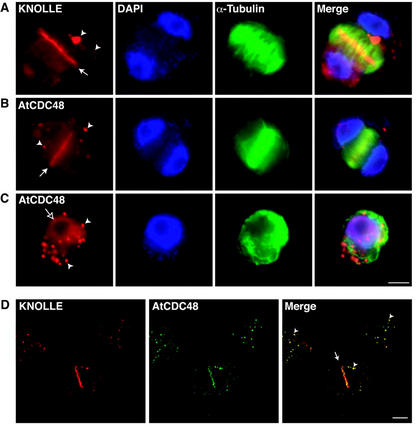
Previously Feiler et al. (1995) provided evidence that mammalian p97 antibodies weakly immunolabeled the phragmoplast midzone of dividing Arabidopsis cells. We have therefore reexamined the localization of CDC48/p97 in dividing and nondividing plant cells using affinity-purified AtCDC48-specific antibodies (Fig. 4). Cells in cytokinesis (Fig. 4, rows A and B) were identified using DAPI to detect binucleated cells, anti-α-tubulin antibodies to visualize phragmoplast microtubules, and the cell plate syntaxin KNOLLE (Fig. 4, row A) to identify the cell plate. KNOLLE antibodies were also found to reproducibly label large punctate organelles in dividing (Fig. 4, row A; see black arrowheads) and nondividing (data not shown) protoplasts derived from Arabidopsis suspension-cultured cells.
Figure 4.
Localization of AtCDC48 in interphase and dividing Arabidopsis cells. Dividing Arabidopsis cells were analyzed by wide-field indirect immunofluorescence microscopy (A–C) and confocal microscopy (D). A through C, Cells were immunolabeled with anti-α-tubulin (green) and affinity-purified anti-KNOLLE (row A; red) or anti-AtCDC48 (rows B and C; red) antibodies and 4′,6-diamino-phenylindole (DAPI; blue). Electronically complied images (merged) were generated from the pseudocolored images. D, Colocalization (yellow) of KNOLLE (red) and AtCDC48 (green) in dividing (center) and nondividing (top left and right) cells were examined by indirect confocal immunofluorescence microscopy. White arrows indicate the location of the cell plate. White arrowheads indicate the position of subcellular membrane compartments containing KNOLLE and AtCDC48 (see text for discussion). The unfilled arrow (C) indicates nuclear localization of AtCDC48. Bar = 50 μm.
By wide-field epifluorescence and confocal microscopy, affinity-purified AtCDC48 antibodies were found to label the equatorial region of phragmoplasts in telophase cells (Fig. 4, rows B and D; black arrows). In addition, AtCDC48 was observed in punctate, cytoplasmically distributed structures in both dividing and interphase cells (Fig. 4, rows B–D; arrowheads). During interphase, AtCDC48 was also found to be associated with the nuclear envelope (Fig. 4C, white arrow), as confirmed by colocalization with the nuclear import receptor, α-importin (Smith and Raikhel, 1998; data not shown). Immunolabeling of the division plane or other intracellular structures was not observed in cells treated with preimmune IgYs or with fluorescently labeled secondary antibodies (data not shown).
Confocal immunofluorescence microscopy analysis (Fig. 4, row D) of cells immunolabeled for both KNOLLE (red) and AtCDC48 (green) showed significant but incomplete colocalization of the two proteins at the plane of division, as shown by the yellow color in the merged image (Fig. 4, row D; merged, black arrow) of the two separate emission channels (red and green) obtained from a single optical section. Complete colocalization, however, was observed in the cytoplasmic punctate structures (Fig. 4D, merged, black arrowheads). Control double immunolabeling experiments with either anti-KNOLLE or anti-AtCDC48 and antibodies directed against Arabidopsis α-mannosidase, a Golgi-resident marker protein (T.G. Falbel and S.Y. Bednarek, unpublished data), demonstrated that the punctate cytoplasmic structures that contain both KNOLLE and AtCDC48 do not correspond to Golgi apparati (data not shown).
AtCDC48 Cofractionates with Membranes Containing KNOLLE, SYP31, and SYP21
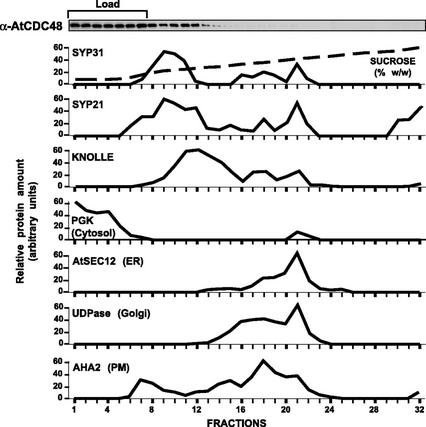
To further characterize the localization of AtCDC48, we analyzed its subcellular distribution by Suc density gradient centrifugation (Fig. 5). The distribution of AtCDC48 present in a postnuclear supernatant (S1) was compared with various compartment-specific marker proteins including PGK (cytosol; Kang et al., 2001), AtSEC12 (ER; Bar-Peled and Raikhel, 1997), UDPase (Golgi; Orellana et al., 1997), AHA2p (plasma membrane; DeWitt and Sussman, 1995), and the SNAREs, KNOLLE (cell plate; Lauber et al., 1997), SYP21 (endosomes; Bassham et al., 2000), and SYP31 (Bassham et al., 2000; Sanderfoot et al., 2000). Cytosolic AtCDC48 that cofractionated with PGK did not enter the gradient and remained in the load (Fig. 5, load, fractions 1–7), whereas membrane-associated AtCDC48 was found exclusively in slowly sedimenting fractions (Fig. 5, fractions 8–32, 17%–25% [w/w] Suc) that coincide with major peaks for SYP31, SYP21, and KNOLLE. Two smaller but significant peaks of KNOLLE, SYP21, and SYP31 were also observed to cosediment with dense fractions containing Golgi (Fig. 5, fractions 16–19) and ER (Fig. 5, fractions 20–22) marker proteins, however, these membranes were devoid of AtCDC48. Suc density gradient flotation analysis confirmed that the pool of AtCDC48 that entered into the Suc sedimentation gradient (Fig. 5, fractions 11–12) was membrane associated (data not shown). Our results demonstrate that membrane-associated AtCDC48 cofractionates with SYP31, SYP21, and KNOLLE.
Figure 5.
Membrane-bound AtCDC48 is primarily associated with a low-density membrane fraction. An Arabidopsis post-nuclear supernatant (S1) was fractionated by velocity centrifugation on a Suc gradient at 150,000g for 2 h at 4°C. The refractive index (expressed as Suc %, w/w) and content of various subcellular marker proteins of each gradient fraction was determined by enzyme activity assays (Golgi UDPase) or immunoblotting.
SYP31 Localizes to the Division Plane during Cytokinesis
The yeast and mammalian SYP31 orthologs, Sed5p/syntaxin 5 have been shown previously to be associated with the ER-to-Golgi branch of the secretory pathway (Banfield et al., 1994; Hui et al., 1997; Hay et al., 1998); however, localization of SYP31 in plant cells has not been reported to date.
As shown in Figure 1, affinity-purified SYP31 antibodies detect a single polypeptide with a molecular mass of approximately 33 kD in protein extracts prepared from Arabidopsis suspension-cultured cells (Fig. 1, SYP31) and Wassilewskija ecotype plant tissue (data not shown). As expected for a SNARE-type protein, the 33-kD polypeptide was found to be associated exclusively with microsomal (P150) membranes in wild-type Arabidopsis protein extracts (Fig. 1) and with an additional 64-kD protein in transgenic plants that express a SYP31-green fluorescent fusion protein (S. Park and S.Y. Bednarek, unpublished data). In addition to SYP31, the Arabidopsis genome encodes a second Sed5p/syntaxin 5 ortholog, SYP32 (Sanderfoot et al., 2000), that displays limited amino acid sequence identity to the N terminus of SYP31 (46% identical, 62% similar). Control immunoblot experiments verified that affinity-purified SYP31 antibodies did not cross-react with an Escherichia coli expressed fusion protein containing the cytoplasmic domain of SYP32 (data not shown).
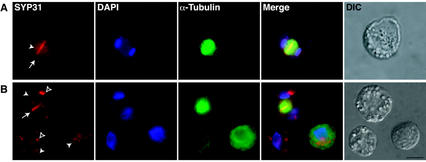
To further assess the subcellular localization of SYP31 in dividing and interphase cells, Arabidopsis suspension-cultured cells were processed for indirect double-immunolabeling using affinity-purified SYP31 antibodies and anti-α-tubulin to visualize cortical and phragmoplast microtubules and analyzed by wide-field epifluorescence microscopy (Fig. 6). In both dividing and interphase cells, SYP31 was found to be associated with both large (Fig. 6A, white arrowheads) and small cytoplasmic punctate structures (Fig. 6A, black arrowheads). The nature of these structures remains to be determined; however, the size and distribution of the small vesicular bodies closely resemble Golgi stacks when viewed by immunolabeling with anti-α-mannosidase (T.G. Falbel and S.Y. Bednarek, unpublished data). Consistent with this idea, we have shown by subcellular fractionation that a significant fraction of SYP31 cofractionates with the Golgi-marker protein, UDPase (Fig. 5). During cytokinesis, the subcellular distribution of SYP31 was found to expand to include the division plane (Fig. 6, A [top cell], and B, see black arrow). In contrast, α-mannosidase was not observed within the division plane (data not shown) as previously demonstrated (Nebenführ et al., 2000). Double immunolabeling with anti-SYP31 and anti-α-mannosidase was not performed because both primary antibodies are of rabbit origin. Our localization studies suggest that in contrast to the cell plate syntaxin KNOLLE, which is expressed only during cell division, SYP31 is present throughout the cell cycle and is recruited to the division plane during cytokinesis.
Figure 6.
Localization of SYP31 in dividing Arabidopsis cells. Arabidopsis cells were double immunolabeled with anti-α-tubulin antibodies (green) and affinity-purified anti-SYP31 antibodies (red) and DAPI. Merged images were generated electronically from the three preceding pseudocolored images and are shown in the indicated panels (merged). Differential interphase contrast images of the cells analyzed are presented (DIC). White arrows indicate the location of the cell plate. White arrowheads indicate the position of large undefined subcellular structures. Unfilled arrowheads indicate the position of small cytoplasmic punctate structures. Bar = 50 μm.
ATP-Dependent Binding of AtCDC48 to SYP31
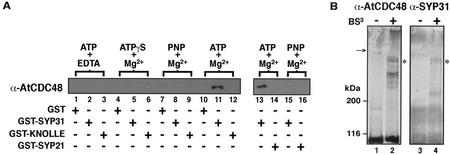
The above observations indicate that AtCDC48 cofractionates with KNOLLE and SYP31 and that all three proteins are targeted to the division plane during cytokinesis. To determine whether AtCDC48 interacts with either SYP31 or KNOLLE, we assessed the in vitro binding of cytosolic AtCDC48 to immobilized glutathione S-transferase (GST) fusion proteins containing the N-terminal cytosolic domains of KNOLLE, SYP31, and the prevacuolar compartment SNARE, SYP21. In addition, the nucleotide requirement for binding of AtCDC48 to the SNAREs was examined. As shown in Figure 7, cytosolic AtCDC48 was found to interact specifically with GST-SYP31ΔTM (Fig. 7A, lanes 11 and 13), whereas binding of AtCDC48 to GST, GST-SYP21ΔTM, and GST-KNOLLEΔTM was not detected under any of the experimental conditions used. E. coli expressed GST-SYP31ΔTM, GST-SYP21ΔTM, and GST-KNOLLEΔTM were found to specifically interact with mammalian α-SNAP, indicating that all of our fusion proteins were active in the in vitro binding assay (data not shown).
Figure 7.
AtCDC48 interacts with SYP31. A, AtCDC48 interacts with SYP31 in vitro. Bacterially expressed GST and GST fusion proteins containing the cytosolic domains of SYP31 (GST-SYP31), SYP21 (GST-SYP21), and KNOLLE (GST-KNOLLE) were incubated with Arabidopsis cytosol in the presence of adenosine nucleotides (1 mm) and either Mg2+ (1 mm) or EDTA (10 mm). Binding of AtCDC48 was assessed by immunoblotting. PNP is an abbreviation for 5′-adenylylimidodiphosphate. B, AtCDC48 interacts with SYP31 in vivo. An Arabidopsis post-nuclear supernatant (S1) was treated with the chemical cross-linker (BS3 +) or prequenched cross-linker (−). Total quenched reactions were solubilized with 5× SDS-PAGE sample buffer, and protein was analyzed by SDS-PAGE and immunoblot of entire discontinuous gel (both stacking and resolving). Blots were first probed with anti-AtCDC48 primary antibodies (lanes 1 and 2) followed by stripping and reprobing with anti-SYP31 antibodies (lanes 3 and 4). Asterisks indicate protein band that colabels with both primary antibodies. The arrow highlights the interface between the stacking and resolving gels.
In the presence of ATPγS, a non-hydrolyzable ATP analog and the adapter protein α-SNAP, the AAA ATPase, NSF/Sec18p, binds stably to SNAREs forming a 20S complex (Söllner et al., 1993; Hay et al., 1997). In contrast, binding of AtCDC48 with SYP31 was only observed in the presence of ATP and under conditions that support nucleotide hydrolysis (Fig. 7A, lanes 11 and 13). Binding of cytosolic AtCDC48 to GST-SYP31ΔTM was inhibited in the presence of two different non-hydrolyzable ATP analogs, ATPγS (Fig. 7A, lanes 4–6) and 5′-adenylylimidodiphosphate (Fig. 7A, lanes 7–9, 15, and 16). Mg2+ is required for ATP hydrolysis by AAA proteins. Addition of EDTA to coordinate the Mg2+ in the presence of ATP also inhibited binding AtCDC48 to GST-SYP31ΔTM (Fig. 7A, lane 2). These data strongly suggest that AtCDC48 interaction with SYP31 is ATP-hydrolysis dependent.
The specificity of the observed in vitro interaction between GST-SYP31ΔTM and cytosolic AtCDC48 was confirmed through chemical cross-linking experiments using mechanically disrupted Arabidopsis suspension-cultured cells (Fig. 7B). Because of our inability to immunoprecipitate using either anti-AtCDC48 or anti-SYP31 antibodies, we determined endogenous Arabidopsis AtCDC48/SYP31 interaction via chemical cross-linking with a non-cleavable cross-linker and immunoblot analysis looking for comigration of high Mr species that were immunoreactive to both anti-AtCDC48 and anti-SYP31 antibodies. To perform this analysis, a protein blot was first probed with anti-AtCDC48 followed by chemical removal of bound antibodies and re-probed with anti-SYP31 antibodies. Upon chemical cross-linking of Arabidopsis protein fractions, several anti-AtCDC48 immunoreactive large complexes were detected when compared with samples not cross-linked (Fig. 7B, compare lanes 2 and 1, respectively). Upon reprobing the stripped blot with anti-SYP31 antibodies, a single band was detected that entered the resolving gel and comigrated with an anti-AtCDC48 immunoreactive band (Fig. 7B, compare lanes 4 to 2, respectively; note the asterisks). In contrast KNOLLE was not found to interact with AtCDC48 under the same cross-linking/immunoblotting conditions (data not shown).
KNOLLE Interacts with NSF and α-SNAP
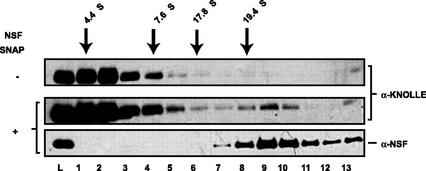
As described above, AtCDC48 was found to interact with SYP31 and not KNOLLE under the binding conditions tested both in vitro and in vivo. To test whether the cell plate syntaxin KNOLLE interacts with Sec18p/NSF, detergent-solubilized Arabidopsis microsomal proteins were incubated in the presence of mammalian α-SNAP and NSF under conditions that block ATP hydrolysis (Fig. 8). Genes encoding NSF/Sec18p and SNAP/Sec17p orthologs are present in the Arabidopsis genome (Sanderfoot et al., 2000), however, E. coli expression constructs for these proteins are currently unavailable. Therefore, mammalian α-SNAP and NSF were used for these experiments because previous studies have shown that the yeast and mammalian NSF/Sec18p and SNAP/Sec17p are interchangeable (Clary et al., 1990). In addition, it has been shown that mammalian α-SNAP binds the Arabidopsis SNARE, SYP21, indicating that the function of these proteins in membrane fusion is conserved between species (Bassham and Raikhel, 1999). In the presence of α-SNAP and NSF, a significant fraction of total cellular KNOLLE was recruited into an approximately 20S complex that cofractionated with NSF (Fig. 8). The efficiency of 20S particle formation observed in Figure 8 is similar to that observed for the formation of 20S particles containing syntaxin 5 from rat liver membranes (Hay et al., 1997). KNOLLE behaved exclusively as a monomer in the absence of α-SNAP and NSF. The results support the model proposed by G. Jürgens and colleagues (Lukowitz et al., 1996; Lauber et al., 1997; Heese et al., 2001) that KNOLLE is likely a syntaxin for a Sec18p/NSF-dependent membrane fusion pathway at the cell plate during plant cytokinesis.
Figure 8.
KNOLLE interacts with NSF and α-SNAP. Detergent-solubilized Arabidopsis microsomal membranes were incubated in the absence (−) or presence (+) of purified E. coli-produced mammalian myc-NSF-His6 and His6-α-SNAP and fractionated by glycerol gradient velocity sedimentation. Fractions were analyzed by immunoblotting to determine the distribution of KNOLLE and mammalian NSF. The relative mobility of protein calibration standards is indicated.
DISCUSSION
Recent evidence suggests that plant cytokinesis is dependent upon interaction between membrane trafficking components that are either expressed in cells in division or those that constitutively play a general role in membrane trafficking and maintenance. The latter group would be recruited during cell division to perform cytokinesis-specific functions. The cell plate SNARE, KNOLLE, which is expressed only in dividing plant cells (Lukowitz et al., 1996), interacts with two constitutively expressed SNAREs, AtSNAP33 (Heese et al., 2001) and NSPN11 (Zheng et al., 2002). Genetic and biochemical data have also suggested that the activity of KNOLLE is regulated by KEULE, a constitutively expressed member of the Sec1p-family (Assaad et al., 2001, 1996). In this paper, we present evidence for the existence of an additional membrane fusion pathway at the plane of division involving AtCDC48 and the SNARE, SYP31. Mammalian and yeast orthologs of AtCDC48, Cdc48p/p97, have been implicated previously in cell division. CDC48 was originally identified as a mutant that displayed defects in the progression of cells through the cell division cycle (Moir et al., 1982). As described above, Cdc48p/p97 is involved various processes including homotypic membrane fusion of ER, nuclear, and mitotic Golgi fragments (Latterich et al., 1995; Rabouille et al., 1995, 1998; Roy et al., 2000; Hetzer et al., 2001), and its recruitment to these various diverse processes is mediated through a set of distinct adapter proteins.
By glycerol gradient sedimentation and gel filtration analysis, we have demonstrated that the majority of cytosolic AtCDC48 fractionated, as predicted, as a hexameric complex. In addition, the broad and skewed profile of the AtCDC48 observed by velocity glycerol gradient sedimentation analysis (Fig. 3B) suggested that a significant proportion of hexameric AtCDC48 is associated with additional factors forming higher order heteroligomeric complexes. In support of this, we have identified several plant-specific proteins that interact with the AtCDC48 complex and SYP31; however, binding of these putative plant adapters to the AtCDC48 complex appears to be labile (D.M. Rancour, S.R. Knight, and S.Y. Bednarek, unpublished data). This weak interaction may account for difference in mass observed for the AtCDC48 complex by gel filtration and velocity sedimentation analysis (Fig. 3). The interaction between the putative plant adapter proteins and AtCDC48 may be stabilized under the conditions used for glycerol gradient sedimentation analysis resulting in a more complex and heterogeneous AtCDC48 fractionation profile.
The current prevailing model for the role of Cdc48p/p97 in homotypic membrane fusion is to function as a mechanochemical complex to disassemble paired SNAREs (e.g. Ufe1p and syntaxin 5) in post-fusion membranes to make them available for subsequent rounds of fusion. This model is largely based upon the Sec18p/NSF-dependent mechanism of membrane fusion and does not reconcile the many biochemical differences between these two AAA ATPases including their interaction with adapter proteins (Wilson et al., 1992; Kondo et al., 1997) and the control of ATP hydrolysis (Morgan et al., 1994; Meyer et al., 1998).
Like other AAA ATPases, it is thought that nucleotide-dependent conformational changes in the hexameric p97 complex are transmitted mechanically to bound substrates to mediate their assembly or disassembly. Recent structural and biochemical studies however reach different conclusions about which step in the ATPase cycle (e.g. ATP binding, hydrolysis, or product release) is critical for the conformational changes in p97 (Rouiller et al., 2000; Zhang et al., 2000b; Lamb et al., 2001). Our findings regarding the ATP-dependent association of AtCDC48 and SYP31 may have significant implications for these models. Similar to the reported observation by Warren and colleagues (Rabouille et al., 1998), binding of cytosolic AtCDC48 to SYP31, the plant ortholog of syntaxin 5, was ATP dependent. Furthermore, we have demonstrated that this interaction, in contrast to the association of NSF with SNAREs, is strictly dependent upon conditions that support nucleotide hydrolysis. These results suggest that nucleotide binding and hydrolysis are required for the interaction of Cdc48p/p97 with its substrate. One model to explain our result is that at least one round of ATP hydrolysis is required to prime and reorient the AtCDC48/SYP31-specific adapter complex to allow for binding to SYP31. As an alternative, ATP hydrolysis may be required to promote the exchange of associated adapter proteins with cytosolic AtCDC48 to facilitate SYP31 interaction. Because our in vitro binding assay used Arabidopsis cytosol as a source of AtCDC48 and adapter proteins, a third possibility is that the observed ATP hydrolysis requirement reflects posttranslational phosphorylation of AtCDC48. It has been previously shown that Cdc48p/p97 activity is regulated by phosphorylation (Madeo et al., 1998; Mayr et al., 1999; Lavoie et al., 2000), however, an ATP requirement for binding of purified Cdc48p/p97 and p47 to syntaxin 5 in vitro (Rabouille et al., 1998) would argue against this model. These models can be tested through the use of our in vitro SYP31 binding assay using purified AtCDC48 adapters and through the in vitro and in vivo analysis of AtCDC48 ATPase mutants.
Analysis of the Arabidopsis genome sequence has revealed two genes that encode Sed5p/syntaxin 5 orthologs, SYP31 and SYP32 (Sanderfoot et al., 2000). Previous localization studies of Sed5p and syntaxin 5 in yeast and mammalian cells, respectively, suggest that the major steady-state localization of Sed5p/syntaxin 5 to be the cis-Golgi and at lower levels in the ER and intermediate compartment (Hardwick and Pelham, 1992; Bennett et al., 1993; Banfield et al., 1994, 1995; Dascher et al., 1994; Hay et al., 1997; Hui et al., 1997). Our localization studies similarly suggest that a fraction of the intracellular pool of SYP31 is associated with the Golgi apparatus during interphase, suggesting that function of the Sed5p/syntaxin 5 family is conserved. However, yeast complementation studies have shown that SYP31 cannot functionally replace Sed5p, indicating that SYP31 and Sed5p may have only partially overlapping functions (A. Sanderfoot, personal communication). Here, we have provided evidence that SYP31 is targeted to the division plane during plant cytokinesis. Its role in plant cell division, however, remains to be determined. Similar to the reassembly of the mammalian Golgi apparatus after mitosis, AtCDC48 and AtSYP31 could function in parallel with the NSF-dependent KNOLLE membrane fusion pathway required for cell plate membrane assembly and maturation.
As an alternative, SYP31 and AtCDC48 may be required for the fusion of “other” secretory membranes at the plane of cell division. EM analysis of dividing plant cells has revealed an extensive array of ER membrane tightly juxtaposed to the developing cell plate (Hepler, 1982). Furthermore live cell imaging has revealed that ER membranes accumulate at the division plane as the cell plate expands (Cutler and Ehrhardt, 2002). This network of ER may mediate direct lipid transfer to the cell plate and/or provide the appropriate ionic environment (e.g. Ca2+) necessary for cell plate membrane fusion. Tubular elements of the cell plate-associated ER network also fuse and become entrapped forming the desmotubule that traverse plasmodesmata (Hepler, 1982). Similar to p97/p47 and syntaxin 5, which are required for homotypic fusion of mammalian t-ER membranes (Roy et al., 2000), SYP31 and AtCDC48 may be required for assembly and/or maintenance of early secretory compartment membranes including t-ER at the plane of cell division to support cytokinesis. Our Suc gradient fractionation data (Fig. 5) showing cofractionation of AtCDC48 with low-density membranes containing SYP31 is consistent with the fractionation properties of low-density membrane fractions used to reconstitute p97-mediated assembly of smooth t-ER tubules in vitro (Lavoie et al., 1996; Roy et al., 2000). Immuno-electron microscopy localization of AtCDC48 and SYP31 will determine the membrane structures with which they associate at the division plane during cytokinesis.
In conclusion, our results indicate that there are at least two distinct membrane fusion pathways involving Cdc48p/p97 and Sec18p/NSF that operate at the division plane to mediate plant cytokinesis. Through a combination of genetic and biochemical approaches, we will further elucidate the roles of each of these membrane fusion pathways in plant cell cytokinesis.
MATERIALS AND METHODS
Antibodies and General Reagents
Antibodies against AHA2, AtSEC12, and SYP21 have previously been described (DeWitt and Sussman, 1995; Bar-Peled and Raikhel, 1997; da Silva Conceicão et al., 1997). Anti-PGK and affinity-purified anti-ADL1 antibodies were kindly provided by J. Thorner (University of California, Berkeley) and W. Lukowitz (Carnegie Institution of Washington, Stanford CA), respectively. Monoclonal mouse anti-mammalian NSF antibodies (2E5; Tagaya et al., 1993) were provided by T. Martin (University of Wisconsin, Madison). Monoclonal rat anti-tubulin antibodies (MAS078p) were purchased from Harlan Sera-Lab Ltd. (Loughborough, UK). Donkey anti-rabbit, sheep anti-mouse, and rabbit anti-chicken horseradish peroxidase conjugates were purchased from Amersham Biosciences Inc. (Piscataway, NJ) and Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA). Secondary antibodies (goat anti-rabbit, donkey anti-chicken, or goat anti-rat) conjugated to Cy3 or FITC were purchased from Jackson Laboratories. All other reagents unless specified were from Sigma-Aldrich (St. Louis) and Fisher Scientific (Pittsburgh).
Oligonucleotides
All oligonucleotides used in this study (Table I) were synthesized by Integrated DNA Technologies Inc. (Coralville, IA).
Table I.
Oligonucleotides used in this study
| Name | Sequence (5′→3′) | Target | Region of Homology |
|---|---|---|---|
| SB-34 | ATAGGATCCATCATATGTGGTTTGGAGAGAGTG | AtCDC48 | nt 1,657–1,675 |
| SB-35 | TATCTCGAGATTGTAGAGATCATCATCGTCGTC | AtCDC48 | nt 2,407–2,427 |
| SB-5a | TCCCCCGGGCCGGATCTCAGTGGTG | pET29B | nt 130–145 |
| SB-1 | ATAGGATCCCATATGAACGACTTGATGACG | KNOLLE | nt 1–18 |
| SB-2 | GACCTCGAGCTTTGCAGTCTTCAGCTCA | KNOLLE | nt 807–825 |
| SB-32 | GAGCCCGGGTCAGCACATCCATTTTCTGCTGTT | KNOLLE | nt 838–858 |
| SB-28 | GAGCTCGAGCTTCATCATGAGCCATCTATTTGAC | SYP31 | nt 933–957 |
| T3 | ATTAACCCTCACTAAAG | pBSSK | nt 774–790 |
| SB-366 | AAAGTACATGGGCTCGACGTTCAG | SYP31 | nt 1–18 |
Nucleotide (nt) sequences in bold correspond to regions of homology between the oligonucleotide and the target template, whereas underlined sequences correspond to artificial extensions containing restriction sites used for the cloning of PCR products.
Plasmid Construction
The C-terminal coding region of AtCDC48(690–809) was amplified by PCR using primers SB-34/SB-35 and subcloned as an NdeI-XhoI fragment into pET29A (Novagen, Madison, WI) to yield AtCDC48 (690–809)-his6. To generate the bacterial GST-AtCDC48 (690–809)-his6 expression construct, AtCDC48 (690–809)-his6 was PCR amplified using primers SB-34/SB5a and subcloned as a BamHI-SmaI fragment into pGEX-5A-2 (Amersham Biosciences Inc.).
KNOLLE (1–275) and KNOLLE (1–294) were PCR amplified using pairs SB-1/SB-2 and SB-1/SB-32, respectively, from the bacterial artificial chromosome F22013 (accession no. AC003981; Arabidopsis Biological Resource Center, Columbus, OH) and cloned as NdeI-XhoI and BamHI-SmaI fragments into pET29A and pGEX-2T to yield the KNOLLE(1–294)ΔTM-His6 and GST-KNOLLE (1–294) expression constructs. SYP31(1–319) was amplified by PCR using the primer pair T3/SB-28 from pBS-His6-SYP31 (Bassham et al., 2000), containing the SYP31 cDNA, and subcloned as an NdeI-XhoI fragment into pET29A to generate pSYP31(1–319)ΔTM-His6. SYP31(1–182) was amplified by PCR using primer pair SB-28/SB-366 from pBS-His6-SYP31 and subcloned as an RsaI fragment into a SmaI-cleaved pGEX4T-3 vector to generate pGST-SYP31(1–182). Isolation of the SYP32 cDNA and generation of a GST-SYP32(1–319)ΔTM expression vector will be described by (S. Park and S.Y. Bednarek, unpublished data) GST expression plasmids containing SYP31ΔTM(46–319) and SYP31ΔTM(4–253) and were provided by N. Raikhel (University of California, Riverside). Escherichia coli expression constructs for mammalian his6-NSF and his6-α-SNAP were kindly provided by T. Martin (University of Wisconsin, Madison).
Production of Bacterially Expressed Fusion Proteins
Plasmids expressing GST- and His-tagged fusion proteins were transformed into E. coli strain BL21(DE3) LysS, and fusion proteins were purified by affinity chromatography on glutathione-Sepharose (Amersham Biosciences Inc.) or Ni2+-NTA agarose (Qiagen, Hilden, Germany) according to the manufacturer's instructions. With the exception of GST-SYP32ΔTM (1–319) and SYP31ΔTM-His6, all E. coli-expressed fusion proteins used in this study were soluble. Protein concentrations and purity of E. coli expressed fusion proteins were determined by Bradford assay (Bio-Rad, Hercules, CA) and SDS-PAGE followed by Coomassie Blue staining and scanning densitometry using bovine serum albumin (BSA) as a standard.
Antisera and Affinity Purification
Chicken IgY antibodies were raised against GST-AtCDC48-His6 and purified as described (Gassmann et al., 1990). Rabbit antibodies against KNOLLE and SYP31 were raised using E. coli expressed and purified KNOLLE(1–275)ΔTM-His6 and SYP31(1–319)ΔTM-His6 as antigens. All antibodies generated in this study were purified by affinity chromatography against their respective immobilized-antigens as described previously (Kang et al., 2001).
Immunofluorescence Microscopy
Immunofluorescence microscopy was performed as described (Kang et al., 2001) with the exception that the fixed Arabidopsis protoplasts were permeabilized in microtubule-stabilizing buffer (Goodbody and Lloyd, 1994; 50 mm PIPES-KOH, pH 6.9, 5 mm MgSO4, and 5 mm EGTA) containing 0.5% (v/v) NP-40 and 10% (v/v) dimethyl sulfoxide for 5 min at 20°C before plating. For double-immunolabeling experiments, the cells were incubated sequentially with affinity-purified primary antibodies of different species followed by a mixture of the corresponding secondary antibodies labeled with FITC and Cy3. Wide-field microscopy and image processing were performed as described (Kang et al., 2001). Confocal microscopy was performed using an Axiovert 100 M inverted microscope (Carl Zeiss, Thornwood, NY) equipped with a Bio-Rad MRC1024 laser scanning unit and a 63× (natural abundance 1.4) PlanAPOChroma oil immersion objective lens. Separate images were collected from each fluorescence emission channel, and images were processed using Photoshop 6.0 (Adobe Systems, San Jose, CA). The labeling pattern observed in fixed protoplasts versus cells that were fixed before enzymatic removal of the cell wall resulted in no observed difference in the distribution of AtCDC48 or other marker proteins relative to the images shown in Figure 4.
Cell Fractionation
Soluble and membrane fractions were prepared from 3-d-old Arabidopsis suspension-cultured cells as described (Kang et al., 2001) with the following modifications. In brief, protoplasts were washed once in chilled 2× concentrated MIB ([1×]; 20 mm HEPES-KOH, pH 7.0, 50 mm KOAc, 1 mm Mg(OAc)2, and 250 mm sorbitol) with DTT (1× = 1 mm) and then diluted 1:5 in chilled MIBDTT + protease inhibitor cocktail (PIC) (1 mm phenylmethylsulfonyl fluoride, 5 μg mL−1 pepstatin A, 1 μg mL−1 chymostatin, 1 mm p-aminobenzamidine, 1 mm ε-aminocaproic acid, 5 μg mL−1 aprotinin, 1 μg mL−1 leupeptin, and 10 μg mL−1 E64) and lysed by passage 7× through a 25-gauge needle. A post-nuclear supernatant designated as S1 was prepared by centrifugation at 1,000g for 10 min at 4°C. Microsomal membranes were prepared by centrifugation of the S1 at 150,000g in a TLA100.3 rotor (Beckman Coulter, Fullerton, CA) for 30 min at 4°C. The supernatant (S150) was transferred to a new tube, and the pellet (P150) was resuspended in MIBDTT + PIC using a glass dounce homogenizer (Kontes Glass Co., Vineland, NJ). Aliquots were made of each fraction, snap frozen in N2(L), and stored at −80°C. Protein content of each fraction was determined using the Bio-Rad DC Protein Assay Kit and BSA as a standard.
For Suc gradient fractionation, an S1 (2.5 mL, 5 × 105 protoplast equivalents) was applied to a continuous 10 mL of 14% to 50% (w/w) Suc gradient in MIBDTT containing a 0.5 mL of 60% (w/w) Suc cushion. Gradients were centrifuged at 150,000g (28,500 rpm) in a SW40 rotor (Beckman Coulter) for 2 h at 4°C. Fractions (32 × 0.4 mL) were collected from the top at 0.5 mL min−1 using a gradient collector (model 640, Isco Inc., Lincoln, NE) and analyzed for their content of latent uridine-5′-diphosphatase UDPase activity (below), refractive index, and by SDS-PAGE and immunoblotting. UDPase activity, a marker of the Golgi apparatus was measured as previously described (Orellana et al., 1997).
Determination of the Size of Cytosolic AtCDC48
For sedimentation velocity analysis, membrane-free cytosol (S150′) was generated by centrifugation of an S1 fraction at 150,000g (45 min, 4°C) onto a 10% (w/w) Suc cushion. One hundred microliters of S150′ (600 μg protein, 3.6 × 106 protoplast equivalents) was layered onto a 4.8-mL continuous 10% to 40% (w/w) glycerol gradient in MIB containing 1 mm DTT (MIBDTT). Gradients were centrifuged at 128,000g in a SW50.1 rotor (Beckman Coulter) for 18 h at 4°C. Fractions (12 × 0.4 mL) were collected from the top using a density fraction collector (model 640, Isco Inc.) and analyzed by immunoblotting. In parallel, gradients were centrifuged containing globular protein standards with known sedimentation coefficients: ovalbumin (3.66S), BSA (4.41S), yeast alcohol dehydrogenase (7.61S), β-amylase (8.9S), catalase (11.3S), apoferritin (17.7S), and thyroglobulin (19.4S).
For gel filtration, 4 × 106 Arabidopsis protoplast equivalents of fresh membrane-free S150 cytosol (0.2 mL volume) was spiked with Mr standards (apoferritin and yeast alcohol dehydrogenase) and loaded onto a MIBDTT-equilibrated Superose-6 HR 10/30 column (Pharmacia, Piscataway, NJ) under the control of an FPLC system. The flow rate was 0.5 mL min−1, and 0.5-mL fractions were collected. The protein elution profile was determined using an in-line UV detector (280 nm), and fractions were analyzed by immunoblotting against AtCDC48. In parallel, the elution profile of known molecular mass standards was determined: ovalbumin (43.5 kD), BSA (66 kD), yeast alcohol dehydrogenase (150 kD), β-amylase (200 kD), catalase (250 kD), apoferritin (480 kD), thyroglobulin (669 kD), and blue dextran (2,000 kD, void volume) that consistently eluted at fraction 17.
Analysis of AtCDC48 Membrane Association
Freshly prepared or N2(L) frozen microsomal membranes (100 μg protein) were diluted (approximately 17-fold) into MIBPIC containing either DTT (1 mm), urea (3 m or 8 m), or TX-100 (1% [w/v]), or alternatively, no dilution was made. Samples were incubated for 30 min at room temperature followed by centrifugation (90,000 rpm, 30 min, 20°C; Beckman Coulter TLA100.1). Supernatants were concentrated by precipitation (Aguilar et al., 1999) and 20-μg protein equivalents of supernatant and pellet fractions were analyzed by immunoblotting. Before immunoblotting, the membrane was analyzed by PonceauS staining to confirm protein recovery and equal loading.
In Vitro Binding Studies
For in vitro AtCDC48 binding studies, 200 μg of Arabidopsis S150 protein was incubated with 0.05 nmol of fusion proteins (GST, GST-SYP21, GST-SYP31, or GST-KNOLLE) immobilized on 6.25 μL of glutathione-Sepharose for 2 h at 4°C in binding buffer (20 mm HEPES-KOH, pH 7.0, 50 mm KOAc, 250 mm sorbitol, 1 mm DTT, and 0.1% [v/v] NP40 plus protease inhibitor cocktail) in the presence of adenosine nucleotides (1 mm final) and either MgOAc (1 mm final) or EDTA (10 mm final). The beads were washed four times with 1 mL of binding buffer, and bound proteins were solubilized in 2× SDS sample buffer and analyzed by immunoblotting.
Chemical Cross-Linking
To determine whether AtCDC48/SYP31 interact in vivo, 100 μg of Arabidopsis S1 protein in buffer (20 mm HEPES-KOH, pH 7.0, 50 mm KOAc, 250 mm sorbitol, 1 mm Mg(OAc)2 1 mm DTT, 1 mm ATP, and protease inhibitor cocktail) was incubated with either Bis[sulfosuccinimidyl]suberate (BS3; 1 mm final; Pierce Inc., Rockford, IL) or BS3 prequenched with ethanolamine (40 mm final) for 30 min at room temperature. Cross-linking reactions were quenched by the addition of 200 mm ethanolamine in reaction buffer (40 mm final) for 10 min at room temperature. Total reactions were then solubilized in 5× SDS sample buffer and analyzed by discontinuous SDS-PAGE (4% [w/v] stacking and 7% [w/v] resolving) and immunoblotting. The entire stacking and resolving gels were used for protein transfer to nitrocellulose. Successive antibody probings of the same nitrocellulose were performed with a blot that was chemically stripped of antibodies by incubation in stripping solution (100 mm 2-mercaptoethanol, 2% [w/v] SDS, and 62.5 mm Tris-HCl, pH 6.7) for 30 min at 50°C according to the enhanced chemiluminescence protocol from Amersham Pharmacia Biotech. The efficiency of stripping was confirmed by re-exposing the nitrocellulose membrane to film. In addition, the immunoblot was treated with azide between probing to inhibit any residual secondary-antibody horseradish peroxidase-conjugate.
NSF and KNOLLE Interaction Experiments
Microsomal membranes (P150) were washed with 1 m KCl, solubilized in 2% (v/v) Triton X-100 and centrifuged at 150,000g for 30 min. Supernatant (900 μg of 150,000g) was preincubated in 200 μL of NSF-binding buffer (20 mm HEPES-KOH, pH 7.0, 100 mm KCl, 2 mm EDTA, O.5% [v/v] Triton X-100, and 100 mm ATP) in the absence or presence of purified E. coli produced myc-NSF-His6 (20 μg) and His6-α-SNAP (20 μg; Söllner et al., 1993) for 30 min at 4°C and fractionated by velocity sedimentation through a 4.8 mL of 16% to 32.5% (v/v) glycerol density gradient at 200,000g in a SW50.1 rotor (Beckman Coulter) for 20.5 h at 4°C. Glycerol gradients in NSF-binding buffer containing 1 mm DTT were prepared and fractionated as described above. Equal volume fractions were analyzed by SDS-PAGE fractionation followed by Coomassie Blue staining for protein standards or immunoblotting against KNOLLE and mammalian NSF. Anti-mammalian NSF antibodies (2E5; Tagaya et al., 1993) do not cross-react with endogenous Arabidopsis NSF (data not shown).
ACKNOWLEDGMENTS
We thank Dr. Mark Cook and Elizabeth Kostic for their help in generating the AtCDC48 antibodies and Dr. Judith Kimble for the use of her confocal microscope. We are also very grateful to Drs. James Bangs and Richard Amasino and to members of our laboratory for discussion and critical comments on the manuscript.
Footnotes
This work was supported by the Department of Energy, Division of Energy Biosciences (project no. DE–FG02–99ER20332), by the U.S. Department of Agriculture-Plant Growth and Development (project no. 98–35304–6671), by the Milwaukee Foundation (award to S.Y.B.), and by the National Science Foundation/Department of Energy/U.S. Department of Agriculture Collaborative Research in Plant Biology Program (grant no. 9602222 to C.E.D. and S.P.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.011742.
LITERATURE CITED
- Acharya U, Jacobs R, Peters JM, Watson N, Farquhar MG, Malhotra V. The formation of Golgi stacks from vesiculated Golgi membranes requires two distinct fusion events. Cell. 1995;82:895–904. doi: 10.1016/0092-8674(95)90269-4. [DOI] [PubMed] [Google Scholar]
- Aguilar RM, Bustamante JJ, Hernandez PG, Martinez AO, Haro LS. Precipitation of dilute chromatographic samples (ng/ml) containing interfering substances for SDS-PAGE. Anal Biochem. 1999;267:344–350. doi: 10.1006/abio.1998.3018. [DOI] [PubMed] [Google Scholar]
- Assaad F, Huet Y, Mayer U, Jürgens G. The cytokinesis gene keule encodes a sec1 protein that binds the syntaxin knolle. J Cell Biol. 2001;152:531–544. doi: 10.1083/jcb.152.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaad FF, Mayer U, Wanner G, Jürgens G. The KEULE gene is involved in cytokinesis in Arabidopsis. Mol Gen Genet. 1996;253:267–277. doi: 10.1007/pl00008594. [DOI] [PubMed] [Google Scholar]
- Banfield DK, Lewis MJ, Pelham HR. A SNARE-like protein required for traffic through the Golgi complex. Nature. 1995;375:806–809. doi: 10.1038/375806a0. [DOI] [PubMed] [Google Scholar]
- Banfield DK, Lewis MJ, Rabouille C, Warren G, Pelham HR. Localization of Sed5, a putative vesicle targeting molecule, to the cis-Golgi network involves both its transmembrane and cytoplasmic domains. J Cell Biol. 1994;127:357–371. doi: 10.1083/jcb.127.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled M, Raikhel NV. Characterization of AtSec12 and AtSAR1 proteins likely involved in ER and Golgi traffic. Plant Physiol. 1997;114:315–324. doi: 10.1104/pp.114.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham DC, Raikhel NV. The pre-vacuolar t-SNARE AtPEP12p forms a 20S complex that dissociates in the presence of ATP. Plant J. 1999;19:599–603. doi: 10.1046/j.1365-313x.1999.00552.x. [DOI] [PubMed] [Google Scholar]
- Bassham DC, Sanderfoot AA, Kovaleva V, Zheng H, Raikhel NV. AtVPS45 complex formation at the trans-Golgi network. Mol Biol Cell. 2000;11:2251–2265. doi: 10.1091/mbc.11.7.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MK, Garcia-Arraras JE, Elferink LA, Peterson K, Fleming AM, Hazuka CD, Scheller RH. The syntaxin family of vesicular transport receptors. Cell. 1993;74:863–873. doi: 10.1016/0092-8674(93)90466-4. [DOI] [PubMed] [Google Scholar]
- Braun S, Matuschewski K, Rape M, Thoms S, Jentsch S. Role of the ubiquitin-selective CDC48(UFD1/NPL4) chaperone (segregase) in ERAD of OLE1 and other substrates. EMBO J. 2002;21:615–621. doi: 10.1093/emboj/21.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT. Structure of proteins involved in synaptic vesicle fusion in neurons. Annu Rev Biophys Biomol Struct. 2001;30:157–171. doi: 10.1146/annurev.biophys.30.1.157. [DOI] [PubMed] [Google Scholar]
- Clary DO, Griff IC, Rothman JE. SNAPs, a family of NSF attachment proteins involved in intracellular membrane fusion in animals and yeast. Cell. 1990;61:709–721. doi: 10.1016/0092-8674(90)90482-t. [DOI] [PubMed] [Google Scholar]
- Cutler S, Ehrhardt D. Polarized cytokinesis in vacuolate cells of Arabidopsis. Proc Natl Acad Sci USA. 2002;99:2812–2817. doi: 10.1073/pnas.052712299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai RM, Li CC. Valosin-containing protein is a multi-ubiquitin chain-targeting factor required in ubiquitin-proteasome degradation. Nat Cell Biol. 2001;3:740–744. doi: 10.1038/35087056. [DOI] [PubMed] [Google Scholar]
- Dascher C, Matteson J, Balch WE. Syntaxin 5 regulates endoplasmic reticulum to Golgi transport. J Biol Chem. 1994;269:29363–29366. [PubMed] [Google Scholar]
- da Silva Conceicão A, Marty-Mazars D, Bassham D, Sanderfoot AA, Marty F, Raikhel NV. The syntaxin homolog AtPep12p resides on a late post-Golgi compartment in plants. Plant Cell. 1997;9:571–582. [PMC free article] [PubMed] [Google Scholar]
- DeWitt ND, Sussman MR. Immunocytological localization of an epitope-tagged plasma membrane proton pump (H+-ATPase) in phloem companion cells. Plant Cell. 1995;7:2053–2067. doi: 10.1105/tpc.7.12.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distel B, Erdmann R, Gould SJ, Blobel G, Crane DI, Cregg JM, Dodt G, Fujiki Y, Goodman JM, Just WW et al. A unified nomenclature for peroxisome biogenesis factors. J Cell Biol. 1996;135:1–3. doi: 10.1083/jcb.135.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiler HS, Desprez T, Santoni V, Kronenberger J, Caboche M, Traas J. The higher plant Arabidopsis thaliana encodes a functional CDC48 homologue which is highly expressed in dividing and expanding cells. EMBO J. 1995;14:5626–5637. doi: 10.1002/j.1460-2075.1995.tb00250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich KU. An AAA family tree. J Cell Sci. 2001;114:1601–1602. doi: 10.1242/jcs.114.9.1601. [DOI] [PubMed] [Google Scholar]
- Gassmann M, Thömmes P, Weiser T, Hübscher U. Efficient production of chicken egg yolk antibodies against a conserved mammalian protein. FASEB J. 1990;4:2528–2532. doi: 10.1096/fasebj.4.8.1970792. [DOI] [PubMed] [Google Scholar]
- Ghislain M, Dohmen RJ, Levy F, Varshavsky A. Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proteolysis in Saccharomyces cerevisiae. EMBO J. 1996;15:4884–4899. [PMC free article] [PubMed] [Google Scholar]
- Golbik R, Lupas AN, Koretke KK, Baumeister W, Peters J. The Janus face of the archaeal Cdc48/p97 homologue VAT: protein folding versus unfolding. Biol Chem. 1999;380:1049–1062. doi: 10.1515/BC.1999.131. [DOI] [PubMed] [Google Scholar]
- Goodbody KC, Lloyd CW. Immunofluorescence techniques for analysis of the cytoskeleton. In: Harris N, Oparka KJ, editors. Plant Cell Biology: A Practical Approach. Oxford: IRL Press; 1994. pp. 221–243. [Google Scholar]
- Gu X, Verma DP. Phragmoplastin, a dynamin-like protein associated with cell plate formation in plants. EMBO J. 1996;15:695–704. [PMC free article] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Morisaki H, Jahn R, Heuser JE. Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell. 1997;90:523–535. doi: 10.1016/s0092-8674(00)80512-7. [DOI] [PubMed] [Google Scholar]
- Hardwick KG, Pelham HR. SED5 encodes a 39-kD integral membrane protein required for vesicular transport between the ER and the Golgi complex. J Cell Biol. 1992;119:513–521. doi: 10.1083/jcb.119.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay JC, Chao DS, Kuo CS, Scheller RH. Protein interactions regulating vesicle transport between the endoplasmic reticulum and Golgi apparatus in mammalian cells. Cell. 1997;89:149–158. doi: 10.1016/s0092-8674(00)80191-9. [DOI] [PubMed] [Google Scholar]
- Hay JC, Klumperman J, Oorschot V, Steegmaier M, Kuo CS, Scheller RH. Localization, dynamics, and protein interactions reveal distinct roles for ER and Golgi SNAREs. J Cell Biol. 1998;141:1489–1502. doi: 10.1083/jcb.141.7.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese M, Gansel X, Sticher L, Wick P, Grebe M, Granier F, Jürgens G. Functional characterization of the KNOLLE-interacting t-SNARE AtSNAP33 and its role in plant cytokinesis. J Cell Biol. 2001;155:239–249. doi: 10.1083/jcb.200107126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler PK. Endoplasmic reticulum in the formation of the cell plate and plasmodesmata. Protoplasma. 1982;111:121–133. [Google Scholar]
- Hetzer M, Meyer HH, Walther TC, Bilbao-Cortes D, Warren G, Mattaj IW. Distinct AAA-ATPase p97 complexes function in discrete steps of nuclear assembly. Nat Cell Biol. 2001;3:1086–1091. doi: 10.1038/ncb1201-1086. [DOI] [PubMed] [Google Scholar]
- Hitchcock AL, Krebber H, Frietze S, Lin A, Latterich M, Silver PA. The conserved npl4 protein complex mediates proteasome-dependent membrane-bound transcription factor activation. Mol Biol Cell. 2001;12:3226–3241. doi: 10.1091/mbc.12.10.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe T, Matuschewski K, Rape M, Schlenker S, Ulrich HD, Jentsch S. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell. 2000;102:577–586. doi: 10.1016/s0092-8674(00)00080-5. [DOI] [PubMed] [Google Scholar]
- Hui N, Nakamura N, Sonnichsen B, Shima DT, Nilsson T, Warren G. An isoform of the Golgi t-SNARE, syntaxin 5, with an endoplasmic reticulum retrieval signal. Mol Biol Cell. 1997;8:1777–1787. doi: 10.1091/mbc.8.9.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Initiative TAG. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Jahn R, Südhof TC. Membrane fusion and exocytosis. Annu Rev Biochem. 1999;68:863–911. doi: 10.1146/annurev.biochem.68.1.863. [DOI] [PubMed] [Google Scholar]
- Jarosch E, Taxis C, Volkwein C, Bordallo J, Finley D, Wolf DH, Sommer T. Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat Cell Biol. 2002;4:134–139. doi: 10.1038/ncb746. [DOI] [PubMed] [Google Scholar]
- Kang BH, Busse JS, Dickey C, Rancour DM, Bednarek SY. The Arabidopsis cell plate-associated dynamin-like protein, ADL1Ap, is required for multiple stages of plant growth and development. Plant Physiol. 2001;126:47–68. doi: 10.1104/pp.126.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo H, Rabouille C, Newman R, Levine TP, Pappin D, Freemont P, Warren G. p47 is a cofactor for p97-mediated membrane fusion. Nature. 1997;388:75–78. doi: 10.1038/40411. [DOI] [PubMed] [Google Scholar]
- Lamb JR, Fu V, Wirtz E, Bangs JD. Functional analysis of the trypanosomal AAA protein TbVCP with trans-dominant ATP hydrolysis mutants. J Biol Chem. 2001;276:21512–21520. doi: 10.1074/jbc.M100235200. [DOI] [PubMed] [Google Scholar]
- Latterich M, Fröhlich KU, Schekman R. Membrane fusion and the cell cycle: Cdc48p participates in the fusion of ER membranes. Cell. 1995;82:885–893. doi: 10.1016/0092-8674(95)90268-6. [DOI] [PubMed] [Google Scholar]
- Lauber MH, Waizenegger I, Steinmann T, Schwarz H, Mayer U, Hwang I, Lukowitz W, Jürgens G. The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J Cell Biol. 1997;139:1485–1493. doi: 10.1083/jcb.139.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie C, Chevet E, Roy L, Tonks NK, Fazel A, Posner BI, Paiement J, Bergeron JJ. Tyrosine phosphorylation of p97 regulates transitional endoplasmic reticulum assembly in vitro. Proc Natl Acad Sci USA. 2000;97:13637–13642. doi: 10.1073/pnas.240278097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie C, Lanoix J, Kan FW, Paiement J. Cell-free assembly of rough and smooth endoplasmic reticulum. J Cell Sci. 1996;109:1415–1425. doi: 10.1242/jcs.109.6.1415. [DOI] [PubMed] [Google Scholar]
- Littleton JT, Barnard RJ, Titus SA, Slind J, Chapman ER, Ganetzky B. SNARE-complex disassembly by NSF follows synaptic-vesicle fusion. Proc Natl Acad Sci USA. 2001;98:12233–12238. doi: 10.1073/pnas.221450198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Mayer U, Jürgens G. Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell. 1996;84:61–71. doi: 10.1016/s0092-8674(00)80993-9. [DOI] [PubMed] [Google Scholar]
- Madeo F, Schlauer J, Zischka H, Mecke D, Fröhlich KU. Tyrosine phosphorylation regulates cell cycle-dependent nuclear localization of Cdc48p. Mol Biol Cell. 1998;9:131–141. doi: 10.1091/mbc.9.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May AP, Whiteheart SW, Weis WI. Unraveling the mechanism of the vesicle transport ATPase NSF, the N-ethylmaleimide-sensitive factor. J Biol Chem. 2001;276:21991–21994. doi: 10.1074/jbc.R100013200. [DOI] [PubMed] [Google Scholar]
- Mayr PS, Allan VJ, Woodman PG. Phosphorylation of p97(VCP) and p47 in vitro by p34cdc2 kinase. Eur J Cell Biol. 1999;78:224–232. doi: 10.1016/S0171-9335(99)80055-7. [DOI] [PubMed] [Google Scholar]
- Meyer HH, Kondo H, Warren G. The p47 co-factor regulates the ATPase activity of the membrane fusion protein, p97. FEBS Lett. 1998;437:255–257. doi: 10.1016/s0014-5793(98)01232-0. [DOI] [PubMed] [Google Scholar]
- Meyer HH, Shorter JG, Seemann J, Pappin D, Warren G. A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J. 2000;19:2181–2192. doi: 10.1093/emboj/19.10.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir D, Stewart SE, Osmond BC, Botstein D. Cold-sensitive cell-division cycle mutants of yeast: isolation, properties, and pseudoreversion studies. Genetics. 1982;300:547–563. doi: 10.1093/genetics/100.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan A, Dimaline R, Burgoyne RD. The ATPase activity of N-ethylmaleimide-sensitive fusion protein (NSF) is regulated by soluble NSF attachment proteins. J Biol Chem. 1994;269:29347–29350. [PubMed] [Google Scholar]
- Nebenführ A, Frohlick JA, Staehelin LA. Redistribution of Golgi stacks and other organelles during mitosis and cytokinesis in plant cells. Plant Physiol. 2000;124:135–152. doi: 10.1104/pp.124.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols BJ, Ungermann C, Pelham HRB, Wickner WT, Haas A. Homotypic vacuolar fusion mediated by t- and v-SNAREs. Nature. 1997;387:199–202. doi: 10.1038/387199a0. [DOI] [PubMed] [Google Scholar]
- Orellana A, Neckelmann G, Norambuena L. Topography and function of Golgi uridine-5′-diphosphatase from pea stems. Plant Physiol. 1997;114:99–107. doi: 10.1104/pp.114.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JM, Kang SG, Pih KT, Jang HJ, Piao HL, Yoon HW, Cho MJ, Hwang I. A dynamin-like protein, ADL1, is present in membranes as a high-molecular-mass complex in Arabidopsis thaliana. Plant Physiol. 1997;115:763–771. doi: 10.1104/pp.115.2.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SK, Indig FE, Olivieri N, Levine ND, Latterich M. Organelle membrane fusion: a novel function for the syntaxin homolog Ufe1p in ER membrane fusion. Cell. 1998;92:611–620. doi: 10.1016/s0092-8674(00)81129-0. [DOI] [PubMed] [Google Scholar]
- Peters JM, Harris JR, Lustig A, Muller S, Engel A, Volker S, Franke WW. Ubiquitous soluble Mg(2+)-ATPase complex: a structural study. J Mol Biol. 1992;223:557–571. doi: 10.1016/0022-2836(92)90670-f. [DOI] [PubMed] [Google Scholar]
- Peters JM, Walsh MJ, Franke WW. An abundant and ubiquitous homo-oligomeric ring-shaped ATPase particle related to the putative vesicle fusion proteins Sec18p and NSF. EMBO J. 1990;9:1757–1767. doi: 10.1002/j.1460-2075.1990.tb08300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich E, Kerem A, Fröhlich K-U, Diamant N, Bar-Nun S. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol Cell Biol. 2002;22:626–634. doi: 10.1128/MCB.22.2.626-634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouille C, Kondo H, Newman R, Hui N, Freemont P, Warren G. Syntaxin 5 is a component of the NSF- and p97-reassembly pathways of Golgi cisternae from mitotic Golgi fragments in vitro. Cell. 1998;92:603–610. doi: 10.1016/s0092-8674(00)81128-9. [DOI] [PubMed] [Google Scholar]
- Rabouille C, Levine TP, Peters JM, Warren G. An NSF-like ATPase, p97, and NSF mediate cisternal regrowth from mitotic Golgi fragments. Cell. 1995;82:905–914. doi: 10.1016/0092-8674(95)90270-8. [DOI] [PubMed] [Google Scholar]
- Rape M, Hoppe T, Gorr I, Kalocay M, Richly H, Jentsch S. Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell. 2001;107:667–677. doi: 10.1016/s0092-8674(01)00595-5. [DOI] [PubMed] [Google Scholar]
- Roggy JL, Bangs JD. Molecular cloning and biochemical characterization of a VCP homolog in African trypanosomes. Mol Biochem Parasitol. 1999;98:1–15. doi: 10.1016/s0166-6851(98)00114-5. [DOI] [PubMed] [Google Scholar]
- Rouiller I, Butel VM, Latterich M, Milligan RA, Wilson-Kubalek EM. A major conformational change in p97 AAA ATPase upon ATP binding. Mol Cell. 2000;6:1485–1490. doi: 10.1016/s1097-2765(00)00144-1. [DOI] [PubMed] [Google Scholar]
- Roy L, Bergeron JJ, Lavoie C, Hendriks R, Gushue J, Fazel A, Pelletier A, Morré DJ, Subramaniam VN, Hong W, Paiement J. Role of p97 and syntaxin 5 in the assembly of transitional endoplasmic reticulum. Mol Biol Cell. 2000;11:2529–2542. doi: 10.1091/mbc.11.8.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels AL, Giddings TH, Staehelin LA. Cytokinesis in tobacco BY-2 and root tip cells: a new model of cell plate formation in higher plants. J Cell Biol. 1995;130:1345–1357. doi: 10.1083/jcb.130.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot AA, Assaad FF, Raikhel NV. The Arabidopsis genome: an abundance of soluble N-ethylmaleimide-sensitive factor adaptor protein receptors. Plant Physiol. 2000;124:1558–1569. doi: 10.1104/pp.124.4.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HM, Raikhel NV. Nuclear localization signal receptor importin alpha associates with the cytoskeleton. Plant Cell. 1998;10:1791–1799. doi: 10.1105/tpc.10.11.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söllner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- Tagaya M, Wilson DW, Brunner M, Arango N, Rothman JE. Domain structure of an N-ethylmaleimide-sensitive fusion protein involved in vesicular transport. J Biol Chem. 1993;268:2662–2666. [PubMed] [Google Scholar]
- Wilson DW, Whiteheart SW, Wiedmann M, Brunner M, Rothman JE. A multisubunit particle implicated in membrane fusion. J Cell Biol. 1992;117:531–538. doi: 10.1083/jcb.117.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Okuhara K, Iwamatsu A, Seo H, Ohta K, Shibata T, Murofushi H. p97 ATPase, an ATPase involved in membrane fusion, interacts with DNA unwinding factor (DUF) that functions in DNA replication. FEBS Lett. 2000;466:287–291. doi: 10.1016/s0014-5793(99)01673-7. [DOI] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- Zhang H, Wang Q, Kajino K, Greene MI. VCP, a weak ATPase involved in multiple cellular events, interacts physically with BRCA1 in the nucleus of living cells. DNA Cell Biol. 2000a;19:253–263. doi: 10.1089/10445490050021168. [DOI] [PubMed] [Google Scholar]
- Zhang X, Shaw A, Bates PA, Newman RH, Gowen B, Orlova E, Gorman MA, Kondo H, Dokurno P, Lally J et al. Structure of the AAA ATPase p97. Mol Cell. 2000b;6:1473–1484. doi: 10.1016/s1097-2765(00)00143-x. [DOI] [PubMed] [Google Scholar]
- Zheng H, Bednarek SY, Sanderfoot AA, Alonso J, Ecker JE, Raikhel NV. NSPN11 is a cell plate associated SNARE protein that interacts with the Syntaxin KNOLLE. Plant Physiol. 2002;129:530–539. doi: 10.1104/pp.003970. [DOI] [PMC free article] [PubMed] [Google Scholar]